Abstract
Antibody-mediated killing of Streptococcus pneumoniae (pneumococcus) by phagocytes is an important mechanism of protection of the human host against pneumococcal infections. Measurement of opsonophagocytic antibodies by use of a standardized opsonophagocytic assay (OPA) is important for the evaluation of candidate vaccines and required for the licensure of new pneumococcal conjugate vaccine formulations. We assessed agreement among six laboratories that used their own optimized OPAs on a panel of 16 human reference sera for 13 pneumococcal serotypes. Consensus titers, estimated using an analysis-of-variance (ANOVA) mixed-effects model, provided a common reference for assessing agreement among these laboratories. Agreement was evaluated in terms of assay accuracy, reproducibility, repeatability, precision, and bias. We also reviewed four acceptance criterion intervals for assessing the comparability of protocols when assaying the same reference sera. The precision, accuracy, and concordance results among laboratories and the consensus titers revealed acceptable agreement. The results of this study indicate that the bioassays evaluated in this study are robust, and the resultant OPA values are reproducible for the determination of functional antibody titers specific to 13 pneumococcal serotypes when performed by laboratories using highly standardized but not identical assays. The statistical methodologies employed in this study may serve as a template for evaluating future multilaboratory studies.
Antibody-mediated killing of Streptococcus pneumoniae (pneumococcus) by phagocytes involving complement components C3b, iC3b, and C3d is generally the primary mechanism of protection of the human host against pneumococcal infections (24). Accurate measurement of antibodies that can efficiently opsonize and fix complement onto the surface of the pneumococcus is desirable when the functional capacity of antibodies in circulation is being measured. Public health strategies for increasing the concentration of circulating antibodies specific to the predominant serotypes causing disease have involved vaccination with polysaccharide and polysaccharide protein conjugate vaccines that have been licensed for use at different stages of life (4, 5, 6). The successful implementation of these vaccination strategies, particularly using protein conjugate vaccines that range in valency from 7 to 13 serotypes for the infant population, have led to a decrease in the incidence of pneumococcal disease caused by serotypes in the vaccine in those countries that have introduced pneumococcal conjugate vaccines into the infant immunization schedule (7, 15, 16). Improved vaccines with wider serotype coverage are being developed, and licensure of these new conjugate vaccine formulations will be based on head-to-head studies including the licensed formulation and using noninferiority and immunogenicity endpoint data (17, 23).
Functional antibody activity can be measured in the laboratory using cultured phagocytic cell lines and a standardized source of complement (baby rabbit complement) in an opsonophagocytosis assay (OPA) (8, 21). A standardized OPA is needed to consistently evaluate functional antibody activity. The measurement of antibodies specific to the capsular polysaccharides by an enzyme-linked immunosorbent assay (ELISA) (antibody binding assay) (12, 22) is the accepted population-based correlate of protection for invasive pneumococcal disease for the licensure of new vaccine formulations for infants. OPA reflects in vivo mechanisms of defense against pneumococcal infection and is recognized as being increasingly important for regulatory purposes, especially for evaluating the serotypes in extended formulations that are not present in PCV7. Several modifications to the OPA first described by Romero-Steiner et al. (19) have been described (9, 11). Perhaps the most significant modification has been the capacity to measure functional antibodies in multiplex formats to evaluate functional responses to four or more serotypes and reduce the number of opsonophagocytic assays for the evaluation of multivalent vaccines (1-3, 14, 15). As experience with this assay progressed, individual laboratories adapted the killing OPA to utilize HL-60 granulocytes as effector cells, replacing peripheral blood leukocytes, and optimized other reagents and protocol steps for their own assays.
Romero-Steiner et al. (20) previously conducted a well-controlled multilaboratory study where five laboratories used the same assay protocol, reagents, and serum samples to evaluate the assay and measure opsonophagocytic antibodies specific to capsular polysaccharides. They concluded that a standardized OPA could be performed in multiple laboratories with a high degree of interlaboratory reproducibility.
In the present study, we assess agreement among six laboratories using their own standardized OPA protocols without any common reagents other than the serum samples to be evaluated. No specific acceptance criteria were preset prior to submission of the data results to the CDC for analysis. Several analytical approaches were used to improve the interpretation and analysis methodologies employed in the first multilaboratory evaluation of the pneumococcal OPA (20). We conclude that there is an acceptable level of agreement among laboratories using standardized but nonuniform OPAs.
MATERIALS AND METHODS
Study design.
OPA titers were measured for a panel of 24 reference sera (19 unique and 5 random repeat serum samples) obtained from D. Goldblatt (UCL Institute of Child Health) and distributed by the National Institute for Biological Standards and Control (NIBSC; Potters Bar, Hertfordshire, United Kingdom), United Kingdom. Five laboratories (laboratories A to E) assayed all 24 reference specimens, and one laboratory (laboratory F) assayed the 19 unique specimens. There were 3 prevaccination and 21 postvaccination serum samples following administration of a licensed 23-valent pneumococcal polysaccharide vaccine (Pneumovax II; Pasteur Mérieux, Lyon, France, or Merck Sharp and Dohme, Ltd.). The three preimmunization sera were removed from the analysis because the majority of the assayed values were at or below the minimum measurable titer (MMT). This resulted in 16 unique postvaccination serum samples to be included in the final analysis. These sera were collected from healthy adults after receipt of informed consent at the Oxford Blood Transfusion Service, Oxford, United Kingdom. The subjects agreed upon the use of their sera for experimental purposes according to good clinical practice and informed consent guidelines. These quality control sera are currently available at the NIBSC for use in pneumococcal assay standardization. Sera were lyophilized in 2-ml aliquots and stored at −20°C until they were used by the participating laboratories. Reconstituted samples were frozen at −70°C until the day of testing. Each laboratory included an internal quality control serum for quality assurance of the assay.
All specimens were assayed in duplicate. Initially, five laboratories (laboratories A to E) assayed the specimens for seven pneumococcal serotypes (serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F). These serotypes are included in the licensed pneumococcal conjugate vaccine (Prevnar; Wyeth-Lederle). Laboratories A, B, C, and D assayed the specimens for six additional pneumococcal serotypes (serotypes 1, 3, 5, 6A, 7F, and 19A). An additional laboratory, laboratory F, entered the study at a later date and assayed the seven serotypes common to laboratories A to E plus five additional serotypes (serotypes 1, 5, 6A, 7F, and 19A). Laboratory F was blinded to specimen-specific assay results from laboratories A to E prior to submitting their data, as were laboratories A to E from all other laboratories. Worksheets with OPA colony counts were sent to the Bacterial Respiratory Pathogen Reference Laboratory at the University of Alabama at Birmingham (Birmingham, AL) for calculation of OPA titers. These results were then sent to the CDC for final statistical analysis.
Opsonophagocytosis assay.
The type of OPA evaluated was the killing assay with specific modifications described previously (summarized in Table 1). Three laboratories performed the MOPA4 assay, which allowed for 4 serotypes to be tested simultaneously by using antibiotic selection and antibiotic-resistant strains (3). None of the antibiotics used for selection are commonly used in pneumococcal treatment. The Radboud University Nijmegen Medical Centre, Netherlands, performed the OPA described by Burton et al. (3), with the following modification: during the preparation of the bacterial working stocks, the strains were grown to optical densities at 600 nm (OD600s) of 0.1 to 0.2, and bacterial counts were determined at the University of Alabama at Birmingham. The minimum measurable titers were 4 for three laboratories (laboratories A, C, and F) and 8 for the remaining three laboratories (laboratories B, D, and E). In addition, laboratories A and F recorded values above their upper limit as “>16,570” and “>8,748,” respectively, and these titers were set to twice those amounts for analysis purposes. Each serum was assayed in duplicate. Replicates with fold differences greater than 8 were excluded from the analysis, as these sera would have been reassayed under a more rigorous quality control protocol. A total of 37 such replicate pairs among all serotypes were excluded, representing 2.6% of the data.
TABLE 1.
Summary of protocol differences for laboratories participating in the multilaboratory pneumococcal OPA studya
| Protocol variable | Protocol for indicated laboratoryb |
|||||
|---|---|---|---|---|---|---|
| I | II | III | IV | V | VI | |
| Multiplex OPA (MOPA4) (M) or singleplex (S) | S | M | S | S | M | M |
| CDC, mixture (M), or other (O) target strains | M | O | M | CDC | O | O |
| Cultured (C) or frozen (F) target bacteria in assay | F | C | C | F | C | C |
| CDC HL60 differentiation protocol | Yes | Yes | Yes | Yes | Yes | Yes |
| HL60 passage number monitored | Yes | Yes | Yes | Yes | Yes | Yes |
| Baby rabbit (BR) or normal rabbit (NR) serum | BR | NR | BR | BR | BR | BR |
| UAB, CDC, or in-house (I) assay utilized | I | UAB | I | CDC | UAB | UAB |
| Assay protocol reference(s) | 9 | 3 | 11, 19 | 19 | 3 | 3 |
The sequence of laboratories does not correspond to their alphabetic identifications in the text, tables, and figures. UAB, University of Alabama at Birmingham.
I, GlaxoSmithKline Biologicals, Belgium; II, Radboud University Nijmegen Medical Centre, Netherlands; III, Pfizer Vaccine Research, Pearl River, NY; IV, National Institute for Health and Welfare, Finland; V, Institute of Child Health, University College London, England; VI, Bacterial Respiratory Pathogen Reference Laboratory, University of Alabama.
Table 1 presents a summarization of the protocol differences for the participating laboratories and reference OPAs utilized in this study. All participating laboratories performed either a validated OPA or a highly qualified OPA. To ensure the anonymity of the laboratories, their sequence in Table 1 does not correspond to their alphabetic identifications in the text, tables, and figures.
Statistical analysis.
All OPA titers were calculated as continuous titers as previously described (3), using the Opsotiter1 software program at the University of Alabama at Birmingham. All OPA titers were transformed to log base 2 values prior to analysis. The 16 unique reference sera do not have known OPA titer assignments, and hence, “consensus” OPA titer values were estimated using an analysis-of-variance (ANOVA) mixed-effects model from the present data and used to quantify accuracy, reproducibility, repeatability, precision, and bias within and among the six laboratories. Accuracy is defined as the closeness of a laboratory-assayed value to the consensus value and is measured using Lin's coefficient of accuracy (Ca) (13). Precision measures how far a set of observations deviates from a straight line and is quantified using Pearson's correlation coefficient (r). Lin's concordance correlation coefficient (rc), which is a combination of Ca and r, was employed to form a single statistic describing both accuracy and precision. Reproducibility is a measure of between-laboratory or interlaboratory variation and represents an estimate of the true error of the assay. Repeatability is a measure of within-assay or intra-assay variation (within a laboratory) and is estimable because assays were performed in duplicate. Bias is a measure of the directional error (consistent offset) of the laboratory titer compared to the consensus OPA titer (10).
Linear mixed-effects ANOVA models were used to estimate consensus values for each serum and serotype. These models also provided estimates of assay repeatability and reproducibility. All models were fit independently by serotype and included the sample as a fixed effect and laboratory and the laboratory-sample interaction as random effects. Since each serum was run in duplicate, a single predicted titer was estimated using the ANOVA models to represent the duplicate values for analysis and comparison of laboratories in the figures and tables.
We used scatter plots, plots of accuracy (box plots), precision, bias, variance components, Pearson correlation coefficients (r), coefficients of accuracy (Ca), and concordance correlation coefficients (rc). Our assessment and evaluation of the ability of the six laboratories to reproduce the titers among themselves and against the consensus OPA titers used the estimated values from the ANOVA models. Laboratory bias was quantified using the random-effect coefficients from the ANOVA models and by comparing the observed laboratory values to the consensus values. We believe that these methods are generalizable and may be used as a template in the analysis of other multilaboratory studies.
RESULTS
For the 16 serum samples included in the final data, the total recorded MMT measurements and the percentages of the total assayed titers for laboratories A to F were 14 (2.6%), 42 (7.7%), 38 (7.0%), 45 (8.2%), 47 (16.0%), and 18 (4.7%), respectively. The total number of OPA titers at or below the MMT were as follows: for serotype 1, 43 (21.5%); for serotype 3, 11 (6.5%); for serotype 4, 19 (7.9%); for serotype 5, 14 (7.0%); for serotype 6A, 11 (5.5%); for serotype 6B, 30 (12.4%); for serotype 7F, 2 (1.0%); for serotype 9V, 6 (2.5%); for serotype 14, 0 (0.0%); for serotype 18, 9 (3.7%); for serotype 19A, 6 (3.0%); for serotype 19F, 16 (6.6%); and for serotype 23F, 37 (15.3%). In addition, laboratory A recorded eight titers as “>16,570” for serotype 14, and these titers were set to twice that amount (33,140) for analysis purposes.
ANOVA model laboratory-predicted and consensus OPA titers.
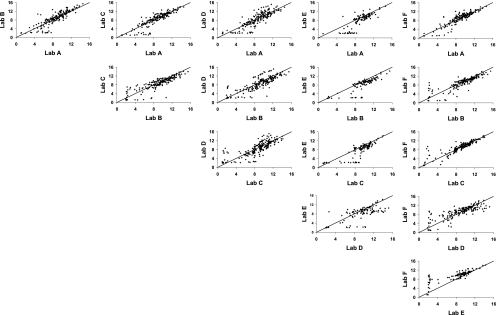
ANOVA models were used to estimate single predicted OPA titers for each serum by laboratory and serotype from the duplicate titers submitted for analysis and for the estimation of the consensus OPA titers for each serum by serotype. Scatter plots comparing these predicted serum OPA titers between laboratories and aggregated over all serotypes are presented in Fig. 1. The line of identity represents perfect agreement (intercept = 0; slope = 1). In general, most laboratory-to-laboratory comparisons yielded clusters of points centered over the line of identity, indicating good agreement. However, titers for laboratory E were lower than those for the other laboratories, as indicated by the shift in the point clusters away from the line. In addition, some laboratories were able to produce measurable titers for samples that other laboratories recorded as MMTs. This is shown by the line of points with titers near 2 along the x and y axes of the comparisons.
FIG. 1.
Scatter plots of pairwise comparison between laboratories aggregated over serotype. Predicted OPA titers derived from ANOVA random-effects models were used for each set of assay duplicates. The solid line indicates perfect agreement (intercept 0 and slope 1).
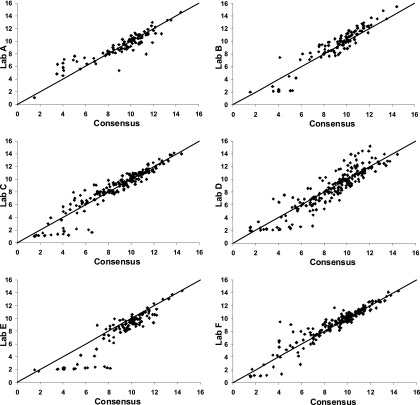
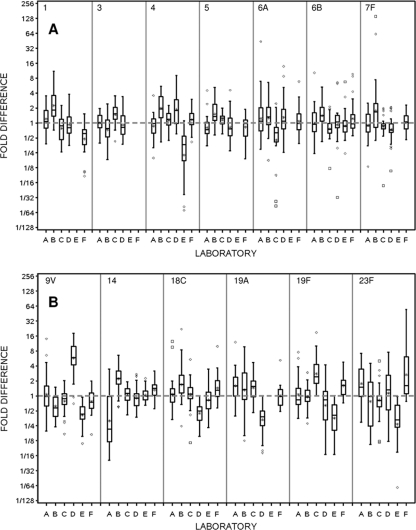
Figure 2 displays the comparison of laboratory-predicted and consensus titers. Laboratory D exhibits slightly more overall variability than the other laboratories, and titers from laboratory E tend to be lower than the consensus values. Box plots displaying the distribution of fold differences between the individual laboratory-reported and consensus OPA titers by serotype are presented in Fig. 3. The distance of the mean (*) from the gray dotted line at a 1-fold difference is a direct measure of the mean bias within a laboratory for each serotype. The size of the box, coupled with the extensions of the vertical lines above and below the box, is a direct indicator of the intralaboratory or within-laboratory variability of the fold differences (repeatability). As an example, small boxes centered about the gray dotted line, with vertical lines extending between 1/2 and 2, indicate a distribution where the laboratory-observed titers were within ±1 titer (2-fold difference) of the consensus value. The positioning of the boxes about the gray dotted line for a given serotype across all laboratories is an indicator of the between-laboratory variability (reproducibility). Laboratory A has a substantial negative mean bias and high variability for serotype 14 and a moderate negative mean bias for serotype 5, but with low variability. In addition, laboratory A has positive mean biases for serotypes 19A and 23F, with moderate variability. Laboratory B has noticeable positive/negative mean biases for all serotypes except 19F and shows increased variability for serotypes 19A and 23F. Laboratory C has a moderate amount of mean bias but generally shows the smallest amount of variability around the consensus titers. Laboratory D exhibits a severe positive bias with serotype 9V and negative biases for serotypes 18C and 19A. Laboratory E has the greatest degree of overall mean bias, underestimating titers for serotypes 4, 9V, 19F, and 23F. Overall, laboratory F exhibits a minor mean bias for serotypes 1 and 23F and, with the exception of serotype 23F, displays small amounts of variability in the fold differences for the remaining serotypes.
FIG. 2.
Scatter plots of pairwise comparisons between laboratories and consensus values aggregated over serotypes. Predicted OPA titers for each laboratory and consensus titers were derived from ANOVA random-effects models. The solid line indicates perfect agreement (intercept 0 and slope 1).
FIG. 3.
Box plots by serotype and laboratory for the fold differences between the consensus and observed OPA titers. Consensus OPA titers were estimated for each sample within a serotype using the random-effects ANOVA model. In these plots, the box is defined by the 25th and 75th percentiles of the distribution; the horizontal line within the box represents the median or 50th percentile, and the asterisk signifies the mean. Vertical lines extend to the most extreme observation that is less than 1.5× the interquartile range (75th to 25th percentiles), and the diamonds and boxes correspond to moderate and severe outlying assay values, respectively.
Interlaboratory and laboratory-to-consensus agreement for OPA titers.
Table 2 presents accuracy (Ca), precision (r), and concordance (rc) measures of agreement between pairs of laboratories and between laboratories and consensus OPA titers. While laboratory E has a definite systematic bias, laboratories A, B, C, D, and F all perform comparably to each other for precision, accuracy, and concordance. Laboratory E consistently underestimated OPA titers compared to the other laboratories, as seen in Fig. 1 and 2, and this is reflected in Table 2, with the lowest values for Ca and rc. The rc value for laboratory E is less than 0.80 (range, 0.67 to 0.78), whereas the rc values are >0.80 for the remaining five laboratories. Similarly, comparison of the results for the laboratories with the consensus OPA titers (Table 2) reveals that laboratory E has the lowest accuracy (0.92) and has the least amount of concordance (0.85) with the consensus values. In contrast, all other laboratories have accuracy values close to 1.0, with concordance values of >0.90.
TABLE 2.
Comparison of OPA titers between laboratories and laboratory-to-consensus OPA titersa
| Laboratory | Statisticb | Value for indicated laboratory |
|||||
|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | ||
| A | Ca | 1.0 | 0.978 | 0.987 | 0.976 | 0.857 | 0.993 |
| r | 1.0 | 0.875 | 0.887 | 0.852 | 0.811 | 0.901 | |
| rc | 1.0 | 0.856 (0.816-0.888) | 0.875 (0.840-0.903) | 0.831 (0.785-0.867) | 0.695 (0.605-0.767) | 0.894 (0.861-0.920) | |
| B | Ca | 1.0 | 0.992 | 0.990 | 0.911 | 0.992 | |
| r | 1.0 | 0.866 | 0.835 | 0.853 | 0.859 | ||
| rc | 1.0 | 0.859 (0.818-0.891) | 0.827 (0.777-0.866) | 0.777 (0.698-0.837) | 0.852 (0.806-0.888) | ||
| C | Ca | 1.0 | 0.998 | 0.921 | 0.996 | ||
| r | 1.0 | 0.805 | 0.833 | 0.911 | |||
| rc | 1.0 | 0.803 (0.749-0.847) | 0.768 (0.689-0.829) | 0.907 (0.878-0.930) | |||
| D | Ca | 1.0 | 0.934 | 0.988 | |||
| r | 1.0 | 0.712 | 0.827 | ||||
| rc | 1.0 | 0.666 (0.553-0.754) | 0.817 (0.763-0.860) | ||||
| E | Ca | 1.0 | 0.839 | ||||
| r | 1.0 | 0.818 | |||||
| rc | 1.0 | 0.686 (0.596-0.759) | |||||
| F | Ca | 1.0 | |||||
| r | 1.0 | ||||||
| rc | 1.0 | ||||||
| Consensus value | Ca | 0.996 | 0.985 | 0.997 | 0.990 | 0.921 | 0.998 |
| r | 0.952 | 0.945 | 0.946 | 0.911 | 0.921 | 0.946 | |
| rc | 0.948 (0.932-0.960) | 0.931 (0.910-0.947) | 0.943 (0.927-0.956) | 0.902 (0.875-0.924) | 0.848 (0.805-0.882) | 0.944 (0.923-0.959) | |
Consensus OPA titers were estimated by sample within a serotype by use of the random-effects ANOVA model. Predicted OPA titers were obtained for each laboratory by sample within a serotype for each of the duplicates by use of the random-effects ANOVA model. Values in parentheses are 95% confidence intervals.
Ca, accuracy; r, precision; rc, concordance correlation coefficient.
Repeatability and reproducibility.
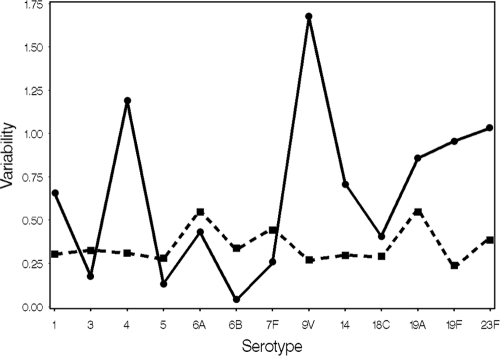
Intralaboratory (repeatability) and interlaboratory (reproducibility) variances are diagramed in Fig. 4. Serotype 9V shows the greatest interlaboratory variability and is influenced by the extreme positive bias in laboratory D for this serotype (Fig. 3). The interlaboratory variance for serotype 9V is reduced by >85% when laboratory D is removed from the analysis, but the intralaboratory variance remains virtually unchanged. If laboratory E is removed from the analysis, then the interlaboratory variability levels are reduced by >82%, >34%, and >58% for serotypes 4, 19F, and 23F, respectively. Repeatability values are similar across serotypes and reflect each laboratory's ability to replicate its results, which are stable across serotypes.
FIG. 4.
Plots of within-laboratory (repeatability [▪]) and between-laboratory (reproducibility [•]) variability by serotype.
Within-laboratory bias per serotype was estimated from the random-effect ANOVA models and illustrated using box plots of titer fold differences (Fig. 3). The mean bias varied greatly across serotypes and laboratories. Bias quantified by serotype and laboratory revealed that for the 71 possible laboratory-serotype combinations, 13 (18.3%) had a mean bias greater than a 2-fold difference and that 2 (2.8%) had a mean bias greater than a 4-fold difference (Fig. 3). There were no systematic patterns, as these represented all laboratories and 8 serotypes. Across all serotypes, laboratories A, C, and D exhibit the lowest degrees of bias, with deviations from consensus titers of 0.01, 0.03, and −0.04, respectively. Laboratories B and F exhibit moderately higher mean bias, with deviations of 0.40 and 0.17, respectively. Laboratory E has a relatively high systematic mean bias compared to the other laboratories, with a deviation of −1.0 compared to the consensus titers. Within a serotype, the expectation is that the mean bias is 0, and this is one of the assumptions of the random-effect ANOVA models. The average absolute bias is generally less than 0.5 deviations from the consensus titers, with a range of 0.25 to 0.88. Serotypes 6B and 9V have the lowest and highest absolute differences, respectively.
Consensus OPA titers and intervals.
Four separate prediction intervals (PIs) were formed about the consensus values for each of the 16 samples and 13 serotypes. These intervals may be used to judge whether future assays generate results comparable to those produced for this study. The 95% and 80% PIs were derived from the ANOVA models and reflect the variability of the titers reported in the present study. We also constructed two nonparametric intervals, representing ±2- and ±4-fold differences from the consensus values, which may also be used as guides for future assays. We calculated the percentages of observed OPA titers that fell within each of the intervals (Table 3). The results illustrate that the ±2-fold-difference interval likely captured less than a desirable percentage of the observed data. Overall, the 80% and 95% prediction intervals and ±4-fold-difference intervals all perform similarly. The 80% prediction interval captured >85% of the observed data for all serotypes and exhibits little variability among the serotypes for the percentages captured (the range is 85.3 to 96.4). In contrast, ±4-fold-difference intervals captured an adequate percentage but exhibited more variability among the serotypes for the percentages, with a range of 72.1 to 98.2.
TABLE 3.
Overall percentages of observed OPA titers that fall within the defined interval aggregated over serotype and sample
| Serotype | % of titers within: |
|||
|---|---|---|---|---|
| 95% PIa | 80% PI | ±2-fold-difference interval | ±4-fold-difference interval | |
| 1 | 96.9 | 90.8 | 68.2 | 92.8 |
| 3 | 98.1 | 92.6 | 79.0 | 98.2 |
| 4 | 95.7 | 92.7 | 63.4 | 89.7 |
| 5 | 97.4 | 87.2 | 80.6 | 97.5 |
| 6A | 97.2 | 90.1 | 63.0 | 89.5 |
| 6B | 95.7 | 91.4 | 79.3 | 93.5 |
| 7F | 97.4 | 96.4 | 76.5 | 96.4 |
| 9V | 99.2 | 85.3 | 54.2 | 80.7 |
| 14 | 97.3 | 90.6 | 75.3 | 92.8 |
| 18C | 97.4 | 89.8 | 69.8 | 91.5 |
| 19A | 98.5 | 89.7 | 59.3 | 86.1 |
| 19F | 98.3 | 87.1 | 56.2 | 84.1 |
| 23F | 96.9 | 87.3 | 52.8 | 72.1 |
PI, prediction interval.
DISCUSSION
To our knowledge, this is the first time a study has been conducted to measure the agreement among a series of laboratories running standardized but nonuniform OPAs that have been optimized in each individual laboratory. In this multilaboratory study, we utilize a comprehensive statistical analysis plan to analyze OPA titers generated by six laboratories to measure the performance of their well-controlled opsonophagocytosic killing assays. Each laboratory used its own established acceptance criteria for reporting titers. We estimated consensus OPA titers for a panel of 16 reference sera for 13 pneumococcal serotypes and evaluated four distinct intervals, which could serve as guides to determine if future assays deliver results comparable to those reported here. This study expands on a previous report by Romero-Steiner et al. (20) by providing a statistical framework which may serve as a template for the analysis of multilaboratory studies. With laboratories using the same assay protocol and reagents, Romero-Steiner et al. report that 88% of sera fell within 2 dilutions of the median titer for seven serotypes. This criterion was previously applied in the report by Romero-Steiner et al. to give an estimate of agreement between laboratories (20). In the present study, laboratories using their own OPAs without any common regents produced results which fell within ±4-fold differences from the consensus values 88% of the time for 12 of 13 serotypes. The results fell within ±4-fold differences from the consensus values 80% of the time for 12 of 13 serotypes (Table 3). As in the previous report, the level of agreement varied by serotype and by participating laboratory. In this study, one laboratory (laboratory E) had a lower level of agreement with the consensus titers by both parametric and nonparametric analyses.
ANOVA mixed modeling is a flexible framework that allows estimation of serum sample titers for each serotype by laboratory. These models may be used to compare and contrast results within and among laboratories. Random-effects ANOVA models allowed us to partition the total variance to measure reproducibility (interassay variability) and repeatability (intra-assay variability). The actual OPA titers for the reference sera used in this study were unknown, so it was not possible to compare experimentally derived titers to a “true” value. The ANOVA mixed model provided a mechanism for estimating consensus values, which served as assigned values for these sera (see Table S1 in the supplemental material).
Our statistical analyses indicated good agreement among the six laboratories participating in this study. Concordance was high among laboratories (Fig. 1 and Table 2) and between results for laboratories and consensus OPA titers (Fig. 2 and Table 2). Concordance is a combined measurement of agreement, i.e., it combines accuracy and precision and in this study was calculated across all serotypes and samples by laboratory. Precision, as measured by the Pearson correlation coefficient and the concordance correlation coefficient, is sensitive to the range and heterogeneity of the data. As the range of measurements increases among a collection of samples, precision, accuracy, and concordance will generally increase. In this study, the range of measurement among sample consensus titers varies by serotype. Serotype-specific coefficients of variation (CVs) using the sample consensus values ranged from 17.3% to 53.2%, with most less than 35.0% (data not shown). This suggests that our data do not suffer from a high degree of sample heterogeneity and that the concordance correlation coefficient adequately reflects the degree to which these laboratories are able to achieve similar values for the reference sera.
Parameter-based prediction intervals can serve as guidance for other laboratories performing OPA validation and trying to assess the variability of their own assays. These intervals differ from the conventional confidence interval, which describes the estimated mean. The use of intervals as a guide for future assay results has been proposed in previous studies using ELISAs to measure antibody concentrations. Plikaytis et al. (18) used a nonparametric acceptance interval of ±40% about the median ELISA concentration value, and Romero-Steiner et al. (20) suggested using ±2- or ±4-fold-difference intervals for OPA. In both of these studies, the assay protocols were uniform and were strictly adhered to by all participating laboratories. No such requirements were present in the current study, which added to the variability of the results. Our data suggest that a 40% range and a ±2-fold-difference interval about the consensus values would be too narrow, given the inherent variability of the OPA assays among these laboratories. The 40% range captured an average of 48% of the measured OPA titers over all serotypes (data not shown). Nonparametric acceptance intervals based on a range of ±2- and ±4-fold differences captured averages of 67.5% and 89.6% of the measurement data, respectively. The 80% and 95% prediction intervals covered 90.1% and 97.4% of the observed data (Table 3). Parameter-based prediction intervals like those estimated by the ANOVA mixed model may be preferred because they account for the variability about the consensus titer and are dependent upon the variability within and between laboratories, samples, and serotypes. In contrast, acceptance intervals based on nonparametric ranges or cutoff values are independent of these different sources of variability. The FDA has created a new panel of 16 reference sera that will supplement and ultimately replace the sera used in this study. Additionally, five laboratories, which include several of the authors in this study, are currently involved in a bridging exercise to replace the pneumococcal reference serum 89SF with a new human reference serum, 007sp (12a). In this process, OPA bioassays will be performed on the new FDA sera, and these data will be used to establish consensus OPA values and derive optimal methodologies to define acceptance criteria.
Our primary study goal was to assess the level of agreement among laboratories using their own standardized and optimized opsonophagocytic killing assays, but not a uniform assay, on a collection of shared specimens from individuals vaccinated with a 23-valent pneumococcal polysaccharide vaccine. A total of 2.6% (37 pairs) of the data were excluded from the analysis because the members of each pair were more than 8-fold apart. However, many replicate pairs included in the analysis still displayed elevated degrees of variability, and the degree of observed agreement among laboratories would improve, as observed in the multilaboratory study by Romero-Steiner et al. (20), if rigorous acceptance criteria were included in the assay protocols requiring repeat testing for those specimens flagged with excessive replicate variability. A secondary goal was to investigate different methodologies to develop prediction intervals for this panel of specimens that could serve as guides for laboratories establishing opsonophagocytic killing assays and to determine if their individual assays are performing in accordance with the reference criteria presented here.
This study was designed to determine whether variations made to the OPA first described by Romero-Steiner et al. (19) compromised interlaboratory agreement. While the laboratories involved in the study by Romero-Steiner et al. used the exact same protocols with shared reagents, the OPAs used in the present study were more disparate, with some laboratories moving substantially away from the original protocol described by Romero-Steiner et al. The results of this study indicate that despite variations made to the original assay, the opsonophagocytic killing assays evaluated in this study are robust and reproducible for the determination of functional antibody titers specific to 13 pneumococcal serotypes when performed by laboratories using highly standardized controlled assays.
Supplementary Material
Acknowledgments
This project was funded in part by NIH contract AI-30021 (NIH/NIAID/DMID) to the University of Alabama at Birmingham (M.H.N.).
We acknowledge the significant contribution to the study concept, design, and analysis by Milan Blake and are deeply saddened by his untimely death.
Footnotes
Published ahead of print on 17 November 2010.
Supplemental material for this article may be found at http://cvi.asm.org/.
REFERENCES
- 1.Bieging, K., et al. 2005. A fluorescent multivalent opsonophagocytic assay for the measurement of functional antibodies to Streptococcus pneumoniae. Clin. Diagn. Lab. Immunol. 12:1238-1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bogaert, D., M. Sluijter, R. de Groot, and P. W. M. Hermans. 2004. Multiplex opsonophagocytosis assay (MOPA): a useful tool for the monitoring of the 7-valent pneumococcal conjugate vaccine. Vaccine 4563:1-7. [DOI] [PubMed] [Google Scholar]
- 3.Burton, R. L., and M. H. Nahm. 2006. Development and validation of a fourfold multiplex opsonization assay (MOPA4) for pneumococcal antibodies. Clin. Vaccine Immunol. 13:1004-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 1997. Prevention of pneumococcal disease: recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend. Rep. 46(RR-8):1-24. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2000. Preventing pneumococcal disease among infants and young children. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recommend. Rep. 49(RR-09):1-38. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2010. Licensure of a 13-valent pneumococcal conjugate vaccine (PCV13) and recommendations for use among children—Advisory Committee on Immunization Practices (ACIP), 2010. MMWR Morb. Mortal. Wkly. Rep. 59(9):258-261. [PubMed] [Google Scholar]
- 7.Centers for Disease Control and Prevention. 2008. Progress in introduction of pneumococcal conjugate vaccine—worldwide, 2000-2008. MMWR Morb. Mortal. Wkly. Rep. 57:1148-1151. [PubMed] [Google Scholar]
- 8.Fleck, R. A., S. Romero-Steiner, and M. H. Nahm. 2005. Use of HL-60 cell line to measure opsonic apacity of pneumococcal antibodies. Clin. Diagn. Lab. Immunol. 12:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Henckaerts, I., N. Durant, N. D. De Grave, L. Schuerman, and J. Poolman. 2007. Validation of a routine opsonophagocytosis assay to predict invasive pneumococcal disease efficacy of conjugate vaccine in children. Vaccine 25:2518-2527. [DOI] [PubMed] [Google Scholar]
- 10.Heyden, Y., and J. Smeyers-Verbeke. 2007. Set-up and evaluation of interlaboratory studies. J. Chromatogr. A 1158:158-167. [DOI] [PubMed] [Google Scholar]
- 11.Hu, B. T., et al. 2005. Approach to validating an opsonophagocytic assay for Streptococcus pneumoniae. Clin. Diagn. Lab. Immunol. 12:287-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jódar, L., et al. 2003. Serological criteria for evaluation and licensure of new pneumococcal conjugate vaccine formulations for use in infants. Vaccine 21:3265-3272. [DOI] [PubMed] [Google Scholar]
- 12a.Lee, L. 2010. Abstr. 7th Int. Symp. Pneum. Pneum. Dis., abstr. 160.
- 13.Lin, L. I. K. 1989. A concordance correlation coefficient to evaluate reproducibility. Biometrics 45:255-268. [PubMed] [Google Scholar]
- 14.Nahm, M. H., D. E. Briles, and X. Yu. 2000. Development of a multi-specificity opsonophagocytic killing assay. Vaccine 18:2768-2771. [DOI] [PubMed] [Google Scholar]
- 15.O'Brien, K. L., et al. 2009. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet 374:893-902. [DOI] [PubMed] [Google Scholar]
- 16.Pilishvili, T., et al. 2010. Sustained reductions in invasive pneumococcal disease in the era of conjugate vaccine. J. Infect. Dis. 201:32-41. [DOI] [PubMed] [Google Scholar]
- 17.Plikaytis, B. D., and G. M. Carlone. 2005. Statistical considerations for vaccine immunogenicity trails. Part 2: non-inferiority and other statistical approaches to vaccine evaluation. Vaccine 23:1606-1614. [DOI] [PubMed] [Google Scholar]
- 18.Plikaytis, B. D., et al. 2000. An analytical model applied to a multicenter pneumococcal ELISA study. J. Clin. Microbiol. 38:2043-2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Romero-Steiner, S., et al. 1997. Standardization of an opsonophagocytic assay for the measurement of functional antibody activity against Streptococcus pneumoniae using differentiated HL-60 cells. Clin. Diagn. Lab. Immunol. 4:415-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Romero-Steiner, S., et al. 2003. Multilaboratory evaluation of a viability assay for measurement of opsonophagocytic antibodies specific to the capsular polysaccharides of Streptococcus pneumoniae. Clin. Diagn. Lab Immunol. 10:1019-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romero-Steiner, S., et al. 2006. Use of opsonophagocytosis for serological evaluation of pneumococcal vaccines. Clin. Vaccine Immunol. 13:165-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Siber, G. R., et al. 2007. Estimating the protective concentration of anti-pneumococcal capsular polysaccharide antibodies. Vaccine 25:3816-3826. [DOI] [PubMed] [Google Scholar]
- 23.World Health Organization. 2009. Recommendations to ensure the quality, safety and efficacy of pneumococcal conjugate vaccines. Proposed replacement of TRS 927, Annex2. http://www.who.int/biologicals/areas/vaccines/pneumo/Pneumo_final_23APRIL_2010.pdf.
- 24.Winkelstein, J. A. 1981. The role of complement in the host's defense against Streptococcus pneumoniae. Rev. Infect. Dis. 3:289-298. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.