Abstract
Saccharomyces cerevisiae cells possess a remarkable capacity to adhere to other yeast cells, which is called flocculation. Flocculation is defined as the phenomenon wherein yeast cells adhere in clumps and sediment rapidly from the medium in which they are suspended. These cell-cell interactions are mediated by a class of specific cell wall proteins, called flocculins, that stick out of the cell walls of flocculent cells. The N-terminal part of the three-domain protein is responsible for carbohydrate binding. We studied the N-terminal domain of the Flo1 protein (N-Flo1p), which is the most important flocculin responsible for flocculation of yeast cells. It was shown that this domain is both O and N glycosylated and is structurally composed mainly of β-sheets. The binding of N-Flo1p to d-mannose, α-methyl-d-mannoside, various dimannoses, and mannan confirmed that the N-terminal domain of Flo1p is indeed responsible for the sugar-binding activity of the protein. Moreover, fluorescence spectroscopy data suggest that N-Flo1p contains two mannose carbohydrate binding sites with different affinities. The carbohydrate dissociation constants show that the affinity of N-Flo1p for mono- and dimannoses is in the millimolar range for the binding site with low affinity and in the micromolar range for the binding site with high affinity. The high-affinity binding site has a higher affinity for low-molecular-weight (low-MW) mannose carbohydrates and no affinity for mannan. However, mannan as well as low-MW mannose carbohydrates can bind to the low-affinity binding site. These results extend the cellular flocculation model on the molecular level.
Saccharomyces cerevisiae flocculation has been defined as the asexual, reversible, and calcium-dependent aggregation of yeast cells to form flocs containing thousands of cells that rapidly sediment to the bottom of the liquid growth substrate (3, 38). Flocculation behavior is of great importance for yeast cells, as it is a way to escape from harsh conditions in the growth medium. It was suggested that cells in the middle of the floc can lyse and act as a source of new nutrients for the other cells (20, 37). Therefore, flocculation enhances the survival rate of yeast cells under starvation conditions. Furthermore, this type of cell adhesion is of considerable importance for the brewing industry, as it provides an environmentally friendly, simple, and cost-effective way to remove yeast cells from beer at the end of fermentation.
In an attempt to elucidate the phenomenon of flocculation, Eddy and Rudin (10) proposed the lectin hypothesis. In the presence of calcium, cells with expressed flocculins are able to bind highly branched mannose polymers located in the cell walls of adjacent cells, leading to cell-cell adhesion (10, 28). Flocculins are specific cell adhesion molecules located on the cell walls of yeast cells. The flocculin family in S. cerevisiae strain S288C includes the FLO1, FLO5, FLO9, FLO10, and FLO11 genes (4, 43). The FLO1, FLO5, FLO9, and FLO10 genes are responsible for flocculation. The expression of FLO11 leads to the adhesion of the cells to substrates such as plastics and agar (14), to diploid pseudohyphal formation, and to haploid invasive growth (26, 27). Some strains carrying the FLO11 gene, such as S. cerevisiae var. diastaticus strains, are flocculent (8).
To further characterize the lectin hypothesis, it was shown that flocculation can be inhibited reversibly by the presence of sugars (29, 30, 35, 39, 41). In particular, the flocculation phenotype governed by FLO1 can be inhibited by mannose but not by glucose, maltose, sucrose, or galactose. Therefore, Flo1p is considered the most specific adhesion protein of the Flo family (12, 25, 44).
The member proteins of the flocculin family have a modular configuration that consists of three domains (the N-terminal, central, and C-terminal domains) and an amino-terminal secretory sequence that is removed when the protein moves to the plasma membrane through the secretory pathway (18, 19, 45). The C-terminal domain contains a glycosylphosphatidylinositol (GPI) attachment site to covalently bind the Flo proteins to β-1,6-glucans of the yeast cell wall via a GPI remnant (4, 6, 7, 15, 16). The central domain contains many tandem repeats and is heavily glycosylated (3, 4, 40, 42). It was suggested that these oligosaccharide side chains enable the flocculins to form a long, semirigid rod-like structure (22). The proline residues, which are common in this region, may also prevent the central domain from forming a compact domain (9). The N-terminal part is the lectin domain, which leads to flocculation by interacting with mannose chains (3, 25).
The open reading frame of the FLO1 gene from S. cerevisiae encodes a protein of 1,537 amino acids, including the N-terminal sugar-binding domain of approximately 240 residues (N-Flo1p) (25, 42, 46). Several studies showed the lectin-like activity of N-Flo1p (3, 25). When N-terminally truncated versions of Flo1 proteins were expressed, the yeast cells lost their flocculation capacity (3). In addition, it was demonstrated indirectly that the N-terminal region of Flo1p contains the sugar recognition domain (25). Until now, no direct interaction between the Flo1p molecule and glycans has been demonstrated.
In this study, the N-terminal domain of Flo1p was expressed in S. cerevisiae, secreted into the medium, and then purified and studied. The secondary structure of the domain was evaluated, as well as the presence of glycans on the protein. The interaction of various mono- and dimannoses with N-Flo1p was characterized using fluorescence spectroscopy and determined quantitatively. Additionally, it was shown that N-Flo1p is able to interact with mannan chains from S. cerevisiae. These results show that the N-terminal domain of Flo1p is indeed responsible for the sugar-binding activity of the protein and confirm the cellular flocculation phenotype on a protein level.
MATERIALS AND METHODS
Construction of N-Flo1p expression plasmid.
Based on alignments of the FLO1 gene and a gene homologous to FLO1 (Lg-FLO1) (13, 25), the N-terminal domain of the Flo1 protein was determined. Two oligonucleotides [Flo1_8, 5′-(TCCTTAGTCAAAAGGACAGAGGCGTGCTTACCAGCA)-3′; and Flo1_9, 5′-(GGAGATCGGAATTCGTCAGTGATGGTGATGGTGATGTGAAGGGTCAGGGACAGTACAGTTAGA)-3′] were designed in order to amplify the N-terminal domain coding sequence from the genomic DNA of S. cerevisiae strain S288C. Primer Flo1_9 contains a sequence for 6 histidines to add a histidine tag to the C-terminal part of the protein to facilitate purification. The yeast-Escherichia coli shuttle vector pYEX-S1 was used for cloning of the N-terminal part of the FLO1 gene. Cloning was performed using the In-Fusion method (Clontech, Mountain View, CA), which is based on recombination between homologous sequences of the vector and the amplified gene (2, 47). The construction of the expression plasmid was confirmed by a combination of PCR and DNA sequencing. In the pYEX-S1 vector, the cloned gene is regulated by the strong constitutive PGK promoter, and this vector also includes the β-lactamase gene for selection in E. coli and the yeast-selectable markers leu2-d and URA3. The Flo1p secretion sequence (amino acids [aa] 1 to 17) was not included during cloning, as the pYEX-S1 vector contains the full-length leader sequence from Kluyveromyces lactis to direct proteins through the secretory pathway for secretion into the growth medium. Transformation into E. coli strain DH5α was carried out as described previously (17). The S. cerevisiae strain S288C BY4741 was transformed using the lithium acetate procedure (21).
Expression and purification of the lectin domain of Flo1p.
S. cerevisiae cells were cultivated in shake flasks. The cells were grown in yeast extract-peptone-dextrose (YPD) medium containing 1% (wt/vol) yeast extract, 2% (wt/vol) peptone, and 2% (wt/vol) d-glucose, according to the following procedure. Overnight cultures were grown by inoculating 90 ml YPD medium with one pYEX-S1-containing S. cerevisiae colony. Cultures were shaken and incubated at 30°C. Subsequently, the overnight cultures were diluted into 2 liters of fresh YPD medium and allowed to grow to an optical density at 600 nm (OD600) of 10 by incubation at 30°C for 64 h with shaking. The cells were separated from the medium by centrifugation (12,000 × g, 20 min, 4°C), and the resulting supernatant was adjusted to pH 7.2 to protonate the histidines of the histidine tag. The lectin domain of Flo1p was captured from the growth medium by affinity chromatography using a nickel-nitrilotriacetic acid (Ni-NTA) Sepharose column (6 ml) and was eluted with 1 M imidazole in phosphate-buffered saline (PBS). Fractions were pooled, concentrated to 2 ml, and subjected to gel filtration chromatography (Superdex 75 HR 10/30; GE Healthcare) in 20 mM Tris, pH 7.5, and 150 mM NaCl. The concentration of pure N-Flo1p was estimated from the absorption at 280 nm.
Detection of proteins.
After purification of the N-terminal domain of Flo1p, sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed to visualize the protein and to estimate its molecular mass. The gel (15% resolving gel) was stained with Coomassie stain (1 g/liter Coomassie brilliant blue R250, 50% [vol/vol] methanol, 10% [vol/vol] acetic acid) for 30 min and destained with 50% (vol/vol) methanol and 10% (vol/vol) acetic acid.
MALDI-MS PMF.
Peptide mass fingerprinting (PMF) was used to identify the purified proteins after SDS-PAGE. The protein bands of interest were excised from the SDS-PAGE gel, washed twice with 50% acetonitrile (ACN)–200 mM ammonium bicarbonate for 20 min at 30°C, and then dried, and 0.1 μg trypsin (Promega) or chymotrypsin (Sigma Aldrich) was added. The gel pieces were kept on ice for 45 min, and 50 mM ammonium bicarbonate was added until they were completely submerged. Digestion was performed by overnight incubation at 37°C. Peptides were extracted by adding 60% ACN–0.1% formic acid twice to the gel spots. The extraction buffer was evaporated in a Speedvac apparatus, and the peptides were redissolved in 8 μl 0.1% formic acid. The peptides then were spotted on a stainless steel matrix-assisted laser desorption ionization (MALDI) target plate and covered with alpha-cyano-4-hydroxycinnamic acid matrix (7 mg/ml in 50% ACN, 0.1% trifluoroacetic acid [TFA], 1 mM ammonium citrate) (1:1 ratio). Identification of the proteins was obtained by measuring the PMF on a MALDI-tandem time of flight mass spectrometry (MALDI-TOF/TOF MS) system (model 4800 proteomic analyzer; Applied Biosystems) in MS mode. MS/MS was performed for verification of the sequences of certain peptides. The obtained spectra were searched against the Swissprot database, using the Mascot platform. The spectra were searched using a 200-ppm peptide mass and a 0.5-Da MS/MS tolerance, with carbamidomethylation (Cys) and oxidation (Met) as variable modification parameters. Proteins were positively identified when at least 2 MS/MS fragmentation spectra were significantly identified using Mascot or when the peptide sequence was assessed manually.
Enzymatic deglycosylation.
Deglycosylation of the purified protein was performed in the presence of EDTA (20 mM Tris, pH 7.5, 150 mM NaCl, 10 mM EDTA) to prevent the released glycans from binding to N-Flo1p during enzymatic deglycosylation. To remove the N-glycans, the protein was treated with endo-β-N-acetylglucosaminidase H (endo H; New England Biolabs) at a concentration of 100 U/μg protein, and the mixture was incubated at 37°C for 4 h. To remove the O-glycans, α-mannosidase from Canavalia ensiformis (Sigma-Aldrich) was used at a concentration of 1 U/μg protein. The mixture was incubated overnight at 30°C to hydrolyze terminal nonreducing α-d-mannose residues. The glycans then were separated from the protein by dialysis to a buffer without EDTA (20 mM Tris, pH 7.5, and 150 mM NaCl).
CD.
Circular dichroism (CD) measurements were performed with a J715 spectropolarimeter (Jasco) in the far-UV (190 to 250 nm) region, using a 0.1-cm cell path length. The protein was diluted in 300 μl phosphate buffer (50 mM at pH 7.5) to obtain a final concentration of 0.2 mg/ml. Data were acquired with a scanning speed of 50 nm/min, a 2-s integration time, and a 1-nm bandwidth at a constant temperature of 25°C. Five spectra were measured and averaged. The mean residual ellipticity, [θ], was calculated as follows: [θ] = (100·θm)/(cL), where θ is the observed ellipticity, m is the mean residual weight, c is the concentration (mg/ml), and L is the path length (cm). A quantitative estimation of the secondary structure contents was made by applying the K2D2 method or by using the Selcon3, ContinLL, and CDsstr programs (1, 23, 31, 33, 36).
Fluorescence spectroscopy.
The binding of various carbohydrate components to N-Flo1p was studied by fluorescence spectroscopy. The N-terminal domain of the Flo1 protein contains 3 tryptophan residues, and one of them is situated in the VSWGT binding motif (25). The measurements were performed at 20°C, using a model LS55 luminescence spectrometer (PerkinElmer, MA). An excitation wavelength of 277 nm, a scan speed of 100 nm/min, and an excitation and emission slit width of 5 nm were used during the experiments. d-(+)-Mannose, α-methyl-d-mannoside, and different dimannoses [α(1-2-, α(1,3)-, and α(1,6)-dimannoses] were tested. Emission spectra were collected from 290 to 450 nm and corrected relative to a buffer blank. The initial sample volume was 500 μl, containing 2 μM N-Flo1p in flocculation buffer (50 mM sodium acetate, pH 4.5, 100 mM NaCl, 7 mM CaCl2). Glycosylated, N-deglycosylated, and both N- and O-deglycosylated N-Flo1p were used at the same concentration and under the same buffer conditions. Additions of carbohydrates were made from concentrated stock solutions containing 50 mM, 500 mM, 1 M, or 2 M sugar. The protein was titrated with appropriate aliquots of the sugar stock solution to final concentrations ranging from 10 μM to 100 mM. At each titration step, the sample was mixed and the fluorescence was measured immediately. The intensity of the tryptophan emission peak at 350 nm was decreased when the carbohydrate was bound to the protein. The quenching of the protein by addition of carbohydrates was followed by measuring the intensity of the emission peak at 350 nm for a series of protein samples with increasing carbohydrate concentrations. The normalized fluorescence signal at 350 nm was plotted versus the carbohydrate concentration and fitted using ligand binding models with Prism 5 (GraphPad, La Jolla, CA). A binding model for a protein with two ligand binding sites was compared to a one-site binding model. The two-ligand binding model is as follows: fluorescence signal = {[carbohydrate]/(KD, Ha + [carbohydrate])} + {[carbohydrate]/(KD, La + [carbohydrate])}, where KD, Ha and KD, La are the dissociation constants at equilibrium of the binding sites of the protein with high and low affinities, respectively, and where [carbohydrate] is the carbohydrate concentration in solution. The one-ligand binding model is as follows: fluorescence signal = [carbohydrate]/(KD + [carbohydrate]). The selection of the best-fitting model was based on the calculated P value. For the d-mannose and α-methyl-d-mannoside binding experiments, measurements were performed in triplicate. Averages and standard deviations were calculated and displayed on the resulting graphs.
For binding tests of various carbohydrates (fucose, galactose, glucose, maltose, rhamnose, sucrose, and d-mannose) to N-Flo1p, only one concentration (100 mM) was added to the protein solution. The decrease of the fluorescence signal was recorded and compared to the signal from the protein without the addition of any carbohydrate. The difference in fluorescence signal resulting from each carbohydrate was converted to a percentage and displayed on a histogram. The experiments were performed in triplicate, and averages and standard deviations were calculated. For galactose, rhamnose, and d-mannose, the measurement was done eight times to obtain better statistics.
The interaction of mannan with N-Flo1p was also evaluated using fluorescence spectroscopy. Therefore, mannan from S. cerevisiae (Sigma-Aldrich) was added to the N-Flo1p solution at different concentrations. The concentrations ranged from 1 mg/ml to 25 mg/ml, obtained by adding appropriate aliquots of a stock solution of 600 mg/ml. Mannan concentrations are expressed in mg/ml instead of as molar concentrations because mannan fragments have a variable length. A titration curve was created and was fitted to the one- and two-binding-site models. The dissociation constant at equilibrium (KD) was determined (expressed in mg/ml).
RESULTS
Expression and purification of N-Flo1p.
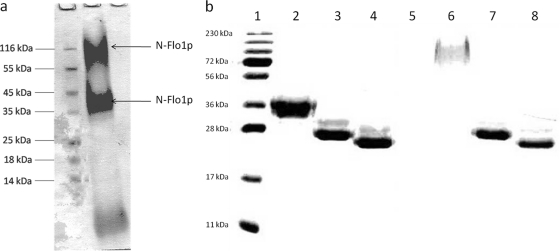
N-Flo1p was purified from the growth medium by use of a nickel column. The eluate was analyzed by SDS-PAGE, and staining of the proteins revealed three bands with different molecular masses (Fig. 1A). N-terminal sequencing and mass spectrometric analysis of the protein bands revealed that the upper two bands correspond to N-Flo1p. Using mass spectrometry, it was shown that Flo1p was present in the visualized bands of approximately 36 kDa and 100 kDa. Three peptides belonging to N-Flo1p were identified after trypsin digestion (KSGMNINFYQYSLK, SGMNINFYQYSLK, and DSSTYSNAAYMAYGYASK), and three peptides were identified after chymotrypsin digestion (GGSLPPNIEGTVY, GTLPISVTLPDGTTVSDDFEGYVY, and GTLPISVTLPDGTTVSDDFEGY).
Fig. 1.
(a) SDS-PAGE of eluted proteins after affinity chromatography using a Ni column. The Coomassie blue-stained gel shows that three protein bands with different molecular masses were purified. Mass spectroscopy revealed that the upper two bands correspond to N-Flo1p, as indicated with arrows. N-Flo1p appears in two different populations, distinguished by their molecular masses. Lane 1, molecular mass marker; lane 2, eluted fraction from Ni column. (b) SDS-PAGE of the two N-Flo1p populations, successively not deglycosylated, N-deglycosylated, and N- and O-deglycosylated. The gel was stained with Coomassie blue. Lane 1, molecular mass marker; lanes 2 and 6, N-Flo1p populations of lower and higher molecular masses, respectively, before enzymatic treatment; lanes 3 and 7, N-Flo1p populations of lower and higher molecular masses, respectively, after endo H treatment; lanes 4 and 8, N-Flo1p populations of lower and higher molecular masses, respectively, after treatment with both endo H and α-mannosidase.
The calculated molecular mass of the N-terminal domain of Flo1p without glycosylation is 26 kDa. On the SDS gel, both N-Flo1p bands correspond to proteins with higher molecular masses, being approximately 36 and 100 kDa (Fig. 1a). Hence, N-Flo1p expressed in S. cerevisiae appears in two different populations, distinguished by their molecular masses. This can be explained by the fact that secreted proteins in S. cerevisiae contain two types of N-linked glycans on their asparagines: core-type glycans and hypermannose-type glycans. The core-type glycans contain approximately 17 mannoses, while the hypermannose-type glycans consist of the core type with hundreds of extra mannoses attached. During protein expression, the core-type or hypermannose glycosylation pathway is selected at random, resulting in the two observed populations (5).
N-Flo1p is both O and N glycosylated.
As observed on the SDS gel, both populations of N-Flo1p had a higher molecular mass than expected (Fig. 1a). To test whether this significant difference in molecular mass was due to glycosylation, the proteins were deglycosylated enzymatically. Therefore, the eluate containing three protein fractions purified on the Ni column was first separated by gel filtration chromatography and then treated with endo H. After endo H treatment, both N-Flo1p populations had the same molecular mass, which was significantly lower than those before N-deglycosylation (Fig. 1b). The N-deglycosylated proteins were further treated with α-mannosidase to remove the O-glycans. The use of both enzymes reduced the molecular mass of N-Flo1p to the expected 26 kDa (Fig. 1b).
These experiments show that both populations of N-Flo1p are both O and N glycosylated and that the glycans can be removed by enzymatic treatment.
N-Flo1p is composed mainly of β-sheets.
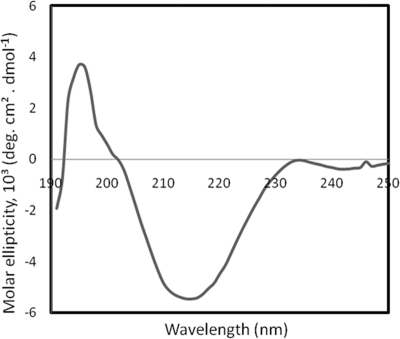
A CD spectrum of the 36-kDa N-Flo1p population was recorded in the far-UV spectrum to investigate its secondary structure (Fig. 2). The observed spectrum is typical for folded proteins, and its shape, exhibiting a maximum around 192 nm and one minimum around 218 nm, indicates a prevalence of β-sheets. The fitting of the CD spectrum, using the method of Perez-Iratxeta and Andrade-Navarro, indicates a β-sheet content of 35.71% and an α-helix content of 13.46% (31). In addition, secondary structure content deconvolution of the spectra, using the Selcon3, ContinLL, and CDsstr programs, suggested that the β-sheet content was 40.4%, 38.2%, and 36.6%, respectively. The α-helix content was also evaluated with the same programs. Values of 4.3%, 5.1%, and 3.2% were found using Selcon3, ContinLL, and CDsstr, respectively, for the α-helix content. In average, the secondary structure of N-Flo1p contains about 37.7% β-sheets and 6.5% α-helices.
Fig. 2.
Far-UV circular dichroism spectrum of N-Flo1p.
N-Flo1p binds specifically to d-mannose.
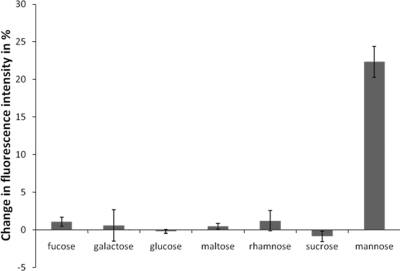
The binding of various carbohydrates to the 36-kDa population of N-Flo1p was studied by fluorescence spectroscopy. The intensity of the emission peak at 350 nm was decreased upon binding of the carbohydrate to the protein. The graph shows the extent of fluorescence signal quenching upon the interaction of various mono- and disaccharides (Fig. 3). It can clearly be observed that N-Flo1p binds specifically to d-mannose. No significant effect on the fluorescence signal was detected when we added other carbohydrates, i.e., fucose, galactose, glucose, maltose, rhamnose, or sucrose.
Fig. 3.
Quenching of N-Flo1p by different carbohydrates, each at a concentration of 100 mM, displayed as the decrease in the intensity of the fluorescence signal at 350 nm.
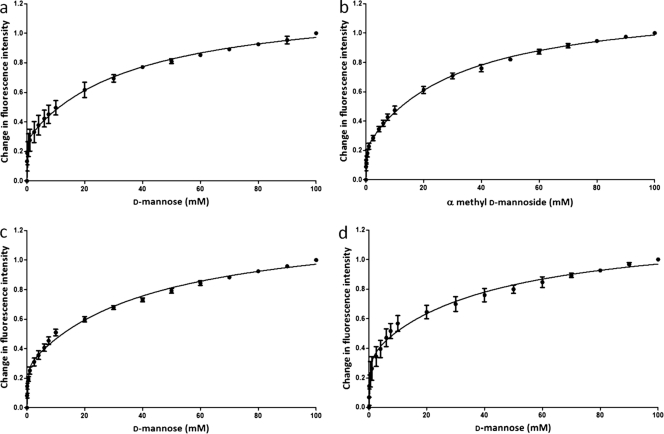
To determine the affinity between N-Flo1p and d-mannose, increasing sugar concentrations were added to the protein. The emission spectra have a maximum at 350 nm, and the intensity decreases with increasing d-mannose concentrations. Using the same technique, the binding of α-methyl-d-mannoside to N-Flo1p was evaluated. Moreover, the interactions of N-deglycosylated N-Flo1p with d-mannose, as well as those of N- and O-deglycosylated N-Flo1p, were tested. For each experiment, a titration curve was obtained by plotting the normalized fluorescence signal at 350 nm versus the sugar concentration (Fig. 4). We observed that the curves have a biphasic nature, which implies the presence of two binding sites. The curves were fitted with a model for a protein with one ligand binding site as well as with a model for a protein with two ligand binding sites, and both models were compared. It was concluded that the preferred model for fitting the data was the model for a protein with two ligand binding sites (P < 0.0001). Therefore, our data suggest that N-Flo1p contains two binding sites for mannose. The values for the dissociation constants at equilibrium for both binding sites are summarized in Table 1. These data indicate that the two binding sites of N-Flo1p have different affinities. The equilibrium dissociation constant is in the micromolar range for the binding site with high affinity, while the site with low affinity has a KD value in the millimolar range.
Fig. 4.
Titration curves of N-Flo1p with increasing concentrations of carbohydrate, ranging from 50 μM to 100 mM. The spheres show the change in fluorescence intensity at each concentration of carbohydrate, and the full lines show the fitting of the data according to the binding model. (a) Glycosylated N-Flo1p was titrated with increasing concentrations of d-mannose. KD, Ha was determined to be 57.77 ± 18.82 μM, and KD, La was determined to be 36.69 ± 6.27 mM. (b) Glycosylated N-Flo1p was titrated with increasing concentrations of α-methyl-d-mannoside. KD, Ha was determined to be 80.56 ± 23.79 μM, and KD, La was determined to be 31.86 ± 3.52 mM. (c) N-deglycosylated N-Flo1p was titrated with increasing concentrations of d-mannose. KD, Ha was determined to be 145.90 ± 37.29 μM, and KD, La was determined to be 43.29 ± 6.55 mM. (d) N- and O-deglycosylated N-Flo1p was titrated with increasing concentrations of d-mannose. KD, Ha was determined to be 283.60 ± 96.85 μM, and KD, La was determined to be 42.49 ± 12.00 mM.
Table 1.
High-affinity (KD, Ha) and low-affinity (KD, La) dissociation constants at equilibrium for the interaction between N-Flo1p and various mannose carbohydrates, based on the two-binding-site model
| N-Flo1p form | Carbohydrate | Mean KD (±SE) |
|
|---|---|---|---|
| KD, Ha (μM) | KD, La (mM) | ||
| Glycosylated | d-Mannose | 57.77 ± 18.82 | 36.69 ± 6.27 |
| Glycosylated | α-Methyl-d-mannoside | 80.56 ± 23.79 | 31.86 ± 3.52 |
| N-deglycosylated | d-Mannose | 145.90 ± 37.29 | 43.29 ± 6.55 |
| N- and O-deglycosylated | d-Mannose | 283.60 ± 96.85 | 42.49 ± 12.00 |
| Glycosylated | α(1,2)-dimannose | 205.70 | 8.47 |
| Glycosylated | α(1,3)-dimannose | 149.50 | 10.99 |
| Glycosylated | α(1,6)-dimannose | 148.20 | 17.55 |
N-Flo1p binds to α-dimannose.
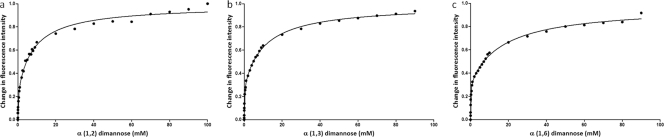
The interaction between N-Flo1p and dimannoses was also evaluated using fluorescence spectroscopy. Therefore, α(1,2)-, α(1,3)-, and α(1,6)-dimannoses were tested as possible ligands for N-Flo1p. A titration curve was generated and the equilibrium dissociation constant calculated for each dimannose (Fig. 5). The titration curves were best fitted using the model for a protein with two ligand binding sites compared to the model with only one binding site (P < 0.0001). The KD values for the binding sites of N-Flo1p with high and low affinities for interaction with dimannoses are summarized in Table 1.
Fig. 5.
Titration curves for glycosylated N-Flo1p with increasing concentrations of dimannoses. The spheres show the change in fluorescence intensity at each concentration of dimannose, ranging from 10 μM to 100 mM for α(1,2)-dimannose and from 10 μM to 90 mM for α(1,3)-dimannose and α(1,6)-dimannose. Determination of the KD values was performed by fitting (full line) the experimental data points with the binding model. (a) KD, Ha = 205.70 μM and KD, La = 8.47 mM for α(1,2)-dimannose. (b) KD, Ha = 149.50 μM and KD, La = 10.99 mM for α(1,3)-dimannose. (c) KD, Ha = 148.20 μM and KD, La = 17.55 mM for α(1,6)-dimannose.
N-Flo1p binds to mannan.
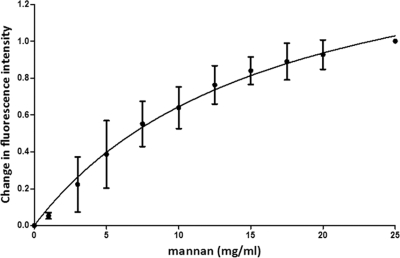
Fluorescence spectroscopy was used to assess the binding of mannan to N-Flo1p. Different concentrations of mannan were added to N-Flo1p, and a titration curve was generated (Fig. 6). In contrast to the titration curves for mannose and the dimannoses, the curve was best fitted using the binding model for a single-site ligand binding protein, and the KD was determined to be 16.53 ± 3.74 mg/ml. The KD value is expressed in mg/ml and cannot be compared to the KD values found for the other binding partners of N-Flo1p. This experiment clearly shows that N-Flo1p binds to mannan.
Fig. 6.
Titration curve for N-Flo1p with increasing concentrations of mannan. The circles show the change in fluorescence intensity at each concentration of mannan, ranging from 1 mg/ml to 25 mg/ml. The full line shows the fitting of the data according to the binding model. KD was determined to be 16.53 ± 3.74 mg/ml.
DISCUSSION
S. cerevisiae flocculation is a result of the interaction of lectin-like proteins on the surfaces of yeast cells with carbohydrates from the cell walls of other yeast cells (28). Flocculins are the cell wall proteins from S. cerevisiae responsible for that interaction (4). The FLO1 gene encodes a protein that leads to the Flo1-type flocculation phenotype, which is characterized by a strong inhibition capacity by mannose. Flo1p is believed to be highly mannose specific (12, 25, 44). However, no direct proof for binding of the Flo1 protein itself to mannose has been reported, since previous results were obtained from experiments with whole cells, not with the purified protein.
In this study, the sugar binding domain of Flo1p was cloned and purified in S. cerevisiae, and it was shown that N-Flo1p does indeed bind to carbohydrates. Using fluorescence spectroscopy, the binding of N-Flo1p to d-mannose was demonstrated. Other carbohydrates, such as fucose, galactose, glucose, maltose, rhamnose, and sucrose, were also tested, but no binding was observed. Recently, inhibition experiments were performed using an S. cerevisiae strain overexpressing Flo1p. The flocculation governed by the overexpressed Flo1p protein could be inhibited only by adding d-mannose to the medium, and it was not inhibited by the other tested sugars, such as maltose, sucrose, glucose, and galactose (44). Hence, our carbohydrate interaction analysis is in line with these results.
The binding of N-Flo1p was also determined by demonstrating its binding to mannose carbohydrates (mannan) present on the cell walls of intact yeast cells. Therefore, yeast cells were added to the purified protein solution. The proteins could be pulled down by the yeast cells, as the protein concentration was considerably (more than 40%) reduced, whereas mannose could inhibit binding to the cells, and no decrease in protein concentration could be observed (data not shown). Additionally, we attempted to demonstrate the binding of N-Flo1p to d-mannose by using other techniques, such as binding of the protein to d-mannose-coated agarose beads, the detection of N-Flo1p bound on yeast cells or on bovine serum albumin-mannose via anti-His antibodies (enzyme-linked immunosorbent assay [ELISA]), the binding of mannose-linked bovine serum albumin to N-Flo1p via surface plasmon resonance, and the binding of N-Flo1p to the 422 different mammalian glycans immobilized on a glycan array (Consortium for Functional Glycomics), where binding is also detected by anti-His antibodies. No single binding event was detected in any of these experiments (data not shown). Considering the fact that N-Flo1p is heavily glycosylated, we can explain the above results by steric hindrance, the poor accessibility of the His tag to anti-His antibodies, and the low binding affinity for mannose carbohydrates. In each experimental setup, mannose was immobilized on a matrix. It is possible that due to its extensive glycosylation, N-Flo1p was prevented from approaching the surface close enough to allow binding with the immobilized mannose. However, we succeeded in detecting an interaction between N-Flo1p and d-mannose carbohydrates by the fluorescence spectroscopy method because the carbohydrate could move freely in solution and find its best position toward the binding site.
The fluorescence titration data show a biphasic course which could best be fit by a two-binding-site model, indicating the presence of two distinct binding sites for mannose. The newly discovered binding site should contain at least one aromatic amino acid that interacts with mannose carbohydrates upon binding, which results in a decrease of fluorescence intensity. In previous reported research, only one binding site was considered. It was proposed that the carbohydrate binding activity of Flo1p is attributed to the VSWGT motif, a pentapeptide encompassing amino acids 226 to 230 (25). By use of this model, carbohydrate recognition by the Flo1 protein was described, and the amino acids that are responsible for this recognition have been identified. It was suggested that the tryptophan residue on position 228 is involved in mannose recognition in the Flo1 flocculation phenotype (25). This was confirmed in the present study by fluorescence spectroscopy, where tryptophan fluorescence was quenched upon binding of mannose.
Furthermore, the affinity of N-Flo1p for mannose carbohydrates was determined. The equilibrium dissociation constant is in the micromolar range for the binding site with high affinity, while the site with low affinity has a KD value in the millimolar range (Table 1). Similar values were found for the interaction of N-Flo1p with α-methyl-d-mannoside. Therefore, it was concluded that N-Flo1p is able to bind α-d-mannose, which is commonly found in nature when mannose chains are formed. For the interactions of N-deglycosylated N-Flo1p and N- and O-deglycosylated N-Flo1p with d-mannose, higher KD values were found (Table 1). This suggests that the glycans present on the protein increase the binding activity of the protein toward d-mannose.
The only quantitative measurement of the affinity between a Flo protein and a carbohydrate has been described for the interaction of a Flo1p homologue, named Lg-Flo1p, with mannose. Only one binding site was assumed, and the value for the dissociation constant was calculated to be 0.77 mM (13). In the past, detailed research about the inhibition of Flo1 phenotype flocculation by sugars has been described on the cellular level. Mannose was added in different concentrations to flocculating yeast cells, inhibiting flocculation at a certain concentration. Very divergent results were obtained. Miki and coworkers quantified the MIC, and they found that up to 500 mM d-mannose was required to completely inhibit flocculation (29). Later, different sugars were ranked according to their inhibition capacity. It was shown that α-d-mannose could disrupt the flocs at a concentration of only 25 mM (35). More recently, this experiment was repeated using a yeast strain overexpressing Flo1p. It was observed that the flocculation mediated by Flo1p was fully inhibited by mannose only at a concentration of 1 M (44).
In addition to the interaction between N-Flo1p and monomannose, the binding of dimannoses [α(1,2)-, α(1,3)-, and α(1,6)-dimannoses] was studied by fluorescence spectroscopy. It was found that dimannoses also bind to two different binding sites of N-Flo1p. The values for the dissociation constant at equilibrium are summarized in Table 1. The KD values for the binding site with high affinity are roughly three times higher than the values obtained for monomannose binding, which suggests that monomannose is a better ligand for the site with high affinity. In contrast, the KD values for the binding site with low affinity show that the affinities of the dimannoses are roughly three times higher than those for monomannose binding. This indicates that the binding site with low affinity has a preference for dimannoses over monomannose.
Moreover, we have shown on the protein level that N-Flo1p is able to interact with mannan (KD = 16.53 ± 3.74 mg/ml), which is part of the cell walls of yeast cells (24). This observation confirms the flocculation model, which assumes that flocculation occurs due to an interaction of cell wall proteins (flocculins) and the mannan layer of the cell walls of neighboring yeast cells (10, 28). Remarkably, only one binding site was detected for the interaction between N-Flo1p and mannan. Taking into account the results for dimannose binding, it was suggested that mannan binds only to the binding site of N-Flo1p with low affinity.
The model suggested by Kobayashi and coworkers (25) is based on only one binding site, comprising the VSWGT binding motif. This pentapeptide allows one mannose residue to enter the binding pocket of Flo1p. We have shown that the binding site with high affinity has a preference for monomannose binding. Therefore, we assume that the Kobayashi model deals only with the binding site with high affinity.
The high-affinity binding site probably has a more enclosed binding pocket where a monomannose just fits in and can interact with the surrounding VSWGT amino acids. The binding affinity of mannose carbohydrates will decrease with their increasing molecular weight due to the reduced probability of fitting into the binding pocket, whereas the low-affinity binding site will have a more open structure with less specificity, since every size of mannose carbohydrates (up to mannan molecules) can bind to this site. It is hypothesized that the initial binding between two interacting yeast cells occurs predominantly by the binding of yeast cell wall high-molecular-weight mannose carbohydrates to the low-affinity binding site. Next, low-molecular-weight mannose carbohydrates (e.g., terminal mannose residues on mannose glycans) bind to the high-affinity binding site. Since the Flo protein is then already fixed in space by binding to the low-affinity binding space, the terminal mannose residues can bind more efficiently to the more enclosed high-affinity binding site. Therefore, the presence of the low-affinity binding site is necessary to increase the efficiency of binding to the high-affinity site. The result is a relatively strong interaction on the single-protein level. Cell-cell binding is further enforced by numerous multiprotein interactions, resulting in a strong cell-cell binding process (9). Indeed, a high concentration of flocculins is found on the surface of a flocculating cell (3).
Until now, no atomic structure of any Flo protein has been determined. However, the N-terminal part of a homologue of Flo1p (Lg-Flo1p) has been crystallized and diffracted to high resolution by X-ray radiation (13), but the structure has not been solved yet. In this study, we have provided more information about the secondary structure of the N-terminal domain of Flo1p, based on CD measurements. We found that N-Flo1p consists mainly of β-sheets (37.7% β-sheets and 6.5% α-helices). This is in accordance with the recent literature about the PA14 domain. Recently, the N-terminal domain of Flo1p was assigned to a new domain family (PF07691) in Pfam (11; http://pfam.sanger.ac.uk), called the PA14 domain. The flocculin N-terminal domain might therefore be considered one of the many PA14 domain variants. The PA14 domain is a conserved domain that has been discovered in a wide variety of bacterial and eukaryotic proteins, which include many glycosidases, glycosyltransferases, proteases, amidases, bacterial toxins such as anthrax protective antigen (PA), and also yeast adhesins. Most of the experimentally characterized PA14-containing proteins are involved in carbohydrate binding and/or metabolism (34). The anthrax toxin PA also contains a PA14 domain. The crystal structure of this toxin indicates that the PA14 domain consists of a series of antiparallel β-strands (32).
In conclusion, the flocculation mechanism was studied at the molecular level by purifying the lectin domain of the Flo1 protein. It was confirmed that the N-terminal domain of Flo1p was the binding part of the flocculin. We demonstrated the binding of N-Flo1p to d-mannose, α-methyl-d-mannoside, and different dimannoses. It was shown that N-Flo1p contains two binding sites, with high and low affinities, in the micromolar and the millimolar range, respectively. These results confirm the cellular flocculation model and extend it to the protein level.
ACKNOWLEDGMENTS
We acknowledge J. E. Edwards (Medicine Department, Division of Infectious Diseases, Harbor-UCLA, CA) for providing us with the pYEX-S1 vector. We thank Lieven Buts and Adinda Wellens for valuable discussions and Khadija Wahni for technical support. We acknowledge the Consortium for Functional Glycomics for the glycan analysis.
This work was supported by the European Space Agency (Prodex program) and the Research Council of the VUB. K.G. acknowledges the Agency for Innovation by Science and Technology (IWT) for her Ph.D. grant.
Footnotes
Published ahead of print on 12 November 2010.
REFERENCES
- 1. Andrade M. A., Chacon P., Merelo J. J., Moran F. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383–390 [DOI] [PubMed] [Google Scholar]
- 2. Benoit R. M., Wilhelm R. N., Scherer-Becker D., Ostermeier C. 2006. An improved method for fast, robust, and seamless integration of DNA fragments into multiple plasmids. Protein Expr. Purif. 45:66–71 [DOI] [PubMed] [Google Scholar]
- 3. Bony M., Thines-Sempoux D., Barre P., Blondin B. 1997. Localization and cell surface anchoring of the Saccharomyces cerevisiae flocculation protein Flo1p. J. Bacteriol. 179:4929–4936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Caro L. H., et al. 1997. In silico identification of glycosyl-phosphatidylinositol-anchored plasma-membrane and cell wall proteins of Saccharomyces cerevisiae. Yeast 13:1477–1489 [DOI] [PubMed] [Google Scholar]
- 5. Dean N. 1999. Asparagine-linked glycosylation in the yeast Golgi. Biochim. Biophys. Acta 1426:309–322 [DOI] [PubMed] [Google Scholar]
- 6. De Groot P. W., Hellingwerf K. J., Klis F. M. 2003. Genome-wide identification of fungal GPI proteins. Yeast 20:781–796 [DOI] [PubMed] [Google Scholar]
- 7. De Nobel H., Lipke P. N. 1994. Is there a role for GPIs in yeast cell-wall assembly? Trends Cell Biol. 4:42–45 [DOI] [PubMed] [Google Scholar]
- 8. Douglas L. M., Li L., Yang Y., Dranginis A. M. 2007. Expression and characterization of the flocculin Flo11/Muc1, a Saccharomyces cerevisiae mannoprotein with homotypic properties of adhesion. Eukaryot. Cell 6:2214–2221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dranginis A. M., Rauceo J. M., Coronado J. E., Lipke P. N. 2007. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol. Mol. Biol. Rev. 71:282–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Eddy A. A., Rudin A. D. 1958. The structure of the yeast cell wall. I. Identification of charged groups at the surface. Proc. R. Soc. Lond. B Biol. Sci. 148:419–432 [DOI] [PubMed] [Google Scholar]
- 11. Finn R. D., et al. 2010. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Govender P., Domingo J. L., Bester M. C., Pretorius I. S., Bauer F. F. 2008. Controlled expression of the dominant flocculation genes FLO1, FLO5, and FLO11 in Saccharomyces cerevisiae. Appl. Environ. Microbiol. 74:6041–6052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Groes M., Teilum K., Olesen K., Poulsen F. M., Henriksen A. 2002. Purification, crystallization and preliminary X-ray diffraction analysis of the carbohydrate-binding domain of flocculin, a cell-adhesion molecule from Saccharomyces carlsbergensis. Acta Crystallogr. D Biol. Crystallogr. 58:2135–2137 [DOI] [PubMed] [Google Scholar]
- 14. Guo B., Styles C. A., Feng Q., Fink G. R. 2000. A Saccharomyces gene family involved in invasive growth, cell-cell adhesion, and mating. Proc. Natl. Acad. Sci. U. S. A. 97:12158–12163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hamada K., Fukuchi S., Arisawa M., Baba M., Kitada K. 1998. Screening for glycosylphosphatidylinositol (GPI)-dependent cell wall proteins in Saccharomyces cerevisiae. Mol. Gen. Genet. 258:53–59 [DOI] [PubMed] [Google Scholar]
- 16. Hamada K., Terashima H., Arisawa M., Kitada K. 1998. Amino acid sequence requirement for efficient incorporation of glycosylphosphatidylinositol-associated proteins into the cell wall of Saccharomyces cerevisiae. J. Biol. Chem. 273:26946–26953 [DOI] [PubMed] [Google Scholar]
- 17. Hanahan D., Jessee J., Bloom F. R. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63–113 [DOI] [PubMed] [Google Scholar]
- 18. Hoyer L. L. 2001. The ALS gene family of Candida albicans. Trends Microbiol. 9:176–180 [DOI] [PubMed] [Google Scholar]
- 19. Hoyer L. L., Payne T. L., Hecht J. E. 1998. Identification of Candida albicans ALS2 and ALS4 and localization of Als proteins to the fungal cell surface. J. Bacteriol. 180:5334–5343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Iserentant D. 1996. Practical aspects of yeast flocculation. Cerevisiae 21:30–33 [Google Scholar]
- 21. Ito H., Fukuda Y., Murata K., Kimura A. 1983. Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153:163–168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jentoft N. 1990. Why are proteins O-glycosylated? Trends Biochem. Sci. 15:291–294 [DOI] [PubMed] [Google Scholar]
- 23. Johnson W. C. 1999. Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins 35:307–312 [PubMed] [Google Scholar]
- 24. Kapteyn J. C., Van Den Ende H., Klis F. M. 1999. The contribution of cell wall proteins to the organization of the yeast cell wall. Biochim. Biophys. Acta 1426:373–383 [DOI] [PubMed] [Google Scholar]
- 25. Kobayashi O., Hayashi N., Kuroki R., Sone H. 1998. Region of FLO1 proteins responsible for sugar recognition. J. Bacteriol. 180:6503–6510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lambrechts M. G., Bauer F. F., Marmur J., Pretorius I. S. 1996. Muc1, a mucin-like protein that is regulated by Mss10, is critical for pseudohyphal differentiation in yeast. Proc. Natl. Acad. Sci. U. S. A. 93:8419–8424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lo W. S., Dranginis A. M. 1998. The cell surface flocculin Flo11 is required for pseudohyphae formation and invasion by Saccharomyces cerevisiae. Mol. Biol. Cell 9:161–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Miki B. L., Poon N. H., James A. P., Seligy V. L. 1982. Possible mechanism for flocculation interactions governed by gene FLO1 in Saccharomyces cerevisiae. J. Bacteriol. 150:878–889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Miki B. L. A., Poon N. H., James A. P., Seligy V. L. 1980. Flocculation in Saccharomyces cerevisiae: mechanism of cell-cell interaction, p. 165–170 In Stewart G. G., Russell I. (ed.), Current developments in yeast research. Pergamon Press, Tokyo, Japan [Google Scholar]
- 30. Nishihara H., Toraya T. 1987. Essential roles of cell surface protein and carbohydrate components in flocculation of a brewer's yeast. Agric. Biol. Chem. 51:2721–2726 [Google Scholar]
- 31. Perez-Iratxeta C., Andrade-Navarro M. A. 2008. K2D2: estimation of protein secondary structure from circular dichroism spectra. BMC Struct. Biol. 8:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Petosa C., Collier R. J., Klimpel K. R., Leppla S. H., Liddington R. C. 1997. Crystal structure of the anthrax toxin protective antigen. Nature 385:833–838 [DOI] [PubMed] [Google Scholar]
- 33. Provencher S. W., Glockner J. 1981. Estimation of globular protein secondary structure from circular dichroism. Biochemistry 20:33–37 [DOI] [PubMed] [Google Scholar]
- 34. Rigden D. J., Mello L. V., Galperin M. Y. 2004. The PA14 domain, a conserved all-beta domain in bacterial toxins, enzymes, adhesins and signaling molecules. Trends Biochem. Sci. 29:335–339 [DOI] [PubMed] [Google Scholar]
- 35. Smit G., Straver M. H., Lugtenberg B. J., Kijne J. W. 1992. Flocculence of Saccharomyces cerevisiae cells is induced by nutrient limitation, with cell surface hydrophobicity as a major determinant. Appl. Environ. Microbiol. 58:3709–3714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Sreerama N., Venyaminov S. Y., Woody R. W. 1999. Estimation of the number of alpha-helical and beta-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 8:370–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Stewart G. G., Russell I. 1981. Yeast flocculation, p. 61–91 In Pollock J. R. A. (ed.), Brewing science, vol. 2 Academic Press, New York, NY [Google Scholar]
- 38. Stratford M. 1989. Evidence for two mechanisms of flocculation in Saccharomyces cerevisiae. Yeast 5:441–445 [PubMed] [Google Scholar]
- 39. Stratford M., Assinder S. 1991. Yeast flocculation: Flo1 and NewFlo phenotypes and receptor structure. Yeast 7:559–574 [DOI] [PubMed] [Google Scholar]
- 40. Straver M. H., Smit G., Kijne J. W. 1994. Purification and partial characterization of a flocculin from brewer's yeast. Appl. Environ. Microbiol. 60:2754–2758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Taylor N. W., Orton W. L. 1978. Aromatic compounds and sugars in flocculation of Saccharomyces cerevisiae. J. Inst. Brewing 84:113–114 [Google Scholar]
- 42. Teunissen A. W., Holub E., van der Hucht J., van den Berg J. A., Steensma H. Y. 1993. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast 9:423–427 [DOI] [PubMed] [Google Scholar]
- 43. Teunissen A. W., Steensma H. Y. 1995. Review: the dominant flocculation genes of Saccharomyces cerevisiae constitute a new subtelomeric gene family. Yeast 11:1001–1013 [DOI] [PubMed] [Google Scholar]
- 44. Van Mulders S. E., et al. 2009. Phenotypic diversity of Flo protein family-mediated adhesion in Saccharomyces cerevisiae. FEMS Yeast Res. 9:178–190 [DOI] [PubMed] [Google Scholar]
- 45. Verstrepen K. J., Reynolds T. B., Fink G. R. 2004. Origins of variation in the fungal cell surface. Nat. Rev. Microbiol. 2:533–540 [DOI] [PubMed] [Google Scholar]
- 46. Watari J., et al. 1994. Molecular cloning and analysis of the yeast flocculation gene FLO1. Yeast 10:211–225 [DOI] [PubMed] [Google Scholar]
- 47. Zhu B., Cai G., Hall E. O., Freeman G. J. 2007. In-fusion assembly: seamless engineering of multidomain fusion proteins, modular vectors, and mutations. Biotechniques 43:354–359 [DOI] [PubMed] [Google Scholar]