Abstract
Atlantic killifish (Fundulus heteroclitus) inhabiting the PCB-contaminated Superfund site in New Bedford Harbor (MA, USA) have evolved genetic resistance to the toxic effects of these compounds. They also lack induction of cytochrome P4501A (CYP1A) and other aryl hydrocarbon receptor (AHR)-dependent responses after exposure to AHR agonists, suggesting an overall down-regulation of the AHR signaling pathway. In this study, we hypothesized that the genetic resistance is due to altered AHR expression resulting from hypermethylation of DNA in the promoter region of AHR genes in fish inhabiting New Bedford Harbor. To test this hypothesis, we cloned and sequenced AHR1 and AHR2 promoter regions and employed bisulfite conversion-polymerase chain reaction (BS-PCR) followed by clonal analysis to compare the methylation status of CpG islands of AHR1 and AHR2 in livers of adult killifish collected from New Bedford Harbor and a reference site (Scorton Creek, MA). No significant differences in methylation profiles were observed in either AHR1 or AHR2 promoter regions between NBH and SC fish. However, hypermethylation of the AHR1 promoter correlated with low expression of transcripts in the liver in both populations. In comparison to AHR1, hepatic mRNA expression of AHR2 is high and its promoter is hypomethylated. Taken together, our results suggest that genetic resistance to contaminants in NBH fish is not due to altered methylation of AHR promoter regions, but that promoter methylation may control tissue-specific expression of AHR genes in killifish.
Introduction
The Atlantic killifish, Fundulus heteroclitus, has become an important model species for studying the mechanisms of evolved resistance to toxicants (Hahn, 1998; Weis et al., 2001; Wirgin and Waldman, 2004; Burnett et al., 2007; Van Veld and Nacci, 2008). Populations of killifish inhabiting highly contaminated estuaries and coastal areas along the North Atlantic U.S. coast have evolved resistance to polynuclear aromatic hydrocarbons (PAHs), polychlorinated biphenyls (PCBs), and 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) (Nacci et al., 1999; Elskus et al., 1999; Powell et al., 2000; Bello et al., 2001; Meyer et al., 2002). One such heavily contaminated site is the Acushnet River estuary located near New Bedford Harbor (NBH), Massachusetts, which is characterized by very high PCB concentrations in the estuarine sediments and in the tissues of resident killifish (Lake et al., 1995; Bello et al., 2001; Nacci et al., 2002). In comparisons of F. heteroclitus populations on the east coast of the United States, NBH killifish are among the least sensitive to the biochemical and toxic effects of dioxin-like compounds (Nacci et al., 1999, 2002, 2010; Bello et al., 2001). The resistant phenotype is seen in embryonic and larval stages as well as in adult fish (Nacci et al., 1999; Bello et al., 2001).
The mechanism of action of certain PAHs, non-ortho-substituted PCBs, TCDD, and other dioxin-like compounds is under the control of the aryl hydrocarbon receptor (AHR) locus. The AHR protein is a ligand-activated transcription factor through which these compounds alter gene expression and cause toxicity. Upon ligand binding, AHR heterodimerizes with AHR nuclear translocator protein (ARNT), binds to the AHR response elements in the target genes, and initiates their transcription (Hankinson, 1995). Killifish express two AHR isoforms (AHR1 and AHR2), the products of distinct loci (Hahn et al., 1997; Karchner et al., 1999). AHR1 is expressed in a tissue-specific manner, whereas AHR2 is ubiquitously expressed (Karchner et al., 1999; Powell et al., 2000). Although the respective functions of these two AHR forms are not well understood (Merson et al., 2009), AHR2 appears to play a major role in mediating the developmental toxicity of PAHs and PCBs (Clark et al., 2010).
To elucidate the mechanistic basis of resistance in killifish, several studies have focused on AHR-dependent regulation of gene expression. One of the widely reported differences exhibited by resistant populations is the significantly reduced inducibility of cytochrome P4501A (CYP1A) as compared to the strong CYP1A inducibility in fish from uncontaminated sites (Van Veld and Westbrook, 1995; Nacci et al., 1999; Elskus et al., 1999; Powell et al., 2000; Bello et al., 2001). Increased CYP1A expression is regarded as a hallmark of AHR pathway activation. The refractory CYP1A phenotype has been observed in killifish populations inhabiting several Superfund sites including NBH, Newark Bay, NJ and Elizabeth River, VA, and resistance to CYP1A induction is highly correlated with resistance to embryotoxicity (Nacci et al., 2010).
The resistant phenotype of NBH killifish is heritable (Nacci et al., 2010). Studies in rodents have provided evidence that environmental toxicant-induced disease states can be transmitted through multiple generations and that the transgenerational effects may involve gene-specific changes in DNA methylation (Anway et al., 2005). This led to the hypothesis that epigenetic modifications such as DNA methylation of CYP1A1 promoter are responsible for stable long term transcriptional silencing of CYP1A gene expression in the resistant populations of killifish (Wirgin and Waldman, 2004; Arzuaga et al., 2004; Timme-Laragy et al., 2005). However, studies to date have found no evidence to support this hypothesis (Arzuaga et al., 2004; Timme-Laragy et al., 2005).
Recently, it has become evident that the resistance to gene induction in PCB- and PAH-resistant killifish populations is not restricted to CYP1A but occurs also for other AHR target genes, such as other CYP1 genes as well as AHRR (Karchner et al 2002; Wills et al., 2010). In addition, microarray-based gene expression profiling suggests that the resistant populations exhibit a genome-wide loss of responsiveness in AHR signaling (Whitehead et al., 2010; Oleksiak, Jenny, Karchner, & Hahn, manuscript in preparation). Thus, epigenetic mechanisms, if they are operating, are more likely to be upstream of AHR target genes, possibly affecting one or both AHRs themselves. The objective of the study described here was to test the hypothesis that fish from PCB-resistant and PCB-sensitive populations of killifish in NBH and Scorton Creek, MA (SC), respectively, exhibit different patterns of DNA methylation in the promoters of AHR genes.
DNA methylation (i.e., cytosine methylation) is defined as a covalent modification in which the 5’ position of cytosine is converted to 5’methylcytosine in a reaction catalyzed by DNA methyltransferases using S-adenosyl-methionine as the methyl donor (Razin and Riggs, 1980). High densities of CpG dinucleotides, commonly called CpG islands, are associated with the promoter regions of genes and are typically unmethylated in active genes (Gardiner-Garden and Frommer, 1987). Methylation of CpG islands located in the 5’ promoter region of genes has been associated with transcriptional inactivation (“silencing”) of genes. Aberrant de novo methylation of CpG islands is seen in several human cancers and silencing of tumor suppressor genes due to hypermethylation of CpG islands has been demonstrated (Feinberg, 2007). AHR expression can be regulated by changes in methylation of its promoter. For example, in human acute lymphoblastic leukemia cells, low constitutive AHR expression was shown to be due to a hypermethylated promoter region and this impaired the binding of transcription factors, such as Sp1, necessary for AHR expression (Mulero-Navarrao et al., 2006).
In this study, we tested the hypothesis that hypermethylation of hepatic AHR promoters is associated with decreased sensitivity to PCBs in NBH killifish compared to fish from a reference site, Scorton Creek, MA, USA. In order to test the hypothesis, we cloned and sequenced AHR1 and AHR2 promoters and then used bisulphite conversion of DNA followed by DNA sequencing of PCR products to analyze the methylation status of CpG islands in the promoter regions of livers of individual killifish from resistant (NBH) and sensitive (SC) populations. We analyzed CpG island methylation in the liver because it is an important organ involved in xenobiotic and energy metabolism, and because previous studies have demonstrated that this tissue is among those of NBH fish that are refractory to effects of dioxin-like compounds (Bello et al., 2001).
Materials and Methods
Animals
Adult Atlantic killifish were collected from NBH and SC in May 2009 using minnow traps as described previously (Karchner et al., 1999). Liver and brain tissues were dissected from 8-10 fish per site and quickly frozen in liquid nitrogen. Tissues were stored at -80°C until further analysis. All the animal husbandry practices followed were according to the regulations of the Animal Care and Use Committee of the Woods Hole Oceanographic Institution.
Genomic DNA isolation
Genomic DNA was isolated using the Nucleospin DNA trace kit (Macherey-Nagel, Germany) following manufacturer’s instructions. It involved proteinase K digestion followed by RNase treatment. The concentration of DNA was determined using a NanoDrop Spectrometer and A260/280 ratios were between 1.9-2.1. The quality of DNA was checked by running an aliquot on a 0.8% agarose gel and visualizing the DNA with ethidium bromide staining under UV light.
AHR promoter sequencing
AHR1 and AHR2 promoters were amplified using the Genome Walker (Clontech, California, USA) kit following manufacturer’s instructions. Briefly, the genomic DNA was digested by blunt end cutting restriction enzymes, DraI, PvuII, EcoRV and StuI and ligated with adaptors. This adaptor ligated DNA was used as a template in a primary PCR reaction with forward adaptor primer (AP1) and gene specific reverse primer (GSP1). The product of the primary PCR reaction was then diluted and used as a template for the secondary PCR with the nested adaptor primer (AP2) and nested gene-specific (GSP2) primer. The major PCR products were gel extracted using the Gene Clean II kit (MP Biomedicals, OH) and cloned into the pGEM-T easy vector (Promega, Wisconsin, USA) before sequencing. This whole procedure was repeated to obtain approximately 2kb of the 5’ promoter region. All the primer sequences used in promoter sequencing are listed in Table 1.
Table 1.
List of primers used to sequence AHR1 and AHR2 promoters using genome walker kit. GSP-gene specific primer; AP- adaptor primer
| Gene | Primer Sequence (5’-3’) | |
|---|---|---|
| AHR1 | GSP1 | CAGCATACATGACTGTTCCTTTTGTGTG |
| GSP2 | CTCTGGACGGGTTTTCTCCTCTTGCGTC | |
| AHR2 | GSP1 | GACGGGCTTCTTCCTCTTCT |
| GSP2 | CCGCTCGGTTCTTCTCAGT | |
| GSP3 | GACCGTTGACACCACAGCAT | |
| GSP4 | AACCTGCCTGCTGTGTTCCT | |
| AP1 | CCATCCTAATACGACTCACTATAGGGC | |
| AP2 | ACTCACTATAGGGCTCGAGCGGC |
Bisulphite conversion of DNA
Bisulphite conversion of DNA was done using the EZ methylation kit (Zymo Research Corporation, CA, USA) following instructions provided. Briefly, one microgram of genomic DNA was denatured by the addition of dilution buffer and incubation at 37°C for 15 minutes. Following denaturation, 100 μL of CT conversion reagent was added to the DNA and incubated in the dark for 3.5 hours at 65°C for bisulphite conversion. Bisulphite converted DNA was purified using spin columns and eluted from the column matrix in a total volume of 10 μL. BS-DNA was stored at -20°C for later use.
Identification of CpG islands in AHR promoter regions
The identification of CpG islands was based on the criteria by Takai and Jones (2002). They define CpG islands as being longer than 500 bp and having a GC content greater than 55% and an [observed CpG]/[expected CpG] ratio ≥ 0.65 (Takai and Jones, 2002). AHR promoter sequences were analyzed for CpG islands using the CpG Island searcher (http://cpgislands.usc.edu) using the above settings.
Bisulphite PCR (BS-PCR)
Methylation analysis of AHR CpG islands was performed by BS-PCR. A 25 μl PCR was carried out in 1X PCR buffer, 5 mM MgCl2, 1 mM dNTP mix, 1 unit of Taq polymerase, 50 pmol each of the forward primer and reverse primer and ~50 ng of bisulfite-treated genomic DNA. BS-PCR primers were designed using the sense strand of the bisulphite-converted DNA; the primer sequences are provided in Table 2. PCR cycling conditions were 94°C for 10 min, followed by 40 cycles of [94°C for 30 s, 55°C for 30 s and 72°C for 30 s], followed by 72°C for 8 min and stored at 4°C. PCR products were electrophoresed on 1% agarose gels, bands excised and gel extracted using the Gene Clean II kit. Purified PCR products were cloned using the pGEM-Teasy cloning kit as per the manufacturer’s protocol. Mini-preps were prepared using Pure Yield plasmid miniprep Kit (Promega). For each sample, a minimum of 5 clones were sequenced. BS-PCR together with sequencing of several clones provides allele-specific methylation profiles. This approach also helps in identifying single nucleotide polymorphisms (SNPs) within the CpG islands.
Table 2.
List of primers used to amplify CpG islands in AHR1 and AHR2 promoters. BS-PCR primers were designed based on bisulphite converted DNA template sequence. Genomic DNA corresponding to CpG islands were amplified using genomic DNA primers.
| Gene | CpG island | Primer Sequence (5’-3’) | |
|---|---|---|---|
| BS-PCR primers | |||
| AHR1 | CpG island I | Forward | GTTATGATGTATTTTTTTAATAAGTTGTTT |
| Reverse | CTAAACAACAAAAACTTTCTAACATAAC | ||
| AHR1 | CpG island II | Forward | GTTTTGTTTTATTTAAGTTGTTAGAGG |
| Reverse | ACAAAACCCAACACATCTCTTCTAC | ||
| AHR2 | CpG island | Forward | TATGTTTTTTTGAATTATGGTAATAG |
| Reverse | AACTTCTTCCTCTTCTTATTAAC | ||
| Genomic DNA primers | |||
| AHR1 | CpG island I | Forward | CAGTTGGCAGAACAGCAGATAG |
| Reverse | GTGAACATAGAGCTCCACAGCA | ||
| AHR1 | CpG island II | Forward | AGACATCTGCTTCCGTGTCTTT |
| Reverse | GAATCTTCCGCCTGTACTCATC | ||
| AHR2 | CpG island | Forward | GCAGCAGTATGCTGTGGTGT |
| Reverse | CCGCTCGGTTCTTCTCAGT | ||
Genomic regions corresponding to the CpG islands were also amplified from untreated genomic DNA for comparison purposes using primers designed based on a genomic DNA template (Table 2). PCR cycling conditions were 94°C for 10 min, 40 cycles of [94°C for 30 s, 60°C for 30 s and 72°C for 45 s], followed by 72°C for 8 min and stored at 4°C. Genomic PCR products were also cloned and a minimum of 5 clones were sequenced as described above. All the clones were sequenced using either SP6 or T7 primers. Sequencing was done on an ABI 3730×l DNA analyzer by Eurofins MWG Operon (Huntsville, AL).
Real-time RT-PCR
Total RNA was isolated using the standard protocol for RNA STAT60 (Tel-Test Inc., Texas, USA). cDNA was synthesized from 1 μg total RNA using random hexamers and the Omniscript cDNA Synthesis Kit (Qiagen, Valencia, CA). Quantitative PCR was performed using the iQ SYBR Green Supermix (Bio-Rad, Hercules, CA) in a MyiQ Single-Color Real-Time PCR Detection System (Bio-Rad). Real-time PCR primers for β-actin, AHR1 and AHR2 are listed in Table 3. The PCR conditions used were 95°C for 3 min and 95°C for 15 s/64°C (AHR1) or 66°C (AHR2 and β-actin) for 1 min (40 cycles). At the end of each PCR run, a melt curve analysis was performed to ensure that only a single product was amplified. Three technical replicates were used for each sample. Relative expression was normalized to that of β-actin (2-ΔCt; where ΔCt = [Ct(AHR) − Ct(β-actin)]. AHR1 and AHR2 mRNA expression levels in liver and brain were compared using paired t-test (GraphPad Prism version 5.3). A probability level of p < 0.01 was considered statistically significant.
Table 3.
Real-time PCR primers for AHR1 and AHR2.
| Gene | Primer Sequence (5’-3’) | |
|---|---|---|
| AHR1 | Forward | CAGGACTCCTCCCAAGAGATGG |
| Reverse | GAAGCTGCTCCGGGTTGTAGG | |
| AHR2 | Forward | GCAGTGATGTACAACCCTGAGC |
| Reverse | CCCGTGGAACTTCAGTGCCAGG | |
| β-actin | Forward | TGGAGAAGAGCTACGAGCTCC |
| Reverse | CCGCAGGACTCCATTCCGAG |
Results
CpG islands in AHR promoter regions
We cloned 2269 base pairs (bp) of the AHR1 promoter (GenBank accession number HQ241280). The AHR1 promoter region has two CpG islands of 489 bp (CpG island I; -1548bp to -2037bp) and 784 bp (CpG island II; -812bp to -28bp). CpG islands I and II have 17 and 27 CpG dinucleotides, respectively, based on the genomic DNA sequences (Fig. 1A).
Figure 1.
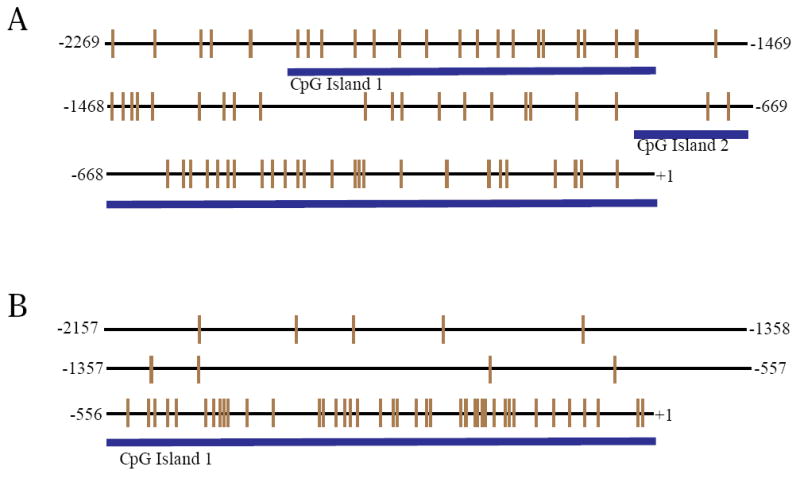
CpG islands in AHR promoters. Representation of CpG islands in the 5’ region of the AHR1 (A) and AHR2 (B) promoters. Each vertical line represents a single CpG dinucleotide. The numbers on the left and right sides indicate the relationship to the translational start site (ATG; +1).
The AHR2 promoter of 2157 bp length was cloned and sequenced (GenBank accession number HQ241281). The AHR2 promoter has only one CpG Island of 662 bp in length (-661bp - +1bp) with 41 CpG dinucleotides (Fig. 1B).
Single nucleotide polymorphisms (SNPs) (C/T and G/A substitutions) were identified in several of the CpG dinucleotides. These SNPs were found in both AHR1 and AHR2 CpG islands in both NBH and SC fish. The locations of SNPs are illustrated in Figs. 2-4.
Figure 2.
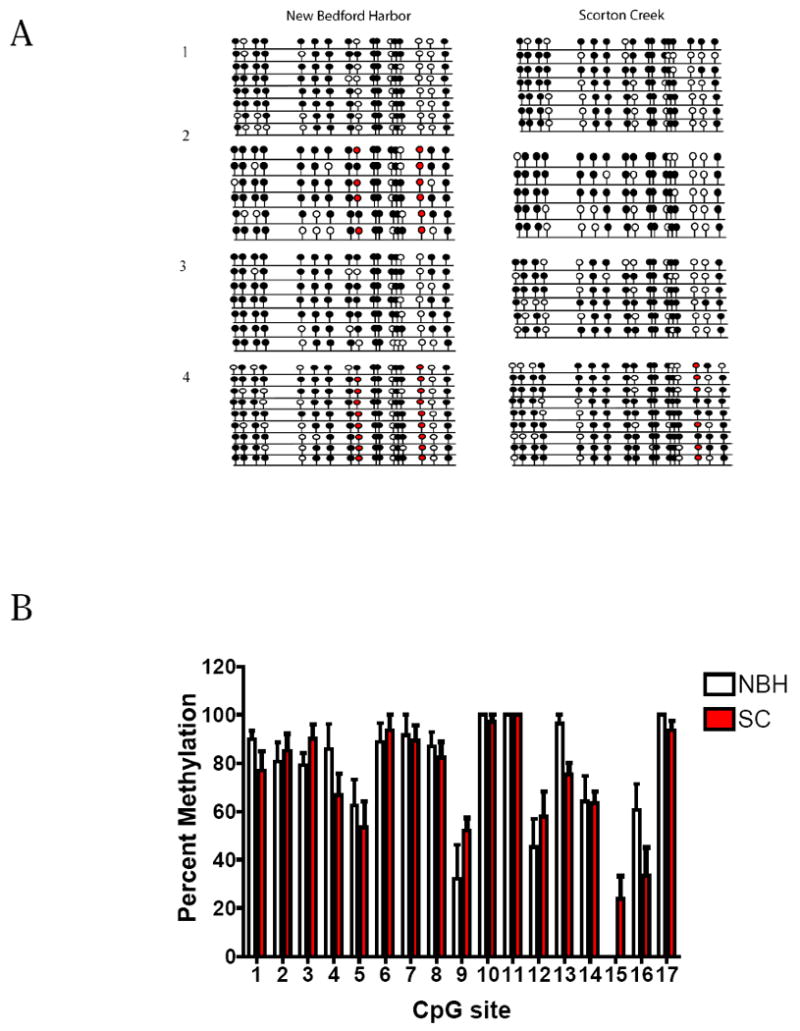
CpG island I in AHR1 promoter: Hepatic methylation patterns in NBH and SC fish. (A) Lollipop diagram showing the differences in methylation status between NBH and SC fish. Numbers 1-4 refer to individual fish from each site. Each line represents one sequenced clone. Each lollipop represents one CpG dinucleotide. Filled and open circles denote methylated and unmethylated sites, respectively. Single nucleotide polymorphisms (SNPs) are represented by red circles. (B) Histogram showing the percentage of methylation for each CpG site in CpG island I. Percent methylation was calculated by dividing the number of clones that were methylated at a particular site by the total number of clones sequenced and multiplying by 100. CpG sites with SNPs were not considered in calculating the percentage of methylation (N = 4 individual fish per site; 5-9 clones per fish). All values represent mean + Standard error of mean (S.E.M.). No significant differences in percentage of methylation were observed.
Figure 4.
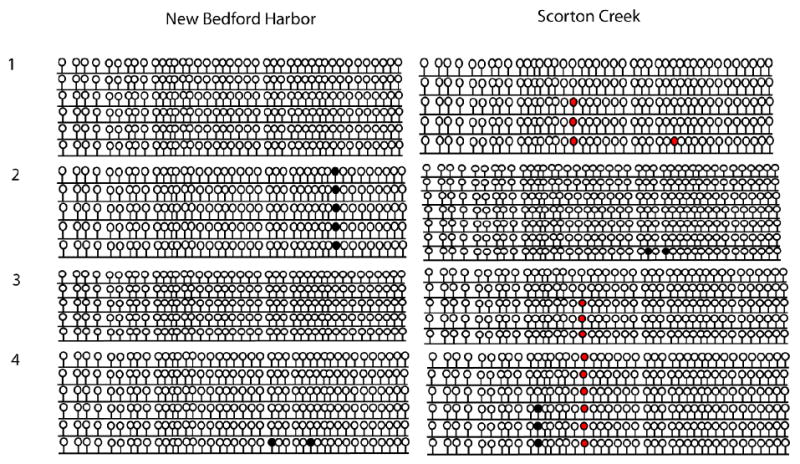
CpG island in AHR2 promoter: Hepatic methylation patterns in NBH and SC fish. Lollipop diagram showing methylation profile of each CpG dinucleotide. Filled and open circles represent methylated sites and unmethylated sites respectively. Red circles are the CpG positions where single nucleotide polymorphisms (SNPs) were observed. Methylation profiles were determined in 4 individual fish from each site and 5-7 clones were sequenced from each fish.
DNA methylation profile of the AHR1 promoter
AHR1 CpG island I was highly methylated, with ≥80% methylation at 11 of 17 CpG sites, with ≥50% methylation at most of the remaining sites (Fig. 2A,B). In contrast, CpG island II of AHR1 was highly unmethylated (Fig. 3). There were no significant differences between NBH and SC killifish in hepatic DNA methylation profiles for CpG islands I or II of AHR1 (Figs. 2,3).
Figure 3.
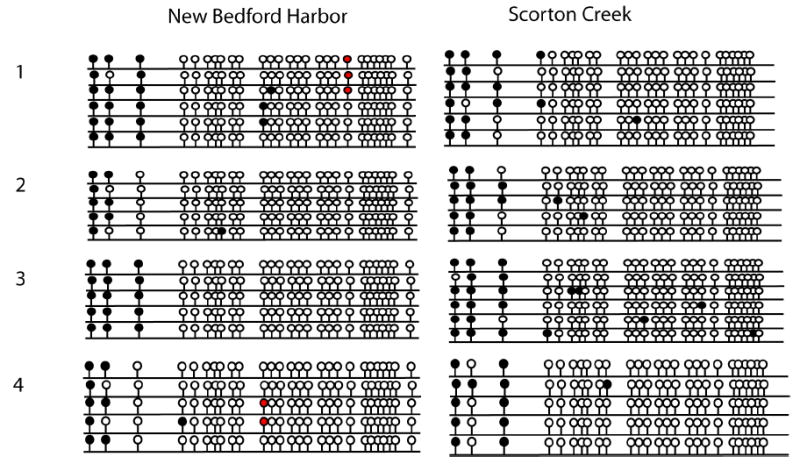
CpG island II in AHR1 promoter: Hepatic methylation patterns in NBH and SC fish. Lollipop diagram showing methylation profile of each CpG dinucleotide. Filled and open circles represent methylated sites and unmethylated sites respectively. Red circles are the CpG positions where single nucleotide polymorphisms (SNPs) were observed. Methylation profiles were determined in 4 individual fish from each site and 5 or 6 clones were sequenced from each fish.
DNA methylation profile of the AHR2 promoter
The CpG island in the AHR2 promoter was highly unmethylated (Fig. 4). There were no significant differences between NBH and SC killifish in hepatic DNA methylation profiles for the CpG island of AHR2 (Fig. 4).
AHR transcript levels
We observed tissue-specific differences in levels of AHR1 transcripts in both NBH and SC fish. AHR1 transcript levels were substantially lower (NBH: 85-fold; SC: 22-fold) in the liver in comparison to the brain (Fig. 5A). No significant differences in hepatic AHR1 transcripts were observed between the sites. In the brain, AHR1 levels were 69% higher in NBH fish than in fish from SC. AHR2 transcripts showed no tissue-specific or site-specific differences (Fig. 5B). A comparison of AHR1 and AHR2 levels in the liver suggests that AHR2 transcripts were more abundant (22-fold) than AHR1 transcripts in this tissue. A similar comparison in the brain revealed no significant differences in the relative abundance of AHR1 transcripts as compared to AHR2 transcripts.
Figure 5.
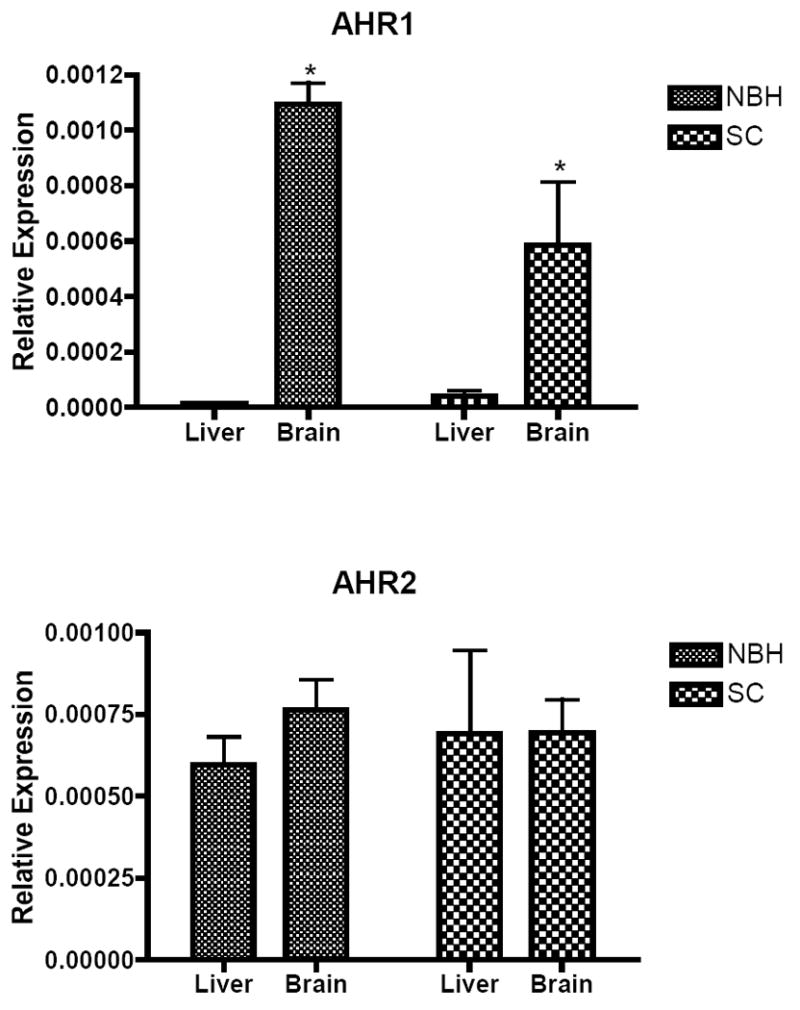
AHR1 and AHR2 transcript levels in liver and brain of NBH and SC fish. The delta Ct method (2 -ΔCt ; ΔCt = (AHR-β-actin)) was used to calculate the relative expression of AHR1 and AHR2 mRNA. All values represent mean + standard error of mean (S.E.M; n = 4-6). * denote statistically significant difference in AHR1 mRNA levels between liver and brain tissues. The same four samples that were used for determining methylation profiles were used to determine the transcript levels.
Discussion
The mechanisms underlying the heritable resistance to environmental chemicals in fish populations are not completely understood. Field and laboratory studies so far have demonstrated that killifish populations from Superfund sites have developed heritable resistance to AHR agonist-induced CYP1A expression (Van Veld and Nacci, 2008). Similarly, the expression of aryl hydrocarbon receptor repressor (AHRR), a key player in repressing AHR-mediated transcription, was not elevated in NBH fish (Karchner et al., 2002) and was also not inducible by PCB126 in the NBH progeny (Jenny M et al., unpublished). Recent microarray studies also suggest that the resistance to gene induction is not specific to CYP1A alone, but also is observed in several other genes likely to be under the control of AHRs (Whitehead et al., 2010).
In addition to aberrant transcriptional profiles of AHR-regulated genes, previous studies have also shown population specific differences in AHR mRNA profiles (Karchner et al., 1999; Powell et al., 2000). In adult killifish from SC, AHR2 was ubiquitously expressed, while AHR1 was expressed only in ovary, brain, heart and testis (Karchner et al., 1999). In contrast, in PCB-resistant NBH fish, AHR1 was expressed in almost all the tissues, suggesting that chronic exposure to contaminants causes altered AHR gene expression. This was hypothesized to be due to differences in the properties of the AHR1 promoter regions between the resistant and sensitive populations (Powell et al., 2000).
In this study we compared the DNA methylation profiles of AHR1 and AHR2 promoters in the livers of adult fish from NBH and SC to determine if differences in the methylation patterns are responsible for differential sensitivity to PCBs and for differences in AHR expression. DNA methylation of CpG islands in the promoter regions can interfere with binding of transcription factors by recruiting various methylated-DNA binding factors and changing the chromatin conformation from an active (euchromatin) to inactive (heterochromatin) state (Razin and Riggs, 1980; Feinberg, 2007). We did not observe any significant differences between NBH and SC fish in methylation patterns of CpG islands in either AHR gene, suggesting that the differential sensitivity is not due to changes in DNA methylation of hepatic AHR promoters.
Our studies focused on possible changes in DNA methylation of AHR promoters, but no population differences were found. Similarly, previous investigations of DNA methylation at killifish CYP1A promoters also showed no significant differences between contaminated (Elizabeth River, VA) and reference fish populations (King’s Creek) (Timme-Laragy et al., 2005). In addition, exposure of developing embryos to the DNA methylation inhibitor, 5-aza-CdR did not change their sensitivity to PCB-induced CYP1A catalytic activity in Newark Bay (NJ) resistant population (Arzuaga et al., 2004). Although we cannot rule out the possible role for AHR methylation in other tissues or life stages, these results suggest that resistance to AHR agonists is not due to aberrant promoter methylation, at least at these loci. Thus, the mechanism of resistance is more likely to be genetically based rather than epigenetic. Killifish from NBH, Newark Bay, and Elizabeth River have been chronically exposed to toxicants for several generations, providing strong selection pressures favoring fish with the resistant phenotype. Killifish are well known for developing genetically based adaptive phenotypic traits in response to environmental changes (Schulte et al., 2000). Population genetic studies currently underway could shed some light on the mechanisms involved in developing resistance (Hahn et al., 2004, 2005; Williams and Oleksiak, 2008; Williams et al., 2010).
Although we found no population-specific differences in methylation, patterns of methylation were AHR gene-specific and showed an inverse relationship with hepatic mRNA levels. The CpG island I in the AHR1 promoter was highly methylated in the liver and this corresponded with low AHR1 mRNA levels in this tissue, whereas the CpG island in the AHR2 promoter was unmethylated and this correlated with high levels of AHR2 transcripts in liver. Our results are in agreement with earlier findings that AHR1 is highly expressed in some extra-hepatic tissues but poorly expressed in the liver of fish from the sensitive fish populations from SC (Karchner et al., 1999; Powell et al., 2000). The present results suggest that these tissue-specific differences in AHR gene expression could be due to the methylation status of their promoter regions.
In an earlier study from our laboratory, Powell et al. (2000) reported that AHR1 mRNA levels in NBH fish showed aberrant widespread expression in several tissues, including liver. We did not observe high hepatic AHR1 expression in the NBH fish in the current study. This discrepancy could reflect differences in sample preparation. In this study, we quantified the mRNA levels using individual fish livers, whereas pooled liver samples were used in the earlier study. We have noticed differences in the expression patterns between individual fish from NBH, with a small percentage of fish expressing high levels of AHR1 (unpublished results) and this could have affected the earlier results obtained using pooled livers.
We identified SNPs within CpG dinucleotides in both AHR promoters; C/T and G/A were the most prevalent substitutions. SNPs in CpG islands have been extensively studied in the context of mutation-induced human diseases, because methylated cytosines are prone to mutations by spontaneous deamination to thymidine (Selker and Stevens, 1985; Cooper et al., 1987; Cooper and Krawczak, 1989). We did not see any population specific differences in these SNPs, but to understand their possible roles a more detailed population genetic analysis would need to be conducted. Previously, several SNPs have been identified within the AHR coding regions in this species, but functional studies revealed no differences in the ligand binding properties (Hahn et al., 2004). However, distinct patterns in the distribution of these alleles among sensitive and resistant populations have been observed (Hahn et al., 2005). Studies are currently in progress to ascertain the role of these SNPs in developing toxicant resistance.
In conclusion, our results demonstrate that there are no differences in hepatic AHR promoter methylation patterns between PCB-resistant (NBH) and PCB-sensitive (SC) fish populations. Our results agree with previous findings on the AHR mRNA expression patterns and we provide evidence that isoform-specific mRNA expression may be related to differences in DNA methylation of AHR promoter regions.
Acknowledgments
This work is funded in part by the Superfund Basic Research Program at Boston University to MEH (NIH Grant P42ES007381) and the postdoctoral scholar program at WHOI, with funding provided by the Dr. George D. Grice Postdoctoral Scholarship Fund to NA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Anway MD, Cupp AS, Uzumcu M, Skinner MK. Epigenetic transgenerational actions of endocrine disruptors and male fertility. Science. 2005;308:1466–1469. doi: 10.1126/science.1108190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arzuaga X, Calcaño W, Elskus A. The DNA de-methylating agent 5-azacytidine does not restore CYP1A induction in PCB resistant Newark Bay killifish (Fundulus heteroclitus) Mar Environ Res. 2004;58:517–520. doi: 10.1016/j.marenvres.2004.03.039. [DOI] [PubMed] [Google Scholar]
- Bello SM, Franks DG, Stegeman JJ, Hahn ME. Acquired resistance to aryl hydrocarbon receptor agonists in a population of Fundulus heteroclitus from a marine superfund site: In vivo and In vitro studies on the induction of xenobiotic metabolizing enzymes. Toxicol Sci. 2001;60:77–91. doi: 10.1093/toxsci/60.1.77. [DOI] [PubMed] [Google Scholar]
- Burnett KG, Bain LJ, Baldwin WS, Callard GV, Cohen S, Di Giulio RT, Evans DH, Gómez-Chiarri M, Hahn ME, Hoover CA, Karchner SI, Katoh F, Maclatchy DL, Marshall WS, Meyer JN, Nacci DE, Oleksiak MF, Rees BB, Singer TD, Stegeman JJ, Towle DW, Van Veld PA, Vogelbein WK, Whitehead A, Winn RN, Crawford DL. Fundulus as the premier teleost model in environmental biology: opportunities for new insights using genomics. Comp Biochem Physiol Part D Genomics Proteomics. 2007;2:257–286. doi: 10.1016/j.cbd.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BW, Matson CW, Jung D, Di Giulio RT. AHR2 mediates cardiac teratogenesis of polycyclic aromatic hydrocarbons and PCB-126 in Atlantic killifish (Fundulus heteroclitus) Aquat Toxicol. 2010;99:232–240. doi: 10.1016/j.aquatox.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Gerber-Huber S, Nardelli D, Schubiger JL, Wahli W. The distribution of the dinucleotide CpG and cytosine methylation in the vitellogenin family. J Mol Evol. 1987;25:107–115. doi: 10.1007/BF02101752. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Human Genet. 1989;83:181–188. doi: 10.1007/BF00286715. [DOI] [PubMed] [Google Scholar]
- Elskus AA, Monosson E, McElroy AE, Stegeman JJ, Woltering DS. Altered CYP1A expression in Fundulus heteroclitus adults and larvae: A sign of pollutant resistance? Aquat Toxicol. 1999;45:99–113. [Google Scholar]
- Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Hahn ME. Mechanisms of innate and acquired resistance to dioxin-like compounds. Rev Toxicol. 1998;2:395–443. [Google Scholar]
- Hahn ME, Karchner SI, Franks DG, Evans BR, Nacci D, Champlin D, Cohen S. Mechanism of PCB- and dioxin-resistance in fish in the Hudson River estuary: Role of receptor polymorphisms. Hudson River Foundation Grant 004/02A report. 2005 July;:33. ( http://www.hudsonriver.org/ls/)
- Hahn ME, Karchner SI, Franks DG, Merson RR. Aryl hydrocarbon receptor polymorphisms and dioxin resistance in Atlantic killifish (Fundulus heteroclitus) Pharmacogenetics. 2004;14:131–143. doi: 10.1097/00008571-200402000-00007. [DOI] [PubMed] [Google Scholar]
- Hahn ME, Karchner SI, Shapiro MA, Perera SA. Molecular evolution of two vertebrate aryl hydrocarbon (dioxin) receptors (AHR1 and AHR2) and the PAS family. Proc Natl Acad Sci USA. 1997;94:13743–13748. doi: 10.1073/pnas.94.25.13743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankinson O. The aryl hydrocarbon receptor complex. Annu Rev Pharmacol Toxicol. 1995;35:307–340. doi: 10.1146/annurev.pa.35.040195.001515. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Franks DG, Powell WH, Hahn ME. Regulatory interactions among three members of the vertebrates aryl hydrocarbon receptor family: AHR repressor, AHR1 and AHR2. J Biol Chem. 2002;277:6949–6959. doi: 10.1074/jbc.M110779200. [DOI] [PubMed] [Google Scholar]
- Karchner SI, Powell WH, Hahn ME. Structural and functional characterization of two highly divergent aryl hydrocarbon receptors (AHR1 and AHR2) in the teleost Fundulus heteroclitus. Evidence for a novel class of ligand-binding basis helix-loop-helix Per-ARNT-Sim(bHLH-PAS) factors. J Biol Chem. 1999;274:33814–33824. doi: 10.1074/jbc.274.47.33814. [DOI] [PubMed] [Google Scholar]
- Lake JL, McKinney R, Lake CA, Osterman FA, Heltshe J. Comparisons of patterns of polychlorinated biphenyl congeners in water, sediment, and indigenous organisms from New Bedford Harbor, Massachusetts. Arch Environ Contam Toxicol. 1995;29:207–220. [Google Scholar]
- Merson RR, Karchner SI, Hahn ME. Interaction of fish aryl hydrocarbon receptor paralogs (AHR1 and AHR2) with the retinoblastoma protein. Aquat Toxicol. 2009;94:47–55. doi: 10.1016/j.aquatox.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer JN, Nacci DE, Di Giulio RT. Cytochrome P4501A (CYP1A) in killifish (Fundulus heteroclitus): heritability of altered expression and relationship to survival in contaminated sediments. Toxicol Sci. 2002;68:69–81. doi: 10.1093/toxsci/68.1.69. [DOI] [PubMed] [Google Scholar]
- Mulero-Navarro S, Carvajal-Gonzalez JM, Herranz M, Ballestar E, Fraga MF, Ropero S, Esteller M, Fernandez-Salguero PM. The dioxin receptor is silenced by promoter hypermethylation in human acute lymphoblastic leukemia through inhibition of Sp1 binding. Carcinogenesis. 2006;27:1099–1104. doi: 10.1093/carcin/bgi344. [DOI] [PubMed] [Google Scholar]
- Nacci D, Coiro L, Champlin D, Jayaraman S, McKinney R, Gleason T, Munns WR, Jr, Specker JL, Cooper K. Adaptation of wild populations of the estuarine fish Fundulus heteroclitus to persistent environmental contaminants. Mar Biol. 1999;134:9–17. [Google Scholar]
- Nacci D, Coiro L, Champlin D, Jayaraman S, McKinney R. Predicting the occurrence of genetic adaptation to dioxin-like compounds in populations of the estuarine fish Fundulus heteroclitus. Environ Toxicol Chem. 2002;21:1525–1532. [PubMed] [Google Scholar]
- Nacci DE, Champlin D, Jayaraman S. Adaptation of the estuarine fish Fundulus heteroclitus (Atlantic killifish) to polychlorinated biphenyls (PCBs) Estuaries and Coasts. 2010;33:853–864. [Google Scholar]
- Powell WH, Bright R, Bello SM, Hahn ME. Developmental and tissue-specific expression of AHR1, AHR2, and ARNT2 in dioxin-sensitive and -resistant populations of the marine fish Fundulus heteroclitus. Toxicol Sci. 2000;57:229–239. doi: 10.1093/toxsci/57.2.229. [DOI] [PubMed] [Google Scholar]
- Razin A, Riggs AD. DNA methylation and gene function. Science. 1980;210:604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Schulte PM, Glemet HC, Fiebig AA, Powers DA. Adaptive variation in lactate dehydrogenase-B gene expression: Role of a stress-responsive regulatory element. Proc Natl Acad Sci USA. 2000;97:6597–6602. doi: 10.1073/pnas.97.12.6597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selker EV, Stevens JN. DNA methylation at asymmetric sites is associated with numerous transition mutations. Proc Natl Acad Sci USA. 1985;82:8114–8118. doi: 10.1073/pnas.82.23.8114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takai D, Jones P. Comprehensive analysis of CpG islands in human chromosomes 21 and 22. Proc Natl Acad Sci USA. 2002;99:3740–3745. doi: 10.1073/pnas.052410099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Timme-Laragy AR, Meyer JN, Waterland RA, Di Giulio RT. Analysis of CpG methylation in the killifish CYP1A promoter. Comp Biochem Physiol C Toxicol Pharmacol. 2005;141:406–411. doi: 10.1016/j.cbpc.2005.09.009. [DOI] [PubMed] [Google Scholar]
- Van Veld PA, Nacci D. Chemical tolerance: Acclimation and adaptations to chemical stress. In: Di Guilio RT, Hinton DE, editors. The Toxicology of Fishes. Washington, D.C.: Taylor and Francis; 2008. pp. 597–644. [Google Scholar]
- Van Veld PA, Westbrook DJ. Evidence for depression of cytochrome P4501A in a population of chemically resistant mummichog (Fundulus heteroclitus) Environ Sci. 1995;3:221–234. [Google Scholar]
- Weis JS, Smith G, Zhou T, Santiago-Bass C, Weis P. Effects of contaminants on behavior: Biochemical mechanisms and ecological consequences. BioScience. 2001;51:209–217. [Google Scholar]
- Whitehead A, Deborah T, Champlin D, Nacci D. Comparative transcriptomics implicates mechanisms of evolved pollution tolerance in a killifish population. Mol Ecol. doi: 10.1111/j.1365-294X.2008.03942.x. [DOI] [PubMed] [Google Scholar]
- Williams LM, Oleksiak MF. Signatures of selection in natural populations adapted to chronic pollution. BMC Evolutionary Biology. 2008;8:282. doi: 10.1186/1471-2148-8-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams LM, Ma X, Boyko AR, Bustamante CD, Oleksiak MF. SNP identification, verification, and utility for population genetics in a non-model genus. BMC Genetics. 2010;11:32. doi: 10.1186/1471-2156-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills LP, Matson CW, Landon CD, Di Giullio RT. Characterization of the recalcitrant CYP1 phenotype found in Atlantic killifish (Fundulus heteroclitus) inhabiting a Superfund site on the Elizabeth River, VA. Aquat Toxicol. 2010;99:33–41. doi: 10.1016/j.aquatox.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirgin I, Waldman JR. Resistance to contaminants in North American fish populations. Mutat Res. 2004;552:73–100. doi: 10.1016/j.mrfmmm.2004.06.005. [DOI] [PubMed] [Google Scholar]


