Abstract
Several factors can influence ortholog replacement between closely related species. We evaluated the transcriptional expression and metabolic performance of ortholog substitution complementing a Sinorhizobium meliloti argC mutant with argC from Rhizobiales (Agrobacterium tumefaciens, Rhizobium etli, and Mesorhizobium loti). The argC gene is necessary for the synthesis of arginine, an amino acid that is central to protein and cellular metabolism. Strains were obtained carrying plasmids with argC orthologs expressed under the speB and argC (S. meliloti) and lac (Escherichia coli) promoters. Complementation analysis was assessed by growth, transcriptional activity, enzymatic activity, mRNA levels, specific detection of ArgC proteomic protein, and translational efficiency. The argC orthologs performed differently in each complementation, reflecting the diverse factors influencing gene expression and the ability of the ortholog product to function in a foreign metabolic background. Optimal complementation was directly related to sequence similarity with S. meliloti, and was inversely related to species signature, with M. loti argC showing the poorest performance, followed by R. etli and A. tumefaciens. Different copy numbers of genes and amounts of mRNA and protein were produced, even with genes transcribed from the same promoter, indicating that coding sequences play a role in the transcription and translation processes. These results provide relevant information for further genomic analyses and suggest that orthologous gene substitutions between closely related species are not completely functionally equivalent.
Synteny, gene neighboring, or conservation of chromosomal gene order has been proposed to be a result of the interdependence between a gene product and its genomic context. Conservation of a syntenic block could be favored by selective pressure because the arrangement allows the sequential or coordinated expression of genes that correctly integrate important metabolic functions (41, 64, 69). By a comparative analysis of four Rhizobiales species (namely, Sinorhizobium meliloti, Agrobacterium tumefaciens, Mesorhizobium loti, and Brucella melitensis), we found that syntenic genes exhibit striking differences compared to nonsyntenic genes, including increased operon and network organization, high sequence conservation, diminished evolutionary rates, essential functional role, and specific phylogenetic associations (32; H. Peralta, G. Guerrero, A. Aguilar, and J. Mora, unpublished data).
Orthologs encode similar functions sharing a common ancestor and exhibiting various degrees of conservation, due in large part to functional adaptation and cellular role in different species (45). We have proposed a new parameter termed “species signature” to extract the amount of amino acid residues specific for a given species, based on multiple sequence alignment. We hypothesized that the species signature represents the proportion of a particular sequence that responds to adaptation (32); it is a useful evolutive measure because we found a high direct correlation with the nonsynonymous substitution rate (Peralta et al., unpublished). Although orthologous genes of Rhizobiales encode proteins with high degrees of identity among several species, the proteins differ in many respects, including isoelectric point. Small differences in protein sequence may be the result of evolutionary adaptation in response to the particular intracellular environment of a given species. In this scenario, specific amino acid changes would be selected for optimal performance in a particular genetic background. Knight et al. (40) carried out a virtual analysis of proteomes of nearly 100 organisms and showed that there was a correlation between theoretical proteomes and ecological niches; conversely, there was no correlation between phylogeny and differences observed in the theoretical proteomes. Our previous work has shown that mutation of a single gene, aniA (a carbon flux regulator), produced a proteomic alteration of approximately 800 proteins (16, 22), indicating that the absence or modification of a single gene can result in complex changes in global gene expression. In this context, syntenic orthologs are ideal to evaluate the functional importance of species signature in related organisms.
The Rhizobiales order is a versatile group of bacteria that present very interesting features, such as a huge amount of genes, abundance of genes acquired by horizontal transfer events, a symbiotic or pathogenic association with higher organisms such as plants or animals, and the ability for nodulation and nitrogen fixation in some species (9, 13, 29, 31, 50, 54, 65). In addition to a circular chromosome, some rhizobia carry secondary chromosomes or plasmids of high molecular size. Arginine is an essential amino acid in bacteria, and its synthesis requires a great deal of energy and reducing power (14, 49, 63). Arginine is the most common nitrogen storage compound and precursor of polyamine synthesis (48, 61). argC genes are commonly organized in operons, exemplified by the Escherichia coli argCBH operon (56). In R. etli, the argC mutation affects growth capacity using ammonium as the sole nitrogen source and results in failure to nodulate the common bean root (23).
We were interested in exploring whether the species signature (particular amino acid sequence variations) in orthologous syntenic genes reflects adaptations to the particular conditions of a given species, both intracellular (interactions with other enzymes and metabolites) and extracellular. The effect of heterologous complementation of an S. meliloti argC mutant with the corresponding ortholog from other Rhizobiales species (A. tumefaciens, R. etli, and M. loti) was compared in terms of growth, excretion of organic compounds, transcriptional and enzymatic activity, and molecular parameters such as mRNA and translation efficiency. An analysis of the putative speB-argC operon was also carried out to define a novel functional argC promoter.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Bacterial strains and plasmids used in the present study are listed in Table 1. The S. meliloti argC mutant was obtained cloning an 1.3-kb argC SmaI-SpeI fragment into the pJQ200SK+ vector (60). The pHP45Ω-Sp plasmid (57) was digested with BamHI to obtain the Ω-Sp interposon, and it was cloned on a BglII site of the argC fragment. The obtained plasmid was conjugated to the S. meliloti 1021 wild-type strain, and double recombinants were selected on minimal medium (MM) (19) (1.2 mM K2HPO4, 0.8 mM MgSO4, 1.5 mM CaCl2, 0.01 mM FeCl3), supplemented with 10 mM succinic acid and 10 mM ammonium chloride (NH4Cl). The wild-type, argC mutant, and argC complemented strains were maintained in PY medium, as described previously (19). When necessary, antibiotics were added at the following concentrations: nalidixic acid, 20 μg/ml; streptomycin, 200 μg/ml; spectinomycin, 100 μg/ml; gentamicin, 15 μg/ml; and tetracycline, 5 μg/ml. MM was used supplemented with 10 mM succinic acid or mannose as a carbon source and 10 mM ammonium chloride (NH4Cl) or potassium nitrate (KNO3) as a nitrogen source. Cultures were grown aerobically at 30°C in a shaker at 200 rpm for all experiments. To confirm that the complementation phenotypes observed were due exclusively to plasmid-encoded sequences, in some cases the plasmids were cured and conjugation repeated. Similar results were obtained (data not shown).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristicsa | Source or reference |
|---|---|---|
| Rhizobiales strains | ||
| Sinorhizobium meliloti 1021T | Wild type (type strain) | 51 |
| Rhizobium etli CFN42T | Wild type (type strain) | 62 |
| Rhizobium etli CE3 | Smr derivative of CFN42 | 53 |
| Agrobacterium tumefaciens C58 | Wild-type strain | ATCC 33970 |
| Mesorhizobium loti MAFF303099 | Wild-type strain | 38 |
| Sinorhizobium meliloti argC mutant | S. meliloti 1021 derivative, argC::ΩSp | This study |
| Escherichia coli strains | ||
| DH5α | supE44ΔlacU169(φ80lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | 34 |
| HB101 | supE44 hsdS20 (rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 6 |
| Plasmids | ||
| pCR2.1 TOPO | Cloning vector for PCR products; Apr Kmr | Invitrogen |
| pJQ200SK+ | Suicide cloning vector; Gmr | 60 |
| pBBR1MCS3 | Broad host range cloning vector; Tcr | 42 |
| pBBMCS53 | ΔplacZ pBBR1MCS5 derivative, carrying the promoterless gus gene; Gmr | 28 |
| pRK2073 | Conjugation helper plasmid; Sm/Spr | 4 |
| pRK2013 | Conjugation helper plasmid; Kmr | 25 |
| pHP45ΩSp | Plasmid containing the Ω-Sp interposon; Spr | 57 |
| pFGP1 | pBBR1MCS3 derivative with S. meliloti speB-argC genes | This study |
| pFGP2 | pBBR1MCS3 derivative with A. tumefaciens speB-argC genes | This study |
| pFGP3 | pBBR1MCS3 derivative with R. etli speB-argC genes | This study |
| pFGP4 | pBBR1MCS3 derivative with M. loti speB-argC genes | This study |
| pFGP5 | pBBMCS53 derivative plac::gus transcriptional fusion | This study |
| pFGP6 | pBBMCS53 derivative pargCSm::gus transcriptional fusion | This study |
| pFGP7 | pBBMCS53 derivative pspeBSm::gus transcriptional fusion | This study |
| pFGP8 | pBBMCS53 derivative plac-argCSm::gus transcriptional fusion | This study |
| pFGP9 | pBBMCS53 derivative plac-argCAt::gus transcriptional fusion | This study |
| pFGP10 | pBBMCS53 derivative plac-argCRe::gus transcriptional fusion | This study |
| pFGP11 | pBBMCS53 derivative plac-argCMl::gus transcriptional fusion | This study |
| pFGP12 | pBBMCS53 derivative pargC-argCSm::gus transcriptional fusion | This study |
| pFGP13 | pBBMCS53 derivative pargC-argCAt::gus transcriptional fusion | This study |
| pFGP14 | pBBMCS53 derivative pargC-argCRe::gus transcriptional fusion | This study |
| pFGP15 | pBBMCS53 derivative pargC-argCMl::gus transcriptional fusion | This study |
| pFGP16 | pBBMCS53 derivative pspeB-argCSm::gus transcriptional fusion | This study |
| pFGP17 | pBBMCS53 derivative pspeB-argCAt::gus transcriptional fusion | This study |
| pFGP18 | pBBMCS53 derivative pspeB-argCRe::gus transcriptional fusion | This study |
| pFGP19 | pBBMCS53 derivative pspeB-argCMl::gus transcriptional fusion | This study |
| pFGP20 | pBBMCS53 derivative carrying the S. meliloti argC gene under its promoter | This study |
| pFGP21 | pBBMCS53 derivative carrying the A. tumefaciens argC gene under the control of the S. meliloti argC promoter. | This study |
| pFGP22 | pBBMCS53 derivative carrying the R. etli argC gene under the control of the S. meliloti argC promoter | This study |
| pFGP23 | pBBMCS53 derivative carrying the M. loti argC gene under the control of the S. meliloti argC promoter | This study |
| pFGP24 | pBBMCS53 derivative pargCAt::gus transcriptional fusion | This study |
| pFGP25 | pBBMCS53 derivative pargCRe::gus transcriptional fusion | This study |
| pFGP26 | pBBMCS53 derivative pargCMl::gus transcriptional fusion | This study |
Tcr, tetracycline resistance; Apr, ampicillin resistance; Smr, streptomycin; Spr, spectinomycin resistance; Kmr, kanamycin resistance; Gmr, gentamicin resistance.
Introduction of overlapping ends by PCR amplification using chimeric primers.
Hybrid fusion genes containing argC genes from S. meliloti 1021, A. tumefaciens C58, R. etli CE3, and M. loti MAFF303099 under the control of different promoters were constructed by using overlap extension PCR methodology (66). Chimeric primers were designed with additional 5′ sequences to introduce homologous ends into the fragments to be fused. We used Oligo 6.0 software to construct the primer sequences, and the primers were purchased from the Unidad de Síntesis Química (IBt-UNAM).
The S. meliloti argC and speB regulatory regions containing overlapping ends corresponding to each argC ortholog were amplified by PCR using genomic DNA from S. meliloti strain 1021 as the template. Eight different PCRs were performed to obtain specific 896- and 500-bp PCR products with distinct overlapping ends. The specific primers used to generate the hybrid genes are listed in Table S1 in the supplemental material Amplification was carried out using a MasterCycler 5330 (Eppendorf, Hamburg, Germany) and AccuPrime Pfx DNA polymerase (Invitrogen). The reaction conditions were as follows: denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 20 s, 62°C for 30 s, and 68°C for 60 s. A final elongation step was carried out at 72°C for 10 min.
The complete argC coding regions of A. tumefaciens C58 (932 bp), R. etli CFN42 (932 bp), and M. loti MAFF303099 (926 bp) were obtained by PCR amplification using chimeric primers containing the S. meliloti argC promoter region overlapping ends. Total DNA from each strain was used as a template. The reaction conditions were as follows: denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 15 s and 68°C for 60 s. A final elongation step was carried out at 72°C for 10 min. For these reactions, Pfx50 DNA polymerase from Invitrogen was used. All PCR products were analyzed by agarose gel electrophoresis and then purified by using a QiaQuick gel extraction kit from Invitrogen.
To join the two DNA fragments, 1 μl of each PCR was mixed and cycled without primers. The reaction conditions were as follows: an initial denaturation step at 95°C for 2 min; followed by 20 cycles of 95°C for 1 min; annealing at 68°C for 10 s, 66°C for 5 s, 64°C for 5 s, and 62°C for 5 s; and extension at 72°C for 90 s. An additional extension was carried out at 72°C for 10 min. Portions (2 μl) of this reaction were mixed with the appropriate outer primers, and the assembled products were recovered. The reaction conditions were as follows: initial denaturation at 95°C for 2 min, followed by 30 cycles of 95°C for 15 s and 70°C for 2 min, followed by an additional extension at 72°C for 10 min. The reactions were carried out by using AccuPrime Taq DNA Polymerase High Fidelity (Invitrogen). The hybrid genes were cloned into pCR2.1 TOPO. All plasmids were sequenced to confirm that the fused gene was obtained with no nucleotide changes. Each set of constructs presented identical flanking sequences, including the regulatory ones, so that the only variant was the argC coding sequence from each organism.
Construction of plasmids for complementation experiments.
Plasmids able to replicate in S. meliloti were constructed by using the cloning vector pBBMCS53, a derivative of pBBR1MCS5 carrying the promoterless gus gene (28). This permitted the complementation and the transcriptional monitoring with the same construction. The sequences of all primers used are listed in Table S1 in the supplemental material. A plasmid, pFGP20, containing a 1,889-bp PCR fragment corresponding to the S. meliloti argC regulatory and coding regions, was constructed. Total DNA isolated from S. meliloti strain 1021 was used as the template for PCR with the primers SmUpXba and Lw_H1_PSm. The resulting fragment was cloned into pCR2.1 TOPO for sequencing (pTOPO::pargCSm-argCSm). An 896-bp regulatory region and the complete argC coding region were cloned into the pBBMCS53 plasmid as a 1,937-bp XbaI fragment. The 1,906-bp EcoRI hybrid fragment from TOPO::pargCSm-argCAt was cloned into pBBMCS53 to generate pFGP21. To generate pFGP22, the 1,924-bp XbaI-BamHI fragment from TOPO::pargCSm-argCRe was cloned into pBBMCS53. The XbaI-BamHI fragment from TOPO::pargCSm-argCMl was cloned into the same vector to generate pFGP23. The resulting recombinant plasmids were analyzed by restriction enzyme digestion and nested PCR to confirm that they contained the proper hybrid gene. Another complete set of plasmids was obtained, containing only argC genes on pBBR1MCS3 vector, under each of the mentioned promoters in order to be used in mRNA and proteomic ArgC detection and quantification.
Construction of β-glucuronidase transcriptional fusions.
To analyze the level of expression of each used promoter and argC orthologs or their segments, we constructed β-glucuronidase (gus) transcriptional gene fusions. The recombinant plasmids described above were used as templates for PCR with the primers SmUpXba and FusC_gus_R. Amplified fragments of the correct size were cloned into pCR2.1 TOPO. Four additional plasmids were constructed carrying the 896-bp regulatory region of S. meliloti argC and the complete argC gene sequence of each species (lacking the last 50 nucleotides) fused with the gus gene. This cassette was inserted into pBBMCS53 as an XbaI-KpnI fragment. Plasmids pFGP12, pFGP13, and pFGP14 contained an 1,828-bp XbaI-KpnI fragment that corresponded to argC genes from S. meliloti, A. tumefaciens, and R. etli under the S. meliloti argC promoter, respectively. A plasmid containing the M. loti argC gene under the control of the S. meliloti argC promoter was constructed by cloning an 1,822-bp XbaI-KpnI fragment in pBBMCS53 (pFGP15).
Conjugative transfer of plasmids from E. coli to Rhizobium was done by triparental mating, using either pRK2013 (25) or pRK2073 (4) as a helper. For determination of plasmid profiles, a modified Eckhardt procedure was used (36).
Determination of plasmid copy number.
The plasmid copy number was assessed in pspeB promoter-plasmids, by PCR, using tetA gene from plasmids and rpoA, a chromosomal gene, to calibrate the results. Complemented strains were grown for 8 h with shaking (200 rpm) at 30°C in MM containing succinate-ammonium. The initial inoculation was at an optical density at 540 nm (OD540) of 0.05. An aliquot of the cell culture (2 ml) was removed, and the cells were washed with sterile, deionized water. The cells were lysed by heat treatment at 95°C for 20 min. The lysate was diluted 500-fold and used as a template for PCR amplification of the tetA gene (for quantification of plasmid) and rpoA gene (as an endogenous, single-copy control). Amplification and detection of DNA by real-time PCR was performed by using the ABI Prism 7300 sequence detection system (Applied Biosystems) in optical-grade 96-well plates. Triplicate samples were routinely used for the determination of DNA by real-time PCR. The reaction conditions for amplification of DNA were as follows: 50°C for 2 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min. The data analysis was carried out by using sequence detection software (version 1.6.3; Applied Biosystems).
RACE.
The transcriptional start site of the S. meliloti argC gene was identified by using the 5′ rapid amplification of cDNA ends (5′RACE) system (version 2.0; Invitrogen). cDNA was synthesized from 1 to 5 μg of total RNA with the complementary primer argCGSP-1 (corresponding to nucleotides 415 to 430 of the argC coding region). After first-strand cDNA synthesis, mRNA was removed by treatment with RNase and then the cDNA was purified by using a SNAP column (Invitrogen). Terminal deoxynucleotidyltransferase and dCTP were used to add homopolymeric tails to the 3′ ends of the cDNA. Nested PCR was carried out by using the gene-specific primer argCGSP-2 (corresponding to nucleotides 340 to 355) and the abridged anchor primer (AAP) GGCCACGCGTCGACTAGTACGGGIIGGGIIGGGIIG provided with the kit. The amplified fragment was cloned into pCR2.1 TOPO for sequence analysis.
RNA isolation, cDNA probe obtention, and real-time RT-PCR assays.
RNA extraction was performed by using an RNeasy kit according to the manufacturer's instructions (Qiagen, Inc., Valencia, CA). Samples were treated with DNase I prior to reverse transcription-PCR (RT-PCR) using the protocol for preparation of DNA-free RNA (Fermentas). cDNA was synthesized from purified total RNA by RT using random primers and reverse transcriptase from the RevertAid H Minus First Strand cDNA synthesis kit (Fermentas). For real-time RT-PCR, TaqMan Universal PCR Mastermix, primer mixes containing a FAM reporter probe, and the gene-specific TaqMan MGB probe (6-FAM dye-labeled) (TaqMan gene expression assay) were obtained from Applied Biosystems. RT-PCR mixtures (25 μl) contained template cDNA, 2× TaqMan Universal PCR Mastermix buffer (12.5 μl), and forward and reverse primers. The optimal primer and probe concentrations and amplification conditions were determined. The forward and reverse argC primers and TaqMan probe for each species were added at concentrations of 5 and 2.5 pmol/μl, respectively.
The S. meliloti rpoA gene encodes the α subunit of RNA polymerase (RNAP) and is stably transcribed throughout different growth stages. Expression of rpoA was used as an endogenous control (normalizer) at concentrations of 1 and 2 pmol/μl. The forward and reverse gus primers and TaqMan gus probe were added at concentrations of 5 pmol/μl each. Reactions were analyzed by using an ABI Prism 7300 sequence detector (7300 System SDS, v1.3.0 software; Applied Biosystems). The reaction conditions were as follows: 2 min at 50°C, 10 min of polymerase activation at 95°C, and then 40 cycles of 95°C for 15 s and 55°C for 60 s. Each assay included (in triplicate) a standard curve of six serial dilutions of cDNA. The baseline was corrected by using the default adaptive baseline algorithm, and the threshold was manually adjusted by visual analysis of log-scale amplification plots.
Linear regression analysis was used to calculate the standard curve, estimate the r2 and slope, and calculate the PCR efficiency (%). After correcting the cycle threshold values (CT) for amplification efficiency, the expression levels were normalized to the endogenous control, and relative quantification of gene expression was obtained by the comparative CT method (2−ΔΔCT) (47).
Measurement of β-glucuronidase activity.
S. meliloti cultures were grown to mid-exponential phase in PY medium. Cells were collected by centrifugation at 6,000 × g in a Sorvall SS34 rotor, washed with sterile MM, and then concentrated 100-fold. Portions (50 ml) of MM containing 10 mM succinate and 10 mM ammonium chloride as carbon and nitrogen sources, respectively, were inoculated with the cell suspension at an initial OD540 of 0.05. Cultures were grown with shaking (200 rpm) for 8 h at 30°C. Quantitative β-glucuronidase activity was measured in a 1.0-ml culture sample using 4-2-nitrophenyl β-d-glucuronide (pNPG) as a substrate (28). The data were normalized to total cell protein concentration obtained by the Lowry method. The specific activities are expressed as nanomoles of p-nitrophenol (pNP) per minute per milligram of protein (nmol/min/mg of protein).
Determination of N-acetyl-γ-glutamyl phosphate reductase (ArgC) activity.
Bacteria were grown in MM. Cell cultures in logarithmic growth phase (8 h for MM with succinate-ammonium or 24 h for MM with mannose-nitrate) were collected by centrifugation and then washed. The cell pellets were resuspended in 5 mM Tris-HCl buffer (pH 7.5), and then the cells were disrupted by sonic oscillation. The supernatant was treated with ammonium sulfate, and the precipitate was discarded. Material that was precipitated by the addition of more ammonium sulfate was collected and dissolved in 5 mM Tris-HCl buffer (pH 7.4), containing 0.6 mM 2-mercaptoethanol. The protein solution was loaded on a Microcon YM-30 column, and the soluble cell extract was collected and used for the measurement of ArgC. The enzymatic activity was measured spectrophotometrically by the disappearance of NADPH in the presence of N-acetyl glutamate and ATP. The assay was initiated by adding ArgB (N-acetyl glutamate kinase, purified in our laboratory) to the reaction mix since N-acetyl glutamyl phosphate, the product of ArgB and substrate of ArgC, is highly unstable and not commercially available. Reaction rates were corrected for NADPH consumption, measured in the absence of added N-acetyl glutamate. The specific activity was expressed as nanomoles of NADP produced per minute per milligram of protein. The protein concentration was determined by the Lowry method.
Determination of SpeB activity.
For the measurement of agmatine ureohydrolase (SpeB) activity, 150-ml cultures were grown (either for 8 h in MM supplemented with succinate-ammonium or for 24 h in MM supplemented with mannose-nitrate), and the cells were collected by centrifugation. The pellets were washed and then resuspended in 1.0 ml of ice-cold SpeB reaction buffer. The cells were disrupted, and then the cell debris was removed by centrifugation. The supernatant was collected and used to assay SpeB activity. The reaction was stopped by the addition of a perchloric acid solution. The ammonia content in the sample was measured by using a diagnostic urea/ammonia determination procedure (Boehringer Mannheim). The specific activity was expressed as micrograms of ammonia produced per milligram of protein. The protein concentration was determined by the Lowry method.
ArgC proteomic identification and quantification.
Bacterial proteins were obtained by sonicating cell cultures (grown 8 h in MM supplemented with succinate-ammonium) for 5 cycles of 1 min each at 4°C in a Vibra Cell (Soniprep150, MSE Sanyo) in the presence of a protease inhibitor (Complete tablets; Roche Diagnostics GmbH, Mannheim, Germany). To further limit proteolysis, protein isolation was performed by using phenol extraction (35). To solubilize and obtain completely denatured and reduced proteins, pellets were dried and resuspended as previously described (21). Prior to electrophoresis, the samples were mixed with 7 M urea, 2 M thiourea, 4% CHAPS {3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate}, 2 mM tributhyl phosphine (TBP), 2% ampholytes, and 60 mM dithiothreitol. Sample preparation, analytical and preparative two-dimensional polyacrylamide gel electrophoresis (2D-PAGE), and image analysis were performed as previously described (20). pH gradients were obtained using a 2D-PAGE standard (Sigma). For separation in the first dimension, ∼500 mg of total protein was loaded. The gels were stained with Coomassie Blue R-250, and protein spots were detected at a 127-by-127-mm resolution using a PDI Image Analysis System and PD-Quest software (Protein Databases, Inc., Huntington Station, NY). Selected spots were excised manually and prepared for mass spectroscopy (MS), as previously described (21). Purified ArgC protein was subjected to enzymatic digestion, and then the resulting peptide fragments were analyzed by complementary MS methods (matrix-assisted laser desorption ionization-time of flight-MS and liquid chromatography-electrospray ionization-tandem MS). ArgC quantification was performed by dot pixel evaluation with PD-Quest software and extrapolated to the total gel load. All experiments were performed three times. Mass spectra were obtained using a Bruker Daltonics Autoflex (Bruker Daltonics, Bellerica, MA) operated in the delayed extraction and reflectron mode. Spectra were externally calibrated using a peptide calibration standard (Bruker Daltonics standard 206095). Peak lists of the tryptic peptide masses were generated and searched against the NCBI nr databases or the Rhizobase (http://bacteria.kazusa.or.jp/rhizo/) using the Mascot search program (Matrix Science, Ltd., London United Kingdom).
Determination of excreted organic acids and amino acids.
For extracellular organic acids and amino acids determination, cells were grown for 8 h in MM with succinate-ammonium or 24 h for MM with mannose-nitrate, and 25-ml cultures were subjected to centrifugation, and then the supernatant, free of bacteria, was lyophilized. Amino acids and organic acids were separated and quantified by using reversed-phase high-performance liquid chromatography with a Symmetry C18 column (5 μm, 3.9 by 150 mm; Waters). The data are expressed as nmol/mg of protein and μmol/mg of protein, respectively.
Bioinformatics methods.
Codon adaptation index (CAI) calculations were performed with the CAI program in the EMBOSS (European Molecular Biology Open Software Suite) package available at www.emboss.org. The species signature was calculated as the number of specific amino acid residues in the encoded protein that were unique or least abundant at each position based on multiple alignments, and was expressed as a percentage of protein length. Species signatures were calculated with a perl custom program (available on request) based on multiple alignments performed with CLUSTAL W (70). For phylogeny, multiple alignment of translated amino acid sequences was carried out by using MUSCLE v3.7 (17) and the approximate-likelihood rate method with PhyML v3.0 (1, 33) on the Phylogeny.fr server (15). A bootstrap was applied with 100 replicates, and a WAG model for amino acid substitution was used. The tree was drawn with TreeView1.6.6 (Roderic Page). ArgC sequences for species signature evaluation and phylogenetic analysis were obtained from GenBank using the following access numbers: A. tumefaciens C58 (NP_354256), A. vitis S4 (YP_002549205), Bradyrhizobium ORS278 (YP_001205564), B. melitensis 16 M (NP_540088), E. coli K-12 W3110 (AP_003852), M. loti MAFF303099 (Q982X3), R. etli CFN42 (ABC90369), and S. meliloti 1021 (NP_385346). Obtained argC sequences were registered in the GenBank and assigned the following provisional access numbers: M. loti strains R88B (HM753558), N86A99 (HM753564), VTI (HM753565), CJ4 (HM753561), CJ2 (HM753562), R12C (HM753559), R7A (HM753560), and CJ6 (HM753563). S. meliloti strains Ts48 (HM753568), Cx2 (HM753573), Ter3 1 (HM753570), Mac3 5 (HM753572), REF21 (HM753574), Cx48 (HM753571), SauI 5 (HM753569), Cx26 (HM753575), Tx 30 (HM753567), and ES2 (HM753566). A. tumefaciens strains IAM12048 (HM753578), IAM13129 (HM753576), IAM13549 (HM753577), and IAM14040 (HM753579). R. etli strains Kim5 (HM753582), CIAT652 (HM753583), GR56 (HM753580), CIAT151 (HM753584), and Brasil5 (HM753581).
RESULTS
Structural analysis of argC genes and their protein sequences. (i) Species signatures and phylogenetic relationships of argC from Rhizobiales.
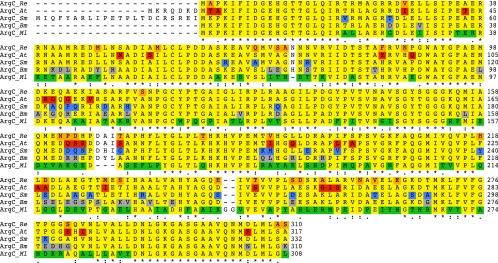
Sequence differences among orthologs could be the result of evolutionary adaptation to a specific cellular environment. The extent of these specific changes has been termed the “species signature” (32). In order to assess the functional importance of species signature in Rhizobiales, we focused on a syntenic gene, argC, which encodes an N-acetyl-γ-glutamyl phosphate reductase that is essential for arginine biosynthesis. Alignment and comparison of the amino acid sequences of ArgC from five species—R. etli, A. tumefaciens, S. meliloti, B. melitensis, and M. loti—showed that there were 133 positions with identical residues, and 32 (10.3%), 30 (9.6%), 30 (9.6%), and 113 (36.4%) residues, respectively, that were unique and are considered species signatures (Fig. 1); the isoelectric points were 5.46, 5.39, 6.0, and 5.4, respectively.
FIG. 1.
Species signature of ArgC from Rhizobiales. A multiple alignment was carried out using CLUSTAL W. Species signatures were determined with a perl custom script. Yellow blocks indicate identical residues in at least two species. Colored residues for each organism denote species signature as follows: orange, R. etli; red, A. tumefaciens; blue, S. meliloti; gray, B. melitensis; and green, M. loti. Residues not in blocks are gaps in some organism or nonhomologous sequences.
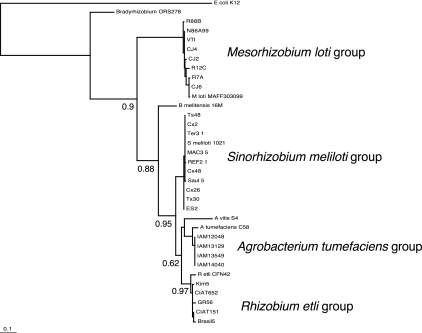
To determine the degree of conservation of the ArgC protein among members of the same species of Rhizobiales, the argC coding regions of different Sinorhizobium, Rhizobium, Agrobacterium, and Mesorhizobium strains were amplified by PCR and then sequenced. The translated sequences showed 98 to 99% similarity among species. A phylogenetic tree was constructed to assess the relationships among Rhizobiales species based on the ArgC sequences (Fig. 2). Rhizobium-Agrobacterium species exhibited the closest relationships, followed by Sinorhizobium strains; M. loti sequences were located on a more distant branch. This species relationship was in general concordance with other vertically inherited markers, such as DnaK, GyrB, and AtpD (data not shown).
FIG. 2.
Phylogenetic analysis of ArgC from Rhizobiales. The alignment was done using the MUSCLE program. Phylogeny was generated by the PhyML method. The tree was drawn with TreeView. Bootstrap analysis was performed with 100 replicates. Supporting values are shown in the main branches. The E. coli sequence was taken as the outgroup. The bar represents substitutions per residue.
(ii) Genomic context analysis and identification of the argC promoter.
The genomic organization of argC in Rhizobiales was analyzed by using a publicly available annotated database (http://www.ncbi.nlm.nih.gov/genomes/MICROBES). In the circular chromosome in each Rhizobiales species analyzed, the argC gene are contiguous to upstream speB (agmatine ureohydrolase), which participates in the synthesis of putrescine, part of the polyamine synthesis pathway (e.g., spermine and spermidine) involved in several cellular and metabolic functions (37). Previous in silico analyses have indicated that argC and speB are organized in an operon (52, 58).
As an initial approach, the complete putative speB-argC operon of each species was cloned and used for complementation analysis of an S. meliloti argC mutant (argC::ΩSp). The enzymatic activity of ArgC and SpeB expressed from the putative speB-argC operons of S. meliloti, A. tumefaciens, R. etli, and M. loti was evaluated in the S. meliloti argC mutant strain. We found differences in ArgC and SpeB activities (data not shown), which conducted us to consider independent promoters for each gene. To identify the putative argC promoter, the speB-argC segment-containing plasmids were modified by deletion of 150 bp of speB gene using XbaI-KpnI restriction sites (data not shown). Disruption of speB did not compromise the ability of the plasmids to complement the mutant phenotype (data not shown).
To verify that there was an independent promoter region for argC located within the speB coding region, we identified the transcriptional start site of S. meliloti argC by 5′RACE using total RNA purified from wild-type strain cells. The argC transcriptional start site was located 125 nucleotides upstream of the putative translational start codon, and a putative −10 box characteristic of sigma 70-dependent promoters was identified (see Fig. S1 in the supplemental material).
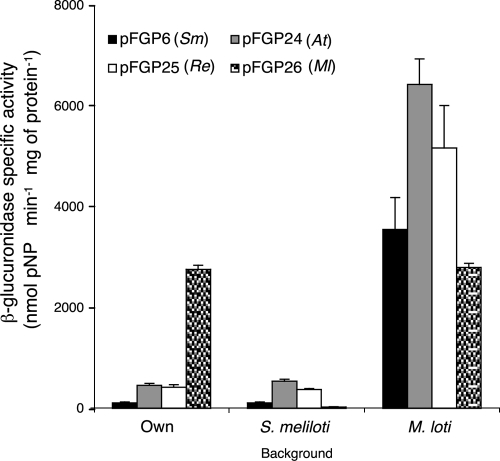
To determine the transcriptional activity of the putative argC promoter of Rhizobiales, argC promoter-β-glucuronidase (gus) gene fusions were constructed, and their activity was evaluated in S. meliloti and M. loti. As a control, each species-specific promoter-gus construct was also evaluated in its native background. As seen in Fig. 3, the activities of the S. meliloti, A. tumefaciens, and R. etli argC promoters were similar in their native backgrounds and in that of S. meliloti. Also, the M. loti argC promoter had very low activity in the latter background. However, the activities were more induced in the M. loti background compared to the M. loti argC promoter activity. Thus, the newly identified argC promoter was active in all strains, and the M. loti argC promoter was weakly active in the S. meliloti background.
FIG. 3.
Transcriptional analysis of argC promoter activity. Expression of the indicated argC promoter-gus (β-glucuronidase) gene fusions is depicted as follows: first group, in the genetic background of the indicated species; second group, in S. meliloti; and third group, in M. loti. Bars: black, S. meliloti; gray, A. tumefaciens; empty, R. etli; patterned, M. loti.
The species signatures of Rhizobiales ArgC orthologs ranged from 10 to 36%, and phylogenetic analysis revealed that R. etli was closely related to A. tumefaciens. Unique to Rhizobiales, the argC gene is linked to speB (involved in putrescine synthesis), and in the case of S. meliloti it was demonstrated that argC has its own active promoter inside the coding sequence of speB. M. loti argC promoter activity was very low in the S. meliloti background compared to its native background.
Expression analysis of argC regulated by different promoters.
The aim of the present study was to determine the complementation efficiency of argC orthologs from related species in an S. meliloti argC mutant. The level of mRNA synthesis is dependent on promoter activity; thus, we were interested in the expression patterns of the Rhizobiales argC genes under the control of the same promoter, either the S. meliloti speB (pspeB) promoter, the S. meliloti argC (pargC) promoter, or the E. coli lac (plac) promoter.
The level of expression of β-glucuronidase under the control of the lac promoter was three times higher than the S. meliloti argC promoter (compare the first and second lines in Table S2 in the supplemental material). In contrast, the S. meliloti speB promoter exhibited very low activity under the same conditions (26- and 8-fold less than plac or pargC, respectively) (third line in Table S2 in the supplemental material). These results indicated that in S. meliloti, plac is a strong promoter, whereas pargC and pspeB are moderate and weak promoters, respectively. For a comparative analysis, we used these promoters to drive the expression of argC from A. tumefaciens, S. meliloti, R. etli, and M. loti. The levels of transcriptional activity from the different promoters were similar to those obtained above (data not shown).
To determine the transcriptional activity through the argC sequences, we analyzed the expression of transcriptional fusions either containing the complete argC sequence, containing a partial argC sequence (either with 880 or 100 bp), or lacking the coding region completely (containing only the ATG codon) under the control of the S. meliloti argC promoter. Constructs were analyzed in an S. meliloti wild-type strain background. Deletion of argC gene portions had variable effects on transcription rates, resulting in increased reporter gene expression in the first 100 to 880 bp of the gene (Table 2). The effect was more drastic with M. loti argC (an ∼2.5-fold increase compared to the start codon alone), followed by S. meliloti (2-fold increase) and A. tumefaciens (50% increase). The exception was the R. etli argC, whose activity was similar for the truncated sequences and the start codon alone. Interestingly, expression was drastically reduced when the complete argC gene sequence was evaluated, indicating sequences with a stalling effect on transcription. The expression of R. etli argC was nearly constant with shorter versions, but with the complete gene sequence the effect was very strong. These results suggested that, at least in plasmid, the argC coding region itself can influence gene expression, in agreement with other recent results about gene sequences modulating their transcription-translation rates (27, 43). Codon adaptation index (CAI) values were calculated for each argC gene in the S. meliloti background and yielded values ranging from 0.76 to 0.8.
TABLE 2.
Effect of argC gene sequences on transcriptional expression
| Sequence in fusion | Mean β-glucuronidase sp act ± SDa |
|||
|---|---|---|---|---|
| S. meliloti | A. tumefaciens | R. etli | M. loti | |
| ATG | 233 ± 15 | 210 ± 28 | 230 ± 39 | 249 ± 17 |
| Partial CDS (100 bp) | 366 ± 23 | 284 ± 30 | 206 ± 25 | 637 ± 131 |
| Partial CDS (880 bp) | 439 ± 51 | 319 ± 48 | 270 ± 48 | 591 ± 103 |
| Complete CDS | 145 ± 6 | 280 ± 30 | 30 ± 3 | 153 ± 7 |
Values are expressed as nmol of p-nitrophenol min−1 mg of protein−1. Data represent the means of two replicates from three independent experiments. The expression under pargCSm was determined.
Plasmid copy number also can play an effect in expression of genes; therefore, we assessed argC gene copy using plasmid with pspeB promoter and chromosomal rpoA as a control. The control plasmid (vector alone) was present in cells at approximately two plasmid copies per chromosome (1.8 ± 0.17), as were the R. etli (2.1 ± 0.18), A. tumefaciens (2.3 ± 0.1), and S. meliloti (2.9 ± 0.11) gene-containing plasmids. In contrast, the M. loti gene-containing plasmid was present at 9.2 ± 0.23 plasmid copies per chromosome.
Based on gus reporter gene expression analysis, we were able to characterize three promoters of various strengths: weak (S. meliloti pspeB), intermediate (S. meliloti pargC), and strong (E. coli plac). Differential transcriptional regulation of complete and partial gene sequences suggested that argC coding sequences may modulate their transcriptional rates. An additional mechanism exists that modulated the plasmid copy number in the case of M. loti argC ortholog complementation.
Complementation analysis of argC from Rhizobiales under different promoters. (i) Physiological analysis: growth, duplication times, and excretion of metabolites.
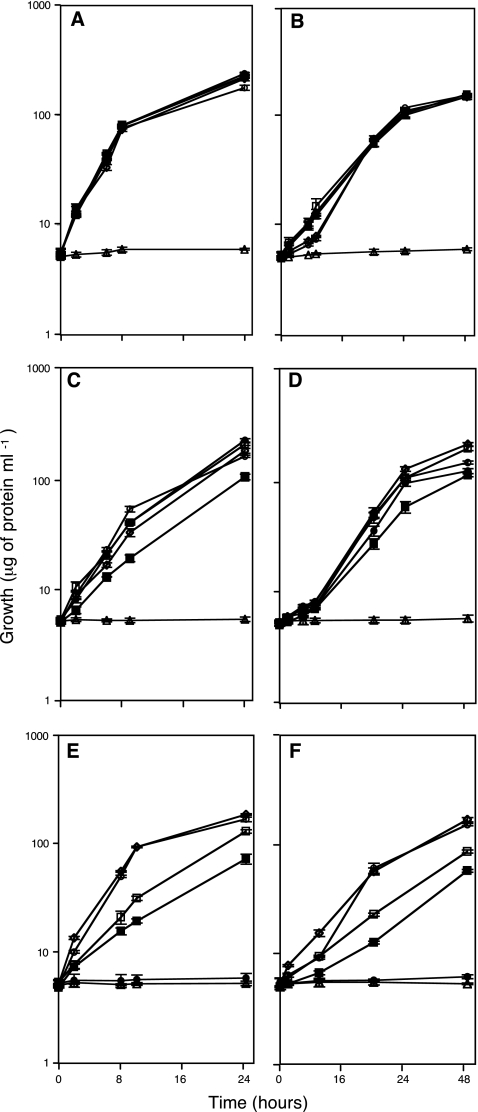
The optimal growth conditions for S. meliloti wild-type strain was MM containing succinate-ammonium, and typical doubling times were 3 h (Fig. 4). We also investigated suboptimal growth conditions in order to differentiate complemented strains. In MM containing mannose-nitrate, the wild-type strain exhibited delayed growth, with a doubling time of 6 h. Under either set of conditions, the argC mutant strain failed to grow. The best complementation was achieved with argC under the control of pargC (Fig. 4A and B). Under that promoter in both MM growth conditions, all complemented strains grew at similar rates and to levels similar to that of the wild-type strain, and similar duplication times were obtained. Under the control of plac, under both sets of conditions, the growth of all complemented strains was similar to that of the S. meliloti wild-type strain (Fig. 4C and D). The A. tumefaciens-complemented strain exhibited delayed growth, with increased duplication times of 4 and 8 h in succinate-ammonium and mannose-nitrate, respectively. Under the control of pspeB (from S. meliloti), the complemented strains exhibited reduced growth under both sets of conditions. The R. etli- and A. tumefaciens-complemented strains exhibited reduced growth, increased duplication times (8 h), and long lag phases (Fig. 4E and F); the M. loti-complemented strain failed to grow. Only S. meliloti-complemented strain growth was similar to that of the wild type.
FIG. 4.
Growth curves of S. meliloti argC-complemented strains. The left panels depict growth in MM supplemented with 10 mM succinic acid and 10 mM ammonium chloride. The right panels depict growth in MM supplemented with 10 mM mannose, 10 mM potassium nitrate. (A and B) argC under the control of the S. meliloti argC promoter; (C and D) argC under the control of the E. coli lac promoter; (E and F) argC under the control of the S. meliloti speB promoter. Each panel represents the argC mutant strain complemented with argC gene from S. meliloti (⧫), A. tumefaciens (▪), R. etli (□), and M. loti (•); the S. meliloti wild-type strain (○); and the argC mutant strain (▴).
Given the limited growth of the complemented strains when argC was placed under the control of pspeB (Fig. 4E and F), we investigated the concentration of metabolites excreted into the culture medium, possibly as a response to a faulty metabolism. In MM containing succinate-ammonium, the argC mutant strain excreted 6 to 15 times more glutamate and glycine compared to the wild-type strain, 15 times more α-ketoglutarate, and consumed only a fraction of the succinate consumed by the wild-type strain (Table 3). These levels were restored to wild-type levels when the mutant strain was grown in MM supplemented with arginine. The S. meliloti-complemented strain exhibited the most wild-type-like metabolic profile, while the R. etli- and A. tumefaciens-complemented strains excreted slightly more α-ketoglutarate and consumed a little less succinate, than the rest of complemented strains (Table 3). The M. loti-complemented strain showed the highest levels of excreted metabolites, even compared to the argC mutant strain (twice the level of glutamate and more than 50% more α-ketoglutarate compared to the argC mutant). In MM containing mannose-nitrate, higher glutamate and glycine levels were observed for the mutant strain, compared to MM containing succinate-ammonium, as well as a reduction in the excreted levels with the addition of arginine. The complemented strains showed differential behavior compared to MM with succinate-ammonium. The S. meliloti- and R. etli-complemented strains showed 10-fold reduction of glutamate excretion, while the A. tumefaciens- and M. loti-complemented strains showed a slight increase. For glycine excretion, a severalfold increase was observed for all of the complemented strains and was drastic with the M. loti gene. In the case of α-ketoglutarate, there was a slight reduction for the S. meliloti gene and the reduction was approximately 50% in the rest of complemented strains.
TABLE 3.
Metabolite excretion of S. meliloti argC complemented strains under the S. meliloti pspeB promoter
| Strain | Metabolite excretion (mean ± SD)a |
||||||
|---|---|---|---|---|---|---|---|
| MM succinate-ammonium (8 h) |
MM mannose-nitrate (24 h) |
||||||
| Glu* | Gly* | α-Kg† | Succ† | Glu* | Gly* | α-Kg† | |
| S. meliloti | |||||||
| 1021 wild type | 1.6 ± 0.7 | 0.1 ± 0.1 | 13.6 ± 1.0 | 226.6 ± 25.2 | 0.1 ± 0.2 | 12.6 ± 2.0 | 10.4 ± 0.8 |
| argC mutant | 7.1 ± 3.3 | 1.5 ± 0.1 | 228.6 ± 74.0 | 3356.6 ± 1201.2 | 24.6 ± 0.5 | 396.4 ± 78.2 | 129.2 ± 36.3 |
| argC mutant (+1 mM Arg) | 0.2 ± 0.1 | 0.4 ± 0.2 | 11.7 ± 0.7 | 176.8 ± 1.2 | 0.4 ± 0.3 | 3.6 ± 0.8 | 9.2 ± 4.8 |
| argC mutant complemented strains | |||||||
| pFGP16 (S. meliloti) | 1.0 ± 0.2 | 0.8 ± 0.1 | 13.3 ± 0.9 | 235.6 ± 23.0 | 0.1 ± 0.5 | 54.4 ± 13.0 | 11.3 ± 1.2 |
| pFGP17 (A. tumefaciens) | 6.5 ± 0.3 | 0.6 ± 0.1 | 68.5 ± 3.9 | 1246.0 ± 93.9 | 11.4 ± 2.4 | 183.4 ± 74.9 | 40.6 ± 4.6 |
| pFGP18 (R. etli) | 1.1 ± 0.3 | 0.5 ± 0.1 | 53.7 ± 3.8 | 854.8 ± 38.7 | 0.1 ± 0.2 | 168.2 ± 17.9 | 19.7 ± 2.7 |
| pFGP19 (M. loti) | 15.9 ± 2.4 | 1.6 ± 0.4 | 328.4 ± 11.6 | 5217.9 ± 1841.0 | 18.4 ± 0.3 | 898.0 ± 10.6 | 151.6 ± 8.4 |
Glu, glutamate; Gly, glycine; α-Kg, α-ketoglutarate; Succ, succinate. *, Expressed as nmol mg of protein−1; †, expressed as μmol mg of protein−1.
These results clearly demonstrated that the argC promoters and gene sequences were important determinants of growth capacity in the complemented strains. The newly identified argC promoter was optimum for all strains, followed by the strong, constitutive lac promoter. There were clear differences among the phenotypes of strains complemented with A. tumefaciens, R. etli, and M. loti argC under the control of pspeB, none of which fully complemented the argC mutant phenotype.
(ii) Biochemical characterization of complemented strains: ArgC and SpeB enzymatic activity.
We evaluated the N-acetyl-γ-glutamyl phosphate reductase (ArgC) of strains containing plasmids with promoter-argC constructions and compared the results to the growth phenotype. The wild-type strain showed reduced activity and, compared to the S. meliloti-complemented strain, the multicopy effect was apparent again, in diverse proportions, with the exception of construct under pspeB promoter (Table 4). Complemented strains grown in MM supplemented with mannose-nitrate showed a reduction in ArgC activity in almost all cases, except for S. meliloti under plac and A. tumefaciens in the speB-argC construct. The tendency toward reduced activity was possibly because these compounds are poor carbon and nitrogen sources for S. meliloti.
TABLE 4.
ArgC and SpeB specific activities of S. meliloti argC complemented strains under the control of different promoters
| Protein and conditions | Mean ArgC and SpeB sp act ± SDa of argC mutant complemented strainsa |
||||
|---|---|---|---|---|---|
| WT | S. meliloti | A. tumefaciens | R. etli | M. loti | |
| ArgC | |||||
| pargC (pFGP12 to pFGP15) | |||||
| Succinate-ammonium | 16.6 ± 0.4 | 134.0 ± 2.6 | 80.1 ± 2.3 | 83.1 ± 1.4 | 13.3 ± 0.8 |
| Mannose-nitrate | 16.3 ± 0.4 | 68.8 ± 0.9 | 28.7 ± 1.4 | 32.3 ± 1.9 | 7.1 ± 0.3 |
| plac (pFGP8 to pFGP11) | |||||
| Succinate-ammonium | 16.7 ± 0.3 | 405.4 ± 3.4 | 49.0 ± 1.5 | 59.5 ± 0.9 | 60.6 ± 0.7 |
| Mannose-nitrate | 16.4 ± 0.1 | 432.4 ± 20.7 | 81.7 ± 2.8 | 32.1 ± 3.1 | 81.9 ± 8.7 |
| pspeB (pFGP16 to pFGP19) | |||||
| Succinate-ammonium | 16.7 ± 0.3 | 20.8 ± 0.4 | 19.3 ± 0.5 | 18.9 ± 1.3 | 0.8 ± 0.2 |
| Mannose-nitrate | 16.1 ± 0.3 | 25.0 ± 0.1 | 16.6 ± 0.1 | 17.7 ± 0.1 | 0.9 ± 0.1 |
| speB-argC (pFGP1 to pFGP4)b | |||||
| Succinate-ammonium | 14.5 ± 0.3 | 102.6 ± 7.4 | 107.5 ± 3.9 | 38.0 ± 6.6 | 0.8 ± 0.2 |
| Mannose-nitrate | 16.2 ± 0.1 | 57.3 ± 10.3 | 115.4 ± 1.7 | 35.2 ± 0.8 | 1.0 ± 0.1 |
| SpeBc | |||||
| speB-argC (pFGP1 to pFGP4) | |||||
| Succinate-ammonium | 6.5 ± 0.1 | 39.9 ± 6.1 | 53.2 ± 2.3 | 55.1 ± 1.9 | 62.2 ± 1.2 |
| Mannose-nitrate | 9.2 ± 0.1 | 46.6 ± 3.4 | 43.7 ± 3.4 | 38.9 ± 1.6 | 21.5 ± 6.6 |
Expressed as nmol of NADP min−1 mg of protein−1, except as noted (see footnote c). WT, S. meliloti 1021 wild-type strain.
That is, constructs containing speB and argC under their own promoters.
Expressed as μg of ammonia mg of protein−1.
Under the control of pargC, in MM containing succinate-ammonium, the S. meliloti-complemented strain had the highest enzymatic activity, and the M. loti-complemented strain had the lowest. In MM containing mannose-nitrate, similar proportions among the strains were observed (Table 4). These activity levels were sufficient for all of the strains to achieve optimal growth (see Fig. 4A and B).
Under both growth conditions, plac conferred high activity levels, most notably in the S. meliloti-complemented strain (Table 4). In this case, the growth delay of the A. tumefaciens-complemented strain did not correlate with diminished activity. Under the control of pspeB, there was reduced ArgC activity in all strains (Table 4). In the M. loti-complemented strain, this level of activity was insufficient to support growth (see Fig. 4E and F). In addition, although both R. etli- and A. tumefaciens-complemented strains exhibited similar activity levels in terms of complementation of the argC mutant phenotype in MM containing succinate-ammonium compared to the S. meliloti-complemented strain, they did not grow well and had long lag phases in MM containing mannose-nitrate.
Strains containing the construct speB-argC (with each gene expressed from its own promoter) showed similar growth compared to wild-type (not shown) and intermediate ArgC activity levels (Table 4)—with the exception of the M. loti-complemented strain, which did not grow in both MM conditions and showed very reduced ArgC activity (Table 4). The activity of SpeB was also evaluated for strains with the speB-argC construct, and similar levels of activity were found among the complemented strains in both growth conditions (Table 4).
Plasmids with S. meliloti and M. loti argC genes also were introduced into the S. meliloti wild-type strain, and no differences in ArgC activity were observed in MM supplemented with succinate-ammonium, with the exception of M. loti-complemented strain under pspeB, that presented one-third reduction in the activity (not shown). This was possibly due to the formation of slightly inefficient ArgC hybrid multimers that reduced native activity.
With the exception of strains carrying the pspeB promoter in MM containing succinate-ammonium, there was a good agreement between growth curves and ArgC activity. Optimal growth and high ArgC activity were obtained with pargC and plac. In the case of pspeB, the M. loti-complemented strain showed the lowest complementation efficiency and a no-growth phenotype. R. etli- and A. tumefaciens-complemented strains under the pspeB promoter exhibited similar enzymatic activities compared to the S. meliloti-complemented strain; however, they showed severe growth delay.
(iii) Molecular analysis: argC transcript levels, direct ArgC quantification, and translational index.
Given the results of the analysis of ArgC activity and growth phenotype, we next investigated important factors influencing the ability of orthologous argC genes to complement the S. meliloti argC mutant phenotype, such as mRNA production and translational efficiency in strains containing the pBBMCS53 vector, carrying the argC and gus genes, which permitted the complementation and monitoring of transcription simultaneously (see Materials and Methods). The mRNA level in the wild-type strain was used as the reference and given a value of 1.0. The ArgC protein was quantified directly by two-dimensional gel electrophoresis and mass spectrometry. The mRNA levels and the amount of protein detected by proteomic identification were used to calculate a translational efficiency index (proteomic protein/mRNA).
The wild-type strain exhibited nearly constant values for mRNA, proteomic protein, and translational index in each set of assays (Table 5). In general, the S. meliloti-complemented strain obtained the best translational efficiency index values in each condition compared to the rest of the strains. Also, despite the fact that argC orthologs were controlled by the same promoter in each set, different amounts of mRNA and protein were produced, confirming the findings regarding the transcriptional effect of argC coding sequences detected with gus fusions (see Table 2).
TABLE 5.
mRNA, protein quantification, and translational efficiency index for S. meliloti argC mutant complemented strains
| Promoter and parametera |
argC mutant complemented strainsb |
||||
|---|---|---|---|---|---|
| WT | S. meliloti | A. tumefaciens | R. etli | M. loti | |
| pargC (pFGP12 to pFGP15) | |||||
| mRNA | 1 | 9.4 ± 0.5 | 51.1 ± 4.8 | 127.6 ± 22.4 | 19.9 ± 0.4 |
| Proteomic protein | 9.8 ± 0.5 | 771.2 ± 91.1 | 442.7 ± 20.8 | 331.6 ± 29.9 | 273.6 ± 58.8 |
| TEI | 9.8 | 82.0 | 8.7 | 2.6 | 13.8 |
| plac (pFGP8 to pFGP11) | |||||
| mRNA | 1 | 121.4 ± 11.8 | 1,759.2 ± 68.2 | 1,596.7 ± 69.7 | 1,598.7 ± 42.9 |
| Proteomic protein | 9.5 ± 0.1 | 1,834.2 ± 100.3 | 1,399.5 ± 213.2 | 91.7 ± 26.9 | 1,696.1 ± 303.1 |
| TEI | 9.5 | 15.1 | 0.8 | 0.1 | 1.1 |
| pspeB (pFGP16 to pFGP19) | |||||
| mRNA | 1 | 4.9 ± 0.6 | 53.1 ± 2.9 | 35.0 ± 1.1 | 162.9 ± 9.5 |
| Proteomic protein | 9.5 ± 0.1 | 25.1 ± 0.1 | 65.5 ± 2.8 | 72.9 ± 7.9 | 67.4 ± 5.2 |
| TEI | 9.5 | 5.1 | 1.2 | 2.1 | 0.4 |
mRNA values were normalized relative to the abundance of mRNA in the wild-type strain. Proteomic protein was quantified by using spectrometry and dot pixels. TEI, translational efficiency index (proteomic protein/mRNA).
WT, S. meliloti 1021 wild-type strain. Cells were grown for 8 h in MM supplemented with 10 mM succinic acid and 10 mM ammonium chloride.
In the case of expression driven by pargC promoter, the S. meliloti-complemented strain had 10 and 78 times more mRNA and ArgC protein production, respectively, than the wild-type strain, and an 8-fold increase in translational index (Table 5). The levels of mRNA in the other complemented strains were in the range of 20- to 140-fold higher than in the wild type, and yet the translational index in these strains was lower, with the R. etli-complemented strain having the lowest translational index. Despite these variations, all strains exhibited similar growth efficiency.
When strains were controlled by plac promoter they exhibited highly elevated levels of mRNA. In the S. meliloti-complemented strain, there was an almost 110-fold increase in mRNA compared to the wild-type strain; mRNA levels in the other complemented strains increased more than 1,500-fold (Table 5). A similar trend in high protein levels was observed, with the exception of the R. etli-complemented strain, which also had the lowest translational index in this set. The S. meliloti-complemented strain demonstrated the highest protein level, but because its mRNA was so much lower, it had the highest translational index.
In the last set, strains with argC expression under the pspeB promoter exhibited the lowest values of mRNA and proteomic protein production. The complemented strains showed mRNA increases of 5-fold (S. meliloti), 7-fold (R. etli), 11-fold (A. tumefaciens), and 30-fold (M. loti) compared to the wild-type strain. However, the last three strains showed similar levels in proteomic protein (a 7-fold increase compared to the wild type) and very low translational index values.
Another complete set of constructs was obtained with pBBR1MCS3 and argC alone (without gus reporter). Similar values and proportions among the strains were found in comparison to the set of strains described above (data not shown). However, strains with pBBR1MCS3 showed slightly increased duplication times, and some reduced ArgC activity (not shown). It is possible that gus fused to argC genes produced some type of stabilization.
The described molecular parameters, together with the translation/transcription index, provided relevant information about the functional performance of the complemented strains, which demonstrated that complementation of ArgC with the S. meliloti argC gene in its own background showed the highest translational index (Table 5), followed by those with the R. etli and A. tumefaciens orthologs.
DISCUSSION
The species signature was intended to extract the amount of particular specific amino acid residues in positions of multiple alignments of the orthologous protein sequences (32). In this context, the species signature denotes the change in physicochemical properties of the proteins and displays certain particular characteristics of the orthologous products. It is relevant because the sequence differences of the orthologs are translated into amino acid residues; we wanted to establish whether these differences can be studied by ortholog complementation and functional performance when compared in the same cellular background and related to wild-type complementation and wild-type strain.
We studied the argC gene because it forms part of an importantly compromised pathway that is central to amino acid and protein synthesis; also, arginine has the highest nitrogen content among the amino acids, its synthesis demands a huge amount of energy, and it is a precursor for polyamine synthesis (14, 48, 63, 68). The S. meliloti argC gene was mutated in the chromosome, and then the complementation was obtained either with its own gene or with the ortholog from closely Rhizobiales species; they were cloned on the same vector and expressed under the same promoter to study the ability to recover the arginine prototrophy.
Three promoters were used in order to achieve diverse expression degrees, and we found strong (plac), medium (pargC), and weak (pspeB) transcriptional levels (see Table S2 in the supplemental material). That from speB was used because the gene is upstream to argC in Rhizobiales and participates in polyamine synthesis (e.g., arginine utilization), which is important in diverse metabolic processes but not so abundant as amino acids (68). With these promoters we searched expression levels similar to those of the wild-type strain. In the case of argC complementation under the pspeB promoter, we obtained the lowest ArgC specific activity and major differences, in regard to the optimal maintenance of cellular growth: the gene sequences from R. etli and A. tumefaciens showed reduced growth ability and that from M. loti showed no growth (Fig. 4E and F).
Interestingly, diverse factors altered the expression of the argC orthologous sequences and their complementation ability in several levels. In the first level, there was a differential multicopy effect due to plasmid vectors, because under the speB promoter we found two to three plasmid copies with S. meliloti, R. etli, and A. tumefaciens sequences, but nine copies with the M. loti gene. The most divergent sequence from S. meliloti was precisely that from M. loti, and the complemented strain carrying this gene presented marked inability to recover arginine synthesis and restore cell growth (Fig. 4E and F). The increased plasmid copy number was possibly related to a gene dosage compensation to favor abundance of the gene, as observed in other bacteria (67) and eukaryotes (18). Lind et al. (46), in a recent report dealing with orthologous replacement of ribosomal proteins in Salmonella enterica serovar Typhimurium, found that gene amplification is a compensatory mechanism to restore fitness decay. In our results, despite increase in gene copy number, growth was not fully recovered. Lind et al. showed that gene amplification is important for horizontally transferred genes and the evolution of new functions from gene duplication.
In the next level—transcription of the orthologous genes—also was affected. Theoretically, different genetic sequences located downstream of the same promoter will have the same level of transcriptional activity. However, the argC coding sequence itself appeared to modulate transcription to a certain extent (Tables 2And 5). This may be due to codon usage (but in this case similar CAI values were obtained, and thus codon usage apparently did not play a role in complementation efficiency), codon distribution (8), interaction of translation factors with RNA polymerase and ribosome (7, 59), and the formation of mRNA secondary structures that can alter the speed of the transcriptional complex. Other authors have made the same observation, at the level of transcription-translation processes (43). In addition, as reported recently, important transcription modulation occurs in the first and last 100 nucleotides of the genes (71). A similar effect was clearly observed in transcriptional fusions with segments of genes (Table 2) and also when expression was directly quantified by RT-PCR (Table 5), because under each of the three promoters different amounts of synthesized mRNA were found for each argC gene.
The previously described effects on transcription-translation processes, altogether with coding sequence differences of the orthologs (the species signature), consequently affected enzymatic activity of ArgC (Table 4), having a physiological repercussion on growth rate (Fig. 4) and metabolite excretion to the culture medium (Table 3). The diminished growth rate is related to intermediary metabolites that cannot be completely assimilated and are excreted in huge amounts (19). To perform an in-depth analysis of what was occurring in the complementation assay, we decided to measure molecular parameters such as direct mRNA and protein quantification (Table 5) to obtain a translational efficiency index; these parameters helped to clarify the transcriptional-translational ability of argC genes cloned on plasmids.
The S. meliloti wild-type strain showed the best translational efficiency, with almost constant values; this reproducibility supported the reliability of our approach (Table 5). In the complementation with the S. meliloti gene under the pspeB promoter, the copy number effect was evident, since a 5-fold increase in the mRNA amount was found, compared to the wild-type strain. However, this value did not correlate directly with the copy number found (i.e., 2.9), so possibly something else influenced the transcription rate, such as the supercoiling state of the replicon (24).
Under the plac promoter, the highest amounts of transcripts were obtained, but not the maximum translational efficiency indexes. It was apparent that there is a translational limit in the cell. In this case, the R. etli-complemented strain obtained the lowest index, however, it was sufficient to promote an adequate growth recovery (Fig. 4C and D). Under the pspeB promoter, despite the fact that the M. loti-complemented strain showed more gene copies, that amount was not enough to sustain cell growth, possibly because cells had a low translation efficiency index (Table 5And Fig. 4E and F). It is important to note the lack of correlation between protein synthesis and enzymatic activity, which was very marked in the case of plac constructs. We believe that there exists a physiological limitation to reaching high levels of enzymatic activity, a limitation that was exerted mainly in the case of Rhizobiales complementing sequences (because the S. meliloti sequence indeed showed a high level of ArgC activity under plac). Several other enzymatic and physiologic factors may participate and alter the correlation between transcription and translation activity (such as stoichiometry, cofactors, regulators, enzymes of the pathway, energy flux, etc.). Another consideration is that activity was measured in vitro and that this does not completely reflect the physiologic alteration of argC ortholog expression. We thus consider that cellular growth was a more faithful measure of what occurred with ortholog complementation.
The ArgC proteins from S. meliloti, R. etli, and A. tumefaciens displayed similar amounts of species signature differences (ca. 10%), which was indicative of their phylogenetic proximity. Sequences from B. melitensis and M. loti were evolutionarily more distant (Fig. 2), and the species signatures were higher (36% for M. loti) (Fig. 1). We sequenced the argC gene from several strains of each species (with the exception of Brucella melitensis), and the phylogenetic relationships between these sequences helped to define clear species clusters (Fig. 2). The genomic organization of argC genes in Rhizobiales was very unusual, since this gene was closely linked to speB (from the polyamine synthesis pathway). Possibly, these genes have independent promoters and mechanisms of transcriptional regulation, with the ability to respond appropriately and effectively to particular regulatory signals.
Transcriptional activation by species-specific argC promoters was also characterized in the native background and in S. meliloti and M. loti. In the native backgrounds, the lowest activity was seen in S. meliloti, the highest was seen in M. loti, and the other species were intermediate (Fig. 3). Interestingly, the argC promoter from M. loti was not active in S. meliloti; in contrast, the argC promoters of all of the strains were highly active in M. loti (Fig. 3). A partial growth defect was also apparent in the A. tumefaciens- and R. etli-complemented strains when the expression of argC was driven by the speB promoter (Fig. 4E and F). This partially defective growth phenotype correlated with a slight increase of α-ketoglutarate excretion to the culture medium and less utilization of succinate in comparison with the wild-type strain (Table 3). A similar phenotype was observed for a GOGAT mutation in R. etli (10).
Orthologous genes encode the same function (although some differences have been described for the HoxA3 ortholog in mice and zebrafish) (11), but they may also contain sequence characteristics that reflect specific adaptations. The specific sequence differences in a gene alter the physicochemical characteristics of the encoded proteins and may affect protein function. Factors such as transcription and translation rates, enzyme kinetics, transcriptional regulation, intracellular environment, availability of substrates, and interacting metabolites, proteins, and ions could also affect performance (30, 39, 55, 72).
Species adaptation is a slow process. Neutral drift (point mutations) and natural selection produce numerous changes in a gradual manner, each change having a small effect on fitness. However, some changes can have vast effects on an organism and tend to outcompete small-effect mutations. It has been generally accepted that any attempt to discern these changes experimentally is futile (12). Despite the fact that “species” differentiation in bacteria is considered imprecise, and its existence is even debatable (26, 44), homologous recombination and point mutations can gradually convert a homogeneous bacterial population into different, nonrecombinable populations. In this way, small (neutral and almost neutral) and large (non-neutral) genetic changes could eventually result in a new species. It remains to be seen whether the elemental mechanisms for adaptation, considered microevolution, can reconstruct the higher processes of macroevolution, including speciation and the emergence of biodiversity (12). The appearance of a new phenotype, such as citrate utilization in the presence of oxygen by an E. coli population in Lenski's long-term experiment, is something close to speciation (2, 5). However, even random variation (bet hedging) can enhance long-term fitness by increasing the likelihood of individuals to express an adaptive phenotype (3). There is a general lack of knowledge of how ortholog replacement may affect cell functioning and fitness. In this regard, the present study is a valid approach to sorting out the diverse consequences of expression in one species of orthologous sequences from other related species, in which specific amino acid changes may have been derived through adaptation.
In the present study we have demonstrated that an in-deep complementation analysis can uncover unusual complexity. We evaluated the ability of ArgC orthologs from closely related Rhizobiales species containing specific sequence differences to physiologically complement the S. meliloti argC mutant phenotype. We observed many factors participating in the complementation efficiency of each ortholog. In addition, we identified a novel promoter for argC in Rhizobiales, the presence of which was not predicted by existing genomic information. In agreement with recent reports (27, 43), we demonstrated that coding sequences participate in the regulation of transcription and translational efficiency, perhaps modulating mRNA stability through the formation of secondary structures. Altogether, these are important findings that open new research perspectives.
Supplementary Material
Acknowledgments
This study was partially supported by the Dirección General de Asuntos del Personal Académico (DGAPA-Universidad Nacional Autónoma de México), grants IN215307 and IN212710, and by the Ph.D. Program in Biomedical Sciences-UNAM.
We thank Mario Alberto Flores for help in the construction of the S. meliloti argC mutant; Michael Dunn for ArgB purification; Sandra Contreras, Miguel Elizalde, Magdalena Hernández, and Gabriel Martínez-Batallar for metabolite and proteome determinations; Gabriela Guerrero and Alejandro Aguilar for preparation of graphs; Ricarda Rivero, Yunuen Acevedo, and Alma Reyes (CCG-UNAM) for technical assistance; and Claudia Silva (IBt-UNAM) and Clive Ronson (Otago University, Otago, New Zealand) for the donation of strains.
Footnotes
Published ahead of print on 12 November 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Anisimova, M., and O. A. Gascuel. 2006. Approximate likelihood ratio test for branches: a fast, accurate and powerful alternative. Syst. Biol. 55:539-552. [DOI] [PubMed] [Google Scholar]
- 2.Barrick, J. E., S. Y. Yu, S. H. Yoon, H. Jeong, T. K. Oh, D. Schneider, R. E. Lenski, and J. F. Kim. 2009. Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature 461:1243-1247. [DOI] [PubMed] [Google Scholar]
- 3.Beaumont, H. J. E., J. Gallie, C. Kost, G. C. Ferguson, and P. B. Rainey. 2010. Experimental evolution of bet hedging. Nature 462:90-93. [DOI] [PubMed] [Google Scholar]
- 4.Better, M., and D. R. Helinski. 1983. Isolation and characterization of the recA gene of Rhizobium meliloti. J. Bacteriol. 155:311-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blount, Z. D., C. Z. Borland, and R. E. Lenski. 2008. Historical contingency and the evolution of a key innovation in an experimental population of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 105:7899-7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 7.Burmann, B. M., K. Schweimer, X. Luo, M. C. Wahl, B. L. Stitt, M. E. Gottesman, and P. Rösch. 2010. A NusE:NusG complex links transcription and translation. Science 328:501-504. [DOI] [PubMed] [Google Scholar]
- 8.Cannarozzi, G., N. N. Schraudolph, M. Faty, P. von Rohr, M. T. Friberg, A. C. Roth, P. Gonnet, G. Gonnet, and Y. Barral. 2010. A role for codon order in translation dynamics. Cell 141:355-367. [DOI] [PubMed] [Google Scholar]
- 9.Carvalho, F. M., R. C. Souza, F. G. Barcellos, M. Hungria, and T. R. Vasconcelos. 2010. Genomic and evolutionary comparisons of diazotrophic and pathogenic bacteria of the order Rhizobiales. BMC Microbiol. 10:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo, A., H. Taboada, A. Mendoza, B. Valderrama, S. Encarnación, and J. Mora. 2000. Role of GOGAT in carbon and nitrogen partitioning in Rhizobium etli. Microbiology 146:1627-1637. [DOI] [PubMed] [Google Scholar]
- 11.Chen, L., P. Zhao, L. Wells, C. T. Amemiya, B. G. Condie, and N. R. Manley. 2010. Mouse and zebrafish Hoxa3 orthologues have nonequivalent in vivo protein function. Proc. Natl. Acad. Sci. U. S. A. 107:10555-10560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chouard, T. 2010. Revenge of the hopeful monster. Nature 463:864-867. [DOI] [PubMed] [Google Scholar]
- 13.Crossman, L. C., S. Castillo-Ramírez, C. McAnnula, L. Lozano, G. S. Vernikos, J. L. Acosta, Z. F. Ghazoui, I. Hernández-González, G. Meakin, A. W. Walker, M. F. Hynes, J. P. Young, J. A. Downie, D. Romero, A. W. Johnston, G. Dávila, J. Parkhill, and V. González. 2008. A common genomic framework for a diverse assembly of plasmids in the symbiotic nitrogen fixing bacteria. PloS One 3:e2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cunin, R., N. Glansdorff, A. Piérard, and V. Stalon. 1986. Biosynthesis and metabolism of arginine in bacteria. Microbiol. Rev. 50:314-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dereeper, A., V. Guignon, G. Blanc, S. Audic, S. Buffet, F. Chevenet, J.-F. Dufayard, S. Guindon, V. Lefort, M. Lescot, J.-M. Claverie, and O. Gascuel. 2008. Phylogeny.fr: robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 36:W465-469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dunn, M. F., G. Araíza, S. Encarnación, M. C. Vargas, and J. Mora. 2002. Effect of aniA (carbon flux regulator) and phaC (poly-β-hydroxybutyrate synthase) mutations on pyruvate metabolism in Rhizobium etli. J. Bacteriol. 184:2296-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar, R. C. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32:1792-1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edger, P. P., and J. C. Pires. 2009. Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chrom. Res. 17:699-717. [DOI] [PubMed] [Google Scholar]
- 19.Encarnación, S., M. Dunn, K. Willms, and J. Mora. 1995. Fermentative and aerobic metabolism in Rhizobium etli. J. Bacteriol. 177:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Encarnación, S., Y. Guzmán, M. F. Dunn, M. Hernández, M. D. C. Vargas, and J. Mora. 2003. Proteome analysis of aerobic and fermentative metabolism in Rhizobium etli CE3. Proteomics 3:1077-1085. [DOI] [PubMed] [Google Scholar]
- 21.Encarnación, S., M. Hernández, G. Martínez-Batallar, S. Contreras, M. D. C. Vargas, and J. Mora. 2005. Comparative proteomics using 2-D gel electrophoresis and mass spectrometry as tools to dissect stimulons and regulons in bacteria with sequenced or partially sequenced genomes. Biol. Proc. Online 7:117-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Encarnación, S., M. C. Vargas, M. F. Dunn, A. Dávalos, G. Mendoza, Y. Mora, and J. Mora. 2002. AniA regulates reserve polymer accumulation and global protein expression in Rhizobium etli. J. Bacteriol. 184:2287-2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ferraioli, S., R. Taté, E. Caputo, A. Lamberti, A. Riccio, and E. J. Patriarca. 2001. The Rhizobium etli argC gene is essential for arginine biosynthesis and nodulation of Phaseolus vulgaris. Mol. Plant-Microbe Interact. 14:250-254. [DOI] [PubMed] [Google Scholar]
- 24.Figueroa, N., and L. Bossi. 1988. Transcription induces gyration of the DNA template in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 85:9416-9420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. U. S. A. 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fraser, C., W. P. Hanage, and B. G. Spratt. 2007. Recombination and the nature of bacterial speciation. Science 315:476-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fredrick, K., and M. Ibba. 2010. How the sequence of a gene can tune its translation. Cell 141:227-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Girard, L., S. Brom, A. Dávalos, O. López, M. Soberón, and D. Romero. 2000. Differential regulation of fixN-reiterated genes in Rhizobium etli by a novel fixL-fixK cascade. Mol. Plant-Microbe Interact. 13:1283-1292. [DOI] [PubMed] [Google Scholar]
- 29.González, V., R. I. Santamaría, P. Bustos, I. Hernández-González, A. Medrano-Soto, G. Moreno-Hagelsieb, S. C. Janga, M. A. Ramírez, V. Jiménez-Jacinto, J. Collado-Vides, and G. Dávila. 2006. The partitioned Rhizobium etli genome: genetic and metabolic redundancy in seven interacting replicons. Proc. Natl. Acad. Sci. U. S. A. 103:3834-3839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goryanin, I. I., G. V. Lebedeva, E. A. Mogilevskaya, E. A. Metelkin, and O. V. Demin. 2006. Cellular kinetic modeling of the microbial metabolism. Methods Biochem. Anal. 49:437-488. [PubMed] [Google Scholar]
- 31.Guerra, H. 2007. The brucellae and their success as pathogens. Crit. Rev. Microbiol. 33:325-331. [DOI] [PubMed] [Google Scholar]
- 32.Guerrero, G., H. Peralta, A. Aguilar, R. Diaz, M. A. Villalobos, A. Medrano-Soto, and J. Mora. 2005. Evolutionary, structural and functional relationships revealed by comparative analysis of syntenic genes in Rhizobiales. BMC Evol. Biol. 5:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guindon, S., and O. Gascuel. 2003. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52:696-704. [DOI] [PubMed] [Google Scholar]
- 34.Hanahan, D., J. Jessee, and F. R. Bloom. 1991. Plasmid transformation of Escherichia coli and other bacteria. Methods Enzymol. 204:63-113. [DOI] [PubMed] [Google Scholar]
- 35.Hurkman, W. J., and C. K. Tanaka. 1986. Solubilization of plant membrane proteins for analysis by two-dimensional gel electrophoresis. Plant Physiol. 81:802-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hynes, M. F., and N. F. McGregor. 1990. Two plasmids other than the nodulation plasmid are necessary for formation of nitrogen-fixing nodules by Rhizobium leguminosarum. Mol. Microbiol. 4:567-574. [DOI] [PubMed] [Google Scholar]
- 37.Igarashi, K., and K. Kashiwagi. 2000. Polyamines: mysterious modulators of cellular functions. Biochem. Biophys. Res. Commun. 271:559-564. [DOI] [PubMed] [Google Scholar]
- 38.Kaneko, T., Y. Nakamura, S. Sato, E. Asamizu, T. Kato, S. Sasamoto, A. Watanabe, K. Idesawa, A. Ishikawa, K. Kawashima, T. Kimura, Y. Kishida, C. Kiyokawa, M. Kohara, M. Matsumoto, A. Matsuno, Y. Mochizuki, S. Nakayama, N. Nakazaki, S. Shimpo, M. Sugimoto, C. Takeuchi, M. Yamada, and S. Tabata. 2000. Complete genome structure of the nitrogen-fixing symbiotic bacterium Mesorhizobium loti. DNA Res. 7:331-338. [DOI] [PubMed] [Google Scholar]
- 39.Khersonsky, O., and D. S. Tawfik. 2010. Enzyme promiscuity: a mechanistic and evolutionary perspective. Annu. Rev. Biochem. 79:11.1-11.35. [DOI] [PubMed] [Google Scholar]
- 40.Knight, C. G., R. Kassen, H. Hebestreit, and P. B. Rainey. 2004. Global analysis of predicted proteomes: functional adaptation of physical properties. Proc. Natl. Acad. Sci. U. S. A. 101:8390-8395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Korbel, J. O., L. J. Jensen, C. von Mering, and P. Bork. 2004. Analysis of genomic context: prediction of functional associations from conserved bidirectionally transcribed gene pairs. Nat. Biotechnol. 22:911-917. [DOI] [PubMed] [Google Scholar]
- 42.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 43.Kudla, G., A. W. Murray, D. Tollervey, and J. B. Plotkin. 2009. Coding-sequence determinants of gene expression in Escherichia coli. Science 324:255-258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lawrence, J. G. 2002. Gene transfer in bacteria: speciation without species? Theoret. Pop. Biol. 61:449-460. [DOI] [PubMed] [Google Scholar]
- 45.Lemoine, F., O. Lespinet, and B. Labedan. 2007. Assessing the evolutionary rate of positional orthologous genes in prokaryotes using synteny data. BMC Evol. Biol. 7:237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lind, P. A., C. Tobin, O. G. Berg, C. G. Kurland, and D. I. Andersson. 2010. Compensatory gene amplification restores fitness after inter-species gene replacements. Mol. Microbiol. 75:1078-1089. [DOI] [PubMed] [Google Scholar]
- 47.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 48.Llácer, J. L., I. Fita, and V. Rubio. 2008. Arginine and nitrogen storage. Curr. Opin. Struct. Biol. 18:673-681. [DOI] [PubMed] [Google Scholar]
- 49.Lu, C. D., Z. Yang, and W. Li. 2004. Transcriptome analysis of the ArgR regulon in Pseudomonas aeruginosa. J. Bacteriol. 186:3855-3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Masson-Boivin, C., E. Giraud, X. Perret, and J. Batut. 2009. Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 7:458-466. [DOI] [PubMed] [Google Scholar]
- 51.Meade, H. M., S. R. Long, G. B. Ruvkun, S. E. Brown, and F. M. Ausubel. 1982. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J. Bacteriol. 149:114-122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Moreno-Hagelsieb, G., and J. Collado-Vides. 2002. A powerful non-homology method for the prediction of operons in prokaryotes. Bioinformatics 18:S329-S326. [DOI] [PubMed] [Google Scholar]
- 53.Noel, K. D., A. Sánchez, L. Fernández, J. Leemans, and M. A. Cevallos. 1984. Rhizobium phaseoli mutants with transposon Tn5 insertions. J. Bacteriol. 158:148-155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pappas, K. M. 2008. Cell-cell signaling and the Agrobacterium tumefaciens Ti plasmid copy number fluctuations. Plasmid 60:89-107. [DOI] [PubMed] [Google Scholar]
- 55.Phair, R. D. 1997. Development of kinetic models in the nonlinear world of molecular cell biology. Metabolism 46:1489-1495. [DOI] [PubMed] [Google Scholar]
- 56.Piette, J., R. Cunin, A. Boyen, D. Charlier, M. Crabeel, F. Van Vliet, N. Glansdorff, C. Squires, and C. L. Squires. 1982. The regulatory region of the divergent argECBH operon in Escherichia coli K-12. Nucleic Acids Res. 10:8031-8048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Prentki, P., and H. M. Krisch. 1984. In vitro insertional mutagenesis with a selectionable DNA fragment. Gene 29:303-313. [DOI] [PubMed] [Google Scholar]
- 58.Price, M., K. Huang, E. Alm, and A. Arkin. 2005. A novel method for accurate operon predictions in all sequenced prokaryotes. Nucleic Acids Res. 33:880-892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Proshkin, S., R. A. Rahmouni, A. Mironov, and E. Nudler. 2010. Cooperation between translating ribosomes and RNA polymerase in transcription elongation. Science 328:504-508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Quandt, J., and M. F. Hynes. 1993. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene 127:15-21. [DOI] [PubMed] [Google Scholar]
- 61.Quintero, M. J., A. M. Muro-Pastor, A. Herrero, and E. Flores. 2000. Arginine catabolism in the cyanobacterium Synechocystis sp. Strain PCC 6803 involves the urea cycle and arginase pathway. J. Bacteriol. 182:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Quinto, C., H. De la Vega, M. Flores, I. Fernández, T. Ballado, G. Soberón, and R. Palacios. 1982. Reiteration of nitrogen fixation gene sequences in Rhizobium phaseoli. Nature 299:724-726. [Google Scholar]
- 63.Randhawa, G. S., and R. Hassani. 2002. Role of rhizobial biosynthetic pathways of amino acids, nucleotide bases and vitamins in symbiosis. Indian J. Exp. Bot. 40:755-764. [PubMed] [Google Scholar]
- 64.Rocha, E. P. 2004. Order an disorder in bacterial genomes. Curr. Opin. Microbiol. 7:519-527. [DOI] [PubMed] [Google Scholar]
- 65.Sallstrom, B., and S. G. E. Andersson. 2005. Genome reduction in alphaproteobacteria. Curr. Opin. Microbiol. 8:579-585. [DOI] [PubMed] [Google Scholar]
- 66.Shevchuk, N. A., A. V. Bryksin, Y. A. Nusinovich, F. C. Cabello, M. Sutherland, and S. Ladisch. 2004. Construction of long DNA molecules using long PCR-based fusion of several fragments simultaneously. Nucleic Acids Res. 32:e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Svenningsen, S. L., K. C. Tu, and B. L. Bassler. 2009. Gene dosage compensation calibrates four regulatory RNAs to control Vibrio cholerae quorum sensing. EMBO J. 28:429-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tabor, C. W., and H. Tabor. 1985. Polyamines in microorganisms. Microbiol. Rev. 49:81-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tamames, J. 2001. Evolution of gene order conservation in prokaryotes. Genome Biol. 2:R0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tuller, T., A. Carmi, K. Vestsigian, S. Navon, Y. Dorfan, J. Zaborske, T. Pan, O. Dahan, I. Furman, and Y. Pilpel. 2010. An evolutionary conserved mechanism for controlling the efficiency of protein translation. Cell 141:344-354. [DOI] [PubMed] [Google Scholar]
- 72.Zhao, J., D. Ridgway, G. Broderick, A. Kovalenko, and M. Ellison. 2008. Extraction of elementary rate constants from global network analysis of Escherichia coli central metabolism. BMC Syst. Biol. 2:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.