Abstract
Human coronaviruses (HCoVs) are responsible for respiratory tract infections ranging from common colds to severe acute respiratory syndrome. HCoV-NL63 and HCoV-229E are two of the four HCoVs that circulate worldwide and are close phylogenetic relatives. HCoV infections can lead to hospitalization of children, elderly individuals, and immunocompromised patients. Globally, approximately 5% of all upper and lower respiratory tract infections in hospitalized children are caused by HCoV-229E and HCoV-NL63. The latter virus has recently been associated with the childhood disease croup. Thus, differentiation between the two viruses is relevant for epidemiology studies. The aim of this study was to develop a double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) as a potential tool for identification and differentiation between HCoV-NL63 and HCoV-229E. The nucleocapsid (N) proteins of HCoV-NL63 and HCoV-229E were expressed in an Escherichia coli system and used to immunize mice in order to obtain monoclonal antibodies (MAbs) specific for each virus. Three specific MAbs to HCoV-NL63, one MAb specific to HCoV-229E, and four MAbs that recognized both viruses were obtained. After their characterization, three MAbs were selected in order to develop a differential DAS-ELISA. The described assay could detect up to 3 ng/ml of N protein and 50 50% tissue culture infective doses/ml of virus stock. No cross-reactivity with other human coronaviruses or closely related animal coronaviruses was found. The newly developed DAS-ELISA was species specific, and therefore, it could be considered a potential tool for detection and differentiation of HCoV-NL63 and HCoV-229E infections.
Coronaviruses (CoVs) are large enveloped positive-strand RNA viruses that belong to the family Coronaviridae (13). On the basis of their serological cross-reactivity and sequence analysis, coronaviruses are classified into three distinct groups (6, 13). The alpha- and betacoronavirus groups harbor various mammalian CoVs, whereas avian viruses cluster in the gammacoronavirus group. There are five different human CoV (HCoV) species, all of them associated with respiratory tract infections ranging from common colds to severe acute respiratory syndrome (SARS) (7, 14).
HCoV-229E and HCoV-NL63 belong to the alphacoronavirus group and are the only two human CoVs that have a relatively close phylogenetic relationship (5). HCoV-229E was discovered in the mid-1960s from persons with the common cold, and HCoV-NL63 was isolated for the first time in 2004 from a 7-month-old infant with bronchiolitis (9, 28). HCoV-229E and HCoV-NL63 infections have a worldwide distribution, having peak activity during the winter months (1, 10, 26). These viruses are associated with both upper and lower respiratory tract diseases and frequently affect young children (19, 27). In most cases, these infections do not lead to severe clinical symptoms, although acute infections in infants, elderly individuals, and immunocompromised patients can cause more serious respiratory disease, which may require hospitalization (3, 17). Globally, approximately 5% of all upper and lower respiratory tract infections in hospitalized children are caused by HCoV-229E and HCoV-NL63 infections (8, 25). The clinical manifestations of HCoV-229E in infected persons are headache, nasal discharge, chills, cough, and sore throat, whereas the symptoms observed in HCoV-NL63-infected patients are more severe, including fever, cough, sore throat, bronchiolitis, and pneumonia. Moreover, HCoV-NL63 has recently been associated with the childhood disease croup (29). Reinfections with HCoV-229E and HCoV-NL63 occur throughout life.
Currently, there are no therapeutic treatments available for any of the HCoVs, and diagnosis is based on virus detection by reverse transcription-PCR technology (2, 16, 30). Several studies described the development of immunoassays using the nucleocapsid (N) protein for detection of antibodies to human coronaviruses (4, 15, 24, 32). The N protein is abundantly expressed during infection and it is highly immunogenic (11, 33). These features support the nucleocapsid protein as a potential source of antigen for detecting CoV infection.
HCoV-NL63 and HCoV-229E are highly related phylogenetically, although they produce different diseases, with that caused by HCoV-NL63 being more severe, and therefore, differentiation between these viruses is an important issue. Due to the close genetic relationship between HCoV-NL63 and HCoV-229E, their serologic differentiation is complex. Therefore, the aim of the study described in the present report was to establish a double-antibody sandwich enzyme-linked immunosorbent assay (DAS-ELISA) capable of detecting and distinguishing between HCoV-NL63 and HCoV-229E. As mentioned in the background, the N protein appears to be the most suitable candidate for use for viral detection. The homology of the amino acid sequence of the HCoV-NL63 N protein with that of the HCoV-229E N protein has been reported to vary from 42 to 49% (15, 20). Therefore, the N proteins of HCoV-NL63 and HCoV-229E were expressed in the Escherichia coli system and used to immunize mice in order to obtain monoclonal antibodies (MAbs) specific for each virus and therefore allow the differentiation between them.
MATERIALS AND METHODS
Viruses and cells.
LLC-MK2 cells were grown and maintained at 34°C in a 5% CO2 atmosphere using a mixture of Hanks and Earle's minimum essential tissue culture medium (Invitrogen) supplemented with 3% fetal calf serum (FCS) and antibiotics (penicillin, streptomycin).
Huh-7 cells were maintained at 37°C in a 5% CO2 atmosphere in Dulbecco's modified Eagles's minimum essential medium (DMEM) supplemented with 10% FCS and antibiotics (penicillin, streptomycin). Vero cells were maintained at 37°C in a 5% CO2 atmosphere in DMEM supplemented with 5% FCS.
HCoV-NL63 (isolate Amsterdam 1, passage 8) was obtained from a virus culture on LLC-MK2 cells as described previously (28). HCoV-229E (isolate Inf-1, passage 4) was obtained from a virus culture on Huh-7 cells for 5 days at 34°C with 5% CO2. The infectious titer of the virus stocks was determined according to the Reed and Muench formula on LLC-MK2 or Huh-7 cell monolayers (21).
Porcine epidemic diarrhea virus (PEDV) was propagated in Vero cells growing in DMEM without fetal bovine serum and supplemented with streptomycin and penicillin G, 12 μg/ml trypsin, 0.3% tryptose phosphate broth, and 0.02% yeast extract. After 24 h at 37°C with 5% CO2, the virus was harvested from the supernatant. After several cycles of freeze-thaw, culture supernatant was clarified by low-speed centrifugation, concentrated by ultracentrifugation, and purified on a sucrose gradient. Mock-infected Vero cells treated in the same way were used as control antigens.
Antigen preparation.
The recombinant N proteins of HCoV-NL63 and HCoV-229E were prepared using the Escherichia coli system as described previously (4). Briefly, the N genes of each virus were amplified and cloned in the pET100/D-Topo vector (Invitrogen). The recombinant N proteins were expressed by transformation of BL21-derived strain Rosetta 2 (Novagen) cells. Overnight cultures of transformed bacteria were inoculated in Luria broth medium supplemented with 1% glucose, carbenicillin, and chloramphenicol. Cultures were grown to exponential phase prior to induction with isopropyl-β-d-thiogalactopyranoside (IPTG) for 5 h. Recombinant proteins were purified with nickel-nitrilotriacetic acid agarose (Qiagen). The C-terminal halves of the N proteins of HCoV-NL63 (amino acids [aa] 215 to 377) and HCoV-229E (aa 213 to 389) were expressed and purified following the same protocol as the one used to prepare the full-length proteins. The recombinant N proteins of HCoV-HKU1 and HCoV-OC43 were prepared following the same protocol.
The virus-infected cell lysates were prepared from stocks of HCoV-NL63 and HCoV-229E by inactivation with 1% Triton X-100 and 0.3% tri-n-butyl phosphate (TNBP) for 2 h at room temperature (RT). Purified PEDV was used as the antigen in the ELISAs.
Immunization of mice.
Four 10-week-old female BALB/c strain mice were injected subcutaneously with 30 μg of purified N protein of HCoV-NL63 or HCoV-229E. The first immunization was done in complete Freund's adjuvant (CFA) (Difco), and two further doses were done in incomplete Freund's adjuvant (IFA) at 2-week intervals. Mice received a final booster injection with 30 μg of antigen in phosphate-buffered saline (PBS) for 3 consecutive days prior to hybridoma fusion.
Production of monoclonal antibodies.
The production of hybridomas secreting virus-specific MAbs was carried out following a modified version of the procedure previously described (23). Briefly, X63/Ag 8.653 myeloma cells were fused with splenocytes from immunized mice using polyethylene glycol (PEG; Sigma-Aldrich). After fusion, hybrid cells were eluted in DMEM (Gibco) containing 15% FCS (Gibco) and coated on 96-well well plates at a concentration of 0.5 × 106 cells/well. After 24 h at 37°C, 2× hypoxanthine-aminopterin-thymidine medium with hybridoma cloning factor (Roche) was added and was renewed every 48 h over a period of 12 days. At day 15, the culture medium of the hybridomas was screened for reactivity with the corresponding recombinant N protein by ELISA. Antibody-producing hybridomas were cloned by limiting dilution at least four times.
Antibody selection was done using the corresponding nucleocapsid protein and an unrelated viral protein prepared in the same way as the N protein and used as the negative-control antigen in the ELISA.
Finally, the selected hybridomas were used to inoculate BALB/c mice. The resulting MAbs were purified from the ascitic fluid by precipitation with caprylic acid and labeled with peroxidase according to the method described by Nakane and Kawaoi for its further characterization (18).
Characterization of monoclonal antibodies.
The specificities of the MAbs were determined by indirect ELISA and Western blot analysis using the recombinant N protein and the corresponding virus-infected cell lysate as antigens. For the indirect ELISA, the purified MAbs were incubated in 96-well microtiter plates coated with 0.2 μg/well of purified recombinant N protein, 0.1 μg/well of the C-terminal half of the N protein, or virus-infected cell lysates. The plates were blocked with Stabilzyme Select (SurModics, Inc., MN) for 1 h at RT. The bound antibody was detected with a 1/10,000 dilution of horseradish peroxidase (HRP)-labeled goat anti-mouse immunoglobulin (Sigma-Aldrich). After 1 h incubation at RT, followed by extensive washing, the plates were incubated with tetramethylbenzidine (TMB) substrate (TMB-MAX; Neogen Corporation, Lexington, KY), and the reaction was stopped by addition of 0.5 M sulfuric acid. The absorbance was measured at 450 nm in a Multiscan Ascent ELISA reader.
Western blot analysis was carried out by electrophoresing the protein preparations on a 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel under reducing conditions. Detection of the antigens was followed by immunoblotting using the selected hybridoma supernatants and a goat anti-mouse immunoglobulin conjugated with peroxidase as a secondary antibody. Specific bands were visualized using TMB as the substrate.
The isotypes of the MAbs were determined by ELISA, using specific anti-mouse subtype antisera (Sigma).
A competition ELISA between the different MAbs was performed in order to detect if they mapped in the same or different antigenic areas. Briefly, 96-well plates were coated with the corresponding recombinant purified N protein (0.2 μg/well) diluted in sodium carbonate buffer (pH 9.6) and incubated overnight (ON) at 4°C. The plates were blocked for 1 h at RT, and a predetermined limiting dilution of the peroxidase-labeled MAb was added to the wells in the presence of serial dilutions of the unlabeled competitor. After 1 h incubation at 37°C, followed by extensive washing, the plates were incubated with the substrate (TMB), and subsequent development of the assay was performed as described for the indirect ELISA. A negative optical density at 450 nm (OD450) was obtained when the antibodies competed, indicating that they mapped in the same antigenic area. In contrast, when the two antibodies mapped in different regions, the resulting OD450 was positive.
DAS-ELISA.
Microtiter plates were coated with 1 μg/well of purified MAb 1E8 (7 mg/ml) diluted in sodium carbonate buffer (pH 9.6), and the plates were incubated ON at 4°C. After the plates were blocked for 1 h at RT, the recombinant N protein, the C-terminal half of the N protein, or the corresponding virus-infected cell lysates diluted in 0.05% Tween 20 in PBS were added to the plates and the plates were incubated for 1 h at RT. An MAb specific for each virus conjugated with peroxidase (MAbs 2D4-PO and 1E7-PO for HCoV-NL63 and HCoV-229E, respectively) was subsequently used for antigen detection by incubation for 1 h at RT. TMB peroxidase substrate was added, and the reaction was stopped by the addition of 0.5 M sulfuric acid. Enzymatic activity was measured by determination of the OD450 in an ELISA plate reader. Washes between consecutive steps were performed with 0.05% Tween 20 in PBS.
RESULTS
Characterization of MAbs.
Three MAbs were obtained from a fusion between myeloma cells and splenocytes from mice immunized with N protein of HCoV-NL63: MAbs 1B12, 2D4, and 2E6. The fusion between splenocytes from mice immunized with N protein of HCoV-229E and myeloma cells resulted in five MAbs: MAbs 1E7, 1E8, 1H11, 5D5, and 5D11.
An indirect ELISA using the recombinant N protein of HCoV-NL63 or HCoV-229E or the corresponding virus-infected cell lysate as the antigen was performed to test the specificity of each MAb. The results showed that the MAbs could be classified in three groups according to their specificity. The first group included MAbs specific for HCoV-NL63 (MAbs 1B12, 2D4, and 2E6), the second group included an MAb specific for HCoV-229E (MAb 1E7), and finally, a third group of MAbs recognized both viruses (MAbs 1E8, 1H11, 5D5, and 5D11).
The isotypes of the MAbs were analyzed; antibodies 1B12, 1E8, 2D4, 2E6, and 5D11 were classified as IgG1; 1H11 was classified as IgG2a; and 1E7 and 5D5 were classified as IgG2b. All the described characteristics of the MAbs are summarized in Table 1.
TABLE 1.
Characterization of HCoV-specific MAbs
| Specificity | MAb | Isotype | MAb reactivity with N protein of: |
|
|---|---|---|---|---|
| HCoV-NL63 | HCoV-229E | |||
| Specific to HCoV-NL63 | 1B12 | IgG1 | + | − |
| 2D4 | IgG1 | + | − | |
| 2E6 | IgG1 | + | − | |
| Specific to HCoV-229E | 1E7 | IgG2b | − | + |
| Common to HCoV-NL63 | 1E8 | IgG1 | + | + |
| and HCoV-229E | 1H11 | IgG2a | + | + |
| 5D5 | IgG2b | + | + | |
| 5D11 | IgG1 | + | + | |
The ability of the MAbs to bind to the recombinant protein was further investigated by Western blot analysis. All MAbs were suitable for use for immunoblotting, and the results were consistent with the ELISA results. Figure 1 shows an example of the reactivity of one MAb representative of each group. MAb 2D4 showed reactivity with a 41-kDa protein, corresponding to the expected molecular mass of recombinant N protein of HCoV-NL63 (Fig. 1A). MAb 1E7 showed reactivity with a 43-kDa protein, corresponding to the expected molecular mass of recombinant N protein of HCoV-229E, and finally, MAb 5D5 showed cross-reactivity with the N proteins of both viruses (Fig. 1B and C, respectively).
FIG. 1.
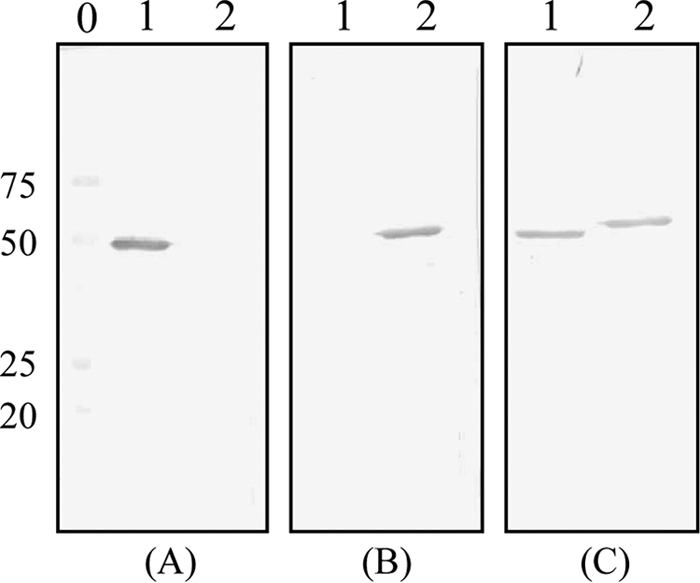
Immunoblot analysis of HCoV N protein. (A) Reactivity with an MAb specific for HCoV-NL63 (MAb 2D4); (B) reactivity with an MAb specific for HCoV-229E (MAb 1E7); (C) reactivity with an MAb that recognizes both viruses (MAb 5D5). Lanes 1, N protein of HCoV-NL63; lanes 2, N protein of HCoV-229E; lane 0, molecular weight markers (numbers on the left are molecular weights, in thousands).
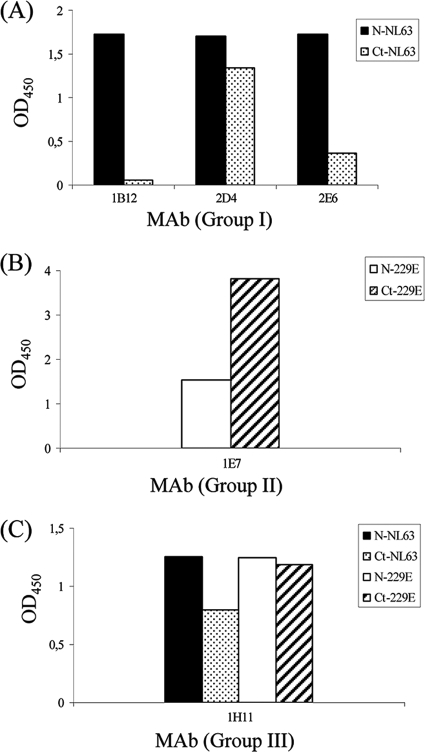
A competition ELISA between antibodies belonging to the same group was done to determine whether they mapped in the same or different antigenic regions of the protein. The results indicated that MAbs specific for the N protein of HCoV-NL63 mapped in different antigenic areas of the protein. When competition between antibodies recognizing both viruses was done, the result showed that all of them competed with each other, indicating that they all mapped in the same region of the protein (data not shown). As a first attempt to localize which regions of the nucleocapsid protein were recognized by the MAbs, a fragment covering the C-terminal half of the N protein (N-HCoV-NL63, aa 215 to 377; N-HCoV-229E, aa 213 to 389) was expressed in the E. coli system and prepared in the same way used for the full-length antigens (described in Materials and Methods). Equimolar amounts of the full-length protein and the corresponding fragment were used to coat the plates. Under these conditions, antibody 2D4 reacted with this region of the protein, while no reactivity was found with either the 1B12 or 2E6 MAb (Fig. 2A). MAb 1E7 bound to the C-terminal end of the protein, and the reactivity with this fragment was even higher than the reactivity observed with the complete molecule (Fig. 2B). Antibodies that recognized both viruses (1E8, 1H11, 5D5, and 5D11) reacted with this region of the protein, and the reactivities of all of them were very similar. Thus, Fig. 2C shows the reactivity of MAb 1H11 as a representative example for the group.
FIG. 2.
Reactivity of MAbs with the N protein and the C-terminal half of the N proteins of HCoVs. (A) Reactivity of MAbs specific for HCoV-NL63 (B); reactivity of MAbs specific for HCoV-229E; (C) reactivity of MAbs that recognize both viruses. ELISA plates were coated with the recombinant N protein of HCoV-NL63 (black bars) or HCoV-229E (white bars) or the corresponding C-terminal fragments (dotted and dashed bars, respectively). The y axis represents the absorbance readings (OD450) obtained in the indirect ELISA.
DAS-ELISA.
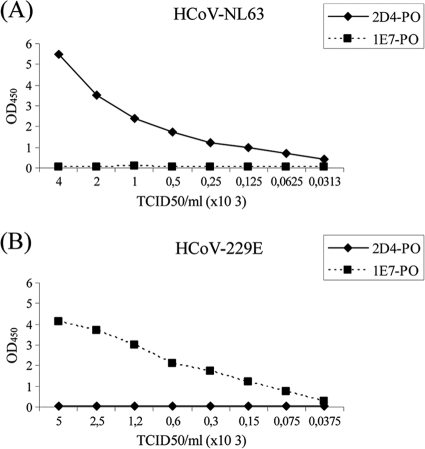
The DAS-ELISA was first set up using the purified recombinant N protein as the antigen, and all the possible combinations of MAbs were tested. Out of the five MAbs common to both viruses, MAb 1E8 was selected as the capture antibody and the optimal specific antibody labeled with peroxidase for each species was chosen: 2D4 for detection of HCoV-NL63 and 1E7 for detection of HCoV-229E. This assay could detect up to 3 ng/ml of recombinant N protein of HCoV-NL63 and 11 ng/ml of recombinant N protein of HCoV-229E. Further experiments were done with virus-infected cell lysates as the antigen, previously inactivated by treatment with 1% Triton X-100 and 0.3% TNBP for 2 h at RT. The specificity of the assay was demonstrated once more, and in this case the detection limit was approximately 50 50% tissue culture infective doses (TCID50s)/ml of viral stock for both viruses. The results of the DAS-ELISA using virus lysates as the antigen are shown in Fig. 3. These results confirm the specificity of the assay.
FIG. 3.
Detection of viral antigen in DAS-ELISA. (A) Detection of HCoV-NL63; (B) detection of HCoV-229E. Purified MAb 1E8 was used as the capture antibody. Serial dilutions of virus-infected cell lysates were added to the plates, and the viral antigen was detected by using a species-specific peroxidase-conjugated MAb (MAbs 2D4-PO and 1E7-PO for HCoV-NL63 and HCoV-229E detection, respectively). The absorbance at 450 nm was plotted against the viral titer, expressed as the numbers of TCID50s/ml.
Cross-reactivity with N protein of other CoVs.
The reactivities of the MAbs to the N proteins of other human coronaviruses belonging to the betacoronavirus group (HCoV-HKU1 and HCoV-OC43) were examined by indirect ELISA and DAS-ELISA. The results of both assays showed that none of the MAbs cross-reacted with any of the other viruses (data not shown).
The cross-reactivity with a CoV of animal origin belonging to the alphacoronavirus group (PEDV) and closely related to HCoV-NL63 and HCoV-229E was also tested. This resulted in a negative OD by indirect ELISA (data not shown) with all the described antibodies, even with those that recognize both viruses.
DISCUSSION
Detection of viral antigens and/or viral genomes is critical for rapid diagnosis of respiratory viral infections, followed by an appropriate treatment. Currently, reverse transcription-PCR is the major technique used for diagnosis of HCoV infection (2, 12, 16). However, this method has some disadvantages, such as possible contamination, instability of the RNA samples, and the need for special equipment that in some cases, in particular, in undeveloped countries, is not available. Other rapid tests are needed in order to facilitate diagnosis. The antigen-capture ELISA using MAbs allows rapid diagnosis of viral infections, and it can be used as an alternative to PCR. On the basis of the low degree of homology among the amino acid sequences of the N proteins of different coronaviruses, together with the high immunogenicity of this protein, the N protein appeared to be a suitable antigenic marker for diagnosis of coronavirus infection. In the work reported here, we described the production and characterization of several species-specific MAbs against the N proteins of human coronaviruses NL63 and 229E and the subsequent development of a differential ELISA that allowed their detection.
The immunization of BALB/c mice with the recombinant N proteins of HCoV-NL63 and HCoV-229E led to the production of several MAbs. Some of these antibodies were species specific, while others were able to recognize both viruses (Table 1). Additionally, the results of the immunoblotting experiments suggested that the epitopes recognized by the MAbs were linear rather than conformational (Fig. 1).
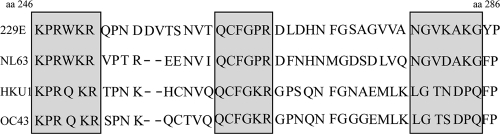
As a first attempt to narrow down the location of the epitopes recognized by the antibodies, a fragment covering the C-terminal half of the N protein was expressed and tested in the ELISA with the different MAbs. It has been previously described that the N-terminal half of all CoV N proteins contains a highly conserved motif (22). However, after two different fusions with the N protein of HCoV-NL63 and the N protein of HCoV-229E, only two out of eight MAbs (1B12 and 2E6) did not bind to the C-terminal half of the N protein (Fig. 2), indicating that the amino region is less immunogenic, at least in mice. This finding agrees with previous studies with SARS coronavirus N protein, where it was described that the most immunoreactive epitopes were located at the C-terminal end of the protein (31). The localization of the developed MAbs in this part of the protein was also supported by the results of the competition ELISA. Thus, MAbs specific for HCoV-NL63 (MAbs 1B12, 2D4, and 1E6) which reacted differently with this part of the protein by indirect ELISA did not compete with each other, suggesting that they mapped in different areas of the protein. In contrast, MAbs recognizing both viruses (MAbs 1E8, 1H11, 5D5, and 5D11) and which reacted in a similar way to the C terminus of the protein in indirect ELISA competed with each other, indicating that they map in the same region or very close regions. A detailed study of the C-terminal end sequence of the N protein revealed that although the overall homology between the different species is low, this fragment includes two regions (KPRWKR and QCFGPR) that are well conserved among all the viruses and one that is conserved only between HCoV-NL63 and HCoV-229E (NGVXAKG) (Fig. 4). This observation, together with our results, leads to the conclusion that the conserved sequence between HCoV-NL63 and HCoV-229E probably includes the epitopes of the selected common MAbs, although further experiments using peptides should be done in order to answer this question.
FIG. 4.
Alignment of the amino acid sequences of the C-terminal half of the nucleocapsid proteins of various HCoVs. Highly conserved regions between the four aligned sequences are in shaded boxes. Sequences used for the alignment included those of the following viruses: HCoV-229E (GenBank accession no. NC_002645), HCoV-NL63 (GenBank accession no. NC_005831), HCoV-HKU1 (GenBank accession no. NC_006577), and HCoV-OC43 (GenBank accession no. NC_005147). The N protein of HCoV-229E was considered the reference sequence (aa 246 to 286), and the alignment was performed with DS Gene (version 1.5) software.
As shown in Fig. 2B, the reactivity of MAb 1E7 (specific for HCoV-229E) with the C-terminal fragment was stronger than the reactivity with the full-length protein. This could be due to the structure of the protein, with the epitope becoming more accessible in the fragment than in the complete protein.
After characterization of the different MAbs, a double-antibody sandwich ELISA was developed for antigen detection. MAb 1E8 was selected as the capture antibody, and MAbs 2D4 and 1E7 conjugated with peroxidase were used as the detecting antibodies for HCoV-NL63 and HCoV-229E, respectively. The results of the DAS-ELISA demonstrated that the MAbs were species specific when the recombinant N protein was used as the antigen. This was also true when virus-infected cell lysates were used (Fig. 3). Besides, the use of one MAb that is able to capture both viruses makes the development of the assay simpler and less expensive, which is an important advantage for commercial purposes.
Finally, no cross-reactivity with viruses belonging to the betacoronavirus group (HCoV-OC43 and HCoV-HKU1) was found, nor was cross-reactivity with a coronavirus of animal origin belonging to the alphacoronavirus group (PDEV) found. This result was somehow expected, since most of the MAbs map in the C-terminal half of the protein, that is, the most variable region (22). In addition, these results strongly confirmed the assay specificity.
Although future experiments using human respiratory samples should be done, the current findings show that the present DAS-ELISA is a potential tool for detection and differentiation between HCoV-NL63 and HCoV-229E infection. This assay could be used as a valid alternative to nucleic acid detection for CoV diagnostic purposes. Moreover, the combined use of DAS-ELISA and PCR may be very helpful, when possible.
Acknowledgments
This work was supported by EU FP6-2005-LIFESCHEALTH-7 (grant 37276 RespViruses).
We thank John N. Barr for critical reading of the manuscript and assistance with the English language.
Footnotes
Published ahead of print on 17 November 2010.
REFERENCES
- 1.Arden, K. E., M. D. Nissen, T. P. Sloots, and I. M. Mackay. 2005. New human coronavirus, HCoV-NL63, associated with severe lower respiratory tract disease in Australia. J. Med. Virol. 75:455-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bastien, N., J. L. Robinson, A. Tse, B. E. Lee, L. Hart, and Y. Li. 2005. Human coronavirus NL-63 infections in children: a 1-year study. J. Clin. Microbiol. 43:4567-4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chiu, S. S., K. H. Chan, K. W. Chu, S. W. Kwan, Y. Guan, L. L. Poon, and J. S. Peiris. 2005. Human coronavirus NL63 infection and other coronavirus infections in children hospitalized with acute respiratory disease in Hong Kong, China. Clin. Infect. Dis. 40:1721-1729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dijkman, R., M. F. Jebbink, N. B. El Idrissi, K. Pyrc, M. A. Muller, T. W. Kuijpers, H. L. Zaaijer, and L. van der Hoek. 2008. Human coronavirus NL63 and 229E seroconversion in children. J. Clin. Microbiol. 46:2368-2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dijkman, R., and L. van der Hoek. 2009. Human coronaviruses 229E and NL63: close yet still so far. J. Formos. Med. Assoc. 108:270-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Drexler, J. F., F. Gloza-Rausch, J. Glende, V. M. Corman, D. Muth, M. Goettsche, A. Seebens, M. Niedrig, S. Pfefferle, S. Yordanov, L. Zhelyazkov, U. Hermanns, P. Vallo, A. Lukashev, M. A. Muller, H. Deng, G. Herrler, and C. Drosten. 2010. Genomic characterization of severe acute respiratory syndrome-related coronavirus in European bats and classification of coronaviruses based on partial RNA-dependent RNA polymerase gene sequences. J. Virol. 84:11336-11349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 8.Gerna, G., E. Percivalle, A. Sarasini, G. Campanini, A. Piralla, F. Rovida, E. Genini, A. Marchi, and F. Baldanti. 2007. Human respiratory coronavirus HKU1 versus other coronavirus infections in Italian hospitalised patients. J. Clin. Virol. 38:244-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hamre, D., and J. J. Procknow. 1966. A new virus isolated from the human respiratory tract. Proc. Soc. Exp. Biol. Med. 121:190-193. [DOI] [PubMed] [Google Scholar]
- 10.Han, T. H., J. Y. Chung, S. W. Kim, and E. S. Hwang. 2007. Human coronavirus-NL63 infections in Korean children, 2004-2006. J. Clin. Virol. 38:27-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hiscox, J. A., D. Cavanagh, and P. Britton. 1995. Quantification of individual subgenomic mRNA species during replication of the coronavirus transmissible gastroenteritis virus. Virus Res. 36:119-130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koetz, A., P. Nilsson, M. Linden, L. van der Hoek, and T. Ripa. 2006. Detection of human coronavirus NL63, human metapneumovirus and respiratory syncytial virus in children with respiratory tract infections in south-west Sweden. Clin. Microbiol. Infect. 12:1089-1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lai, M. M. C., S. Perlman, and J. L. Anderson. 2006. Coronaviridae, p. 1305-1335. In D. M. Knipe and P. M. Howley (ed.), Fields virology. Lippincott Williams & Wilkins, Philadelphia, PA.
- 14.Larson, H. E., S. E. Reed, and D. A. Tyrrell. 1980. Isolation of rhinoviruses and coronaviruses from 38 colds in adults. J. Med. Virol. 5:221-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lehmann, C., H. Wolf, J. Xu, Q. Zhao, Y. Shao, M. Motz, and P. Lindner. 2008. A line immunoassay utilizing recombinant nucleocapsid proteins for detection of antibodies to human coronaviruses. Diagn. Microbiol. Infect. Dis. 61:40-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leung, T. F., C. Y. Li, W. Y. Lam, G. W. Wong, E. Cheuk, M. Ip, P. C. Ng, and P. K. Chan. 2009. Epidemiology and clinical presentations of human coronavirus NL63 infections in Hong Kong children. J. Clin. Microbiol. 47:3486-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Milano, F., A. P. Campbell, K. A. Guthrie, J. Kuypers, J. A. Englund, L. Corey, and M. Boeckh. 2010. Human rhinovirus and coronavirus detection among allogeneic hematopoietic stem cell transplantation recipients. Blood 115:2088-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakane, P. K., and A. Kawaoi. 1974. Peroxidase-labeled antibody. A new method of conjugation. J. Histochem. Cytochem. 22:1084-1091. [DOI] [PubMed] [Google Scholar]
- 19.Principi, N., S. Bosis, and S. Esposito. 2010. Effects of coronavirus infections in children. Emerg. Infect. Dis. 16:183-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pyrc, K., M. F. Jebbink, B. Berkhout, and L. van der Hoek. 2004. Genome structure and transcriptional regulation of human coronavirus NL63. Virol. J. 1:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty per cent endpoints. Am. J. Epidemiol. 27:493-497. [Google Scholar]
- 22.Rota, P. A., M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli, J. P. Icenogle, S. Penaranda, B. Bankamp, K. Maher, M. H. Chen, S. Tong, A. Tamin, L. Lowe, M. Frace, J. L. DeRisi, Q. Chen, D. Wang, D. D. Erdman, T. C. Peret, C. Burns, T. G. Ksiazek, P. E. Rollin, A. Sanchez, S. Liffick, B. Holloway, J. Limor, K. McCaustland, M. Olsen-Rasmussen, R. Fouchier, S. Gunther, A. D. Osterhaus, C. Drosten, M. A. Pallansch, L. J. Anderson, and W. J. Bellini. 2003. Characterization of a novel coronavirus associated with severe acute respiratory syndrome. Science 300:1394-1399. [DOI] [PubMed] [Google Scholar]
- 23.Sanz, A., B. Garcia-Barreno, M. L. Nogal, E. Vinuela, and L. Enjuanes. 1985. Monoclonal antibodies specific for African swine fever virus proteins. J. Virol. 54:199-206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Severance, E. G., I. Bossis, F. B. Dickerson, C. R. Stallings, A. E. Origoni, A. Sullens, R. H. Yolken, and R. P. Viscidi. 2008. Development of a nucleocapsid-based human coronavirus immunoassay and estimates of individuals exposed to coronavirus in a U.S. metropolitan population. Clin. Vaccine Immunol. 15:1805-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vabret, A., J. Dina, S. Gouarin, J. Petitjean, V. Tripey, J. Brouard, and F. Freymuth. 2008. Human (non-severe acute respiratory syndrome) coronavirus infections in hospitalised children in France. J. Paediatr. Child Health 44:176-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van der Hoek, L. 2007. Human coronaviruses: what do they cause? Antivir. Ther. 12:651-658. [PubMed] [Google Scholar]
- 27.van der Hoek, L., G. Ihorst, K. Sure, A. Vabret, R. Dijkman, M. de Vries, J. Forster, B. Berkhout, and K. Uberla. Burden of disease due to human coronavirus NL63 infections and periodicity of infection. J. Clin. Virol. 48:104-108. [DOI] [PMC free article] [PubMed]
- 28.van der Hoek, L., K. Pyrc, M. F. Jebbink, W. Vermeulen-Oost, R. J. Berkhout, K. C. Wolthers, P. M. Wertheim-van Dillen, J. Kaandorp, J. Spaargaren, and B. Berkhout. 2004. Identification of a new human coronavirus. Nat. Med. 10:368-373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Hoek, L., K. Sure, G. Ihorst, A. Stang, K. Pyrc, M. F. Jebbink, G. Petersen, J. Forster, B. Berkhout, and K. Uberla. 2005. Croup is associated with the novel coronavirus NL63. PLoS Med. 2:e240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Elden, L. J., A. M. van Loon, F. van Alphen, K. A. Hendriksen, A. I. Hoepelman, M. G. van Kraaij, J. J. Oosterheert, P. Schipper, R. Schuurman, and M. Nijhuis. 2004. Frequent detection of human coronaviruses in clinical specimens from patients with respiratory tract infection by use of a novel real-time reverse-transcriptase polymerase chain reaction. J. Infect. Dis. 189:652-657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang, J., J. Wen, J. Li, J. Yin, Q. Zhu, H. Wang, Y. Yang, E. Qin, B. You, W. Li, X. Li, S. Huang, R. Yang, X. Zhang, L. Yang, T. Zhang, Y. Yin, X. Cui, X. Tang, L. Wang, B. He, L. Ma, T. Lei, C. Zeng, J. Fang, J. Yu, J. Wang, H. Yang, M. B. West, A. Bhatnagar, Y. Lu, N. Xu, and S. Liu. 2003. Assessment of immunoreactive synthetic peptides from the structural proteins of severe acute respiratory syndrome coronavirus. Clin. Chem. 49:1989-1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Woo, P. C., S. K. Lau, B. H. Wong, H. W. Tsoi, A. M. Fung, K. H. Chan, V. K. Tam, J. S. Peiris, and K. Y. Yuen. 2004. Detection of specific antibodies to severe acute respiratory syndrome (SARS) coronavirus nucleocapsid protein for serodiagnosis of SARS coronavirus pneumonia. J. Clin. Microbiol. 42:2306-2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao, J., W. Wang, W. Wang, Z. Zhao, Y. Zhang, P. Lv, F. Ren, and X. M. Gao. 2007. Comparison of immunoglobulin G responses to the spike and nucleocapsid proteins of severe acute respiratory syndrome (SARS) coronavirus in patients with SARS. Clin. Vaccine Immunol. 14:839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]