Abstract
To improve the enzymatic hydrolysis (saccharification) of lignocellulosic biomass by Trichoderma reesei, a set of genes encoding putative polysaccharide-degrading enzymes were selected from the coprophilic fungus Podospora anserina using comparative genomics. Five hemicellulase-encoding genes were successfully cloned and expressed as secreted functional proteins in the yeast Pichia pastoris. These novel fungal CAZymes belonging to different glycoside hydrolase families (PaMan5A and PaMan26A mannanases, PaXyn11A xylanase, and PaAbf51A and PaAbf62A arabinofuranosidases) were able to break down their predicted cognate substrates. Although PaMan5A and PaMan26A displayed similar specificities toward a range of mannan substrates, they differed in their end products, suggesting differences in substrate binding. The N-terminal CBM35 module of PaMan26A displayed dual binding specificity toward xylan and mannan. PaXyn11A harboring a C-terminal CBM1 module efficiently degraded wheat arabinoxylan, releasing mainly xylobiose as end product. PaAbf51A and PaAbf62A arabinose-debranching enzymes exhibited differences in activity toward arabinose-containing substrates. Further investigation of the contribution made by each P. anserina auxiliary enzyme to the saccharification of wheat straw and spruce demonstrated that the endo-acting hemicellulases (PaXyn11A, PaMan5A, and PaMan26A) individually supplemented the secretome of the industrial T. reesei CL847 strain. The most striking effect was obtained with PaMan5A that improved the release of total sugars by 28% and of glucose by 18%, using spruce as lignocellulosic substrate.
Lignocellulosic biomass is a desirable feedstock for the supply of second-generation bioethanol, being the largest renewable source of carbohydrates, but the recalcitrance of raw materials makes biotechnological conversion complex and costly (34).
The main components of plant cell walls are cellulose, lignin, and hemicellulose, which form a tight complex with varying proportions between plants (for a review, see reference 45). Cellulose microfibrils are formed by crystalline-like combination of linear and highly ordered polymer chains of β-1,4-linked d-glucose residues, and lignin consists of an amorphous aromatic heteropolymer with hydrophobic properties. Hemicellulose is a combined designation of a diverse set of noncrystalline carbohydrate polymers from plant tissues that form closely associated networks with cellulose microfibrils and lignin (6, 24). Typically, the most abundant component of hemicellulose in the cell walls of monocots (e.g., cereals) is β-1,4-xylan, which consists of β-1,4-linked d-xylose residues substituted with l-arabinosyl, 4-O-methyl-glucuronosyl and acetyl side chains (46, 53). Arabinofuranosyl residues of arabinoxylan may also be esterified with hydroxycinnamic acid residues, e.g., ferulic and p-coumaric acids (53). In softwoods (gymnosperms), the main hemicellulose component is β-1,4-mannan, which consists of a backbone formed by β-1,4-linked d-mannose and d-glucose residues substituted with α-1,6-linked d-galactose residues (39). Linear mannan and glucomannan chains containing more than 5% (wt/wt) d-galactose are called galactomannans and galactoglucomannans, respectively (39).
The proportion of monosaccharide residues in plant biomass degradation products depends on the raw material and the hydrolysis procedure, but significantly, pentose sugars such as d-xylose, constitute the second most abundant carbohydrate in plant raw materials after glucose (26). Thus, it is considered now that the use of all sugar monomers is one of the possible strategies to provide cost-efficient biorefinery processes (28), which is partly due to the recent progress made with pentose-fermenting yeast strains (36). Therefore, bioconversion of hemicelluloses into fermentable sugars is essential to enable the commercial production of lignocellulosic ethanol (24).
One of the crucial steps in the bioconversion of LCB to fuel-grade ethanol is the enzymatic degradation of this recalcitrant biomass (28). The ascomycete Trichoderma reesei (teleomorph Hypocrea jecorina) is one of the most widely used industrial fungi, able to produce cellulase-rich enzyme cocktails with up to 100 g of extracellular protein per liter of culture (13). Such high yields enable a wide application of T. reesei enzyme preparations in different fields of white biotechnology, in the bioconversion of biomass to fuel and other useful products (34). Recent genetic and biochemical technologies have improved our knowledge of the full set of T. reesei enzymes. Although analysis of the secretome of T. reesei CL847 industrial strain revealed only 22 secreted enzymes, most of them involved in biomass degradation (27), complete genome sequencing of the QM6a strain (35) identified about 200 genes encoding glycoside hydrolases as defined by the CAZy classification (10). Despite this high number, the T. reesei genome is surprisingly poor in terms of number and diversity of carbohydrate-degrading enzymes, including GHs, polysaccharide lyases, and carbohydrate esterases potentially able to participate in LCB degradation, compared to other fungi known for their carbohydrate-degrading capacity (35). In addition, the T. reesei genome has the smallest number of carbohydrate-binding modules of all of the Sordariomycetes sequenced to date (35). The greatest current challenge facing the development of second generation biofuels is enzyme cost reduction, the main strategy to meet this goal being the improvement of the efficiency of T. reesei enzymatic cocktail for the saccharification of LCB (34).
Possible strategies to circumvent T. reesei weaknesses in LCB hydrolysis include the search for enzymatic activities able to supplement those of T. reesei industrial enzyme preparations. The identification of novel glycosidases that are either distantly related to or absent from the T. reesei genome is a promising option for enhancing enzymatic hydrolysis of lignocellulosic materials. The increasing number of sequenced genomes annotated for their content of genes encoding for CAZymes (i.e., CAZomes) is a potential source of novel enzymes that warrants exploration. Among the fungal genomes already sequenced, that of the coprophilic ascomycete Podospora anserina, a late grower in the droppings of grass herbivores, encodes for a large, highly specialized set of genes involved in the utilization of carbon sources commonly found in its natural biotope, together with the highest number of CBMs of all of the fungal genomes sequenced to date (19). Interestingly, its CAZome has also revealed an increased potential for the digestion of resistant components of hemicellulose, presenting abundant enzyme sets dedicated to the hydrolysis of cross-linked xylan (19), from which no CAZymes have yet been characterized experimentally.
Owing to the heterogeneity of the hemicellulose fraction of LCB, complete hydrolysis requires a broad variety of cooperatively acting enzymes, such as xylanases, xylosidases, arabinofuranosidases, glucuronidases, acetylxylan esterases, ferulic acid esterases, and p-coumaric acid esterases for the xylan fraction and mannanases, galactosidases, glucosidases, and acetylmannan esterases for the mannan fraction. Until now, most of the characterization and applied studies on hemicellulases have focused on xylanases found in CAZy families GH10 and GH11. Accessory enzymes, such as arabinose-debranching enzymes, are found in six families (GH3, GH43, GH51, GH54, GH62, and GH93), whose members display different specificities. Although mannan is the principal hemicellulose component in softwood, studies of mannan-hydrolyzing enzymes found in families GH5 and GH26 have been largely neglected compared to xylanases.
The objective of the present study was to investigate the potential of single enzyme candidates from the hemicellulolytic enzymatic arsenal of P. anserina for improving the degradation of LCB by an industrial strain of T. reesei. By means of genome analysis using CAZy database tools, we identified genes encoding enzymes that degrade carbohydrates distantly related to or absent from T. reesei. We thus selected and cloned a set of genes from P. anserina encoding putative hemicellulases, which were heterologously expressed and purified to give insight into their biochemical characteristics and enzymatic specificities. The T. reesei enzyme cocktail was further used as a reference to investigate the contribution made by each specific P. anserina auxiliary enzyme on wheat straw and/or spruce, taking advantage of the recently developed automated saccharification method (40).
MATERIALS AND METHODS
Abbreviations.
Abf, α-l-arabinofuranosidase; AFMB, autoclaved fraction of maize bran; AX, arabinoxylan; CAZy, carbohydrate active enzyme; CBM, carbohydrate binding module; CE, carbohydrate esterase; DNS, dinitrosalycilic acid; d.m., dry matter; GH, glycoside hydrolase; HPAEC-PAD, high-performance anion exchange chromatography coupled with pulsed amperometric detection; IEF, isoelectric focusing; LBG, locust bean gum; LCB, lignocellulosic biomass; Man, endo-β-1,4-mannanase; PL, polysaccharide lyase; PX, mixture of pectin, birchwood, and larchwood xylans; Xyn, endo-β-1,4-xylanase.
Bioinformatics-guided sequence selection.
Previous analysis of the T. reesei (anamorph of Hypocrea jecorina) strain QM6a genome had revealed weaknesses in hemicellulolytic capacity (35). Candidate hemicellulolytic CAZymes from the P. anserina genome were subjected to BLASTp analysis against all sequences present in CAZy (10) supplemented by T. reesei CAZyme sequences (35). A number of P. anserina candidates that were (i) absent or distant from those found in the T. reesei genome, (ii) distant from known characterized enzymes from other sources, and/or (iii) displayed an unusual combination of catalytic and carbohydrate-binding modules, were selected for cloning and characterization. For each individual target, the enzyme activity was predicted from the sequence comparisons with BLASTp against a specific library of characterized enzyme modules derived from CAZy. As a complement, the signal peptide cleavage sites were predicted using the signalP 3.0 server (http://www.cbs.dtu.dk/services/SignalP/).
Cloning of genes encoding hemicellulases.
The homokaryotic P. anserina strain S mat+ used in the present study was kindly provided by P. Silar (CNRS, Paris, France). Currently used methods and culture media can be accessed at the P. anserina Genome Project web site (http://podospora.igmors.u-psud.fr). For the induction of enzymes, P. anserina was grown on M2 medium supplemented with AFMB (autoclaved fraction of maize bran at 15 g liter−1) or PX (pectin, birchwood xylan, and larchwood xylan from Sigma-Aldrich (Lyon, France), each at 5 g liter−1. AFMB was prepared as described previously (7). Growth was carried out in baffled flasks at 120 rpm and 27°C. The mycelium was recovered by sterile filtration using miracloth after 3, 4, and 5 days of culture and directly frozen at −80°C. Total RNA was extracted by using RNeasy plant kit (Qiagen, Courtaboeuf, France), and first-strand cDNA synthesis was performed with SuperScript reverse transcriptase (Invitrogen, Cergy-Pontoise, France) and oligo(dT18) primer according to the manufacturer's instructions. DNA was amplified by PCR using Pwo Super yield DNA polymerase (GE Healthcare, Buc, France) and the specific primers (including restriction sites) designated in Table 1 . The amplified fragments were subcloned into pCRII-TOPO vector (Invitrogen) and subjected to sequencing, and the corresponding genes were deposited in GenBank (for accession numbers, see Table 2 and the legend to Fig. 1). Amplified fragments were inserted at the corresponding sites into vector pPICZαA or pPICZαC (Table 1) in frame with both the yeast α-secretion factor and the C-term-His6 tag encoding sequences. The resulting recombinant expression plasmids were linearized with PmeI and transformed into P. pastoris X-33 (Invitrogen) by electroporation. P. pastoris transformants were isolated on plates containing increasing concentrations of zeocin ranging from 100 to 1,000 μg ml−1. Zeocin-resistant P. pastoris transformants were then screened for protein expression in 10 ml of BMGY (in 50-ml tubes) at 30°C in an orbital shaker (200 rpm) for 16 h to an optical density at 600 nm of 2 to 6, and expression was induced by transferring cells into 2 ml of BMMY, followed by growth for another 3 days. Each day the medium was supplemented with 3% (vol/vol) methanol. The supernatant was then analyzed by SDS-PAGE to determine the transformant with the best secretion yield for each enzyme.
TABLE 1.
Oligonucleotide sequences used in this study
| Orientation and primer | Sequence (5′-3′)a | Destination vector |
|---|---|---|
| Forward | ||
| PaMan5AEcoF | TTTGAATTCCTCCCCCAAGCACAAGGTG | pPiCZαA |
| PaXyn11AEcoF | TTTGAATTCCGTCCTTTCGACTTTCTCG | pPiCZαA |
| PaMan26AClaF | TTTATCGATAAAGCCTTGTAAGCCTCGTG | pPiCZαC |
| PaAbf51AEcoF | TTTTTTGAATTCATTGATTTGTTTGTCAAGTC | pPiCZαA |
| PaAbf62AEcoF | TTGAATTCTCTTCCCTGCCAGCTGGGC | pPiCZαA |
| Reverse | ||
| PaMan5AXbaR | TTTTCTAGACCCGCCGGGAGAGCATTGATAG | pPiCZαA |
| PaXyn11AXbaR | TTTTCTAGACCGTGTGTCTGGACATAAATGTC | pPiCZαA |
| PaMan26AXbaR | TTTTCTAGACCACTCCTCCACCCCTGAATCTC | pPiCZαC |
| PaAbf51AXbaR | TTTTCTAGACCGTTACCCTTTCCCTTGCCCTTG | pPiCZαA |
| PaAbf62AXbaR | TTTTCTAGACCACAAGCAGGGTTGTTGTGTGCG | pPiCZαA |
The restriction sites added to insert genes into pPICZαA or pPICZαC are underlined.
TABLE 2.
Characteristics of the P. anserina hemicellulase-encoding genes selected
| P. anserina gene | Enzyme family | CBM family | GenBank accession no. (locus) | Putative activity | % Identity to T. reesei characterized GH (amino acid sequence) | Condition of inductiona |
|---|---|---|---|---|---|---|
| PaMan5A | GH5 | HM357135 (Pa_6_490) | Endo-β-1,4-mannanase | 62% with TrMan5A | AFMB/PX | |
| PaMan26A | GH26 | CBM35 (N-term) | HM357136 (Pa_5_1950) | Endo-β-1,4-mannanase | NAb | AFMB/PX |
| PaXyn11A | GH11 | CBM1 (C-term) | HM357137 (Pa_2_13730) | Endo-β-1,4-xylanase | 47% with TrXyn1, 64% with TrXyn2 | AFMB/PX |
| PaAbf51A | GH51 | HM357138 (Pa_5_11670) | α-l-Arabinofuranosidase | NA | AFMB/PX | |
| PaAbf62A | GH62 | HM357139 (Pa_0_1370) | α-l-Arabinofuranosidase | 35% with TrAbf2 | AFMB |
mRNA expression was induced using AFMB or pectin and xylan (PX) (see Materials and Methods).
NA, not applicable. That is, no member of this family was present in T. reesei based on genomic data.
FIG. 1.
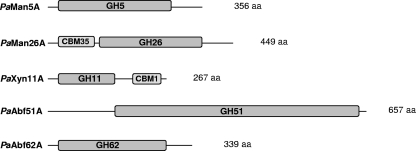
Modular representation of hemicellulases used in the present study. GH, glycoside hydrolases; CBM, carbohydrate-binding modules. The corresponding GenBank accession numbers are HM357135 (PaMan5A), HM357136 (PaMan26A), HM357137 (PaXyn11A), HM357138 (PaAbf51A), and HM357139 (PaAbf62A).
Recombinant enzymes production and purification.
For each enzyme, the best-producing transformant was grown in 200 ml of BMGY in a 1-liter baffled flask. The cells were then transferred to 40 ml of BMMY in 200-ml flasks stirred at 200 rpm and 30°C. The suspension was recovered after 3 days induction by centrifugation (10 min, 4,000 × g), and the supernatant was concentrated 10 times by using Amicon centrifugal units (10-kDa cutoff, 4,000 × g; Millipore, Molsheim, France). A nickel chelate His-Bind Resin (GE Healthcare) column (0.7 by 5 cm) was connected to a fast-performance liquid chromatography Äkta apparatus (GE Healthcare) and equilibrated with 50 mM Tris-HCl (pH 7.8), 150 mM NaCl, and 10 mM imidazole. The concentrated supernatant was diluted into the equilibration buffer and loaded onto the column at 4°C. The column was washed with equilibration buffer, and the enzyme was eluted with the same buffer containing 150 mM imidazole. The eluate was concentrated in the centrifugal units and repeatedly washed with sodium acetate buffer (50 mM, pH 5).
Biochemical characterization.
Protein concentration was determined by using the Bio-Rad protein assay kit with bovine serum albumin as the standard (Bio-Rad, Marnes-la-Coquette, France). SDS-PAGE was performed in 12% (wt/vol) polyacrylamide gel (Bio-Rad) using a Pharmacia LMW electrophoresis calibration kit (GE Healthcare). Native IEF was carried out at 4°C in the Bio-Rad gel system, using pI standards ranging from 4.45 to 8.2 (Bio-Rad). Proteins were visualized with either Coomassie staining or IEF gel staining solution (0.04% [wt/vol] Coomassie blue R250, 0.05% [wt/vol] Cocrein Scarlet, 10% [vol/vol] acetic acid, 27% [vol/vol] isopropanol).
Enzyme assays.
Unless otherwise indicated, assay mixtures contained substrate and suitably diluted enzyme in McIlvaine's buffer (pH 5). Reactions were carried out at 37°C for 5 to 30 min, and the enzyme activity was determined spectrophotometrically. The activity of enzymes toward xylan, mannan, or arabinan was measured by using the 3,5-dinitrosalicylic acid method described earlier (2). Briefly, 10 μl of enzyme was mixed with 190 μl of substrate (xylan or mannan at 10 mg ml−1) in McIlvaine's buffer (pH 5) at 37°C. The reaction was terminated by the addition of 300 μl of dinitrosalicylic acid reagent at 1% (wt/vol) and boiled for 5 min. The reaction mixtures were cooled and centrifuged for 5 min at 13,000 rpm, and 200 μl was transferred to a microtiter plate. The absorbance at 550 nm was measured relative to a xylose or mannose standard curve (0 to 180 μg ml−1). One unit of endo-glycosidase activity was defined as the amount of protein that released 1 μmol of sugar monomer per min. Low-viscosity wheat arabinoxylan, konjac glucomannan, carob galactomannan, ivory nut mannan, sugar beet arabinan, and debranched arabinan were purchased from Megazyme International (Wicklow, Ireland). Birchwood xylan, carboxymethyl cellulose, and locust bean gum galactomannan were purchased from Sigma-Aldrich. Activity toward pNP-α-l-arabinofuranoside (Sigma-Aldrich) was determined by measuring the release of 4-nitrophenol from p-nitrophenyl-α-l-arabinofuranoside (pNP-Ara; 10 mM; Sigma-Aldrich) in 50 mM sodium phosphate buffer (pH 5) at 37°C in a reaction volume of 100 μl. The reaction was stopped by adding 900 μl of 1 M sodium carbonate (pH 9), and the release of 4-nitrophenol was quantified at 410 nm with a molar extinction coefficient for 4NP of 18,300 M−1 cm−1. One unit of enzyme activity was defined as the amount of protein that released 1 μmol of arabinose per min.
The apparent optimal pH was estimated by using wheat arabinoxylan or LBG galactomannan at 10 mg ml−1 in McIlvaine's buffer in the pH range from 2.6 to 7.5 (11 pHs tested). The apparent optimal temperature was estimated in the range 30 to 70°C (nine temperatures tested) under the conditions used. For determination of apparent Michaelis-Menten constants, the initial velocities of the enzymes were measured at 37°C in McIlvaine's buffer (pH 5.0), with substrate concentrations ranging from 2 to 20 mg ml−1 for polymeric substrates and from 0.1 to 20 mM for pNP substrates. The molar concentrations of polymeric substrates could not be calculated because of their heterogeneous nature. Consequently, only an apparent value for the Michaelis constant, Km(app), was determined. The kinetic parameters were estimated by using weighted nonlinear squares regression analysis with Grafit software (Erithacus Software, Horley, United Kingdom).
Determination of the PaMan26A N-terminal CBM35 dissociation constant and the carbohydrate binding capacity.
For the binding assay, the reaction mixture contained 1 mg of insoluble wheat arabinoxylan or insoluble ivory nut mannan (Megazyme) or Avicel PH101 (Sigma-Aldrich) and 10 to 50 μg of PaMan26A ml−1 in a final volume of 225 μl of sodium citrate buffer (pH 5). For each assay, the mixture was incubated at 4°C for 1 h with vertical rotation (20 rpm). Polysaccharide was removed by centrifugation, and the concentration of unbound protein ([UB], μM) in the supernatant was measured by the Bradford method. The bound protein concentration ([B], μmol per g of polysaccharide) was determined by subtracting [UB] from the total protein concentration. All of the assays were carried out in triplicate. Adsorption parameters were determined as described previously (41) based on typical double-reciprocal plots using the equation [B] = [UB] × [B]max/(Kd + [UB]), where Kd (μM) and [B]max (μmol per g of polysaccharide) are the equilibrium dissociation constant and the maximum amount of protein bound, respectively.
Supplementation of T. reesei.
The lignocellulosic raw materials, wheat straw (Triticum aestivum cv. Apache; Aveyron, France) and spruce (Picea abies, Holmen plant; Braviken, Sweden) were kindly prepared by X. Rouau (INRA Montpellier, France). They were reduced to particles of roughly 100 μm by a multistep micronization procedure as previously described (40). Suspensions were made by adding 1 g of wheat straw or 2 g of spruce to 100 ml of 50 mM sodium acetate (pH 5) in the presence of 30 μg of cycloheximide ml−1 and 40 μg of tetracycline ml−1. Using the handling robot (Tecan, Männedorf, Switzerland), 96-well microplates were filled with micronized wheat straw or spruce suspensions, heat sealed, and stored frozen at −20°C until use; then, 30 μg per well of T. reesei industrial xylanase-rich enzyme cocktail (E508) produced from T. reesei CL847 strain (49) was used as a reference in the assay as described previously (40). T. reesei enzyme cocktail contained 0.12 U of filter paper activity, 0.33 U of CMCase, 0.2 U of β-glucosidase, 1.6 U of xylanase, 0.02 U of mannanase, and 0.02 U of arabinofuranosidase per mg of total protein at 37°C (pH 5.0). To investigate the contribution of each specific P. anserina auxiliary enzyme to the saccharification of micronized wheat straw and/or spruce, 30 μg of PaMan5A, 30 μg of PaMan26A, 10 μg of PaXyn11A, 30 μg of PaAbf51A, and 30 μg of PaAbf62A were individually added to the 30 μg of T. reesei enzymatic cocktail. A total of 30 μg of equal amounts of PaMan5A and PaMan26A was also assayed in combination on spruce. The plate was heat sealed and transferred to a shaking incubator set at 40°C. The enzymatic reaction was left to proceed for 24 h, and the plate was filtered. The total reducing sugars were quantified by using the DNS assay as described previously (40). Monosaccharides (glucose, xylose, and mannose) were quantified by HPAEC-PAD (see below). Saccharification results were expressed in μmol of sugar released per mg of d.m.
Analysis of sugar release by HPAEC-PAD.
Monosaccharides generated after hydrolysis of micronized LCB were analyzed by HPAEC coupled with PAD (ICS 3000; Dionex, Sunnyvale, CA) equipped with a Carbo-Pac PA-1 analytical column (250 by 4 mm). Enzymatic reactions were stopped by the addition of 50 mM NaOH before injection (10 μl) into the HPAEC system. Elution (1 ml min−1) was carried out in 18 mM NaOH. Calibration curves were plotted using arabinose, glucose, xylose, mannose, and cellobiose standards (Sigma-Aldrich), from which response factors were calculated (Chromeleon program; Dionex) and used to estimate the amount of product released in test incubations. All of the assays were carried out in triplicate. The results are expressed in μmol of sugar released per mg of d.m. End products generated after hydrolysis of polymeric substrates (ivory nut mannan and wheat arabinoxylan) were analyzed as described earlier (4). The reference β-1,4-xylo-oligosaccharides and β-1,4-manno-oligosaccharides were obtained from Megazyme International.
RESULTS
Selection of genes encoding fungal hemicellulases and sequence analysis.
Genes encoding polysaccharide-degrading enzymes were selected based on (i) the structural features of lignocellulosic raw materials selected for the present study, i.e., wheat straw and spruce; (ii) BLASTp searches using CAZy database by comparison of T. reesei and P. anserina genomes; (iii) the presence and diversity of CBMs; and (iv) the predicted specificity of putative exo- and endo-acting CAZymes. The search yielded the sequences corresponding to the loci Pa_6_490 (designated PaMan5A following characterization), Pa_5_1950 (PaMan26A), Pa_2_13730 (PaXyn11A), Pa_5_11670 (PaAbf51A), and Pa_0_1370 (PaAbf62A).
One of our main goals was to select targets that would contribute to β-mannan degradation. Sequence comparison showed that PaMan5A exhibited 62% identity to the catalytic module of T. reesei β-mannosidase TrMan5A, a well-characterized enzyme (48). Unlike TrMan5A, which bears a CBM1 module at C-term, PaMan5A bears no additional module. Since genome analysis had revealed that likely orthologs from different Trichoderma bear the C-terminal CBM, while a variety of other fungi bear none (data not shown), PaMan5A was selected as a target. The second selected enzyme degrading β-mannan was PaMan26A, which has no equivalent in the T. reesei genome. Interestingly, only a very reduced number of fungal genomes present closely related catalytic modules (unpublished data). The closest characterized enzyme is the β-mannanase from Humicola insolens (30), with 79% identity at the catalytic module, followed by a number of bacterial β-mannanases. All closely related enzymes have CBM35 modules adjacent to the N-terminal side of the GH26 catalytic module, but the substrate affinity of the corresponding binding modules has never been studied.
The next hemicellulolytic target PaXyn11A was selected, although its catalytic module presented, respectively, 47 and 64% sequence identity to T. reesei xylanases TrXyn1 and TrXyn2, two long-established enzymes. (51). The main singularity of this sequence is the fact that it carries a C-term CBM1 module. The number of characterized GH11 enzymes bearing a C-term CBM1 module is relatively small. They include xylanases from Penicillium funiculosum XynB (9, 25), Neocallimastix patriciarum XynS20 (33), Lentinula edodes Xyn11A (32), and Phanerochaete chrysosporium XynB/XynB-1 (17). Although the substrate specificities of these fungal CBM1-containing enzymes have been described, none was chosen for that feature.
Finally, two enzyme targets for arabinofuranose-containing substrates were considered, PaAbf51A and PaAbf62A. The first has no equivalent in the T. reesei genome, and the closest characterized sequences are those of the α-l-arabinofuranosidases Afq1 from Penicillium chrysogenum (43) and Abf2 from Penicillium purpurogenum (21), both at ca. 50% identity. The second target had 35% identity to a still (to our knowledge) uncharacterized TrAbf2 from T. reesei, although a number of transcription studies have identified its presence upon induction by a number of saccharides (1, 20). The closest characterized α-l-arabinofuranosidase to PaAbf62A is CjAbf62A from Cellvibrio japonicus at 43% identity (31).
Cloning and expression of novel hemicellulases.
Five genes (PaMan5A, PaMan26A, PaXyn11A, PaAbf51A, and PaAbf62A) encoding putative CAZymes were cloned by reverse transcription-PCR using mRNA purified from P. anserina grown on different natural inducers (i.e., AFMB or PX). Although both inducers used were found to induce the expression of PaMan5A, PaMan26A, PaXyn11A, and PaAbf51A genes, only AFMB was able to induce the expression of PaAbf62A gene (Table 2). After sequencing, all of the coding sequences were in agreement with those predicted from genomic data except for the PaAbf51A coding sequence (HM357138), which presented an insertion of 54 nucleotides, leading to 18 extra amino acids with no change in the open reading frame. The coding sequence of each gene was inserted into the P. pastoris expression vector in frame with sequences encoding the yeast α-factor secretion peptide and a His6 tag located at the C terminus. Recombinant genes were then introduced into the Pichia genome under the control of the methanol-inducible promoter. Multicopy transformants were screened on small cultures to select transformants that exhibited satisfactory levels of production by the analysis of each culture supernatant by means of SDS-PAGE. All targeted hemicellulases were successfully detected in the supernatant after induction, indicating correct processing of the α-factor signal sequence. Secretion levels were between 0.1 and 1 g liter−1, facilitating further purification of recombinant enzymes taking advantage of His6 tags. In addition, only trace amounts of endogenous proteins were present in the culture supernatant of transformants (data not shown). Purification yields ranged from 1.2 to 17.6 mg of recombinant protein from 40 ml of supernatant after 3 days induction (Table 3).
TABLE 3.
Expression yield and biochemical characterization of P. anserina hemicellulolytic CAZymes
| CAZyme | Expression yield (mg/liter)b | Molecular mass (kDa) |
pI |
Apparent optimum pH and tempa |
|||
|---|---|---|---|---|---|---|---|
| Theoretical | Experimentalc | Theoretical | Experimentald | pH | Temp (°C) | ||
| PaMan5A | 440 | 39.4 | 46 | 7.8 | 7.5 | 3-6 (4) | 50-65 (60) |
| PaMan26A | 100 | 49.8 | 62 | 4.7 | 4.3 | 4-6 (4.5) | 40 (40) |
| PaXyn11A | 30 | 28.5 | 41 | 8.3 | 6.2-6.3 | 5-7 (6) | 50-60 (50) |
| PaAbf51A | 90 | 71.7 | 86 | 6.4 | 6.1 | 4-7 (4.5) | 5-75 (70) |
| PaAbf62A | 300 | 37.7 | 48 | 4.8 | 4.6 | 4-6 (4) | 50-60 (55) |
Values indicate that the relative enzyme activity remaining was above 50%. Values in parentheses indicate the apparent optimum pH and temperature under the conditions used.
Expression yield is expressed in mg of recombinant protein purified from 1 liter of culture.
Estimated from SDS-PAGE.
Estimated from an IEF gel.
Characterization and kinetic properties of individual enzymes.
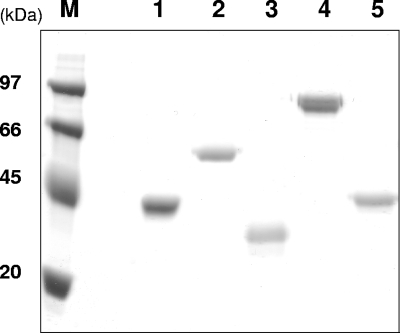
First, P. anserina hemicellulolytic CAZymes purified to homogeneity were biochemically characterized in terms of apparent molecular mass on SDS-PAGE and pI on IEF (Fig. 2 and Table 3). The recombinant proteins displayed apparent molecular masses slightly higher that the theoretical ones (Fig. 2A), probably as a result of posttranslational modification (e.g., glycosylation). Experimental pIs were in close agreement with pIs deduced from the amino acid sequences (Fig. 2B; Table 3). Apparent optimum pH and temperature were first determined on each enzyme, together with temperature stability (Table 3). All of the enzymes were active in the pH 4 to 6 range at temperatures up to 60°C and were stable at 40°C for 1 h with >90% residual activity. P. anserina glycosidases were characterized in depth in terms of activity toward various polysaccharides. Kinetic parameters were further determined at pH 5 and 40°C on the best substrates of each enzyme.
FIG. 2.
SDS-PAGE analysis of P. anserina hemicellulases: 3 μg of purified recombinant was loaded onto a 12% Tris glycine SDS-PAGE, and the proteins were stained with Coomassie blue. PaMan5A (lane 1), PaMan26A (lane 2), PaXyn11A (lane 3), PaAbf51A (lane 4), and PaAbf62A (lane 5) are shown. The molecular masses of marker proteins (M) are indicated on the left.
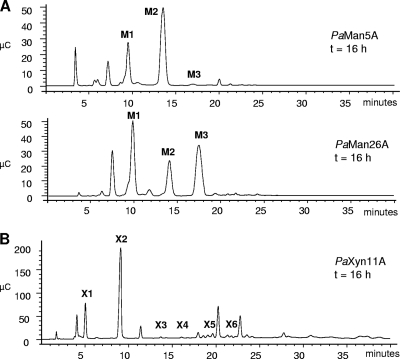
PaMan5A and PaMan26A enzymes efficiently hydrolyzed structurally different mannan polysaccharides, such as galactomannans, glucomannans, and β-1,4-mannans from different sources with specific activities ranging from 35 to 86 U mg−1 (Table 4). Neither of the mannanases was active on carboxymethyl cellulose or Avicel. Determination of kinetic parameters showed that PaMan5A and PaMan26A displayed the highest specificity toward konjac glucomannan, with Km values of 0.7 and 0.8 mg ml−1 and catalytic efficiencies (kcat/Km) of 3,142 and 5,750 mg−1 min−1 ml, respectively. PaMan5A and PaMan26A enzymes also acted efficiently on unsubstituted mannan from ivory nut with Km values of 1.6 and 1.0 mg.ml−1 and catalytic efficiencies (kcat/Km) of 1,044 and 1,928 mg−1 min−1 ml, respectively. The PaXyn11A enzyme efficiently hydrolyzed xylans from birchwood and wheat with a preference for wheat arabinoxylan (Km = 5.8 mg ml−1 and kcat/Km = 2,443 mg−1 min−1 ml, Table 4). To evaluate the mode of action of these P. anserina glycosidases, the products generated by the action of PaMan5A and PaMan26A on unsubstituted mannan and PaXyn11A on arabinoxylan were analyzed by HPAEC-PAD. All three enzymes tested displayed typical endo-activity against polymeric substrates, i.e., a mixture of oligosaccharides was generated during the initial stages of hydrolysis (not shown). As the reaction continued, the oligosaccharides were progressively hydrolyzed, yielding principally mannobiose and xylobiose toward the end of the reaction for PaMan5A and PaXyn11A, respectively, and a mixture of mannose, mannobiose, and mannotriose for PaMan26A (Fig. 3).
TABLE 4.
Enzymatic properties of P. anserina CAZymes acting on hemicellulases from different sources
| CAZyme | Activity | Substrate | Enzymatic property |
|||
|---|---|---|---|---|---|---|
| SAa ± SEM (U mg−1) | Kmb ± SEM (mg ml−1 or mM) | Vmax ± SEM (U mg−1) | kcat/Kmc (mg−1 min−1 ml or M−1 min−1) | |||
| PaMan5A | Endo-β-1,4-mannanase | Konjac glucomannan | 45 ± 0.9 | 0.7 ± 0.1 | 56.4 ± 1.7 | 3,142 |
| Carob galactomannan | 65 ± 1.4 | 1.7 ± 0.1 | 75.5 ± 1.3 | 1,732 | ||
| LBG galactomannan | 68 ± 3.2 | 4.7 ± 1.4 | 107 ± 14 | 899 | ||
| Ivory nut mannan | 35 ± 1.8 | 1.6 ± 0.4 | 42.3 ± 2.4 | 1,044 | ||
| PaMan26A | Endo-β-1,4-mannanase | Konjac glucomannan | 56 ± 1.9 | 0.8 ± 0.0 | 74.2 ± 1.8 | 5,750 |
| Carob galactomannan | 60 ± 3.4 | 2.6 ± 0.6 | 79.5 ± 5.8 | 1,895 | ||
| LBG galactomannan | 86 ± 4.1 | 5.5 ± 1.2 | 126 ± 13 | 904 | ||
| Ivory nut mannan | 46 ± 2.7 | 1.0 ± 0.1 | 48.8 ± 1.7 | 1,928 | ||
| PaXyn11A | Endo-β-1,4-xylanase | Wheat arabinoxylan | 249 ± 7.4 | 5.8 ± 0.4 | 405 ± 12 | 2,443 |
| Birchwood xylan | 95 ± 4.2 | 3.7 ± 0.5 | 124 ± 5 | 1,172 | ||
| PaAbf51A | α-l-Arabinofuranosidase | pNP-Ara | 22.8 ± 0.6 | 1.2 ± 0.1 | 26.3 ± 0.5 | 1,571 |
| Wheat arabinoxylan | 6.9 ± 0.4 | NDd | ND | ND | ||
| Sugar beet arabinan | 3.1 ± 0.3 | ND | ND | ND | ||
| Debranched arabinan | 0.9 ± 0.0 | ND | ND | ND | ||
| PaAbf62A | α-l-Arabinofuranosidase | pNP-ara | 0.38 ± 0.01 | 6.1 ± 0.3 | 0.61 ± 0.02 | 3.7 |
| Wheat arabinoxylan | 1.0 ± 0.1 | ND | ND | ND | ||
| Sugar beet arabinan | 0.11 ± 0.02 | ND | ND | ND | ||
| Debranched arabinan | 0.22 ± 0.03 | ND | ND | ND | ||
SA was determined at 10 mg ml−1 for polysaccharides and at 10 mM for pNP sugars at 40°C and pH 5 in McIlvaine's buffer.
Units are in mg ml−1 for polysaccharides and in mM for pNP substrates.
Units are in mg−1 min−1 ml for polysaccharides and in mM−1 min−1 for pNP substrates.
ND, not detected. Although enzyme activity was present, it was too low to measure the individual kinetic constant.
FIG. 3.
HPAEC analysis of end products released by endo-acting P. anserina hemicellulases. (A) Ivory nut mannan hydrolysis by PaMan5A and PaMan26A: mannose (M1), mannobiose (M2), and mannotriose (M3). (B) Wheat arabinoxylan hydrolysis by PaXyn11A: xylose (X1), xylobiose (X2), xylotriose (X3), xylotetraose (X4), xylopentaose (X5), and xylohexaose (X6).
To investigate the specificity of PaAbf51A and PaAbf62A enzymes, their capacity to hydrolyze pNP-α-l-arabinofuranoside, as well as arabinose-containing polysaccharides, was evaluated. Although PaAbf51A and PaAbf62A displayed similar Km values in the micromolar range (1.2 and 6.1 mM, respectively) toward pNP-α-l-arabinofuranoside, PaAbf51A was approximately 60 times more active than PaAbf62A in terms of specific activity (Table 4). These arabinose-releasing enzymes exhibited low activity against polymeric substrates, preventing accurate determination of the individual kinetic parameters. HPAEC analysis of the reaction products generated by PaAbf51A and PaAbf62A revealed that they released only arabinose from polymeric substrates (wheat arabinoxylan, sugar beet arabinan, and debranched arabinan), thus demonstrating that they cleaved terminal units only (not shown).
Substrate-binding capacities of PaMan26A.
The ability of CBM35 harbored by PaMan26A to bind various insoluble polysaccharides was tested. Although under our experimental conditions PaMan26A did not bind to Avicel (crystalline cellulose), binding to mannan and xylan was observed. Based on typical double-reciprocal plots, binding parameters of PaMan26A were determined for mannan and xylan, yielding Kd values of 6.25 and 1.85 μM and Bmax values of 19 and 12 mg of PaMan26A per g of mannan and xylan, respectively (Table 5).
TABLE 5.
Binding parameters of P. anserina PaMan26A
| Condition | Binding parametera |
|
|---|---|---|
| Kd (μM) | Bmax (mg bound per g of substrate) | |
| Insoluble wheat xylan | 1.85 | 12 |
| Ivory nut mannan | 6.25 | 19 |
| Avicel PH101 | NB | NB |
The binding parameters were determined in triplicate, and the Kd and Bmax values were calculated using double-reciprocal plots. NB, no binding.
Supplementation of T. reesei enzyme cocktail with P. anserina hemicellulases.
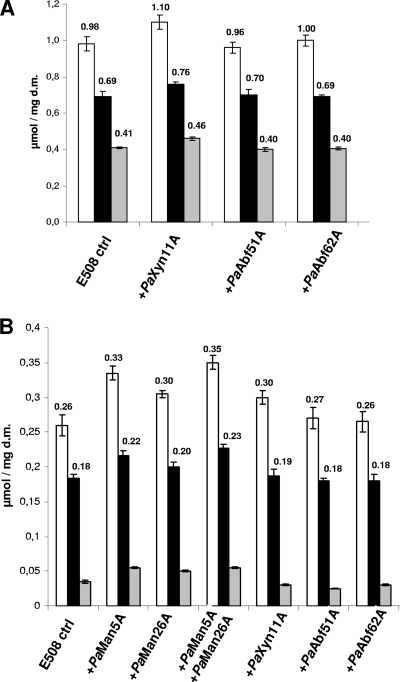
Each enzyme was added to the T. reesei enzymatic cocktail, E508, for the saccharification of micronized wheat straw and spruce. The release of both reducing sugars and individual sugar monomers was quantified by using the DNS assay and HPAEC-PAD, respectively, and compared to E508 (Fig. 4). The addition of PaXyn11A resulted in an increase in reducing sugars, glucose, and xylose of ca. 10% from micronized wheat straw. However, the addition of exo-acting enzymes did not affect the release of sugar monomers compared to T. reesei alone. The combination of PaAbf51A and PaAbf62A exo-acting enzymes with PaXyn11A did not improve the results obtained with the xylanase alone on micronized wheat straw (data not shown). Using spruce as lignocellulosic substrate, a 15% increase in total sugars was observed when PaXyn11A was added, with no effect on the glucose release. Supplementation of E508 with either PaMan5A or PaMan26A significantly increased the release of total sugars by 28 and 17%, respectively, and the release of glucose by 18 and 10%, respectively. Supplementation of E508 with a combination of PaMan5A and PaMan26A improved the release of total sugars by 34% and the release of glucose by 23%. Also, the addition of mannanase alone or in combination improved the release of xylose and mannose by 40 to 50% (Fig. 4).
FIG. 4.
Saccharification of lignocellulosic micronized substrates (wheat straw and spruce) by T. reesei enzymatic cocktail (E508) supplemented with P. anserina hemicellulases. Saccharification was carried out on wheat straw (A) and spruce (B) for 24 h at 40°C as described in Materials and Methods. The release of sugars was expressed in μmol per mg of dry matter (d.m.). White bars indicate the release of reducing sugars determined by using the DNS assay. Black bars indicate the release of glucose measured using HPAEC. Gray bars indicate the release of xylose from wheat straw (A) and the release of xylose plus mannose from spruce (B) measured using HPAEC. Values above each bar are means of sextuplicate measures, and error bars represent standard errors of the mean.
DISCUSSION
In the present study, we characterized five novel hemicellulases from different glycoside hydrolase families from P. anserina and assessed their individual contributions to the saccharification of LCB. P. pastoris was used as a heterologous expression system to express the set of P. anserina hemicellulases, since it is capable of secreting large amounts of correctly folded fungal proteins into the medium, with very few additional proteins (16), making it ideal for the production of recombinant fungal enzymes. It was recently used successfully to express a suite of polysaccharide-degrading enzymes from Aspergillus (3). Using this expression host, we efficiently produced and easily purified five fungal hemicellulases in one step, thus allowing their in-depth characterization.
PaMan5A and PaMan26A enzymes were both active on structurally different mannans. Determination of the kinetic parameters enabled us to demonstrate that their best substrate was the unsubstituted glucomannan in terms of affinity and catalytic efficiency, suggesting a negative impact of d-galactosyl side groups with hydrolysis for the galactomannan substrates as described for T. reesei mannanase (50). Although PaMan5A and PaMan26A displayed similar specificities toward a range of mannan substrates, they differed in terms of end product formation. PaMan26A released mannobiose and mannotriose from mannan polymer (ivory nut mannan), as reported for the mannanase from T. reesei (47). PaMan5A released mainly mannobiose without accumulation of mannotriose, as described for the recently characterized Aspergillus niger GH5 mannanase (18). These results indicate differences in the mode of action of substrate binding within the active sites of the two characterized P. anserina mannanases.
Our data strongly suggest that the N-terminal CBM35 module harbored by PaMan26A displays specificity toward hemicellulose polymers, mannan and xylan, with a similar constant Kd in the μM range. A CBM35 module is conserved in three Cellvibrio xylan-degrading enzymes and has been shown to bind uronic acid of xylan with a calcium-dependent interaction (38), while a module from a Cellvibrio mannanase binds to decorated soluble mannans and manno-oligosaccharides (52). Subtle changes in amino acid chemistry and conformation within the binding site of these bacterial CBM35s have radical impacts on their specificity (14). In fungi, only one CBM35 module binding to β-galactan has been characterized to date in a Phanerochaete chrysosporium galactan β-1,3-galactosidase (29).
Although family GH11 glycoside hydrolases are one of the best-characterized CAZy families, containing exclusively endo-xylanases, PaXyn11A was selected because it harbors a CBM1 module. This family module, formerly designated cellulose-binding domain family 1 (CBD1), is a well-characterized module specific to cellulose and exclusively in fungi (12). PaXyn11A xylanase exhibited an “endo” mode of action on polymeric substrates, releasing mainly xylobiose as major end product with a high catalytic efficiency, a common characteristic of fungal GH11 xylanases (4, 11).
Fungal species have been found to produce several arabinofuranosidase isoenzymes with different modes of action and substrate specificity (21). The PaAbf51A and PaAbf62A arabinofuranosidases characterized in the present study followed Michaelis-Menten kinetics toward pNP-Ara. Since the activity of these enzymes is of the “exo” type, very low activity was observed toward natural substrates. The kinetic properties of PaAbf51A toward pNP-Ara were comparable to the GH51 fungal enzymes characterized to date with a Km value in the mM range, an optimum pH around 5, and an optimum temperature above 50°C (21). P. anserina PaAbf62A displayed a 400-fold decrease in catalytic efficiency compared to its PaAbf51A arabinofuranosidase counterpart, mainly due to a low kcat value. This observed difference suggests that these two enzymes may act on different substitutions.
Genome sequencing of T. reesei has revealed the relatively small number of genes encoding enzymes involved in the breakdown of LCB in terms of diversity compared to other fungi. With only 16 genes encoding putative hemicellulases, T. reesei has the smallest set of hemicellulases of all of the saprophytic fungi analyzed (35). Among ascomycetes, the genome of P. anserina revealed the potential of this coprophilic fungus to hydrolyze recalcitrant lignocellulosic residues (19). It contains a significantly higher number of putative cellulases and xylanases compared to A. niger, A. nidulans and A. oryzae, reflected by its growth profile, which is better than Aspergillus species on cellulose and xylan (15). These observations fit well with the natural biotope of P. anserina, which is a late colonizer of herbivore dung, in which lignocellulose is the main remaining organic component. To our knowledge, none of the numerous putative glycoside hydrolases from different CAZy families encoded by the P. anserina genome have been characterized to date.
Since the T. reesei cocktail is xylanase-rich, the positive effect observed by adding PaXyn11A enzyme on the increase of both reducing sugars and glucose from wheat straw could be due to the presence of a CBM1 module linked to the enzyme that may contribute to the overall xylanase efficiency. To our knowledge, the T. reesei genome contains no gene encoding a GH11 xylanase harboring a CBM1. A possible explanation for the capacity of CBMs to increase the activity of glycoside hydrolases against insoluble substrates is that they reduce the “accessibility problem” by bringing the appended catalytic modules into intimate, prolonged association with their target substrate, thereby enhancing catalytic efficiency (23). Another hypothesis is that xylanases drastically change the structure and physicochemical properties of the AX population. Xylanases in the same GH family, i.e., with similar specificities, differ substantially in substrate selectivity, and the classification of GH family alone is not sufficient to predict the breakdown of insoluble AXs by xylanases (8). Although mechanistic information on the catalytic and substrate specificity of xylanases is helpful for comparing xylanase efficiencies, other factors, such as their degree of selectivity toward soluble versus insoluble substrate, may play a role in determining the functionality of these enzymes in the degradation of hemicellulose from raw materials (5). Although T. reesei has the smallest set of arabinofuranosidases (two GH43, no GH51, one GH62, and no GH93) of all plant cell wall-degrading ascomycetes (35), PaAbf51A and PaAbf62A did not potentiate the action of T. reesei enzymes in our experimental conditions.
In the present study, a positive effect on the release of reducing sugars and glucose from spruce was observed when T. reesei was supplemented with individual and combined addition of PaMan5A and PaMan26A mannanases. Galactoglucomannans are the main hemicellulose components in softwood (10 to 15% dry mass), with concentrations varying with the type of softwood and the location in the plant cell wall (39). Glucomannans can be found incorporated into aggregates of cellulose, as well as parallel to the cellulose fibrils, to which they are tightly connected (44). Our hypothesis is that PaMan5A and PaMan26A mannanases can facilitate the action of the T. reesei enzymatic cocktail by removing mannans that are found around cellulose fibrils. Also, T. reesei mannanase (TrMan5a) is expressed at low levels of secretion with only ∼0.02 U mg−1 detected in the E508 enzymatic cocktail. Although PaMan5A does not carry a CBM, the supplementation effect was higher than that found for PaMan26A. Further fusion of PaMan5A to a CBM specific for cellulose (CBM1) could be way to further enhance the positive effect observed in the present study. We recently demonstrated that the fusion of the Aspergillus niger mannanase (AnMan5) to a CBM1 module potentiates the action of the enzyme toward mannan-containing lignocellulosic substrates (42).
Helper enzymes can loosen up the lignocellulosic structure via degradation of hemicellulosic substrates (mannan/xylan), thus increasing the surface area of cellulose attacked by T. reesei cellulases. Because of the heterogeneity of hemicellulose between lignocellulosic substrates, enzymes with different characteristics are required in industrial applications, but the choice of the enzyme is still largely empirical (5). In the present study, hemicellulose-acting enzymes selected using comparative genomics brought a clearly marked improvement in both the liquefaction and the solubilization of LCB and the release of glucose and xylose/mannose when supplemented to T. reesei. The fact that this effect was more striking with the addition of mannanase might be due to the close association of softwood glucomannan with cellulose microfibrils, whereas xylan is interconnected to lignin. Since pretreatement of lignocellulose is also responsible for the high process cost of bioethanol production from LCB, helper enzymes (e.g., hemicellulases) could be an alternative to chemical and/or physical pretreatments. With addition of xylan- or mannan-acting enzymes to the actual cocktail, we could consider lowering the dosage of the T. reesei cocktail to achieve the same glucose yield (22, 37), since cellulose would be more accessible to cellulases. Tailor-made enzymatic cocktails need to be developed according to the lignocellulosic structure of each biomass for optimal hydrolysis.
The analysis of P. anserina genome has recently revealed a large and highly specialized set of genes involved in utilization of natural carbon sources (e.g., lignocellulose) commonly found in its natural biotope (19). The present findings pave the way for further studies to give more insights into P. anserina's enzymatic machinery. Among this unexpected unique enzymatic equipment, its genome presents a number of other interesting targets potentially acting on the recalcitrant components of plant cell wall. Finding complementary auxiliary enzymes displaying novel specificities will be required to deconstruct efficiently LCB for the production of biofuels and high-value products.
Acknowledgments
This study was funded by the French National Research Agency (ANR, program E-TRICEL ANR-07-BIOE-006).
We thank X. Rouau and G. Ghizzi for the preparation of micronized substrates, P. Silar for advice on P. anserina culturing methods, and D. Navarro for helpful discussions.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Akel, E., B. Metz, B. Seiboth, and C. P. Kubicek. 2009. Molecular regulation of arabinan and l-arabinose metabolism in Hypocrea jecorina (Trichoderma reesei). Eukaryot. Cell 8:1837-1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bailey, M. J., P. Biely, and K. Poutanen. 1992. Interlaboratory testing of methods for assay of xylanase activity. J. Biotechnol. 23:257-270. [Google Scholar]
- 3.Bauer, S., P. Vasu, S. Persson, A. J. Mort, and C. R. Somerville. 2006. Development and application of a suite of polysaccharide-degrading enzymes for analyzing plant cell walls. Proc. Natl. Acad. Sci. U. S. A. 103:11417-11422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berrin, J. G., H. el Ajandouz, J. Georis, F. Arnaut, and N. Juge. 2007. Substrate and product hydrolysis specificity in family 11 glycoside hydrolases: an analysis of Penicillium funiculosum and Penicillium griseofulvum xylanases. Appl. Microbiol. Biotechnol. 74:1001-1010. [DOI] [PubMed] [Google Scholar]
- 5.Berrin, J. G., and N. Juge. 2008. Factors affecting xylanase functionality in the degradation of arabinoxylans. Biotechnol. Lett. 30:1139-1150. [DOI] [PubMed] [Google Scholar]
- 6.Biely, P. 1993. Enzymological aspects of the production of microbial hemicellulases, p. 29-51. In M. P. Coughlan and G. P. Hazelwood (ed.), Hemicellulose and hemicellulases. Portland Press, London, England.
- 7.Bonnin, E., M. Brunel, Y. Gouy, L. Lesage-Meessen, M. Asther, and J.-F. Thibault. 2001. Aspergillus niger I-1472 and Pycnoporus cinnabarinus MUCL39533, selected for the biotransformation of ferulic acid to vanillin, are also able to produce cell wall polysaccharide-enzymes and feruloyl esterases. Enzyme Microb. Technol. 28:70-80. [DOI] [PubMed] [Google Scholar]
- 8.Bonnin, E., S. Daviet, J. F. Sorenson, O. Sibbesen, A. Goldson, N. Juge, and L. Saulnier. 2006. Behaviour of family 10 and 11 xylanases toward arabinoxylans with varying structure. J. Sci. Food Agric. 86:1618-1622. [Google Scholar]
- 9.Brutus, A., C. Villard, A. Durand, T. Tahir, C. Furniss, A. Puigserver, N. Juge, and T. Giardina. 2004. The inhibition specificity of recombinant Penicillium funiculosum xylanase B toward wheat proteinaceous inhibitors. Biochim. Biophys. Acta 1701:121-128. [DOI] [PubMed] [Google Scholar]
- 10.Cantarel, B. L., P. M. Coutinho, C. Rancurel, T. Bernard, V. Lombard, and B. Henrissat. 2009. The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37:D233-D238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cervera-Tison, M., G. Andre-Leroux, M. Lafond, J. Georis, N. Juge, and J. G. Berrin. 2009. Molecular determinants of substrate and inhibitor specificities of the Penicillium griseofulvum family 11 xylanases. Biochim. Biophys. Acta 1794:438-445. [DOI] [PubMed] [Google Scholar]
- 12.Chen, X. A., N. Ishida, N. Todaka, R. Nakamura, J. Maruyama, H. Takahashi, and K. Kitamoto. 2010. Promotion of efficient saccharification of crystalline cellulose by Aspergillus fumigatus swo1. Appl. Environ. Microbiol. 76:2556-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cherry, J. R., and A. L. Fidantsef. 2003. Directed evolution of industrial enzymes: an update. Curr. Opin. Biotechnol. 14:438-443. [DOI] [PubMed] [Google Scholar]
- 14.Correia, M. A., D. W. Abbott, T. M. Gloster, V. O. Fernandes, J. A. Prates, C. Montanier, C. Dumon, M. P. Williamson, R. B. Tunnicliffe, Z. Liu, J. E. Flint, G. J. Davies, B. Henrissat, P. M. Coutinho, C. M. Fontes, and H. J. Gilbert. 2010. Signature active site architectures illuminate the molecular basis for ligand specificity in family 35 carbohydrate binding module. Biochemistry 49:6193-6205. [DOI] [PubMed] [Google Scholar]
- 15.Coutinho, P. M., M. R. Andersen, K. Kolenova, P. A. vanKuyk, I. Benoit, B. S. Gruben, B. Trejo-Aguilar, H. Visser, P. van Solingen, T. Pakula, B. Seiboth, E. Battaglia, G. Aguilar-Osorio, J. F. de Jong, R. A. Ohm, M. Aguilar, B. Henrissat, J. Nielsen, H. Stalbrand, and R. P. de Vries. 2009. Post-genomic insights into the plant polysaccharide degradation potential of Aspergillus nidulans and comparison to Aspergillus niger and Aspergillus oryzae. Fungal Genet. Biol. 46:S161-S169. [DOI] [PubMed] [Google Scholar]
- 16.Daly, R., and M. T. Hearn. 2005. Expression of heterologous proteins in Pichia pastoris: a useful experimental tool in protein engineering and production. J. Mol. Recognit. 18:119-138. [DOI] [PubMed] [Google Scholar]
- 17.Decelle, B., A. Tsang, and R. K. Storms. 2004. Cloning, functional expression and characterization of three Phanerochaete chrysosporium endo-1,4-β-xylanases. Curr. Genet. 46:166-175. [DOI] [PubMed] [Google Scholar]
- 18.Do, B. C., T. T. Dang, J. G. Berrin, D. Haltrich, K. A. To, J. C. Sigoillot, and M. Yamabhai. 2009. Cloning, expression in Pichia pastoris, and characterization of a thermostable GH5 mannan endo-1,4-β-mannosidase from Aspergillus niger BK01. Microb. Cell Fact. 8:59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Espagne, E., O. Lespinet, F. Malagnac, C. Da Silva, O. Jaillon, B. M. Porcel, A. Couloux, J. M. Aury, B. Segurens, J. Poulain, V. Anthouard, S. Grossetete, H. Khalili, E. Coppin, M. Dequard-Chablat, M. Picard, V. Contamine, S. Arnaise, A. Bourdais, V. Berteaux-Lecellier, D. Gautheret, R. P. de Vries, E. Battaglia, P. M. Coutinho, E. G. Danchin, B. Henrissat, R. E. Khoury, A. Sainsard-Chanet, A. Boivin, B. Pinan-Lucarre, C. H. Sellem, R. Debuchy, P. Wincker, J. Weissenbach, and P. Silar. 2008. The genome sequence of the model ascomycete fungus Podospora anserina. Genome Biol. 9:R77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Foreman, P. K., D. Brown, L. Dankmeyer, R. Dean, S. Diener, N. S. Dunn-Coleman, F. Goedegebuur, T. D. Houfek, G. J. England, A. S. Kelley, H. J. Meerman, T. Mitchell, C. Mitchinson, H. A. Olivares, P. J. Teunissen, J. Yao, and M. Ward. 2003. Transcriptional regulation of biomass-degrading enzymes in the filamentous fungus Trichoderma reesei. J. Biol. Chem. 278:31988-31997. [DOI] [PubMed] [Google Scholar]
- 21.Fritz, M., M. C. Ravanal, C. Braet, and J. Eyzaguirre. 2008. A family 51 α-l-arabinofuranosidase from Penicillium purpurogenum: purification, properties and amino acid sequence. Mycol. Res. 112:933-942. [DOI] [PubMed] [Google Scholar]
- 22.Gao, D., S. P. Chundawat, C. Krishnan, V. Balan, and B. E. Dale. 2009. Mixture optimization of six core glycosyl hydrolases for maximizing saccharification of ammonia fiber expansion (AFEX) pretreated corn stover. Bioresour. Technol. 101:2770-2781. [DOI] [PubMed] [Google Scholar]
- 23.Gilbert, H. J. 2010. The biochemistry and structural biology of plant cell wall deconstruction. Plant Physiol. 153:444-455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girio, F. M., C. Fonseca, F. Carvalheiro, L. C. Duarte, S. Marques, and R. Bogel-Lukasik. 2010. Hemicelluloses for fuel ethanol: a review. Bioresour. Technol. 101:4775-4800. [DOI] [PubMed] [Google Scholar]
- 25.Guais, O., G. Borderies, C. Pichereaux, M. Maestracci, V. Neugnot, M. Rossignol, and J. M. François. 2008. Proteomics analysis of “Rovabio Excel”, a secreted protein cocktail from the filamentous fungus Penicillium funiculosum grown under industrial process fermentation. J. Ind. Microbiol. Biotechnol. 35:1659-1668. [DOI] [PubMed] [Google Scholar]
- 26.Hendriks, A. T., and G. Zeeman. 2009. Pretreatments to enhance the digestibility of lignocellulosic biomass. Bioresour. Technol. 100:10-18. [DOI] [PubMed] [Google Scholar]
- 27.Herpoel-Gimbert, I., A. Margeot, A. Dolla, G. Jan, D. Molle, S. Lignon, H. Mathis, J. C. Sigoillot, F. Monot, and M. Asther. 2008. Comparative secretome analyses of two Trichoderma reesei Rut-C30 and CL847 hypersecretory strains. Biotechnol. Biofuels 1:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Himmel, M. E., S. Y. Ding, D. K. Johnson, W. S. Adney, M. R. Nimlos, J. W. Brady, and T. D. Foust. 2007. Biomass recalcitrance: engineering plants and enzymes for biofuels production. Science 315:804-807. [DOI] [PubMed] [Google Scholar]
- 29.Ichinose, H., M. Yoshida, T. Kotake, A. Kuno, K. Igarashi, Y. Tsumuraya, M. Samejima, J. Hirabayashi, H. Kobayashi, and S. Kaneko. 2005. An exo-β-1,3-galactanase having a novel β-1,3-galactan-binding module from Phanerochaete chrysosporium. J. Biol. Chem. 280:25820-25829. [DOI] [PubMed] [Google Scholar]
- 30.Kauppinen, M. S., M. Schulein, K. Schnorr, L. N. Andersen, and M. E. Bjørnvad. 2003. Novel mannanases. U.S. patent 6566114.
- 31.Kellett, L. E., D. M. Poole, L. M. Ferreira, A. J. Durrant, G. P. Hazlewood, and H. J. Gilbert. 1990. Xylanase B and an arabinofuranosidase from Pseudomonas fluorescens subsp. cellulosa contain identical cellulose-binding domains and are encoded by adjacent genes. Biochem. J. 272:369-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, C. C., D. W. Wong, and G. H. Robertson. 2005. Cloning and characterization of the xyn11A gene from Lentinula edodes. Protein J. 24:21-26. [DOI] [PubMed] [Google Scholar]
- 33.Liu, J. R., C. H. Duan, X. Zhao, J. T. Tzen, K. J. Cheng, and C. K. Pai. 2008. Cloning of a rumen fungal xylanase gene and purification of the recombinant enzyme via artificial oil bodies. Appl. Microbiol. Biotechnol. 79:225-233. [DOI] [PubMed] [Google Scholar]
- 34.Margeot, A., B. Hahn-Hagerdal, M. Edlund, R. Slade, and F. Monot. 2009. New improvements for lignocellulosic ethanol. Curr. Opin. Biotechnol. 20:372-380. [DOI] [PubMed] [Google Scholar]
- 35.Martinez, D., R. M. Berka, B. Henrissat, M. Saloheimo, M. Arvas, S. E. Baker, J. Chapman, O. Chertkov, P. M. Coutinho, D. Cullen, E. G. Danchin, I. V. Grigoriev, P. Harris, M. Jackson, C. P. Kubicek, C. S. Han, I. Ho, L. F. Larrondo, A. L. de Leon, J. K. Magnuson, S. Merino, M. Misra, B. Nelson, N. Putnam, B. Robbertse, A. A. Salamov, M. Schmoll, A. Terry, N. Thayer, A. Westerholm-Parvinen, C. L. Schoch, J. Yao, R. Barabote, M. A. Nelson, C. Detter, D. Bruce, C. R. Kuske, G. Xie, P. Richardson, D. S. Rokhsar, S. M. Lucas, E. M. Rubin, N. Dunn-Coleman, M. Ward, and T. S. Brettin. 2008. Genome sequencing and analysis of the biomass-degrading fungus Trichoderma reesei (syn. Hypocrea jecorina). Nat. Biotechnol. 26:553-560. [DOI] [PubMed] [Google Scholar]
- 36.Matsushika, A., H. Inoue, T. Kodaki, and S. Sawayama. 2009. Ethanol production from xylose in engineered Saccharomyces cerevisiae strains: current state and perspectives. Appl. Microbiol. Biotechnol. 84:37-53. [DOI] [PubMed] [Google Scholar]
- 37.Meyer, A. S., L. Rosgaard, and H. R. Sørensen. 2009. The minimal enzyme cocktail concept for biomass processing. J. Cereal Sci. 50:337-344. [Google Scholar]
- 38.Montanier, C., A. L. van Bueren, C. Dumon, J. E. Flint, M. A. Correia, J. A. Prates, S. J. Firbank, R. J. Lewis, G. G. Grondin, M. G. Ghinet, T. M. Gloster, C. Herve, J. P. Knox, B. G. Talbot, J. P. Turkenburg, J. Kerovuo, R. Brzezinski, C. M. Fontes, G. J. Davies, A. B. Boraston, and H. J. Gilbert. 2009. Evidence that family 35 carbohydrate binding modules display conserved specificity but divergent function. Proc. Natl. Acad. Sci. U. S. A. 106:3065-3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Moreira, L. R., and E. X. Filho. 2008. An overview of mannan structure and mannan-degrading enzyme systems. Appl. Microbiol. Biotechnol. 79:165-178. [DOI] [PubMed] [Google Scholar]
- 40.Navarro, D., M. Couturier, G. Ghizzi Damasceno da Silva, J.-G. Berrin, X. Rouau, M. Asther, and C. Bignon. 2010. Automated assay for screening the enzymatic release of reducing sugars from micronized biomass. Microb. Cell Fact. 9:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Okazaki, F., Y. Tamaru, S. Hashikawa, Y. T. Li, and T. Araki. 2002. Novel carbohydrate-binding module of β-1,3-xylanase from a marine bacterium, Alcaligenes sp. strain XY-234. J. Bacteriol. 184:2399-2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pham, T. A., J. G. Berrin, E. Record, K. A. To, and J. C. Sigoillot. 2010. Hydrolysis of softwood by Aspergillus mannanase: role of a carbohydrate-binding module. J. Biotechnol. 148:163-170. [DOI] [PubMed] [Google Scholar]
- 43.Sakamoto, T., and H. Kawasaki. 2003. Purification and properties of two type-B α-l-arabinofuranosidases produced by Penicillium chrysogenum. Biochim. Biophys. Acta 1621:204-210. [DOI] [PubMed] [Google Scholar]
- 44.Salmen, L. 2004. Micromechanical understanding of the cell-wall structure. C. R. Biol. 327:873-880. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez, C. 2009. Lignocellulosic residues: biodegradation and bioconversion by fungi. Biotechnol. Adv. 27:185-194. [DOI] [PubMed] [Google Scholar]
- 46.Saulnier, L., P.-E. Sado, G. Branlard, G. Charmet, and F. Guillon. 2007. Wheat arabinoxylans: exploiting variation in amount and composition to develop enhanced varieties. J. Cereal Sci. 46:261-281. [Google Scholar]
- 47.Stålbrand, H., MattiSika-aho, M. Tenkanen, and L. Viikari. 1993. Purification and characterization of two β-mannanases from Trichoderma reesei. J. Biotechnol. 29:229-242. [Google Scholar]
- 48.Stålbrand, H., A. Saloheimo, J. Vehmaanperä, B. Henrissat, and M. Penttilä. 2005. Cloning and expression in Saccharomyces cerevisiae of a Trichoderma reesei beta-mannanase gene containing a cellulose binding domain. Appl. Environ. Microbiol. 61:1090-1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tabka, M. G., I. Herpoel-Gimbert, F. Monod, M. Asther, and J. C. Sigoillot. 2006. Enzymatic saccharification of wheat straw for bioethanol production by a combined cellulase xylanase and feruloyl esterase treatment. Enzyme Microb. Technol. 39:897-902. [Google Scholar]
- 50.Tenkanen, M., M. Makkonen, M. Perttula, L. Viikari, and A. Teleman. 1997. Action of Trichoderma reesei mannanase on galactoglucomannan in pine kraft pulp. J. Biotechnol. 57:191-204. [DOI] [PubMed] [Google Scholar]
- 51.Törrönen, A., R. L. Mach, R. Messner, R. Gonzalez, N. Kalkkinen, A. Harkki, and C. P. Kubicek. 1992. The two major xylanases from Trichoderma reesei: characterization of both enzymes and genes. Biotechnology 10:1461-1465. [DOI] [PubMed] [Google Scholar]
- 52.Tunnicliffe, R. B., D. N. Bolam, G. Pell, H. J. Gilbert, and M. P. Williamson. 2005. Structure of a mannan-specific family 35 carbohydrate-binding module: evidence for significant conformational changes upon ligand binding. J. Mol. Biol. 347:287-296. [DOI] [PubMed] [Google Scholar]
- 53.Waldron, K., and C. Faulds. 2007. Cell wall polysaccharides: composition and structure, p. 181-201. In J. Kamerling, G. J. Boons, Y. Lee, A. Suzuki, N. Taniguchi, and A. G. J. Voragen (ed.), Comprehensive glycoscience: analysis of glycans/polysaccharide functional properties, vol. 2. Elsevier B.V., Amsterdam, Netherlands. [Google Scholar]