Abstract
Three paralogous transcriptional activators MarA, SoxS, and Rob, activate >40 Escherichia coli promoters. To understand why MarA does not activate certain promoters as strongly as SoxS, we compared MarA, MarA mutants, and SoxS for their abilities to activate 16 promoters and to bind their cognate marbox binding sites. Replacement of the MarA glutamic acid residue 89 with alanine greatly increased the marbox binding and activation of many class I promoters. Like cells constitutive for SoxS, cells expressing the MarA with the E89A mutation were more resistant to superoxides than those harboring WT MarA. The activities of several other E89 substitutions ranked as follows: E89A > E89G > E89V > WT > E89D. Increased binding and activation occurred only at class I promoters when the 12th base of the promoter's marbox (a position at which there is no known interaction between the marbox and MarA) was not a T residue. Furthermore, WT MarA binding to a synthetic marbox in vitro was enhanced when the phosphate group between positions 12 and 13 was eliminated on one strand. The results demonstrate that relatively minor changes in a single amino acid side chain (e.g., alanine to valine or glutamic acid to aspartic acid) can strongly influence activity despite any evidence that the side chain is involved in positive interactions with either DNA or RNA polymerase. We present a model which attributes the differences in binding and activation to the interference between the β- and γ-carbons of the amino acid at position 89 and the phosphate group between positions 12 and 13.
The three paralogous Escherichia coli AraC/XylS family activators MarA, SoxS, and Rob are regulated by three different systems (marRAB, soxRS, and rob, respectively) in response to different stresses (29). These activators transcriptionally activate the same set of >40 promoters (the MarA-SoxS-Rob regulon) but to different extents (2, 26, 33, 43). This work was undertaken to discern structural differences between MarA and SoxS that might be responsible for this “promoter discrimination.”
Treatment of E. coli with phenolic derivatives, such as salicylate, inactivates MarR, leading to derepression of the marRAB operon (7, 24). The resultant increase in cellular resistance to low levels of diverse antibiotics and organic solvents is due primarily to activation of the efflux pump genes acrAB and tolC (3, 32).
Treatment with superoxide-generating compounds, such as paraquat, activates SoxR, which, in turn, transcriptionally activates the expression of soxS (5, 10, 31, 46). Upregulation of soxS renders the cells resistant to the same levels of antibiotics as but to greater levels of superoxides than does marRAB derepression (23). This is likely due to the greater extent of activation by SoxS of promoters involved in superoxide defense, e.g., acnA, fpr, zwf, fumC, and sodA. The Rob protein is primarily regulated posttranslationally (37, 38) and was not studied further here.
The basis for the activation of these promoters is the presence of a 20-bp binding site for MarA and SoxS (the marbox or soxbox). The consensus sequence for this site is highly degenerate, and the marbox must be in one of several configurations relative to the binding signals for RNA polymerase (RNAP) in order to be functional (17, 21, 22, 45). From X-ray crystallographic analysis of the cocrystal of MarA with the marbox from the marRAB promoter, two helix-turn-helix (HTH) motifs were identified that make 34 contacts with the DNA and bend it by 35° (36). While no physical structure is available for SoxS, it is likely to resemble MarA in how it binds DNA: at the MarA positions where amino acid side chains make DNA contact, the identity between MarA and SoxS is 60% (87% homology) even though the overall identity between the two proteins (1) is only 40% (56% homology). The structure of Rob bound to the micF marbox (19) is not pertinent here as it appears to be, in part, an artifact of crystal packing.
Among the promoters activated by MarA, the correlation between the strength of binding and the extent of activation is poor (23). For example, MarA binds tightly to the marRAB promoter in vitro yet stimulates transcription by only ∼3-fold; no significant binding to inaA can be demonstrated by gel shift experiments, yet transcription is stimulated >5-fold. This finding may be related to the fact that the basal transcription of marRAB is high, whereas that of inaA is low. A similar lack of correlation between binding strength and activation is found for SoxS (23).
In contrast, there is a correlation between the relative strengths of binding of MarA and SoxS to the marbox of a particular promoter and the relative abilities of these activators to stimulate that promoter (23). Since MarA activates some promoters more effectively and SoxS activates others more effectively, we wished to determine what structural differences might account for this promoter discrimination (23) and whether these differences were related to DNA binding at the corresponding marboxes.
In this report, we extend our earlier alanine-scanning mutagenesis studies (13) to a large number of promoters. Our principal finding is that MarA with the mutation E89A [MarA(E89A)] behaves more like SoxS than MarA in exhibiting greater activation and binding at class I promoters. We propose a model for the role of E89 in promoter discrimination.
MATERIALS AND METHODS
Bacterial strains.
Table S1 in the supplemental material provides a list of the strains used in this study. The lacZ transcriptional fusions have been described previously (12, 13, 20, 26, 27) except for acrAB::lacZ. This fusion contains the acrAB promoter from nucleotides 485037 to 484920 fused to lacZ and was constructed as described previously (39); the transcription start site is at 484922 (11). Most of the fusions were introduced into, and assayed in, strain M3997 (ΔmarRAB rob::kan Δlon clpP::cat [F′ proAB+ Tn10 lacIq]). The lon mutation prevents degradation of the proteolysis-sensitive MarA and SoxS proteins (15). However, the mdtG::lacZ fusion was introduced into, and assayed in, the wild-type (WT) strain GC4468, and the acrAB::lacZ fusion was introduced into, and assayed in, M4436, an acrR::cat derivative of GC4468. In experiments shown in Fig. 3, fpr::lacZ and mdtG::lacZ fusions were analyzed in strain GC4468. Because of the instability of the strain carrying E89A in GC4468, each strain (see Fig. 3) carrying the MarA wild type or mutant or the SoxS plasmid was reconstructed and purified immediately prior to assay. Since we have demonstrated that the MarA concentrations attained here (20) exceed the Km for the Lon protease, the levels of MarA in lon+ clpP+ cells are ∼65% of those in lon clpP cells and should not affect the results significantly. Both M3997 and N8452 have null mutations in marRAB and rob to eliminate the basal levels of expression of the activators; the WT soxS present on the chromosome is negligibly expressed (our unpublished observations).
The plasmids carrying marA, soxS, and most of the marA mutations used in this study are derivatives of pUC19 and have been described previously (13, 23). However, MarA(E89A) was reconstructed because plasmid recovered from frozen stocks (13) did not have the correct sequence. Strains carrying this plasmid were found to be highly unstable in the absence of lacIq. Other plasmids containing mutants of MarA or SoxS not previously described were constructed in a manner analogous to that of Gillette et al. (13). These include MarA mutants E25R, E77R, Y81D, E89D, E89G, E89V, R85D, E25R R85D, and E77R R85D and the SoxS mutant V83E. All mutations were verified by sequence analysis of both DNA strands.
β-Galactosidase assays.
β-Galactosidase activity was assayed using the SDS-CHCl3 method described by Miller (30). Cells containing a plasmid that expresses wild-type or mutant MarA or SoxS under lacIq control were grown to mid-log phase and derepressed for 1 h with 500 μM isopropyl-β-d-thiogalactopyranoside (IPTG) prior to assay. The poxB promoter, which requires RpoS function, was assayed in stationary-phase cultures. See Materials and Methods in the supplemental material for a discussion of the statistical analysis of the data. The data presented in Tables 2 and 3 and in Fig. 1Are normalized to the activity of the promoter-lacZ fusion carrying the WT MarA plasmid according to the following calculation: (activity of the mutant MarA − activity of the no-activator control)/(activity of WT MarA − activity of the no-activator control).
FIG. 1.
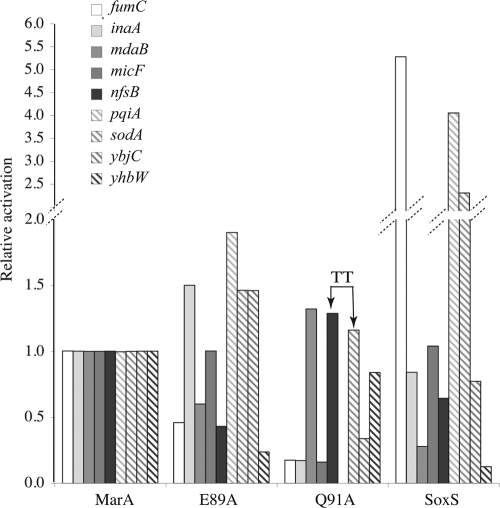
Activation of nine class II promoters by MarA, MarA mutants E89A and Q91A, and SoxS (Table 3 gives details.) The bases present in the sodA and nfsB marboxes at positions 17 and 18 of the consensus marbox are both Ts rather than As (indicated as TT at the top of the bars) (26). The relative activities were calculated as described in Materials and Methods. The WT MarA β-galactosidase activities (Miller units), control and induced, respectively, for the indicated promoters were as follows: for fumC, 75 (control) and 220 (induced); inaA, 100 and 1,400; mdaB, 150 and 3,000; micF, 80 and 800; nfsB, 80 and 1,000; pqiA, 25 and 100; sodA, 1,200 and 3,000; ybjC 150 and 3,800; and yhbW, 400 and 1,800.
The data for Fig. 3 are from overnight cultures diluted 100-fold and allowed to grow 2 h to mid-logarithmic phase before assay.
DNA binding assays.
DNA binding assays were performed as described previously (25). Briefly, MarA, MarA mutants, and SoxS containing the N-terminal fragment encoded in pET15b were purified as previously described (13, 18) and serially diluted 1.67-fold in buffer containing 50 mM HEPES, pH 8.0, 25% glycerol, 0.5 M NaCl, 100 μg/ml bovine serum albumin, and 500 nM poly(dA-dT) (20 bp in length). One microliter of activator was mixed with 9 μl of 5′ 32P end-labeled double-stranded DNA (dsDNA) fragment (20 bp in length; ∼5 pmol/ml, one strand of which is listed in Table 1) in Tris-acetate-EDTA (TAE) buffer with 25% glycerol and subjected to electrophoresis at 150 V on 6% gels for 35 min. Gels were dried and analyzed with a Molecular Dynamics PhosphorImager as previously described (21). Assays were performed in duplicate and had variances of <1 dilution, i.e., ± 1.67-fold. For the experiments employing DNA with one chain interrupted, the complementary 36-nucleotide (nt) fragment (including the marbox of fpr flanked by the 7 nt to its 5′ end and 9 nt to its 3′ end as listed in Table 1, i.e., CCTCTGATTGATTTGATCGATTGAGCCTTCCAGTCC) was end labeled and annealed either to a single unlabeled fragment having the sequence GGACTGGAAGGCTCAATCGATCAAATCAATCAGAGG (marbox underlined) or to two unlabeled fragments (both 5′ and 3′ ends dephosphorylated) together containing the appropriate sequence (e.g., the two oligonucleotides GGACTGGAAGGCTCAATCG and ATCAAATCAATCAGAGG).
TABLE 1.
DNA fragments used in binding studiesa
| Marbox type | Sequence of one strand of the marbox fragment at the indicated position |
|||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | 18 | 19 | 20 | |
| Consensus | A | Y | N | G | C | A | C | N | N | W | N | N | R | Y | Y | A | A | A | C | N |
| Promoter | ||||||||||||||||||||
| acnA | A | A | C | C | C | A | A | A | T | T | G | A | T | A | A | A | A | G | A | G |
| acrAB | A | T | G | G | C | A | C | G | A | A | A | A | A | C | C | A | A | A | C | A |
| fpr | A | A | G | G | C | T | C | A | A | T | C | G | A | T | C | A | A | A | T | C |
| fumC | A | T | G | G | C | A | C | G | A | A | A | G | A | C | C | A | A | A | C | A |
| inaA | A | C | G | A | C | A | C | G | T | T | T | C | A | T | T | A | A | G | A | T |
| marRAB | A | T | G | C | C | A | C | G | T | T | T | T | G | C | T | A | A | A | T | C |
| mdaB | T | T | T | G | C | A | C | A | T | T | T | T | G | C | T | A | A | T | T | T |
| mdtG | A | G | A | G | C | T | T | T | T | A | T | C | G | C | T | A | A | A | T | C |
| micF | A | C | A | G | C | A | C | T | G | A | A | T | G | T | C | A | A | A | A | C |
| nfsB | A | G | C | G | C | A | T | T | T | T | T | C | T | C | G | C | T | T | A | C |
| pqiA | A | A | A | G | C | A | G | A | A | A | C | T | G | T | A | A | A | A | C | G |
| poxB | G | A | G | G | C | A | C | T | A | A | C | G | G | T | T | A | A | A | T | A |
| sodA | A | C | G | G | C | A | T | T | G | A | T | A | A | T | C | A | T | T | T | T |
| ybjC | A | A | A | G | C | T | A | T | A | A | C | T | G | T | T | A | A | A | C | A |
| yhbW | A | T | A | G | C | T | C | A | C | T | T | T | G | T | T | A | A | C | A | A |
| zwf | A | T | C | G | C | A | C | G | G | G | T | G | G | A | T | A | A | G | C | G |
Both strands of the 20-mer marbox from each promoter were synthesized and annealed and used in the binding experiments. Each marbox is shown in the forward orientation (22). Although there is no base preference at position 20, which is why we refer to the marbox as being 19 bp in length, binding in vitro requires a 20-bp fragment (22). At position 12, 6 of the 16 marbox sequences have a T at this position (boldface). At positions 17 and 18, 9 of the 16 marboxes have an A at both positions; underlining shows where there is no A; only the sodA and nfsB marboxes have no A at either position.
Superoxide sensitivity assays.
Bacteria were assayed for superoxide sensitivity on gradient plates as described previously (23, 41).
RESULTS
Detailed examination of the crystal structure of MarA suggested that the two regions of the molecule containing the two helix-turn-helix motifs were likely to be less flexible than those of SoxS (see Fig. S1 in the supplemental material). We therefore considered the possibility that the existence of bridging contacts formed between these two regions by E25 and R85 but absent in SoxS might account in part for the differences between MarA and SoxS with regard to the activation of different promoters. Thirteen MarA mutants (seven single-alanine substitutions, four charge inversion mutations [acidic to basic side chains or vice versa] and two double-charge inversions) were compared with MarA and SoxS for activation of 16 MarA-SoxS-Rob regulon promoters. We reasoned that class I promoters were more likely to reveal a correlation between activity and flexibility, if it existed, since they depend on only a single interaction of MarA with the marbox and a single interaction with RNAP, whereas class II promoters involve additional interactions with RNAP. (The amino acids chosen for this study were known not to involve the interaction of MarA with RNAP [8]).
Only the E89A variant showed a consistent increase in activation of class I promoters that paralleled the activation by SoxS (Table 2). This was surprising since E89A is unlikely to have any direct effect on the flexibility of MarA and was intended only as a control. As expected, there was no correlation between the activation of class II promoters by MarA(E89A) and by SoxS (Fig. 1).
TABLE 2.
Activation of class I promoters by MarA, 13 MarA mutants, and SoxSa
| Activator | Activity relative to that of MarA promoter-lacZ fusionb |
||||||
|---|---|---|---|---|---|---|---|
| marRAB | acnA | acrAB | fpr | mdtG | poxB | zwf | |
| MarA | |||||||
| WT | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| E25R | 1.03 | 0.71 | 1.2 | 0.95 | 1.10 | 1.2 | 0.70 |
| E77A | 0.98 | 0.80 | 1.02 | 0.91 | 1.08 | 0.78 | 1.03 |
| E77R | 1.02 | 0.39 | 0.81 | 0.59 | 1.03 | 1.2 | 0.53 |
| L80A | 0.97 | 1.3 | 3.7 | 1.3 | 1.2 | 0.36 | 1.2 |
| Y81A | 1.01 | 1.4 | 9.5 | 1.4 | 1.5 | 0.24 | 1.2 |
| Y81D | 0.96 | 0.83 | 1.2 | 0.95 | 1.2 | 0.13 | 0.70 |
| E84A | 0.99 | 1.10 | 1.5 | 0.98 | 1.81 | 0.55 | 1.9 |
| R85A | 1.00 | 0.89 | 0.98 | 0.39 | 0.57 | 1.5 | 0.61 |
| R85D | 0.17 | 0.19 | 0.51 | 1.3 | 0.54 | 0.42 | 0.19 |
| E89A | 1.10 | 5.4 | 1.8 | 4.9 | 4.1 | 0.16 | 2.5 |
| Q91A | 0.92 | 0.10 | 1.03 | 0.45 | 0.57 | 0.52 | 0.85 |
| E25R R85D | 0.98 | 0.92 | 0.93 | 0.55 | 0.89 | 0.44 | 0.52 |
| E77R R85D | 1.00 | 0.85 | 0.81 | 0.67 | 0.90 | 1.00 | 0.53 |
| SoxS | 1.10 | 3.2 | 1.8 | 23. | 3.7 | 0.22 | 2.3 |
All strains are derivatives of M3997 and have null mutations in marRAB, rob, lon, and clpP except that those containing the acrAB::lacZ or mdtG::lacZ transcriptional fusion are derivatives of M4435 (acrR) or GC4468 (wild type), respectively, and are wild type for lon and clpP. The wild-type MarA β-galactosidase activities (Miller units), control and induced, respectively, for each of the promoters were as follows: for acnA, 50 and 140; acrAB, 80 and 450; fpr, 80 and 180; marRAB, 750 and 2000; mdtG, 7 and 37; poxB, 35 and 85; and zwf, 200 and 1.200. For a discussion of the statistical significance of these values see the supplemental material.
Numbers in bold show activation >2-fold. See Table S1 for promoter-lacZ fusion references.
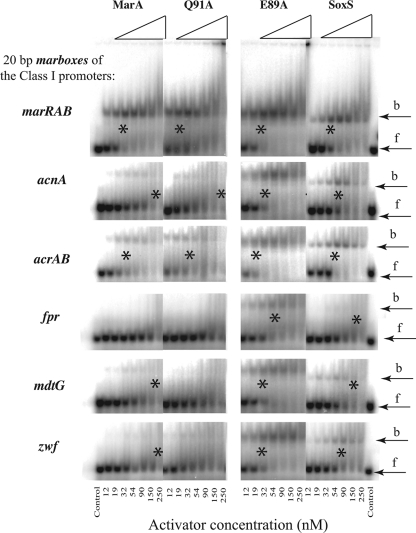
To ascertain whether the greater activation of the acnA, acrAB, fpr, mdtG, and zwf promoters by MarA(E89A) was related to binding, SoxS, MarA, MarA(E89A), and MarA(Q91A) proteins were purified and assayed by gel retardation for their ability to bind the 20 bp marboxes (listed in Table 1). In general, marbox binding (Fig. 2) paralleled promoter activation for the class I promoters (Table 3). Both SoxS and MarA(E89A) showed considerable binding to the marboxes of all of these promoters, whereas MarA and MarA(Q91A) showed significant binding to only the marRAB and acrAB marboxes. MarA(E89A) showed large increases in binding (relative to WT MarA), concomitant with large increases in relative activation for the acnA, fpr, mdtG, and zwf promoters, and showed modest increases in both binding and activation of the acrAB promoter. All four activators bound the marRAB marbox with similar affinities and activated marRAB to similar extents. SoxS bound the acrAB marbox with similar affinity to MarA but, like MarA(E89A), marginally increased acrAB transcription (∼1.8-fold). Binding of the poxB marbox was seen only for MarA(E89A) (data not shown). We note that, of these promoters, acnA, fpr, and zwf are necessary for optimal superoxide resistance (1, 2, 5, 26, 31, 33).
FIG. 2.
Autoradiographs of gel electrophoretic mobility assays using MarA, MarA mutants E89A and Q91A, and SoxS with 32P-labeled 20-bp DNA fragments (Table 1) corresponding to the marbox binding sites at different class I promoters. Partially purified (∼80%) MarA, MarA(E89A), MarA(Q91A), or SoxS starting at a concentration of 250 nM was serially diluted 3:2 and mixed with the DNA, and the amount of protein that bound 50% of the DNA (*) was used to estimate the dissociation complex (KD). When only a weak band was seen at the highest concentration of protein used, the KD was estimated as >250. The positions of the free (f) and bound (b) DNA are indicated. The calculated KDs from these gels are listed in Table 3.
TABLE 3.
Relative activation compared with relative binding for activators and mutants at MarA-SoxS-Rob regulon promotersa
| Promoter class and name | WT MarA |
MarA(Q91A) |
Position 12c | MarA(E89A) |
SoxS |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| RA | RB | KD (nM)b | RA | RB | KD (nM) | RA | RB | KD (nM) | RA | RB | KD (nM) | ||
| Class I | |||||||||||||
| marRAB | 1.0 | 1.0 | 25 | 0.9 | ∼0.8 | 30 | T | 1.1 | 1.0 | 25 | 1.1 | 0.8 | 30 |
| acnA | 1.0 | 1.0 | 180 | 0.1 | 0.7 | 250 | A | 5.4 | 6.0 | 30 | 3.2 | 3.0 | 60 |
| acrAB | 1.0 | 1.0 | 30 | 1.0 | 0.6 | 50 | A | 1.8 | 1.5 | 20 | 1.8 | 0.8 | 40 |
| fpr | 1.0 | — | >250 | 0.5 | — | >250 | G | 4.9 | ∼5.0 | 50 | 23.0 | >2.0 | 200 |
| mdtG | 1.0 | 1.0 | 200 | 0.6 | <0.8 | >250 | C | 4.1 | 6.7 | 30 | 3.7 | 1.3 | 150 |
| zwf | 1.0 | 1.0 | 200 | 0.9 | <0.8 | >250 | G | 2.5 | 6.7 | 30 | 2.3 | 2.7 | 75 |
| Class II | |||||||||||||
| fumC | 1.0 | 1.0 | 50 | 0.2 | <0.3 | >150 | G | 0.5 | ∼1.7 | 30 | 5.3 | ∼1.0 | 50 |
| inaA | 1.0 | — | ≫150 | 0.2 | — | ≫150 | C | 1.5 | ∼3.0 | 50 | 0.8 | — | ≫150 |
| mdaB | 1.0 | 1.0 | 50 | 1.3 | 0.8 | 63 | T | 0.6 | 1.4 | 35 | 0.3 | 0.5 | 100 |
| micF | 1.0 | 1.0 | 25 | 0.2 | 1.0 | 25 | T | 1.0 | 1.0 | 25 | 1.0 | ∼2.0 | 50 |
| nfsB | 1.0 | 1.0 | 200 | 1.3 | ∼.8 | ∼250 | C | 0.4 | 2.7 | 75 | 0.6 | 1.3 | 150 |
| pqiA | 1.0 | — | ≫150 | 0.0 | — | ≫150 | T | 1.9 | — | ≫150 | 4.0 | — | ≫150 |
| sodA | 1.0 | — | ≫150 | 1.2 | >1.0 | >150 | A | 1.5 | ∼1.0 | >150 | 2.3 | — | ≫150 |
| ybjC | 1.0 | — | ≫150 | 0.3 | — | ≫150 | T | 1.5 | ∼1.5 | 100 | 0.8 | — | ≫150 |
| yhbW | 1.0 | — | >150 | 0.8 | <1.0 | ≫150 | T | 0.2 | ∼1.5 | 100 | 0.1 | — | >150 |
The data for the activation relative to that of WT MarA are from Table 2 and Fig. 1. RA, relative activation; RB, relative binding constant. Where values are shown in boldface, the relative activation did not parallel the relative binding. —, value could not be calculated.
The dissociation constant (KD) was calculated from gel shift assays similar to those shown in Fig. 2.
The nucleotide at position 12 for this marbox sequence.
The binding of MarA, the two MarA mutants, and SoxS to the marbox sequences of nine class II promoters was also determined by gel mobility assays and is summarized in Table 3. In every case, MarA(E89A) bound as well as or more tightly (i.e., had a lower KD [equilibrium dissociation constant) than WT MarA. For several promoter marboxes this change was dramatic: the KD dropped from ≫150 to 50 for inaA, from 200 to 75 for nfsB, and from ≫150 nM to ∼100 nM for both ybjC and yhbW. In spite of this, MarA(E89A) activation was greater than that of WT MarA for inaA, sodA, and ybjC; it was comparable to that of WT MarA for micF and less than that of WT MarA for fumC, mdaB, nfsB, and ybhW (Table 3And Fig. 1). We conclude that the WT glutamic acid of MarA at position 89 is an inhibitor of MarA binding to many marboxes.
In an effort to understand why E89 is inhibitory, we examined the sequences of the 14 marboxes for which we have data (Table 3). We noticed that five have a T at position 12 and that the binding to none of these sequences is increased by the E89A substitution (less than 1.7-fold). In contrast, of the remaining nine sequences that do not have a T at this position, the binding of E89A was increased for six of them by >2.5-fold and for the seventh by 1.7-fold; only two do not show increased binding. Although the cocrystal structure of MarA with the marRAB marbox DNA (with a T at position 12) indicates no interaction between the two molecules at this position (36), we considered the possibility that steric hindrance between the marbox DNA and MarA could limit activation by MarA when position 12 is not a T residue (see the Discussion for a fuller treatment).
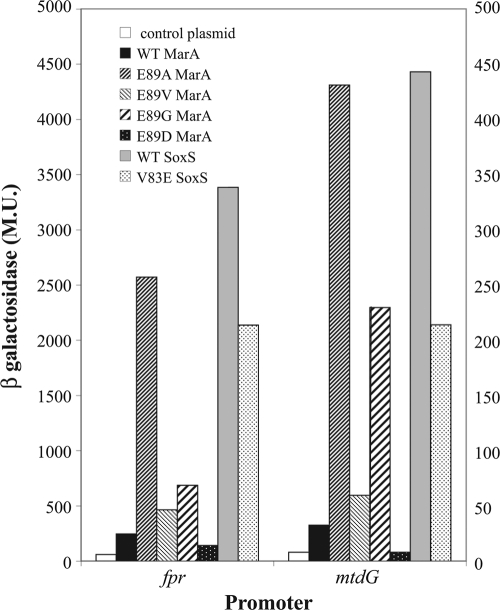
To examine one facet of this possibility, namely, that the glutamic acid side chain of E89 sterically inhibits interaction with marbox DNAs lacking a T at position 12, we tested the effects of several amino acid substitutions at residue 89 on the fpr::lacZ fusion and the mdtG::lacZ fusion, the two fusions that showed the greatest effects of E89A on activation (see above). Plasmids carrying WT MarA, the MarA variant MarA(E89A) (with a nonpolar single methyl group side chain), MarA(E89G) (with no side chain), MarA(E89D) (with a side chain one methylene group shorter than glutamic acid), MarA(E89V) (with the amino acid present at the corresponding position of SoxS and having a dimethyl methylene side chain,), WT SoxS, and SoxS(V83E) were introduced into these fusions and assayed for β-galactosidase. Again (Table 2), activation of these promoters by SoxS was much greater than that by MarA (Fig. 3). Similarly, MarA(E89A) was considerably more active than WT MarA. In contrast, the E89D variant was less active than WT MarA for fpr and completely inactive (indistinguishable from the control plasmid) for mdtG. E89G was approximately twice as active as WT MarA for fpr and 4-fold more active for mdtG although for both promoters it was considerably less active than E89A. The E89V variant was marginally more active than the WT for both promoters but substantially less so than E89A. SoxS(V83E) reduced the activation of these promoters relative to WT SoxS, but SoxS(V83E) was still more active than WT MarA. (The activations shown here are greater than those apparent in Table 2 because the expression of the plasmids carrying MarA, SoxS, and their mutants is not entirely shut off by lacIq so that the increases expressed in Table 2Appear smaller.) As outlined in the Discussion, these results, namely, (i) that variant E89D is virtually inactive, (ii) that E89G is very active although to a lesser extent than E89A, (iii) that E89V is only marginally more active than the WT, and (iv) that SoxS(V83E) has reduced activation although not to the low levels expressed by WT MarA, are consistent with the possibility that that the side chain of E89 sterically inhibits interaction with the DNA for marbox sequences lacking a T at position 12.
FIG. 3.
Activation of class I promoters by MarA and SoxS and their mutants at position E89 (MarA) or V83 (SoxS). The absolute β-galactosidase values (Miller units [MU]) for the fpr::lacZ fusion (left-hand scale) and the mdtG::lacZ and acrAB::lacZ fusions (right-hand scale) are plotted for the plasmid control, MarA, MarA(E89A), MarA(E89V), SoxS, and SoxS(V83E).
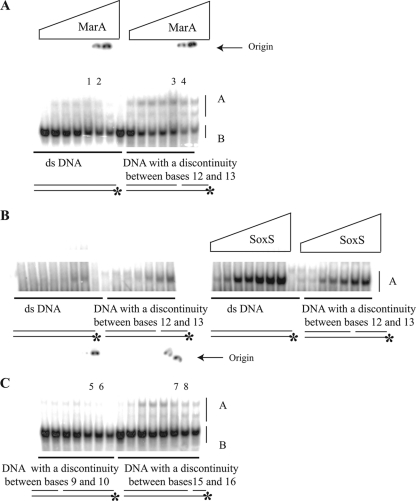
If, as outlined above and presented in greater detail in the Discussion, steric interference with the phosphate between positions 12 and 13 and the glutamic acid side chain at position 89 is responsible for the very poor activation of promoters such as fpr, then it would be predicted that binding of WT MarA to the fpr marbox would be enhanced if that phosphate were absent. We therefore compared the binding affinity of WT MarA to either a 36-bp double-stranded DNA containing the marbox sequence of fpr or with dsDNA of the same sequence and length but prepared so that the phosphate linkage between positions 12 and 13 of the marbox was eliminated (see Materials and Methods). Again (Fig. 2And Table 3), MarA bound very poorly to the fpr marbox (Fig. 4). However, binding increased significantly when the fpr DNA lacked the phosphate group between positions 12 and 13 (Fig. 4A and B). In contrast, SoxS bound more tightly to dsDNA than to the discontinuous DNA (Fig. 4B). When the phosphate located 3 nt farther upstream (between positions 9 and 10) was absent, no significant alteration in binding was observed (Fig. 4) although a small increase was observed when the phosphate between positions 15 and 16 was absent. We conclude that the phosphate group between nt 12 and 13 of the consensus sequence inhibits the ability of MarA to bind.
FIG. 4.
Autoradiographs of gel retardation assays as in Fig. 2 except that the DNA fragment was 36 nt long and corresponds to the 7 nt upstream and 9 nt downstream of the native fpr marbox (GGACTGGAAGGCTCAATCGATCAAATCAATCAGAGG; the marbox is in boldface). Gels A and C were run at the same time using the same preparation of highly purified MarA with the His6 tag removed. We have no explanation for the slower-moving band seen only with this preparation of MarA. The ratios of the bound to unbound DNA (intensity of the A/B bands) in the indicated lanes were as follows: lane 1, 0.19; lane 2, 0.21; lane 3, 0.34; lane 4, 0.44; lane 5, 0.05; lane 6, 0.08; lane 7, 0.20; and lane 8, 0.22. Gel B (showing only the bound material) employed a different preparation of MarA from which the His6 tag had not been removed (the same as in Fig. 2) and the comparable preparation of SoxS. The concentrations of MarA and SoxS are as described in the legend of Fig. 2.
Activation and marbox binding by MarA(Q91A).
The only other MarA mutation found here to differ significantly from WT MarA in the activation of class I promoters was Q91A (Table 2). Of the seven class I promoters examined, MarA(Q91A) activated the acrAB, marRAB, and zwf promoters to similar extents as WT MarA but activated acnA, fpr, mdtG, and poxB to only 60% or less of MarA WT levels (Table 2; see also below).
For the nine class II promoters, MarA(Q91A) significantly reduced the activation of fumC, inaA, micF, pqiA, and ybjC but had no significant effect on nfsB, mdaB, sodA, or yhbW. MarA- (Q91) has been identified as forming van der Waals interactions with the methyl groups of the two thymidines that are complementary to the adenines at positions 17 and 18 of the consensus sequence (36). Thus, it would be expected that the Q91A substitution might reduce activation of the seven class II promoters that have at least one A at position 17 or 18 but not the two promoters, nfsB and sodA, that have no A residues at these positions. Indeed, nfsB and sodA are among the promoters that Q91A activated to the same extent as WT MarA. Since Q91A reduced the expression of four of the seven class I promoters and four of the nine class II promoters, our results are inconsistent with the proposition that Q91 is principally required for interactions at class II promoters, as has been proposed for the analogous site (Q85) in SoxS (16).
For the majority of these promoters, the gel mobility assay for binding of MarA to marboxes was insufficiently precise to determine whether there is a correlation between binding and activation by MarA(Q91A) relative to MarA (Table 3). Among the class I promoters that exhibited measurable binding to MarA, MarA(Q91A) showed no greater binding or activation for acrAB, a modest reduction in activation and binding for mdtG and zwf, and a reduction in binding but not in activation for marRAB. A modest reduction in binding with a small increase in activation was seen for the class II mdaB promoter. Although not observed in these experiments, a more detailed analysis of the binding of MarA(Q91A) to the micF marbox (using protein without the His6 tag) showed a very small reduction in binding concomitant with the reduced ability of MarA(Q91A) to activate the class II micF promoter (13). Thus, relative to MarA, there may be a correlation between activation and binding for Q91A at class I promoters, but none is obvious with regard to the class II promoters.
Activation of superoxide resistance by MarA(E89A).
If the glutamic acid at position 89 of MarA is a major determinant in vivo of the reduced activity of MarA at many promoters where SoxS is more active, we would expect cells carrying MarA(E89A)to be more resistant to superoxides than cells carrying WT MarA. This was tested with gradient plate assays of sensitivity to two superoxide-generating compounds, phenazine methosulfate (PMS) and menadione (Table 4). Cells constitutively expressing MarA(E89A)were more resistant than WT MarA to PMS (1.7-fold) and, to a lesser extent, to menadione (1.3-fold). Comparable MICs with SoxS for PMS were 1.6-fold and for menadione 2.0-fold. Clearly, MarA(E89A)activates superoxide resistance to a greater extent than WT MarA. Curiously, the Q91A substitution had no effect on resistance to the superoxide generator PMS (MIC of 35 μM for both the Q91A mutant and the WT MarA) but lowered resistance to the superoxide generator menadione (MIC of 0.9 mM for the Q91A mutant and 1.8 mM for the WT).
TABLE 4.
Superoxide resistance of strains carrying MarA, MarA mutations, or SoxS
| Plasmid | Strain no. | Resistance to the indicated superoxide-generating compounda |
|||
|---|---|---|---|---|---|
| Phenazine methosulfate |
Menadione |
||||
| MIC (μM) | Relative increase in MIC | MIC (mM) | Relative increase in MIC | ||
| Vector | M5390 | 12 | 0.5 | ||
| WT MarA | M5391 | 35 | 1.0 | 1.8 | 1.0 |
| MarA(E25R) | M5392 | 31 | 0.89 | 1.6 | 0.91 |
| MarA(E77A) | M5393 | 22 | 0.65 | 1.2 | 0.68 |
| MarA(L80A) | M5394 | 42 | 1.2 | 1.4 | 0.76 |
| MarA(E89A) | M5395 | 57 | 1.7 | 2.4 | 1.3 |
| MarA(Q91A) | M5396 | 35 | 1.0 | 0.9 | 0.49 |
| WT SoxS | M5397 | 55 | 1.6 | 3.5 | 2.0 |
The MIC of each chemical for each strain was estimated from gradient plates that were performed two times. To determine the relative increase, the MIC for each strain was divided by that of the strain carrying the WT MarA plasmid.
DISCUSSION
The MarA-SoxS-Rob regulon of E. coli consists of a variety of genes that enable cells to adapt to multiple stresses. It contains genes that render the cell multidrug and organic-solvent resistant (acrAB, tolC, and micF) and that defend against superoxide stress (e.g., acnA, fpr, fumC, nfsA, sodA, and zwf) (1, 2, 5, 26, 31, 33). Not surprisingly, there are quantitative differences in the extents of activation of particular promoters by the paralogous activators so that the phenotypic outcomes depend on which activator is upregulated (2, 26, 34, 43).
E89 inhibition.
We have shown here that the MarA glutamic acid residue E89 is responsible for decreasing the binding of WT MarA relative to SoxS for the class I marbox promoters, acnA, mdtG, fpr, and zwf, thereby decreasing the relative activation of these promoters and hence the resistance engendered to superoxides by MarA. We think the following may explain these results.
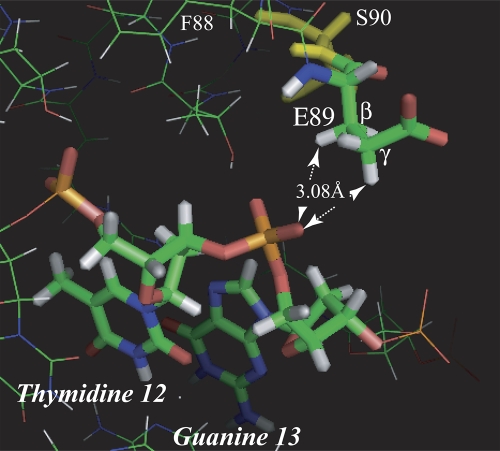
A detail of the MarA structure (Fig. 5) shows that E89 is oriented in the cocrystal with its acidic groups exposed to the solvent away from the DNA backbone. In so doing, methyl hydrogens on the β- and γ-carbons are positioned restrictively close to an oxygen of the phosphate group between bases T12 and G13 of the marRAB marbox. Even this orientation of E89 is only possible as the result of a small “clash” (as predicted by the MolProbity program [http://molprobity.biochem.duke.edu/]) between E89 and S90 that permits the rotation of E89 into the MarA core and away from the DNA backbone. This suggests that any further displacement of the phosphate group closer to MarA would be unfavorable. Indeed, Dangi et al. (9) have shown that there are no differences detectable (shift difference of <0.45 ppm) by nuclear magnetic resonance (NMR) in the backbone chemical shifts for E89 of MarA when it binds to the marboxes of marRAB, fumC, fpr, or micF although such shifts are detectable at other positions.
FIG. 5.
Detail of the MarA DNA cocrystal structure (36) showing the close proximity of hydrogens of the β- and γ-carbons of E89 to one of the oxygens of the phosphate in the DNA backbone between positions 12 and 13 of the MarA marbox. The other γ-hydrogen is 3.20 Å from the same oxygen. The MolProbity program (http://molprobity.biochem.duke.edu/) indicates a clash between the oxygen of the peptide bond of E89 with a β-hydrogen of S90 (in yellow) of 0.445Å, which has the effect of allowing E89 to rotate slightly into the MarA structure, thereby moving the β- and γ-hydrogens to permissible separations from the phosphate oxygen. At the same time, the oxygens of E89 can face away from the DNA into the solvent.
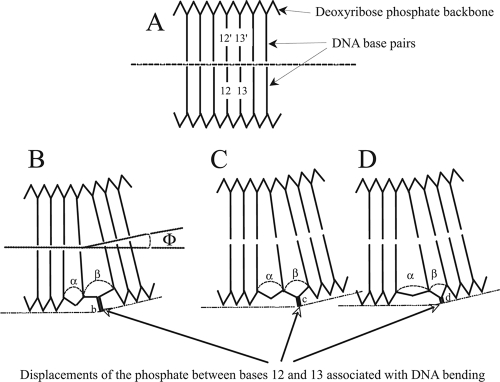
We suggest that a thymidine at position 12 (see Results) might optimize the location of the phosphate group so as to minimize its interference with E89 (Fig. 6). Rhee et al. (36) have noted that the displacement of the DNA to achieve the 35° bend required for binding is not uniform and is primarily limited to the regions of the DNA between bases 7 and 9 and bases 11 and 13 of the marbox. Hydrogen bonds between the bases are not significantly disrupted at these points (i.e., there is no melting of the DNA), but significant stacking energy must be lost between bases on the convex face of the DNA since they are separated to a greater extent than those on the concave face. Thus, it is not simply the base pair at this position that is critical but the specific base on the convex surface. While there is still disagreement as to how to estimate base stacking energies, there is general agreement on the following: that stacking energies may vary from one base to another by several kcal, that there are energetic differences between which base is to the 5′ side, and that there are additional “many-body effects” (i.e., effects on the stacking energy between bases 1 and 2 by base 3) and nonadditive effects (40, 47).
FIG. 6.
(A) Schematic representation of a portion of a marbox DNA sequence showing the bases stacked at a 3.2-Å separation, the ribose-phosphate backbone connecting them, and a dotted line to indicate that the DNA is linear. The numbers (12 and 13) indicate the positions of the bases in the consensus marbox sequence, and 12′ and 13′ are their complements. (B, C, and D) To allow for a bend of angle 2Φ (35°) in the marbox DNA, Rhee et al. (36) have pointed out that the DNA does not distribute the necessary distortion over the entire 20-bp length but, rather, limits the distortion to the regions between bases 7 to 9 and 11 to 13 as illustrated in these three diagrams. The distortion of the backbone between the bases at positions 11 to 13 results in separations of α and β. The relative sizes of these displacements and the absolute energy required to generate them will depend on the stacking energies of the particular bases occupying positions 11, 12, and 13. As a consequence, the position of the phosphate between bases 12 and 13 will be altered relative to the surface of the DNA. We postulate that the presence of T at position 12 of the consensus sequence in general results in the most favorable of these structures to accommodate the glutamic acid at position 89.
Consistent with this model, we find that, first, WT MarA binds more tightly to DNA of the fpr marbox lacking a phosphate between positions 12 and 13 than to uninterrupted dsDNA. This is not simply the result of compensation for the energy lost on bending normal DNA with this sequence since an interruption in the ribose-phosphate backbone at the other stress point (position 9) does not enhance DNA binding. Furthermore, SoxS binds more tightly to the dsDNA if it has the phosphate between positions 12 and 13. Thus, the model offers an explanation as to why there is a strong correlation between E89A activation and the lack of a T at position 12 of the marboxes, namely, to prevent interference with the phosphate. Second, for two of the promoters most restricted by E89 (fpr and mdtG), there is a significant difference between different MarA substitutions. As expected from the model, since E89A and E89G lack γ-carbons, they are the most effective in binding and activation. E89V, with two γ-carbons that could clash with the phosphate between 12 and 13 but are still somewhat free to rotate, is less effective. WT E89 is even less effective since, if the model were correct, the rotation of its γ-carbon would be severely limited by the hydrophilicity of its acidic group. Finally, E89D is almost inactive since its γ-carbon is part of the acidic group, and we would predict a significant clash with the phosphate leading to distortions of the structure.
Implications of these results for the mechanism of activation by MarA.
The differences in behavior of the E89A and Q91A mutants relative to WT MarA at class I and class II promoters is instructive with regard to the mechanism of transcriptional activation. MarA(E89A) exhibited increased binding relative to WT MarA for 13 of the 16 promoter marboxes examined although the increase was small for acrAB (Table 3). Only two promoters, marRAB and micF, showed no increase (no binding to pqiA was found at even the highest activator concentrations). In the case of the six rpoD-stimulated class I promoters, the increase in binding by all except marRAB was associated with increased activation. The implication of this is that an increase in the interaction between activator and binding site is associated with increased RNAP activity at these five promoters. This, in turn, implies that an important part of the mechanism for activation is recruitment of RNAP by the bound activator (35). A further conclusion is that recruitment is not an element of the mechanism for activation of the marRAB promoter, as has been demonstrated elsewhere (42).
Furthermore, MarA may have a special role at the marRAB promoter since it appears to be a competitive inhibitor of MarR (at least in solution [28]), thereby freeing the −10 and −35 signals for RNAP binding. This effect would not have been seen here or in the experiments of Wall et al. (42) since they were carried out in the absence of MarR. Similarly, since the activation experiments with the acrAB promoter were carried out in an acrR null mutant, the small increases in binding and activation of acrAB by MarA(E89A) compared to levels of the WT may have masked any competition between MarA and AcrR for the acrAB promoter.
The lack of correlation between marbox binding by MarA(E89A) and activation at class II promoters (Table 3) is consistent with a large body of information indicating that additional interactions between activator and RNAP are essential at class II promoters (4, 6, 14, 44). Like the E89A variant, MarA(Q91A) reduced the activation of and, to a limited extent, the binding to about half of both the class I and class II promoters. Thus, we see no indication of a specific role for Q91 in class II promoter activation. This is contrary to the finding that the corresponding amino acid in SoxS, Q85, interacts with the σ subunit of RNAP (16).
Finally, we note that discrimination between SoxS and MarA is not entirely the result of increased binding of SoxS at class I promoters. While a number of the principal functions required for superoxide resistance are controlled at class I promoters, others are at class II promoters (e.g., fumC and sodA). The greater ability of SoxS over MarA to activate these class II promoters appears to have a different basis and will be the subject of a future communication.
Supplementary Material
Acknowledgments
We thank Michael Gleghorn for modeling the SoxS coordinates and David Davies, Frederick Dyda, Alison B. Hickman, and Jane Richardson for helpful discussions.
This research was supported by the Intramural Research Program of the NIH, NIDDK.
Footnotes
Published ahead of print on 19 November 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ariza, R. R., Z. Li, N. Ringstad, and B. Demple. 1995. Activation of multiple antibiotic resistance and binding of stress-inducible promoters by Escherichia coli Rob protein. J. Bacteriol. 177:1655-1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa, T., and S. Levy. 2000. Differential expression of over 60 chromosomal genes in Escherichia coli by constitutive expression of MarA. J. Bacteriol. 182:3467-3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baucheron, S., et al. 2004. AcrAB-TolC directs efflux-mediated multidrug resistance in Salmonella enterica serovar Typhimurium DT104. Antimicrob. Agents Chemother. 48:3729-3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bhende, P., and S. Egan. 2000. Genetic evidence that transcription activation by RhaS involves specific amino acid contacts with sigma 70. J. Bacteriol. 182:4959-4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blanchard, J., W. Wholey, E. Conlon, and P. Pomposiello. 2007. Rapid changes in gene expression dynamics in response to superoxide reveal SoxRS-dependent and independent transcriptional networks. PLoS One 2:e1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Busby, S., and R. Ebright. 1999. Transcription activation by catabolite activator protein (CAP). J. Mol. Biol. 293:199-213. [DOI] [PubMed] [Google Scholar]
- 7.Cohen, S. P., S. B. Levy, J. Foulds, and J. L. Rosner. 1993. Salicylate induction of antibiotic resistance in Escherichia coli: activation of the mar operon and a mar-independent pathway. J. Bacteriol. 175:7856-7862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dangi, B., A. M. Gronenborn, J. L. Rosner, and R. G. Martin. 2004. Versatility of the carboxy-terminal domain of the α subunit of RNA polymerase in transcriptional activation: use of the DNA contact site as a protein contact site for MarA. Mol. Microbiol. 54:45-59. [DOI] [PubMed] [Google Scholar]
- 9.Dangi, B., et al. 2001. Structure and dynamics of MarA-DNA complexes: an NMR investigation. J. Mol. Biol. 314:113-127. [DOI] [PubMed] [Google Scholar]
- 10.Demple, B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53-57. [DOI] [PubMed] [Google Scholar]
- 11.Eguchi, Y., et al. 2003. Transcriptional regulation of drug efflux genes by EvgAS, a two-component system in Escherichia coli. Microbiology 149:2819-2828. [DOI] [PubMed] [Google Scholar]
- 12.Fàbrega, A., R. Martin, J. Rosner, M. Tavio, and J. Vila. 2010. Constitutive SoxS expression in a fluoroquinolone-resistant strain with a truncated SoxR protein and identification of a new member of the marA-soxS-rob regulon, mdtG. Antimicrob. Agents Chemother. 54:1218-1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gillette, W., R. Martin, and J. Rosner. 2000. Probing the Escherichia coli transcriptional activator MarA using alanine-scanning mutagenesis: residues important for DNA binding and activation. J. Mol. Biol. 299:1245-1255. [DOI] [PubMed] [Google Scholar]
- 14.Grainger, D. C., C. L. Webster, T. A. Belyaeva, E. I. Hyde, and S. J. Busby. 2004. Transcription activation at the Escherichia coli melAB promoter: interactions of MelR with its DNA target site and with domain 4 of the RNA polymerase sigma subunit. Mol. Microbiol. 51:1297-1309. [DOI] [PubMed] [Google Scholar]
- 15.Griffith, K., I. Shah, and R. J. Wolf. 2004. Proteolytic degradation of Escherichia coli transcription activators SoxS and MarA as the mechanism for reversing the induction of the superoxide (SoxRS) and multiple antibiotic resistance (Mar) regulons. Mol. Microbiol. 51:1801-1816. [DOI] [PubMed] [Google Scholar]
- 16.Griffith, K. L., and R. E. Wolf, Jr. 2002. A comprehensive alanine scanning mutagenesis of the Escherichia coli transcriptional activator SoxS: identifying amino acids important for DNA binding and transcription activation. J. Mol. Biol. 322:237-257. [DOI] [PubMed] [Google Scholar]
- 17.Griffith, K. L., and R. E. Wolf, Jr. 2001. Systematic mutagenesis of the DNA binding sites for SoxS in the Escherichia coli zwf and fpr promoters: identifying nucleotides required for DNA binding and transcription activation. Mol. Microbiol. 40:1141-1154. [DOI] [PubMed] [Google Scholar]
- 18.Jair, K. W., W. P. Fawcett, N. Fujita, A. Ishihama, and R. E. Wolf, Jr. 1996. Ambidextrous transcriptional activation by SoxS: requirement for the C-terminal domain of the RNA polymerase alpha subunit in a subset of Escherichia coli superoxide-inducible genes. Mol. Microbiol. 19:307-317. [DOI] [PubMed] [Google Scholar]
- 19.Kwon, H. J., M. H. Bennik, B. Demple, and T. Ellenberger. 2000. Crystal structure of the Escherichia coli Rob transcription factor in complex with DNA. Nat. Struct. Biol. 7:424-430. [DOI] [PubMed] [Google Scholar]
- 20.Martin, R., E. Bartlett, J. Rosner, and M. Wall. 2008. Activation of the Escherichia coli marA/soxS/rob regulon in response to transcriptional activator concentration. J. Mol. Biol. 380:278-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin, R., W. Gillette, N. Martin, and J. Rosner. 2002. Complex formation between activator and RNA polymerase as the basis for transcriptional activation by MarA and SoxS in Escherichia coli. Mol. Microbiol. 43:355-370. [DOI] [PubMed] [Google Scholar]
- 22.Martin, R., W. Gillette, S. Rhee, and J. Rosner. 1999. Structural requirements for marbox function in transcriptional activation of mar/sox/rob regulon promoters in Escherichia coli: sequence, orientation and spatial relationship to the core promoter. Mol. Microbiol. 34:431-441. [DOI] [PubMed] [Google Scholar]
- 23.Martin, R., W. Gillette, and J. Rosner. 2000. Promoter discrimination by the related transcriptional activators MarA and SoxS: differential regulation by differential binding. Mol. Microbiol. 35:623-634. [DOI] [PubMed] [Google Scholar]
- 24.Martin, R., and J. Rosner. 1995. Binding of purified multiple antibiotic-resistance repressor protein (MarR) to mar operator sequences. Proc. Natl. Acad. Sci. U. S. A. 92:5456-5460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin, R., and J. Rosner. 1997. Fis, an accessorial factor for transcriptional activation of the mar (multiple antibiotic resistance) promoter of Escherichia coli in the presence of the activator MarA, SoxS, or Rob. J. Bacteriol. 179:7410-7419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin, R., and J. Rosner. 2002. Genomics of the marA/soxS/rob regulon of Escherichia coli: identification of directly activated promoters by application of molecular genetics and informatics to microarray data. Mol. Microbiol. 44:1611-1624. [DOI] [PubMed] [Google Scholar]
- 27.Martin, R., and J. Rosner. 2004. Transcriptional and translational regulation of the marRAB multiple antibiotic resistance operon in Escherichia coli. Mol. Microbiol. 53:183-191. [DOI] [PubMed] [Google Scholar]
- 28.Martin, R. G., K. W. Jair, R. E. Wolf, Jr., and J. L. Rosner. 1996. Autoactivation of the marRAB multiple antibiotic resistance operon by the MarA transcriptional activator in Escherichia coli. J. Bacteriol. 178:2216-2223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Martin, R. G., and J. L. Rosner. 2005. Structure and function of MarA and its homologs, p. 235-246. In D. G. White, M. N. Alekshun, and P. F. McDermott (ed.), Frontiers in antimicrobial resistance: a tribute to Stewart B. Levy. ASM Press, Washington, DC.
- 30.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 31.Nunoshiba, T., E. Hidalgo, C. Amábile Cuevas, and B. Demple. 1992. Two-stage control of an oxidative stress regulon: the Escherichia coli SoxR protein triggers redox-inducible expression of the soxS regulatory gene. J. Bacteriol. 174:6054-6060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Okusu, H., D. Ma, and H. Nikaido. 1996. AcrAB efflux pump plays a major role in the antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J. Bacteriol. 178:306-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pomposiello, P., M. Bennik, and B. Demple. 2001. Genome-wide transcriptional profiling of the Escherichia coli responses to superoxide stress and sodium salicylate. J. Bacteriol. 183:3890-3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pomposiello, P. J., and B. Demple. 2001. Redox-operated genetic switches: the SoxR and OxyR transcription factors. Trends Biotechnol. 19:109-114. [DOI] [PubMed] [Google Scholar]
- 35.Ptashne, M., and A. Gann. 1997. Transcriptional activation by recruitment. Nature 386:569-577. [DOI] [PubMed] [Google Scholar]
- 36.Rhee, S., R. G. Martin, J. L. Rosner, and D. R. Davies. 1998. A novel DNA-binding motif in MarA: the first structure for an AraC family transcriptional activator. Proc. Natl. Acad. Sci. U. S. A. 95:10413-10418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rosenberg, E. Y., D. Bertenthal, M. L. Nilles, K. P. Bertrand, and H. Nikaido. 2003. Bile salts and fatty acids induce the expression of Escherichia coli AcrAB multidrug efflux pump through their interaction with Rob regulatory protein. Mol. Microbiol. 48:1609-1619. [DOI] [PubMed] [Google Scholar]
- 38.Rosner, J., B. Dangi, A. Gronenborn, and R. Martin. 2002. Posttranscriptional activation of the transcriptional activator Rob by dipyridyl in Escherichia coli. J. Bacteriol. 184:1407-1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Simons, R., F. Houman, and N. Kleckner. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene 53:85-96. [DOI] [PubMed] [Google Scholar]
- 40.Sponer, J., H. Gabb, J. Leszczynski, and P. Hobza. 1997. Base-base and deoxyribose-base stacking interactions in B-DNA and Z-DNA: a quantum-chemical study. Biophys. J. 73:76-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Szybalski, W., and V. Bryson. 1952. Genetic studies on microbial cross resistance to toxic agents. I. Cross resistance of Escherichia coli to fifteen antibiotics. J. Bacteriol. 64:489-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wall, M. E., D. A. Markowitz, J. L. Rosner, and R. G. Martin. 2009. Model of transcriptional activation by MarA in Escherichia coli. PLoS Comput. Biol. 5:e1000614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.White, D. G., M. N. Alekshun, and P. F. McDermott (ed.). 2005. Frontiers in antimicrobial resistance: a tribute to Stewart B. Levy. ASM Press, Washington, DC.
- 44.Wickstrum, J., and S. Egan. 2004. Amino acid contacts between sigma 70 domain 4 and the transcription activators RhaS and RhaR. J. Bacteriol. 186:6277-6285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wood, T. I., et al. 1999. Interdependence of the position and orientation of SoxS binding sites in the transcriptional activation of the class I subset of Escherichia coli superoxide-inducible promoters. Mol. Microbiol. 34:414-430. [DOI] [PubMed] [Google Scholar]
- 46.Wu, J., and B. Weiss. 1992. Two-stage induction of the soxRS (superoxide response) regulon of Escherichia coli. J. Bacteriol. 174:3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yakovchuk, P., E. Protozanova, and M. Frank-Kamenetskii. 2006. Base-stacking and base-pairing contributions into thermal stability of the DNA double helix. Nucleic Acids Res. 34:564-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.