Abstract
The Candida albicans plasma membrane plays important roles in interfacing with the environment, morphogenesis, and cell wall synthesis. The role of the Sur7 protein in cell wall structure and function was analyzed, since previous studies showed that this plasma membrane protein is needed to prevent abnormal intracellular growth of the cell wall. Sur7 localizes to stable patches in the plasma membrane, known as MCC (membrane compartment occupied by Can1), that are associated with eisosome proteins. The sur7Δ mutant cells displayed increased sensitivity to factors that exacerbate cell wall defects, such as detergent (SDS) and the chitin-binding agents calcofluor white and Congo red. The sur7Δ cells were also slightly more sensitive to inhibitors that block the synthesis of cell wall chitin (nikkomycin Z) and β-1,3-glucan (caspofungin). In contrast, Fmp45, a paralog of Sur7 that also localizes to punctate plasma membrane patches, did not have a detectable role in cell wall synthesis. Chemical analysis of cell wall composition demonstrated that sur7Δ cells contain decreased levels of β-glucan, a glucose polymer that confers rigidity on the cell wall. Consistent with this, sur7Δ cells were more sensitive to lysis, which could be partially rescued by increasing the osmolarity of the medium. Interestingly, Sur7 is present in static patches, whereas β-1,3-glucan synthase is mobile in the plasma membrane and is often associated with actin patches. Thus, Sur7 may influence β-glucan synthesis indirectly, perhaps by altering the functions of the cell signaling components that localize to the MCC and eisosome domains.
Candida albicans is the most common human fungal pathogen. Although it is a commensal organism of humans, under certain conditions it causes life-threatening infections in susceptible individuals (18, 27). One feature that underlies the pathogenesis of C. albicans is its ability to grow in different morphologies, including buds, elongated pseudohyphae, and long filamentous hyphal cells (35). Induction of the hyphal form correlates with increased expression of virulence factors, including adhesin proteins that promote biofilm formation and attachment to human cells, secreted hydrolytic enzymes that facilitate invasive growth and inactivate components of the complement pathway, and antioxidant enzymes that counteract the immune system (5, 7, 20, 40). The role of the plasma membrane has been under investigation in C. albicans pathogenesis because of the critical roles that it plays in the production of virulence factors and in transducing information and nutrients from the extracellular environment. The plasma membrane is also involved in dynamic cellular processes, such as secretion, endocytosis, and cell wall biogenesis, that are essential for proper morphogenesis and viability. The critical role of the plasma membrane is highlighted by the fact that it is the target of the most commonly used antifungal drugs (28).
The Saccharomyces cerevisiae plasma membrane is composed of at least two different subdomains (24, 25). One domain, termed MCP (membrane compartment occupied by Pma1), contains proteins that readily diffuse, such as the plasma membrane ATPase Pma1. Another domain appears as 300-nm patches that are immobile in the membrane. The latter domain was termed MCC (membrane compartment occupied by Can1), since it contains the Can1 arginine permease (24, 25). These static MCC domains are distinct from the mobile cortical actin patches or the finger-like invaginations that form at sites of endocytosis. Subsequent studies identified other symporters in the MCC and also two families of proteins that are thought to contain four transmembrane domains (TMDs) (15, 17). The best-known examples of these two different families of tetra-spanning proteins are Nce102 and Sur7. Nce102 has been implicated in sphingolipid signaling that regulates the formation of the MCC domains (15). The function of Sur7 in S. cerevisiae is not clear, perhaps because of genetic redundancy, since there are at least three paralogs of Sur7 (Fmp45, Ylr414c, and Pun1/Ynl194c) (2, 44). Mutation of the SUR7 family members does not cause strong phenotypes under standard growth conditions (2, 44), although Pun1 was recently shown to be important for proper response to nitrogen stress (43). Pun1/Ynl194c is more divergent than the other members of the Sur7 family in S. cerevisiae, including differences in the Cys-containing motif in extracellular loop 1 of Sur7 family proteins and at the analogous position in the mammalian Claudin proteins found at tight junctions (2). In contrast, C. albicans contains just two members of this family, Sur7 and Fmp45. This suggests that the functions of Sur7 family members may have diverged between S. cerevisiae and C. albicans.
The structure of the MCC domains was broadened by identification of a complex of cytoplasmic proteins known as eisosomes that associate with the inner surface of the plasma membrane at MCC sites (39). Eisosomes have been implicated in endocytosis, but it is not clear how these static domains relate to sites of endocytosis mediated by actin patches, since these regions appear to be distinct in the plasma membrane (39, 44). However, there is likely to be at least an indirect link. Two proteins that localize to eisosomes, Pil1 and Lsp1, are needed for proper rates of endocytosis (39). Pil1 and Lsp1 associate with the Pkh1/2 protein kinases (23, 38) that influence endocytosis and a variety of other processes, including cell wall integrity, actin localization, and response to heat stress (10, 23). Pkh1/2 also mediate the effects of sphingolipids on the regulation of the assembly and disassembly of eisosomes by phosphorylating Pil1 and Lsp1 (23, 38).
Analysis of a C. albicans sur7Δ (Casur7Δ) mutant implicated the MCC domains in the regulation of cell wall synthesis. In contrast to S. cerevisiae, a Casur7Δ mutant displayed obvious defects in bud and hyphal morphogenesis (1–3). Interestingly, the mutant cells showed an unusual phenotype in that there were large intracellular regions of cell wall growth. The abnormal cell wall synthesis was suggested to be due in part to a defect in β-1,3-glucan synthesis, since the sur7Δ mutant showed changes in gene expression that overlapped with those observed when cells are treated with caspofungin, an inhibitor of cell wall β-1,3-glucan synthase (2). In addition, septins were mislocalized in sur7Δ cells, analogous to the altered localization of septins caused by treatment of cells with caspofungin (4). Therefore, C. albicans sur7Δ and fmp45Δ mutants were analyzed in this study to determine how the structure and function of the cell wall were altered. The results demonstrated that Sur7 is important for resistance to conditions that perturb cell wall function and also for the proper synthesis of the β-1,3-glucan component of the cell wall.
MATERIALS AND METHODS
Strains and media.
The C. albicans strains (Table 1) were propagated on rich YPD medium (2% glucose, 1% peptone, and 2% yeast extract) or on SD (yeast nitrogen base synthetic medium with dextrose), essentially as described previously (34). Uridine (80 mg/liter) was added to cultures of ura3Δ cells. Hyphal morphogenesis was monitored by growing cells at 37°C in medium containing 10% bovine calf serum. Invasive growth was assayed by spotting 3 × 103 cells onto agar medium containing 4% bovine calf serum and then incubating the plates for 7 days at 37°C.
Table 1.
Strains used in this study
| C. albicans strain | Parent | Genotype |
|---|---|---|
| BWP17 | Sc5314 | ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| DIC185 | BWP17 | ura3Δ::λimm434/URA3 his1::hisG/HIS1 arg4::hisG/ARG4 |
| YJA10 | BWP17 | sur7Δ::ARG4/sur7Δ::HIS1 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA11 | YJA10 | sur7Δ::ARG4/sur7Δ::HIS1 URA3/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YJA12 | YJA10 | sur7Δ::ARG4/sur7Δ::HIS1 SUR7::URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW1 | BWP17 | fmp45Δ::ARG4/FMP45 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW2 | YHXW1 | fmp45Δ::ARG4/fmp45Δ::HIS1 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW3 | YHXW2 | fmp45Δ::ARG4/fmp45Δ::HIS1 URA3/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW4 | BWP17 | BWP17, except SUR7-GFPγ::URA3 |
| YHXW5 | BWP17 | BWP17, except FMP45-GFPγ::URA3 |
| YHXW6 | YJA10 | sur7Δ strain YJA10, except FMP45-GFPγ::URA3 |
| YHXW7 | YHXW2 | fmp45Δ strain YHXW2, except SUR7-GFPγ::URA3 |
| YHXW8 | YHXW2 | fmp45Δ::NAT/fmp45Δ::HIS1 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW9 | YHXW8 | fmp45Δ::NAT/fmp45Δ::HIS1 sur7Δ::UAU/SUR7 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW10-UAU1 | YHXW9 | fmp45Δ::NAT/fmp45Δ::HIS1 sur7Δ::UAU/sur7Δ::URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW10-UAU2 | YHXW9 | fmp45Δ::NAT/fmp45Δ::HIS1 sur7Δ::UAU/sur7Δ::URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
| YHXW10-UAU3 | YHXW9 | fmp45Δ::NAT/fmp45Δ::HIS1 sur7Δ::UAU/sur7Δ::URA3 ura3Δ::λimm434/ura3Δ::λimm434 his1::hisG/his1::hisG arg4::hisG/arg4::hisG |
The C. albicans sur7Δ homozygous deletion mutant and the sur7Δ- plus SUR7-complemented strain were constructed previously in C. albicans strain BWP17 (2). The fmp45Δ homozygous deletion mutant was constructed using similar methods. In brief, PCR primers that contained ∼70 bp of sequence homologous to the 5′ and 3′ ends of the FMP45 open reading frame were used to amplify a cassette containing either the ARG4 or the HIS1 selectable marker gene (41). The sur7Δ fmp45Δ double mutants were constructed using the UAU method (13). The starting strain for this procedure was YHXW8 (fmp45::HIS1 fmp45::NAT), which was constructed in part by using the SAT-flipper (32). One allele of SUR7 was then deleted by homologous recombination with the UAU cassette and identified after selection on medium lacking arginine. The UAU cassette consists of an ARG4 gene flanked by the 5′ and 3′ portions of the URA3 gene (13). The mutant cells were then grown nonselectively to permit the endogenous recombination pathways to replace the other allele of SUR7 to generate a sur7::UAU/sur7::UAU homozygous deletion. Segregants were then isolated on medium lacking Arg and Ura to identify cells that underwent a further recombination event that converts one UAU allele into a functional URA3 gene by looping out the ARG4 sequences. More than 30% of the Arg+ Ura+ colonies contained a homozygous deletion, sur7::UAU/sur7::URA3, and lacked the wild-type SUR7 gene, which is similar to the frequency reported for other nonessential genes (13). Integration of the deletion cassettes at the appropriate sites was verified by PCR using combinations of primers that flanked the integration site and also primers that annealed within the introduced cassettes.
The 3′ ends of the SUR7 and FMP45 open reading frames were fused to green fluorescent protein (GFP) via homologous recombination using previously described methods (45). PCR primers containing ∼70 bp of sequence homologous to the 3′ end of the SUR7 or FMP45 open reading frames were used to amplify a cassette containing a more photostable version of enhanced GFP (CaGFPγ) and a URA3 selectable marker (45). The Ura+ colonies resulting from the transformation of the cassette into C. albicans were then screened for GFP-positive cells by fluorescence microscopy and confirmed by PCR.
Microscopy.
Cells used for microscopic analysis of Sur7-GFP and Fmp45-GFP were grown overnight to log phase. Fluorescence microscopy was used to detect GFP, and differential interference contrast (DIC) optics was used to detect cell morphology. Chitin staining was carried out by incubating cells with 40 ng/ml calcofluor white (Sigma-Aldrich) as described previously (29). In order to visualize the intracellular growth of the cell wall, cells were first permeabilized by washing them with methanol and acetone and then stained with calcofluor white. Covalent labeling of the cell walls with the fluorescent dye fluorescein was carried out by washing the cells 3 times in phosphate-buffered saline (PBS), resuspending them at 108 cells/ml in PBS, and then adding fluorescein-5-maleimide (FM) (Invitrogen) to a final concentration of 0.2 μg/ml. The cells were incubated for 15 min, washed 3 times with PBS, and then resuspended in YPD medium. Labeling the cells with FM in this manner did not have detectable effects on growth. At different times after FM labeling, the cells were permeabilized as described above and then stained with 4 μg/ml calcofluor white. Cell integrity was analyzed by mixing equal volumes of cell culture and a solution of 0.4% trypan blue (Sigma-Aldrich), incubating them for 5 min, and then visualizing them by light microscopy to identify lysed cells that stained blue. Images were captured using an Olympus BH2 microscope equipped with a Zeiss AxioCam digital camera.
Drug and chemical sensitivity.
Sensitivity to SDS, Congo red, calcofluor white, and hygromycin was tested by spotting dilutions of cells onto YPD plates prepared with the indicated concentration of the corresponding chemical. Three microliters of a 10-fold dilution series of cells was spotted on the surfaces of the plates. The plates were then incubated at 37°C for the indicated time and photographed. The sensitivity of cells to nikkomycin Z, rapamycin, and tunicamycin was assayed by testing the effects of a serial dilution series of drug concentrations in 96-well plates. The cells were adjusted to 1 × 104 cells/ml in SD medium and plated in the wells, and then a 2-fold dilution series of the indicated drug was applied. The plates were incubated at 37°C for 2 days, and the extent of growth was quantified with a spectrophotometer. Sensitivities to fluconazole, amphotericin, and caspofungin were assayed using Etest strips according to the manufacturer's directions (bioMérieux, Inc., Durham, NC). In brief, 1 × 106 cells were spread on an agar plate containing RPMI 1640 medium (Sigma-Aldrich), 0.165 M MOPS (morpholinepropanesulfonic acid), pH 7, and 2% dextrose; an Etest strip was applied; and the plates were incubated at 37°C for 48 h.
Chemical analysis of cell wall composition.
Strains were adjusted to an optical density at 600 nm (OD600) of 0.01 and then grown in YPD at 30°C for 15 h. The cells were washed twice with water and boiled twice in 50 mM Tris-HCl buffer (pH 7.5) containing 50 mM EDTA, 2% SDS, and 40 mM β-mercaptoethanol for 1 h. The sediment obtained after centrifugation (3,600 rpm; 10 min) was washed five times with water and then incubated twice in 1 M NaOH containing 0.5 M NaBH4 at 65°C for 1 h. The insoluble pellet obtained upon centrifugation (3,600 rpm; 10 min) was washed with water to neutrality and freeze-dried (alkali-insoluble [AI] fraction). The supernatant fraction was then neutralized, dialyzed against water, and freeze-dried (alkali-soluble [AS] fraction). The monosaccharide composition was determined by gas-liquid chromatography (GLC) (Perichrom PR2100 instrument; Perichrom, Saulx-les-Chartreux, France) equipped with a flame ionization detector (FID) and a fused silica capillary column (30 m by 0.32-mm inside diameter; filled with BP1 matrix composed of dimethyl polysiloxane) using mesoinositol as the internal standard. Derivatized hexoses (alditol acetates) were obtained after hydrolysis (4 N trifluoroacetic acid/8 N hydrochloric acid; 100°C for 4 h) of the AI and AS fractions, followed by reduction and peracetylation. The monosaccharide composition (percent) was calculated from the peak areas with respect to that of the internal standard (14).
RESULTS
Sur7-GFP and Fmp45-GFP localize to punctate plasma membrane domains in C. albicans.
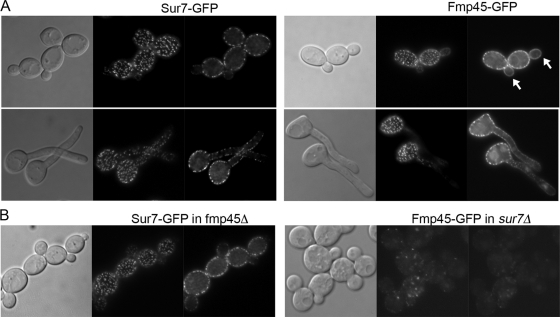
Analysis of the C. albicans genome detected only one paralog of Sur7, named Fmp45, which showed 37% identity and 54% similarity to the Sur7 sequence. In contrast, S. cerevisiae contains at least three paralogs of Sur7 (Fmp45, Ylr414c, and Pun1/Ynl194c). Although the C. albicans paralog of Sur7 was previously named Fmp45, it shows approximately equal amino acid sequence relatedness to S. cerevisiae Fmp45 and Ynl194c (31% identity). To determine if CaFmp45 was present in punctate patches in the plasma membrane, we examined the localization of proteins that were fused to GFP. For this analysis, a recently described version of GFP (GFPγ) that is more photostable (45) was fused to the C-terminal coding sequences of SUR7 and FMP45. As expected, Sur7-GFP was detected in a punctate pattern in the plasma membrane in mother cells but was only weakly detected in new buds (Fig. 1A). Fmp45-GFP was also detected in punctate structures in mother cells, in agreement with previous reports for Fmp45-GFP in S. cerevisiae and C. albicans (3, 44). In contrast to Sur7-GFP, however, we also detected Fmp45-GFP in new buds. The Fmp45-GFP present in small buds was distributed in a more homogeneous fashion and was not concentrated into punctate domains as it was in mature cells (Fig. 1A). Thus, Sur7-GFP and Fmp45-GFP give similar, but not identical, localization patterns.
Fig. 1.
Punctate plasma membrane localization of Sur7-GFP and Fmp45-GFP. (A) C. albicans strain BWP17 engineered to express SUR7-GFP or FMP45-GFP fusion genes. (Top) Budding cells. (Bottom) Cells induced with 10% bovine calf serum to form hyphae. In each series of images, the image on the left is a light microscope image to show cell morphology. The two fluorescence microscope images on the right demonstrate the localization of Sur7-GFP and Fmp45-GFP in punctate patches in focal planes that correspond to the top of the cell and to the medial portion. The arrows indicate bud localization of Fmp45-GFP. (B) Localization of Sur7-GFP in fmp45Δ cells and Fmp45-GFP in sur7Δ cells. The microscope images are as described for panel A. All of the fusion genes were constructed with GFPγ, a more photostable version of GFP (45). The strains used in these experiments included YHXW4 (SUR7-GFP), YHXW5 (FMP45-GFP), YHXW6 (sur7Δ FMP45-GFP), and YHXW7 (fmp45Δ SUR7-GFP).
Analysis of cell wall localization in sur7Δ and fmp45Δ cells.
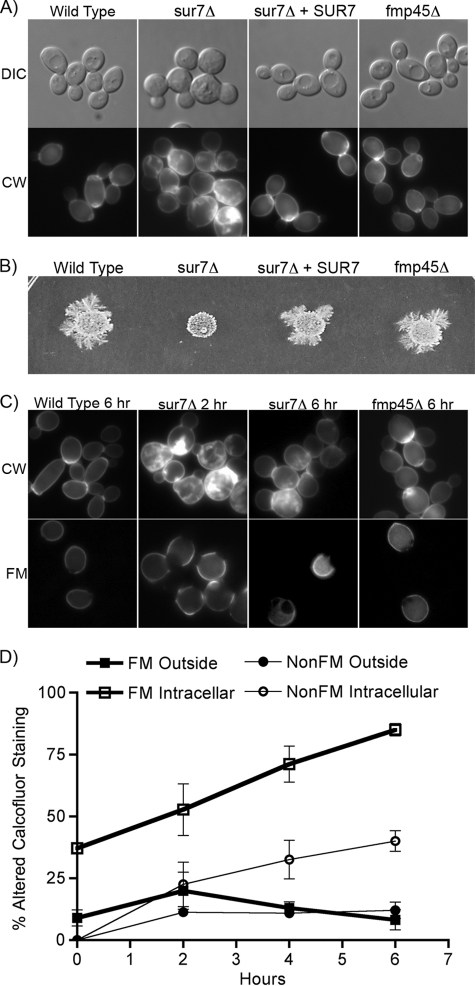
To determine whether Fmp45 functions in cell wall morphogenesis, a homozygous fmp45Δ deletion mutant was constructed in the C. albicans BWP17 strain. Microscopic analysis of live cells did not reveal obvious morphological defects for the fmp45Δ cells; they formed buds (Fig. 2A) and hyphae (data not shown) that were similar to those of the wild-type control strain. The fmp45Δ cells also did not contain the intracellular growth of the cell wall seen in the sur7Δ mutant (Fig. 2A, bottom), and Sur7-GFP localized properly in the fmp45Δ cells (Fig. 1B). The fmp45Δ mutant also was not defective in invasive growth into agar, as was the sur7Δ mutant (Fig. 2B). In contrast, as reported previously (2), about half the sur7Δ mutant cells showed obvious morphological defects, including enlarged cells and bud necks, and many cells displayed ectopic intracellular growth of the cell wall (Fig. 2A). Fmp45-GFP was detected at greatly reduced levels in sur7Δ cells, demonstrating that Sur7 is required for normal organization of MCC domains (Fig. 1B).
Fig. 2.
Analysis of the cell wall and morphogenesis phenotypes of the sur7Δ and fmp45Δ mutants. (A) The morphology of live cells of the indicated genotypes is shown in the upper images. The lower images show a different set of cells, viewed by fluorescence microscopy, that were permeabilized and stained with calcofluor white to detect cell wall structures. (B) Invasive-growth assay of the indicated C. albicans strains. Equal numbers of cells were spotted onto an agar plate containing 4% serum and then incubated at 37°C for 7 days. (C) Comparison of cell wall structures in older and newer cells. Cells of the indicated genotype were cross-linked to fluorescein-maleimide and then grown for 2 or 6 h at 30°C in YPD. The cells were then permeabilized and stained with calcofluor white. Fluorescence microscope images of cells stained with calcofluor white are shown above; the older cells that were cross-linked with fluorescein are shown below. (D) Quantitation of calcofluor white-stained cells after labeling with FM at time zero versus new cells that were not labeled (NonFM). Outside, cells that showed altered calcofluor white staining of the outer cell wall; Intracellular, altered calcofluor white staining within the cell due to ectopic growth of the cell wall. The error bars indicate SD.
Older cells were examined to search more sensitively for a weak phenotype caused by fmp45Δ. The rationale for this was based on a previous suggestion that older sur7Δ mutant cells may have more severe defects than younger cells, since most of the new buds appeared to be relatively normal (2). To identify older cells in the population, the cells were covalently labeled with FM so that they could be detected by fluorescence microscopy, whereas any new cells that formed subsequent to the labeling procedure would not be fluorescent. The FM-labeled cells were then grown for 6 h at 30°C, permeabilized, and analyzed by staining the cell wall with calcofluor white. The older FM-labeled fmp45Δ cells did not show any obvious differences from the wild-type control strain (Fig. 2C). In contrast, nearly all of the older sur7Δ mutant cells showed altered calcofluor white staining of the outer cell wall and intracellular growth of the cell wall (Fig. 2C). Time course studies demonstrated that the percentage of FM-labeled sur7Δ cells with cell wall abnormalities increased over time from ∼45% to ∼90% at the 6-h time point (Fig. 2D). Interestingly, the newer cells that were not fluorescent developed altered cell wall structures as early as 2 h after FM labeling (Fig. 2C and D). The altered cell wall localization in the newer (non-FM-labeled) cells increased over time to a steady-state level of about 45% by 6 h. Thus, the sur7Δ mutant accumulates cell wall abnormalities over time, indicating defects in both the spatial and temporal regulation of wall synthesis.
The sur7Δ mutant is more sensitive to agents that exacerbate cell wall defects.
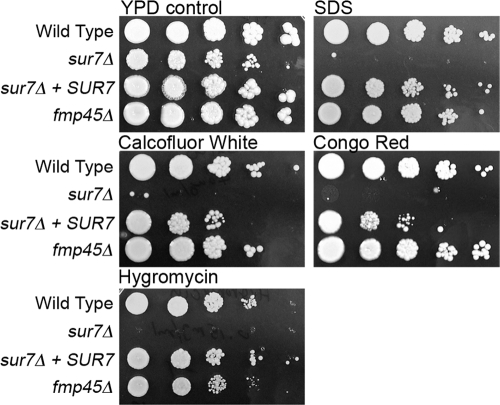
The sur7Δ mutant forms extra ectopic cell wall, but it is not clear whether this affects the function of the cell wall. To investigate this, the sur7Δ and fmp45Δ mutants were analyzed for the ability to grow on agar medium containing compounds that exacerbate the growth defects of cell wall synthesis mutants, including SDS, calcofluor white, Congo red, and hygromycin (9). Interestingly, the sur7Δ mutant showed greatly increased sensitivity to all four of these reagents, whereas the fmp45Δ mutant did not (Fig. 3). Calcofluor white and Congo red are thought to perturb cell wall synthesis by binding to the chitin component of the cell wall. The basis for the sensitivity to SDS is not known, but it may be due to enhanced sensitivity of cell wall synthesis components or a general effect on the plasma membrane. Hygromycin is an aminoglycoside antibiotic whose ability to inhibit protein synthesis is increased in a wide range of cell wall mutants for unknown reasons. Altogether, these results indicate that the sur7Δ mutant forms defective cell walls.
Fig. 3.
The sur7Δ mutant is more sensitive to chemicals that exacerbate the growth defects of cell wall mutants. Ten-fold dilutions of the wild-type control strain DIC185, YJA11 (sur7Δ), YJA12 (sur7Δ plus SUR7), and YHXW3 (fmp45Δ) were spotted onto rich-medium YPD plates containing the following concentrations of the indicated additive: 0.06% SDS, 0.04 mg/ml calcofluor white, 0.1 mg/ml Congo red, and 0.15 mg/ml hygromycin. The plates were incubated at 37°C for 2 days and then photographed.
Sensitivity to antifungal drugs.
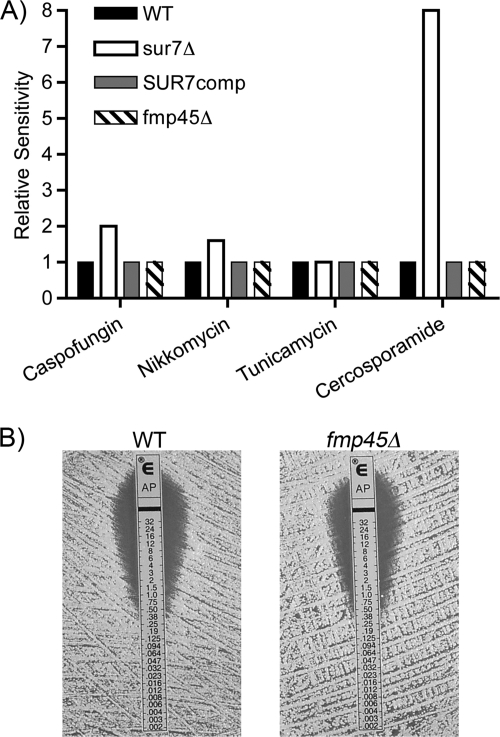
The mutant cells were analyzed further by testing their sensitivity to drugs that perturb cell wall synthesis. The Pkc1 cell wall stress pathway is expected to play an important role in cells that form abnormal cell walls. To test this, broth dilution assays in 96-well plates were used to examine sensitivity to the drug cercosporamide, which is a selective inhibitor of the C. albicans Pkc1 protein kinase (36). The results demonstrated that sur7Δ cells were 8-fold more sensitive, consistent with the mutant cells forming defective cell walls (Fig. 4A). The sur7Δ mutant was also about 50% more sensitive to nikkomycin Z, an inhibitor of chitin synthase (Fig. 4A), but showed normal sensitivity to tunicamycin, an inhibitor of N-linked glycosylation that is expected to perturb a wide range of glycosylated cell wall proteins. As reported previously, Etest assays showed that the sur7Δ mutant was 2-fold more sensitive to caspofungin (2). Thus, the sur7Δ mutant is slightly more sensitive to inhibitors of both chitin and β-glucan synthases. The sur7Δ mutant was not hypersensitive to all compounds that exacerbate the defects of cell wall mutants. For example, it showed only a slight difference in sensitivity to rapamycin (≤2-fold) and was not sensitive to a high calcium concentration (0.5 M) (data not shown).
Fig. 4.
Altered sensitivity of the sur7Δ mutant to antifungal drugs. (A) C. albicans strains were tested for sensitivity to nikkomycin Z, tunicamycin, and cercosporamide by assaying their ability to grow in 96-well plates containing a 2-fold dilution series of each drug. Sensitivity to caspofungin was tested with Etest strips, as described previously for the sur7Δ mutant (2). The results represent the average of at least two independent experiments. (B) The fmp45Δ mutant displays a wild-type level of sensitivity to amphotericin. Cells were spread on a plate containing RPMI 1640 medium, the Etest strip was applied, and then the plates were incubated at 37°C for 48 h. The Etest strips release a gradient of drug, causing a zone of growth inhibition. The strains used in these assays included DIC185, YJA11 (sur7Δ), YJA12 (sur7Δ plus SUR7), and YHXW3 (fmp45Δ).
The fmp45Δ mutant did not display increased sensitivity to cercosporamide or inhibitors of cell wall synthesis. Etest strip assays also showed that fmp45Δ did not show increased sensitivity to fluconazole (data not shown), as does sur7Δ (2). Furthermore, we did not detect increased sensitivity of the fmp45Δ homozygous deletion mutant to amphotericin (Fig. 4B), which contrasts with a previous report from a high-throughput study that an fmp45Δ/FMP45 heterozygote is more sensitive to amphotericin (42).
Altered cell wall composition of sur7Δ cells.
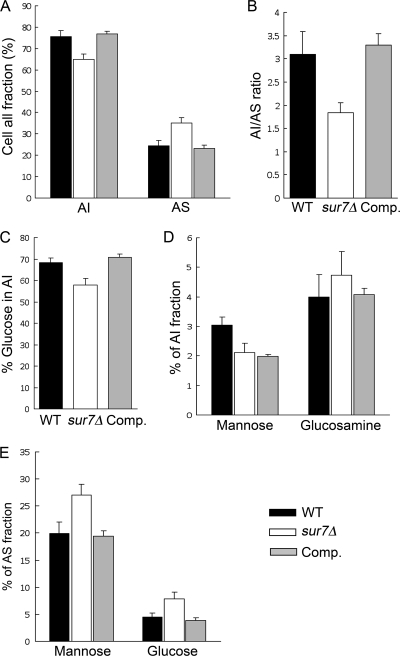
The carbohydrate composition of the sur7Δ cell wall was analyzed to determine if the constituents were altered compared to the wild-type control strain. The cell walls were divided into AI and AS fractions (see Materials and Methods). Interestingly, the sur7Δ mutant showed a decrease in the AI fraction of the cell wall that includes the important structural polymers, chitin and β-glucan (Fig. 5A). The AI/AS ratio decreased from 3.2 ± 0.5 for the wild-type control (DIC185) to 1.9 ± 0.2 for the sur7Δ strain (Fig. 5B). Analysis of the monosaccharide composition in the AI fraction from the wild-type control strain DIC185 showed that glucose was the major component, consistent with β-glucan being the most abundant fraction of the cell wall. There was a significant decrease (P < 0.01) in the glucose content in the sur7Δ cell wall AI fraction compared to that of the wild-type control strain (Fig. 5C). The levels of glucosamine detected in the AI fractions, which correspond to N-acetylglucosamine (GlcNAc) in the cell wall, were not significantly different for the wild-type and sur7Δ mutant strains (Fig. 5D), even though increased GlcNAc is often detected in cell wall mutants due to a compensatory mechanism that increases chitin levels. In contrast, the AS fraction was increased in the sur7Δ cells, mainly due to an elevated mannose content (Fig. 5E). The reduced AI/AS ratio and the low level of β-glucan suggest that the sur7Δ cells are likely to form weaker cell walls.
Fig. 5.
Decreased alkali-insoluble fraction in sur7Δ cell walls. The chemical compositions of the cell walls for the wild-type (DIC185), sur7Δ (YJA11), and sur7Δ plus SUR7 complemented (YJA12) strains were compared. (A) Relative amounts of AS and AI fractions as a percentage of the total dry weight of the cell wall. (B) AI/AS ratio for each strain. (C) Glucose compositions of the AI fractions. (D) Mannose and glucosamine compositions of the AI fractions. (E) Mannose and glucose compositions of the AS fractions. Cell wall fractions were analyzed as described in Materials and Methods. The results represent the average of 4 independent experiments plus SD.
Osmotic rescue of the sur7Δ cell lysis defect.
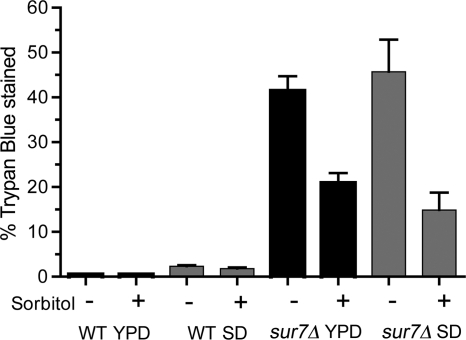
The altered cell wall composition suggested that sur7Δ cells would be more sensitive to lysis. To test this, cells were grown at 42°C, since elevated temperature promotes cell wall stress. As expected, only a small percentage of the cells of the wild-type control strain DIC185 stained with trypan blue, a vital stain used to determine cell viability. In contrast, the sur7Δ cells grew more slowly than the wild type at 42°C, and trypan blue staining demonstrated that there was a large fraction of lysed cells. More than 40% of the sur7Δ cells stained with trypan blue at 42°C in either rich YPD or synthetic SD medium, indicating that the cells had lysed and the trypan blue dye could gain access to the interior of the cell (Fig. 6). Increasing the osmolarity by adding 1 M sorbitol to the medium decreased the percentage of trypan blue staining from 41.5% to 20.9% in rich medium and 45.7% to 14.9% in synthetic SD medium. Similar results were observed for medium with 1 M NaCl (data not shown). The observation that increased osmolarity decreased the number of trypan blue-stained sur7Δ cells confirms that the lysis defect is due at least in part to weakened cell walls.
Fig. 6.
Increased osmolarity suppresses the cell lysis defect of the sur7Δ mutant at elevated temperatures. Wild-type (DIC185) and sur7Δ (YJA11) strains were grown overnight at 42°C in the presence or absence of 1 M sorbitol to increase osmolarity. The fraction of lysed cells was determined by staining them with trypan blue. For comparison, cells were grown in either rich YPD medium or synthetic SD medium. The results represent the average of at least two independent experiments plus SD.
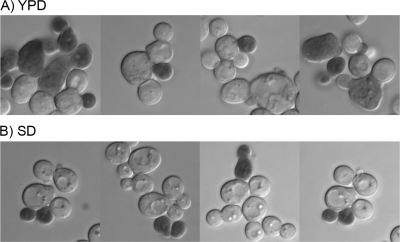
Microscopic analysis of the cells also revealed that both large and small sur7Δ cells stained with trypan blue. When the sur7Δ mutant was grown in rich YPD medium at 30°C, most of the cells that stained with trypan blue were larger. It is not surprising that the larger cells observed in the sur7Δ mutant cultures would lyse more readily, since their increased volume places extra demands on the cell wall. However, it was interesting that some smaller cells also stained with trypan blue (Fig. 7A). The lysis of smaller cells was more obvious when the cells were grown in synthetic SD medium, since the sur7Δ mutant formed fewer large cells under these conditions but still showed lysis of smaller cells (Fig. 7B). These results indicate that the sur7Δ mutation causes defects in cell wall synthesis in younger cells, as well as the older, larger cells that have accumulated more of the aberrant ectopic cell wall structures.
Fig. 7.
Spontaneous lysis of both large and small sur7Δ cells. The sur7Δ strain YJA11 was grown at 30°C in either rich YPD (A) or synthetic SD (B) medium and then stained with trypan blue to detect lysed cells.
Deletion of FMP45 does not exacerbate the phenotype of sur7Δ cells.
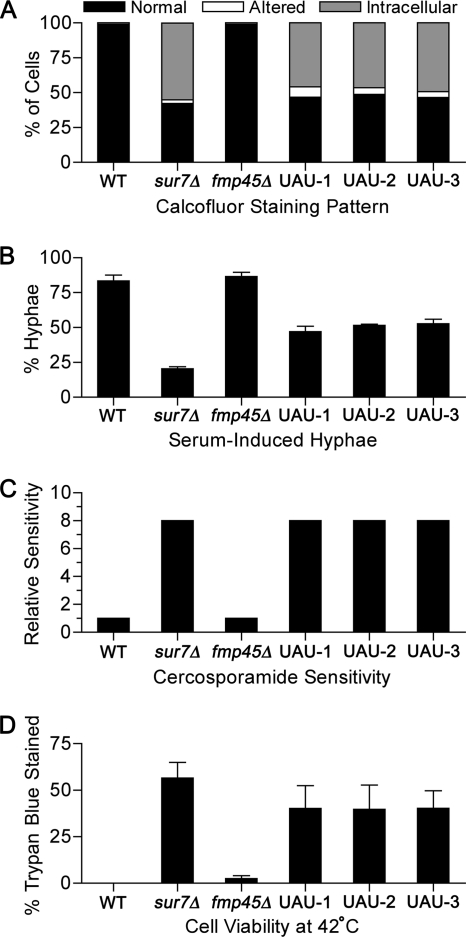
Double-mutant sur7Δ fmp45Δ cells were analyzed to determine whether Fmp45 plays a significant role in the absence of Sur7. In view of the possibility that the double mutation may cause lethality, we used the UAU method to create a sur7Δ fmp45Δ mutant strain (13). The UAU method entails deleting one copy of a gene with the UAU cassette (an ARG4 gene flanked by the 5′ and 3′ portions of a URA3 gene) and then selecting on medium lacking arginine and uridine for cells in which both copies of the targeted gene are deleted (see Materials and Methods). This approach was successful in yielding sur7Δ fmp45Δ mutants, three of which were picked for analysis. The double mutants showed ectopic staining of cell wall chitin (Fig. 8A), increased cercosporamide sensitivity (Fig. 8C), and lysis at 42°C (Fig. 8D) that was similar to that of the sur7Δ single mutant, but not worse. The three sur7Δ fmp45Δ mutants were also better able to form hyphae in response to serum than sur7Δ alone (Fig. 8B), suggesting the possibility that production of Fmp45 in the absence of Sur7 may have negative effects. Altogether, these results indicate that Fmp45 does not contribute a significant function in the absence of Sur7, which is consistent with the observation that Fmp45-GFP is not stably produced in sur7Δ cells (Fig. 1).
Fig. 8.
Phenotypes of sur7Δ fmp45Δ mutants. Three sur7Δ fmp45Δ double mutants (UAU-1, UAU-2, and UAU-3) were generated by the UAU method as described in Materials and Methods. The mutants were compared to the following strains: wild-type control (DIC185), sur7Δ (YJA11), and fmp45Δ (YHXW3). (A) Permeabilized cells stained with calcofluor white and then examined microscopically to determine the fractions of cells displaying a normal cell wall, an altered cell wall, or an intracellular cell wall pattern (n = >200). (B) Cells induced with 10% serum and then examined for hyphal formation (n = >200). (C) Sensitivity to cercosporamide tested by broth dilution assays in 96-well plates. (D) Cells grown overnight at 42°C and then stained with trypan blue to determine viability (n = >200). The error bars indicate the SD.
DISCUSSION
Cell wall synthesis plays a key role in C. albicans pathogenesis. The essential role of the cell wall for virulence is highlighted by the success of the drug caspofungin in treating infections by C. albicans and A. fumigatus (28). The cell wall also plays important roles by interacting with the immune system during infection (26). Many of the key steps of cell wall synthesis occur at the plasma membrane, where Sur7 is localized. This includes the action of β-glucan and chitin synthases that extrude the elongating polymers of glucose and GlcNAc, respectively, through the plasma membrane (19, 33). β-Glucan and chitin fibrils form the inner layer of the cell wall that gives it strength and rigidity. The outer layer is composed primarily of different types of mannoproteins. These various cell wall components must be regulated in concert to efficiently coordinate cell wall growth.
Cell wall function.
Previous studies demonstrated that Sur7 was needed for proper spatial regulation of cell wall growth (2). This unusual ectopic cell wall phenotype of sur7Δ cells also raised the question of whether the mutant cell walls could function properly. Several lines of evidence now confirm that the sur7Δ mutant produces defective cell walls. One is that the sur7Δ mutant was more sensitive to agents that exacerbate cell wall defects, including SDS, calcofluor white, and Congo red (Fig. 3) (3). However, the sur7Δ cells were not sensitive to all conditions that can affect cell wall mutants. For example, they were not more sensitive to elevated calcium and were only weakly sensitive to rapamycin (data not shown). Furthermore, the sur7Δ mutant was only slightly more sensitive to caspofungin and nikkomycin Z, inhibitors of β-glucan and chitin synthases, respectively (Fig. 4) (2). One possible reason for the relatively small effect of caspofungin and nikkomycin Z is that the sur7Δ mutation might alter the coordination of cell wall synthesis but might not directly affect the enzymatic activities of the β-glucan and chitin synthases. Altogether, these results indicate that mutation of SUR7 affects an important subset of cell wall functions.
As a test of cell wall strength, the sur7Δ mutant was examined for a cell lysis defect that could be suppressed by increased osmolarity. Growing the sur7Δ cells at 42°C resulted in a high proportion of lysed cells (>40%), consistent with previous observations that growth at elevated temperatures stresses the cell wall and enhances the defects of many cell wall mutants. Increasing the osmolarity of the medium by adding 1 M sorbitol partially rescued this lysis defect for the sur7Δ cells (Fig. 6). This confirms that the sur7Δ mutation causes defects in the functional properties of the cell wall, as well as in the spatial regulation of cell wall synthesis.
In contrast to Sur7, the Fmp45 protein did not have a detectable role in cell wall synthesis. The fmp45Δ mutant did not display altered morphogenesis or cell wall synthesis, and it was not more sensitive to a variety of conditions that were expected to exacerbate cell wall defects. Furthermore, the phenotype of a sur7Δ fmp45Δ mutant was not worse than that of the single sur7Δ mutant (Fig. 8). Thus, although Sur7 and Fmp45 show ∼37% identity, they have distinct roles.
Cell wall synthesis.
Chemical analysis demonstrated that the cell wall composition was significantly altered in sur7Δ cells (Fig. 5). In particular, the ratio of the alkali-insoluble to alkali-soluble fractions was greatly decreased, due to both a decrease in the AI fraction and an increase in the AS fraction. The increase in the AS fraction was primarily due to elevated mannan levels, which is consistent with the observation from microarray studies that the expression of the mannosyl transferase genes MNN1 and MNN4-4 was increased cells ∼8-fold and ∼5-fold, respectively, in sur7Δ (2). Altered mannosylation may also account for the increased sensitivity of the sur7Δ mutant to hygromycin, since defects in mannosylation have been linked to hypersensitivity to the drug (21). The biggest change in the AI fraction for the sur7Δ mutant was a decrease in β-glucan, the glucose polymer that constitutes the major component of the cell wall (19, 33). This decrease in β-glucan content is consistent with previous studies, which showed that the sur7Δ mutant displayed changes in gene expression and altered septin localization that were similar to effects cause by caspofungin (2, 4, 6). Furthermore, the reduced β-glucan levels are consistent with the formation of weaker cell walls in sur7Δ cells, since β-glucan plays a major role in cell wall strength and rigidity (22).
The next major question that needs to be resolved is whether Sur7 plays a direct or indirect role in cell wall synthesis. An indirect role is suggested by the very different patterns of subcellular localization for the static Sur7 patches and the mobile actin patches (2, 44). In S. cerevisiae, β-glucan synthase associates with mobile actin patches, which is thought to facilitate proper expansion of bud growth (11, 30, 37). In addition, Sur7-GFP is not readily detected in new buds (Fig. 1) (44), which are very active sites of cell wall synthesis. These differences in subcellular localization suggest that Sur7 is not directly involved in cell wall synthesis. However, other evidence raises the possibility that Sur7 or other MCC/eisosome proteins may directly regulate at least some aspects of cell wall synthesis. For example, an echinocandin class inhibitor of β-1,3-glucan synthesis was found to chemically cross-link primarily to the C. albicans Pil1 and Lsp1 proteins (12, 31), which are components of eisosomes that associate with Sur7 (39). In addition, a high-throughput study of S. cerevisiae proteins identified the Fks1 subunit of β-1,3-glucan synthase in a complex with Pil1 (16).
Further support for the idea that MCC/eisosomes are involved in cell wall regulation comes from analysis of the S. cerevisiae Pkh1/2 protein kinases, which localize to eisosomes (23, 38). In S. cerevisiae, Pkh1/2 regulate the Pkc1, Sch9, and Ypk1 protein kinases that act in part to control cell wall synthesis (8). Interestingly, the C. albicans sur7Δ cells were 8-fold more sensitive to the Pkc1 inhibitor cercosporamide (Fig. 4), similar to C. albicans cells with reduced Pkc1p activity (36). Since Pkc1 regulates many genes involved in cell wall synthesis, this raised the possibility that the cell wall defect of sur7Δ cells is due to decreased Pkc1 activity. However, other data indicate that Pkc1 is active in sur7Δ cells. For one, PKC1 mutations cause the opposite effect of mutating SUR7. The pkc1 mutant cells produce decreased cell wall and are not known to cause extra ectopic growth of the cell wall. Also, targets of the Pkc1-activated Rlm1 transcription factor (6) were not decreased in sur7Δ cells but were instead highly expressed. PGA13 was induced 16-fold, and CRH11 was induced 4-fold (2). Expression of cell wall synthesis genes was also not decreased in the sur7Δ cells. GSC1 (β-1,3-glucan synthase), as well as KRE1 and KRE7 (β-1,6-glucan synthases), were expressed at slightly increased levels, and the expression of the chitin synthase genes was not changed significantly in sur7Δ cells (2). In contrast, there were extensive changes in the expression of genes that encode a variety of other putative cell wall proteins, especially predicted glycosylphosphatidylinositol (GPI)-anchored proteins that are likely to directly interact with the cell wall. Many of these genes are thought to be activated by the Cas5 transcription factor, which is important for response to caspofungin treatment (6). These data indicate that the cell wall defects of the sur7Δ cells are not due to a general failure of cells to sense and respond to cell wall damage. Future studies on MCC/eisosomes will help to define the links between these novel domains and cell wall synthesis.
ACKNOWLEDGMENTS
We thank the members of our laboratories for their helpful advice and comments on the manuscript, Francisco Javier Alvarez for providing strains, and Yoshikazu Ohya for advice.
This research was supported by a grant awarded to J.B.K. from the National Institutes of Health (RO1 AI47837) and a grant awarded to J.-P.L. from ERA-NET PathoGenoMics ANTIFUN.
Footnotes
Published ahead of print on 29 November 2010.
REFERENCES
- 1. Alvarez F. J., Douglas L. M., Konopka J. B. 2009. The Sur7 protein resides in punctate membrane subdomains and mediates spatial regulation of cell wall synthesis in Candida albicans. Commun. Integr. Biol. 2:76–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Alvarez F. J., Douglas L. M., Rosebrock A., Konopka J. B. 2008. The Sur7 protein regulates plasma membrane organization and prevents intracellular cell wall growth in Candida albicans. Mol. Biol. Cell 19:5214–5225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bernardo S. M., Lee S. A. 2010. Candida albicans SUR7 contributes to secretion, biofilm formation, and macrophage killing. BMC Microbiol. 10:133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Blankenship J. R., Fanning S., Hamaker J. J., Mitchell A. P. 2010. An extensive circuitry for cell wall regulation in Candida albicans. PLoS Pathog. 6:e1000752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Blankenship J. R., Mitchell A. P. 2006. How to build a biofilm: a fungal perspective. Curr. Opin. Microbiol. 9:588–594 [DOI] [PubMed] [Google Scholar]
- 6. Bruno V. M., et al. 2006. Control of the C. albicans cell wall damage response by transcriptional regulator Cas5. PLoS Pathog. 2:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Calderone R. A., Fonzi W. A. 2001. Virulence factors of Candida albicans. Trends Microbiol. 9:327–335 [DOI] [PubMed] [Google Scholar]
- 8. Casamayor A., Torrance P. D., Kobayashi T., Thorner J., Alessi D. R. 1999. Functional counterparts of mammalian protein kinases PDK1 and SGK in budding yeast. Curr. Biol. 9:186–197 [DOI] [PubMed] [Google Scholar]
- 9. de Groot P. W., et al. 2001. A genomic approach for the identification and classification of genes involved in cell wall formation and its regulation in Saccharomyces cerevisiae. Comp. Funct. Genomics 2:124–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Dickson R. C., Sumanasekera C., Lester R. L. 2006. Functions and metabolism of sphingolipids in Saccharomyces cerevisiae. Prog. Lipid Res. 45:447–465 [DOI] [PubMed] [Google Scholar]
- 11. Drgonová J., et al. 1996. Rho1p, a yeast protein at the interface between cell polarization and morphogenesis. Science 272:277–279 [DOI] [PubMed] [Google Scholar]
- 12. Edlind T. D., Katiyar S. K. 2004. The echinocandin “target” identified by cross-linking is a homolog of Pil1 and Lsp1, sphingolipid-dependent regulators of cell wall integrity signaling. Antimicrob. Agents Chemother. 48:4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Enloe B., Diamond A., Mitchell A. P. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730–5736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fontaine T., et al. 2000. Molecular organization of the alkali-insoluble fraction of Aspergillus fumigatus cell wall. J. Biol. Chem. 275:27594–27607 [DOI] [PubMed] [Google Scholar]
- 15. Fröhlich F., et al. 2009. A genome-wide screen for genes affecting eisosomes reveals Nce102 function in sphingolipid signaling. J. Cell Biol. 185:1227–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gavin A. C., et al. 2002. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415:141–147 [DOI] [PubMed] [Google Scholar]
- 17. Grossmann G., et al. 2008. Plasma membrane microdomains regulate turnover of transport proteins in yeast. J. Cell Biol. 183:1075–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Heitman J., Filler S. G., Edwards J. E. J., Mitchell A. P. 2006. Molecular principles of fungal pathogenesis. ASM Press, Washington, DC [Google Scholar]
- 19. Klis F. M., de Groot P., Hellingwerf K. 2001. Molecular organization of the cell wall of Candida albicans. Med. Mycol. 39(Suppl. 1):1–8 [PubMed] [Google Scholar]
- 20. Kumamoto C. A., Vinces M. D. 2005. Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell Microbiol. 7:1546–1554 [DOI] [PubMed] [Google Scholar]
- 21. Lambou K., Perkhofer S., Fontaine T., Latge J. P. 2010. Comparative functional analysis of the OCH1 mannosyltransferase families in Aspergillus fumigatus and Saccharomyces cerevisiae. Yeast 27:625–636 [DOI] [PubMed] [Google Scholar]
- 22. Lesage G., Bussey H. 2006. Cell wall assembly in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:317–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Luo G., Gruhler A., Liu Y., Jensen O. N., Dickson R. C. 2008. The sphingolipid long-chain base-Pkh1/2-Ypk1/2 signaling pathway regulates eisosome assembly and turnover. J. Biol. Chem. 283:10433–10444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Malinská K., Malinsky J., Opekarova M., Tanner W. 2003. Visualization of protein compartmentation within the plasma membrane of living yeast cells. Mol. Biol. Cell 14:4427–4436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Malinska K., Malinsky J., Opekarova M., Tanner W. 2004. Distribution of Can1p into stable domains reflects lateral protein segregation within the plasma membrane of living S. cerevisiae cells. J. Cell Sci. 117:6031–6041 [DOI] [PubMed] [Google Scholar]
- 26. Netea M. G., Brown G. D., Kullberg B. J., Gow N. A. 2008. An integrated model of the recognition of Candida albicans by the innate immune system. Nat. Rev. Microbiol. 6:67–78 [DOI] [PubMed] [Google Scholar]
- 27. Odds F. C. 1988. Candida and candidosis. Bailliere Tindall, Philadelphia, PA [Google Scholar]
- 28. Odds F. C., Brown A. J., Gow N. A. 2003. Antifungal agents: mechanisms of action. Trends Microbiol. 11:272–279 [DOI] [PubMed] [Google Scholar]
- 29. Pringle J. R. 1991. Staining of bud scars and other cell wall chitin with calcofluor. Methods Enzymol. 194:732–735 [DOI] [PubMed] [Google Scholar]
- 30. Qadota H., et al. 1996. Identification of yeast Rho1p GTPase as a regulatory subunit of 1,3-beta-glucan synthase. Science 272:279–281 [DOI] [PubMed] [Google Scholar]
- 31. Radding J. A., Heidler S. A., Turner W. W. 1998. Photoaffinity analog of the semisynthetic echinocandin LY303366: identification of echinocandin targets in Candida albicans. Antimicrob. Agents Chemother. 42:1187–1194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Reuss O., Vik A., Kolter R., Morschhauser J. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119–127 [DOI] [PubMed] [Google Scholar]
- 33. Ruiz-Herrera J., Elorza M. V., Valentin E., Sentandreu R. 2006. Molecular organization of the cell wall of Candida albicans and its relation to pathogenicity. FEMS Yeast Res. 6:14–29 [DOI] [PubMed] [Google Scholar]
- 34. Sherman F. 1991. Getting started with yeast. Methods Enzymol. 194:3–21 [DOI] [PubMed] [Google Scholar]
- 35. Sudbery P., Gow N., Berman J. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317–324 [DOI] [PubMed] [Google Scholar]
- 36. Sussman A., et al. 2004. Discovery of cercosporamide, a known antifungal natural product, as a selective Pkc1 kinase inhibitor through high-throughput screening. Eukaryot. Cell 3:932–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Utsugi T., et al. 2002. Movement of yeast 1,3-beta-glucan synthase is essential for uniform cell wall synthesis. Genes Cells 7:1–9 [DOI] [PubMed] [Google Scholar]
- 38. Walther T. C., et al. 2007. Pkh-kinases control eisosome assembly and organization. EMBO J. 26:4946–4955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Walther T. C., et al. 2006. Eisosomes mark static sites of endocytosis. Nature 439:998–1003 [DOI] [PubMed] [Google Scholar]
- 40. Whiteway M., Oberholzer U. 2004. Candida morphogenesis and host-pathogen interactions. Curr. Opin. Microbiol. 7:350–357 [DOI] [PubMed] [Google Scholar]
- 41. Wilson R. B., Davis D., Mitchell A. P. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu D., et al. 2007. Genome-wide fitness test and mechanism-of-action studies of inhibitory compounds in Candida albicans. PLoS Pathog. 3:e92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Xu T., et al. 2010. A profile of differentially abundant proteins at the yeast cell periphery during pseudohyphal growth. J. Biol. Chem. 285:15476–15488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Young M. E., et al. 2002. The Sur7p family defines novel cortical domains in Saccharomyces cerevisiae, affects sphingolipid metabolism, and is involved in sporulation. Mol. Cell. Biol. 22:927–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhang C., Konopka J. B. 2010. A photostable green fluorescent protein variant for analysis of protein localization in Candida albicans. Eukaryot. Cell 9:224–226 [DOI] [PMC free article] [PubMed] [Google Scholar]