Abstract
Bacterial protein secretion is a highly orchestrated process that is essential for infection and virulence. Despite extensive efforts to predict or experimentally detect proteins that are secreted, the characterization of the bacterial secretome has remained challenging. A central event in protein secretion is the type I signal peptidase (SPase)-mediated cleavage of the N-terminal signal peptide that targets a protein for secretion via the general secretory pathway, and the arylomycins are a class of natural products that inhibit SPase, suggesting that they may be useful chemical biology tools for characterizing the secretome. Here, using an arylomycin derivative, along with two-dimensional gel electrophoresis and liquid chromatography-tandem mass spectrometry (LC-MS/MS), we identify 11 proteins whose secretion from stationary-phase Staphylococcus epidermidis is dependent on SPase activity, 9 of which are predicted to be translated with canonical N-terminal signal peptides. In addition, we find that the presence of extracellular domains of lipoteichoic acid synthase (LtaS) and the β-lactam response sensor BlaR1 in the medium is dependent on SPase activity, suggesting that they are cleaved at noncanonical sites within the protein. In all, the data define the proteins whose stationary-phase secretion depends on SPase and also suggest that the arylomycins should be valuable chemical biology tools for the study of protein secretion in a wide variety of different bacteria.
Secreted proteins play a central role in the interaction of bacteria with their environment (64). For the human pathogen Staphylococcus epidermidis, protein secretion facilitates commensal growth on skin as well as the invasive biofilm growth associated with implanted devices (41). Generally, the secretion of many of proteins, in both Gram-positive bacteria like S. epidermidis as well as Gram-negative bacteria, is mediated by the evolutionarily conserved general secretory (Sec) system. In Gram-negative bacteria, the Sec system exports proteins to the periplasm, where they may then be recognized by other specialized systems for transport across the outer membrane, while in Gram-positive bacteria, Sec translocates proteins directly into the extracellular environment. Proteins are targeted to the Sec system via an N-terminal signal peptide that is removed after translocation by a type I signal peptidase (SPase).
While there is much interest in defining the bacterial secretome, the essentiality of SPase, as well as other components of the Sec system, precludes the characterization of secretion in their absence. Instead, much effort has been directed toward the use of sequence analysis to predict potential SPase signal peptides and, thus, proteins that may be targeted for secretion. The consensus signal peptide recognized by SPase comprises on average the first 25 to 32 amino acids of the preprotein, with the sequences typically being longer in Gram-positive bacteria (42), and has a tripartite organization, with a positively charged N terminus, followed by a hydrophobic portion that commonly terminates with a Gly or Pro and then a C terminus that includes the SPase recognition site. The recognition site itself consists of small, aliphatic residues at positions −1 and −3 relative to the cleaved bond, and these correspondingly bind in the S1 and S3 binding sites of SPase. While Ala is most common at both the −1 and −3 positions, Gly, Ser, Cys, and Thr are also found at the −1 position, and Gly, Ser, Cys, Ile, Val, and Leu are also found at the −3 position. The surrounding residues at the +1, −2, −4, and −5 positions appear to be less critical, with most residues tolerated, with the exception of Pro at the +1 position (42), and although a role for the residues beyond the +1 position have been noted (14), their contribution is not well understood. Thus, while many trends have been identified, the prediction of signal peptides recognized by SPase based on sequence alone remains challenging and is further complicated by signal peptides that only poorly conform to the consensus rules (2, 3, 25, 33, 44, 59). Proteins processed by SPase may also have cell wall retention signals, which are themselves difficult to predict (8, 25, 61). Moreover, this approach predicts only which proteins have the potential to be secreted, as secretion will depend on growth and environmental factors.
In principle, the proteins secreted under a given set of conditions may be determined experimentally by identifying the proteins found in the media (13, 16, 21, 53, 60, 63, 67). However, this approach does not determine whether the presence of the protein in the medium is due to secretion or instead due to small levels of unavoidable cell lysis; in fact, known cytoplasmic proteins may comprise the majority of the proteins detected (2). While the detection of extracellular proteins that have been cleaved at likely SPase sites mitigates this complication (46), it does not rule out cleavage by a protease other than SPase and is complicated by further proteolysis, which is common and can remove the evidence of SPase activity.
The arylomycins are a class of natural products that were isolated from Streptomyces strain Tü 6075 based on their ability to inhibit Escherichia coli SPase in vitro (26, 50). Previously, we synthesized one member, arylomycin A2, and a synthetic derivative, arylomycin C16 (Fig. 1), and found that they are both potent and selective inhibitors of S. epidermidis SPase in vivo (47, 55a). While the arylomycins potently kill S. epidermidis (MICs of 1.0 and 0.25 μg/ml for arylomycin A2 and arylomycin C16, respectively), cell death is minimal when stationary-phase cells are treated for only brief periods of time, suggesting that these compounds may be useful chemical biology probes of protein secretion. Here, we use arylomycin C16 to identify the S. epidermidis proteins that are processed by SPase and secreted into the medium during stationary-phase growth via two-dimensional gel electrophoresis in combination with liquid chromatography (LC)-tandem mass spectrometry (MS) (MS/MS). The results reveal that only 11 proteins appear to be secreted in an appreciable quantity via the general Sec pathway during stationary-phase growth. However, we observed different levels of inhibition among the secreted proteins, suggesting in turn that they have different affinities for SPase, possibly revealing a previously unappreciated level of control over the secretome. Moreover, these proteins represent important virulence factors, and thus, the results shed light on the survival strategies of S. epidermidis and suggest that an arylomycin antibiotic might act to reduce virulence during therapy. Finally, we identify two proteins that appear to have noncanonical SPase recognition sites located internally within the protein that are generally difficult to detect using other approaches.
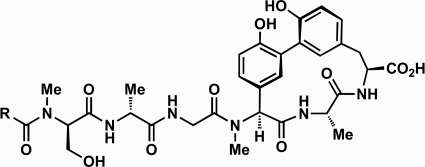
FIG. 1.
Arylomycin A2 (R = iso-C12) and arylomycin C16 (R = iso-C16).
MATERIALS AND METHODS
Materials.
Components for growth media were manufactured by Difco Laboratories (Detroit, MI). Arylomycin C16 was synthesized as described previously (47). S. epidermidis RP62A was obtained from the ATCC (ATCC 35984).
Two-dimensional gel electrophoresis.
S. epidermidis ATCC 35984 (RP62A) was grown on tryptic soy agar (TSA) overnight at 37°C. A single colony was inoculated into 3 ml of tryptic soy broth (TSB) and grown overnight at 37°C with vigorous shaking. An aliquot (1 ml) of the culture grown overnight was then inoculated into 500 ml TSB and also grown overnight under the same conditions. Saturated cultures were centrifuged (10,000 × g for 5 min) and washed with TSB (200 ml) to remove any residual secreted proteins and then resuspended in 500 ml TSB to a density of ∼3 × 109 CFU/ml. The resulting saturated 500-ml culture was then separated into 100-ml aliquots, and the following concentrations of arylomycin C16 were added in a fixed volume of dimethyl sulfoxide (DMSO): 0×, 0.5×, 1.0×, and 4.0× the MIC (corresponding to 0, 0.25, 0.5, and 2 μg/ml, respectively). After incubation with shaking for 2 h at 37°C, the optical density was recorded, and aliquots were plated to determine the number of viable cells for each culture. Cells were then collected by centrifugation (10,000 × g for 5 min) and discarded. Supernatants containing the secreted proteins were clarified by filtration using a 0.22-μm low-protein-binding membrane. Proteins were then precipitated by the addition of 10% (wt/vol) trichloroacetic acid (TCA) followed by an overnight incubation on ice and then harvested by centrifugation (18,600 × g for 1 h). The resulting pellets were washed four times with 100% ethanol, dried, resuspended in 100 μl of 4% 3-[(3-cholamidopropyl)-dimethylammonio]-1-propanesulfonate (CHAPS) plus 8 M urea, and assayed for total protein (Bio-Rad protein assay).
Protein samples for electrophoresis were normalized by the number of viable cells, which were found to be ∼2-fold decreased in all arylomycin-treated samples compared to the DMSO-only control. To further increase our confidence that changes in viable cell numbers did not contribute to the observed protein levels, we also quantified total protein levels, which were also found to be decreased by ∼2-fold. For the first dimension of electrophoresis, linear immobilized pH gradient strips, pH 3 to 10 (Ready-Strip; Bio-Rad, Hercules, CA), were passively rehydrated with protein sample overnight at room temperature according to the protocol supplied by the manufacturer. Isoelectric focusing of the strips was run to 24,000 V-h. For the second dimension, 10% to 14.5% precast SDS-polyacrylamide gradient gels (Criterion; Bio-Rad) were run at 200 V for 54 min. After the second dimension, each gel was fixed for 30 min in 10% methanol-7% acetic acid and then stained overnight with SYPRO ruby protein gel stain (Invitrogen), washed in 10% methanol-7% acetic acid for an additional 30 min, and finally rehydrated in water for subsequent MS analysis. Sets of gels corresponding to secretion in the presence of 0×, 0.5×, 1.0×, and 4.0× the MIC of arylomycin C16 were independently reproduced three times.
Protein digestion.
From each set of two-dimensional (2D) gels, protein spots whose intensity decreased with increasing arylomycin C16 concentrations were identified by visual inspection and were excised from the arylomycin C16-free gel for characterization by LC-MS/MS. The excised gel bands were destained with a mixture of a 50:50 (vol/vol) dilution of acetonitrile-25 mM ammonium bicarbonate before being dried with a SpeedVac system. The proteins contained in the gel were then reduced in 25 μl of 10 mM d,l-dithiothreitol (Sigma) for 1 h and alkylated with 25 μl of 55 mM iodoacetamide (Sigma) for 30 min in the dark prior to an 18-h trypsin digestion at 37°C using a 1:30 (wt/wt) enzyme-to-substrate ratio. The resulting peptides were extracted twice with a 50:45:5 (vol/vol) volume of acetonitrile-water-formic acid and concentrated to 30 μl before being analyzed.
Separation of peptides and mass spectrometry.
Peptides were analyzed by reverse-phase chromatography prior to mass spectrometry analysis using the following method. Nanoelectrospray capillary column tips were made in-house by using a P-100 laser puller (Sutter Instruments). The columns were packed with Zorbax SB-C18 stationary phase (Agilent) purchased in bulk (5-mm particles, with a 15-cm length and a 75-mm inner diameter). The reverse-phase gradient separation was performed by using water and acetonitrile (0.1% formic acid) as the mobile phases. The gradient began at 5% acetonitrile and was ramped to 8% acetonitrile over 10 min. The acetonitrile was ramped to 35% acetonitrile over 20 min and then increased to 90% acetonitrile for another 20 min and maintained for another 10 min prior to reequilibration to 5% acetonitrile.
Data-dependent MS/MS data were obtained with an LTQ linear ion trap mass spectrometer using a home-built nanoelectrospray source at 2 kV at the tip. One MS spectrum was followed by 4 MS/MS scans on the most abundant ions after the application of the dynamic exclusion list. Tandem mass spectra were extracted by use of Xcalibur software. All MS/MS samples were analyzed by using Mascot (version 2.1.04; Matrix Science, London, United Kingdom) with S. epidermidis proteins contained in the NCBInr protein database, assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0.80 Da and a parent ion tolerance of 2.0 Da; identification was done at the 95% confidence level with a calculated false-positive rate of <1% as determined by using a reversed concatenated protein database. Scaffold (version Scaffold-01_06_03; Proteome Software Inc., Portland, OR) was used to additionally validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater than 95.0% probability as specified by the Peptide Prophet algorithm (28). Protein identifications were accepted if they could be established at a greater than 99.0% probability and contained at least 2 identified peptides as specified by the Protein Prophet algorithm (28). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
RT-PCR.
S. epidermidis cells for total RNA preparation were grown and treated with arylomycin as described above for two-dimensional gel electrophoresis and then lysed by using RNAprotect bacterial reagent (Qiagen) and the enzymatic lysis, proteinase K digestion, and mechanical disruption method supplied by the manufacturer. Lysates were subjected to total RNA isolation with the RNeasy minikit (Qiagen) according to the manufacturer's protocols. One microgram of total RNA was used for reverse transcription (RT) using the Superscript III first-strand synthesis kit (Invitrogen) according to the manufacturer's protocol. Subsequent first-strand cDNA (50 ng) was combined with SYBR green and 10 pmol primers (Table 1) in a total volume of 20 μl and subjected to the following cycling conditions with Taq polymerase (NEB) in a Bio-Rad iCycler: 94°C for 3 min followed by 30 cycles of 94°C for 15 s, 50°C for 30 s, and then 68°C for 1.5 min, followed by a melt curve to check that each primer set had only a single peak. Data were analyzed automatically with iCycler iQ software (Bio-Rad) using the comparative threshold cycle (CT) method. The transcription of 16S rRNA was used as a control, and samples were analyzed in duplicate.
TABLE 1.
Primers used for RT-PCR
| Primer | Sequence |
|---|---|
| SERP0318-5′ | 5′-GCCTTTGCAATTACAGCCGCTTCAGGCGC |
| SERP0318-3′ | 5′-CCATCATTCGCTGCATTTGACGCCCAG |
| SERP0379-5′ | 5′-CCGGTGACTCAACAGTAGACGG |
| SERP0379-3′ | 5′-GTCACCATTTAACACGCTGTCACTC |
| SERP0636-5′ | 5′-CGATGATGAAGACGATGATAGCTCAACAGACC |
| SERP0636-3′ | 5′-GCCATCAACAAATGCGTGTACGAATGC |
| SERP1397-5′ | 5′-GCAGCGTTAGCAACTACTACAATGG |
| SERP1397-3′ | 5′-CCCACATTGTGGCTAATGGTTTGTCACCAGGG |
| SERP1428-5′ | 5′-GCCTTTCCGTTTTCACATGTCGC |
| SERP1428-3′ | 5′-CGTCTACACCTTCACCTATCGTCACGC |
| SERP1460-5′ | 5′-GCACTGTTTGGACTTGACCG |
| SERP1460-3′ | 5′-CGTAACCTACAAACCACCC |
| SERP2138-5′ | 5′-CAGGACATGAAGCACACGCTTCAG |
| SERP2138-3′ | 5′-CTGAACCAGTTGAACCCCAACCTGGC |
| SERP2263-5′ | 5′-CCAAGCAGGTTTCCGTAAATACCCAAG |
| SERP2263-3′ | 5′-GCCCAGCTTCAAACACAACTGCAG |
| SERP2297-5′ | 5′-GCGTAGGTGCATCATCCATTCTGATTGCAGC |
| SERP2297-3′ | 5′-GAGAGTCTTCTTGTTGGGCAGTTG |
| SERP2388-5′ | 5′-GGAACGCCTCATAATGGCACACCTGCTGC |
| SERP2388-3′ | 5′-CGGGTCTCACTTGCCAGATACCCTTGTCAG |
| SERP2390-5′ | 5′-GCAGTTCGCAAACTTGCTGAAG |
| SERP2390-3′ | 5′-GCCGGCACACCAACTATTATACCC |
| 16S-rRNA-5′ | 5′-GCCTAATACATGCAAGTCGAGCGAACAGACG |
| 16S-rRNA-3′ | 5′-CCCCAATCATTTGTCCCACCTTCGACG |
Protein sequence analysis.
Molecular masses and average pI values were calculated by using the ExPASy proteomics server (http://expasy.org/cgi-bin/pi_tool). SPase cleavage sites, membrane anchor sequences, and transmembrane (TM) segments were predicted by using SMART (34) (http://smart.embl-heidelberg.de/), TMHMM (31) (http://www.cbs.dtu.dk/services/TMHMM/), LocateP (65) (http://www.cmbi.ru.nl/locatep-db/cgi-bin/locatepdb.py), SignalP 3.0 neural networks (SignalP-NN) and hidden Markov model (SignalP-HMM) algorithms (9) (http://www.cbs.dtu.dk/services/SignalP/), and PrediSi (http://www.predisi.de). Default settings (for Gram-positive bacteria) were used for these Web-based programs.
RESULTS
To identify proteins in S. epidermidis whose secretion relies on SPase, we compared protein spots on 2D gels of the extracellular fraction isolated from strain RP62A exposed during stationary-phase growth to various concentrations of arylomycin C16 (0×, 0.5×, 1.0×, and 4.0× the MIC) (Fig. 2). Roughly 125 protein spots were identified in the 2D gels of untreated samples. Visual inspection of the gels revealed 18 spots whose intensity showed a dose-dependent decrease with arylomycin treatment (Fig. 2). The corresponding spots were excised from the gels of untreated samples and subjected to LC-MS/MS analysis, resulting in the identification of 11 different proteins (Table 2). In comparison, intracellular levels of the corresponding mRNA were virtually unaffected (Fig. 3), confirming that inhibition occurred at the level of secretion and not transcription.
FIG. 2.
Representative set of 2D gels from 0× (a), 0.5× (b), 1.0× (c), 4.0× (d) the MIC of arylomycin-treated samples. Labeled spots correspond to proteins whose extracellular levels decrease with arylomycin treatment.
TABLE 2.
Identification of S. epidermidis proteins whose secretion is inhibited in a dose-dependent fashion by arylomycin C16
| Locus | Protein | Signal peptideb | Spot intensitya |
|||
|---|---|---|---|---|---|---|
| 0× | 0.5× | 1× | 4× | |||
| SERP0318 | SsaA-like | 1MKKLAFAITAASGAAAVLSHHDAEA | +++ | − | − | − |
| SERP0379 | LtaS | 144WLDNRAFSKKFVPVVMATSVALFFLNLAFA | ++++ | +++ | ++ | + |
| SERP0636 | AtlE | 1MAKKFNYKLPSMVALTLFGTAFTAHQANA | +++ | − | − | − |
| SERP1397 | SspA | 1MKKRFLSICTMTIAALATTTMVNTSYA | +++ | ++ | ++ | − |
| SERP1460 | BlaR1 | 302NLKKQSKLILIFICIFTFFIMIIQSQFLMG | +++ | − | − | − |
| SERP1428 | IsaB | 1MKIFKLTSLTLAALTLAFPFSHVAQA | +++ | ++ | + | − |
| SERP2138 | IsaA | 1MKKTVIASTLAVSLGIAGYGLSGHEAHA | ++++ | −/++/++++ | −/+/+++ | −/+/++ |
| SERP2263 | 1MKKNKFLVYLLSTALITPTFATQTAFA | +++ | ++ | − | − | |
| SERP2297 | GehC | 1MKTRQNKYSIRKFSVGASSILIAALLFMGGGSAQA | +++ | − | − | − |
| SERP2388 | GehD | 1MKNNNETRRFSIRKYTVGVVSIITGITIFVSGQHAQA | ++++c | −/++ | −/+ | −c |
| SERP2390 | SspB | 1MKKKLSYMITIMLAFTLSLALGLFFNSAHA | +++c | ++c | +c | +c |
Qualitative spot intensity as a function of added arylomycin C16, determined by visual inspection of 2D gels. The intensities for multiple spots are separated by slashes (see text).
Numbers correspond to the translated sequence. The sequences of LtaS and BlaR1 were arbitrarily started at residues 144 and 302, respectively.
All spots showed roughly the same sensitivity to inhibition.
FIG. 3.
Intracellular levels of mRNA of identified proteins detected after treatment with different arylomycin dosages (first bar, 0 μM; second bar, 0.125 μM [0.25× MIC]; third bar, 0.25 μM [0.5× MIC]; fourth bar, 0.5 μM [1.0× MIC]; fifth bar, 2.0 μM [4.0× MIC]). (A) SERP0318; (B) SERP0379; (C) SERP0636; (D) SERP1397; (E) SERP1428; (F) SERP1460; (G) SERP2138; (H) SERP2263; (I) SERP2297; (J) SERP2388; (K) SERP2390. See Materials and Methods for details.
The SsaA-like protein (SERP0318, 100% identical to SE_0433) was detected as a single spot in the samples without arylomycin treatment that was absent in all treated samples, and each prediction methodology employed identified the same 25-amino-acid signal peptide (Table 2). The single spot migrated with an apparent mass of ∼25 kDa and a pI of ∼6.5, consistent with those predicted for the SPase-processed, full-length protein (calculated mass of 25.8 kDa and pI of 6.0). Correspondingly, peptides identified by mass spectrometry correspond to sequences throughout the protein, minus the signal peptide.
The autolysin AtlE (SERP0636) was detected as two spots, one at ∼30 kDa with a pI of ∼9.5 and another at ∼75 kDa with a pI of ∼4.5, neither of which was observed for any of the arylomycin-treated samples. AtlE is predicted to have a well-defined, 29-amino-acid signal peptide (Table 2), and the secreted protein is known to be further processed to yield a 120-kDa protein and then cleaved again to generate a 60-kDa alanine amidase fragment and a 52-kDa glucosaminidase fragment, which also appear to be subject to further degradation (22). The peptides isolated from both spots correspond to the alanine amidase domain, with the 75-kDa spot comprised of peptides covering amino acids 41 to 461 and the 30-kDa spot comprised of peptides covering amino acids 318 to 461.
SERP2263 is annotated as an autolysin and is predicted to be targeted for secretion by a 27-residue signal peptide (Table 2). The protein was detected as a single spot with an apparent molecular mass of ∼25 kDa and an apparent pI of ∼4.5 whose intensity decreased until it was not visible at 4× the MIC. The observed mass is only a fraction of that predicted for the secreted protein (70.6 kDa), and the only peptides detected were derived from residues between residues 154 and 365, which are just N terminal to the predicted glucosaminidase and C-terminal cysteine-, histidine-dependent aminohydrolase/peptidase (CHAP) domains, suggesting that the protein may undergo proteolysis after secretion.
The protease SspA (SERP1397) is predicted to be translated with a 28-residue signal peptide (Table 2) and was detected as a single spot with an apparent mass of ∼24 kDa and a pI of ∼7.5 that was not observed in the presence of higher inhibitor concentrations. The mass and pI are in good and fair agreement, respectively, with those predicted for the fully processed protein (calculated mass of 23.6 kDa and pI of 6.2). Correspondingly, peptides from most of the protein minus the predicted signal sequence (amino acids 45 to 282) were detected.
In the case of SERP1428, which is an apparent homolog of Staphylococcus aureus immunodominant antigen B (IsaB) (17% identity and 26% similarity) and which is predicted to be translated with a 26-residue signal peptide (Table 2), we detected a single spot with an apparent molecular mass of ∼17.5 kDa and pI of ∼5.5, in agreement with the mass and pI predicted for the full-length secreted protein (17.4 kDa and pI 5.7, respectively). However, only peptides from the C-terminal half of the protein (between amino acids 91 and 178) were detected, possibly due to poor N-terminal fragment detection. The intensity of the spot decreased as a function of added arylomycin, and none was detectable at the highest concentration.
Our analysis also identified the lipases GehC and GehD, which each appear to be synthesized as preproenzymes with signal peptides of 35 and 37 amino acids, respectively (Table 2). In the case of GehC, we observed a single spot in the untreated control sample that was absent in all arylomycin-treated samples. While the apparent mass of the protein (∼80 kDa) is slightly above the calculated mass for the processed proprotein (73.7 kDa), it is significantly larger than the mass of the mature lipase (44 kDa) (54). Correspondingly, peptides from regions throughout the proprotein were detected, and the apparent pI agrees with the calculated value of 5.5 for the full-length proprotein. In the case of GehD, we observed two spots in the 2D gels of all samples, both of which decreased with added arylomycin. Like GehC, the slower-migrating spot had an apparent mass (∼75 kDa) that was slightly greater than that calculated for the proprotein (68.1 kDa) and an apparent pI (∼8.5) that was higher than expected (pI 6.6). The slower-migrating spot had an apparent mass of ∼45 kDa, consistent with that predicted for the mature lipase (44 kDa), although the apparent pI of ∼8 is slightly higher than that predicted (pI 6.5). Peptides from the entire length of the proprotein were detected in the 75-kDa spot, and peptides corresponding to only the mature lipase were detected in the 45-kDa spot. While the intensity of both GehD spots decreased with added arylomycin, the heavier protein spot was not observed at or above a concentration of 1× the MIC, and the second protein spot persisted until a concentration of 4× the MIC, possibly reflecting the various proteolyses of the differently processed proteins.
The cysteine protease SspB (SERP2390) is predicted to be translated with a 30-amino-acid signal peptide (Table 2) and was detected in four different spots with apparent masses varying from approximately 18 to 45 kDa and apparent isoelectric points of approximately 9 to 10. Peptides corresponding to regions starting just beyond the predicted SPase cleavage site and spanning the entire protein were detected in the 45-kDa spot, suggesting that it corresponds to the full-length secreted protein, which is further supported by its predicted mass of 41.4 kDa and pI of 9.4 (with the faster-migrating spots assumed to correspond to proteolytic fragments). While the secretion of the protease decreased with added arylomycin, it remained detectable at all inhibitor concentrations examined.
IsaA (SERP2138) is a likely transglycosylase and virulence factor that is predicted to be translated with a 28-residue signal peptide (Table 2) that was previously shown to be recognized by SPase in vitro (45). Two spots corresponding to IsaA were observed at ∼23 and ∼25 kDa, with apparent isoelectric points of ∼5.5 and ∼6, respectively. The two spots may correspond to variously processed forms of the secreted proteins, and both appear to migrate slower than expected (the predicted mass of the secreted protein is 21.8 kDa). Only peptides corresponding to a central region of the protein between amino acids 126 and 216 were detected. The heavier fragment was observed only in the absence of arylomycin; while the amount of the smaller fragment decreased with added inhibitor, it was still present at the highest arylomycin concentration examined.
A canonical N-terminal signal peptide was not detected with two of the identified proteins, LtaS (SERP0379), which is involved in lipoteichoic acid synthesis (36), and BlaR1 (SERP1460), which is a component of a two-component response regulator involved in the sensing of β-lactam antibiotics (30). However, LtaS is predicted by sequence analysis to have five N-terminal transmembrane helices and a C-terminal, extracellular domain (which has been annotated as a sulfatase [UniProt accession number Q5HR16]). When the sequences surrounding each of the predicted transmembrane helices were examined individually, a high-probability SPase recognition site was detected between amino acids 173 and 174, which is located just C terminal to the last transmembrane helix (Table 2). The protein was detected as a single spot at ∼50 kDa and pI ∼8, which is consistent with the values predicted for the C-terminal domain after cleavage between residues 173 and 174 (36, 66), and all of the detected peptides were from the extracellular domain. The intensity of the spot decreased with added arylomycin, although it still persisted at the highest arylomycin C16 concentrations examined.
BlaR1 was detected as a single spot with a mass of ∼25 kDa and a pI of ∼9 that was not present in the arylomycin-treated samples. While BlaR1 does not appear to have an obvious Sec-type signal peptide, as with LtaS, sequence analysis indicates that it does have a series of N-terminal transmembrane helices (19) and a high-probability SPase recognition sequence predicted just C terminal to the last transmembrane helix (between amino acids 331 and 332) (Table 2). Correspondingly, the apparent mass and pI of the protein are consistent with those predicted for the extracellular domain (30 kDa and pI 9.1). Moreover, all of the peptides detected corresponded to fragments from within the extracellular domain (between amino acids 341 and 562).
DISCUSSION
During the early growth phase, bacteria are thought to export surface proteins that function primarily as adhesins, but as growth proceeds toward the late exponential phase, the expression of these proteins is suppressed and extracellular proteins are upregulated and secreted into the environment (15, 64). These secreted proteins are thought to include important virulence factors, such as proteases, nucleases, lipases, and autolysins, which facilitate persistence and/or dissemination (1, 64). However, the determination of exactly which proteins are processed by SPase has proven difficult.
To develop a general approach for identifying proteins that are secreted in an SPase-dependent manner, we synthesized arylomycin C16, a derivative of the arylomycin class of natural products, which potently and selectively inhibit S. epidermidis SPase in vivo (47, 55a). S. epidermidis is the most common cause of nosocomial infections (41), most notably with implanted devices, and costs the United States alone an estimated $28.4 to $33.8 billion annually (51). Based on the available sequence of the S. epidermidis RP62A genome, it is estimated that more than 600 proteins are synthesized with potential Sec-type signal peptides, although most are predicted to remain associated with the cell membrane, and canonical SPase recognition sequences, in the absence of any cell wall retention signals, are obvious in only 29 of these proteins (65). We detected roughly 125 protein spots in medium prepared from stationary-phase cultures of S. epidermidis by 2D PAGE. However, in the presence of increasing amounts of arylomycin C16, we observed a dose-dependent decrease in the intensity of spots corresponding to only 11 proteins, suggesting that only these proteins are recognized by SPase and actually secreted via the general Sec pathway (note that S. epidermidis appears to lack a twin-arginine translocation pathway, which in other organisms also relies on SPase [11, 42]).
Of the 29 S. epidermidis proteins that are predicted by LocateP (65) to be translated with SPase cleavage sites and which lack identifiable cell wall retention signals, we identified 7 whose presence in the medium is indeed dependent on SPase. Six of these were also identified previously by Batzilla et al. (6), including AtlE, GehC, GehD, IsaA, SspA, and the SsaA-like protein. We also identified five proteins that were not detected by Batzilla et al. (6), including three with canonical signal peptides, SspB, IsaB, and SERP2263, as well as two that appear to have noncanonical SPase cleavage sites, LtaS and BlaR1.
It is also interesting to consider the proteins that are predicted to be secreted but which were not detected as SPase substrates. For example, despite detecting the SsaA-like protein encoded by SERP0318, we did not detect SsaA itself (SERP1880/SERP2136), which is predicted to have a signal peptide and which has been detected immunologically in sera from some patients with S. epidermidis infections (32). In addition, while we detected several proteases, we did not detect the thermolysin-like zinc metalloprotease SepA, despite its synthesis with an apparent Sec-type signal peptide and its secretion into the medium as the processed protein lacking the signal peptide sequence when expressed in Staphylococcus carnosus (58). SepA was also detected in the extracellular fraction of clinical, biofilm-forming S. epidermidis strain 567 previously by Batzilla et al. (6) but not by Ohara-Nemoto et al. in their studies of non-biofilm-forming S. epidermidis strain ATCC 14990 (40). Despite detecting the lipases GehC and GehD, we did not detect several other enzymes with similar proposed functions that are predicted to have signal peptides, including the putative lipases SERP0018 and SERP2336 as well as the putative esterase SERP2109. Other proteins predicted to be translated with a signal peptide but not detected in our experiments include a lysozyme domain protein (SERP0043), a putative transcriptional regulator (SERP0640), the transglycosylase SceD, the UTP-glucose-1-phosphate uridylyltransferase GalU, the adhesion protein IcaB, the ATPase subunit KdpC, as well as eight hypothetical proteins. While it is possible that some of these proteins are secreted under other conditions, we cannot exclude the possibility that some are secreted at undetectable concentrations and/or are obscured by other proteins in the 2D gels. Regardless, the nature of the 11 proteins that were detected provides important information about the growth and virulence of S. epidermidis.
Peptidoglycan hydrolases are essential for reorganizing the cell wall during vegetative growth and cell division and are also thought to be important virulence factors (62). We found that arylomycin C16 inhibited the secretion of four peptidoglycan hydrolases: AtlE, the SsaA-like protein encoded by SERP0318, SERP2263, and IsaA, making this the major type of protein secreted by S. epidermidis. The autolysin-adhesin AtlE is the major autolysin of S. epidermidis, and it has a well-defined role in virulence (22, 49, 55) and is also thought to play a role in biofilm formation through its hydrophobicity (22) and its ability to bind various plasma and matrix proteins (22, 23, 43). The SsaA-like protein encoded by SERP0318 has a C-terminal cysteine- and histidine-dependent aminohydrolase/peptidase (CHAP) domain and two N-terminal LysM peptidoglycan binding domains, which are commonly associated with bacterial cell wall degradation. It is homologous to the secretory antigen SsaA-like protein of S. aureus (80% identity), which is a surface antigen that is upregulated by sigB and is important for biofilm formation (7, 10). SERP0318 is also similar to S. aureus Sle1 (46% identical), which is an autolysin with N-acetylmuramyl-l-alanine amidase activity (27). SERP2263 also appears to contain an N-acetylmuramoyl-l-alanine amidase as well as a transglycosylase domain, while IsaA is annotated as a transglycosylase, and both are known virulence factors (35, 56).
Secreted proteases play an important role in virulence by scavenging nutrients in the form of short peptides, processing other extracellular bacterial proteins, and degrading host proteins. Commonly, serine proteases, cysteine proteases, and metalloproteases are secreted (17), but only the serine protease SspA (SERP1397, also known as Esp, GseA, and GluSE) and the cysteine protease SspB (also known as Ecp) were observed to be secreted in S. epidermidis. SspA and SspB are transcribed on a single polycistronic mRNA that is regulated by the Agr and Sar systems (6, 17). SspA is homologous to the S. aureus V8 protease (65% identity), which is an important virulence factor (17, 38, 39) that possibly interferes with complement activation and blood coagulation (17, 38). SspB is homologous to both of the S. aureus staphopain cysteine proteases SspB1 and SspB2 (61% and 75% identical, respectively) and has been shown to degrade human immunoglobulins, serum albumin, fibronectin, and all three subunits of fibrinogen, which might contribute to immune system avoidance and the establishment and/or dissemination of infection (52). The extracellular localization of the S. aureus homologs was demonstrated previously and was also shown to be dependent on the alternative sigma factor σB (24).
Secreted lipases are thought to be important for the colonization of lipid-rich environments, such as the skin habitat of S. epidermidis (12, 54). As with secreted proteases, secreted lipases are synthesized as preproproteins, and we detected two, GehC and GehD, whose presence in the medium was dependent on SPase activity. In addition to a signal peptide, GehC and GehD each contain the conserved YSIRK-G/S motif, which has been detected in surface proteins of other bacteria and is thought to target these proteins to a specialized mechanism of secretion (4, 48, 57). However, our data reveal that at least the majority of their secretion is mediated by the general Sec pathway and SPase. Consistent with this conclusion, neither GehC nor GehD has a C-terminal LPXTG sorting signal for cell wall attachment as was observed with other YSIRK-G/S motif-containing proteins (4).
The secretion of SERP1428, an apparent homolog of S. aureus IsaB, was also inhibited by arylomycin C16. IsaB is an S. aureus virulence factor, and its secretion and nonspecific DNA and RNA binding were demonstrated previously (37). Given the contribution of extracellular DNA to biofilm formation, Mackey-Lawrence et al. explored the contribution of IsaB to S. aureus biofilm formation (37), and although they did not detect any in their plastic adherence assay, those authors pointed out that this does not rule out a role for IsaB under more physiological conditions.
An interesting observation from the arylomycin dose dependencies of the secreted proteins is that the secretion of different proteins appears to show different sensitivities to inhibition (Table 2). For example, among the peptidoglycan hydrolases, the secretion of AtlE and the SsaA-like protein encoded by SERP0318 was strongly inhibited by arylomycin C16, while that of SERP2263 and IsaA was only moderately inhibited. The secretions of the two lipases GehC and GehD also showed very different dose dependencies, with the former being more inhibited than the latter. This finding suggests that different Sec-type signal peptides are recognized by SPase with different affinities (although the origins of the differential recognition need not reside entirely within the signal peptide sequence itself [14]). This in turn suggests that under conditions of limiting SPase activity, SPase specificity may contribute a previously unappreciated level of control over the secretome.
From the perspective of potential drug development, it is interesting that much recent interest has focused on the targeting of virulence mechanisms (5, 18, 29). Nonetheless, approaches based on inhibiting virulence will likely suffer from a limited spectrum, as most proteins that contribute to virulence are not essential and thus free to diverge. In this regard, SPase is unique in that it is essential and its active site is highly conserved. Thus, the demonstration that SPase activity is required for the secretion of multiple virulence factors suggests that the arylomycins might be developed as broad-spectrum antibiotics that inhibit virulence while also killing the bacteria.
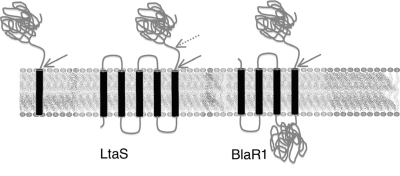
A surprising result of the current study is that levels of the cytoplasmic domains of the LtaS and BlaR1 transmembrane proteins found in the media decrease with added arylomycin. This result suggests that both proteins are SPase substrates, despite their apparent lack of N-terminal signal peptides, which are the only generally accepted SPase substrates. However, an internal site that is topologically identical to a canonical N-terminal signal peptide was identified in each protein (Fig. 4). The finding that LtaS is cleaved at an internal Ala-X-Ala site, suggestive of SPase recognition, was recently reported by Lu and coworkers for the S. aureus protein, but the participation of SPase was not confirmed (36). In this case, N-terminal sequencing revealed cleavage at 217ALA-SE (cleavage at the hyphen). However, this site is not predicted by SignalP (SignalP-HMM probability of 0.0), in contrast with the site that we detected at 173AFA-ET (SignalP-HMM probability of 1.0). It is thus possible that additional processing occurs after the initial SPase-mediated cleavage or that SignalP is unable to recognize the site in this case. Moreover, Bacillus subtilis encodes five SPases, and the double mutant bereft of two SPases showed decreased levels of the LtaS homolog YfiI (2). Finally, there are a variety of reports in the literature suggesting that SPase may act at internal sites, analogous to that proposed here with LtaS and BlaR1 (2, 3, 25, 33, 44, 59). This, and the fact that 2 out of 11 SPase substrates detected in S. epidermidis appear to be cleaved internally, suggests that internal cleavage sites are not uncommon and that SPase may have a variety of other previously unappreciated regulatory roles, such as in cell wall biogenesis and the regulation of inherent resistance to β-lactam antibiotics.
FIG. 4.
Topology of SPase substrates. (Left) General representation of a signal peptide protein showing a transmembrane region and an extracellular domain that is released via the indicated SPase cleavage site (arrow). (Center) Proposed topology of LtaS showing five TM domains and two proposed cleavage sites, one (solid arrow) predicted by SignalP 3.0 (9) and a second (hashed arrow) predicted by proteomic data (36, 66). (Right) Proposed topology of BlaR1 (20) showing four TM domains, an intracellular zinc binding domain, and the extracellular domain released via the indicated cleavage site.
It is likely that the arylomycins will provide a general chemical biology tool for the study of SPase-mediated secretion in a variety of bacteria and under a variety of conditions. However, it should be noted that the approach identifies only proteins that are synthesized and secreted under the experimental conditions employed. In this study, we examined stationary-phase cells in order to minimize the effects of antibiotic-mediated cell lysis, although any cell lysis associated with the arylomycin treatment of exponentially growing bacteria would be expected to increase protein levels in the medium and thus should not interfere with the identification of SPase-mediated export (which is identified by decreased levels of a given protein in the medium). Regarding generality, we have recently identified a diverse range of Gram-positive and Gram-negative bacteria whose SPases are potently inhibited by arylomycin C16 in vivo (55a), and thus, arylomycin C16 should be useful for the characterization of protein secretion in these bacteria as well. While the SPases of E. coli, Pseudomonas aeruginosa, and S. aureus are resistant to the arylomycins, we have identified strains of each bacterium with SPase mutants that are also potently inhibited by arylomycin C16, making it possible to profile secretion in these important pathogens as well. However, the approach on its own cannot distinguish between proteins whose levels are decreased as a direct result of SPase inhibition and proteins whose levels are decreased due to the indirect effects of inhibiting an essential process. In this study RT-PCR was used to demonstrate that the decreased levels of the identified proteins do not result from altered transcription, and all of the identified proteins have Sec-type leader sequences; therefore, the data support the conclusion that the proteins are indeed secreted in an SPase-dependent manner. In a similar fashion, arylomycin C16 and the chemical biology approach utilized here should prove valuable for studying protein secretion in a wide range of bacteria.
Acknowledgments
Funding for this work was provided by the Office of Naval Research (grant N000140810478) and the National Institutes of Health (grant AI081126).
Footnotes
Published ahead of print on 12 November 2010.
REFERENCES
- 1.An, Y. H., and R. J. Friedman (ed.). 2000. Handbook of bacterial adhesion: principles, methods, and applications. Humana Press, Totowa, NJ.
- 2.Antelmann, H., H. Tjalsma, B. Voigt, S. Ohlmeier, S. Bron, J. M. van Dijl, and M. Hecker. 2001. A proteomic view on genome-based signal peptide predictions. Genome Res. 11:1484-1502. [DOI] [PubMed] [Google Scholar]
- 3.Antelmann, H., R. C. Williams, M. Miethke, A. Wipat, D. Albrecht, C. R. Harwood, and M. Hecker. 2005. The extracellular and cytoplasmic proteomes of the non-virulent Bacillus anthracis strain UM23C1-2. Proteomics 5:3694-3695. [DOI] [PubMed] [Google Scholar]
- 4.Bae, T., and O. Schneewind. 2003. The YSIRK-G/S motif of staphylococcal protein A and its role in efficiency of signal peptide processing. J. Bacteriol. 185:2910-2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baron, C. 2010. Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 13:100-105. [DOI] [PubMed] [Google Scholar]
- 6.Batzilla, C. F., S. Rachid, S. Engelmann, M. Hecker, J. Hacker, and W. Ziebuhr. 2006. Impact of the accessory gene regulatory system (Agr) on extracellular proteins, codY expression and amino acid metabolism in Staphylococcus epidermidis. Proteomics 6:3602-3613. [DOI] [PubMed] [Google Scholar]
- 7.Beenken, K. E., P. M. Dunman, F. McAleese, D. Macapagal, E. Murphy, S. J. Projan, J. S. Blevins, and M. S. Smeltzer. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665-4684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bendtsen, J. D., L. Kiemer, A. Fausboll, and S. Brunak. 2005. Non-classical protein secretion in bacteria. BMC Microbiol. 5:58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bendtsen, J. D., H. Nielsen, G. von Heijne, and S. Brunak. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783-795. [DOI] [PubMed] [Google Scholar]
- 10.Bischoff, M., P. Dunman, J. Kormanec, D. Macapagal, E. Murphy, W. Mounts, B. Berger-Bachi, and S. Projan. 2004. Microarray-based analysis of the Staphylococcus aureus σB regulon. J. Bacteriol. 186:4085-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Biswas, L., R. Biswas, C. Nerz, K. Ohlsen, M. Schlag, T. Schafer, T. Lamkemeyer, A. K. Ziebandt, K. Hantke, R. Rosenstein, and F. Gotz. 2009. Role of the twin-arginine translocation pathway in Staphylococcus. J. Bacteriol. 191:5921-5929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bowden, M. G., L. Visai, C. M. Longshaw, K. T. Holland, P. Speziale, and M. Hook. 2002. Is the GehD lipase from Staphylococcus epidermidis a collagen binding adhesin? J. Biol. Chem. 277:43017-43023. [DOI] [PubMed] [Google Scholar]
- 13.Cash, P., and E. Argo. 2009. Analysis of bacterial proteins by 2DE. Methods Mol. Biol. 519:131-144. [DOI] [PubMed] [Google Scholar]
- 14.Choo, K. H., and S. Ranganathan. 2008. Flanking signal and mature peptide residues influence signal peptide cleavage. BMC Bioinformatics 9(Suppl. 12):S15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman, G., C. M. Jakeman, and N. Martin. 1978. Patterns of total extracellular protein secretion by a number of clinically isolated strains of Staphylococcus aureus. J. Gen. Microbiol. 107:189-192. [DOI] [PubMed] [Google Scholar]
- 16.Cravatt, B. F., G. M. Simon, and J. R. Yates III. 2007. The biological impact of mass-spectrometry-based proteomics. Nature 450:991-1000. [DOI] [PubMed] [Google Scholar]
- 17.Dubin, G. 2002. Extracellular proteases of Staphylococcus spp. Biol. Chem. 383:1075-1086. [DOI] [PubMed] [Google Scholar]
- 18.Felise, H. B., H. V. Nguyen, R. A. Pfuetzner, K. C. Barry, S. R. Jackson, M. P. Blanc, P. A. Bronstein, T. Kline, and S. I. Miller. 2008. An inhibitor of Gram-negative bacterial virulence protein secretion. Cell Host Microbe 4:325-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hanique, S., M. L. Colombo, E. Goormaghtigh, P. Soumillion, J. M. Frere, and B. Joris. 2004. Evidence of an intramolecular interaction between the two domains of the BlaR1 penicillin receptor during the signal transduction. J. Biol. Chem. 279:14264-14272. [DOI] [PubMed] [Google Scholar]
- 20.Hardt, K., B. Joris, S. Lepage, R. Brasseur, J. O. Lampen, J. M. Frere, A. L. Fink, and J. M. Ghuysen. 1997. The penicillin sensory transducer, BlaR, involved in the inducibility of beta-lactamase synthesis in Bacillus licheniformis is embedded in the plasma membrane via a four-alpha-helix bundle. Mol. Microbiol. 23:935-944. [DOI] [PubMed] [Google Scholar]
- 21.Hecker, M., D. Becher, S. Fuchs, and S. Engelmann. 2010. A proteomic view of cell physiology and virulence of Staphylococcus aureus. Int. J. Med. Microbiol. 300:76-87. [DOI] [PubMed] [Google Scholar]
- 22.Heilmann, C., M. Hussain, G. Peters, and F. Gotz. 1997. Evidence for autolysin-mediated primary attachment of Staphylococcus epidermidis to a polystyrene surface. Mol. Microbiol. 24:1013-1024. [DOI] [PubMed] [Google Scholar]
- 23.Heilmann, C., G. Thumm, G. S. Chhatwal, J. Hartleib, A. Uekotter, and G. Peters. 2003. Identification and characterization of a novel autolysin (Aae) with adhesive properties from Staphylococcus epidermidis. Microbiology 149:2769-2778. [DOI] [PubMed] [Google Scholar]
- 24.Hempel, K., J. Pane-Farre, A. Otto, S. Sievers, M. Hecker, and D. Becher. 2010. Quantitative cell surface proteome profiling for SigB-dependent protein expression in the human pathogen Staphylococcus aureus via biotinylation approach. J. Proteome Res. 9:1579-1590. [DOI] [PubMed] [Google Scholar]
- 25.Hirose, I., K. Sano, I. Shioda, M. Kumano, K. Nakamura, and K. Yamane. 2000. Proteome analysis of Bacillus subtilis extracellular proteins: a two-dimensional protein electrophoretic study. Microbiology 146(Pt. 1):65-75. [DOI] [PubMed] [Google Scholar]
- 26.Holtzel, A., D. G. Schmid, G. J. Nicholson, S. Stevanovic, J. Schimana, K. Gebhardt, H. P. Fiedler, and G. Jung. 2002. Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by Streptomyces sp. Tü 6075. II. Structure elucidation. J. Antibiot. 55:571-577. [DOI] [PubMed] [Google Scholar]
- 27.Kajimura, J., T. Fujiwara, S. Yamada, Y. Suzawa, T. Nishida, Y. Oyamada, I. Hayashi, J. Yamagishi, H. Komatsuzawa, and M. Sugai. 2005. Identification and molecular characterization of an N-acetylmuramyl-L-alanine amidase Sle1 involved in cell separation of Staphylococcus aureus. Mol. Microbiol. 58:1087-1101. [DOI] [PubMed] [Google Scholar]
- 28.Keller, A., A. I. Nesvizhskii, E. Kolker, and R. Aebersold. 2002. Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74:5383-5392. [DOI] [PubMed] [Google Scholar]
- 29.Keyser, P., M. Elofsson, S. Rosell, and H. Wolf-Watz. 2008. Virulence blockers as alternatives to antibiotics: type III secretion inhibitors against Gram-negative bacteria. J. Intern. Med. 264:17-29. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi, T., Y. F. Zhu, N. J. Nicholls, and J. O. Lampen. 1987. A second regulatory gene, blaR1, encoding a potential penicillin-binding protein required for induction of beta-lactamase in Bacillus licheniformis. J. Bacteriol. 169:3873-3878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krogh, A., B. Larsson, G. von Heijne, and E. L. Sonnhammer. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305:567-580. [DOI] [PubMed] [Google Scholar]
- 32.Lang, S., M. A. Livesley, P. A. Lambert, W. A. Littler, and T. S. Elliott. 2000. Identification of a novel antigen from Staphylococcus epidermidis. FEMS Immunol. Med. Microbiol. 29:213-220. [DOI] [PubMed] [Google Scholar]
- 33.Lequette, Y., E. Lanfroy, V. Cogez, J. P. Bohin, and J. M. Lacroix. 2008. Biosynthesis of osmoregulated periplasmic glucans in Escherichia coli: the membrane-bound and the soluble periplasmic phosphoglycerol transferases are encoded by the same gene. Microbiology 154:476-483. [DOI] [PubMed] [Google Scholar]
- 34.Leutunic, I., T. Doerks, and P. Bork. 2008. SMART 6: recent updates and new developments. Nucleic Acids Res. 37:D229-D332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenz, U., K. Ohlsen, H. Karch, M. Hecker, A. Thiede, and J. Hacker. 2000. Human antibody response during sepsis against targets expressed by methicillin resistant Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 29:145-153. [DOI] [PubMed] [Google Scholar]
- 36.Lu, D., M. E. Wormann, X. Zhang, O. Schneewind, A. Grundling, and P. S. Freemont. 2009. Structure-based mechanism of lipoteichoic acid synthesis by Staphylococcus aureus LtaS. Proc. Natl. Acad. Sci. U. S. A. 106:1584-1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mackey-Lawrence, N. M., D. E. Potter, N. Cerca, and K. K. Jefferson. 2009. Staphylococcus aureus immunodominant surface antigen B is a cell-surface associated nucleic acid binding protein. BMC Microbiol. 9:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moon, J. L., A. Banbula, A. Oleksy, J. A. Mayo, and J. Travis. 2001. Isolation and characterization of a highly specific serine endopeptidase from an oral strain of Staphylococcus epidermidis. Biol. Chem. 382:1095-1099. [DOI] [PubMed] [Google Scholar]
- 39.Ohara-Nemoto, Y., Y. Ikeda, M. Kobayashi, M. Sasaki, S. Tajika, and S. Kimura. 2002. Characterization and molecular cloning of a glutamyl endopeptidase from Staphylococcus epidermidis. Microb. Pathog. 33:33-41. [DOI] [PubMed] [Google Scholar]
- 40.Ohara-Nemoto, Y., T. Oho, Y. Shimoyama, S. Kimura, and T. K. Nemoto. 2008. Homologous and heterologous expression and maturation processing of extracellular glutamyl endopeptidase of Staphylococcus epidermidis. Biol. Chem. 389:1209-1217. [DOI] [PubMed] [Google Scholar]
- 41.Otto, M. 2009. Staphylococcus epidermidis—the ‘accidental’ pathogen. Nat. Rev. Microbiol. 7:555-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paetzel, M., A. Karla, N. C. Strynadka, and R. E. Dalbey. 2002. Signal peptidases. Chem. Rev. 102:4549-4580. [DOI] [PubMed] [Google Scholar]
- 43.Pourmand, M. R., S. R. Clarke, R. F. Schuman, J. J. Mond, and S. J. Foster. 2006. Identification of antigenic components of Staphylococcus epidermidis expressed during human infection. Infect. Immun. 74:4644-4654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pugsley, A. P. 1993. The complete general secretory pathway in Gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rao, S., K. Bockstael, S. Nath, Y. Engelborghs, J. Anné, and N. Geukens. 2009. Enzymatic investigation of the Staphylococcus aureus type I signal peptidase SpsB—implications for the search for novel antibiotics. FEBS J. 276:3222-3232. [DOI] [PubMed] [Google Scholar]
- 46.Ravipaty, S., and J. P. Reilly. 2010. Comprehensive characterization of methicillin-resistant Staphylococcus aureus subsp. aureus COL secretome by two-dimensional liquid chromatography and mass spectrometry. Mol. Cell. Proteomics 9:1898-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberts, T. C., P. A. Smith, R. T. Cirz, and F. E. Romesberg. 2007. Structural and initial biological analysis of synthetic arylomycin A2. J. Am. Chem. Soc. 129:15830-15838. [DOI] [PubMed] [Google Scholar]
- 48.Rosenstein, R., and F. Gotz. 2000. Staphylococcal lipases: biochemical and molecular characterization. Biochimie 82:1005-1014. [DOI] [PubMed] [Google Scholar]
- 49.Rupp, M. E., J. S. Ulphani, P. D. Fey, K. Bartscht, and D. Mack. 1999. Characterization of the importance of polysaccharide intercellular adhesin/hemagglutinin of Staphylococcus epidermidis in the pathogenesis of biomaterial-based infection in a mouse foreign body infection model. Infect. Immun. 67:2627-2632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schimana, J., K. Gebhardt, A. Holtzel, D. G. Schmid, R. Sussmuth, J. Muller, R. Pukall, and H. P. Fiedler. 2002. Arylomycins A and B, new biaryl-bridged lipopeptide antibiotics produced by Streptomyces sp. Tü 6075. I. Taxonomy, fermentation, isolation and biological activities. J. Antibiot. 55:565-570. [DOI] [PubMed] [Google Scholar]
- 51.Scott, R. D., III. 2009. The direct medical costs of healthcare-associated infections in U.S. hospitals and the benefits of prevention. Centers for Disease Control and Prevention, Atlanta, GA.
- 52.Shaw, L., E. Golonka, J. Potempa, and S. J. Foster. 2004. The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150:217-228. [DOI] [PubMed] [Google Scholar]
- 53.Sibbald, M. J., A. Ziebandt, S. Engelmann, M. Hecker, A. de Jong, H. Harmsen, G. Raangs, I. Stokroos, J. Arends, J.-Y. Dubois, and J. M. van Dijl. 2006. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70:755-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Simons, J. W., M. D. van Kampen, S. Riel, F. Götz, M. R. Egmond, and H. M. Verhij. 1998. Cloning, purification and characterization of the lipase from Staphylococcus epidermidis—comparison of the substrate selectivity with those of other microbial lipases. Eur. J. Biochem. 253:675-683. [DOI] [PubMed] [Google Scholar]
- 55.Sivadon, V., M. Rottman, J. C. Quincampoix, E. Prunier, P. de Mazancourt, L. Bernard, A. Lortat-Jacob, P. Piriou, T. Judet, and J. L. Gaillard. 2006. Polymorphism of the cell wall-anchoring domain of the autolysin-adhesin AtlE and its relationship to sequence type, as revealed by multilocus sequence typing of invasive and commensal Staphylococcus epidermidis strains. J. Clin. Microbiol. 44:1839-1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55a.Smith, P. A., T. C. Roberts, and F. E. Romesberg. Broad spectrum antibiotic activity of the arylomycin natural products is masked by natural target mutations. Chem. Biol., in press. [DOI] [PMC free article] [PubMed]
- 56.Stapelton, M. R., M. J. Horsburgh, E. J. Hayhurst, L. Wright, I. M. Jonsson, A. Tarkowski, J. F. Kokai-Kun, J. J. Mond, and S. J. Foster. 2007. Characterization of IsaA and SceD, two putative lytic transglycosylases of Staphylococcus aureus. J. Bacteriol. 189:7316-7325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tettelin, H., K. E. Nelson, I. T. Paulsen, J. A. Eisen, T. D. Read, S. Peterson, J. Heidelberg, R. T. DeBoy, D. H. Haft, R. J. Dodson, A. S. Durkin, M. Gwinn, J. F. Kolonay, W. C. Nelson, J. D. Peterson, L. A. Umayam, O. White, S. L. Salzberg, M. R. Lewis, D. Radune, E. Holtzapple, H. Khouri, A. M. Wolf, T. R. Utterback, C. L. Hansen, L. A. McDonald, T. V. Feldblyum, S. Angiuoli, T. Dickinson, E. K. Hickey, I. E. Holt, B. J. Loftus, F. Yang, H. O. Smith, J. C. Venter, B. A. Dougherty, D. A. Morrison, S. K. Hollingshead, and C. M. Fraser. 2001. Complete genome sequence of a virulent isolate of Streptococcus pneumoniae. Science 293:498-506. [DOI] [PubMed] [Google Scholar]
- 58.Teufel, P., and F. Gotz. 1993. Characterization of an extracellular metalloprotease with elastase activity from Staphylococcus epidermidis. J. Bacteriol. 175:4218-4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Thony-Meyer, L., P. James, and H. Hennecke. 1991. From one gene to two proteins: the biogenesis of cytochromes b and c1 in Bradyrhizobium japonicum. Proc. Natl. Acad. Sci. U. S. A. 88:5001-5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tjalsma, H., H. Antelmann, J. D. H. Jongbloed, P. G. Braun, E. Darmon, R. Dorenbos, J.-Y. Dubois, H. Westers, G. Zanen, W. J. Quax, O. P. Kuipers, S. Bron, M. Hecker, and J. M. van Dijl. 2004. Proteomics of protein secretion by Bacillus subtilis: separating the “secrets” of the secretome. Microbiol. Mol. Biol. Rev. 68:207-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tjalsma, H., and J. M. Dijl. 2005. Proteomics-based consensus prediction of protein retention in a bacterial membrane. Proteomics 7:73-81. [DOI] [PubMed] [Google Scholar]
- 62.Vollmer, W., B. Joris, P. Charlier, and S. Foster. 2008. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32:259-286. [DOI] [PubMed] [Google Scholar]
- 63.Wang, P., and R. E. Dalbey. 2010. In vitro and in vivo approaches to studying the bacterial signal peptide processing. Methods Mol. Biol. 619:21-37. [DOI] [PubMed] [Google Scholar]
- 64.Wooldridge, K. (ed.). 2009. Bacterial secreted proteins: secretory mechanisms and role in pathogenesis. Caister Academic Press, Norwich, United Kingdom.
- 65.Zhou, M., J. Boekhorst, C. Francke, and R. J. Siezen. 2008. LocateP: genome-scale subcellular-location predictor for bacterial proteins. BMC Bioinformatics 9:173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ziebandt, A. K., D. Becher, K. Ohlsen, J. Hacker, M. Hecker, and S. Engelmann. 2004. The influence of agr and σB in growth phase dependent regulation of virulence factors in Staphylococcus aureus. Proteomics 4:3034-3047. [DOI] [PubMed] [Google Scholar]
- 67.Ziebandt, A. K., H. Kusch, M. Degner, S. Jaglitz, M. J. Sibbald, J. P. Arends, M. A. Chlebowicz, D. Albrecht, R. Pantucek, J. Doskar, W. Ziebuhr, B. M. Broker, M. Hecker, J. M. van Dijl, and S. Engelmann. 2010. Proteomics uncovers extreme heterogeneity in the Staphylococcus aureus exoproteome due to genomic plasticity and variant gene regulation. Proteomics 10:1634-1644. [DOI] [PubMed] [Google Scholar]