Abstract
Iron acquisition in aerobic habitats is complicated by the low solubility of ferric hydroxides. Siderophores that bind ferric iron with high affinity are used to mobilize iron. The reduction of ferric iron to the ferrous form can be coupled to the release of iron from siderophores. Iron is also stored intracellularly as a ferric mineral in proteins, such as ferritin, and must be reduced during release. In Escherichia coli, the yqjH gene encodes a putative ferric siderophore reductase that is also part of the Fur regulon. Here we show that YqjH has ferric reductase activity and is required for iron homeostasis in E. coli. Divergently transcribed from yqjH is the yqjI gene, which encodes a novel member of the winged-helix family of transcriptional regulators and also contains an N-terminal extension similar to the Ni2+-binding C-terminal tail of SlyD. Deletion of yqjI leads to constitutive high-level activity of the yqjH and yqjI promoters. Purified YqjI binds inverted repeat target sequences within the yqjH and yqjI promoters. We also observed that YqjI-dependent transcriptional repression is reduced when cells are exposed to elevated nickel levels, resulting in increased expression of yqjH and yqjI. YqjI binding to nickel or iron reduces YqjI DNA-binding activity in vitro. Furthermore, we found that elevated nickel stress levels disrupt iron homeostasis in E. coli and that deletion of yqjH increases nickel toxicity. Our results suggest that the YqjI protein controls expression of yqjH to help maintain iron homeostasis under conditions (such as elevated cellular nickel levels) that disrupt iron metabolism.
Iron is an essential transition metal required for critical cellular pathways, including respiration and photosynthesis. However, maintenance of iron homeostasis is a daunting task due to the low solubility of iron in aerobic environments and to the spurious redox chemistry catalyzed by iron in the presence of oxygen. It is also clear that other transition metals, such as copper and cobalt, can effectively displace and/or compete with iron during metalloenzyme assembly if they are present in excess in the intracellular environment (32, 33, 45, 50).
To circumvent the difficulties of iron acquisition and trafficking, complex iron homeostasis systems have evolved in most organisms. Bacterial iron homeostasis pathways include high-affinity extracellular chelators (siderophores) for extraction of ferric iron from the environment, membrane iron transporters for a variety of iron chelates, intracellular iron storage proteins, and dedicated iron metalloenzyme assembly systems (1). In E. coli, transcriptional expression of these various iron homeostasis pathways, collectively referred to as the iron stimulon, is largely controlled by the iron metalloregulatory protein Fur (20).
Iron coordination by the Fur protein controls its DNA-binding activity. The Fe2+-Fur homodimer binds to a 19-bp sequence (the “iron box” or “Fur box”) to repress transcription from target promoters, while apo-Fur dissociates from target promoters, leading to upregulation of iron acquisition systems to increase cellular iron levels (21). Fur also controls expression of the RyhB small RNA that acts as a posttranscriptional regulator of mRNA transcripts from the iron stimulon (35).
Despite extensive study of iron homeostasis in E. coli, the full picture of iron homeostasis is incomplete. For example, a recent DNA macroarray analysis of iron-dependent gene regulation in E. coli showed that approximately one-third of the 101 genes regulated by the Fe2+-Fur complex are hypothetical open reading frames with no known function (37). Establishing the biochemical roles of these uncharacterized genes is critical to gain a complete understanding of in vivo iron metabolism and homeostasis.
In this study, we examine the function and regulation of yqjH, which was shown to be part of the Fur regulon (37). We show here that YqjH is a NADPH-dependent ferric reductase that plays a role in iron homeostasis. Interestingly, we discovered that yqjH transcription is controlled primarily by a second regulator encoded by the divergent yqjI gene. YqjI represses yqjH transcription and is a nickel-binding protein. Our results suggest that YqjI may regulate yqjH and other target genes to protect iron homeostasis from disruption by environmental stresses, such as elevated intracellular nickel levels.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Escherichia coli wild-type strain MG1655 was the parent strain for all studies (Table 1). The gene deletion strains were constructed as described previously (14). Briefly, a kanamycin resistance (Kanr) cassette was amplified from pKD4 using primer pairs containing approximately 35 bp of sequence homologous to regions upstream and downstream of the target genes. The PCR products were transformed into NM400 expressing the λ Red recombinase system, resulting in replacement of the target gene with the Kanr cassette. Mutations were then moved by P1 transduction into wild-type MG1655. In some cases, the Kanr cassette was removed from a single mutant strain after transformation with the pCP20 plasmid so that double mutant strains could be constructed by P1 transduction (14). Promoter-lacZ fusions from the 287-bp intergenic region between yqjH and yqjI were constructed. The yqjH promoter was amplified by yqjH_287_up and yqjH_upstr_L, and the yqjI promoter was amplified by yqjI_287_up and yqjI_upstr_L (see Table S1 in the supplemental material). The promoter fragments were digested with BamHI and XhoI and cloned into the corresponding sites in the pPK7035 plasmid (28). A fragment, containing part of lacI, the Kanr gene, and the entire 287-bp intergenic sequence up to but not including the ATG start codons of yqjH or yqjI fused to the region upstream of the lacZ start codon, was amplified by the primer pair 5′-lacI-Kn and 3′-lacZ and then transformed into NM400 as previously described (28). Recombination resulted in replacement of the native lacZ promoter with either the yqjH or yqjI promoter. Fusions were then moved into MG1655 by P1 transduction. For protein expression and complementation studies, the open reading frames of yqjH and yqjI were amplified by PCR using the primers yqjH_pET21a_up/dn and yqjI_pET21a_up/dn. The fragments were digested and cloned into the NdeI and BamHI sites of the pET21a vector (Novagen). In both cases, the fully wild-type sequence of each open reading frame containing no mutations was used. All plasmids were confirmed by DNA sequencing.
TABLE 1.
Strains used in this study
| Strain | Genotype and/or characteristicsa | Source or reference |
|---|---|---|
| MG1655 | Wild-type E. coli K-12 | Laboratory strain |
| NM400 | MG1655 mini-λ Cmr Ts | Laboratory strain |
| BL21(DE3) | Laboratory strain | |
| DJ480 | MG1655 ΔX74lac | Laboratory strain |
| WO452 | MG1655 Φ(yqjH-lacZ) | This study |
| WO453 | MG1655 Φ(yqjI-lacZ) | This study |
| WO454 | MG1655 ΔyqjI Φ(yqjH-lacZ) | This study |
| WO455 | MG1655 ΔyqjI Φ(yqjI-lacZ) | This study |
| WO456 | MG1655 Δfur Φ(yqjH-lacZ) | This study |
| WO457 | MG1655 ΔrcnA Φ(yqjH-lacZ) | This study |
| WO458 | MG1655 ΔyqjI Φ(yqjH-lacZ)/pET21a_yqjI | This study |
| WO19 | MG1655 ΔsufABCDSE | 41 |
| WO460 | MG1655 ΔyqjH::Kanr | This study |
| WO461 | MG1655 ΔyqjI::Kanr | This study |
| WO462 | MG1655 Δfes::Kanr | This study |
| WO463 | MG1655 ΔyqjH/Δfes::Kanr | This study |
| WO464 | MG1655 ΔyqjH/ΔfhuF::Kanr | This study |
| WO465 | MG1655 ΔrcnA | This study |
| WO466 | BL21(DE3) pET21a_yqjH | This study |
| WO472 | BL21(DE3) pET21a_yqjI | This study |
Ts, temperature sensitive.
Growth assays.
For growth on LB plates, overnight cultures of wild-type or mutant derivatives of MG1655 in LB were normalized to an optical density at 600 nm (OD600) of 2.0 and then serially diluted from 2 × 10−1 to 2 × 10−6 in sterile LB. A total of 5 μl of each dilution was plated on LB agar plates with the appropriate concentrations of 2,2′-dipyridyl or NiCl2. Plates were incubated for 14 to 20 h at 37°C. Final growth was recorded by photography. For growth in minimal medium, overnight cultures of wild-type or mutant derivatives of MG1655 in LB were normalized to an OD600 of 2.0 and then washed with M9 minimal medium with 0.2% gluconate twice before diluting cells to a starting OD600 of 0.02 (1:100 dilution) in the same medium with various concentrations of NiCl2. Cell growth was evaluated as the OD600 after 1 day (22 to 24 h) of incubation at 37°C, with shaking at 225 rpm.
β-Galactosidase assays.
Wild-type or mutant derivatives of MG1655, containing ΦyqjH-lacZ or ΦyqjI-lacZ, were grown in LB to mid-exponential phase (OD600 = 0.4 to 0.6) at 37°C and then assayed for β-galactosidase activity. For complementation studies, some strains also carried plasmid pET21a or pYqjI. For metal regulation experiments, strains containing ΦyqjH-lacZ were grown in LB overnight and then diluted to a starting OD600 of 0.02 into M9 minimal medium supplemented with 0.2% glucose and containing various concentrations of divalent metals (Ni2+, Fe2+, Co2+, Cu2+, or Mn2+). Cells were grown aerobically with vigorous shaking at 37°C. β-Galactosidase activity was measured when cells reached mid-exponential phase.
Protein expression and purification.
Plasmids were expressed in BL21(DE3). Bacteria were grown to an OD600 of 0.5 in LB with 100 μg/ml ampicillin at 37°C and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) at 37°C for 3 h for YqjH or at 30°C for 16 h for YqjI. In order to obtain Ni-YqjI, 500 μM NiCl2 was added into the LB medium at the time of inoculation. For YqjH purification, harvested cell pellets were resuspended in binding buffer (20 mM HEPES, pH 7.5, and 10% glycerol), lysed by sonication for 5 min, and cleared by centrifugation for 20 min at 16,000 × g. The supernatant was filtered through a 0.22-μm syringe filter (Millipore MCE membrane) before being loaded on a HiTrap DEAE column (GE Healthcare). YqjH was eluted using a linear gradient from 0 to 1 M NaCl. Bright yellow fractions were pooled, concentrated to 2 ml, and then further purified using a Superdex 75 gel filtration column (HiLoad 16/60; GE Healthcare) run with 20 mM HEPES at pH 7.5, 100 mM NaCl, and 10% glycerol. Yellow fractions containing YqjH were combined and further purified to homogeneity on a HiTrap phenyl FF column (GE Healthcare) using a linear gradient of 1 to 0 M ammonium sulfate. Pure YqjH was dialyzed overnight against 20 mM HEPES, pH 7.5, and 10% glycerol at 4°C to remove ammonium sulfate. The purity of final product was estimated by 12.5% Tris-glycine sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to be ≥95%.
Apo-YqjI and Ni-YqjI were purified using the same protocol. Cell pellets were resuspended in 20 mM HEPES, pH 7.5, with 10 mM β-mercaptoethanol and 5% glycerol and lysed by sonication for 2 min. After centrifugation, cleared lysate was loaded onto a HiTrap heparin HP column (GE Healthcare) and eluted with a linear gradient of 0 to 1 M NaCl. The last two elution peaks were found by SDS-PAGE to contain YqjI. The fractions that correspond to YqjI peak 1 were pooled, concentrated, and loaded onto a Superdex 75 gel filtration column to purify the monomer form of YqjI, while the YqjI peak 2 fractions were further purified by a Superdex 200 gel filtration column (HiLoad 16/60; GE Healthcare) to obtain the oligomer form of YqjI. Both columns were run with 20 mM HEPES at pH 7.5, 10 mM β-mercaptoethanol, 500 mM NaCl, and 5% glycerol. Protein purity was estimated by SDS-PAGE to be ≥90%, and protein concentration was determined by the Bradford assay. In some cases, the oligomeric states of YqjI were confirmed by analytical gel filtration on a Superdex 200 column (Superdex 10/300 GL; GE Healthcare). The column was calibrated using cytochrome c (12.4 kDa), carbonic anhydrase (29 kDa), ovalbumin (44 kDa), bovine serum albumin (66 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa), and thyroglobulin (669 kDa). The native oligomeric states were also confirmed by 4 to 16% native bis-Tris PAGE (NativePAGE Novex; Invitrogen).
Ferric reductase assay.
A ferric reductase assay was performed anaerobically in 500 μl of 50 mM Tris-HCl, pH 7.5, containing NADPH (0 to 1 mM) and a ferric iron substrate, ferric-EDTA (0 to 2 mM), ferric citrate (0 to 2 mM), or ferric enterobactin (0 to 0.7 mM), and 2 mM ferrozine to detect ferrous iron formation. Various concentrations of pure YqjH were added to initiate the reaction. The absorbance at 562 nm was recorded every 15 s for up to 20 min, and the extinction coefficient at 562 nm of the ferrous-ferrozine complex (ɛ562 = 27.9 mM−1 cm−1) was used to quantify ferrous iron generation. To determine the Km for ferric-EDTA, NADPH concentration was fixed at 0.25 mM, and YqjH was added at a 0.73 μM final concentration. To determine the Km for NADPH, the ferric-EDTA concentration was fixed at 2 mM, and YqjH was added at a 0.73 μM final concentration. Ferric enterobactin was purified as previously described (42). Iron release from ferric enterobactin was monitored by the decrease of the absorbance at 495 nm for ferric enterobactin in the reaction.
In vitro nickel reconstitution of the apo-YqjI oligomer.
The apo-YqjI concentration was determined by Bradford assay and diluted with 20 mM HEPES at pH 7.5, 500 mM NaCl, 10 mM β-mercaptoethanol, and 5% glycerol to 20 μM. A total of 200 μM NiCl2 (>99.99%) was prepared from 100 mM NiCl2 solution by serial dilutions. Changes in the YqjI UV-visible absorption spectrum upon Ni2+ addition were monitored after stepwise addition of 2 μl of 200 μM NiCl2 to a quartz cuvette containing 200 μl of 20 μM apo-YqjI. Protein precipitation began to occur when the molar ratio of Ni2+ to YqjI monomers increased to above 4:1. For standard preparation of reconstituted Ni-YqjI, a 3-fold molar excess of Ni2+ was added to apo-YqjI, and the sample was incubated on ice for 1 h. Excess nickel was then removed by concentration and dilution three times using a YM-10 Microcon (Millipore). Use of this protocol typically resulted in a 2:1 molar ratio of Ni2+ to YqjI monomers.
Metal analysis.
For protein metal analysis, apo-YqjI and holo-YqjI were diluted to ∼0.5 to 1.0 mg/ml in 4 ml buffer. Samples were analyzed on a Varian Liberty Series II inductively coupled plasma atomic emission spectrometer (ICP-AES). Ni, Zn, and Fe standards were prepared as 0, 0.3, 0.6, 0.9, and 1.2 ppm concentrations in ultrapure water. A buffer-only control was also measured to control for background contamination of nickel and zinc in buffer components. Alternatively, the 4-(2-pyridylazo)resorcinol (PAR) assay was used to measure Ni content in apo- and holo-YqjI. The method was modified from a previously published protocol (24). YqjI was prepared in 50 mM HEPES, pH 7.3, and 6 M guanidine hydrochloride. A total of 60 μl of PAR solution (1 mg/ml PAR dissolved in buffer) was added to 300 μl of standards or protein samples. Ni concentrations were measured after 20 min at 505 nm using a calibration calculated from Ni standards of 0, 3, 6, 9, 12, and 15 μM.
DNA binding using fluorescence anisotropy.
5′-fluorescein-labeled single-stranded DNA oligos, corresponding to the predicted YqjI binding sites (PyqjH and PyqjI), the predicted Fur binding site (Pfur), or a randomly selected sequence (random) in the yqjH-yqjI promoter region, were obtained from Fisher Scientific. Single-stranded oligos were annealed to their unlabeled complementary strands in 20 mM HEPES at pH 7.5 and 50 mM NaCl by being heated at 95°C for 5 min, followed by being slow cooled to room temperature. Equilibrium binding of apo-YqjI to 10 nM 5′-fluorescein-labeled double-stranded DNA was measured in 20 mM HEPES at pH 7.8, 100 mM NaCl, and 10 mM β-mercaptoethanol at 26°C after 30 s of equilibration using a SpectraMax M5 microplate reader (Molecular Devices) under the “fluorescence polarization” mode. The excitation wavelength (ex) was 494 nm, the emission wavelength (em) was 525 nm, and the cutoff was 515 nm. Photomultiplier tube (PMT) sensitivity was set to the highest level in order to obtain accurate readings. Each data point was the average result from three readings. To characterize the DNA-protein interactions, 1, 5, 10, 20 μM protein stocks were prepared and titrated into the DNA sample at up to 400 nM. Concentration response data from titration of labeled PyqjH and PyqjI oligonucleotides with apo-YqjI was fit with a sigmoidal (Boltzmann) function by iterative nonlinear least-squares regression using the SOLVER function in Microsoft Excel 2004 for Mac as previously described (6). The coefficients of determination (R2) for fitting of the binding data were 0.979 for PyqjH and 0.984 for PyqjI. To determine the effects of divalent metals on apo-YqjI DNA binding, Ni2+, Zn2+, or Fe2+ was titrated into 10 nM PyqjH oligonucleotide preequilibrated with 40 nM apo-YqjI oligomer. The change in anisotropy was measured after 30 s. The Fe2+ solution was freshly prepared in anaerobic buffer under anaerobic conditions from ferrous ammonium sulfate immediately prior to use.
RESULTS
A key step in iron homeostasis is the mobilization of ferric iron from both extracellular and intracellular sources. Ferric iron reduction may help release iron from siderophores (47) and is required for mobilization of ferric iron from intracellular iron storage proteins like ferritin (26, 49). While there are a number of potential mechanisms to drive intracellular ferric iron reduction, only a few proteins in E. coli have been directly or indirectly linked to this process (36, 39).
YqjH is a homologue of ViuB that is required for siderophore utilization in Vibrio cholerae (7, 37). A recently published crystal structure of YqjH shows that it is structurally related to the NAD(P)H:flavin oxidoreductase superfamily (4). Purified YqjH binds flavin adenine dinucleotide (FAD) and may utilize this cofactor to transfer electrons from NADH or NADPH to a ferric chelate to reduce iron to the ferrous form. However, the in vivo substrate for YqjH and its exact role in the iron starvation response are unknown.
To further characterize the biochemical function of YqjH, we overexpressed and purified the recombinant protein. As previously reported, YqjH was purified in a bright yellow fraction, with UV-visible absorption maxima at 384 and 451 nm and shoulders at 428 and 475 nm, consistent with the presence of oxidized FAD (see Fig. S1A in the supplemental material) (4). We determined the flavin extinction coefficient at 451 nm by using published protocols and found it to be 10,482 M−1cm−1 (31). Using this extinction coefficient, we conclude that purified YqjH contains 0.93 FAD per protein monomer.
Fluorescence measurement of YqjH after separation by SDS-PAGE and soaking in 7% acetic acid showed the presence of a fluorescent band at the same molecular weight as that of YqjH, demonstrating that FAD is covalently linked to and comigrates with YqjH (see Fig. S1B in the supplemental material). Pretreatment of the gel with 5% performic acid increased the YqjH-FAD fluorescence, indicating that the flavin is bound to YqjH via an S-cysteinyl flavin linkage consisting of a thioether bond between a cysteine side chain and the FAD (see Fig. S1B) (48). The UV-visible absorption spectrum of purified YqjH, with two absorption maxima at 384 and 451 nm (see Fig. S1A), indicates an 8α-S-cysteinyl flavin linkage rather than a 6-S-cysteinyl linkage, which typically shows only a single absorption peak around 437 nm (48). Based on our analysis, it appears that YqjH uses FADH2/FAD as a covalently bound cofactor rather than a substrate. Thus, YqjH is distinct from other flavin reductases (such as Fre in E. coli) that release free reduced flavins as products that are then separately used to reduce ferric iron in vivo (47).
Next, we determined if purified YqjH can reduce ferric iron in vitro. Purified YqjH reduced ferric iron from the ferric-EDTA complex, with a specific activity of 22.0 nmol Fe2+/min/mg and a Km of 33 μM. YqjH showed no ferric reductase activity toward ferric chloride. YqjH activity was dependent on the presence of NADPH, with a Km for NADPH of 43 μM. No YqjH ferric reductase activity was detected using NADH as an electron donor, indicating that the enzyme is specific for NADPH. YqjH showed weak activity (1.0 nmol Fe2+/min/mg) toward purified ferric enterobactin. This in vitro analysis indicates that purified YqjH can function as a NADPH-dependent ferric reductase, although its in vivo substrate does not appear to be enterobactin due to its low activity toward that substrate. However, it is possible that a linear ferric enterobactin complex or iron bound to an enterobactin precursor, such as 2,3-dihydroxybenzoic acid, might be a substrate for YqjH ferric reductase activity. These possible substrates await further testing.
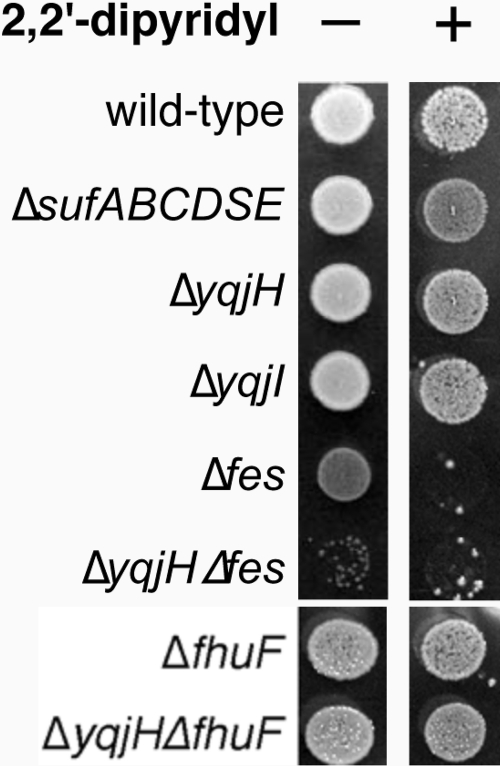
Based on its homology to viuB, we also tested if a yqjH deletion mutant is sensitive to iron starvation. A sufABCDSE deletion strain was monitored under the same conditions. The Suf pathway is a stress-responsive Fe-S cluster assembly system, and deletion of the suf operon renders E. coli sensitive to conditions that disrupt iron homeostasis, such as iron starvation and oxidative stress (41). However, on LB plates, a yqjH deletion strain was as resistant to the ferrous iron chelator 2,2′-dipyridyl as the wild-type strain (Fig. 1).
FIG. 1.
Sensitivity of various strains of E. coli to 200 μM 2,2′-dipyridyl. Overnight cultures of strains were normalized to the same OD600 and spotted on LB agar plates. Final growth was recorded after 14 h at 37°C.
Next, we tested if a yqjH deletion shows synthetic phenotypes when combined with mutations in siderophore utilization systems in E. coli. Release of ferric iron from the native E. coli siderophore enterobactin precedes through hydrolysis of the siderophore backbone by the Fes esterase, followed by an ill-defined iron reduction step and release of the iron (30). The FhuF ferric reductase is required for utilization of the nonnative iron source ferric ferrioxamine B (36). We constructed the Δfes and ΔfhuF mutations alone and in combination with ΔyqjH and examined the resulting phenotypes.
Deletion of fes leads to a slow-growth phenotype even on rich media, such as LB (Fig. 1), and produces small pink colonies due to the accumulation of ferric enterobactin. This slow-growth phenotype increases in the ΔyqjH Δfes deletion strain, suggesting that YqjH and Fes work in parallel but separate pathways. Furthermore, both Δfes and ΔyqjH Δfes strains were extremely sensitive to 2,2′-dipyridyl (Fig. 1). In contrast, neither the ΔfhuF deletion strain nor the ΔyqjH ΔfhuF deletion strain showed any growth phenotypes on rich medium and were as resistant to 2,2′-dipyridyl as the wild-type control strain. We also attempted to repeat these experiments in chemically defined minimal medium. Unfortunately, the Δfes single mutant strain grew poorly or not at all in M9 medium, making it difficult to compare relative growth phenotypes among the single and double deletion mutants of fes and yqjH (data not shown). The synthetic phenotypes observed in the ΔyqjH Δfes strain support a role for YqjH in adaptation to iron starvation but argue against a direct role for YqjH in ferric enterobactin utilization.
YqjI and Fur regulate yqjH.
A putative binding site for the iron metalloregulatory protein Fur overlaps the predicted −10 RNA polymerase binding site upstream of yqjH (Fig. 2A) (11, 37). Two repetitive extragenic palindromic (REP) elements are also present in the yqjH-yqjI intergenic region (2). The functional roles of the REP elements are unclear, although they may be able to form DNA stem-loops or serve as novel protein binding sites. It was previously shown by DNA macroarray studies that yqjH is repressed by the iron-dependent regulator Fur and that Fur repression is lost under iron starvation conditions (37). In good agreement with previous studies, we observed that a yqjH-lacZ promoter fusion construct shows 2- to 3-fold induction in LB medium in the Δfur strain or upon addition of the ferrous iron chelator dipyridyl in the wild-type strain (data not shown).
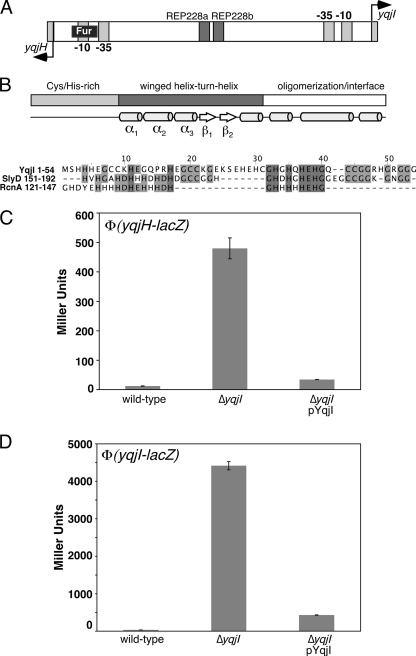
FIG. 2.
Regulation of yqjH by the YqjI transcription factor. (A) Diagram of the yqjH-yqjI intergenic region showing the putative Fur binding site and REP elements (not to scale). (B) Diagram of the YqjI protein domain organization with predicted secondary structural elements shown for each domain (43). Below the diagram is an amino acid alignment of the Cys/His-rich YqjI N terminus with SlyD and RcnA Cys/His-rich regions. The shading indicates the degree of conservation, with darker colors indicating more conserved residues. (C) Activity of the yqjH-lacZ promoter fusion in various strains of E. coli grown in LB medium. (D) Activity of the yqjI-lacZ promoter fusion in various strains of E. coli grown in LB medium. “pYqjI” is the pET21a-yqjI plasmid expressing a low level of the YqjI protein.
The previously uncharacterized gene yqjI is divergently transcribed from yqjH. The N-terminal region 1 to 54 of YqjI includes a number of potential metal-binding amino acids (12 histidines, 7 cysteines, and 7 glutamates) and is similar to the SlyD C-terminal metal-binding tail (E value of less than 0.01). The SlyD peptidyl-prolyl cis/trans isomerase is required for [NiFe] hydrogenase maturation during anaerobic growth in E. coli (53). E. coli SlyD has a C-terminal metal-binding tail containing 15 histidines, 6 cysteines, and 7 aspartates or glutamates. The YqjI N terminus is also similar to residues 121 to 147 of the RcnA Ni2+/Co2+ efflux transporter that are located on a loop between predicted transmembrane domains 3 and 4 and are part of the histidine-rich signature motif that characterizes major facilitator superfamily (MFS) transporters that efflux nickel (Fig. 2B) (46). At present, the exact functional role of this region in RcnA-mediated nickel efflux is unclear.
Residues 57 to 207 of YqjI are similar to those of the PadR family of winged helix-turn-helix (wHTH) transcription factors (InterPro accession no. IPR005149) (34). PadR is a transcriptional repressor that controls genes involved in the phenolic acid stress response in some microorganisms (19). A model three-dimensional structure of YqjI was calculated using the crystal structure of AphA, a PadR homologue from V. cholerae, as a template (16, 44). The comparison to AphA suggests that residues 62 to 152 of YqjI constitute a winged helix fold DNA-binding domain, while residues 153 to 206 form an oligomerization and/or ligand-binding domain (Fig. 2B) (16).
Based on the close proximity of the potential YqjI regulator, we measured the transcriptional activity of the yqjH-lacZ promoter fusion in wild-type and ΔyqjI strains. Deletion of yqjI resulted in a 30-fold increase in the basal expression of yqjH over wild-type levels in LB (Fig. 2C). Introduction of a plasmid expressing a low basal level of yqjI (pYqjI) (Fig. 2C) in the ΔyqjI strain reduced transcriptional expression of yqjH to much closer to wild-type levels, confirming that the YqjI protein is required to fully repress yqjH expression. Together, these results confirm that both Fur and YqjI repress transcription of yqjH.
Since many regulators can autoregulate their own expression, we also measured the activity of a yqjI-lacZ promoter fusion in the ΔyqjI strain. We found that basal expression of yqjI-lacZ in LB is increased by 130-fold in the ΔyqjI strain (Fig. 2D). YqjI provided in trans on a plasmid was able to reduce transcription from 130-fold to 12-fold over wild-type levels in the ΔyqjI strain (Fig. 2D). The residual induction of yqjI in the complementation experiment may be due to low expression of YqjI from the plasmid construct (which is noninducible in this strain). This result indicates that YqjI also represses its own transcription.
Metal-dependent regulation of the yqjH promoter.
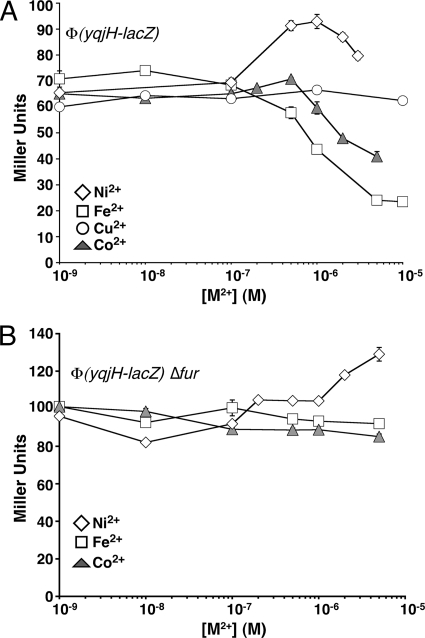
Based on the observed regulation by Fur, the presence of a potential nickel-binding region within the YqjI transcriptional regulator, and the observed regulation of yqjH and yqjI by YqjI, we tested if expression of yqjH is altered by transition metal ions in vivo. We conducted these experiments in chemically defined M9 minimal medium, with glucose as the carbon source. For cells grown in M9/glucose minimal medium, the basal expression of yqjH-lacZ increased 6-fold (from 10 to 60 Miller units) compared to that of cells grown in LB. This difference in basal expression is likely due to the larger amount of iron present in LB (about 5 μM) than in minimal medium (below 300 nM) (40), which leads to lower yqjH expression levels in cells grown in LB due to increased repression by Fe2+-Fur.
Addition of Fe2+ decreased yqjH expression starting at 100 nM, with a maximum repression of 4-fold occurring at 10 μM Fe2+ (Fig. 3A). Addition of CoCl2 concentrations of more than 500 nM also repressed yqjH expression by about 50% (Fig. 3A). The iron- and cobalt-dependent repression of yqjH in M9/glucose medium was abolished in the Δfur strain (Fig. 3B), indicating that the Fur metalloregulatory protein mediates iron and cobalt regulation of yqjH. Although Fur primarily regulates iron homeostasis, Fur can respond to other divalent metals, including Co2+, in vivo and in vitro (3, 15).
FIG. 3.
Metal-responsive regulation of yqjH. Activity of the yqjH-lacZ promoter fusion was measured in triplicate in various strains of E. coli grown in M9/glucose medium at 37°C with increasing amounts of various divalent metals. (A) Ni2+, Fe2+, Cu2+, or Co2+ was added to the wild-type strain. (B) Fe2+, Co2+, or Ni2+ was added to the Δfur strain. Lines are for emphasis only and are not fits.
Upon addition of NiCl2, yqjH-lacZ expression increased starting at 100 nM NiCl2, with maximum induction by 1 μM NiCl2 (Fig. 3A). NiCl2 induced yqjH expression by 42% over basal levels. In contrast, CuCl2 did not induce yqjH expression even at levels of up to 100 μM (Fig. 3A). The nickel-dependent induction of yqjH still occurred in the Δfur strain, demonstrating that Fur does not provide nickel-dependent regulation of yqjH (Fig. 3B).
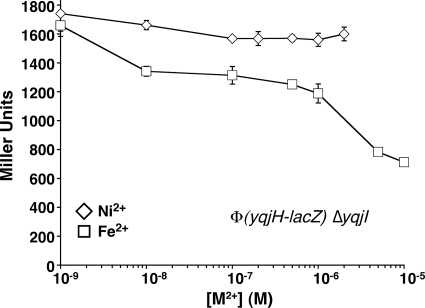
Deletion of yqjI led to constitutive expression levels of yqjH that were 30-fold higher than wild-type basal expression levels in M9/glucose medium (Fig. 4). Furthermore, deletion of yqjI abolished nickel-responsive regulation of yqjH (Fig. 4). In contrast, the iron-dependent repression of yqjH still occurred in the ΔyqjI strain over the same concentration range of added iron (Fig. 4). Together, our results indicate that YqjI is required for nickel-dependent regulation of yqjH, while Fur is required for iron- and cobalt-dependent regulation of yqjH. Since YqjI was seen to repress its own expression (Fig. 2D), we also tested to see if nickel addition alters expression of yqjI in vivo. Addition of nickel led to a maximum 6-fold increase in yqjI expression (see Fig. S2 in the supplemental material). This result further indicates that YqjI repression of target promoters is somehow responsive to elevated nickel in vivo.
FIG. 4.
Role of YqjI in yqjH nickel regulation. Activity of the yqjH-lacZ promoter fusion was measured in triplicate in the ΔyqjI strain grown in M9/glucose medium with increasing amounts of Ni2+ or Fe2+. Lines are for emphasis only and are not fits.
Deletion of rcnA, which encodes the RcnA nickel efflux transporter, leads to increased nickel sensitivity in E. coli (46). To determine if disruption of nickel efflux alters yqjH nickel regulation, we monitored yqjH expression upon nickel addition in both wild-type and ΔrcnA strains (see Fig. S3 in the supplemental material). Basal expression of yqjH in the ΔrcnA strain in minimal medium did not change. However, yqjH expression was more highly induced at lower nickel concentrations in the ΔrcnA strain than that observed in the wild-type strain (see Fig. S3). This result confirms that yqjH expression is responsive to physiologically relevant changes in E. coli nickel homeostasis. Based on the altered regulation in the ΔrcnA strain, it seems likely that YqjI, which provides nickel-responsive regulation of yqjH, responds to the same intracellular pool of nickel transported by RcnA (25).
YqjI binds to target sequences in the yqjH-yqjI intergenic region.
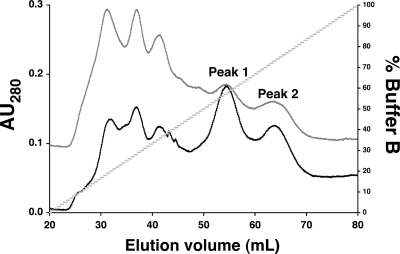
To characterize the DNA-binding activity of YqjI, we overexpressed and purified wild-type YqjI in E. coli. Recombinant YqjI protein, expressed in E. coli, eluted on a heparin column in two peaks (Fig. 5). The first peak to elute on the heparin column was separately retained and analyzed by gel filtration chromatography. Approximately 90% of YqjI from the first heparin peak eluted at an apparent molecular weight of 28,000. About 10% of YqjI eluted as a higher-molecular-weight species at above 100,000. Since YqjI has a theoretical molecular weight of 23,401, the first heparin peak likely contained predominantly the monomer form of YqjI. The second YqjI peak to elute from the heparin column was similarly analyzed by gel filtration analysis. Approximately 90% of YqjI from the second heparin peak eluted at an apparent molecular weight of 179,000. While this apparent molecular weight would be most consistent with a hexamer of YqjI, most PadR family members form dimer or tetramer species. The gel filtration data do not allow us to determine the exact stoichiometry of the YqjI oligomer. Based on YqjI similarity to other PadR family members, we used the theoretical molecular weight of the YqjI tetramer for all molar calculations but refer to this complex as an YqjI oligomer.
FIG. 5.
Purification of YqjI. Overexpressed YqjI eluted as two peaks during heparin column chromatography (gray trace). The addition of 500 μM NiCl2 to LB during cell growth and protein expression did not alter the subsequent YqjI elution profile (black trace). The first YqjI elution peak contained monomeric YqjI, while the second elution peak contained an oligomeric form of YqjI that is likely tetrameric or a higher quaternary structure (as subsequently determined by gel filtration chromatography). AU280, absorbance at 280 nm.
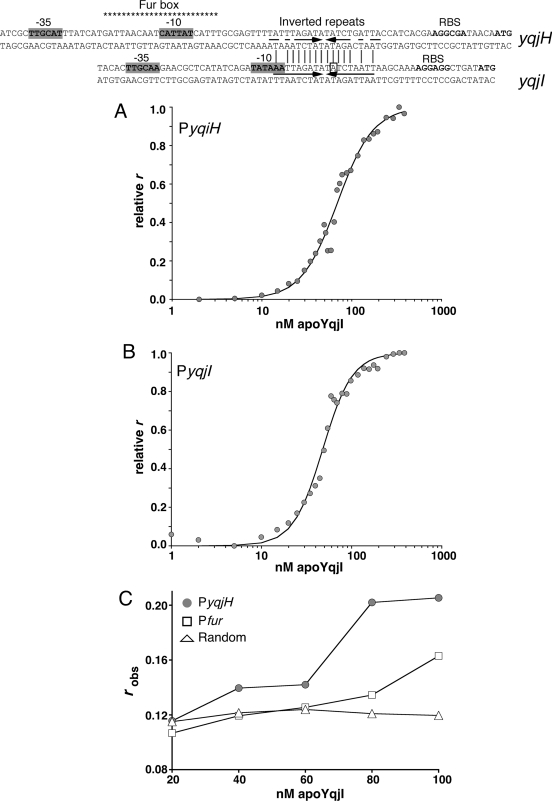
Close examination of the yqjH-yqjI intergenic region revealed two highly similar inverted repeats located near the predicted −10 RNA polymerase binding sites for both the yqjH and yqjI promoters (referred to as PyqjH and PyqjI) (Fig. 6). Since both the yqjH and yqjI promoters are repressed by YqjI, we tested if these inverted repeats act as binding sites for YqjI. Both monomer and oligomer forms of apo-YqjI were incubated with fluorescent oligonucleotides matching the inverted repeat sequences, and the change in fluorescence anisotropy (FA) was measured (Fig. 6). The oligomer form of apo-YqjI bound the PyqjH and PyqjI oligonucleotides, with Kd (dissociation constant) values of 68.9 nM for PyqjH (Fig. 6A) and 47.8 nM for PyqjI (Fig. 6B). In contrast, the monomer form of apo-YqjI showed only weak binding to the oligonucleotides (data not shown). As controls for nonspecific DNA binding, we also incubated oligomer apo-YqjI with a random oligonucleotide with the same length as PyqjH or with an oligonucleotide matching the Fur binding site upstream of yqjH. Apo-YqjI showed only weak interaction with these control oligonucleotides (Fig. 6C). Since the apo-YqjI oligomer binds PyqjH and PyqjI, these inverted repeat sequences are strong candidates for the apo-YqjI binding sites within the yqjH and yqjI promoters.
FIG. 6.
(Top) Alignment of inverted repeats found in the yqjH and yqjI promoters (indicated by arrows). The putative Fur box in the yqjH promoter is shown with asterisks. The previously mapped +1 transcriptional start site of yqjI is indicated with a box (38). (A, B) Oligomer apo-YqjI binding to 10 nM PyqjH (A) or PyqjI (B) oligonucleotides, as measured by DNA fluorescence anisotropy. Boldface lines indicate fits to binding data calculated as described in Materials and Methods. (C) Binding of oligomer apo-YqjI to 10 nM PyqjH oligonucleotide (gray circle), random-sequence oligonucleotide (open triangle), or Pfur oligonucleotide (open square). Lines are for emphasis only and are not fits.
Divalent metals can regulate YqjI DNA-binding activity in vitro.
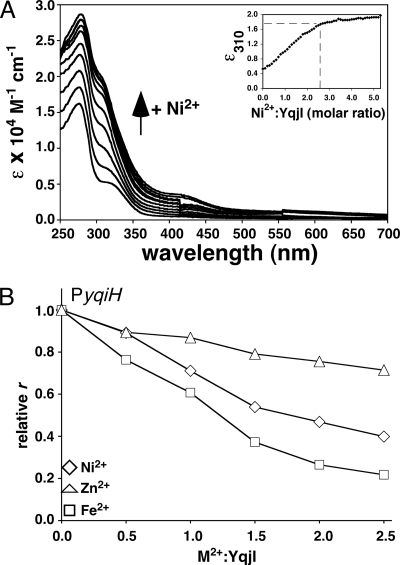
The potential nickel-binding N terminus of YqjI and the in vivo nickel-responsive regulation of yqjH suggest that YqjI activity may be directly or indirectly regulated by divalent metals. The metal content of as-purified YqjI was measured to determine if YqjI is a metalloprotein. The Ni2+/YqjI ratio was 0.06 for the monomer form and 0.15 for the oligomer form if YqjI was expressed in cells grown in standard LB, which contains approximately 120 nM Ni2+ (40). If cells were grown in LB supplemented with 500 μM nickel chloride, the Ni2+/YqjI ratio increased to 1.1 for the monomer form and 1.6 for the oligomer form of YqjI. All forms of YqjI (monomer and oligomer) also contained zinc at a Zn2+/YqjI ratio of 0.5 ± 0.1, regardless of the nickel content of the medium. Attempts to remove all bound metals by overnight incubation of YqjI with EDTA resulted in protein precipitation. Addition of nickel to the LB medium during cell growth did not alter the separation of YqjI into monomer and oligomer peaks during purification on the heparin column (Fig. 5), and stoichiometric nickel addition to the apo-YqjI oligomer did not alter its oligomeric state, as monitored by gel filtration chromatography (data not shown).
To better define the maximum nickel stoichiometry of YqjI, 20 μM of the apo-YqjI oligomer (containing 0.15 Ni2+ and 0.5 Zn2+ per monomer) was titrated with increasing nickel concentrations. Upon nickel addition, the UV-visible absorption spectrum of YqjI showed peaks at 280 nm and 310 nm, consistent with S− → Ni(II) or imidazole− → Ni(II) charge transfer transitions from Cys-Ni or His-Ni coordination (Fig. 7A). The absorption intensity at 310 nm began to saturate as 2 to 3 molar equivalents of nickel were added (Fig. 7A). Similar spectra were observed for nickel titration of the apo-YqjI monomer (data not shown). After completion of the nickel titration, excess nickel was removed from YqjI by thorough buffer exchange. The buffer-exchanged, Ni-reconstituted YqjI retained 2.1 ± 0.02 Ni2+ per YqjI monomer. Based on the UV-visible absorption spectra and the nickel content of buffer-exchanged YqjI, the YqjI protein can accommodate approximately 2 Ni2+ per monomer in vitro. In comparison, the nickel stoichiometry for full-length SlyD (as measured by equilibrium dialysis) is 4.2 per monomer (27).
FIG. 7.
(A) UV-visible absorption spectra for apo-YqjI titrated with up to 5 molar equivalents of nickel. The inset shows the change in absorbance at 310 nm (ɛ310) with increasing NiCl2 addition. Metal-to-protein ratios were calculated using the monomer molecular weight of YqjI. (B) Effects of divalent metal addition on binding of 40 nM apo-YqjI to 10 nM PyqjH. Lines are not fits and are for emphasis only.
The UV-visible absorption spectra of Ni-YqjI in the 270- to 400-nm region could imply tetrahedral Ni(II) coordination (10, 24). Although the absence of significant absorbance in the far visible region (beyond 600 nm) would seem to argue against tetrahedral Ni coordination, this characteristic absorbance can be masked in some tetrahedral Ni complexes (51). In addition, the presence of a shoulder at around 420 nm is attributed to d-d transitions that are similar to published square planar Ni(II) complexes (10). The complexity of the UV-visible absorption spectra for Ni-YqjI suggests multiple nickel-binding sites with different ligands and/or geometries. Recent X-ray absorption spectroscopy (XAS) and extended X-ray absorption fine-structure (EXAFS) spectroscopy of Ni-SlyD also show a mixture of nickel sites with different geometries and coordination numbers, indicating that the Ni-binding motifs of YqjI and SlyD are flexible in their ability to accommodate nickel (27).
We also tested if excess zinc is capable of displacing nickel bound to YqjI. Ni-reconstituted YqjI containing 2 Ni2+ per monomer was titrated with increasing concentrations of zinc. We observed no change in the UV-visible spectrum of Ni-reconstituted YqjI, as zinc was added at up to a 4-fold molar excess over YqjI protein (equivalent to a 2-fold molar excess over bound nickel), indicating excess zinc was unable to displace the bound nickel (data not shown). We were unable to continue the zinc titration above a 4-fold molar excess over the YqjI protein, as we observed protein precipitation at these levels of zinc.
Based on the in vitro nickel binding by apo-YqjI, we determined if the presence of nickel alters oligomer YqjI binding to PyqjH or PyqjI binding sites. As increasing levels of NiCl2 were titrated into samples containing apo-YqjI and the PyqjH oligonucleotide, the FA decreased, indicating that nickel causes the dissociation of YqjI from PyqjH (Fig. 7B). A 50% decrease in FA occurs at a Ni2+/YqjI ratio of 2:1, consistent with the nickel stoichiometry of 2:1 obtained after in vitro nickel reconstitution of apo-YqjI. We also observed that addition of ZnCl2 had a weak effect on the FA, reducing it by approximately 20% (Fig. 7B). However, Fe2+ addition (as anaerobically prepared ferrous ammonium sulfate) decreased the FA by nearly 80%, indicating it was more effective than nickel at reducing the YqjI affinity for DNA (Fig. 7B). These results indicate that divalent metals such as Ni2+ and Fe2+ have a similar negative effect on YqjI DNA-binding activity in vitro, although the observed nickel regulation parallels the in vivo nickel-specific YqjI-dependent regulation of the yqjH promoter.
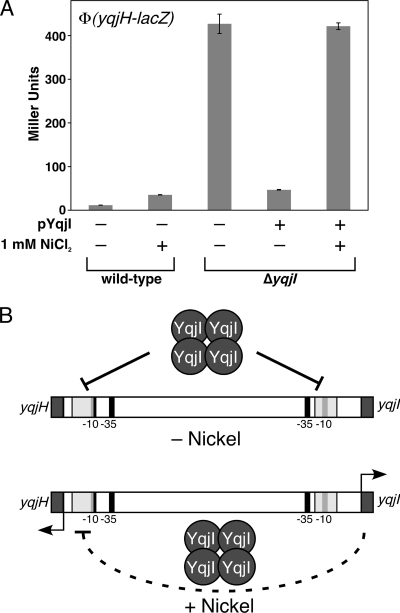
YqjI autoregulation may lead to dampening of yqjH nickel induction.
Although nickel can regulate YqjI DNA-binding activity in vitro, the nickel induction of the yqjH and yqjI promoters is rather mild in vivo, especially compared to the high constitutive expression observed in the ΔyqjI strain (Fig. 2D). However, the autoregulation of yqjI indicates that increased nickel levels block YqjI repression of yqjI transcription, thereby causing levels of the YqjI apoprotein to increase. Increased apo-YqjI then counteracts the loss of YqjI repression at the yqjH promoter (Fig. 8 B). To test if this regulatory loop is occurring in vivo, one must uncouple expression of YqjI from its native promoter so that the addition of nickel will no longer lead to increased YqjI expression. Nickel induction of yqjH can then be measured without the added complication of YqjI autoregulation.
FIG. 8.
Nickel-responsive autoregulation of yqjI. (A) Activity of the yqjH-lacZ promoter fusion was measured in triplicate in wild-type or ΔyqjI strains of E. coli with or without pYqjI grown in LB medium at 37°C with or without 1 mM nickel. “pYqjI” is the pET21a-yqjI plasmid expressing a low level of the YqjI protein. (B) Proposed negative feedback regulation of yqjH under nickel stress.
To accomplish this, we utilized the ΔyqjI strain carrying a plasmid that expresses a low basal level of yqjI (pYqjI) (Fig. 2). In this strain, yqjI expression is uncoupled from its native promoter and is driven only by leaky transcription from the plasmid-borne T7 promoter upstream of yqjI, which is adequate to repress yqjH and yqjI transcription nearly to the levels observed in the wild-type strain (Fig. 2). To match earlier complementation studies, these experiments were carried out in LB medium, which has an excess capacity to chelate divalent metals compared to that of minimal medium. Larger amounts of nickel were required to observe nickel induction of yqjH (see Fig. S4 in the supplemental material). Upon addition of 1 mM nickel to the wild-type strain (without the pYqjI plasmid), yqjH expression increased approximately 3-fold (Fig. 8A). In the ΔyqjI strain carrying the pYqjI plasmid, addition of 1 mM nickel increased yqjH expression to the same high level that was observed in the complete absence of yqjI (compare Fig. 8A to 2C). This result indicates that nickel addition is able to completely block YqjI repression of yqjH so long as YqjI protein levels are not allowed to increase due to the YqjI autoregulatory circuit (Fig. 8B) or differential regulation of YqjI by other factors.
YqjH helps maintain iron homeostasis under excess nickel conditions.
To determine if yqjH is required for resistance to nickel toxicity, we analyzed the growth phenotype of a ΔyqjH strain in response to increasing nickel in M9/gluconate minimal medium. Cells grown on gluconate are sensitized to environmental stresses that perturb iron homeostasis due to the resulting block in the [4Fe-4S] enzyme phosphogluconate dehydratase, making this a useful growth medium for testing if nickel directly disrupts iron homeostasis (17, 41). For comparison, we also characterized the phenotype of a ΔrcnA strain under the same conditions, since the ΔrcnA strain is known to be sensitive to elevated nickel when grown in minimal medium (46). In addition, we also characterized a sufABCDSE deletion strain under the same conditions. Deletion of suf makes E. coli more sensitive to excess levels of copper and cobalt that appear to disrupt iron metabolism by competing with iron for incorporation into metalloenzymes (32, 33, 45).
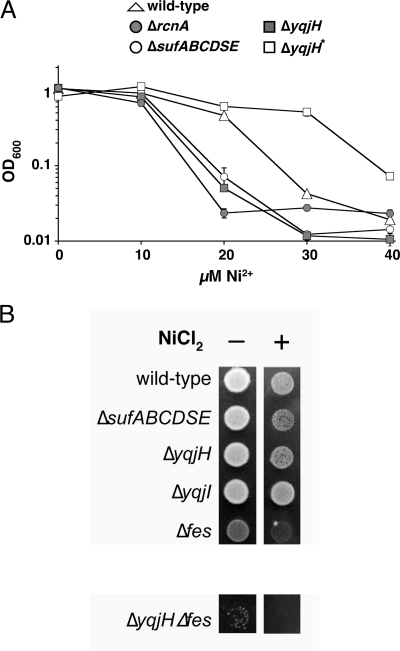
The ΔrcnA strain was more sensitive to elevated nickel in M9/gluconate minimal medium than the wild-type strain (Fig. 9A). The ΔsufABCDSE strain also was sensitive to elevated nickel compared to the wild-type strain (Fig. 9A), indicating that iron homeostasis and/or Fe-S cluster metabolism is disrupted in response to elevated nickel in E. coli. During our attempts to characterize the nickel sensitivity of the ΔyqjH strain, we encountered a highly variable phenotype. In approximately half of the independent growth assays, the ΔyqjH strain was as sensitive to elevated nickel as the ΔrcnA strain (Fig. 9A). However, in the other half of the assays, the ΔyqjH strain was more nickel resistant than the wild-type strain. We refer to this apparent suppressor phenotype as ΔyqjH* (Fig. 9A). When ΔyqjH* cells were isolated from the high-nickel medium and retested for nickel sensitivity, they uniformly grew the same or better than the wild-type control strain in response to elevated nickel, suggesting the selection of a stable suppressor mutation(s) (data not shown). We confirmed that both ΔyqjH and ΔyqjH* still contained the actual yqjH deletion using colony PCR with primers flanking the site of the deletion (data not shown). The variable nickel sensitivity phenotype was always observed, even when we independently reconstructed and retested the ΔyqjH strain. The variable nickel sensitivity phenotype indicates a high level of suppressor mutations in the ΔyqjH strain in response to nickel toxicity in M9/gluconate minimal medium. At present, the exact nature of the suppressor mutation(s) is unclear and is the subject of ongoing studies.
FIG. 9.
(A) Sensitivity of various strains of E. coli to nickel. Strains were normalized to the same OD600 and inoculated in triplicate into M9/gluconate minimal medium with increasing levels of NiCl2. Final growth was recorded after 24 h at 37°C. (B) Overnight cultures of various strains of E. coli were normalized to the same OD600 and spotted on LB agar plates containing 1 mM NiCl2. Final growth was recorded after 14 h at 37°C. The ΔyqjH* strain contains unknown secondary mutations that suppress the ΔyqjH phenotype and render it nickel resistant.
We also examined the phenotypes of the various gene deletion strains under high-nickel conditions on LB medium. Although the nickel sensitivity phenotypes of all strains grown on LB were reduced compared to those of strains grown on gluconate minimal medium, we observed that the sufABCDSE deletion strain was sensitive to high-nickel concentrations, indicating the disruption of Fe-S cluster biosynthesis or increased turnover of Fe-S enzymes (Fig. 9B). A Δfes strain was also sensitive to high-nickel conditions (Fig. 9B). The yqjH deletion strain showed a degree of nickel sensitivity similar to that of ΔsufABCDSE. However, the ΔyqjH Δfes double mutant strain showed a higher sensitivity to high-nickel stress than either the yqjH or fes single deletion strains (Fig. 9B). The severe phenotype of the ΔyqjH Δfes double mutant strain is consistent with parallel but separate physiological roles for YqjH and Fes during nickel stress. The yqjI gene deletion strain grew as well as the wild-type control strain.
DISCUSSION
YqjI is the main regulator of yqjH and yqjI transcription.
This study shows that YqjI is the primary regulator of yqjH transcription, since deletion of yqjI leads to a 40-fold increase in yqjH basal expression while deletion of fur derepresses yqjH by only 3-fold. A close regulatory connection between yqjH and yqjI is also supported by phylogenetic analysis. The yqjI and yqjH genes are found in close proximity in the Gammaproteobacteria and Betaproteobacteria subclasses of the Proteobacteria (see Fig. S5 in the supplemental material). In most Enterobacteriaceae, the genes are oriented divergently to each other, just as in E. coli. However, in Pseudomonas and Xanthomonas Gammaproteobacteria as well as in Ralstonia and Burkholderia spp., yqjI and yqjH are transcribed in the same direction rather than divergently (see Fig. S5). The N-terminal, SlyD-like region of E. coli YqjI is conserved mainly in species closely related to E. coli. However, Ralstonia metallidurans and Pseudomonas syringae each contain a YqjI homologue with a N-terminal extension that is histidine rich but lacks the cysteine residues present in E. coli YqjI (data not shown).
What are the key regulatory signals for YqjI in vivo?
The nickel-dependent regulation of yqjH and yqjI by YqjI is physiologically consistent with the nickel homeostasis systems in E. coli that are regulated by the NikR and RcnR transcription factors. The nikABCDE nickel import system is repressed by Ni-NikR as nickel concentrations rise above 100 nM in M63/glucose minimal medium (25). Repression of the Ni2+/Co2+ efflux transporter RcnA by apo-RcnR is relieved in M63/glucose minimal medium as nickel levels increase above 500 nM (25). Together, these previously published expression profiles indicate that nickel-responsive gene regulation in E. coli occurs over a concentration range of 100 to 500 nM nickel in minimal medium. Nickel-dependent regulation of yqjH by YqjI in minimal medium occurs beginning at nickel concentrations above 100 nM, with maximum induction between 500 nM and 1 μM. Since yqjH induction occurs at the high end of the regulatory concentration range, this suggests that YqjH could play a role in cellular adaptation to nickel toxicity.
The in vivo YqjI-dependent nickel-responsive regulation of yqjH and the in vitro nickel binding by YqjI suggest a model in which YqjI may regulate yqjH directly in response to nickel levels in order to integrate iron and nickel homeostasis. Such cross-regulation has been observed in species like Helicobacter pylori, which utilizes the NikR regulator to coordinate expression of both iron and nickel metal homeostasis systems (13). This cross-regulation may be important in H. pylori due to the high levels of nickel required to activate the highly abundant urease enzyme needed for cell survival at low pH (5). Previous studies in E. coli also have shown that rcnA may be regulated by Fur in response to iron and that the RcnR protein (that represses rcnA expression) is itself induced by iron via a Fur-independent pathway (29). Presumably, this cross-regulation of rcnA and rcnR by iron provides a careful balance between iron and nickel uptake and efflux to maintain enzyme maturation without poisoning metal homeostasis pathways.
However, full nickel induction of yqjH to the same expression levels observed in the ΔyqjI strain could be achieved only if YqjI expression was uncoupled from autoregulation (Fig. 8A). This autoregulatory circuit could indicate that nickel is not the only regulatory signal that controls YqjI DNA-binding activity in vivo. Multiple signal inputs may be required to fully reverse YqjI repression of yqjH in vivo in the wild-type background. Possibly the regulation of yqjI by other regulators could act indirectly to control yqjH transcription. It was previously reported that deletion of the IscR Fe-S regulatory protein leads to upregulation of yqjI, suggesting that IscR may normally repress yqjI expression (18). It is likely that the loss of IscR repression under nickel stress (when iron and Fe-S homeostasis are disrupted) leads to increased expression of apo-YqjI. It was also shown by DNA microarray experiments that YqjI expression is lower in the Δfnr strain, although the significance of this result under aerobic conditions is unclear (12).
We clearly observed that the DNA-binding activity of YqjI was significantly diminished in the presence of Ni2+ or Fe2+ (and mildly altered by Zn2+), indicating that YqjI can be regulated in vitro by multiple metals. However, the in vivo results show that YqjI regulates yqjH specifically (although somewhat weakly) in response to nickel. Zinc addition of up to 100 μM did not alter yqjH expression (data not shown), while iron and cobalt regulation of yqjH was mediated solely through Fur (Fig. 3B). The metal-binding domain of SlyD, which is similar to the YqjI N terminus, also can bind multiple metals in vitro, but the ΔslyD phenotype is specifically rescued by nickel addition in vivo (22, 27, 52). The exact biochemical function of the SlyD C-terminal tail is unknown, but the C terminus is necessary for metal-dependent inhibition of SlyD peptidyl-prolyl isomerase (PPIase) activity, suggesting that it could function as a metal-responsive switch to control SlyD activity or interactions with substrate proteins (22, 52, 53).
One regulatory model, suggested by the in vivo regulation, in vivo phenotype, and in vitro metal-binding studies, shows that nickel or other ligands act as a molecular switch to negatively regulate YqjI DNA-binding activity by directly binding the N-terminal SlyD-like domain. However, at present we cannot conclude that nickel directly binds YqjI to regulate DNA-binding activity in vivo. It is possible that disruption of iron homeostasis by elevated nickel indirectly affects YqjI activity through an unknown mechanism.
Nickel disrupts iron homeostasis in E. coli.
Strains lacking yqjH are sensitive to nickel toxicity (Fig. 9). In addition, we observed that a ΔsufABCDSE strain, lacking a stress-response Fe-S biosynthesis pathway, is also sensitive to elevated nickel levels (Fig. 9). Together, these results indicate that iron homeostasis can be disrupted by elevated nickel levels, providing a physiological rationale for cross-regulation of iron and nickel homeostasis in E. coli. It is not surprising that nickel can disrupt iron homeostasis since the Irving-Williams series (Mn < Fe < Co < Ni < Cu > Zn) predicts that nickel, cobalt, and copper should bind more strongly to potential cellular ligands than ferrous iron. Indeed, cobalt and copper have already been shown to disrupt iron metabolism in bacteria (23, 32, 45, 50).
Previous studies in higher eukaryotes have clearly shown that nickel competes directly with iron for cell entry through the DMT1 divalent metal transporter, leading to decreased cellular iron accumulation (8). More recent work indicates that nickel can compete directly with iron for incorporation into 2-oxoglutarate-dependent histone demethylases, suggesting that misincorporation of nickel into iron metalloproteins is a another facet of nickel toxicity (9). The exact mechanism of nickel toxicity in E. coli remains to be elucidated and may involve disrupted iron transport and/or direct inhibition of iron metalloproteins by nickel.
Regardless of the mechanism of nickel toxicity, the YqjH ferric reductase appears to play a role in maintaining iron homeostasis under nickel stress. Combination of the yqjH deletion with a fes deletion leads to a synthetic phenotype in LB and under high nickel stress. The weak activity of YqjH toward purified enterobactin and the synthetic phenotype with Δfes argues against a direct role for YqjH in the enterobactin utilization pathway. YqjH may be used to reduce ferric iron from another iron siderophore or to release iron from ferric iron storage proteins, such as ferritin, under low-iron conditions. Further studies are needed to clarify the in vivo substrate for the YqjH ferric reductase.
These studies establish that the PadR family member YqjI represses the transcription of yqjH and yqjI in vivo. YqjI repression of these target promoters is weakly responsive to elevated nickel in vivo, although YqjI DNA-binding activity is clearly altered by transition metals in vitro. Future experiments will focus on identifying other signals and/or regulators that may control YqjI DNA-binding activity in vivo.
Supplementary Material
Acknowledgments
We thank Caryn E. Outten for helpful comments and reading of the manuscript.
This work was supported by the University of South Carolina Research Foundation and grant MCB 1022288 from the National Science Foundation (to F.W.O.).
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. [DOI] [PubMed] [Google Scholar]
- 2.Bachellier, S., J. M. Clement, and M. Hofnung. 1999. Short palindromic repetitive DNA elements in enterobacteria: a survey. Res. Microbiol. 150:627-639. [DOI] [PubMed] [Google Scholar]
- 3.Bagg, A., and J. B. Neilands. 1987. Ferric uptake regulation protein acts as a repressor, employing iron (II) as a cofactor to bind the operator of an iron transport operon in. Escherichia coli. Biochemistry 26:5471-5477. [DOI] [PubMed] [Google Scholar]
- 4.Bamford, V. A., et al. 2008. Preliminary X-ray diffraction analysis of YqjH from Escherichia coli: a putative cytoplasmic ferri-siderophore reductase. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 64:792-796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bauerfeind, P., R. Garner, B. E. Dunn, and H. L. Mobley. 1997. Synthesis and activity of Helicobacter pylori urease and catalase at low pH. Gut 40:25-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown, A. M. 2001. A step-by-step guide to non-linear regression analysis of experimental data using a Microsoft Excel spreadsheet. Comput. Methods Programs Biomed. 65:191-200. [DOI] [PubMed] [Google Scholar]
- 7.Butterton, J. R., and S. B. Calderwood. 1994. Identification, cloning, and sequencing of a gene required for ferric vibriobactin utilization by Vibrio cholerae. J. Bacteriol. 176:5631-5638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen, H., T. Davidson, S. Singleton, M. D. Garrick, and M. Costa. 2005. Nickel decreases cellular iron level and converts cytosolic aconitase to iron-regulatory protein 1 in A549 cells. Toxicol. Appl. Pharmacol. 206:275-287. [DOI] [PubMed] [Google Scholar]
- 9.Chen, H., et al. 2010. Nickel ions inhibit histone demethylase JMJD1A and DNA repair enzyme ABH2 by replacing the ferrous iron in the catalytic centers. J. Biol. Chem. 285:7374-7383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, X., M. Chu, and D. P. Giedroc. 2000. Spectroscopic characterization of Co(II)-, Ni(II)-, and Cd(II)-substituted wild-type and non-native retroviral-type zinc finger peptides. J. Biol. Inorg. Chem. 5:93-101. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Z., et al. 2007. Discovery of Fur binding site clusters in Escherichia coli by information theory models. Nucleic Acids Res. 35:6762-6777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Constantinidou, C., et al. 2006. A reassessment of the FNR regulon and transcriptomic analysis of the effects of nitrate, nitrite, NarXL, and NarQP as Escherichia coli K12 adapts from aerobic to anaerobic growth. J. Biol. Chem. 281:4802-4815. [DOI] [PubMed] [Google Scholar]
- 13.Contreras, M., J. M. Thiberge, M. A. Mandrand-Berthelot, and A. Labigne. 2003. Characterization of the roles of NikR, a nickel-responsive pleiotropic autoregulator of Helicobacter pylori. Mol. Microbiol. 49:947-963. [DOI] [PubMed] [Google Scholar]
- 14.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lorenzo, V., S. Wee, M. Herrero, and J. B. Neilands. 1987. Operator sequences of the aerobactin operon of plasmid ColV-K30 binding the ferric uptake regulation (fur) repressor. J. Bacteriol. 169:2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.De Silva, R. S., et al. 2005. Crystal structure of the virulence gene activator AphA from Vibrio cholerae reveals it is a novel member of the winged helix transcription factor superfamily. J. Biol. Chem. 280:13779-13783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gardner, P. R., and I. Fridovich. 1991. Superoxide sensitivity of the Escherichia coli 6-phosphogluconate dehydratase. J. Biol. Chem. 266:1478-1483. [PubMed] [Google Scholar]
- 18.Giel, J. L., D. Rodionov, M. Z. Liu, F. R. Blattner, and P. J. Kiley. 2006. IscR-dependent gene expression links iron-sulphur cluster assembly to the control of O2-regulated genes in Escherichia coli. Mol. Microbiol. 60:1058-1075. [DOI] [PubMed] [Google Scholar]
- 19.Gury, K., L. Barthelmebs, N. P. Tran, C. Divies, and J. F. Cavin. 2004. Cloning, deletion, and characterization of PadR, the transcriptional repressor of the phenolic acid decarboxylase-encoding padA gene of Lactobacillus plantarum. Appl. Environ. Microbiol. 70:2146-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hantke, K. 1984. Cloning of the repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol. Gen. Genet. 197:337-341. [DOI] [PubMed] [Google Scholar]
- 21.Hantke, K. 2001. Iron and metal regulation in bacteria. Curr. Opin. Microbiol. 4:172-177. [DOI] [PubMed] [Google Scholar]
- 22.Hottenrott, S., T. Schumann, A. Pluckthun, G. Fischer, and J. U. Rahfeld. 1997. The Escherichia coli SlyD is a metal ion-regulated peptidyl-prolyl cis/trans-isomerase. J. Biol. Chem. 272:15697-15701. [DOI] [PubMed] [Google Scholar]
- 23.Irving, H., and R. J. P. Williams. 1948. Order of stability of metal complexes. Nature 162:746-747. [Google Scholar]
- 24.Iwig, J. S., S. Leitch, R. W. Herbst, M. J. Maroney, and P. T. Chivers. 2008. Ni(II) and Co(II) sensing by Escherichia coli RcnR. J. Am. Chem. Soc. 130:7592-7606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Iwig, J. S., J. L. Rowe, and P. T. Chivers. 2006. Nickel homeostasis in Escherichia coli—the rcnR-rcnA efflux pathway and its linkage to NikR function. Mol. Microbiol. 62:252-262. [DOI] [PubMed] [Google Scholar]
- 26.Jones, T., R. Spencer, and C. Walsh. 1978. Mechanism and kinetics of iron release from ferritin by dihydroflavins and dihydroflavin analogues. Biochemistry 17:4011-4017. [DOI] [PubMed] [Google Scholar]
- 27.Kaluarachchi, H., et al. 2009. The Ni(II)-binding properties of the metallochaperone SlyD. J. Am. Chem. Soc. 131:18489-18500. [DOI] [PubMed] [Google Scholar]
- 28.Kang, Y., K. D. Weber, Y. Qiu, P. J. Kiley, and F. R. Blattner. 2005. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J. Bacteriol. 187:1135-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Koch, D., D. H. Nies, and G. Grass. 2007. The RcnRA (YohLM) system of Escherichia coli: a connection between nickel, cobalt and iron homeostasis. Biometals 20:759-771. [DOI] [PubMed] [Google Scholar]
- 30.Lin, H., M. A. Fischbach, D. R. Liu, and C. T. Walsh. 2005. In vitro characterization of salmochelin and enterobactin trilactone hydrolases IroD, IroE, and Fes. J. Am. Chem. Soc. 127:11075-11084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Macheroux, P. 1999. UV-visible spectroscopy as a tool to study flavoproteins, p. 1-8. In S. K. Chapman and G. A. Reid (ed.), Flavoprotein protocols, vol. 131. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 32.Macomber, L., and J. A. Imlay. 2009. The iron-sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U. S. A. 106:8344-8349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Macomber, L., C. Rensing, and J. A. Imlay. 2007. Intracellular copper does not catalyze the formation of oxidative DNA damage in Escherichia coli. J. Bacteriol. 189:1616-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Marchler-Bauer, A., et al. 2009. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 37:D205-D210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Masse, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matzanke, B. F., S. Anemuller, V. Schunemann, A. X. Trautwein, and K. Hantke. 2004. FhuF, part of a siderophore-reductase system. Biochemistry 43:1386-1392. [DOI] [PubMed] [Google Scholar]
- 37.McHugh, J. P., et al. 2003. Global iron-dependent gene regulation in Escherichia coli. A new mechanism for iron homeostasis. J. Biol. Chem. 278:29478-29486. [DOI] [PubMed] [Google Scholar]
- 38.Mendoza-Vargas, A., et al. 2009. Genome-wide identification of transcription start sites, promoters and transcription factor binding sites in E. coli. PLoS One 4:e7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Miethke, M., and M. A. Marahiel. 2007. Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71:413-451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Outten, C. E., and T. V. O'Halloran. 2001. Femtomolar sensitivity of metalloregulatory proteins controlling zinc homeostasis. Science 292:2488-2492. [DOI] [PubMed] [Google Scholar]
- 41.Outten, F. W., O. Djaman, and G. Storz. 2004. A suf operon requirement for Fe-S cluster assembly during iron starvation in Escherichia coli. Mol. Microbiol. 52:861-872. [DOI] [PubMed] [Google Scholar]
- 42.Payne, S. M. 1994. Detection, isolation, and characterization of siderophores. Methods Enzymol. 235:329-344. [DOI] [PubMed] [Google Scholar]
- 43.Petersen, B., T. N. Petersen, P. Andersen, M. Nielsen, and C. Lundegaard. 2009. A generic method for assignment of reliability scores applied to solvent accessibility predictions. BMC Struct. Biol. 9:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pieper, U., et al. 2009. MODBASE, a database of annotated comparative protein structure models and associated resources. Nucleic Acids Res. 37:D347-D354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ranquet, C., S. Ollagnier-de-Choudens, L. Loiseau, F. Barras, and M. Fontecave. 2007. Cobalt stress in Escherichia coli—the effect on the iron-sulfur proteins. J. Biol. Chem. 282:30442-30451. [DOI] [PubMed] [Google Scholar]
- 46.Rodrigue, A., G. Effantin, and M. A. Mandrand-Berthelot. 2005. Identification of rcnA (yohM), a nickel and cobalt resistance gene in Escherichia coli. J. Bacteriol. 187:2912-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schroder, I., E. Johnson, and S. de Vries. 2003. Microbial ferric iron reductases. FEMS Microbiol. Rev. 27:427-447. [DOI] [PubMed] [Google Scholar]
- 48.Scrutton, N. S. 1999. Identification of covalent flavoproteins and analysis of the covalent link, p. 181-194. In S. K. Chapman and G. A. Reid (ed.), Flavoprotein protocols, vol. 131. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 49.Sirivech, S., E. Frieden, and S. Osaki. 1974. The release of iron from horse spleen ferritin by reduced flavins. Biochem. J. 143:311-315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Thorgersen, M. P., and D. M. Downs. 2007. Cobalt targets multiple metabolic processes in Salmonella enterica. J. Bacteriol. 189:7774-7781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vasak, M., J. H. R. Kagi, B. Holmquist, and B. L. Vallee. 1981. Spectral studies of cobalt(II)-metallothionein and nickel(II)-metallothionein. Biochemistry 20:6659-6664. [DOI] [PubMed] [Google Scholar]
- 52.Wulfing, C., J. Lombardero, and A. Pluckthun. 1994. An Escherichia coli protein consisting of a domain homologous to FK506-binding proteins (FKBP) and a new metal binding motif. J. Biol. Chem. 269:2895-2901. [PubMed] [Google Scholar]
- 53.Zhang, J. W., G. Butland, J. F. Greenblatt, A. Emili, and D. B. Zamble. 2005. A role for SlyD in the Escherichia coli hydrogenase biosynthetic pathway. J. Biol. Chem. 280:4360-4366. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.