Abstract
Iron is an essential element for the survival of living cells. However, excess iron is toxic, and its uptake is exquisitely regulated by the ferric uptake regulator, Fur. In Salmonella, the Salmonella pathogenicity island 1 (SPI-1) encodes a type three secretion system, which is required for invasion of host epithelial cells in the small intestine. A major activator of SPI-1 is HilA, which is encoded within SPI-1. One known regulator of hilA is Fur. The mechanism of hilA regulation by Fur is unknown. We report here that Fur is required for virulence in Salmonella enterica serovar Typhimurium and that Fur is required for the activation of hilA, as well as of other HilA-dependent genes, invF and sipC. The Fur-dependent regulation of hilA was independent of PhoP, a known repressor of hilA. Instead, the expression of the gene coding for the histone-like protein, hns, was significantly derepressed in the fur mutant. Indeed, the activation of hilA by Fur was dependent on 28 nucleotides located upstream of hns. Moreover, we used chromatin immunoprecipitation to show that Fur bound, in vivo, to the upstream region of hns in a metal-dependent fashion. Finally, deletion of fur in an hns mutant resulted in Fur-independent activation of hilA. In conclusion, Fur activates hilA by repressing the expression of hns.
Salmonella enterica serovar Typhimurium is a Gram-negative facultative anaerobe capable of causing food-borne gastroenteritis. Two serovars, S. Typhimurium and S. Enteritidis, are reported as the primary human isolates of nontyphoid Salmonella infections (8). S. Typhimurium, like many pathogens, contains genetic material acquired through horizontal transfer, usually phage mediated, which enhance virulence within the host (reviewed in reference 6). These horizontal gene transfer events culminated in the acquisition of Salmonella pathogenicity islands (SPIs). SPI-1 is a DNA segment of ∼40 kb and encodes regulators of SPI-1, a type three secretion system (T3SS), chaperone proteins, and effector proteins that Salmonella secretes into host cells and are required for initiating host cytoskeletal rearrangement and the eventual uptake of the pathogen (19, 32, 61).
Regulation of SPI-1 is complex. Several environmental signals regulate the expression of SPI-1 (i.e., low O2, high osmolarity, and acidic pH) (3, 36). Presumably, these environmental signals are similar to the conditions in the ileum of the small intestine, the preferred site for invasion by S. Typhimurium (32, 51). Oxygen tension is a critical signal that promotes invasion of epithelial cells (36), and in a previous study we demonstrated that a mutation in the oxygen-sensitive transcriptional regulator fumarate nitrate reduction (Fnr) is fully attenuated in mice and is unable to survive in macrophages (24). In addition, microarray data revealed reduced expression of several genes located within SPI-1 in the fnr mutant (24). Whether the two observations (i.e., increased invasion in low O2 and avirulence of the fnr mutant) are connected requires further work.
Three AraC/XylS-type regulators, HilC, HilD, and RtsA (also known as STM4315) are important for the activation of SPI-1 in S. Typhimurium. These three regulators are capable of activating each other independently, though HilD appears to be critical following per oral inoculation (20). In addition, these three regulators (i.e., HilC, HilD, and RtsA) activate a regulator within SPI-1, HilA, which shows homology with the OmpR/ToxR family of DNA-binding proteins and induces transcriptional activation of SPI-1 genes in response to a variety of stimuli (1, 59). Specifically, HilA activates prgH and invF, driving expression of the prg/org and inv/spa genes (34, 37). Like HilC and HilD proteins, InvF belongs to the AraC/XylS family of transcriptional regulators, and its gene lies within SPI-1 (28, 33). When bound with SicA, InvF is capable of activating expression of downstream sic/sip genes, the effector gene, sigD, and the chaperone gene, sigE, which lies outside of SPI-1 (16-18). In addition to indirectly activating sic/sip expression through InvF, HilA is capable of directly activating sic/sip expression via readthrough transcription of invF (16, 19). This indicates that regulation of HilA is important for determining the activation of SPI-1.
HilA expression was shown to be repressed by addition of an intracellular iron chelator, deletion of the ferric uptake regulator (Fur), or by phosphorylated PhoP (3, 22). Deletion of phoP also renders the cells susceptible to iron-mediated oxidative stress and profound attenuation in vivo (10, 41). Because Fur controls iron uptake in many bacteria and there is fierce competition between the pathogen and the host for free iron, an understanding of how iron homeostasis regulates hilA is crucial to our knowledge of the role of Fur in the activation of SPI-1 and virulence in Salmonella.
In a recent transcriptome analysis of S. Typhimurium growing in anaerobiosis, we found that the expression of hns is significantly upregulated in a fur mutant (64). A putative Fur binding site was identified upstream of hns, but no evidence was presented for Fur binding to this region (64). In the present study, we tested the hypothesis that derepression of hns in the fur mutant is responsible for the reduced expression of hilA previously noted under conditions of iron chelation or Fur deletion (22, 62). We also tested whether Fur was required for systemic infection in mice. Our data indicated that Fur was required for full virulence in mice regardless of the presence or absence of the natural resistance-associated macrophage protein 1, Nramp1. In addition, we determined that under anaerobic conditions the expression of hilA was dependent on Fur, and we demonstrated that PhoP was not involved in this regulation. We also identified 28 nucleotides upstream of hns that contained the Fur binding motif responsible for the Fur-dependent activation of hilA. Moreover, using a modified chromatin immunoprecipitation (ChIP) assay, we determined that binding of Fur upstream of hns is dependent on metal. Finally, we demonstrated that in the absence of H-NS the expression of hilA was independent of Fur.
MATERIALS AND METHODS
Strains, plasmids, and growth conditions.
The strains and plasmids used in the present study are listed in Table 1. P22-mediated transduction transferred mutations into the appropriate genetic backgrounds and transductants were purified on Evans blue-uranine agar plates. S. enterica serovar Typhimurium 14028s and its isogenic strains were used throughout the present study. Transductants containing the hilA-lacZ transcriptional fusion were selected for tetracycline resistance and a lac+ on 0.4% lactose MacConkey agar plates. hilA-lacZ was also transduced into strains AV0474 (phoP::FRT), WN153, which contained an in-frame deletion of amino acids 61 to 65 of RpoS, conferring reduced function of RpoS (rpoSlow) (46), and WN341 (rpoSlow hns::kan) (46). Constructed strains were confirmed by PCR, lac+ phenotype, and antibiotic resistance of linked markers. Strains with mutation in Fur were checked for siderophore secretion on Tris-buffered 0.3% xylose chrome azurol (CAS) plates (54).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| S. Typhimurium strains | ||
| 14028s | Wild type | ATCCa |
| RM5938 | hilA-lacZY | 3 |
| KLM001 | fur::bla | 65 |
| NC1091 | fur::bla hilA-lacZY | This study |
| NC1137 | hilA-lacZY pACYC184 | This study |
| NC1153 | hilA-lacZY pfur-ha | This study |
| NC1141 | fur::bla hilA-lacZY pACYC184 | This study |
| NC1154 | fur::bla hilA-lacZY pfur-ha | This study |
| NC1038 | fur::bla pfur-ha | This study |
| AV0474 | phoP::FRT | 5 |
| NC1092 | phoP::FRT hilA-lacZY | This study |
| NC1119 | fur::bla phoP:FRT hilA-lacZY | This study |
| NC1148 | hilA-lacZY phns−385 | This study |
| NC1156 | hilA-lacZY phns−357 | This study |
| NC1149 | hilA-lacZY phns−190 | This study |
| NC1147 | fur::bla hilA-lacZY phns−385 | This study |
| NC1150 | fur::bla hilA-lacZY phns−357 | This study |
| NC1140 | fur::bla hilA-lacZY phns−190 | This study |
| WN153 | rpoSlow | 46 |
| WN341 | rpoSlowhns::kan | 46 |
| NC1133 | rpoSlowhilA-lacZY | This study |
| NC1132 | rpoSlowhns::kan hilA-lacZY | This study |
| NC1131 | rpoSlowfur::bla hilA-lacZY | This study |
| NC1130 | rpoSlowhns::kan fur::bla hilA-lacZY | This study |
| Plasmids | Source | |
| pACYC184 | cat ori p15A tet | 12 |
| pfur-ha | fur with in-frame expression of hemagglutinin epitope in AvaI HindIII sites of pACYC184 (cat) | This study |
| phns−385 | −385 bp from ATG of hns to 20 bp after TAA cloned into AvaI HindIII sites of pACYC184 (cat) | This study |
| phns−357 | −357 bp from ATG of hns to 20 bp after TAA cloned into AvaI HindIII sites of pACYC184 (cat) | This study |
| phns−190 | −190 bp from ATG of hns to 20 bp after TAA cloned into AvaI HindIII sites of pACYC184 (cat) | This study |
ATCC, American Type Culture Collection.
Truncations of the hns promoter were constructed using the primers listed in Table 2. Forward primers contained a 5′ AvaI site (underlined), and the reverse primer contained a 5′ HindIII site (underlined). Genomic DNA of 14028s was used as a template for PCR amplification of fragments that contained the hns promoter and the open reading frame. PCR products were extracted from agarose gels and digested with AvaI and HindIII. Purified inserts were ligated to AvaI/HindIII-digested pACYC184 and cloned into 14028s with selection on LB agar plates containing chloramphenicol. Plasmids were confirmed by restriction enzyme digest and tetracycline-sensitive (Tets) phenotype. Plasmid DNA and genomic DNA was purified with miniprep and DNA extraction kits (Qiagen, Valencia, CA), respectively. The pfur-ha plasmid contained the fur open reading frame expressed in-frame with the hemagglutinin (HA) epitope. This plasmid restored Fur+ wild-type (WT) siderophore production to the fur mutant; as phenotypically confirmed on CAS plates (54) (see Fig. S1 in the supplemental material).
TABLE 2.
Primers
| Primer | Sequence (5′-3′) | Relevance |
|---|---|---|
| AvaIhns−385 fwd | AATAAATCCCGAGTTCATCAACAATGCTTATCA | phns−385 |
| AvaIhns−357 fwd | AATAAATCCCGAGTTACAATATGAAAACCTTGTAC | phns−357 |
| AvaIhns−190 fwd | AATAAATCCCGAGGAGTATCCCCCCTGCCAA | phns−190 |
| HindIIIhns rev | ATATAAAGCTTTTAAGCATCCAGGAAGTAAA | All phns constructs |
| hns fwd | TTGTTACCGTTACTCATACCC | ChIP |
| hns rev | TACGCTTTATTAAGCACACG | ChIP |
| hns fwd | TAAAATTCTGAACAACATCC | qRT-PCR |
| hns rev | GATACTGTTGCAGTTTACG | qRT-PCR |
| rpoD fwd | TGCTGCTGGCTGAAAATACC | ChIP |
| rpoD rev | TTCAGGGTCAATGCTGTTGT | ChIP |
| rpoD fwd | CGATGTCTCTGAAGAAGTGC | qRT-PCR |
| rpoD rev | TTCAACCATCTCTTTCTTCG | qRT-PCR |
| dinP fwd | GCTTGTTCCTTGATAAAACG | ChIP |
| dinP rev | GACATATTCGAAGGTATGACG | ChIP |
Anaerobic cultures were maintained in a Coy Anaerobic chamber filled with anaerobic gas mixture (10% H2, 5% CO2, 85% N2). Growth was monitored by measuring changes in optical density at 600 nm (OD600) over time. The medium used throughout was buffered LB medium containing 100 mM morpholinepropanesulfonic acid and 20 mM xylose at pH 7.4 (LB-MOPS-X) and equilibrated in the anaerobic chamber for at least 48 h prior to experiments (24). Tetracycline, chloramphenicol, ampicillin, and kanamycin were used at 20, 20, 120, and 55 μg/ml, respectively.
Pathogenicity assays.
Immunocompetent 6- to 8-week-old C3H/HeN (Nramp+/+) or C57BL/6 (Nramp−/−) mice purchased from Charles River were tested at the animal facility of the University of Colorado Denver according to Institutional Animal Care and Use Committee guidelines. Stationary-phase serovar Typhimurium (WT and fur mutant) cultures grown aerobically in LB-MOPS-X broth were used. The bacteria were diluted in phosphate-buffered saline (PBS). Intraperitoneal (i.p.) challenge consisted of groups of five C3H/HeN mice inoculated with approximately 3,000 CFU in 500 μl of PBS/mouse or groups of five C57BL/6 mice inoculated with 250 CFU in 500 μl of PBS/mouse, and mortalities were scored over 21- and 30-day periods, respectively.
β-Galactosidase assays.
Reporter assays of the transcriptional hilA-lacZ fusion were determined as described previously (40). Initial experiments were conducted by a 200-fold dilution of cultures into medium under anaerobic conditions containing the appropriate antibiotics. After ∼14 h of anaerobic growth, reporter activity was determined from at least four independent cultures. Empty vector controls were included when plasmid constructs were used in experiments. In order to determine the growth phase-dependent regulation (i.e., kinetics) and the rate of expression of gene fusions, we used differential plots as previously described (14, 15, 31, 44). This was accomplished by monitoring the growth of the culture (from an initial OD600 of 0.02 to an OD600 of ∼1) and the reporter activity (U ml−1) in the growing cultures. The data were plotted in the form of differential plots, where reporter activity (U ml−1) was plotted versus growth (OD600) of the culture, and the slopes of the linear portion of these plots were used to determine the rate of expression of the reporter gene. This is beneficial for the identification of the precise OD600 at which point the induction of the hilA-lacZ reporter gene occurred. This also allowed for determining enzyme induction of the reporter as defined by the differential rate of synthesis in proportion of the increase in total protein measured as OD600 (42, 47). A paired Student t test was used to determine significance and graphs were compiled by using GraphPad Prism 4.0.
qRT-PCR and ChIP assays.
For quantitative reverse transcription-PCR (qRT-PCR) experiments, three independent cultures were grown to an OD600 of ca. 0.35 to 0.4 and treated with RNAlater. Total RNA was extracted from each culture with TRIzol reagent (Invitrogen, Carlsbad, CA) and treated with RNase free DNase (Promega, Madison, WI) according to the manufacturer's specifications. DNase-treated RNA was purified by using an RNeasy miniprep kit (Qiagen), the RNA quality was determined by agarose gel electrophoresis, and the RNA was quantified by determining the absorbance at 260 nm. qRT-PCR analyses were performed in triplicate using a Quantitect SYBR green RT-PCR kit (Qiagen) that contained 50 ng of total RNA in an iCycler (Bio-Rad, Hercules, CA). Reaction conditions were 49°C for 50 min for cDNA synthesis, followed by 95°C for 15 min with 45 cycles of 95°C for 15 s, 50°C for 30 s, and 72°C for 30 s. Melting-curve analysis was performed beginning at 72°C and ending at 95°C and confirmed a single PCR product. Primers were used at 0.5 μM. The data were analyzed by using Bio-Rad Optical System Software, version 3.1, according to the manufacturer's specifications. The absence of genomic DNA was confirmed by PCR using primers targeting within the coding gene of hns. A standard curve of rpoD was used to determine transcript copy number. The primers are listed in Table 2.
Antibodies for ChIP included rabbit polyclonal antibody (pAb) to HA tag (ab9110-100; Abcam, Cambridge, MA) and an IgG isotype-matched control (ab6721-1; Abcam). For ChIP analyses, nucleoprotein complexes from anaerobic samples in LB-MOPS-X at an OD600 of ∼0.25 in the presence or absence of 2,2′ dipyridyl (dip; 200 μM) were reversibly cross-linked in 1% formaldehyde for 10 min at room temperature. Cross-linked reactions were neutralized in glycine (125 mM) for 5 min at room temperature. The cells were pelleted and washed in 1× PBS and sonicated extensively on ice to produce DNA fragments 300 to 500 bp in size.
ChIP was performed as previously described (55). Prior to ChIP, each antibody (1 μg) was incubated (1 h at 4°C) with protein A- and protein G-coupled Dynabeads (Invitrogen) at 5 μl/IP in 50 μl of radioimmunoprecipitation assay (RIPA) buffer (10 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS], 0.1% sodium deoxycholate) supplemented with 0.5 mg of sheared salmon sperm DNA/ml, 1× protease inhibitor cocktail (Sigma-Aldrich, St. Louis, MO), and 1 mM phenylmethylsulfonyl fluoride. Conjugated antibodies were washed and combined with equivalent chromatin amounts in 100 μl of supplemented RIPA buffer. Protein-DNA complexes were immunoprecipitated 2 h at 4°C and then washed three times in RIPA buffer and twice in Tris-EDTA (pH 8.0). Protein-DNA complexes were eluted for 15 min (100 mM NaHCO3, 1% SDS). and cross-links were reversed for 15 min at 95°C in the presence of 200 mM NaCl. Protein was digested with 10 μg of proteinase K/ml 1 h at 37°C, and DNA was purified using QIAquick nucleotide removal columns (Qiagen) according to the manufacturer's instructions.
For real-time PCR, 3-μl IP and input control samples were amplified by using 1× SensiMix Plus (Quantace, London, United Kingdom) and primers specific for rpoD, dinP, and hns (Table 2). The cycling parameters for 20-μl reactions were as follows: 95°C for 10 min, followed by 40 to 50 cycles of 95°C for 20 s, 53°C for 30 s, and 72°C for 30 s. Fold enrichment in the bound fractions relative to input was calculated as previously described (13). Binding signals using anti-HA pAbs were normalized to ChIP signals using control IgG and are expressed as the fold enrichment over those obtained from cells cultured in the presence of 200 μM dip.
RESULTS
Fur is required for systemic infection of mice.
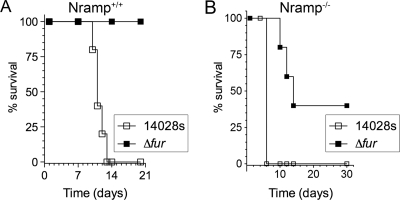
The importance of Fur in virulence was determined in a systemic model of infection. Two strains of mice: Nramp+/+ (C3H/HeN) and Nramp−/− (C57BL/6) were inoculated via i.p. with 14028s or the fur mutant. This approach allowed us to determine the contribution of a functional Nramp1 (for natural resistance-associated macrophage protein 1) to S. Typhimurium with or without the Fur protein. Mutations in Nramp1 increase host susceptibility to intracellular pathogens (57). Nramp1 is a transporter of Mn2+, Fe2+, and Co2+ that also enhances acidification of the phagosome (25, 29). Figure 1A demonstrates that Fur is required for virulence in Nramp+/+ (C3H/HeN) mice. The mice succumbed to infection with 14028s in less than 2 weeks, but mice infected with the fur mutant survived the entire study. On the other hand, the fur mutant was partially attenuated in Nramp−/− (C57BL/6) mice (Fig. 1B). Thus, all mice infected with 14028s succumbed in <1 week, but 40% of mice infected with the fur mutant survived the 30-day experiment. Our findings indicated that Fur-dependent regulation was important to the virulence of S. Typhimurium and are in agreement with a previous report in the C3H/HeN mice (65).
FIG. 1.
Fur is required for virulence in mice. (A) C3H/HeN (Nramp+/+) mice were inoculated i.p. with 3,000 CFU of WT 14028s or fur mutant. Mice were observed for 21 days after infection. (B) C57BL/6 (Nramp−/−) mice were inoculated i.p. with 250 CFU of WT 14028s or fur mutant. Mice were observed for 30 days after infection.
Fur is required for expression of hilA.
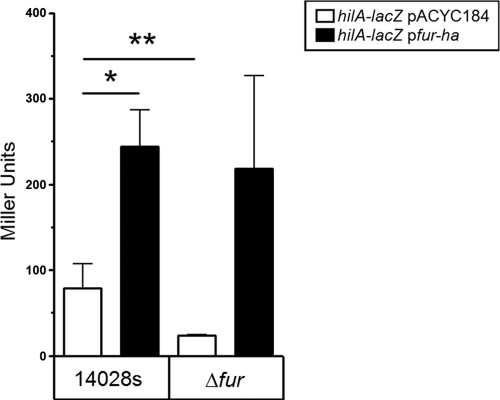
Previous studies have suggested that activation of hilA is Fur dependent (22, 62). Inspection of our Fur microarray data revealed that the expression of genes within SPI-1 (prgK, prgH, sipADCB, sicA, and invJICBA) was significantly (1.4- to 2-fold) reduced in the fur mutant (64). However, in that study a 2.5-fold change was arbitrarily set to identify genes regulated by Fur. Thus, under this 2.5-fold constraint, the microarray data did not identify hilA as differentially regulated by Fur. To confirm the regulatory role of Fur in the regulation of hilA in anaerobically cultured cells, we assessed the impact of a deletion in fur on the expression of a hilA-lacZ reporter. The data in Fig. 2, show that the expression of hilA-lacZ in the fur mutant was >3-fold lower than that in the parent strain. Moreover, introduction of a low-copy-number plasmid that contained a functional fur gene increased the activity of hilA-lacZ by ∼3-fold in the parent strain and by ∼10-fold in the fur mutant (Fig. 2). This confirmed previous findings and demonstrated that the copy number of fur has an impact on the expression of hilA.
FIG. 2.
Fur is required for the expression of hilA-lacZ. WT and the fur mutant strains carrying a single-chromosomal copy of hilA-lacZ were used. Expression of hilA-lacZ was determined in cells harboring empty vector (pACYC184) and compared to cells harboring pfur-ha. Cultures were grown anaerobically for ∼14 h before β-galactosidase was determined. A paired Student t test was used to determine significance (*, P ≤ 0.05; **, P ≤ 0.01; n = 5). Mean values ± the standard deviations (SD) are shown.
Effect of Fur on hilA is independent of PhoP.
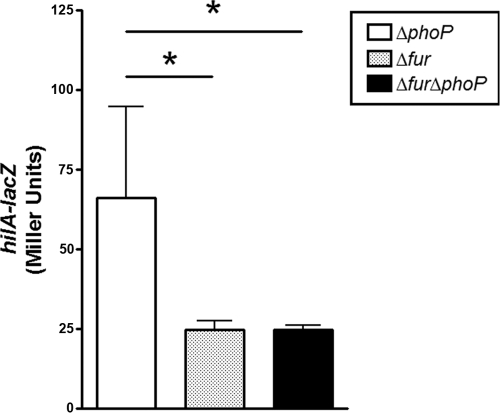
Several pathways regulate hilA expression (reviewed in references 2And 21) and a major regulator of SPI-1 is PhoP. PhoP is a possible candidate for the observed Fur-dependent regulation of hilA because a deletion of phoP renders the cell sensitive to iron-mediated oxidative stress (10), suggesting a potential role for Fur in mediating this sensitivity. Therefore, we examined how PhoP affects Fur regulation of hilA-lacZ. The data in Fig. 3 show that deletion of phoP in the fur mutant background did not restore expression of hilA-lacZ. Therefore, PhoP-dependent regulation was not involved in the observed activation of hilA-lacZ by Fur. This is not surprising because activation of PhoP by its cognate histidine kinase, PhoQ, is dependent on a low [Mg2+], and it is likely that Mg2+ is plentiful in the LB media used here (9, 58).
FIG. 3.
Fur regulates hilA-lacZ independent of the response regulator PhoP. β-Galactosidase activity was determined in the ΔphoP, Δfur, or ΔphoP Δfur mutant strains with single-copy hilA-lacZ. Activity was determined as in Fig. 2. A paired Student t test was used to determine significance (*, P ≤ 0.05; **, P ≤ 0.01; n = 4). Means ± the SD are shown.
Previous reports indicated that induction of hilA is dependent on intracellular divalent cations, especially ferrous iron (22). Indeed, we found that growth in the presence of the ferrous iron chelator 2,2′-dipyridyl resulted in reduced expression of hilA-lacZ (data not shown). To rule out the possibility that 2,2′-dipyridyl may scavenge Mg2+ from the cell, we assessed the effect of the chelator on the expression of a PhoP activated gene (pagP) using a pagP-lacZ fusion, where PhoP binds pagP and is essential for induction of this gene (4, 52). Unlike with the hilA fusion, the expression of pagP-lacZ was not differentially affected by the addition of 200 μM 2, 2′dipyridyl to the growth media (see Fig. S2 in the supplemental material). This clearly demonstrated that PhoP is not involved in the iron- and Fur-dependent activation of hilA.
Fur regulation of hns.
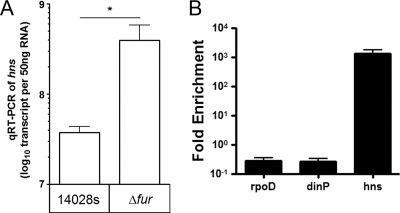
The microarray data (64) identified hns, a gene encoding the histone-like protein H-NS, as part of the Fur regulon in anaerobically grown S. Typhimurium. The expression of hns was significantly upregulated in the fur mutant, and a putative Fur binding site was identified upstream of the gene (64). To confirm the role of Fur in regulating hns, we used qRT-PCR to quantify the expression of hns in the presence or absence of Fur. The data showed that the expression of hns was >14-fold higher in the fur mutant than in the parent strain (Fig. 4A). We next assessed the binding of Fur to site(s) upstream of hns using a modified ChIP assay. Cross-linked nucleoprotein complexes were isolated from independent, anaerobic cultures of the fur mutant complemented with a plasmid expressing an HA-tagged Fur protein. This plasmid expressed an ∼18-kDa protein that cross-reacted with the HA antibodies and is capable of restoring the WT siderophore secretion phenotype on CAS plates (see Fig. S1 in the supplemental material). The data in Fig. 4B show that the HA-tagged Fur was not detected at the control genes, rpoD or dinP, but was enriched >103-fold at hns. Importantly, Fur enrichment to the upstream region of hns was metal dependent, noting that the data in Fig. 4B were normalized to 2,2′-dipyridyl-treated controls.
FIG. 4.
Fur binds upstream of hns in a metal-dependent manner. (A) Expression of hns was significantly elevated in the fur mutant determined by qRT-PCR. A paired Student t test was used to determine significance (*, P ≤ 0.05; n = 3). (B) Fur enrichment upstream of hns in S. Typhimurium. ChIP was performed on chromatin from the fur mutant that contained the pfur-ha plasmid. Resultant DNAs were analyzed by qPCR for antibody-mediated enrichment of Fur-HA at rpoD, dinP, and hns. Amplicon enrichment with anti-HA pAb was normalized to matched signal in nonspecific IgG control immunoprecipitations and expressed as the fold enrichment over that obtained from cells treated with dip (200 μM). Bars indicate means ± the SD of triplicate qPCRs and are representative of four independent ChIP experiments.
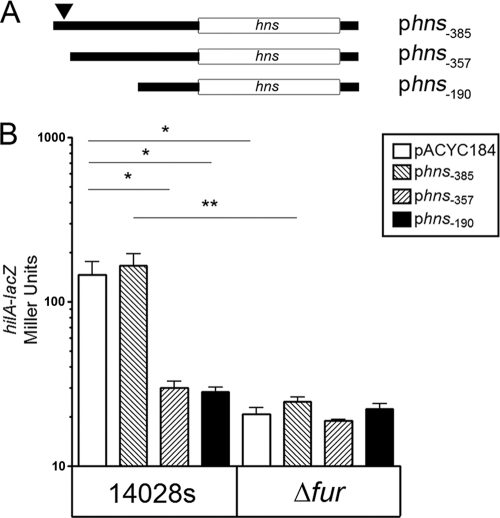
Activation of hilA-lacZ by Fur is dependent on the putative Fur-binding site upstream of hns.
H-NS has been shown to repress the transcription of hilA, a number of its activators, and other horizontally acquired SPIs (38, 46, 48, 49, 53). We hypothesized that the upregulation of hns in the fur-deficient cells may explain the observed Fur-dependent expression of hilA (Fig. 2) (22, 62). To test this hypothesis, we examined the physiological role of the putative Fur binding site in the hns promoter on the expression of hilA-lacZ. For the present study, we constructed a set of low-copy-number plasmids that contained the full-length hns gene downstream of progressive 5′ promoter deletions (Fig. 5A). The predicted Fur binding site is located within 385 and 357 nucleotides upstream of hns (signified by a filled triangle in Fig. 5A). The different hns plasmids were transformed into the WT and fur mutant strains carrying hilA-lacZ reporters. The data in Fig. 5B, show that the expression of hilA-lacZ in the WT background was the same in cells carrying the empty vector control and in cells with a construct that contained 385 nucleotides upstream of hns (relative to the ATG codon). The expression of hilA-lacZ in the WT background decreased in cells carrying a construct that contained the upstream 357- or 190-nucleotide segments (Fig. 5B). In the Δfur background, the expression of hilA-lacZ in cells carrying the empty vector or the construct with the 385 nucleotides upstream of hns was much lower than in the WT background, and shorter constructs had no further effect on the expression of hilA-lacZ in the mutant (Fig. 5B). This indicated that the DNA segment within 385 and 357 nucleotides upstream of hns was important for Fur-dependent activation of hilA-lacZ.
FIG. 5.
Fur regulates hilA-lacZ through nucleotides upstream of hns. (A) The truncated constructs used to determine the role of the upstream region of the hns promoter in the Fur-dependent regulation of hilA-lacZ. A filled triangle depicted a predicted Fur binding site. (B) The β-galactosidase activity was determined in the parent strain and in the fur mutant that contained single-copy hilA-lacZ and harbored the empty vector (pACYC184) or in the different truncated constructs listed in panel A. β-Galactosidase activity was determined as in Fig. 2. A paired Student t test was used to determine significance (*, P ≤ 0.05; **, P ≤ 0.01; n = 6). Means ± the SD are shown.
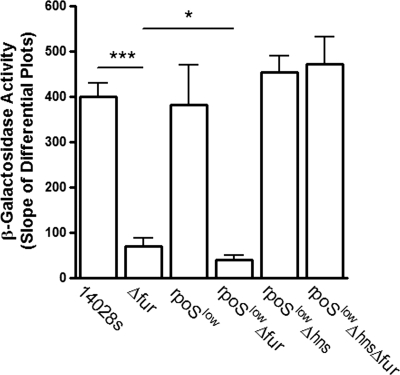
H-NS reduces hilA-lacZ expression in the fur mutant.
Next, we wanted to examine the effect of Fur on the expression of hilA-lacZ in a strain that lacked H-NS. It has been reported that, in S. Typhimurium, a deletion of hns is lethal, and certain mutations in the alternative sigma factor (rpoSlow) can compensate for this lethality (38, 46, 53). Therefore, we used P22 transduction to move hilA-lacZ into strains WN153 (rpoSlow) and WN341 (rpoSlowΔhns) that were derived from 14028s (38, 46). We then introduced the Δfur mutation into each strain to eliminate Fur expression. We monitored the expression of hilA-lacZ throughout anaerobic growth (see Fig. S3 in the supplemental material). These data are presented in the form of a differential plot (47), where the slopes were used to determine the rate of expression of the reporter (Fig. 6). Using this approach, we determined that hilA-lacZ induction occurs across a cell density that corresponds to the mid-log to the onset of stationary phase (i.e., an optical density at 600 nm [OD600] of ∼0.25 to ∼1 [under our conditions]; see Fig. S3 in the supplemental material).
FIG. 6.
Deletion of hns ablates Fur-dependent regulation of hilA-lacZ. β-Galactosidase activity was determined under anaerobic conditions. The data are presented as slopes from the differential plots (see Fig. S3 in the supplemental material). Means ± the SD are shown (*, P ≤ 0.05; ***, P ≤ 0.001; n = 6). The expression of hilA-lacZ in rpoSlow was biphasic (see the text and Fig. S3 for details).
The requirement of Fur for the activation of hilA was apparent as the rate of synthesis of hilA-lacZ was significantly reduced in Δfur compared to 14028s (P ≤ 0.001, Fig. 6), and lack of Fur resulted in decreased expression of HilA-activated genes (i.e., invF and sipC) (see Fig. S4 in the supplemental material). Interestingly, reporter activity exhibited biphasic expression in rpoSlow genetic background (see Fig. S3 in the supplemental material). Thus, during the early phase of growth the expression of hilA-lacZ in the rpoSlow background was similar to that of the wild type (Fig. 6) but was significantly reduced during the later phase compared to 14028s (i.e., 146.2 ± 54.7 U OD600−1 versus 399.8 ± 29.6 U OD600−1, P ≤ 0.01; see Fig. S3 in the supplemental material). In addition, reporter activity was significantly reduced in rpoSlowΔfur relative to Δfur (P ≤ 0.05; Fig. 6). Together, these findings strongly indicate a role for RpoS in hilA-lacZ induction during stationary phase, under anaerobic conditions. As expected, deletion of hns in the rpoSlow background restored reporter activity to wild-type expression (Fig. 6). Reporter activity was similar to wild type when fur was deleted in the rpoSlowΔhns background (Fig. 6). Collectively, these data demonstrate that Fur-dependent activation of hilA-lacZ during anaerobic growth is due to the repression of hns by Fur.
DISCUSSION
In Salmonella, virulence genes are clustered in the SPIs that are acquired via horizontal gene transfer. H-NS has been shown to serve as a silencer of horizontally transferred genes (46, 49). SPI-1 is an important facet of Salmonella virulence because it facilitates the invasion and penetration of the gastrointestinal epithelium. A major regulator of SPI-1 is the DNA-binding protein HilA, which activates all components necessary for a T3SS. Indeed, in vivo competition experiments indicate deletion of either hilA or SPI-1 result in the same phenotype (20). A previous study showed that the fur mutant of S. Typhimurium was avirulent in C3H/HeN (Nramp+/+) mice infected i.p. (65), and other work showed that Fur is required for the activation of hilA (22, 62). However, the identity of the Fur-regulated gene(s) responsible for the positive effect of Fur on the regulation of hilA and virulence in S. Typhimurium remained unknown. In the present study, we report that Fur regulates the expression of H-NS and this in-turn regulates the expression of hilA and virulence in mice.
Fur and HilA.
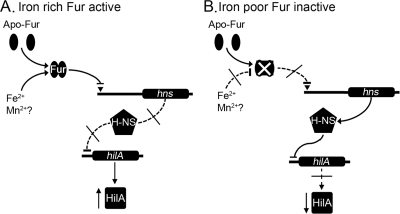
Normally, Fur acts as a repressor of genes required for iron homeostasis (30). In E. coli, Fur has been shown to positively affect the expression of few genes (e.g., sodB) via its negative regulation of a small RNA, RyhB (39). The possibility that the Salmonella sRNAs (RfrA and RfrB) or the RNA-binding protein (Hfq) were involved in the activation of HilA by Fur was ruled out by the excellent work of Ellermeier and Slauch (22). We confirmed that Hfq has no role in the activation of hilA-lacZ under anaerobic conditions (see Fig. S4 in the supplemental material). The nature of the factor required for the Fur activation of HilA remained elusive until the present study. Thus, we showed that Fur negatively regulates the expression of hns and that the positive effect of Fur on the expression of hilA is dependent on an upstream sequence that contained the putative Fur-binding site in hns. In addition, a deletion in hns abrogated the effect of Fur on the expression of hilA. These results are summarized in the diagram depicted in Fig. 7, where the presence of active Fur (i.e., Fur-Fe2+) results in decrease in [H-NS] and increased the expression of hilA (Fig. 7A). On the other hand, under conditions where Fur is inactive (i.e., via deletion of fur or iron deprivation) [H-NS] increases and results in repression of hilA (Fig. 7B). In addition, the growth-phase-dependent regulation of hilA (Fig. S3) may explain the small change (i.e., <2.5-fold) observed in our Δfur microarray analysis (64).
FIG. 7.
Model depicting the role of Fur in the regulation of hilA. (A) Conditions that activate Fur (i.e., the presence of iron) repress the expression of hns (solid arrow), and allow the expression of hilA (blocked dotted arrows) and increased HilA. (B) Mutation in Fur or lack of the cofactor (i.e., Fe2+) results in the expression of hns and the H-NS protein (blocked dotted arrows) and the reduced expression of hilA (solid arrows) and HilA. Implicit in this model is the role of ferrous iron transport and iron homeostasis in the activation of hilA/HilA and virulence.
Earlier work determined that Fur activation of hilA operated through HilD and that overexpression of HilD resulted in Fur-independent induction of hilA (22). This may be due to HilD countersilencing H-NS repression. H-NS displacement is needed to allow access of RNA polymerase to hilA. This distinguishes H-NS from other regulators, for instance, those that require an appropriate metal for DNA binding, e.g., Fur, which can be displaced from DNA-binding sites by removal of the metal. However, the identity of the factor(s) required for H-NS displacement is currently unknown. It has recently been shown that HilD does countersilence H-NS repression of another SPI (SPI-2) during in vitro growth (7). This suggests the removal of H-NS by HilD may also be responsible for activation of hilA in a mutated fur background. Thus, countersilencing of H-NS repression appears to be a common aspect of its regulation.
Fur and virulence.
We found the fur mutant to be avirulent in the C3H/HeN (Nramp+/+)-infected mice, in agreement with (65), but it was attenuated in C57BL/6 (Nramp−/−) mice (Fig. 1). This result could be attributed to the metal restriction imposed on intracellular pathogens in the Nramp+/+ background. However, as expected, we detected upregulation of Mn2+, Fe2+, and Fe3+ acquisition genes (64) and increased siderophore production in the fur mutant (see Fig. S1B in the supplemental material). This indicates that metal restriction in the Nramp+/+ may not be the cause for the avirulent phenotype of the fur mutant. Instead, the difference in the severity of the infection in the two types of mice may be due to the Nramp1-dependent acidification of the phagosome in the Nramp+/+ mice instead of the contribution to metal restriction from the pathogen (29). Indeed, Fur is essential during acid stress in Salmonella (26, 27). However, it could be suggested that reduced virulence of the fur mutant is related to the increased concentration of free iron in the cells and oxidative stress (50, 63, 65). In the present study, we demonstrated the role of Fur in activating a regulator of SPI-1, HilA, which may contribute to the reduced virulence of the Fur mutant. However, one may argue that in the present and the previous (65) studies mice were infected via i.p. routes that bypass the need for invasion genes found in SPI-1. Although the classical role of SPI-1 is traversing the host epithelial cell layer, several lines of evidence show that genes within SPI-1 are necessary for survival in phagocytic cells (11, 24, 35, 56). In addition, since H-NS is a major silencer of horizontally acquired genes (46), we postulate that overexpression of H-NS in the fur mutant results in the reduced expression of genes within SPI-1 (i.e., HilA-dependent genes), as well as genes within other SPIs, and collectively contribute to the reduced virulence of the Fur mutant in mice.
Regulation of H-NS by Fur.
The nucleoid-associated protein, H-NS, is important for the selective silencing and regulation of horizontally acquired genes in enteric bacteria (23, 46, 60). Indeed, expression of horizontally acquired genes at the right time and in response to the appropriate environmental signals is essential for peak fitness of the organism (23, 46, 60). There are many reports on the antisilencing mechanisms deployed by enteric bacteria (for a review, see reference 60). We reported here, for the first time, on the regulation of H-NS by Fur. Thus, suggesting that the regulation of horizontally acquired genes in Salmonella is indeed integrated into the preexisting regulatory network for iron homeostasis. However, evidence presented here raises two major questions regarding H-NS regulation.
First, to what extent does H-NS interact with iron metabolism? The metal-dependent regulation of hns by Fur suggests that iron concentrations (possibly other metals such as manganese) within the cell may be key signals in H-NS regulation. Indeed, evidence in Escherichia coli suggests that H-NS represses a major iron-storing ferritin (ftnA) and that Fur counteracts this repression by physically removing H-NS from the repressive site (45). Also, in E. coli, the effect of H-NS on iron acquisition is divergent; it acts as a repressor of salmochelin (IroN) and activator of the hemin receptor (ChuA), while having no effect on the expression of enterobactin receptor (FebA) (43). In addition, in S. Typhimurium, activation of ftnA depends on iron in a Fur-dependent fashion (65). It is also noteworthy that Fur is required for induction of the least studied ferritin-like protein in S. Typhimurium, FtnB (64). H-NS binds the region of ftnB and represses transcription (46), which indicates a possible mechanism for Fur activation of ftnB. H-NS also binds to the promoter of the ferrous iron transport genes, feoAB, as well as the activator of feoAB transcription, RstA (46). Given that RstA (STM1475) is a member of the OmpR family of response regulators, it may not be surprising that H-NS regulation is iron dependent and indicates that H-NS may reduce ferrous iron acquisition and transport, thereby reducing Fur activity. We speculate that a primary role of RstA activation of Fur in acidic conditions is to reduce ferrous iron transport, repress hns, and promote Fur-mediated hydroperoxidase activity (64, 66). Collectively, our data and the reports cited above suggest an interaction between H-NS and Fur in iron homeostasis.
Second, how does Fur regulate expression of hns at such a distant regulatory site? The predicted −35 and −10 sites of hns are located 70 and 50 nucleotides upstream of ATG, respectively, 309 and 292 nucleotides downstream from our putative Fur binding site. This indicates that Fur may repress hns expression by a novel mechanism, perhaps in combination with other, yet-to-be discovered regulators. The recent evidence that Fur displaces H-NS from regulatory sites of ftnA and not vice versa (45) indicates that once Fur is activated the cell is committed to countersilencing and repression of H-NS until iron concentrations are reduced enough to inhibit Fur binding. Subsequent work should determine the exact mechanism of Fur regulation of hns. For instance, does Fur binding to hns require additional regulators? Can Fur bind to multiple regulatory sites of hns? The benefit of our ChIP assays was the determination of Fur binding to the regulatory region of hns, in vivo. However, in vitro work is needed to determine the extent of Fur affinity to our predicted Fur binding site and perhaps other sites located upstream of hns.
In conclusion, we provided, for the first time, evidence that Fur represses hns expression. In addition, Fur-controlled activation of hilA through 28 bp located at a distant site upstream of hns. Our findings advance the current model of SPI-1 regulation (2, 21) and identified the previously unexplained relationship between iron and Fur in the regulation of hilA (signified as “X” in reference 22). Also, our data provide a plausible explanation of the reduced virulence of the fur mutant. Further studies are needed to define the exact mechanism of H-NS regulation via iron homeostasis.
Supplementary Material
Acknowledgments
This study was supported in part by the North Carolina Agricultural Research Service (to HM.H.). A.V.-T. and J.J. were supported by NIH grant AI054959 and the Burroughs Wellcome Fund.
We are grateful to C. Altier, F. C. Fang, and S. J. Libby for strains.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Albright, L. M., E. Huala, and F. M. Ausubel. 1989. Prokaryotic signal transduction mediated by sensor and regulator protein pairs. Annu. Rev. Genet. 23:311-336. [DOI] [PubMed] [Google Scholar]
- 2.Altier, C. 2005. Genetic and environmental control of Salmonella invasion. J. Microbiol. 43(Spec. No.):85-92. [PubMed] [Google Scholar]
- 3.Bajaj, V., R. L. Lucas, C. Hwang, and C. A. Lee. 1996. Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol. Microbiol. 22:703-714. [DOI] [PubMed] [Google Scholar]
- 4.Belden, W. J., and S. I. Miller. 1994. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect. Immun. 62:5095-5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bourret, T. J., S. Porwollik, M. McClelland, R. Zhao, T. Greco, H. Ischiropoulos, and A. Vazquez-Torres. 2008. Nitric oxide antagonizes the acid tolerance response that protects Salmonella against innate gastric defenses. PLoS One 3:e1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brussow, H., C. Canchaya, and W. D. Hardt. 2004. Phages and the evolution of bacterial pathogens: from genomic rearrangements to lysogenic conversion. Microbiol. Mol. Biol. Rev. 68:560-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bustamante, V. H., L. C. Martinez, F. J. Santana, L. A. Knodler, O. Steele-Mortimer, and J. L. Puente. 2008. HilD-mediated transcriptional cross-talk between SPI-1 and SPI-2. Proc. Natl. Acad. Sci. U. S. A. 105:14591-14596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Centers for Disease Control and Prevention. 2008. Salmonella surveillance: annual summary, 2006, p. 5-27. U.S. Department of Health and Human Services, Atlanta, GA.
- 9.Chamnongpol, S., M. Cromie, and E. A. Groisman. 2003. Mg2+ sensing by the Mg2+ sensor PhoQ of Salmonella enterica. J. Mol. Biol. 325:795-807. [DOI] [PubMed] [Google Scholar]
- 10.Chamnongpol, S., and E. A. Groisman. 2002. Mg2+ homeostasis and avoidance of metal toxicity. Mol. Microbiol. 44:561-571. [DOI] [PubMed] [Google Scholar]
- 11.Chan, K., C. C. Kim, and S. Falkow. 2005. Microarray-based detection of Salmonella enterica serovar Typhimurium transposon mutants that cannot survive in macrophages and mice. Infect. Immun. 73:5438-5449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ciccone, D. N., K. B. Morshead, and M. A. Oettinger. 2004. Chromatin immunoprecipitation in the analysis of large chromatin domains across murine antigen receptor loci. Methods Enzymol. 376:334-348. [DOI] [PubMed] [Google Scholar]
- 14.Costanzo, A., and S. E. Ades. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, sigmaE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Costanzo, A., H. Nicoloff, S. E. Barchinger, A. B. Banta, R. L. Gourse, and S. E. Ades. 2008. ppGpp and DksA likely regulate the activity of the extracytoplasmic stress factor sigmaE in Escherichia coli by both direct and indirect mechanisms. Mol. Microbiol. 67:619-632. [DOI] [PubMed] [Google Scholar]
- 16.Darwin, K. H., and V. L. Miller. 1999. InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J. Bacteriol. 181:4949-4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Darwin, K. H., and V. L. Miller. 2000. The putative invasion protein chaperone SicA acts together with InvF to activate the expression of Salmonella typhimurium virulence genes. Mol. Microbiol. 35:949-960. [DOI] [PubMed] [Google Scholar]
- 18.Darwin, K. H., and V. L. Miller. 2001. Type III secretion chaperone-dependent regulation: activation of virulence genes by SicA and InvF in Salmonella typhimurium. EMBO J. 20:1850-1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichelberg, K., and J. E. Galan. 1999. Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and hilA. Infect. Immun. 67:4099-4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ellermeier, C. D., J. R. Ellermeier, and J. M. Slauch. 2005. HilD, HilC, and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 57:691-705. [DOI] [PubMed] [Google Scholar]
- 21.Ellermeier, J. R., and J. M. Slauch. 2007. Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr. Opin. Microbiol. 10:24-29. [DOI] [PubMed] [Google Scholar]
- 22.Ellermeier, J. R., and J. M. Slauch. 2008. Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J. Bacteriol. 190:476-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fang, F. C., and S. Rimsky. 2008. New insights into transcriptional regulation by H-NS. Curr. Opin. Microbiol. 11:113-120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fink, R. C., M. R. Evans, S. Porwollik, A. Vazquez-Torres, J. Jones-Carson, B. Troxell, S. J. Libby, M. McClelland, and H. M. Hassan. 2007. FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J. Bacteriol. 189:2262-2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forbes, J. R., and P. Gros. 2003. Iron, manganese, and cobalt transport by Nramp1 (Slc11a1) and Nramp2 (Slc11a2) expressed at the plasma membrane. Blood 102:1884-1892. [DOI] [PubMed] [Google Scholar]
- 26.Foster, J. W. 1991. Salmonella acid shock proteins are required for the adaptive acid tolerance response. J. Bacteriol. 173:6896-6902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foster, J. W., and H. K. Hall. 1992. Effect of Salmonella typhimurium ferric uptake regulator (fur) mutations on iron- and pH-regulated protein synthesis. J. Bacteriol. 174:4317-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gallegos, M. T., R. Schleif, A. Bairoch, K. Hofmann, and J. L. Ramos. 1997. Arac/XylS family of transcriptional regulators. Microbiol. Mol. Biol. Rev. 61:393-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hackam, D. J., O. D. Rotstein, W. Zhang, S. Gruenheid, P. Gros, and S. Grinstein. 1998. Host resistance to intracellular infection: mutation of natural resistance-associated macrophage protein 1 (Nramp1) impairs phagosomal acidification. J. Exp. Med. 188:351-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hantke, K. 1981. Regulation of ferric iron transport in Escherichia coli K-12: isolation of a constitutive mutant. Mol. Gen. Genet. 182:288-292. [DOI] [PubMed] [Google Scholar]
- 31.Hassan, H. M., and H. C. Sun. 1992. Regulatory roles of Fnr, Fur, and Arc in expression of manganese-containing superoxide dismutase in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 89:3217-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones, B. D., N. Ghori, and S. Falkow. 1994. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer's patches. J. Exp. Med. 180:15-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaniga, K., J. C. Bossio, and J. E. Galan. 1994. The Salmonella typhimurium invasion genes invF and invG encode homologues of the AraC and PulD family of proteins. Mol. Microbiol. 13:555-568. [DOI] [PubMed] [Google Scholar]
- 34.Klein, J. R., T. F. Fahlen, and B. D. Jones. 2000. Transcriptional organization and function of invasion genes within Salmonella enterica serovar Typhimurium pathogenicity island 1, including the prgH, prgI, prgJ, prgK, orgA, orgB, and orgC genes. Infect. Immun. 68:3368-3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lawley, T. D., K. Chan, L. J. Thompson, C. C. Kim, G. R. Govoni, and D. M. Monack. 2006. Genome-wide screen for Salmonella genes required for long-term systemic infection of the mouse. PLoS Pathog. 2:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee, C. A., and S. Falkow. 1990. The ability of Salmonella to enter mammalian cells is affected by bacterial growth state. Proc. Natl. Acad. Sci. U. S. A. 87:4304-4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lostroh, C. P., V. Bajaj, and C. A. Lee. 2000. The cis requirements for transcriptional activation by HilA, a virulence determinant encoded on SPI-1. Mol. Microbiol. 37:300-315. [DOI] [PubMed] [Google Scholar]
- 38.Main-Hester, K. L., K. M. Colpitts, G. A. Thomas, F. C. Fang, and S. J. Libby. 2008. Coordinate regulation of Salmonella pathogenicity island 1 (SPI1) and SPI4 in Salmonella enterica serovar Typhimurium. Infect. Immun. 76:1024-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Miller, S. I., A. M. Kukral, and J. J. Mekalanos. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 86:5054-5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Monod, J. 1958. An outline of enzyme induction. Recueil Des Travaux Chimiques Des Pays-Bas. J. R. Netherlands Chem. Soc. 77:569-585. [Google Scholar]
- 43.Muller, C. M., U. Dobrindt, G. Nagy, L. Emody, B. E. Uhlin, and J. Hacker. 2006. Role of histone-like proteins H-NS and StpA in expression of virulence determinants of uropathogenic Escherichia coli. J. Bacteriol. 188:5428-5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mutalik, V. K., G. Nonaka, S. E. Ades, V. A. Rhodius, and C. A. Gross. 2009. Promoter strength properties of the complete sigma E regulon of Escherichia coli and Salmonella enterica. J. Bacteriol. 191:7279-7287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nandal, A., C. C. Huggins, M. R. Woodhall, J. McHugh, F. Rodriguez-Quinones, M. A. Quail, J. R. Guest, and S. C. Andrews. 2010. Induction of the ferritin gene (ftnA) of Escherichia coli by Fe2+-Fur is mediated by reversal of H-NS silencing and is RyhB independent. Mol. Microbiol. 75:637-657. [DOI] [PubMed] [Google Scholar]
- 46.Navarre, W. W., S. Porwollik, Y. Wang, M. McClelland, H. Rosen, S. J. Libby, and F. C. Fang. 2006. Selective silencing of foreign DNA with low GC content by the H-NS protein in Salmonella. Science 313:236-238. [DOI] [PubMed] [Google Scholar]
- 47.Neidhardt, F. C., J. L. Ingraham, and M. Schaechter. 1990. Physiology of the bacterial cell: a molecular approach, p. 331. Sinauer Associates, Sunderland, MA.
- 48.Olekhnovich, I. N., and R. J. Kadner. 2006. Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J. Mol. Biol. 357:373-386. [DOI] [PubMed] [Google Scholar]
- 49.Olekhnovich, I. N., and R. J. Kadner. 2007. Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J. Bacteriol. 189:6882-6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Park, S., X. You, and J. A. Imlay. 2005. Substantial DNA damage from submicromolar intracellular hydrogen peroxide detected in Hpx- mutants of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 102:9317-9322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Penheiter, K. L., N. Mathur, D. Giles, T. Fahlen, and B. D. Jones. 1997. Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer's patches. Mol. Microbiol. 24:697-709. [DOI] [PubMed] [Google Scholar]
- 52.Perez, J. C., D. Shin, I. Zwir, T. Latifi, T. J. Hadley, and E. A. Groisman. 2009. Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 5:e1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schechter, L. M., S. Jain, S. Akbar, and C. A. Lee. 2003. The small nucleoid-binding proteins H-NS, HU, and Fis affect hilA expression in Salmonella enterica serovar Typhimurium. Infect. Immun. 71:5432-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 55.Sikes, M. L., J. M. Bradshaw, W. T. Ivory, J. L. Lunsford, R. E. McMillan, and C. R. Morrison. 2009. A streamlined method for rapid and sensitive chromatin immunoprecipitation. J. Immunol. Methods 344:58-63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva-Herzog, E., and C. S. Detweiler. 2010. Salmonella enterica replication in hemophagocytic macrophages requires two type three secretion systems. Infect. Immun. 74:3369-3377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skamene, E., E. Schurr, and P. Gros. 1998. Infection genomics: Nramp1 as a major determinant of natural resistance to intracellular infections. Annu. Rev. Med. 49:275-287. [DOI] [PubMed] [Google Scholar]
- 58.Soncini, F. C., E. Garcia-Vescovi, F. Solomon, and E. A. Groisman. 1996. Molecular basis of the magnesium deprivation response in Salmonella typhimurium: identification of PhoP-regulated genes. J. Bacteriol. 178:5092-5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stock, J. B., A. J. Ninfa, and A. M. Stock. 1989. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol. Rev. 53:450-490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stoebel, D. M., A. Free, and C. J. Dorman. 2008. Anti-silencing: overcoming H-NS-mediated repression of transcription in Gram-negative enteric bacteria. Microbiology 154:2533-2545. [DOI] [PubMed] [Google Scholar]
- 61.Takeuchi, A. 1967. Electron microscope studies of experimental Salmonella infection. I. Penetration into the intestinal epithelium by Salmonella typhimurium. Am. J. Pathol. 50:109-136. [PMC free article] [PubMed] [Google Scholar]
- 62.Thompson, A., M. D. Rolfe, S. Lucchini, P. Schwerk, J. C. Hinton, and K. Tedin. 2006. The bacterial signal molecule, ppGpp, mediates the environmental regulation of both the invasion and intracellular virulence gene programs of Salmonella. J. Biol. Chem. 281:30112-30121. [DOI] [PubMed] [Google Scholar]
- 63.Touati, D., M. Jacques, B. Tardat, L. Bouchard, and S. Despied. 1995. Lethal oxidative damage and mutagenesis are generated by iron in delta fur mutants of Escherichia coli: protective role of superoxide dismutase. J. Bacteriol. 177:2305-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Troxell, B. 2010. The role of ferric uptake regulator in regulation of metal homeostasis, metabolism, virulence, and protection against hydrogen peroxide in Salmonella enterica serovar Typhimurium. Dissertation. North Carolina State University, Raleigh, NC.
- 65.Velayudhan, J., M. Castor, A. Richardson, K. L. Main-Hester, and F. C. Fang. 2007. The role of ferritins in the physiology of Salmonella enterica sv. Typhimurium: a unique role for ferritin B in iron-sulphur cluster repair and virulence. Mol. Microbiol. 63:1495-1507. [DOI] [PubMed] [Google Scholar]
- 66.Zaid, T., T. S. Srikumar, and L. Benov. 2003. Growth of Escherichia coli in iron-enriched medium increases HPI catalase activity. J. Biochem. Mol. Biol. 36:608-610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.