Abstract
Because most fungi have evolved to be free-living in the environment and because the infections they cause are usually opportunistic in nature, it is often difficult to identify specific traits that contribute to fungal pathogenesis. In recent years, there has been a surge in the number of sequenced genomes of human fungal pathogens, and comparison of these sequences has proved to be an excellent resource for exploring commonalities and differences in how these species interact with their hosts. In order to survive in the human body, fungi must be able to adapt to new nutrient sources and environmental stresses. Therefore, genes involved in carbohydrate and amino acid metabolism and transport and genes encoding secondary metabolites tend to be overrepresented in pathogenic species (e.g., Aspergillus fumigatus). However, it is clear that human commensal yeast species such as Candida albicans have also evolved a range of specific factors that facilitate direct interaction with host tissues. The evolution of virulence across the human pathogenic fungi has occurred largely through very similar mechanisms. One of the most important mechanisms is gene duplication and the expansion of gene families, particularly in subtelomeric regions. Unlike the case for prokaryotic pathogens, horizontal transfer of genes between species and other genera does not seem to have played a significant role in the evolution of fungal virulence. New sequencing technologies promise the prospect of even greater numbers of genome sequences, facilitating the sequencing of multiple genomes and transcriptomes within individual species, and will undoubtedly contribute to a deeper insight into fungal pathogenesis.
Despite the myriad yeasts and fungal spores in the environment, relatively few directly impact human health. However, those fungi that do cause disease in humans continue to represent a significant challenge for infectious disease physicians and clinical microbiologists. Of the 100 or so fungal species that have been implicated in human disease, species belonging to the genera Candida and Aspergillus are widely recognized as the most important human fungal pathogens. Aside from the human costs associated with these infections, it was estimated in 2002 that the direct costs associated with systemic fungal infections in the United States alone amounted to $2.6 billion (60). For the most part, these infections are opportunistic in nature, usually occurring in patients who are hospitalized and immunocompromised, although cases of community-acquired candidemia have recently been reported (49). The incidence of these infections continues to increase (43), and mortality rates remain high due to difficulties associated with the early diagnosis of fungal infections, resulting in failure to provide appropriate antifungal therapy in time to treat fungal infections effectively.
Since most fungal infections are opportunistic in nature, it is predominantly host factors that lead to the establishment of infection. However, not all fungi have the capacity to cause disease in even the most immunocompromised patients, and human pathogenic fungi must be able to express specific traits that allow them to grow in humans. While it is sometimes relatively easy to identify virulence factors directly associated with human disease in many bacterial and viral pathogens, the identification of virulence factors or, more correctly, virulence-associated factors in fungal pathogens is usually less clear-cut. Because of this complexity, our understanding of the pathogenesis of fungal infections lags behind that for other classes of microbial infections, and we still have much to learn regarding how fungi cause disease. Fortunately, new genome sequencing technologies offer future hope for improving our understanding of fungal pathogenesis. Over the past decade, the genomes of many of the most important human fungal pathogens have been sequenced, and the world of “-omics” has been opened up to medical mycologists. In this minireview, we discuss how analysis of these genome sequences, particularly comparative genomic analysis (CGA) of related species, has contributed to our understanding of the pathogenesis of some of the most common human fungal pathogens, i.e., Aspergillus spp., Candida spp., Coccidioides spp., and Cryptococcus spp., and how virulence has evolved in these organisms.
IDENTIFICATION OF LINEAGE-SPECIFIC GENES IN HUMAN PATHOGENIC FUNGI
The overriding goal of CGA with human pathogenic fungi is the identification of pathogen-specific genes required to efficiently colonize and infect the human host. CGA can produce large volumes of data, but implicating specific genetic differences between pathogenic and nonpathogenic fungi in the disease process can be problematic. Initial CGA studies on pathogenic fungi focused mainly on comparisons with previously sequenced and distantly related nonpathogenic model organisms. Such studies provided broad insights into genome evolution within the relevant genera but rarely pinpointed specific genetic differences associated with virulence. In order to maximize the potential power of CGA to provide clues to identify virulence-associated genes, it is essential to choose species that are taxonomically close yet distinct in the phenotype of interest, in this case the capacity of an organism to cause human infection. CGA with the genus Aspergillus demonstrates how analyses of organisms with various degrees of relatedness have provided different perspectives into the evolution of the genus and, in particular, into the evolution of virulence traits in the opportunistic pathogen Aspergillus fumigatus. Aspergilli are free-living filamentous saprophytic fungi that are ubiquitous in the environment, growing mainly in soil and decaying vegetation (21). These include Aspergillus nidulans, a model species long used in genetic studies, the industrially important species Aspergillus niger and Aspergillus oryzae, and A. fumigatus, one of the most important human fungal pathogens. Of all environmental filamentous fungi, A. fumigatus is by far the most common cause of invasive human disease (11). Although this saprophyte has evolved specifically to grow in compost, some of the traits that permit growth in this environmental niche also, by happenstance, allow it to grow in humans with compromised immune systems. In an attempt to identify A. fumigatus genes specifically associated with the capacity to cause human infection, CGA was used to compare its genome sequence with the genome sequences of A. oryzae and A. nidulans. This analysis revealed the presence of nine A. fumigatus-specific allergens (including ribotoxin, an enzyme that cleaves ribosomal 28S RNA in two) and more than a dozen clusters of genes encoding secondary metabolites (e.g., fumagillin) that have been suggested to play a role in virulence (21). In total, more than 500 genes were found to be A. fumigatus specific, although most of these have no known function. It is important to realize, however, that these three species are in fact distantly related (orthologous proteins share approximately 70% identity, on average, which is similar to the relationship between mammals and fish [15]) and that A. fumigatus is the only one of the three species that can be described as pathogenic in humans. So while comparison of these genomes has been informative in terms of general physiology and genome evolution (e.g., the detection of a set of genes involved in a sexual mating cycle in A. fumigatus), there are other species that are more appropriate for comparative genomic analysis in relation to pathogenicity. Subsequent analyses applied CGA to two of the species most closely related to A. fumigatus, namely, Neosartorya fischeri and Aspergillus clavatus (18). The former is a species that has only very rarely been associated with infection, while A. clavatus, which is known to produce allergens and mycotoxins, is a rare cause of alveolar inflammation, particularly in malt workers (6, 10). At a gross level, significant differences in genome size were observed, which may be explained by the greater number of transposable elements in the N. fischeri genome. Closer analysis confirmed the taxonomic relatedness of the three species and revealed a high level of synteny, with more than 7,500 orthologous core genes present in the three genomes. However, despite the relatively close relatedness of these species, a significant number of species-specific genes were identified. Relative to A. nidulans and A. oryzae, the genomes of these three organisms were enriched for genes involved in ion transport, choline transport, and carbohydrate metabolism as well as for genes involved in the production of secondary metabolites. Specific analysis of A. fumigatus also revealed the presence of species-specific genes involved in the catabolism of carbohydrates, polysaccharides, and amino-sugars and in amine transport (18). Although the production of specific secondary metabolites may play a role in virulence in these organisms, it is clear that the capacity to grow in vivo is associated with the presence of genes with roles in nutrient transport and catabolism, suggesting that growth in humans may require a high degree of catabolic flexibility. Interestingly, many of these A. fumigatus-specific genes were smaller, on average, than core shared Aspergillus genes, had fewer introns, and displayed a significant telomeric bias, whose significance is described in greater detail below.
The genus Candida, which is comprised of more than 150 disparate yeast species, has also been the subject of several noteworthy CGA studies. The majority of Candida spp. are environmental saprophytes, and only a dozen or so of these have been associated with human colonization or infection. Candida albicans is a common human commensal and is often referred to as the most pathogenic fungal species, as it is responsible for most cases of candidiasis, although other species, such as Candida glabrata, Candida tropicalis, and Candida parapsilosis, can also cause infection (40). The most common form of candidiasis, referred to as superficial candidiasis or thrush, occurs on the mucosal surfaces of the mouth or vagina as a result of immunosuppression or disturbances in the normal microbial flora. In severe cases of immunosuppression (e.g., following chemotherapy or neutropenia), especially if the integrity of the gut wall is compromised, endogenous Candida cells can invade and penetrate into the bloodstream, resulting in candidemia and systemic infection (42). Unlike the vast majority of Candida species and the Aspergillus species described earlier, these pathogenic yeast species are members of the normal human flora and have evolved to colonize the oral cavity, the gastrointestinal tract, and the vagina. Initial comparative analysis of the C. albicans genome with that of the model yeast Saccharomyces cerevisiae revealed striking differences in metabolic capabilities and the size and expansion of a range of gene families in C. albicans (7, 58). Many of these gene families have roles in nutrient acquisition (e.g., secreted aspartyl proteinases [SAPs] and lipases) and nutrient uptake (e.g., ferric reductases, iron transporters, and oligopeptide and amino acid permeases) and may play a role in host colonization and infection. Candida albicans also differs from S. cerevisiae in having additional genes required for respiratory catabolism, including acyl-coenzyme A (acyl-CoA) oxidase, fatty acid-CoA synthase, and numerous oxidoreductase genes, all of which could be expected to play a role in nutrient acquisition and energy production in vivo. Indeed, it has already been demonstrated that genes involved in the metabolism of alternative carbohydrate sources are required for virulence in C. albicans (46). Further catabolic diversity is provided by the presence of several genes encoding amino acid oxidases (7). As in the case of the pathogenic filamentous fungus A. fumigatus, the enhanced nutrient uptake and catabolic capacity of C. albicans suggest that growth in vivo requires greater metabolic flexibility and that the genes involved in these processes were either acquired by C. albicans and related species or lost by S. cerevisiae since the two species diverged.
Subsequently, a broader comparative analysis of Candida genomes provided a deeper insight into the evolution of pathogenic mechanisms (8). This study included several members of the so-called CTG clade, a group of yeasts that translate the CUG codon as serine and includes yeasts that are pathogenic (Candida albicans, Candida tropicalis, and Candida parapsilosis), moderately pathogenic (Candida lusitaniae and Candida guilliermondii), and nonpathogenic (Lodderomyces elongisporus and Debaryomyces hansenii). Despite the taxonomic distance separating these species, CGA identified Candida-specific gene families, with some of these families specifically enriched in the genomes of the most pathogenic species (see below) (8). These included the ALS gene family, which encodes a group of well-characterized proteins proposed to act primarily as adhesins. Other identified gene families encode the rapidly evolving Hyr/Iff protein family, most of whose members possess glycosylphosphatidylinositol (GPI) anchor sites, suggesting that they are cell wall attached and possibly involved in host-pathogen interaction. A third gene family that is overrepresented in the Candida clade encodes Pga30-like proteins, but the role of these proteins in virulence has yet to be assessed. Although these families are enriched in the pathogenic species, they are found in most of the Candida genomes examined so far. For instance, while there are 8 members of the ALS family in C. albicans, there are 16 ALS genes in the less pathogenic species C. tropicalis and 4 in the exceedingly rare pathogen L. elongisporus (8). Consequently, while these gene family expansions may explain why the Candida clade is generally more pathogenic than Saccharomyces species, they do not fully explain why C. albicans is by far the most pathogenic member of the clade.
More recently, CGA was applied to the closely related species C. albicans and Candida dubliniensis. First identified in 1995, C. dubliniensis is by far the species most closely related to C. albicans (22, 53, 54), and even though the two species diverged from a common ancestor approximately 20 million years ago (39), they are sufficiently closely related to mate in vitro (45). Consequently, C. dubliniensis shares many phenotypic characteristics with C. albicans, to the point where the two species are very difficult to distinguish using phenotypic methods. Despite the very close phenotypic and phylogenetic relatedness of C. albicans and C. dubliniensis, epidemiological data suggest that the former is a far more effective pathogen. The significantly lower virulence level of C. dubliniensis has also been confirmed using a range of experimental infection models, including murine systemic (2, 22, 59) and orogastric (52) models of system infection and the ex vivo oral reconstituted human epithelial (RHE) infection model of superficial oral infection (50, 52). On this basis, C. albicans might be expected to exhibit a more extensive array of virulence factors than C. dubliniensis. However, many of the virulence factors proposed to play a role in Candida pathogenicity, such as adherence, dimorphism, phenotypic switching, and the ability to produce SAPs, are shared by both species (13, 22, 24, 25, 37). The ability to switch between yeast and true hyphal forms has long been recognized as an important C. albicans virulence trait. However, although C. dubliniensis is capable of producing true hyphae, it does so less efficiently than C. albicans, both in vivo and under a wide range of in vitro conditions (2, 41, 52, 59). As observed in all of the other comparative analyses of fungal genomes described earlier, the main disparity between C. albicans and C. dubliniensis is due primarily to differences in specific gene families in C. albicans, but these differences are often quite subtle. Differences include genes with a known role in virulence, including a number of genes that are expressed only by C. albicans hyphae. Some of the most notable genes absent from the C. dubliniensis genome are two hypha-specific SAP genes and ALS3, which is also hypha specific and appears to have arisen in C. albicans by a unique transposition event. Als3 is believed to play an important role in C. albicans virulence, as it has been demonstrated to have invasin-like properties (44) and has been proposed to play a role in iron acquisition from ferritin in vivo (1). In addition to the subtle differences in the sizes of gene families known to be involved in virulence, the main disparities in gene family size occur in families with no known function, such as the IFA genes, which are proposed to encode a family of transmembrane proteins, and the TLO gene family. While there are 14 telomere-associated (TLO) genes in C. albicans (58), there are only 2 in C. dubliniensis, representing the most notable difference in gene content between the two species. The function of the TLO genes is unknown, but the presence of a conserved Med2 domain suggests that they may encode a novel family of transcriptional regulators. Disruption of the C. dubliniensis TLO genes results in defective morphogenesis, suggesting that they may play a role in Candida pathogenesis (27).
Studies to compare the genomes of the human pathogens Coccidioides immitis and Coccidioides posadasii with the closely related nonpathogenic species Uncinocarpus reesii and the more distantly related pathogenic species Histoplasma capsulatum have also revealed pathogen-specific adaptations (48). Coccidioides spp. are fungi which are endemic in the southwestern United States and arid areas of northern Mexico and are the causative agents of “valley fever,” a pulmonary disease that affects 100,000 to 300,000 people in the United States each year. The fungus grows in the soil as filaments that produce arthroconidia (asexual spores), which when inhaled into the mammalian lung develop into multinucleate spherical structures, called spherules, which are filled with endospores (43). CGA identified 93 genes specific to the Coccidioides lineage since its divergence from U. reesii that exhibited spherule-specific expression, including genes involved in energy metabolism and a gene required for the use of allantoin as a nitrogen source. This gene set also included a range of integral membrane and cell surface protein genes. Additionally, Coccidioides spp. have retained or acquired a set of heme-binding proteins, suggesting that as with other pathogenic fungi, acquisition of iron in vivo is crucial for virulence. As with Aspergillus species, many of the species-specific genes in Coccidioides spp. are found in “genomic islands” in subtelomeric regions, although the role of these genes in virulence has yet to be established (48). Comparison of Onygenales genome sequences with those of members of the sister order Eurotiales (fungi that are associated primarily with plants, including the aspergilli) revealed that Onygenales species either lack or have reduced numbers of genes associated with growth on plant matter, e.g., plant cell wall-degrading enzymes, such as cellulases, cutinase, pectate lyase, and pectin esterases, as well as genes required for carbohydrate metabolism. In contrast, the Coccidioides spp. and U. reesii have larger families of protease genes, encoding extracellular serine proteases, such as keratinase, and the deuterolysin metalloprotease family, some of whose members are found only in Coccidioides spp. These data suggest that the Coccidioides spp. might not be soil saprophytes; instead, it has been proposed that they have evolved to associate specifically with animal hosts, probably rodents (48).
Unlike the pathogenic fungi described thus far, which are all members of the phylum Ascomycota, the pathogenic yeast Cryptococcus neoformans belongs to the phylum Basidiomycota. Cryptococcus neoformans is the causative agent of cryptococcal meningitis, an infection of immunocompromised patients, particularly those with HIV infection. Two varieties of C. neoformans are known to cause infection in humans, namely, serotype A (C. neoformans var. grubii) and serotype D (C. neoformans var. neoformans), with serotype A accounting for the majority of infections in AIDS patients (34). Comparison of the C. neoformans genome with that of ascomycetous yeasts revealed considerable differences in genomic architecture, with the cryptococcal genome being rich in introns and antisense messages (35). Comparison of the whole genomes of C. neoformans var. grubii and C. neoformans var. neoformans strains revealed that an ancient, nonreciprocal transfer of 40 kb occurred approximately 2 million years ago, from C. neoformans var. grubii to C. neoformans var. neoformans, and is widespread in the natural population. This event was likely the result of the creation of a serotype AD hybrid intermediate. Such hybrids can be created in the laboratory and are found in nature, but they are genetically unstable and often aneuploid. This region, referred to as an identity island, is widespread in naturally occurring strains of C. neoformans var. neoformans and may have imparted a selective advantage which has allowed it to become fixed in the population (29).
EVOLUTION OF VIRULENCE-ASSOCIATED GENES IN PATHOGENIC FUNGI
The application of comparative genomics to human pathogenic fungi has revealed that several different genetic processes have played important roles in the acquisition of virulence-associated genes. These processes are largely driven by gene duplication to provide the raw material for new open reading frames (ORFs), although horizontal gene transfer (HGT) may also play a minor role. Specialization for life as a commensal may also lead to gene loss, as costly genetic material associated with previous lifestyles is lost to increase reproductive fitness. Although these processes are not unique to pathogenic fungi, their importance in virulence gene evolution has been highlighted repeatedly in CGA studies of pathogenic fungi. The transfer of fungi from the environment to a living mammalian host introduces them to a range of challenging new environments. As the subsequent sections outline, fungi have used a variety of evolutionary mechanisms to facilitate growth in these potentially stressful environments.
Gene family expansion.
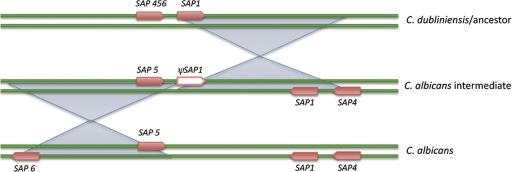
Gene duplication is an important force in evolution. Duplicated genes can increase fitness in an environment where gene dosage is important, or duplicated genes may diversify to take on new functions. Gene duplication and the formation of gene clusters or families may therefore lead to specialization for specific environmental conditions. Expansion of tandem gene arrays often occurs under selective pressure when increased gene dosage is required. Comparative analysis of the sequenced Saccharomyces and Candida genomes revealed that three cell wall-associated protein gene families are particularly enriched in the pathogenic species. These families, which encode the Als, Iff, and Pga30 proteins, show evidence of tandem duplications and subsequent divergence and have been proposed to play important roles in host interactions (8). For C. glabrata, several large gene clusters have been described that exhibit evidence of functional diversification. One is a cluster of six YPS genes, encoding the yapsins, which are extracellular GPI-linked aspartyl proteinases, and another is a cluster of 8 alpha-1,3-mannosyltransferase genes (14, 15). One of the most important C. glabrata virulence factors is the Epa family, which is a family of GPI-anchored cell wall proteins that facilitate host recognition and adhesion by C. glabrata. The number of EPA genes differs from strain to strain (up to 23 paralogs have been found in one strain), but they all are located in clusters adjacent to telomeres, where they are subject to transcriptional silencing (12, 47). Evidence of tandem gene duplication also occurs in C. albicans; however, most gene families in this species appear to be dispersed, with subgroups of related genes on the same chromosome. This model of duplication and dispersion is well exemplified by the LIP family of lipase genes. Two related clusters of LIP genes can be identified, on chromosome 1 (LIP1, -2, -3, -6, and -10) and chromosome 7 (LIP5, -8, and -9), with an orphan gene, LIP4, on chromosome 6, indicating that at least two translocations between chromosomes have occurred, followed by expansion on chromosomes 1 and 7 (26, 58). Interestingly, the less pathogenic species C. parapsilosis has only two LIP genes, only one of which appears to be functional (20). Although the functional LIP gene in C. parapsilosis has been shown to be required for virulence, the large size of the LIP gene family in C. albicans may contribute in some way to enhancing the ability of this species to colonize and infect humans. Comparison of the SAP gene clusters on chromosome 6 in C. albicans and C. dubliniensis provides insight into the mechanisms of gene family expansion along a single chromosome. Candida albicans possesses three closely related genes, SAP4, -5, and -6, on chromosome 6, whereas C. dubliniensis possesses one gene (CdSAP456) orthologous to this subfamily. It appears that in C. albicans, the ancestral SAP456 gene underwent two separate duplications and segmental inversions in the region that dispersed the cluster along chromosome 6 (Fig. 1). However, the origin of SAP1 in this region is more difficult to explain, as SAP1 is more closely related to SAP2 on chromosome R, indicating that proximity is not always a reliable indicator of relatedness. SAP1 was likely derived from a separate translocation event, perhaps involving homologous flanking sequences (27, 58).
Fig. 1.
Cartoon depicting how the SAP gene family may have expanded on chromosome 6 in C. albicans. It is proposed that C. dubliniensis represents the ancestral state (top) and that a single inversion event led to the duplication of the tandem pair of SAP456 and SAP1 to create an intermediate strain (middle) with SAP5, SAP1, and SAP4, but which has lost one of the copies of SAP1 (ψSAP1). It is proposed that another inversion resulted in the duplication of SAP5 to create SAP6, representing the current complement of genes in C. albicans (bottom). (Adapted from reference 27 with permission of the publisher.)
Several gene family expansions have also been identified in Coccidioides. Both Coccidioides spp. and the nonpathogenic species U. reesii harbor an expanded family of extracellular serine proteases, which are perhaps required to acquire nutrients from mammalian tissues. This expansion most likely occurred before divergence of U. reesii from Coccidioides and may have evolved to allow growth on decaying animal matter rather than virulence within a living host. However, another family of metalloprotease genes homologous to the known virulence factor MEP1 was identified in these organisms, and in this case, three additional members could be identified in the pathogenic Coccidioides spp. relative to U. reesii (48).
Telomeric “gene factories.”
In many pathogenic fungi, the telomere-proximal regions appear to be locations where pathogenic species have acquired novel genes that are often absent from closely related, nonpathogenic relatives. This phenomenon has been particularly well described for A. fumigatus. In contrast to C. albicans, where novel duplicated genes are usually dispersed along a chromosome, almost 50% of the A. fumigatus-specific genes can be clustered together in blocks or genomic islands of 10 or more genes (18). Many of these clusters of genes are involved in the production of secondary metabolites such as mycotoxins and fumigaclavine. These genomic islands show a strong telomeric bias, with the majority located within 300 kb of telomere ends. Although initial studies suggested that these clusters may have arisen by horizontal gene transfer, subsequent studies identified paralogous genes in other aspergilli, allowing the evolution of these genes to be traced within the genus and suggesting that they were recently duplicated and translocated to telomere-proximal locations (18). Unexpectedly, a tendency was found for these A. fumigatus lineage-specific genes to be shorter than core Aspergillus genes and to contain fewer introns. This may be due to reduced selective pressures on these duplicated genes that may have allowed loss of introns and the accumulation of premature stop codons. The tendency of these genes to cluster at telomere-proximal regions may be linked to their rapid evolution due to the relaxed selective constraints at these regions. Telomeric loci have been associated with accelerated evolution in protozoa (4). Fedorova et al. (18) speculated that the telomere-proximal regions of Aspergillus spp. may act as gene factories where duplicated genes may undergo significant divergence (or pseudogenization) in the absence of pressure to maintain function and subsequently be translocated to other areas of the genome. This theory has been termed the duplication, differentiation, and differential gene loss (DDL) hypothesis. Alternatively, clustering of these genes at subtelomeric loci may be a strategy for coordinated regulation of virulence gene expression. Movement of these clusters into subtelomeric regions would place them under the regulatory control of LaeA, a factor that regulates gene expression through chromatin remodeling (9). Microarray data support the idea that clustering facilitates the coordinated epigenetic regulation of virulence gene expression, as ∼30% of clustered genes are induced during initiation of invasive aspergillosis in a mouse infection model (38).
Genes belonging to expanded gene families have also been identified at telomere-proximal regions in Candida spp. The EPA genes, mentioned above, occur in tandem arrays at the telomeres of C. glabrata (12), a location that may have promoted their functional divergence and facilitated their regulation by transcriptional silencing. As described above, a family of TLO genes has also been identified in C. albicans (58). Comparative genomic analysis with the less pathogenic species C. dubliniensis revealed that while 14 copies are present in C. albicans SC5314, only 2 copies are present in the sequenced type strain of C. dubliniensis (i.e., Cd36) (27). The specific expansion of this family in C. albicans suggests that this family may play a specific role in commensalism or virulence of C. albicans. The telomere-specific location of these genes suggests a method of duplication and dispersal distinct from that for the previously described gene families of C. albicans. Dispersal of this family in C. albicans may have occurred by telomeric recombination. In addition, each C. albicans TLO gene is flanked at the 5′ end by long terminal repeat (LTR) kappa, possibly implicating the movement of retrotransposons in their evolution. Furthermore, comparison of the sequences of the encoded C. albicans Tlo proteins with those of the C. dubliniensis proteins and the single C. tropicalis ortholog suggests that the C. albicans family has diverged significantly from the ancestral state (27). The sequences of the C. albicans TLO genes are highly similar, suggesting a recent expansion or one that occurred to increase gene dosage rather than functional diversity.
HGT.
HGT, whereby genes are exchanged between individual strains and species, is a significant force in prokaryotic evolution and plays a major role in the spread of genes involved in bacterial virulence and antibiotic resistance (28). For fungi, the role of horizontal gene transfer in evolution, either from bacteria or from other fungi, has been considered marginal. However, the relative dearth of evidence for HGT in fungi may be due to the relatively small number of fungal genomes available for analysis and because these events may be very ancient and difficult to identify. Indeed, numerous good candidates for HGT events in fungi have been described recently. For example, the S. cerevisiae URA1 gene appears to have been acquired from a Lactobacillus species and may have facilitated adaptation to anaerobic growth (23). Evidence also exists for the transfer of a whole cluster of genes involved in secondary metabolism from the plant pathogen Magnaporthe grisea to A. clavatus (31). There is also evidence suggesting that the reason that the A. oryzae genome contains more than 1,000 extra genes compared with A. fumigatus and A. nidulans is because of HGT from a range of fungi, including Sodariomycetes. This suggests that A. oryzae may be more competent to take up foreign DNA than other fungal species (32). Recently, genome sequence analyses provided evidence for the interkingdom transfer of a gene encoding a proline racemase and a gene involved in the metabolism of phenazine from Burkholderia spp. to C. parapsilosis (19). The paucity of examples of recent HGT in the Candida CTG clade may be due to the unique codon usage of these organisms, which may act to restrict HGT. Therefore, although there are examples of interkingdom and intergenus HGT in fungi, there is little evidence thus far to suggest that this phenomenon has played a significant role in the evolution of virulence in human pathogenic fungi.
Adaptive evolution of gene sequences.
Positive selection can lead to high amino acid substitution rates in particular genes, and this can often be detected by determining the ratio of the rate of nonsynonymous substitutions (Ka) to the rate of synonymous substitutions (Ks) between homologous genes. Genes with high Ka/Ks ratios are usually said to be evolving rapidly under positive selection. Four virulence-associated genes of A. fumigatus were shown to exhibit evidence of accelerated evolution within the Aspergillus clade (18). This may be due to a relaxation of selection or, more likely, to a positive selection process leading to rapid functional diversification. The four genes (PabaA, fos-1, pes1, and pksP) are involved in nutrient acquisition and the oxidative stress response. Comparison of Coccidioides spp. with the closely related species U. reesii identified 67 Coccidioides genes that exhibit rapid evolution. This group includes the well-characterized immunization antigen 1 gene, which can confer protective immunity in mice (48).
Several gene families that exhibit positive selection were identified in C. albicans, including the IFF-HYR1, ALS, and PGA30-like families of cell surface protein genes that are enriched in the most pathogenic Candida species (8). Recombination may also play a role in the functional diversification of cell surface proteins. Phylogenetic comparison of the C. albicans and C. dubliniensis ALS genes reveals that syntenic genes at the corresponding genomic locations often lack the expected sequence similarities. Evidence exists for ALS mosaicism, indicating a high degree of intergenic recombination, which may play a major role in generating functional diversity in this gene family (27).
Gene loss.
Although it is a member of the genus Candida, C. glabrata is more closely related to S. cerevisiae than to C. albicans (15); comparison of the S. cerevisiae and C. glabrata genomes confirmed the shared presence of sister chromosomal regions or blocks, thought to be the result of an ancestral whole-genome duplication event. However, the number and size of duplicated blocks in C. glabrata are smaller than those for S. cerevisiae, which is indicative of significant gene loss in the former species since its divergence. When its gene content was compared with those of S. cerevisiae and Kluyveromyces lactis, C. glabrata was found to have lost genes involved in galactose metabolism, phosphate metabolism, and nitrogen and sulfur metabolism (14, 15). This level of difference is not unexpected considering the evolutionary distance between these species, and it may reflect metabolic specialization of C. glabrata for life in the human gastrointestinal tract.
The impact of gene loss or pseudogenization on genome content is also clearly evident when the genomes of C. albicans and C. dubliniensis are compared. It is apparent that C. dubliniensis is undergoing reductive evolution, with many genes potentially involved in morphogenesis and virulence (e.g., HYR1, a member of the virulence-associated IFF gene family [3] which confers neutrophil resistance on C. albicans [36]) having been lost entirely or in the process of being lost through pseudogenization. Candida dubliniensis was found to possess 78 pseudogenes which possessed intact orthologs in C. albicans, including 16 genes designated filamentous growth regulators (FGRs) (27, 57). Another gene family, the IFA family, encoding putative transmembrane proteins, is unique to C. albicans, C. dubliniensis, and C. tropicalis. In C. albicans and C. dubliniensis, there is evidence of widespread expansion of this gene family, with 31 loci in C. albicans and 21 loci in C. dubliniensis. However, a substantial component of the C. dubliniensis IFA gene repertoire appears to be in a state of mutational decay. Many of the IFA ORFs are heavily decayed gene relics, while others contain a few point mutations or frameshifts, suggesting that gene loss is an ongoing process (27). Clearly, the evolutionary pressure on C. albicans and C. dubliniensis is to become successful commensals in the human host. However, given the opportunity (e.g., damage to the gut wall, neutropenia due to immunosuppressive therapy, etc.), both species have the capacity to become pathogens when they overgrow and invade tissue, ultimately leading to systemic candidiasis. Taken together, the CGA data suggest that C. dubliniensis may be undergoing reductive evolution for specialized growth in an as yet unidentified anatomic niche, while the larger repertoire of C. albicans genes may allow this species to adapt to and thrive under a more diverse range of environmental conditions, including those found in a wider range of sites in the body. Therefore, the presence of colonizing C. albicans in greater numbers in more anatomic sites results in this species being more pathogenic than C. dubliniensis, and therefore responsible for a greater number of cases of invasive infection. Further evidence from the pathogenic fungi that genome reduction is an important evolutionary mechanism that has led to niche specialization is found in the genomes of Coccidioides spp., which lack many of the genes necessary for growth on plant matter that are present in species such as the aspergilli (48).
Functional genomics.
While CGA has proved to be a very helpful tool in investigating fungal virulence, it is clear that analysis of gene content alone is not sufficient to explain differences between, or indeed within, fungal species. For example, comparison of two strains of C. albicans that differ in the ability to invade tissue and cause infection in vivo by use of comparative genomic hybridization suggested that the gene contents of the two species are identical. However, global transcription comparisons revealed significant differences in gene expression profiles that are suggestive of transcriptional rewiring, which could contribute to differences in growth rate between strains in vivo (55). These data indicate that functional genomic approaches will be very helpful in identifying differences in gene expression and function between strains and closely related species. For example, cross-species forward genetic screens have been used successfully for whole-genome functional comparisons between C. albicans and C. dubliniensis (17, 51). Analysis of transcriptional networks in C. albicans and other ascomycete species, using transcript profiling and chromatin immunoprecipitation-microarray (ChIP-chip) analysis, showed that some transcriptional pathways have been modified specifically in Candida species. In C. albicans, these changes are associated with a high turnover of transcription factor binding sites, as has been shown for MCM1, and with changes in the regulatory networks controlling glycolysis and ribosome biogenesis (56). These types of changes are likely to have played a significant role in the evolution of C. albicans from an ancestral, environmental yeast and were recently reviewed by Lavoie et al. (33). In terms of host adaptation, C. albicans has been shown to possess numerous transcriptional programs activated in response to conditions commonly encountered in vivo, including alkaline pH, oxidative stresses, and morphogenetic signals (5). A recent transcript profiling analysis of the less virulent species C. dubliniensis suggested that many of these transcriptional programs are conserved (41). However, the nature of the stimuli required to trigger these transcriptional responses appears to differ in the two species and may help to account for the differences in pathogenicity of these two organisms (41, 50).
CONCLUSIONS
Fungi have evolved through several specific evolutionary mechanisms. CGA has revealed that virulence and virulence-associated factors have evolved through the same mechanisms in primary and opportunistic human pathogens (summarized in Fig. 2). One of the most important of these mechanisms involves gene duplication and subsequent expansion of specific gene families and clusters. Many of the genes expressed by pathogenic fungi confer flexibility in nutrient acquisition and metabolic diversity (e.g., A. fumigatus) as well as in host recognition and adhesion (e.g., C. albicans). Expanded gene clusters and gene islands implicated in virulence are often present in subtelomeric regions (e.g., the EPA genes of C. glabrata), with families undergoing expansion adjacent to telomeres and being exchanged between the telomeres of individual chromosomes (e.g., the TLO genes of C. albicans). Lineage-specific genes have also been identified in the subtelomeric regions of nonpathogenic fungi belonging to the genus Saccharomyces, suggesting that the chromatin structure adjacent to telomeres facilitates accelerated evolution of genes located there and that these evolutionary hot spots are a common feature of fungi in general (30). In addition to gene “gain” through expansion, there are also clear cases of gene “loss” occurring in pathogenic fungi, particularly in species that are specifically adapted for life as mammalian commensals, which is suggestive of a process of reductive evolution (e.g., C. glabrata). Genomic analysis also indicates that evolution of virulence in fungi has taken a different path from that in bacterial pathogens. In bacteria, one of the most important means of acquiring virulence is through HGT of virulence factor genes (16). However, although there is evidence for HGT between different fungal species (23, 31) and between bacteria and fungi (19), it appears to be relatively rare, and there have been no examples detected of the transfer of genes that might contribute directly to virulence in humans.
Fig. 2.
Summary of the genomic processes responsible for the evolution of human fungal pathogens.
Although the genomes of the most important fungal pathogens have become available only recently, they have already provided very useful insight into how fungi cause disease in humans and how fungal virulence has evolved. Up to now, comparative genomics has been hampered by the lack of publicly available completed fungal genome sequences. However, new rapid and high-throughput DNA and RNA sequencing technologies offer the potential to generate genomic and transcriptomic data for multiple strains and species, allowing the comparison of any number of strains that differ in the capacity to cause disease or in their host or geographic range. Comparison of the genome sequences of multiple strains will facilitate the functional analysis of the many genes of unknown function in all of the fungal pathogens and will facilitate the identification of rapidly evolving genes, which are most likely to play a role in host-pathogen interaction. Similar comparative studies using whole-transcriptome shotgun sequencing (also referred to as RNA-seq) have the potential to revolutionize fungal pathogenomics and identify new virulence-associated genes. We are now on the brink of an explosion of genomic information, and while a major bottleneck in the past was the lack of sequence data, the coming tsunami of genomic data will provide a new challenge to bioinformaticians and evolutionary biologists.
ACKNOWLEDGMENTS
Work in our laboratory is supported by the Board of the Dublin Dental Hospital, Science Foundation Ireland (04/IN3/B463), and the Health Research Board (RP/2004/235 and HRA/2009/3).
Footnotes
Published ahead of print on 12 November 2010.
REFERENCES
- 1. Almeida R. S., et al. 2008. The hyphal-associated adhesin and invasin Als3 of Candida albicans mediates iron acquisition from host ferritin. PLoS Pathog. 4:e1000217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Asmundsdóttir L. R., Erlendsdóttir H., Agnarsson B. A., Gottfredsson M. 2009. The importance of strain variation in virulence of Candida dubliniensis and Candida albicans: results of a blinded histopathological study of invasive candidiasis. Clin. Microbiol. Infect. 15:576–585 [DOI] [PubMed] [Google Scholar]
- 3. Bates S., et al. 2007. Candida albicans Iff11, a secreted protein required for cell wall structure and virulence. Infect. Immun. 75:2922–2928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Berriman M., et al. 2002. The architecture of variant surface glycoprotein gene expression sites in Trypanosoma brucei. Mol. Biochem. Parasitol. 122:131–140 [DOI] [PubMed] [Google Scholar]
- 5. Biswas S., Van Dijck P., Datta A. 2007. Environmental sensing and signal transduction pathways regulating morphopathogenic determinants of Candida albicans. Microbiol. Mol. Biol. Rev. 71:348–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Blyth W., Grant I. W., Blackadder E. S., Greenberg M. 1977. Fungal antigens as a source of sensitization and respiratory disease in Scottish maltworkers. Clin. Allergy 7:549–562 [DOI] [PubMed] [Google Scholar]
- 7. Braun B. R., et al. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1:36–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Butler G., et al. 2009. Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459:657–662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cairns T., Minuzzi F., Bignell E. 2010. The host-infecting fungal transcriptome. FEMS Microbiol. Lett. 307:1–11 [DOI] [PubMed] [Google Scholar]
- 10. Chim C. S., Ho P. L., Yuen K. Y. 1998. Simultaneous Aspergillus fischeri and herpes simplex pneumonia in a patient with multiple myeloma. Scand. J. Infect. Dis. 30:190–191 [DOI] [PubMed] [Google Scholar]
- 11. Dagenais T. R., Keller N. P. 2009. Pathogenesis of Aspergillus fumigatus in invasive aspergillosis. Clin. Microbiol. Rev. 22:447–465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. De Las Peñas A., et al. 2003. Virulence-related surface glycoproteins in the yeast pathogen Candida glabrata are encoded in subtelomeric clusters and subject to RAP1- and SIR-dependent transcriptional silencing. Genes Dev. 17:2245–2258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. de Repentigny L., Aumont F., Bernard K., Belhumeur P. 2000. Characterization of binding of Candida albicans to small intestinal mucin and its role in adherence to mucosal epithelial cells. Infect. Immun. 68:3172–3179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dujon B. 2010. Yeast evolutionary genomics. Nat. Rev. Genet. 11:512–524 [DOI] [PubMed] [Google Scholar]
- 15. Dujon B., et al. 2004. Genome evolution in yeasts. Nature 430:35–44 [DOI] [PubMed] [Google Scholar]
- 16. Ehrlich G., Hiller N. L., Hu F. 2008. What makes pathogens pathogenic. Genome Biol. 9:225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Enjalbert B., et al. 2009. Genome-wide gene expression profiling and a forward genetic screen show that differential expression of the sodium ion transporter Ena21 contributes to the differential tolerance of Candida albicans and Candida dubliniensis to osmotic stress. Mol. Microbiol. 72:216–228 [DOI] [PubMed] [Google Scholar]
- 18. Fedorova N. D., et al. 2008. Genomic islands in the pathogenic filamentous fungus Aspergillus fumigatus. PLoS Genet. 4:e1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fitzpatrick D., Logue M., Butler G. 2008. Evidence of recent interkingdom horizontal gene transfer between bacteria and Candida parapsilosis. BMC Evol. Biol. 8:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gácser A., Trofa D., Schäfer W., Nosanchuk J. D. 2007. Targeted gene deletion in Candida parapsilosis demonstrates the role of secreted lipase in virulence. J. Clin. Invest. 117:3049–3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galagan J. E., et al. 2005. Sequencing of Aspergillus nidulans and comparative analysis with A. fumigatus and A. oryzae. Nature 438:1105–1115 [DOI] [PubMed] [Google Scholar]
- 22. Gilfillan G. D., et al. 1998. Candida dubliniensis: phylogeny and putative virulence factors. Microbiology 144:829–838 [DOI] [PubMed] [Google Scholar]
- 23. Hall C., Brachat S., Dietrich F. S. 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot. Cell 4:1102–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hannula J., et al. 2000. Comparison of virulence factors of oral Candida dubliniensis and Candida albicans isolates in healthy people and patients with chronic candidosis. Oral Microbiol. Immunol. 15:238–244 [DOI] [PubMed] [Google Scholar]
- 25. Hoyer L. L., et al. 2001. Characterization of agglutinin-like sequence genes from non-albicans Candida and phylogenetic analysis of the ALS family. Genetics 157:1555–1567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hube B., et al. 2000. Secreted lipases of Candida albicans: cloning, characterisation and expression analysis of a new gene family with at least ten members. Arch. Microbiol. 174:362–374 [DOI] [PubMed] [Google Scholar]
- 27. Jackson A. P., et al. 2009. Comparative genomics of the fungal pathogens Candida dubliniensis and C. albicans. Genome Res. 19:2231–2244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jain R., Rivera M. C., Moore J. E., Lake J. A. 2003. Horizontal gene transfer accelerates genome innovation and evolution. Mol. Biol. Evol. 20:1598–1602 [DOI] [PubMed] [Google Scholar]
- 29. Kavanaugh L. A., Fraser J. A., Dietrich F. S. 2006. Recent evolution of the human pathogen Cryptococcus neoformans by intervarietal transfer of a 14-gene fragment. Mol. Biol. Evol. 23:1879–1890 [DOI] [PubMed] [Google Scholar]
- 30. Kellis M., Patterson N., Endrizzi M., Birren B., Lander E. S. 2003. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423:241–254 [DOI] [PubMed] [Google Scholar]
- 31. Khaldi N., Collemare J., Lebrun M. H., Wolfe K. H. 2008. Evidence for horizontal transfer of a secondary metabolite gene cluster between fungi. Genome Biol. 9:R18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Khaldi N., Wolfe K. H. 2008. Elusive origins of the extra genes in Aspergillus oryzae. PLoS One 3:e3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lavoie H., Hogues H., Whiteway M. 2009. Rearrangements of the transcriptional regulatory networks of metabolic pathways in fungi. Curr. Opin. Microbiol. 12:655–663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Lin X. 2009. Cryptococcus neoformans: morphogenesis, infection, and evolution. Infect. Genet. Evol. 9:401–416 [DOI] [PubMed] [Google Scholar]
- 35. Loftus B. J., et al. 2005. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science 307:1321–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Luo G., et al. 2010. Candida albicans Hyr1p confers resistance to neutrophil killing and is a potential vaccine target. J. Infect. Dis. 201:1718–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McCullough M., Ross B., Reade P. 1995. Characterization of genetically distinct subgroup of Candida albicans strains from oral cavities of patients infected with human immunodeficiency virus. J. Clin. Microbiol. 33:696–700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. McDonagh A., et al. 2008. Sub-telomere directed gene expression during initiation of invasive aspergillosis. PLoS Pathog. 4:e1000154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mishra P. K., Baum M., Carbon J. 2007. Centromere size and position in Candida albicans are evolutionarily conserved independent of DNA sequence heterogeneity. Mol. Genet. Genomics 278:455–465 [DOI] [PubMed] [Google Scholar]
- 40. Moran G. P., Sullivan D. J., Coleman D. C. 2002. Emergence of non-Candida albicans Candida species as pathogens, p. 37–53In Calderone R. A. (ed.), Candida and candidiasis. ASM Press, Washington, DC [Google Scholar]
- 41. O'Connor L., Caplice N., Coleman D. C., Sullivan D. J., Moran G. P. 2010. Differential filamentation of Candida albicans and C. dubliniensis is governed by nutrient regulation of UME6 expression. Eukaryot. Cell 9:1383–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perlroth J., Choi B., Spellberg B. 2007. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med. Mycol. 45:321–346 [DOI] [PubMed] [Google Scholar]
- 43. Pfaller M. A., Diekema D. J. 2010. Epidemiology of invasive mycoses in North America. Crit. Rev. Microbiol. 36:1–53 [DOI] [PubMed] [Google Scholar]
- 44. Phan Q. T., et al. 2007. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 5:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pujol C., et al. 2004. The closely related species Candida albicans and Candida dubliniensis can mate. Eukaryot. Cell 3:1015–1027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ramirez M. A., Lorenz M. C. 2007. Mutations in alternative carbon utilization pathways in Candida albicans attenuate virulence and confer pleiotropic phenotypes. Eukaryot. Cell 6:280–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rosas-Hernandez L. L., et al. 2008. yKu70/yKu80 and Rif1 regulate silencing differentially at telomeres in Candida glabrata. Eukaryot. Cell 7:2168–2178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Sharpton T. J., et al. 2009. Comparative genomic analyses of the human fungal pathogens Coccidioides and their relatives. Genome Res. 19:1722–1731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Sofair A. N., et al. 2006. Epidemiology of community-onset candidemia in Connecticut and Maryland. Clin. Infect. Dis. 43:32–39 [DOI] [PubMed] [Google Scholar]
- 50. Spiering M. J., et al. 2010. Comparative transcript profiling of Candida albicans and Candida dubliniensis identifies SFL2, a C. albicans gene required for virulence in a reconstituted epithelial infection model. Eukaryot. Cell 9:251–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Staib P., Morschhauser J. 2005. Differential expression of the NRG1 repressor controls species-specific regulation of chlamydospore development in Candida albicans and Candida dubliniensis. Mol. Microbiol. 55:637–652 [DOI] [PubMed] [Google Scholar]
- 52. Stokes C., et al. 2007. Lower filamentation rates of Candida dubliniensis contribute to its lower virulence in comparison with Candida albicans. Fungal Genet. Biol. 44:920–931 [DOI] [PubMed] [Google Scholar]
- 53. Sullivan D. J., Moran G. P., Coleman D. C. 2005. Candida dubliniensis: ten years on. FEMS Microbiol. Lett. 253:9–17 [DOI] [PubMed] [Google Scholar]
- 54. Sullivan D. J., Westerneng T. J., Haynes K. A., Bennett D. E., Coleman D. C. 1995. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology 141:1507–1521 [DOI] [PubMed] [Google Scholar]
- 55. Thewes S., et al. 2008. Phenotypic screening, transcriptional profiling, and comparative genomic analysis of an invasive and non-invasive strain of Candida albicans. BMC Microbiol. 8:187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Tuch B. B., Galgoczy D. J., Hernday A. D., Li H., Johnson A. D. 2008. The evolution of combinatorial gene regulation in fungi. PLoS Biol. 6:e38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Uhl M. A., Biery M., Craig N., Johnson A. D. 2003. Haploinsufficiency-based large-scale forward genetic analysis of filamentous growth in the diploid human fungal pathogen C. albicans. EMBO J. 22:2668–2678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. van het Hoog M., et al. 2007. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol. 8:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vilela M. M., et al. 2002. Pathogenicity and virulence of Candida dubliniensis: comparison with C. albicans. Med. Mycol. 40:249–257 [DOI] [PubMed] [Google Scholar]
- 60. Wilson L. S., et al. 2002. The direct cost and incidence of systemic fungal infections. Value Health 5:26–34 [DOI] [PubMed] [Google Scholar]