Abstract
The ability to isolate fetal nucleated red blood cells (NRBCs) from the maternal circulation makes possible prenatal genetic analysis without the need for diagnostic procedures that are invasive for the fetus. Such isolation requires antibodies specific to fetal NRBCs. To generate a panel of antibodies to antigens present on fetal NRBCs, a new type of nonimmune phage antibody library was generated in which multiple copies of antibody fragments are displayed on each phage. Antibody fragments specific for fetal NRBCs were isolated by extensive predepletion of the phage library on adult RBCs and white blood cells (WBCs) followed by positive selection and amplification on fetal liver erythroid cells. After two rounds of selection, 44% of the antibodies analyzed bound fetal NRBCs, with two-thirds of these showing no binding of WBCs. DNA fingerprint analysis revealed the presence of at least 16 unique antibodies. Antibody specificity was confirmed by flow cytometry, immunohistochemistry, and immunofluorescence of total fetal liver and adult RBCs and WBCs. Antibody profiling suggested the generation of antibodies to previously unknown fetal RBC antigens. We conclude that multivalent display of antibodies on phage leads to efficient selection of panels of specific antibodies to cell surface antigens. The antibodies generated to fetal RBC antigens may have clinical utility for isolating fetal NRBCs from maternal circulation for noninvasive prenatal genetic diagnosis. Some of the antibodies may also have possible therapeutic utility for erythroleukemia.
Keywords: antibody phage display, monoclonal antibody, single chain Fv, fetal erythroid antibodies
It has long been known that fetal red blood cells (RBCs) routinely leak into the maternal circulation during normal pregnancy (1, 2). More recently, it has been established that a very small number of fetal nucleated RBCs (NRBCs) are also routinely present in the maternal circulation (3, 4). These cells are considered the ideal target for noninvasive DNA prenatal diagnosis, but presently they cannot be readily isolated from the maternal circulation in high enough numbers and purity for routine clinical use. Because the isolation methods for purifying fetal NRBCs from maternal circulation rely on antibody-based separation and detection techniques, progress in this area has been hampered by the relative lack of antibodies to unique fetal erythroid antigenic determinants (5). Well characterized antigens expressed on fetal erythroid cells but not adult RBCs, such as CD71 and CD36, are also expressed on a number of adult white blood cells (WBCs) resulting in contamination by many WBCs in purification techniques relying on these antibodies.
Fetal erythroid lineage antigens classically have been identified by massive screening of mAbs produced by conventional murine hybridoma technology using mice immunized with human fetal NRBCs. The majority of antibodies generated by this method are nonspecific and react with irrelevant epitopes present on all human cells. Conventional murine hybridoma technology also tends to produce antibodies only to immunogenic antigens, because it relies on natural immune response in an animal. Thus, antibodies to antigens that are strongly evolutionarily conserved tend not to be produced by this technology.
To overcome these limitations, we applied antibody phage display technology to isolate new fetal erythroid lineage specific antibodies. In antibody phage display, large nonimmune libraries are created and display single-chain variable antibody fragments (scFv) on the surface of filamentous bacteriophage virions (refs. 6 and 7; reviewed in ref. 8). The gene for the displayed antibody is carried in the phage genome, thus linking genotype with phenotype. Antigen specific antibodies are selected from the library by a variety of different affinity chromatography techniques. Because this approach does not depend on a natural immune response and uses entirely in vitro selection techniques, antibodies can be isolated to any antigens, including nonimmunogenic and conserved antigens (9–11). Antibodies to cell surface antigens can be directly isolated from phage antibody libraries by panning on cells, including blood cells (12, 13). In fact, RBCs were the first cell type used to demonstrate the feasibility of cell surface selection by antibody phage display (12). Such cell selections, however, have not proven generally successful for generation of panels of cell-type specific antibodies. Here we describe the generation of a new type of nonimmune phage antibody library in which multiple copies of antibody fragments are displayed on each phage and report its successful application to generate a panel of antibodies to unique fetal erythroid cell surface markers.
Methods
Blood Cell Preparations.
Buffy coats containing peripheral blood leukocytes were obtained from the Irwin Memorial Blood Bank (San Francisco). Fetal livers of gestational ages ranging from 14–24 weeks were obtained from San Francisco General Hospital with the approval of the University of California, San Francisco Committee for the Protection of Human Subjects. For phage antibody selection and immunocytochemistry, fetal erythroid cells were isolated from the human fetal liver by straining through 70 μm nylon mesh (Becton Dickinson Labware, Franklin Lakes, NJ) to remove fetal hepatocytes and clumped cells, followed by panning on polystyrene plates coated with anti-glycophorin A (GPA) antibodies (Beckman Coulter, Westbrook, ME) at 10 μg/ml in 0.5 M Tris⋅HCl (pH 9.5) as follows: fetal cells were resuspended in 3 ml of PBS supplemented with 5% FCS at a concentration of 107 cells/ml and allowed to attach for 2 h at 4°C. Cells that did not attach were removed by washing four times with PBS/1% FCS.
For flow cytometry, light-density fetal liver cells, containing a high proportion of immature erythroid progenitors, were isolated by first homogenizing the liver through a wire mesh and washing the cells in PBS containing 0.5% fraction-V ethanol-extracted BSA (Boehringer Mannheim), and 50 μg/ml gentamicin (GIBCO/BRL). The fetal liver cells were next layered on a 1.077 g/ml solution of Nycoprep (GIBCO/BRL) and centrifuged at 1,000 × g for 25 min at room temperature. The cells were washed and resuspended in PBS/0.5% BSA for phenotypic analysis. Light-density fetal liver cells depleted of GPA+ cells were prepared by immunomagnetic bead depletion as described (14).
Phage Display Library Construction.
To generate phage displaying multiple copies of antibody fragment, an scFv phage antibody library was constructed in fd phage. The fd phage display library (B.B., Dave O'Connell, and J.D.M., unpublished work) was derived from a 7 × 109 member phagemid library (11) by subcloning the SfiI/NotI scFv insert from pHEN1 into fd-SfiI/NotI (15), (provided by Andrew Griffiths, Medical Research Council, Cambridge, U.K.). Ligation mixtures were used to transform Escherichia coli TG1 and the transformation mixture plated on TYE plates containing 15 μg/ml tetracycline. Library size was calculated by counting the number of tetracycline-resistant colonies. Library quality was verified by determining the percentage of clones with inserts of appropriate size for an scFv gene, performed by colony PCR screening using the primers fdseq (7) and fd2, 5′-TTTTTGGAGATTTTCAAC-3′. Library diversity was confirmed by BstN1 fingerprinting the amplified scFv genes (16) as described in ref. 7. The library was stored in 2× TY containing 15 μg/ml tetracycline and 15% glycerol at −80°C.
Phage Library Preparation and Selection.
Phage were prepared by inoculation in 1 liter of 2× TY containing 15 μg/ml tetracycline with an aliquot of library glycerol stock, and the culture was grown overnight at 30°C with shaking at 250 rpm. Phage were harvested by centrifugation, concentrated by precipitation with polyethylene glycol (PEG) (7), and purified by CsCl gradient centrifugation as described (17). Phage concentration was determined by titering on E. coli TG1.
Before selection, the phage library was extensively depleted against a mixture of adult RBCs and WBCs. A total of 1012 phage particles were incubated with 109 adult RBCs and 108 adult WBCs in PBS/1% BSA in a total volume of 1 ml for 15 min at room temperature with rotation. After incubation, phage binding adult RBCs and WBCs were removed by centrifugation and collection of the supernatant. The supernatant was used to resuspend fresh adult RBCs and WBCs. This procedure was repeated six times each with adult RBCs and WBCs The supernatant was further depleted of phage binding adult RBCs by incubation for 60 min at 4°C with adult RBCs attached to 15-cm polystyrene plates coated with 10 μg/ml anti-GPA antibodies. This supernatant was further depleted of phage binding WBCs by incubation for 60 min at 4°C with adult human WBCs attached to 15-cm polystyrene plates coated with 10 μg/ml anti-CD45 and anti-CD13 (Caltag, Burlingame, CA).
After the depletion steps, supernatant containing phage was incubated at 4°C for 60 min with fetal NRBCs attached to 10-cm polystyrene plates coated with 10 μg/ml anti-GPA antibodies. Plates were washed 10 times with 10 ml of ice-cold PBS/0.5% BSA. After washing, the fetal RBCs were scraped off the plates, washed twice with PBS/0.5% BSA and collected by centrifugation. After washing, fetal RBCs were lysed with 1 ml of 100 mM Triethlyamine (Sigma). The lysate was neutralized with 0.5 ml of 1 M Tris⋅HCl (pH 6.8) and then used to infect 10 ml of exponentially growing E. coli TG1 as described (7). E. coli was grown at 37°C for 1 h with shaking at 250 rpm after which time the culture was plated on TYE plates containing 15 μg/ml tetracycline. After overnight growth, colonies were scraped from the plates and used to generate phage for a second round of selection using depletion and positive selection steps as described above. The number of unique scFv was estimated by PCR fingerprinting of the scFv genes with the restriction enzyme BstNI.
Flow Cytometry.
For flow cytometry, phage was prepared from individual colonies. Phage was prepared by inoculation of 500 ml of 2× TY containing 15 μg/ml tetracycline with individual clones and grown overnight at 30°C. Phage were concentrated by PEG precipitation and resuspended in 2 ml of PBS/1% BSA. FACS analysis was performed on a FACScan or FACSCalibur flow cytometer (Becton Dickinson Immunchemistry Systems).
Blood cells were prepared as described above, washed with PBS/1% BSA and then incubated with phage. Approximately 1–5 × 105 cells and 1012 phage were incubated on ice for 1 h in 100 μl of PBS/1% BSA. After washing twice, cells were resuspended in 100 μl of biotinylated polyclonal anti-M13 (5 Prime → 3 Prime) diluted 1:2,000 in PBS/1% BSA and incubated on ice for 30 min. After being washed twice, cells were resuspended in either streptavidin-R-phycoerythrin (PE) conjugate (Molecular Probes) or streptavidin-fluorescein isothiocyanate (FITC) (Molecular Probes) diluted 1:200 in PBS/1% BSA and incubated on ice for 30 min. DNA was stained by incubating cells with Hoescht 33342 (Molecular Probes) at 10 μg/ml on ice for 30 min. Cells were washed twice and analyzed by flow cytometry. Dead cells were stained with 1 μg/ml propidium iodide (Sigma).
Total light-density fetal liver cells and GPA− light-density fetal liver cells were analyzed for binding of FITC-labeled phage and PE-labeled GPA or CD34 (Beckman Coulter), respectively. Binding of FITC-labeled phage was directly compared with binding of FITC-labeled CD36 (Beckman Coulter) and CD71 (Becton Dickinson Immunchemistry Systems) mAbs. Phage were directly labeled with FITC by incubating on ice, for 90 min, 1012 phage in 500 μl of 100 mM NaHCO3 (pH 8.5) with 5 μl of 6-[fluorescein-5-(and -6)-carboxamido]hexanoic acid, succinimidyl ester [5(6)-SFX] (Molecular Probes) suspended at 10 mg/ml in N,N-dimethyl formamide. Phage was precipitated with PEG and resuspended three times. After labeling with FITC, the phage were resuspended in 500 μl of PBS/1% BSA and then used for cell staining. Approximately 1–5 × 105 cells were stained, in a volume of 20–50 μl in 96-well V-bottom plates (Costar), with saturating levels of labeled phage or mAbs for 30 min on ice. Thereafter, the cells were washed twice with washing buffer consisting of PBS containing 0.5% BSA and 0.01% NaN3. The cells were resuspended in the same washing buffer supplemented with propidium iodide for the analysis of live cells.
Immunohistochemisty and Immunofluorescence Microscopy.
For cell staining, biotinylated phage was used. Phage was biotinylated by incubating 1012 phage in 500 μl of 100 mM NaHCO3 (pH 8.5) with 40 μl of 2 mg/ml Sulfo-NHS-LC-biotin (Pierce) for 30 min on ice. Phage was precipitated with PEG and resuspended three times. For staining, 106 cells were first blocked in a total volume of 100 μl of PBS/1% BSA containing the helper phage M13K07 at a concentration of 1013/ml for 20 min. Ten microliters of biotinylated phage antibody was added to the cells and incubated for 30 min on ice. Cells were washed with PBS/1% BSA and used to prepare slides for immunohistochemistry and immunofluorescence using a cytocentrifuge (Sakura Finetek, Torrance, CA). Slides were air dried overnight and fixed with 2% formaldehyde at room temperature for 20 min. After washing, phage staining was detected by alkaline phosphatase conjugated streptavidin (DAKO) diluted 1:120 with PBS at room temperature for 20 min. Slides were washed twice in PBS and then developed with Fast Red (DAKO) at room temperature for 20 min. For immunofluorescence, phage binding was detected by incubation with Alexa fluor 546-conjugated streptavidin (Molecular Probes) diluted 1:400 with PBS. Fetal hemoglobin was also detected by incubation with anti-γ (Hb F) mAb (Perkin-Elmer Wallace) diluted 1:400, followed by staining with a goat anti-mouse FITC-conjugated antibody (Caltag) (18). DNA was stained with 4′,6-diamidino-2-phenylindole mounting media (Vector Laboratories). The stained cells were evaluated by using Zeiss Axiophot fluorescence microscope with filters (UV, blue, or green excitation) for 4′,6-diamidino-2-phenylindole, FITC, or Alexa fluor 546, respectively, and a dual band filter (blue and green excitation) for combined FITC and Alexa 546 detection.
Results
To generate mAbs to fetal erythroid antigens, a scFv phage antibody library was constructed in the phage vector fdDOG (6). Unlike antibody libraries constructed in phagemid vectors, use of a phage vector yields three to five copies of scFv-pIII fusion per phage, resulting in multivalent display. Such multivalent display confers an increase in the functional affinity constant of the phage antibodies and should theoretically result in both more efficient depletion of antibodies to antigens in common with adult cells, and more efficient positive selection of antibodies to fetal antigens. To construct a nonimmune phage antibody library, an scFv gene repertoire was subcloned as SfiI–NotI gene fragments from a large nonimmune phage antibody library in the phagemid vector pHEN1 into the phage vector fd. After transformation, a library of 3.2 × 108 tetracycline-resistant clones was obtained. PCR screening revealed that 95% of clones had an scFv size insert, yielding a functional library size of 3.04 × 108 members. To verify that scFv display levels were higher in phage vs. phagemid vectors, phage were prepared from both libraries, and subjected to SDS/PAGE followed by Western blotting with an anti-pIII antibody. Blots indicated that for the phage library, approximately 50% of pIII consisted of scFv fusion. In contrast, only 1–5% of pIII consisted of scFv fusion for the phagemid library (data not shown).
To generate a panel of mAbs to fetal NRBC antigens, the phage library was panned on fetal nucleated erythroid cells obtained from fetal liver. Because fetal erythroid cells express antigens that are also present on some adult WBCs and RBCs, the phage library was first extensively depleted against both adult RBCs and adult WBCs. After the first round of selection, the titer of the recovered phage was 3 × 104. The recovered phage were amplified by growth in bacteria and used for a second round of selection. After the second round, the titer of the recovered phage was 5 × 105.
To determine the outcome of the selection strategy, polyclonal phage were prepared after the second round of selection and analyzed for binding to adult buffy coat WBCs and total fetal liver cells by flow cytometry. Only a very small amount of binding was detected on adult WBCs, whereas the majority of cells from the total fetal liver bound phage (Fig. 1). To identify phage that specifically bound fetal erythroid antigens, phage were prepared from individual colonies from the second round of selection and analyzed for fetal erythroid cell binding and adult WBC binding by flow cytometry. Of 95 random clones analyzed, 42 bound fetal erythroid cells by flow cytometry. Two-thirds of the antibodies showed no evidence of WBC binding (for example, mAb fd-H7 in Fig. 2). One-third of the mAbs bound a small percentage of WBC from the buffy coat as determined by flow cytometry (for example, mAb fd-B6 in Fig. 2). To determine the number of unique antibodies generated, the scFv gene was amplified by PCR and the PCR product was digested with the frequently cutting restriction enzyme BstN1 (PCR fingerprinting). From the 42 fetal erythroid binding scFv, 16 unique fingerprints were observed, indicating the presence of 16 unique antibodies (Fig. 3).
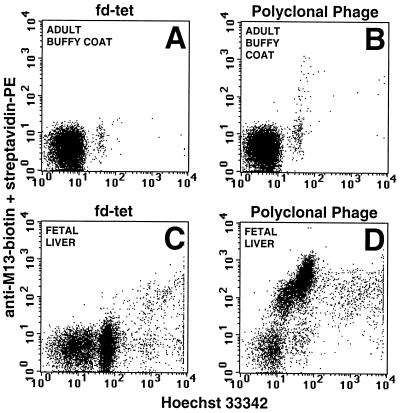
Figure 1.
Binding of polyclonal phage antibodies to adult buffy coat WBCs and total fetal liver cells. After the second round of selection, polyclonal phage were prepared and analyzed for their ability to bind either adult buffy coat WBCs (B) or total fetal liver (D). There is a large shift in the fetal liver cells compared with a minimal shift of a small population of adult buffy coat WBCs. In contrast, control wild-type fd phage did not significantly shift either cell population (A and C).
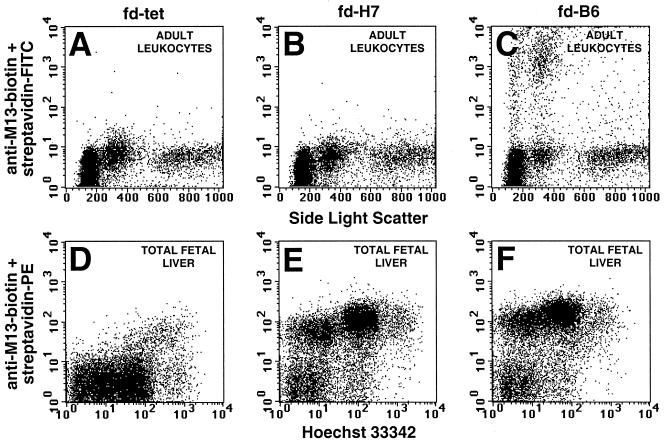
Figure 2.
Binding of monoclonal phage antibodies to adult buffy coat WBCs and total fetal liver cells. Phage were prepared from individual colonies after the second round of selection and analyzed for their ability to bind adult buffy coat WBCs or total fetal liver cells. The representative antibody fd-H7 (B and E) bound only fetal liver cells, whereas the antibody fd-B6 (C and F) bound fetal liver and a small subpopulation of WBCs consistent with monocytes. Staining with wild-type fd phage (A and D) is included as a control.
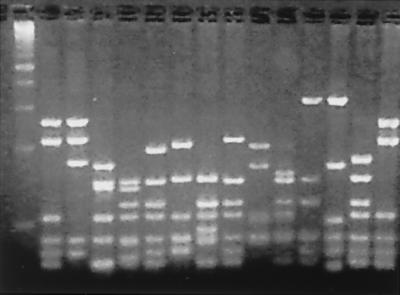
Figure 3.
DNA fingerprint analysis of the scFv genes of individual antibodies from the second round of selection. scFv DNA was amplified by PCR directly from colonies and digested with the frequently cutting restriction enzyme BstN1. A diverse banding pattern was observed, with each unique pattern representing a unique antibody sequence. First lane is a 100-bp DNA marker.
These 16 phage antibodies were analyzed according to their binding to fetal liver derived light-density cells that are comprised predominantly of NRBCs but also of hematopoietic progenitors as well as mature leukocytes (Table 1). The phage antibodies bound 70–99% of the GPA+ population of these light-density cells. The binding to the GPA− population was more variable, ranging from 7.5–71.5% cells, suggesting that they were binding to different antigens on the more primitive erythroid cells. These phage antibodies were then further analyzed for their binding to CD34+ or CD34− cells. Their binding properties were also compared with those of anti-CD36 and anti-CD71 mAbs. By these criteria, as well as by judging the FACS analysis patterns, the phage antibodies could be grouped into five clusters. Two of them that had the binding characteristics similar to antibodies against CD36 or CD71 also demonstrated appreciable binding to adult peripheral blood cells. The other three clusters, A, B, and C, bound fetal liver-derived, GPA+ light-density cells but not mature RBCs. They differ from one another in that clusters A and C bound more GPA+ cells than cluster B did, and cluster C bound more CD34+ cells from the GPA− fraction than clusters A and B did. The antigen detected by cluster B also appeared to be more restricted to GPA+ cells. These results indicate that the phage antibodies that we have generated are binding to different antigens on immature erythroid and hematopoietic cells other than CD36 and CD71.
Table 1.
Binding of phage antibodies by subsets of fetal liver cells
| Antibody fragment/mAb* | Light-density
fetal liver
|
GPA− light-density fetal liver
|
||
|---|---|---|---|---|
| GPA+ | GPA− | CD34+ | CD34− | |
| Cluster A | ||||
| FSH8 | 90.9% | 16.9% | 6.6% | 5.3% |
| FSH7 | 92.1% | 39.6% | 2.0% | 3.9% |
| FSG5 | 98.7% | 66.7% | 7.0% | 6.7% |
| FSG9 | 86.2% | 45.7% | 3.1% | 4.2% |
| Cluster B | ||||
| C7 | 71.6% | 7.5% | 5.5% | 4.8% |
| F4 | 81.7% | 9.8% | 5.4% | 4.6% |
| C3 | 84.1% | 7.9% | 7.1% | 6.5% |
| B5 | 88.0% | 8.0% | 9.9% | 8.7% |
| Cluster C | ||||
| FSE2 | 99.2% | 46.9% | 18.4% | 8.2% |
| A6 | 81.6% | 11.3% | 14.8% | 6.2% |
| CD36* | 98.4% | 34.6% | 43.2% | 25.7% |
| FSH3 | 90.7% | 22.0% | 22.9% | 18.9% |
| FSD8 | 99.2% | 53.6% | 53.7% | 51.5% |
| CD71* | 99.2% | 47.8% | 52.0% | 13.7% |
| FSA7 | 97.9% | 30.3% | 42.8% | 20.0% |
| FSD10 | 99.4% | 51.3% | 45.0% | 18.0% |
| H7 | 99.3% | 71.5% | 61.9% | 23.0% |
| G4 | 99.1% | 34.0% | 39.0% | 13.8% |
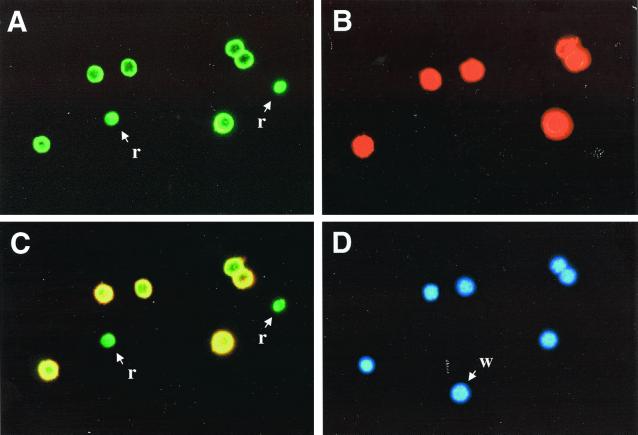
To confirm further which of these antibodies were binding specifically to fetal NRBCs, immunohistochemistry was performed on total fetal liver and on adult WBC buffy coats. About two-thirds of the antibodies stained only the erythroid cells in fetal liver but not the buffy coat WBCs (Fig. 4 A and C). The other one-third stained erythroid cells in fetal liver and also adult monocytes (Fig. 4 B and D). These results confirm the specificity demonstrated by flow cytometry. As an additional demonstration of specificity, immunofluorescence was performed on fetal liver cells, and a representative result is shown in Fig. 5. Only fetal hemoglobin containing cells with nuclei (Fig. 5D) were stained by the FITC-tagged anti-fetal hemoglobin mAb (Fig. 5A) and by the Alexa 546-tagged phage antibody (Fig. 5B). When visualized with a dual band filter, the fetal NRBCs yielded yellow fluorescence (Fig. 5C). The two enucleated fetal RBCs stained only green with FITC, whereas the WBC stained with neither.
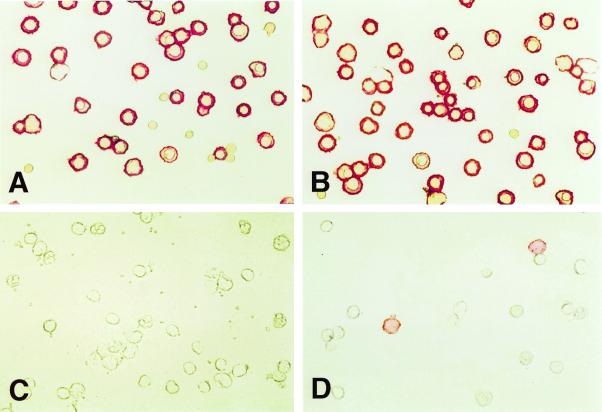
Figure 4.
Staining of adult buffy coat WBCs and total fetal liver by phage antibodies. Adult buffy coat WBCs or fetal liver cells were stained with biotinylated phage antibodies and the cells applied to microscope slides, fixed, and stained with streptavidin-alkaline phosphatase and Fast Red. The phage antibody FSA7 stains fetal erythroid cells (A) but not WBCs (C), whereas the phage antibody FSH3 stains both fetal liver (B) and a subset of WBCs (D).
Figure 5.
Staining of total fetal liver by a fetal NRBC specific phage antibody. Fetal liver cells were incubated with biotinylated phage antibody FSG9, applied to microscope slides, and then stained with FITC anti-fetal hemoglobin antibody (A), streptavidin Alexa fluor 546 to detect phage binding (B), or 4′,6-diamidino-2-phenylindole (D). Fetal hemoglobin and phage binding were visualized together with a dual band filter (C). Only fetal NRBCs stained with both. r, enucleated RBC; w, WBC.
Discussion
Production of mAbs by xenotypic immunization of mice with intact human cells usually results in the production of antibodies against immunodominant epitopes found on more than one cell type. As a result, it is impossible to generate a complete set of antibodies to surface receptors by using hybridoma technology. Phage antibody libraries represent a potential solution to this problem, and successful cell surface selections have been reported by using fixed (19) or living cells (12, 13, 20–23). However, because of the heterogeneity of the starting material, such selections require elaborate subtraction protocols to avoid the selection of irrelevant antibodies. Although there are several reports of successful selections on cells using large nonimmune libraries (10, 12, 13), this approach has been most successful by using small libraries from immunized sources. This limits the spectrum of antibody specificities obtainable from any single library to those present on the immunizing cell, and does not completely overcome the problem of immunodominant epitopes. Furthermore, the antibodies obtained are usually murine.
The step limiting the selection of binders from large naïve libraries by cell panning seems to be the relatively high background binding of nonspecific phage and relatively low binding of specific phage (24–26). The low binding of specific phage is partially related to the low concentration of a given binding phage in the polyclonal preparation (approximately 1.6 × 10−17 M for a single member of a 109 library in a phage preparation of 1 × 1013 particles per ml). The low concentration simultaneously limits the efficiency of both subtraction of common binders and enrichment of specific binders. To overcome this limitation, we generated a large nonimmune phage library in the phage vector fd. Compared with existing nonimmune libraries constructed in phagemid vectors, such phage vectors will display an antibody fragment on each of the five copies of pIII, leading to multivalent antibody display. Multivalent display leads to an increase in the functional affinity constant of the phage antibody, resulting in both increased efficiency of depletion and positive selection. In contrast, phagemid libraries display less than one antibody fragment per phage because of the presence of wild-type pIII from the helper phage.
For the purposes of prenatal genetic diagnosis, there are several advantages of targeting fetal erythroid cells. Fetal NRBCs are already found in the mother's blood during the first trimester and are present throughout the entire pregnancy. They have a limited lifespan and do not persist from prior pregnancies as it has been found with fetal lymphocytes (4). Although fetal NRBCs are an ideal source of fetal genetic material, they are present at an extremely low frequency in the absence of fetomaternal hemorrhage. It is estimated that the frequency is one fetal NRBC per 106–108 maternal nucleated cells (4). Hence, antibodies specific for fetal cells will be invaluable for their enrichment and confirmation.
At present, antibodies to fetal and embryonic hemoglobin are being used to identify fetal RBCs. However, Hb F in the maternal RBC may be elevated in some pregnancies, and although anti-ζ antibodies are more specific, embryonic hemoglobin has a narrower temporal window of expression (27). Additionally, targeting intracellular antigens makes the purification steps more subject to cell loss, as the fragile erythroid cells need to be permeabilized to make the hemoglobin accessible to these antibodies. Thus, cell surface markers are preferred for enrichment. Cell surface antibodies to CD71, CD36, and I/i antigens are most commonly used for purification. However, they have the disadvantage of being expressed on subpopulations of maternal WBCs. Because of this fact, additional erythroid markers are likely to be useful for fetal cell enrichment or verification of fetal RBCs. Preliminary results of phage antibodies generated by panning on fetal liver cells have also been reported by others (27, 28). The panel of antibodies we have generated are likely to span a wide range of fetal RBC antigens other than CD71, CD36, and I/i. Further characterization of the specificity and utility of these phage antibodies for erythroid and hematopoietic cells of different lineage in the normal and disease states is in progress. If, for example some can be found to be specific for primitive erythroid cells, they may prove useful not only for prenatal diagnosis, but also for treatment of polycythemia or erythroleukemia.
Acknowledgments
This work was supported by grants from the Dermatology Foundation, Avon Products, Inc., Department of Defense (DAMD-17–98-1–8189), and the National Institutes of Health/National Cancer Institute (5 P50 CA 58207 and DK16666).
Abbreviations
- scFv
single-chain variable antibody fragment
- RBC
red blood cell, WBC, white blood cell
- NRBC
nucleated RBC
- GPA
glycophorin A
- PEG
polyethylene glycol
- FITC
fluorescein isothiocyanate
- PE
phycoerythrin
References
- 1.Zipursky A, Hull A, White F D, Israels L G. Lancet. 1959;i:451–452. doi: 10.1016/s0140-6736(59)92264-0. [DOI] [PubMed] [Google Scholar]
- 2.Schroder J. J Med Genet. 1975;12:230–242. [PMC free article] [PubMed] [Google Scholar]
- 3.Bianchi D, Flint A F, Pizzimenti M F, Knoll J H M, Latt S A. Proc Natl Acad Sci USA. 1990;87:3279–3283. doi: 10.1073/pnas.87.9.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchi D. Br J Haematol. 1999;105:574–583. doi: 10.1046/j.1365-2141.1999.01383.x. [DOI] [PubMed] [Google Scholar]
- 5.Adinolfi M. Nat Genet. 1992;1:316–318. doi: 10.1038/ng0892-316. [DOI] [PubMed] [Google Scholar]
- 6.McCafferty J, Griffiths A D, Winter G, Chiswell D. Nature (London) 1990;348:552–554. doi: 10.1038/348552a0. [DOI] [PubMed] [Google Scholar]
- 7.Marks J D, Hoogenboom H R, Bonnert T P, McCafferty J, Griffiths A D, Winter G. J Mol Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 8.Marks C, Marks J D. N Engl J Med. 1996;335:730–733. doi: 10.1056/NEJM199609053351008. [DOI] [PubMed] [Google Scholar]
- 9.Griffiths A D, Malmqvist M, Marks J D, Bye J M, Embleton M J, McCafferty J, Baier M, Holliger K P, Gorick B D, Hughes-Jones N C, Hoogenboom H R, Winter G. EMBO J. 1993;12:725–734. doi: 10.1002/j.1460-2075.1993.tb05706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vaughan T J, Williams A J, Pritchard K, Osbourn J K, Pope A R, Earnshaw J C, McCafferty J, Hodits R A, Wilton J, Johnson K S. Nat Biotech. 1996;14:309–314. doi: 10.1038/nbt0396-309. [DOI] [PubMed] [Google Scholar]
- 11.Sheets M D, Amersdorfer P, Finnern R, Sargent P, Lindqvist E, Schier R, Hemingsen G, Wong C, Gerhart J C, Marks J D. Proc Natl Acad Sci USA. 1998;95:6157–6162. doi: 10.1073/pnas.95.11.6157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marks J D, Ouwehand W H, Bye J M, Finnern R, Gorick B D, Voak D, Thorpe S J, Hughes-Jones N C, Winter G. Bio/Technology. 1993;11:1145–1149. doi: 10.1038/nbt1093-1145. [DOI] [PubMed] [Google Scholar]
- 13.de Kruif J, Terstappen L, Boel E, Logtenberg T. Proc Natl Acad Sci USA. 1995;92:3938–3942. doi: 10.1073/pnas.92.9.3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barcena A, Muench M O, Song K S, Ohkubo T, Harrison M R. Exp Hematol. 1999;27:1428–1439. doi: 10.1016/s0301-472x(99)00080-6. [DOI] [PubMed] [Google Scholar]
- 15.Poul M A, Marks J D. J Mol Biol. 1999;288:203–211. doi: 10.1006/jmbi.1999.2678. [DOI] [PubMed] [Google Scholar]
- 16.Gussow D, Clackson T. Nucleic Acids Res. 1989;17:4000. doi: 10.1093/nar/17.10.4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith G P, Scott J K. Methods Enzymol. 1993;217:228–257. doi: 10.1016/0076-6879(93)17065-d. [DOI] [PubMed] [Google Scholar]
- 18.Cheung M-C, Goldberg J D, Kan Y W. Nat Genet. 1996;14:264–268. doi: 10.1038/ng1196-264. [DOI] [PubMed] [Google Scholar]
- 19.Van Ewijk W, de Kruif J, Germeraad W, Berendes P, Ropke C, Platenburg P, Logtenberg T. Proc Natl Acad Sci USA. 1997;94:3903–3908. doi: 10.1073/pnas.94.8.3903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Andersen P, Stryhn A, Hansen B, Fugger L, Engberg J, Buus S. Proc Natl Acad Sci USA. 1996;93:1820–1824. doi: 10.1073/pnas.93.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cai X, Garen A. Proc Natl Acad Sci USA. 1995;92:6537–6541. doi: 10.1073/pnas.92.14.6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Osbourn J, Derbyshire E, Vaughan T, Field A, Johnson K. Immunotechnology. 1998;3:293–302. doi: 10.1016/s1380-2933(97)10007-0. [DOI] [PubMed] [Google Scholar]
- 23.Siegel D, Chang T, Russell S, Bunya V. J Immunol Methods. 1997;206:73–85. doi: 10.1016/s0022-1759(97)00087-2. [DOI] [PubMed] [Google Scholar]
- 24.Becerril B, Poul M A, Marks J D. Biochem Biophys Res Commun. 1999;255:386–393. doi: 10.1006/bbrc.1999.0177. [DOI] [PubMed] [Google Scholar]
- 25.Pereira S, Maruyama H, Siegel D, Van Belle P, Elder D, Curtis P, Herlyn D. J Immunol Methods. 1997;203:11–24. doi: 10.1016/s0022-1759(97)00005-7. [DOI] [PubMed] [Google Scholar]
- 26.Watters J, Telleman P, Junghans R. Immunotechnology. 1997;3:21–29. doi: 10.1016/s1380-2933(96)00056-5. [DOI] [PubMed] [Google Scholar]
- 27.Zheng Y-L, Zhen D K, Farina A, Berry S M, Wapner R J, Williams J M, Bianchi D W. Am J Obstet Gynecol. 1999;180:1234–1239. doi: 10.1016/s0002-9378(99)70622-8. [DOI] [PubMed] [Google Scholar]
- 28.Zheng Y-L, Zhen D K, DeMaria M A, Berry S M, Wapner R J, Evans M I, Copeland D, Williams J M, Bianchi D W. Hum Genet. 1997;100:35–42. doi: 10.1007/s004390050462. [DOI] [PubMed] [Google Scholar]