Abstract
CsgD and cyclic-3′,5′-di-guanylate are key regulators of biofilm formation in Salmonella enterica serovar Typhimurium. Our results show that polynucleotide phosphorylase and NlpI oppositely altered expression of CsgD. Polynucleotide phosphorylase and NlpI also had opposite effects on the expression of yjcC, which codes for a cyclic-3′,5′-di-guanylate phosphodiesterase affecting CsgD expression.
The ability to create multicellular communities in the form of a biofilm has gained increased appreciation as a prokaryotic response to selected environmental cues (3, 6, 8). In Salmonella enterica serovar Typhimurium and Escherichia coli, biofilm formation is initiated upon growth at a low temperature on solid media of low osmolarity (5, 7, 13). In the presence of the dye Congo red, this biofilm is characterized by a red, dry and rough (rdar) colony appearance (6). The rdar morphotype involves production of at least four extracellular matrix components, including CsgAB fimbriae (2, 6, 7).
Biofilm development in S. Typhimurium is a genetically highly regulated process that includes an array of key regulatory proteins (8) and the secondary messenger cyclic-di-guanylic acid monophosphate (c-di-GMP) (6, 8, 12). The intracellular level of c-di-GMP is spatially and temporally regulated by the activities of GGDEF and EAL domain proteins (8, 12). GGDEF and EAL domain proteins synthesize and degrade c-di-GMP, respectively (8, 12).
Polynucleotide phosphorylase (PNPase) (encoded by pnp) is an evolutionarily conserved enzyme affecting gene expression in bacteria, plants, and mammals (10). In E. coli, PNPase assists acclimatization to cold through its ability to degrade selected cold-induced transcripts (14). We have previously demonstrated that PNPase affects around 4% of the coding sequences in S. Typhimurium during growth in rich medium (15). Recently we noted that PNPase also contributes to growth at decreased temperatures and motility of S. Typhimurium. The expression of these phenotypes also required the membrane protein NlpI (S. F. Rouf et al., unpublished data). Here we demonstrate that PNPase and NlpI, in contrast, regulate rdar morphotype development in S. Typhimurium SR-11 oppositely.
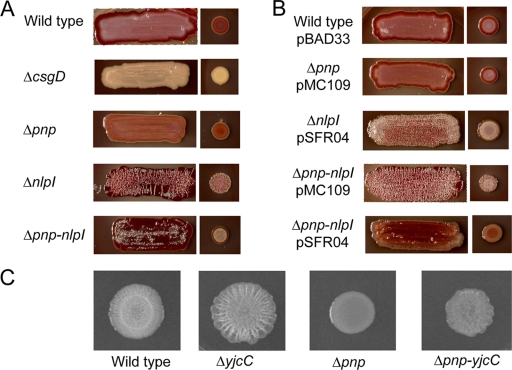
rdar morphotypes of pnp and nlpI mutants.
rdar morphotype development was assayed on Congo red Luria agar plates without salt at 28°C (12). Plates were inoculated with a 10-μl drop of overnight cultures grown in Luria broth suspended in phosphate-buffered saline (PBS) to an optical density at 600 nm (OD600) of 0.5. The rdar morphotype development was followed for up to 72 h. When necessary, plates were supplemented with antibiotics (ampicillin, 100 μg/ml; chloramphenicol, 10 μg/ml; and kanamycin sulfate, 50 μg/ml; Sigma Aldrich). For induction of NlpI from the recombinant plasmid pSFR04 (cloned into pBAD33 under the araC promoter; S. F. Rouf et al., unpublished data), media were supplemented with 0.1% l-(+)-arabinose (Sigma).
Compared to that of the wild type, development of the rdar morphotype was delayed in the pnp mutant and enhanced in the nlpI mutant (Fig. 1A). The pnp nlpI double mutant appeared more rugous than the wild type but less rugous than the nlpI mutant (Fig. 1A). Complementation with the cloned pnp gene (pMC109) (1) restored the rdar morphotype in the pnp mutant. The cloned nlpI gene (pSFR04) suppressed biofilm development in the nlpI mutant (Fig. 1B).
FIG. 1.
Formation of the rdar morphotype in S. Typhimurium on Congo red plates. Plates were seeded as either streaks or drop-on lawns. Incubation was carried out at 28°C for 72 h, except for noncomplemented drop-on lawns, which were read after 48 h.
Expression of CsgD and CsgA in pnp and nlpI mutants.
CsgD is a master activator of biofilm formation through its ability to induce expression of the CsgA and CsgB fimbrial subunits and cellulose (3, 5, 9). We therefore compared the expression of CsgD and CsgA in the wild-type and mutant strains. CsgD was assayed by immunoblotting (12). Equal numbers of wild-type and mutant bacteria grown in LB without salt at 28°C for 20 h were solubilized in SDS-PAGE sample buffer. The proteins were separated on 12% SDS-PAGE gels (4) and transferred to polyvinylidene difluoride membranes (Hybond P; Amersham). Polyclonal rabbit anti-CsgD peptide antiserum (1:5,000) and horseradish peroxidase-conjugated goat anti-rabbit IgG antibodies (1:5,000; Pierce) were used as primary and secondary antibodies (7). Chemoluminescence signal intensities of CsgD were analyzed using the Quantity One software program (Bio-Rad). CsgA was extracted from bacterial cultures grown at 28°C for 20 h on LB agar without salt (12). CsgA was depolymerized in formic acid, and the CsgA monomers were analyzed on 12% SDS-PAGE gels (4) after staining with Coomassie blue.
In agreement with rdar morphotype development, expression of CsgD and CsgA was substantially reduced in the pnp mutant (Fig. 2, left panels). In contrast, the nlpI mutant showed increased expression of both CsgD and CsgA (Fig. 2, left panels). Quantification of immunoblot signals revealed a more than 10-fold difference in CsgD expression when the pnp and nlpI mutants were compared (Fig. 2, top left). The pnp nlpI mutant showed an intermediate level of CsgD expression (Fig. 2, top left).
FIG. 2.
Effects of pnp and nlpI mutations on CsgD and CsgA expression in S. Typhimurium. CsgD and CsgA expression was analyzed by immunoblotting with Coomassie blue-stained 12% SDS-PAGE gels. Relative signal intensities and the ratios thereof are shown for CsgD.
To estimate csgD and csgA mRNA levels, total bacterial RNA was isolated using the TRI Reagent (Sigma) from cultures grown in LB without salt at 28°C. RNA samples were reverse transcribed for quantitative real-time PCR (qRT-PCR) (SYBR green Jumpstart; Sigma) (Table 1). The transcripts were quantified on an ABI Prism 7000 sequence detection system (Applied Biosystems). To assess qRT-PCR primer efficiencies, the dissociation curve was analyzed for each primer pair. Each of these showed a single peak. Relative signals of the transcripts were analyzed using the rfaH gene as an endogenous control, since the expression of rfaH is unaffected by pnp (15). RNA samples that had not been reversed transcribed were used as negative controls for DNA contamination. In these analyses, the csgD and csgA mRNA levels closely followed the corresponding protein expression levels (Fig. 3).
TABLE 1.
Primers used for qRT-PCR
| Primer | Sequence | Target gene |
|---|---|---|
| FadrA | TCGCTGGAAGTCACGCTCT | adrA |
| RadrA | CGCTTATGTTCCGCTAATTTAATG | adrA |
| FcsgA | CATCGACCAGTGGAACGCTA | csgA |
| RcsgA | TTACGCTGGAATCAGATGCG | csgA |
| FcsgD | CCCTGACGATTATCCCTACCG | csgD |
| RcsgD | CCCTGTAATCCGCTGACCAC | csgD |
| FrfaH | GGCGTTGTCGATCCTGAAAC | rfaH |
| RrfaH | CGTGATGATGACGCTATCGC | rfaH |
| FyciR | GCGAATACAGACACCGTCACC | yciR |
| RyciR | ACGATACCCACCTGCGACTC | yciR |
| FyjcC | AAAAACGCGTCAGCATTCAGT | yjcC |
| RyjcC | CCGCGCTCAGTCACTTCAA | yjcC |
| FyhjH | CTGCTGCTGCAACTGATGAAC | yhjH |
| RyhjH | CTCCACGCCCTCGACAAT | yhjH |
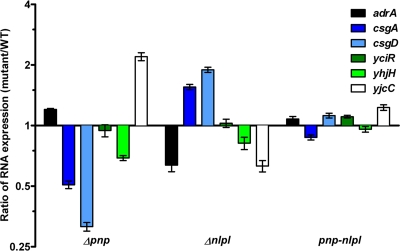
FIG. 3.
Effect of pnp and nlpI mutations on gene expression in S. Typhimurium. Transcript expression ratios for adrA, csgA, csgD, yciR, yhjH, and yjcC mRNA levels as given for the pnp, nlpI, and pnp nlpI mutants in relation to those for the wild type at 8 h postinoculum.
Biofilm formation in yjcC mutants.
The c-di-GMP synthetase AdrA and the phosphodiesterases YhjH, YciR, and YjcC all have an impact on rdar morphotype development (3, 5, 12). YjcC and YciR in particular suppress expression of CsgD (12). We therefore extended the qRT-PCR analysis to include adrA, yciR, yjcC, and yhjH. The mRNA coding for the biofilm suppressor YjcC was markedly increased in the pnp mutant and was decreased in the nlpI mutant (Fig. 3). Furthermore, pnp and nlpI had minor, but inverse, effects on the expression of adrA. The expression levels of yjcC, yhjH, and adrA were restored to wild-type levels in the pnp nlpI mutant (Fig. 3).
To establish a connection between pnp, yjcC, and biofilm formation, we constructed a yjcC mutant and a pnp yjcC double mutant by P22 int phage transduction (11). Compared to that of the wild type, biofilm development was enhanced in the yjcC and pnp yjcC mutants (Fig. 1C). When CsgD expression was quantified by immunoblotting, the yjcC mutant revealed a 2.8-fold enhancement in CsgD expression (Fig. 2, top right). When the yjcC mutation was introduced into the pnp mutant, we noted a 4.6-fold enhancement in the expression of CsgD (Fig. 2).
Conclusion.
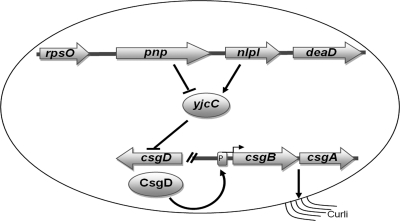
Here we have demonstrated that PNPase and NlpI have opposite effects on the expression of CsgD, a key activator of biofilm formation. This observation is associated with a subsequent divergent effect of PNPase and NlpI on the expression of CsgA and the biofilm-associated rdar morphotype development. YjcC is a major phosphodiesterase suppressing CsgD expression (12). Expression of yjcC was increased in the pnp mutant and decreased in the nlpI mutant. The altered expression of yjcC became restored in the pnp nlpI double mutant. In parallel, we noted that the impact of YjcC on the expression of CsgD was more pronounced in the absence of PNPase. Therefore, the effect of PNPase and NlpI on biofilm formation could relate to their opposite effects on the expression of YjcC (Fig. 4). These results also bring forth an unrecognized gene regulatory connection between PNPase and c-di-GMP metabolism.
FIG. 4.
Model for the concerted activity of pnp, nlpI, and yjcC in relation to the expression of csgD.
Acknowledgments
This study was supported by the Swedish Medical Research Council. S.F.R. is a Ph.D. fellow from International Research Training Group 1273, funded by the German Research Foundation, N.A. and I.A. are Ph.D. fellows of the Higher Education Commission, Pakistan, whereas A.K. was funded by Elitforskartjänst (KI).
Footnotes
Published ahead of print on 12 November 2010.
REFERENCES
- 1.Clements, M. O., S. Eriksson, A. Thompson, S. Lucchini, J. C. Hinton, S. Normark, and M. Rhen. 2002. Polynucleotide phosphorylase is a global regulator of virulence and persistency in Salmonella enterica. Proc. Natl. Acad. Sci. U. S. A. 99:8784-8789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Collinson, S. K., S. C. Clouthier, J. L. Doran, P. A. Banser, and W. W. Kay. 1996. Salmonella enteritidis agfBAC operon encoding thin, aggregative fimbriae. J. Bacteriol. 178:662-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonas, K., Ö. Melefors, and U. Römling. 2009. Regulation of c-di-GMP metabolism in biofilms. Future Microbiol. 4:341-358. [DOI] [PubMed] [Google Scholar]
- 4.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 5.Pesavento, C., G. Becker, N. Sommerfeldt, A. Possling, N. Tschowri, A. Mehlis, and R. Hengge. 2008. Inverse regulatory coordination of motility and curli-mediated adhesion in Escherichia coli. Genes Dev. 22:2434-2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Römling, U. 2005. Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell. Mol. Life Sci. 62:1234-1246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Römling, U., Z. Bian, M. Hammar, W. D. Sierralta, and S. Normark. 1998. Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J. Bacteriol. 180:722-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Römling, U., D. Pesen, and S. Yaron. 2007. Biofilms in Salmonella enterica, p. 127-146. In M. Rhen, D. Maskell, P. Mastroeni, and J. Therfall (ed.), Salmonella: molecular biology and pathogenesis. Horizon Bioscience, Norwich, United Kingdom.
- 9.Römling, U., M. Rohde, A. Olsén, S. Normark, and J. Reinköster. 2000. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 36:10-23. [DOI] [PubMed] [Google Scholar]
- 10.Sarkar, D., and P. B. Fisher. 2006. Polynucleotide phosphorylase: an evolutionary conserved gene with an expanding repertoire of functions. Pharmacol. Ther. 112:243-263. [DOI] [PubMed] [Google Scholar]
- 11.Schmieger, H. 1972. Phage P22-mutants with increased or decreased transduction abilities. Mol. Gen. Genet. 119:75-88. [DOI] [PubMed] [Google Scholar]
- 12.Simm, R., A. Lusch, A. Kader, M. Andersson, and U. Römling. 2007. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar Typhimurium. J. Bacteriol. 189:3613-3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sukupolvi, S., R. G. Lorenz, J. I. Gordon, Z. Bian, J. D. Pfeifer, S. J. Normark, and M. Rhen. 1997. Expression of thin aggregative fimbriae promotes interaction of Salmonella typhimurium SR-11 with mouse small intestinal epithelial cells. Infect. Immun. 12:5320-5325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yamanaka, K., and M. Inoune. 2001. Selective mRNA degradation by polynucleotide phosphorylase in cold shock adaptation in Escherichia coli. J. Bacteriol. 183:2808-2816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ygberg, S. E., M. O. Clements, A. Rytkönen, A. Thompson, D. W. Holden, J. C. Hinton, and M. Rhen. 2006. Polynucleotide phosphorylase negatively controls spv virulence gene expression in Salmonella enterica. Infect. Immun. 74:1243-1254. [DOI] [PMC free article] [PubMed] [Google Scholar]