Abstract
Hph1 and Hph2 are homologous integral endoplasmic reticulum (ER) membrane proteins required for Saccharomyces cerevisiae survival under environmental stress conditions. To investigate the molecular functions of Hph1 and Hph2, we carried out a split-ubiquitin-membrane-based yeast two-hybrid screen and identified their interactions with Sec71, a subunit of the Sec63/Sec62 complex, which mediates posttranslational translocation of proteins into the ER. Hph1 and Hph2 likely function in posttranslational translocation, as they interact with other Sec63/Sec62 complex subunits, i.e., Sec72, Sec62, and Sec63. hph1Δ hph2Δ cells display reduced vacuole acidification; increased instability of Vph1, a subunit of vacuolar proton ATPase (V-ATPase); and growth defects similar to those of mutants lacking V-ATPase activity. sec71Δ cells exhibit similar phenotypes, indicating that Hph1/Hph2 and the Sec63/Sec62 complex function during V-ATPase biogenesis. Hph1/Hph2 and the Sec63/Sec62 complex may act together in this process, as vacuolar acidification and Vph1 stability are compromised to the same extent in hph1Δ hph2Δ and hph1Δ hph2Δ sec71Δ cells. In contrast, loss of Pkr1, an ER protein that promotes posttranslocation assembly of Vph1 with V-ATPase subunits, further exacerbates hph1Δ hph2Δ phenotypes, suggesting that Hph1 and Hph2 function independently of Pkr1-mediated V-ATPase assembly. We propose that Hph1 and Hph2 aid Sec63/Sec62-mediated translocation of specific proteins, including factors that promote efficient biogenesis of V-ATPase, to support yeast cell survival during environmental stress.
The synthesis of proteins destined for the secretory pathway begins with their translocation into the endoplasmic reticulum (ER). Once in the ER, these proteins are folded, modified, and assembled into complexes so that they can function in their target compartments within the secretory pathway. Proteins can be translocated into the ER cotranslationally as they exit the ribosome or posttranslationally once polypeptide synthesis is complete (50). During cotranslational translocation, the signal recognition particle (SRP) binds the signal sequence on the nascent polypeptide and targets the polypeptide/ribosome complex to the ER translocation pore by interacting with the SRP receptor (SR) (37). The translocation pore itself is made up of the heterotrimeric Sec61 complex (Sec61, Sbh1, and Sss1), and translocation additionally requires the activity of Sec63 and BiP (13, 14, 16, 17, 41, 44). In contrast, posttranslational translocation is SRP independent. In this case, Hsp40- and Hsp70-type chaperones maintain fully synthesized peptides in a soluble state, competent for translocation (2, 6). These polypeptides are delivered to the translocation pore through their interaction with the Sec63/Sec62 complex, comprised of Sec63, Sec62, Sec71, and Sec72 (18–20, 23). Interestingly, Saccharomyces cerevisiae cells lacking Sec71 and Sec72 are viable, demonstrating that their function in protein translocation is not essential. Nascent polypeptides are targeted to either the co- or posttranslational translocation machinery based primarily on the hydrophobicity of their emerging signal sequence. Proteins with signal sequences of high hydrophobicity bind SRP and undergo cotranslational translocation; polypeptides that contain fewer hydrophobic signal sequences interact less efficiently with SRP and are targeted to the posttranslational translocation pathway (34). In yeast, the two pathways seem to be equally important, and some proteins use both mechanisms for translocation. In mammals, however, the majority of proteins translocate cotranslationally (50). Although the core components of the translocation machinery are well defined, additional modulators likely remain to be identified. For example, Yet1, Yet3, and Ylr301w interact with the Sec63/Sec62 complex in yeast, although their functions in protein translocation are unclear (47, 48). In addition, phosphorylation of Sec63 enhances its binding to Sec62, suggesting that the activity of this complex is regulated in vivo (46). In this study, we show that Hph1 and Hph2, two integral ER membrane proteins with unknown molecular functions, are novel interacting partners of the Sec63/Sec62 complex that may function in translocation.
Hph1 and Hph2 are homologous proteins required for cell survival during environmental stress (3, 5, 8, 24, 38). They have overlapping functions, and cells lacking both are viable but display growth defects during alkaline, saline, and cell wall stress (24). Hph1, but not Hph2, interacts with and is a substrate of calcineurin, a Ca2+/calmodulin-dependent serine/threonine protein phosphatase that activates specific stress responses (10, 24). Calcineurin positively modulates Hph1. An Hph1 mutant (Hph1ΔPVIAVN) that neither interacts with nor is dephosphorylated by calcineurin does not fully rescue the growth defect of hph1Δ hph2Δ cells under alkaline pH or high-salt conditions (24). Thus, Hph1 and Hph2 are required for stress survival; however, their molecular functions remain unknown. Here, we report on further characterization of hph1Δ hph2Δ cells, which revealed phenotypes resembling those of mutants defective for vacuolar proton ATPase (V-ATPase) activity.
The yeast V-ATPase is a multisubunit complex whose function, structure, and assembly have been well characterized. Cells with impaired V-ATPase activity (Vma−) fail to acidify the vacuole, cannot grow at alkaline pH, and are sensitive to high concentrations of extracellular calcium. In addition, Vma− mutants cannot use nonfermentable carbon sources and are sensitive to high concentrations of heavy metals, oxidative stress, and cell wall-damaging agents (29, 32). The yeast V-ATPase is made up of the peripherally associated V1 complex and the integral membrane V0 complex (29). The V0 complex is comprised of the proteolipid c ring (Vma3, Vma11, and Vma16), Vma6, Vma9, and the a subunit (either Vph1 or Stv1). The Vph1-containing ATPase acidifies the vacuole, whereas Stv1-containing V-ATPase localizes to endosomes and vesicles (30, 31). A set of dedicated protein factors assembles V0 within the ER, and loss of any of these (Vma12, Vma21, Vma22, Voa1, or Pkr1) severely compromises V-ATPase function (11, 22, 27, 28, 40). After V0 is assembled, Vma21 and Voa1 export it from the ER in COPII vesicles destined for the Golgi apparatus (40). One of the hallmarks of cells defective in V0 assembly is the rapid degradation of unassembled Vph1 via the ER-associated degradation (ERAD) pathway (26). The instability of unassembled Vph1 is apparent in cells that lack either an assembly factor or another subunit of the V0 complex. In this study, we show that defects in the Sec63/Sec62 complex cause Vph1 instability, indicating that the substrate(s) of this translocation complex contributes to V0 production. We further demonstrate that Hph1 and Hph2 act together with the Sec63/Sec62 complex in this process.
MATERIALS AND METHODS
Growth media and general methods.
Yeast media and culture conditions were essentially as previously described, except that twice the levels of amino acids and nucleotides were used in synthetic complete medium (43). Yeast transformations were carried out by the lithium acetate method (1). Yeast strains are listed in Table 1; strains from the yeast deletion collection were purchased from Open Biosystems (Huntsville, AL).
Table 1.
Strains used in this study
| Strain | Genotype | Reference |
|---|---|---|
| BY4741 (YSC1048) | MATaleu2Δ ura3Δ his3Δ met15Δ | Yeast deletion collection (Open Biosystems) |
| BY4742 (YSC1049) | MATα leu2Δ ura3Δ his3Δ lys2Δ | Yeast deletion collection (Open Biosystems) |
| VHY70 | MATaleu2Δ ura3Δ his3Δ | 24 |
| VHY60 | MATaleu2Δ ura3Δ his3Δ met15Δ hph1Δ::KanMX4 hph2Δ::KanMX4 | 24 |
| hph1Δ (no. 1621) | Isogenic to BY4741 | Yeast deletion collection (Open Biosystems) |
| hph2Δ (no. 380) | Isogenic to BY4741 | Yeast deletion collection (Open Biosystems) |
| pkr1Δ (no. 6564) | Isogenic to BY4741 | Yeast deletion collection (Open Biosystems) |
| sec71Δ (no. 3311) | Isogenic to BY4741 | Yeast deletion collection (Open Biosystems) |
| hph1Δ hph2Δ sec71Δ | MATaleu2Δ ura3Δ his3Δ hph1Δ::KanMX4 hph2Δ::KanMX4 sec71Δ::KanMX4 | This study |
| hph1Δ hph2Δ pkr1Δ | MATa leu2Δ ura3Δ his3Δ hph1Δ::KanMX4 hph2Δ::KanMX4 pkr1Δ::KanMX4 | This study |
| vma21Δ (no. 4735) | Isogenic to BY4741 | Yeast deletion collection (Open Biosystems) |
| BY4743-SEC71-TAP | MATa/α leu2Δ/leu2Δ ura3Δ/ura3Δ met15Δ/MET15 lys2Δ/LYS2 SEC71::TAP::HIS3MX6/SEC71 | This study |
| BY4743-SEC72-TAP | MATa/α leu2Δ/leu2Δ ura3Δ/ura3Δ met15Δ/MET15 lys2Δ/LYS2 SEC72::TAP::HIS3MX6/SEC72 | This study |
| BY4743-SEC61-TAP | MATa/α leu2Δ/leu2Δ ura3Δ/ura3Δ met15Δ/MET15 lys2Δ/LYS2 SEC61::TAP::HIS3MX6/SEC61 | This study |
| BY4743-SEC62-TAP | MATa/α leu2Δ/leu2Δ ura3Δ/ura3Δ met15Δ/MET15 lysΔ2/LYS2 SEC62::TAP::HIS3MX6/SEC62 | This study |
| BY4743-SEC63-TAP | MATa/α leu2Δ/leu2Δ ura3Δ/ura3Δ met15Δ/MET15 lysΔ2/LYS2 SEC63::TAP::HIS3MX6/SEC63 | This study |
The plasmids used in this study are described in Table 2. All genes were initially cloned by amplification with Taq High-Fidelity polymerase (Invitrogen, Beverly, MA) with the indicated restriction sites. The glutathione S-transferase (GST) coding sequence was amplified from plasmid pES298-9 using the forward primer 5′-CTAGTCTAGAATGTCCCCTATACTAGGTTATTGG-3′ and the reverse primer 5′-CTAGTCTAGACAGATCCGATTTTGGAGGATGGTC-3′, flanked by XbaI restriction sites, and cloned into pUG34 or pUG36 (CEN URA3 pMET25-yEGFP or CEN HIS3 pMET25-yEGFP; gifts of U. Güldener and J. H. Hegemann), which had been digested with XbaI and gel purified to remove the green fluorescent protein (GFP) coding sequence, thus generating pFJP10 and pFJP13, respectively. HPH1 was subcloned into pFJP10 or pFJP13 as a BamHI/HindIII fragment from pVH1 to generate pFJP11 and pFJP14. hph11ΔPVIAVN was subcloned from pVH12 into pFJP10 as a BamHI/XhoI fragment to generate pFJP12. GST-HPH2 was generated by in vivo recombination in VHY70 yeast cells. The GST coding sequence was amplified, flanked by the MET25 promoter sequence on the 5′ end and the HPH2 sequence on the 3′ end, for homologous recombination between pVH2 (digested with XbaI and gel purified) and the GST PCR product in VHY70 to generate pFJP20 (forward primer, CATCTACTATTTCCTTCGTGTAATACAGGGTCGTCAGATACATAGATACAATTCTAT TACCCCCATCCATACTCTAGAATGTCCCCTATACTAGGTTATTGG; reverse primer, CCATCTTTGCTATTGCGATCTGTACCATCTATTCCGCTGCCTTTAGAAGAGCTCTTTATTTGAGCATTTTGCATGGATCCACTAGT TCTAGACAGATCCGATTTTGGAGGATGG). Alternatively, HPH2 was PCR amplified from pVH2, flanked by GST coding sequence on the 5′ end and CYC terminator sequence on the 3′ end, for homologous recombination between pFJP10 (digested with BamHI and gel purified) and the HPH2 PCR product (forward primer, GCATGGCCTTTGCAGGGCTGGCAAGCCACGTTTGGTGGTGGCGACCATCCTCCAAAATCGGATCTGTCTAGAACTAGTGGAT CCATGCAAAATGCTCAAATAAAGAGC; reverse primer, CGGTTAGAGCGGATGTGGGGGGAGGGCGTGAATGTAAGCGTGACATAACTAATTA CATGACTCGAGGAGCTCGTCTAGATTATTTATGCGATACTAGATGC) in VHY70 to generate pFJP21. All constructs were confirmed by sequence analysis and complementation.
Table 2.
Plasmids used in this study
| Plasmid | Vector | Insert | Reference |
|---|---|---|---|
| pFJP10 | Modified pUG34 lacking GFP | GST | This study |
| pFJP11 | pFJP10 | HPH1 | This study |
| pFJP12 | pFJP10 | hph1ΔPVIAVN | This study |
| pFJP13 | pUG36 without GFP | GST | This study |
| pFJP14 | pFJP13 | HPH1 | This study |
| pFJP20 | pFJP13 | GST-HPH2 | This study |
| pFJP21 | pFJP10 | GST-HPH2 | This study |
| pVH1 | pUG36 | HPH1 | 24 |
| pVH2 | pUG36 | HPH2 | 24 |
| pUG36 | 24 |
The hph1Δ::KanMX4 hph2Δ::KanMX4 pkr1Δ::KanMX4 mutant was generated by mating MATa hph1Δ::KanMX4 to MATα pkr1Δ::KanMX4 and MATa hph2Δ::KanMX4 to MATα pkr1Δ::KanMX4 mutants. The resulting diploids were sporulated to generate MATα hph1Δ::KanMX4 pkr1Δ::KanMX4 (spore 3D) and MATa hph2Δ::KanMX4 pkr1Δ::KanMX4 (spore 5C) mutants, respectively. The hph1Δ::KanMX4 pkr1Δ::KanMX4 (3D) mutant was mated to the hph2Δ::KanMX4 pkr1Δ::KanMX4 (5C) mutant, and the resulting diploid was sporulated to generate the hph1Δ::KanMX4 hph2Δ::KanMX4 pkr1Δ::KanMX4 (spore 3D) mutant. Three independent hph1Δ::KanMX4 hph2Δ::KanMX4 pkr1Δ::KanMX4 mutants (spores 1C, 2D, and 3D) were generated, and all behaved similarly in growth assays.
The hph1Δ::KanMX4 hph2Δ::KanMX4 sec71Δ::KanMX4 mutant was generated by mating the MATa hph1Δ::KanMX4 mutant to the MATα sec71Δ::KanMX4 mutant and the MATa hph2Δ::KanMX4 mutant to the MATα sec71Δ::KanMX4 mutant. Diploids were sporulated to generate the MATa hph1Δ::KanMX4 sec71Δ::KanMX4 (spore 18C) mutant and the MATα hph2Δ::KanMX4 sec71Δ::KanMX4 (spore 17D) mutant, respectively. The MATa hph1Δ::KanMX4 sec71Δ::KanMX4 (18C) mutant was mated to the MATα hph2Δ::KanMX4 sec71Δ::KanMX4 (17D) mutant to generate the MATa hph1Δ::KanMX4 hph2Δ::KanMX4 sec71Δ::KanMX4 (spore 4D) mutant. Three independent hph1Δ::KanMX4 hph2Δ::KanMX4 sec71Δ::KanMX4 mutants (spores 4D, 19A, and 22D) were generated, and all behaved similarly in growth assays.
Appropriate segregation was monitored for four independent marker loci to verify tetrads for all mutants that were generated. Double and triple mutants were verified by PCR.
To confirm the interaction between Hph1 and Hph2 and the Sec63/Sec62 complex, strains from the yeast TAP-tagged collection (catalog number YSC1178; Open Biosystems, Huntsville, AL) were mated to BY4742 to generate a SEC-TAP/SEC heterozygous diploid strain (Table 1). Heterozygous strains were transformed with pFJP13, pFJP14, or pFJP20.
Quinacrine staining.
Yeast cells were grown to mid-log phase in yeast-peptone-dextrose (YPD) medium buffered to pH 5.5 with 50 mM morpholineethanesulfonic acid (MES) and 50 mM morpholinepropanesulfonic acid (MOPS). Cells were collected by centrifugation; washed once with water; grown for 2 h in YPD, pH 7.6, with 100 mM HEPES-KOH; collected; resuspended in fresh YPD, pH 7.6, with 100 mM HEPES-KOH with 200 μM quinacrine (Sigma-Aldrich, St. Louis, MO); and incubated for 5 min at 30°C with shaking. The cells were collected and washed three times with cold 100 mM HEPES (pH 7.6) buffer with 2% glucose. After the third wash, the cells were concentrated 10-fold, and 2 μl was placed on a microscope slide. The cells were imaged by differential interference contrast (DIC) or with a fluorescein isothiocyanate (FITC) filter to detect quinacrine fluorescence with a Zeiss Axio Imager M1 microscope (Carl Zeiss, Jena, Germany) and Hamamatsu Orca-R digital camera coupled to Openlab Software 5.0.1 (Perkin Elmer, Waltham, MA). To quantify quinacrine fluorescence, cells were treated as described above. After the third wash, 200 μl was transferred to a 96-well microplate (Greiner PS; F-bottom) for analysis (TECAN Safire; excitation, 470 nm; emission, 507 nm). The values on the graphs (see Fig. 4A and 5A) represent the averages of three independently grown, stained, and analyzed cultures. Values were normalized to the average of the wild-type (WT) strain (BY4741) in all cases. The error bars represent the standard deviations of the mean (SDM).
Growth assays.
Growth was assayed by plating serial dilutions of stationary-phase yeast cultures grown in YPD (50 mM MES, 50 mM MOPS, pH 5.5) on YPD, YPD with 100 mM Tris-HCl (pH 7.6), YP-2% ethanol, or YPD containing NaCl, LiCl, H2O2, CaCl2, ZnCl2, CoCl2, CdCl2, or CsCl2 at the indicated concentrations. The first culture dilution was at 2.5 × 106 cells/ml, and each subsequent spot was a 7-fold serial dilution. The plates were incubated at room temperature for the indicated number of days (or at 30°C for 2 days [see Fig. 5]).
GST pulldown assays from yeast extracts.
Copurification studies were carried out by transforming BY4742 with pFJP10, pFJP11, or pFJP12 (Fig. 1A); VHY70 cells with pUG36, pVH1, or pVH2 and pFJP10 or pFJP11 (Fig. 1B); or BY4743-Sec71-TAP, BY4743-Sec72-TAP, BY4743-Sec61-TAP, BY4743-Sec62-TAP, and BY4743-Sec63-TAP cells with pFJP13, pFJP14, or pFJP20 (Fig. 1C). The cells were grown overnight in synthetic complete (SC) medium without uracil and with methionine (3 g/liter) and diluted to 2 × 106 cells/ml in SC without uracil or methionine to induce expression of GST, GST-Hph1, or GST-Hph2 from the MET25 promoter. The cells were grown to mid-log phase, treated with 8 mM dithiothreitol (DTT) for 2 h (DTT treatment increased the stability of GST-Hph1 and GST-Hph2 and yielded more protein in the GST pulldown [data not shown]), and collected by centrifugation (1,000 × g for 5 min). The cells were washed once with water, flash frozen in liquid nitrogen, and stored at −80°C. Cell protein extracts were generated by glass bead lysis in breaking buffer (20 mM HEPES-KOH [pH 6.8], 250 mM sorbitol, 250 mM NaCl, 70 mM potassium acetate, 1 mM magnesium acetate, 1 mM DTT) and protease inhibitors at 4°C. The cells were centrifuged at 1,000 × g for 5 min at 4°C to remove unbroken cells and the glass beads. n-Dodecyl maltoside was added to the supernatant at a 0.8% final concentration and incubated for 20 min at room temperature with end-over-end rotation and then centrifuged at 13,000 × g for 15 min at 4°C. The supernatant (S13) was centrifuged at 100,000 × g for 10 min at 4°C, and 5 mg of protein extract (S100) in 1 ml of breaking buffer was incubated with 20 μl of glutathione-Sepharose beads (GE Healthcare, Buckinghamshire, United Kingdom) for 2 h at 4°C with end-over-end rotation. The beads were washed 3 times with 800 μl breaking buffer containing 300 mM NaCl and 0.1% n-dodecyl maltoside, dried by aspiration, resuspended in SDS-PAGE buffer, and heated at 37°C for 15 min. Samples were analyzed by SDS-PAGE and Western blotting using standard procedures. The immunoblots were probed with mouse monoclonal anti-GST (Covance, Emeryville, CA), mouse monoclonal anti-GFP (Covance, Emeryville, CA), rabbit polyclonal anti-TAP (Open Biosystems, Huntsville, AL), or rabbit polyclonal antiserum to Cna2 (39). Anti-mouse and anti-rabbit secondary antibodies conjugated to horseradish peroxidase (HRP) (GE Healthcare, Buckinghamshire, United Kingdom) and a SuperSignal West Dura Extended Duration Substrate kit (Thermo Scientific, Rockford, IL) were used to detect immunoreactive bands on Kodak Biomax XAR film (Kodak, Rochester, NY).
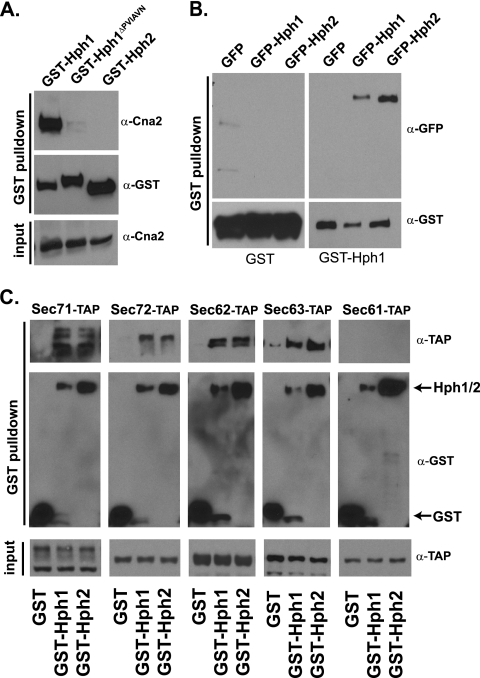
Fig. 1.
Hph1 interacts with calcineurin, itself, and Hph2. Hph1 and Hph2 also interact with components of the posttranslational translocation machinery, i.e., the Sec63/Sec62 complex. (A) Calcineurin copurifies with GST-Hph1, but not with GST-Hph1ΔPVIAVN or GST-Hph2. GST fusion proteins were purified from extracts of wild-type cells (BY4742) expressing GST-Hph1, GST-Hph2, or GST-Hph1ΔPVIAVN and analyzed by SDS-PAGE and immunoblotting with anti-GST and anti-Cna2 antisera. (B) GST-Hph1 or GST was purified from extracts of hph1Δ hph2Δ cells (VHY70) coexpressing GFP, GFP-Hph1, or GFP-Hph2 and analyzed by SDS-PAGE and immunoblotting with anti-GST anti-GFP antisera. (C) GST, GST-Hph1, or GST-Hph2 was purified from extracts of cells containing a genomically integrated TAP-tagged allele of the Sec71, Sec72, Sec62, Sec63, or Sec61 gene (21); analyzed by SDS-PAGE; and immunoblotted with anti-GST and anti-TAP antibodies.
Pulse-chase analysis of Vph1.
Vph1 stability was quantified by pulse-chase analysis as described previously (36). Vph1 was immunoprecipitated either with a monoclonal Vph1 antibody (10D7; Molecular Probes) or using a polyclonal Vph1 antibody provided by Randy Schekman (University of California, Berkeley, CA).
RESULTS
Hph1 and Hph2 interact with components of the posttranslational translocation machinery.
Hph1 and Hph2 are important mediators of the cellular stress response; however, their molecular functions are undetermined (24). To better understand their functions, we carried out a membrane-based yeast two-hybrid screen (MbY2H) assaying both Hph1 and Hph2 for interaction against a library of integral membrane proteins (33). This screen identified 24 candidates as interacting with both Hph1 and Hph2. Sixteen of these candidates interacted with Hph1 and Hph2 by MbY2H in a secondary screen, and 12 of these were further examined for copurification with Hph1 and Hph2 (see Table S1 in the supplemental material). N-terminal GST fusions of Hph1, Hph1ΔPVIAVN (an Hph1 mutant that lacks the calcineurin binding domain), and Hph2, all expressed under the control of the MET25 promoter, were used for these studies. The Hph1 and Hph2 GST fusion proteins were functional and rescued the growth defects of hph1Δ hph2Δ cells (data not shown). Since Hph1 and Hph2 are integral membrane proteins, we identified extraction conditions that solubilized Hph1 and Hph2 while preserving protein-protein interactions. We first examined the interaction of Hph1 with calcineurin, which was previously identified using the yeast two-hybrid technique (24). Calcineurin copurified with GST-Hph1, but not with GST-Hph1ΔPVIAVN or GST-Hph2 (Fig. 1A), validating our purification method. In addition, we examined the interaction between Hph1 and Hph2, which colocalize at the ER membrane and interact with each other in yeast two-hybrid assays. To perform this experiment, we coexpressed GST-Hph1 with N-terminal GFP fusions of Hph1 or Hph2 (24). GFP-Hph1 and GFP-Hph2 both copurified with GST-Hph1 (Fig. 1B), which is consistent with previous findings (24) and establishes that Hph1 and Hph2 make higher-order complexes with each other and possibly other proteins.
The same method was used to test the interaction of GST-Hph1 with candidates identified in the MbY2H screen (see Table S1 in the supplemental material). Only Sec71, a nonessential subunit of the Sec63/Sec62 complex, reproducibly interacted with GST-Hph1 and GST-Hph2 (Fig. 1C and data not shown) (18, 20). We asked whether Hph1 and Hph2 interact with other components of this posttranslational translocation complex. Specifically, we examined the interaction of GST-Hph1 and GST-Hph2 with genomically expressed, TAP-tagged alleles of Sec72, Sec62, Sec63, and Sec61 (21). These TAP-tagged alleles were functional, as untranslocated CPY (preprocarboxypeptidase Y) did not accumulate in the tagged haploid strains (data not shown). GST-Hph1 and GST-Hph2, but not GST, copurified with Sec71-TAP, Sec72-TAP, Sec62-TAP, and Sec63-TAP (Fig. 1C). No interaction was detected with the TAP-tagged version of Sec61, one of the subunits of the translocation pore. The association of Hph1 and Hph2 with the Sec63/Sec62 complex suggests they may function in posttranslational protein translocation at the ER. Surprisingly, hph1Δ hph2Δ cells showed no defects in processing of CPY and prepro-alpha-factor, well-characterized substrates for the Sec63/Sec62 complex (data not shown). Thus, Hph1 and Hph2 interact with the Sec63/Sec62 complex and, while not essential for Sec63/Sec62 function, may selectively promote the translocation of specific proteins.
hph1Δ hph2Δ cells are sensitive to a variety of stress conditions.
To gain insight into proteins whose function might depend on Hph1 and Hph2, we examined phenotypes of hph1Δ hph2Δ cells. Previous studies established that cells lacking both HPH1 and HPH2 (hph1Δ hph2Δ) grow poorly at alkaline pH, under high-NaCl conditions, or in the presence of cell wall-damaging agents (Congo red and calcofluor white), unlike hph1Δ or hph2Δ single mutants, which show no growth defects under these conditions (Fig. 2) (24). In addition to these phenotypes, we found that hph1Δ hph2Δ cells were sensitive to a variety of environmental stress conditions (Fig. 2). Specifically, hph1Δ hph2Δ cells were sensitive to high concentrations of the divalent heavy metal ions zinc, cadmium, cobalt, and cesium. In addition, hph1Δ hph2Δ cells were sensitive to oxidative stress (hydrogen peroxide) and grew poorly on media containing a nonfermentable carbon source (ethanol) (Fig. 2). They also displayed mild sensitivity to calcium chloride (Fig. 2). Under most of these conditions, the growth of hph1Δ and hph2Δ single mutants was indistinguishable from that of wild-type cells, indicating functional overlap of these genes. In contrast, hph1Δ cells grew poorly on media containing elevated amounts of zinc chloride (Fig. 2), although the increased sensitivity of hph1Δ hph2Δ suggested partial redundancy of Hph1 and Hph2 under these conditions, as well. The wide array of hph1Δ hph2Δ phenotypes, together with the interaction of Hph1 and Hph2 proteins with the posttranslational translocation machinery, suggests that Hph1 and Hph2 promote the translocation of one or more proteins that allow cells to cope with various types of environmental stress. hph1Δ hph2Δ phenotypes were strikingly similar to those of cells deficient for vacuolar acidification, such as vma21Δ (Fig. 3A) (29). However, in each case, the hph1Δ hph2Δ growth defect was less severe than that observed for vma21Δ cells, in which failure to assemble and export the V0 complex of the V-ATPase from the ER leads to a severe disruption in V-ATPase activity (27, 40).
Fig. 2.
hph1Δ hph2Δ cells are sensitive to growth under conditions of high concentrations of heavy metals (ZnCl2, CdCl2, CoCl2, and CsCl2), CaCl2, oxidative stress (H2O2), and a nonfermentable carbon source (ethanol). hph1Δ cells are sensitive to growth on ZnCl2. Wild-type (BY4741), hph1Δ (number 1621), hph2Δ (number 380), and hph1Δ hph2Δ (VHY60) strains were grown to stationary phase in YPD. The cultures were diluted to 2.5 × 106 cells/ml, and 7-fold serial dilutions were plated on YPD plates containing the indicated additions. The plates were incubated at room temperature for 2 days (YPD, pH 7.6, CaCl2, and YP-ethanol), 4 days (ZnCl2, CdCl2, CoCl2, CsCl2, and H2O2), and 5 days (NaCl and LiCl).
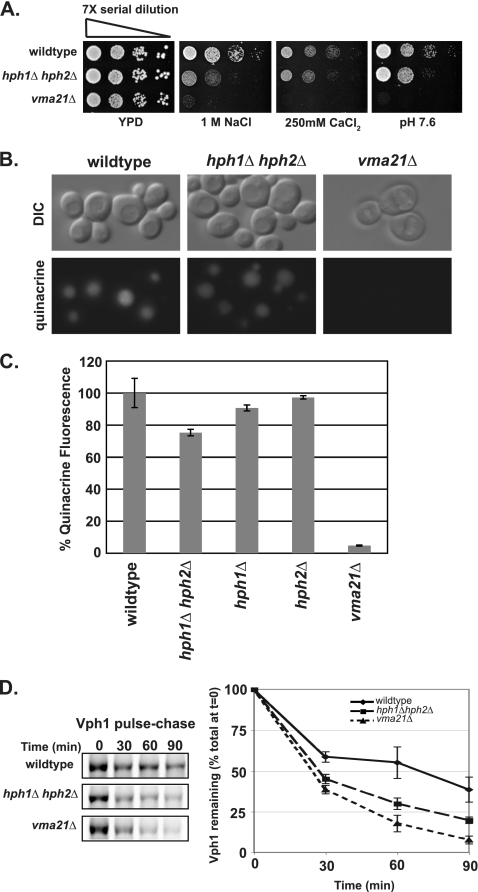
Fig. 3.
hph1Δ hph2Δ cells exhibit a spectrum of growth defects similar to that of vma21Δ cells, a partial defect in vacuolar acidification, and increased Vph1 turnover. (A) Wild-type (BY4741), hph1Δ hph2Δ (VHY60), and vma21Δ (number 4735) cells were grown to stationary phase. Cultures were diluted to 2.5 × 106 cells/ml, and 7-fold serial dilutions were spotted on plates containing YPD, pH 5.5, with the indicated additions. (B) Yeast strains (same as in panel A) were stained with quinacrine and imaged immediately after being stained. (C) The fluorescence of quinacrine-stained cells was quantified as described in the text and normalized to the average fluorescence of wild-type cells. The mean fluorescence intensity of three replicates, normalized to the WT, is presented with SDM. (D) The half-life of Vph1 was determined in the indicated strains by pulse-chase analysis.
hph1Δ hph2Δ cells display reduced vacuolar acidification and increased Vph1 turnover.
We assayed vacuolar acidification in hph1Δ hph2Δ cells using quinacrine, a fluorescent dye whose accumulation in the vacuole depends on acidification of the organelle. When viewed by fluorescence microscopy, vacuoles in WT cells were intensely fluorescent, whereas those of vma21Δ cells were barely visible (Fig. 3B). hph1Δ hph2Δ cells displayed vacuolar fluorescence (Fig. 3B) that was reduced 25% ± 2.0% compared to that of wild-type cells (Fig. 3C), indicating a small but significant defect in vacuolar acidification (42). Neither hph1Δ nor hph2Δ single mutants displayed a decrease in quinacrine accumulation, consistent with the functional overlap observed for the HPH1 and HPH2 genes (Fig. 3C). To investigate the cause of this decreased acidification, we examined the stability of Vph1, one of the subunits of the V0 complex of the V-ATPase, which is degraded via the ERAD pathway in cells disrupted for V0 synthesis or assembly (26). As previously described, Vph1 turnover is increased in vma21Δ compared to wild-type cells. The Vph1 half-life was also significantly shorter in hph1Δ hph2Δ than in wild-type cells (Fig. 3D), suggesting that reduced stability of the V-ATPase compromises vacuolar acidification and causes the growth defects observed in these cells.
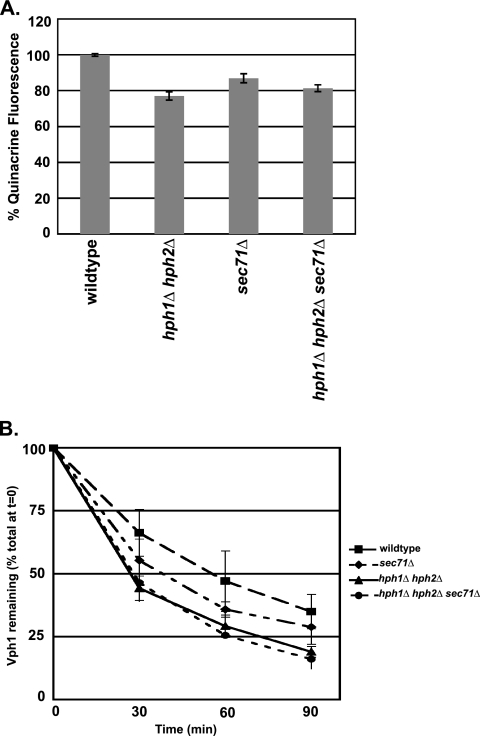
Vacuolar acidification defects of hph1Δ hph2Δ and sec71Δ cells are not additive.
Since Hph1 and Hph2 interact with the Sec63/Sec62 complex, we examined whether Sec71, a nonessential component of the complex, was required for Vph1 stability. sec71Δ cells are deficient in posttranslational translocation (18, 20). Similar to hph1Δ hph2Δ cells, sec71Δ cells exhibited a 20% ± 2.3% decrease in quinacrine fluorescence (Fig. 4A) and a shorter Vph1 half-life than wild-type cells (Fig. 4B). These observations suggest that the posttranslocation machinery is required for optimal biogenesis of the V-ATPase. To test whether Hph1/Hph2 and the Sec63/Sec62 complex alter V-ATPase function through the same or independent mechanisms, we examined the phenotypes of hph1Δ hph2Δ sec71Δ cells. Vacuolar acidification, as observed by quinacrine accumulation, was reduced to similar extents in hph1Δ hph2Δ, sec71Δ, and hph1Δ hph2Δ sec71Δ cells (Fig. 4A). In addition, the half-life of Vph1 in hph1Δ hph2Δ sec71Δ cells was slightly shorter than in sec71Δ but equivalent to that of hph1Δ hph2Δ cells. Thus, the decrease in vacuolar acidification and Vph1 stability observed in hph1Δ hph2Δ cells was not exacerbated by disruption of the Sec63/Sec62 complex. These results are consistent with the hypothesis that Hph1 and Hph2 act in concert with the Sec63/Sec62 complex to translocate one or more proteins that promote V0 biogenesis.
Fig. 4.
sec71Δ cells exhibit reduced vacuolar acidification and Vph1 stability that is not additive with hph1Δ hph2Δ. (A) Wild-type (BY4741), hph1Δ hph2Δ (VHY60), sec71Δ (number 3311), and hph1Δ hph2Δ sec71Δ cells were stained with quinacrine, and the vacuolar fluorescence was quantified. The mean fluorescence intensity of three replicates, normalized to the WT, is presented with the SDM. (B) The half-life of Vph1 was determined in wild-type (BY4741), hph1Δ hph2Δ (VHY60), sec71Δ (number 3311), and hph1Δ hph2Δ sec71Δ cells by pulse-chase analysis.
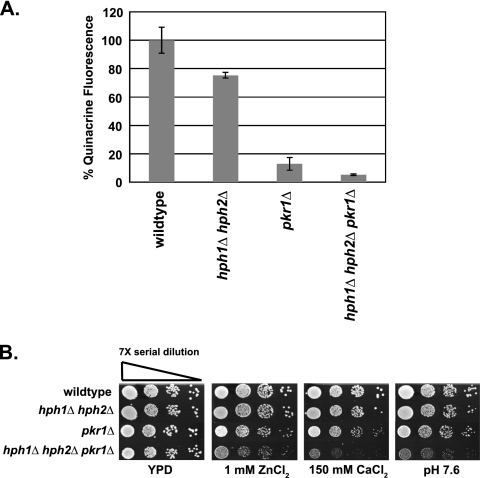
Vacuolar acidification defects caused by hph1Δ hph2Δ and pkr1Δ are additive.
After its translocation into the ER, Vph1 is assembled with the rest of the V0 complex by several factors, including Pkr1 (11, 29); disruption of V0 assembly results in degradation of Vph1 via ERAD (26). To examine a potential role for Hph1 and Hph2 in Vph1 assembly and export, we examined the phenotypes of hph1Δ hph2Δ pkr1Δ cells. Cells lacking PKR1 displayed an 87% ± 4.5% reduction in quinacrine accumulation compared to wild-type cells (Fig. 5A). These cells also displayed growth defects on alkaline media and with high concentrations of calcium chloride and zinc chloride (reference 11 and data not shown). Examination of triple mutants (hph1Δ hph2Δ pkr1Δ) revealed that deletion of PKR1 exacerbated the defects of hph1Δ hph2Δ cells. Quinacrine accumulation was reduced by 95% ± 0.5% in hph1Δ hph2Δ pkr1Δ compared to wild-type cells, whereas hph1Δ hph2Δ displayed only a 25% ± 2.0% reduction. Furthermore, hph1Δ hph2Δ pkr1Δ cells displayed significant growth defects under conditions (1 mM ZnCl2, 150 mM CaCl2, pH 7.6) that had little effect on either hph1Δ hph2Δ or pkr1Δ cells (Fig. 5B). Thus, the effects of hph1Δ hph2Δ and pkr1Δ are additive. These findings suggest that Hph1 and Hph2 act independently of Pkr1 and are consistent with the hypothesis that Hph1 and Hph2, together with the Sec63/Sec62 complex, act upstream of Pkr1-mediated V0 assembly to promote efficient biogenesis of the V0 complex.
Fig. 5.
pkr1Δ exacerbates growth defects and the vacuolar acidification defect of hph1Δ hph2Δ cells. (A) The fluorescence of wild-type (BY4741), hph1Δ hph2Δ (VHY60), pkr1Δ (number 6564), and hph1Δ hph2Δ pkr1Δ cells was determined after incubation with quinacrine and normalized to the average fluorescence of wild-type cells. The error bars represent SDM. (B) Wild-type (BY4741), hph1Δ hph2Δ (VHY60), pkr1Δ (number 6564), and hph1Δ hph2Δ pkr1Δ cells were plated as described for Fig. 3A and grown at 30°C for 2 days.
DISCUSSION
Hph1 and Hph2 interact with the Sec63/Sec62 complex.
Previous characterization showed that Hph1 and Hph2 act redundantly to promote yeast cell growth during saline, alkaline, and cell wall stress, although their molecular functions were not identified (24). Our findings that Hph1 and Hph2 copurify with Sec71, Sec72, Sec62, and Sec63 from yeast extracts suggest that these proteins function in posttranslational ER translocation. However, translocation of preprocarboxypeptidase Y and prepro-alpha-factor, two substrates whose translocation is Sec63/Sec62 dependent, is unperturbed in hph1Δ hph2Δ cells, indicating more specialized roles for Hph1 and Hph2 in the translocation of select substrates. Although the core machinery required for posttranslational translocation has been elucidated, putative accessory proteins, such as Yet1, Yet3, and Ylr301w, have been identified that interact with the Sec63/Sec62 complex but whose roles in translocation are not understood (47, 48). It is likely that such factors promote the translocation of particular substrates and/or regulate the activity of the translocon. However, little is known about such modulatory activities.
Hph1 and Hph2 are low-abundance proteins, estimated at 250 molecules/cell compared to the >7,000 molecules/cell for the rest of the subunits of the Sec63/Sec62 complex (21). This difference in stoichiometry also suggests that Hph1 and Hph2 couple to a subset of Sec63/Sec62 complexes. Hph1 and Hph2 each contain a single C-terminal transmembrane domain and are inserted into the ER via the GET complex (7); thus, the bulk of each protein is cytosolic and available to interact with the cytosolic domains of Sec63/Sec62 complex components. Hph1, Hph2, and Sec71 each contain a predicted coiled-coil motif, providing a plausible interaction mechanism. However, further studies are required to elucidate direct physical contacts between Hph1 and Hph2 and Sec63/Sec62 complex members. Interestingly, association between Hph1 or Hph2 and Sec61, one of the subunits of the translocation pore, was not observed. While this interaction may simply not have been preserved under the experimental conditions used, a more interesting possibility is that Hph1 and Hph2 serve as receptors for translocation substrates, acting upstream of translocation per se to confer substrate specificity but making little or no contact with the translocation pore.
Hph1 and Hph2 are novel factors that contribute to V-ATPase biogenesis.
Investigation of the growth defects of hph1Δ hph2Δ cells revealed striking parallels with those displayed by Vma− mutants, which compromise V-ATPase. V-ATPase acidifies the vacuole, Golgi apparatus, and endosomes and is critical for cellular pH and ion homeostasis (29). When V-ATPase activity is deficient, cells are unable to grow at neutral pH and display sensitivity to CaCl2 and a variety of metal ions (29). While hph1Δ hph2Δ cells share many of these properties, their growth defects are consistently less severe than those of Vma− mutants. Direct examination of vacuolar acidification, as measured by quinacrine accumulation, revealed a small but significant decrease in vacuolar acidification in hph1Δ hph2Δ cells that led us to investigate a possible function for Hph1 and Hph2 in V-ATPase biogenesis. The synthesis and assembly of the V-ATPase V0 complex is highly regulated. Vma12, Vma21, and Vma22 are required for V0 assembly, and Vma21 plays an additional role in the export of V0 from the ER (11, 22, 27, 28, 40). In cells lacking any one of these assembly factors, no V0 is produced, leading to a complete loss of V-ATPase activity and increased turnover of Vph1 in the ER via the ERAD pathway (26). Pkr1 increases the efficiency of V0 assembly, and the amount of functional V0 that is correctly targeted to the vacuolar membrane is severely reduced in pkr1Δ cells. Voa1 functions early in V0 assembly. voa1Δ cells display a modest decrease in V-ATPase activity and no Vma− growth phenotypes; however in combination with a mutant allele of vma21, V-ATPase assembly is dramatically compromised (26). Thus, proteins that contribute to, but are not strictly required for, V-ATPase synthesis and assembly can be difficult to identify. We observed increased turnover of Vph1 in hph1Δ hph2Δ cells, and this finding, together with decreased vacuolar acidification and growth defects of these cells, suggests that Hph1 and Hph2 contribute to V-ATPase biogenesis. Furthermore, we found that the phenotypes of hph1Δ hph2Δ cells were greatly exacerbated when combined with pkr1Δ; hph1Δ hph2Δ pkr1Δ mutants showed a larger reduction in vacuolar acidification and more severe growth defects than either hph1Δ hph2Δ or pkr1Δ cells, suggesting that Hph1/Hph2 act independently of Pkr1-mediated V0 assembly and likely affect a distinct step in V-ATPase biogenesis.
In studying Hph1 and Hph2, we focused on an early step in V-ATPase production, ER translocation, which has not been previously investigated. We found that vacuolar acidification and Vph1 stability are decreased in sec71Δ cells, which are deficient in posttranslational translocation. Surprisingly, sec72Δ cells displayed normal levels of quinacrine accumulation (data not shown). However, Sec72 is destabilized in sec71Δ cells (20), so the defects we observed in those cells may result from deficiencies in both Sec71 and Sec72. Significantly, the loss of Sec71 in hph1Δ hph2Δ cells does not further decrease vacuolar acidification or exacerbate Vph1 instability; thus, we propose that Hph1 and Hph2 function together with the Sec63/Sec62 complex to promote posttranslational translocation of one or more protein(s) required for efficient biogenesis of V0. Currently, the identities of such proteins are unknown; they could be either V0 components or previously identified or novel V0 assembly factors, and their translocation may be fully or partially dependent on Hph1/2 and Sec63/62. We did not detect copurification of Vph1 with GST-Hph1 or GST-Hph2 (data not shown). However, it is unlikely that Vph1 itself is a direct substrate of the Sec63/Sec62 complex, as it would be difficult to maintain this large, polytopic membrane protein in a soluble state posttranslationally prior to translocation. With the exception of Vma22, the known V0 assembly factors, although smaller, are also integral membrane proteins. Interestingly, MbY2H experiments identified a putative interaction between Sec72 and Vma12 (33). Therefore, we isolated Sec72-TAP and Sec71-TAP but did not observe copurification with either Vma12 or Vph1 (data not shown). Future studies will aim to elucidate the specific molecular functions of Hph1 and Hph2 in the translocation process and to identify their targets.
Physiological functions and regulation of Hph1 and Hph2.
Hph1 and Hph2 are required for cell survival under many stress conditions, and Hph1 is positively regulated by the Ca2+/calmodulin-regulated protein phosphatase calcineurin (24). Calcineurin is activated under conditions of environmental stress and dephosphorylates several proteins, including Hph1, Slm1, Slm2, and the Crz1 transcription factor to promote cell survival (4, 10, 24, 35). Many of the genes transcriptionally activated by Crz1/calcineurin encode membrane proteins (49), and by activating Hph1, calcineurin may also promote translocation of these proteins. Surprisingly, we observed identical interactions of Hph1 and Hph1ΔPVIAVN, which is hyperphosphorylated due to its inability to be dephosphorylated by calcineurin, with Sec63/Sec62 components (data not shown). This suggests that some other aspect of Hph1 function, such as its proposed interaction with translocation substrates, is regulated by its phosphorylation state. Further studies are required to examine this possibility.
HPH1 and HPH2 expression increases when cells are exposed to DTT or tunicamycin, which causes ER stress through the accumulation of unfolded proteins in the ER (45). However, Hph1 and Hph2 are not essential under these conditions, nor is the unfolded protein response, as measured by expression of the UPRE-LacZ reporter gene (pJC005 [9]), constitutively induced in hph1Δ hph2Δ cells (data not shown). Still, the Hph1 regulator calcineurin is required for cell survival during ER stress (15), and upregulation of Hph1 and Hph2 under these conditions may contribute to cell survival by enhancing the translocation of proteins that allow cells to cope with ER stress. Alternatively, an increase in unfolded proteins under these conditions may decrease the availability of cytosolic chaperones and increase accumulation of untranslocated precursors (12); Hph1 and Hph2 could compensate for such stress-induced translocation defects.
Hph1, calcineurin, and Hsp90 provide one mechanism by which yeasts acquire resistance to azole antifungals (8, 38). Our studies suggest a possible explanation for Hph1-mediated azole resistance. Overexpression of HIS3 abrogates the azole sensitivity of an erg3Δ hph1Δ hph2Δ strain, and this finding prompted the proposal that Hph1 and Hph2 promote the sensing or import of amino acids in response to azole stress (38). It will be interesting to examine, in light of their interaction with the Sec63/Sec62 complex, if Hph1 and Hph2 promote the translocation of amino acid transporters or factors that aid in the biogenesis of these transporters.
The machinery that mediates protein translocation is well studied; however, we still know relatively little about how this critical and potentially rate-limiting step in protein biogenesis is modulated in vivo (25). Hph1 and Hph2 promise to provide novel insights into the mechanisms and physiological conditions that regulate the ability of proteins to transit the ER membrane.
Supplementary Material
ACKNOWLEDGMENTS
We thank Tom Stevens (University of Oregon) for providing reagents and sharing unpublished data and Wolf Frommer (Carnegie Institute of Plant Biology) and Clara Bermejo for use of the TECAN. We thank members of the Cyert laboratory for technical advice and useful discussion, especially J. Roy for critically reading the manuscript. We are grateful to Randy Schekman for discussion and antibodies.
Funding for this work was provided by NIH research grant GM-48729 to M.S.C. and an Agilent Technologies Foundation Grant to M.S.C and A.F.O. F.J.P was funded by Departmental NIH training grant T32-GM007276, an NSF Graduate Research Fellowship, and an NIH-NRSA predoctoral fellowship (5F31GM084690). Work performed in E.A.M.'s laboratory is supported by NIH grants GM078186 and GM085089. Work in S.F.'s laboratory was supported by NIH grant P41 RR11823. S.F. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
Published ahead of print on 19 November 2010.
REFERENCES
- 1. Ausubel F. M. 1991. Current protocols in molecular biology. John Wiley & Sons Inc: New York, NY [Google Scholar]
- 2. Becker J., Walter W., Yan W., Craig E. A. 1996. Functional interaction of cytosolic hsp70 and DnaJ-related protein, Ydj1, in protein translocation in vivo. Mol. Biol. Cell 16:4378–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Beilharz T., Egan B., Silver P. A., Hofmann K., Lithgow T. 2003. Bipartite signals mediate subcellular targeting of tail-anchored membrane proteins in Saccharomyces cerevisiae. J. Biol. Chem. 278:8219–8223 [DOI] [PubMed] [Google Scholar]
- 4. Bultynck G., et al. 2006. Slm1 and Slm2 are novel substrates of the calcineurin phosphatase required for heat stress-induced endocytosis of the yeast uracil permease. Mol. Cell. Biol. 26:4729–4745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Burri L., Lithgow T. 2004. A complete set of SNAREs in yeast. Traffic 5:45–52 [DOI] [PubMed] [Google Scholar]
- 6. Caplan A. J., Cyr D. M., Douglas M. G. 1992. YDJ1p facilitates polypeptide translocation across different intracellular membranes by a conserved mechanism. Cell 71:1143–1155 [DOI] [PubMed] [Google Scholar]
- 7. Copic A., et al. 2009. Genomewide analysis reveals novel pathway affecting endoplasmic reticulum homeostasis, protein modification and quality control. Genetics 182:757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cowen L. E., Carpenter A. E., Matangkasombut O., Fink G. R., Lindquist S. 2006. Genetic architecture of Hsp90-dependent drug resistance. Eukaryot. Cell 5:2184–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cox J. S., Shamu C. E., Walter P. 1993. Transcriptional induction of genes encoding endoplasmic reticulum resident proteins requires a transmembrane protein kinase. Cell 73:1197–1206 [DOI] [PubMed] [Google Scholar]
- 10. Cyert M. S. 2003. Calcineurin signaling in Saccharomyces cerevisiae: how yeast go crazy in response to stress. Biochem. Biophys. Res. Commun. 311:1143–1150 [DOI] [PubMed] [Google Scholar]
- 11. Davis-Kaplan S. R., et al. 2006. PKR1 encodes an assembly factor for the yeast V-type ATPase. J. Biol. Chem. 281:32025–32035 [DOI] [PubMed] [Google Scholar]
- 12. Deshaies R. J., Koch B. D., Werner-Washburne M., Craig E. A., Schekman R. 1988. A subfamily of stress proteins facilitates translocation of secretory and mitochondrial precursor polypeptides. Nature 332:800–805 [DOI] [PubMed] [Google Scholar]
- 13. Deshaies R. J., Sanders S. L., Feldheim D., Schekman R. 1991. Assembly of the yeast Sec proteins involved in the translocation into the endoplasmic reticulum into a membrane-bound multisubunit complex. Nature 349:806–808 [DOI] [PubMed] [Google Scholar]
- 14. Deshaies R. J., Schekman R. 1987. A yeast mutant defective at an early stage in import of secretory protein precursors into the edoplasmic reticulum. J. Cell Biol. 105:633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dudgeon D. D., Zhang N., Ositelu O. O., Kim H., Cunningham K. W. 2008. Nonapoptotic death of Saccharomyces cerevisiae cells that is stimulated by Hsp90 and inhibited by calcineurin and Cmk2 in response to endoplasmic reticulum stresses. Eukaryot. Cell 7:2037–2051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Esnault Y., Blondel M. O., Deshaies R. J., Schekman R., Kepes F. 1993. The yeast SSS1 gene is essential for secretory protein translocation and encodes a conserved protein of the endoplasmic reticulum. EMBO J. 12:4083–4093 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Esnault Y., Feldheim D., Blondel M. O., Schekman R., Kepes F. 1994. SSS1 encodes a stabilizing component of the Sec61 subcomplex of the yeast protein translocation apparatus. J. Cell Biol. 269:27478–27485 [PubMed] [Google Scholar]
- 18. Fang H., Green N. 1994. Nonlethal sec71-1 and sec72-1 mutations eliminate proteins associated with the Sec63p-BiP complex from S. cerevisiae. Mol. Biol. Cell 5:933–942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Feldheim D., Schekman R. 1994. Sec72p contributes to the selective recognition of signal peptides by the secretory polypeptide translocation complex. J. Cell Biol. 126:935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Feldheim D., Yoshimura K., Admon A., Schekman R. 1993. Structural and functional characterization of Sec66p, a new subunit of the polypeptide translocation apparatus in the yeast endoplasmic reticulum. Mol. Biol. Cell 4:931–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ghaemmaghami S., et al. 2003. Global analysis of protein expression in yeast. Nature 425:737–741 [DOI] [PubMed] [Google Scholar]
- 22. Graham L. A., Hill K. J., Stevens T. H. 1998. Assembly of the yeast vacuolar H+-ATPase occurs in the endoplasmic reticulum and requires a Vma12p/Vma22p assembly complex. J. Cell Biol. 142:39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Green N., Fang H., Walter P. 1992. Mutants in three novel complementation groups inhibit membrane protein insertion into and soluble protein translocation across the endoplasmic reticulum membrane of Saccharomyces cerevisiae. J. Cell Biol. 116:597–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Heath V. L., Shaw S. L., Roy S., Cyert M. S. 2004. Hph1p and Hph2p, novel components of calcineurin-mediated stress responses in Saccharomyces cerevisiae. Eukaryot. Cell 3:695–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Hegde R. S., Kang S. W. 2008. The concept of translocation regulation. J. Cell Biol. 182:225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hill K. C., Anthony A. 2000. Degradation of unassembled Vph1p reveals novel aspects of the yeast ER quality control system. EMBO J. 19:550–561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hill K. J., Stevens T. H. 1994. Vma21p is a yeast membrane protein retained in the endoplasmic reticulum by a di-lysine motif and is required for the assembly of the vacuolar H(+)-ATPase complex. Mol. Biol. Cell 5:1039–1050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jackson D. D., Stevens T. H. 1997. Vma12 encodes a yeast endoplasmic reticulum protein required for vacuolar H+-ATPase assembly. J. Biol. Chem. 272:25928–25934 [DOI] [PubMed] [Google Scholar]
- 29. Kane P. M. 2006. The where, when, and how of organelle acidification by the yeast vacuolar H+-ATPase. Microbiol. Mol. Biol. Rev. 70:177–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawasaki-Nishi S., Bowers K., Nishi T., Forgac M., Stevens T. H. 2001. The amino-terminal domain of the vacuolar proton-translocating ATPase a subunit controls targeting and in vivo dissociation, and the carboxyl-terminal domain affects coupling of proton transport and ATP hydrolysis. J. Biol. Chem. 276:47411–47420 [DOI] [PubMed] [Google Scholar]
- 31. Manolson M. F., et al. 1994. STV1 gene encodes functional homologue of 95-kDa yeast vacuolar H(+)-ATPase subunit Vph1p. J. Biol. Chem. 269:14064–14074 [PubMed] [Google Scholar]
- 32. Milgrom E., Diab H., Middleton F., Kane P. M. 2007. Loss of vacuolar proton-translocatin ATPase activity in yeast results in chronic oxidative stress. J. Biol. Chem. 282:7125–7136 [DOI] [PubMed] [Google Scholar]
- 33. Miller J. P., et al. 2005. Large-scale identification of yeast integral membrane protein interactions. Proc. Natl. Acad. Sci. U. S. A. 102:12123–12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ng D. T., Brown J. D., Walter P. 1996. Signal sequences specify the targeting route to the endoplasmic reticulum membrane. J. Cell Biol. 134:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. O'Donnell A. F., Apffel A., Gardner R. G., Cyert M. S. 2010. {alpha}-Arrestins Aly1 and Aly2 regulate intracellular trafficking in response to nutrient signaling. Mol Biol Cell 21:3552–3566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pagant S., Kung L., Dorrington M., Lee M. C. S., Miller E. A. 2007. Inhibiting endoplasmic reticulum (ER)-associated degradation of misfolded Yor1p does not permit ER export despite the presence of a diacidic sorting signal. Mol. Biol. Cell 18:3398–3413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pool M. R. 2005. Signal recognition particles in chloroplasts, bacteria, yeast, and mammals. Mol. Membr. Biol. 22:3–15 [DOI] [PubMed] [Google Scholar]
- 38. Robbins N., Collins C., Morhayim J., Cowen L. E. 2010. Metabolic control of antifungal drug resistance. Fungal Genet. Biol. 47:81–93 [DOI] [PubMed] [Google Scholar]
- 39. Rodríguez A., et al. 2009. A conserved docking surface on calcineurin mediates interaction with substrates and immunosuppressants. Mol. Cell 33:616–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ryan M., Graham L. A., Stevens T. H. 2008. Voa1p functions in V-ATPase assembly in the yeast endoplasmic reticulum. Mol. Biol. Cell 19:5131–5142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sanders S. L., Whitfield K. M., Vogel J. P., Rose M. D., Schekman R. W. 1992. Sec61p and BiP directly facilitate polypeptide translocation into the ER. Cell 69:353–365 [DOI] [PubMed] [Google Scholar]
- 42. Seol J. H., Shevchenko A., Shevchenko A., Deshaies R. J. 2001. Skp1 forms multiple protein complexes, including RAVE, a regulator of V-ATPase assembly. Nat. Cell Biol. 3:384–391 [DOI] [PubMed] [Google Scholar]
- 43. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 44. Toikkanen J., et al. 1996. Yeast protein translocation complex: isolation of two genes SEB1 and SEB2 encoding proteins homologous to the Sec61 beta subunit. Yeast 12:425–438 [DOI] [PubMed] [Google Scholar]
- 45. Travers K. J., et al. 2000. Functional and genomic analyses reveal an essential coordination between the unfolded protein response and ER-associated degradation. Cell 101:249–258 [DOI] [PubMed] [Google Scholar]
- 46. Wang X., Johnsson N. 2005. Protein kinase CK2 phosphorylates Sec63p to stimulate the assembly of the endoplasmic reticulum protein translocation apparatus. J. Cell Sci. 118:723–732 [DOI] [PubMed] [Google Scholar]
- 47. Willer M., Jermy A. J., Young B. P., Stirling C. J. 2003. Identification of novel protein-protein interactions at the cytosolic surface of the Sec63 complex in the yeast ER membrane. Yeast 20:133–148 [DOI] [PubMed] [Google Scholar]
- 48. Wilson J. D., Barlowe C. 2010. Yet1p and Yet3p, the yeast homologs of BAP29 and BAP31, interact with the endoplasmic reticulum translocation apparatus and are required for inositol prototrophy. J. Biol. Chem. 285:18252–18261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Yoshimoto H., et al. 2002. Genome-wide analysis of gene expression regulated by the calcineurin/Crz1p signaling pathway in Saccharomyces cerevisiae. J. Biol. Chem. 277:31079–31088 [DOI] [PubMed] [Google Scholar]
- 50. Zimmermann R., Eyrisch S., Ahmad M., Helms V. 2010. Protein translocation across the ER membrane. Biochim. Biophys. Acta. doi:10.1016/j.bbamem.2010.06.016 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.