Abstract
The Pseudomonas aeruginosa transcription factor QscR responds to a variety of fatty acyl-homoserine lactones (HSLs), including N-3-oxododecanoyl-HSL (3OC12-HSL), which is produced and detected by the P. aeruginosa quorum-sensing circuit LasI and LasR. As is true for LasR and many other acyl-HSL-dependent transcription factors, production of soluble QscR in sufficient amounts for purification requires growth of recombinant bacteria in the presence of an appropriate acyl-HSL. QscR is thought to bind 3OC12-HSL relatively weakly compared to LasR, and unlike LasR, binding of purified QscR to target DNA was shown to strongly depend on exogenously added 3OC12-HSL. We show that purified QscR is dimeric at sufficiently high concentrations and monomeric at lower concentrations. Furthermore, QscR bound 3OC12-HSL more tightly than previously believed. Purified QscR retained 3OC12-HSL, and at sufficiently high concentrations, it bound target DNA in the absence of added 3OC12-HSL. We also obtained soluble QscR from recombinant Escherichia coli grown in the presence of N-3-oxohexanoyl-HSL (3OC6-HSL) instead of 3OC12-HSL, and because 3OC6-HSL bound much more loosely to QscR than other acyl-HSLs tested, we were able to exchange 3OC6-HSL with other acyl-HSLs in vitro and then estimate binding affinities of QscR for different acyl-HSLs and for target DNA. Our data support a model whereby QscR polypeptides fold properly in the absence of an acyl-HSL, but soluble, acyl-HSL-free QscR does not accumulate because it is subject to rapid aggregation or proteolysis.
Quorum sensing controls expression of hundreds of genes, including genes for production of many secreted virulence factors in the opportunistic human pathogen Pseudomonas aeruginosa (10, 28, 31, 41). There are two acyl-homoserine lactone (HSL) quorum-sensing signals produced by P. aeruginosa, N-3-oxododecanoyl-homoserine lactone (3OC12-HSL), the product of an acyl-HSL synthase called LasI, and N-butanoyl-homoserine lactone (C4-HSL), which is generated by RhlI. The receptors for these quorum-sensing signals are the transcription factors LasR and RhlR, which bind target promoters in their signal-bound forms. The lasI and lasR genes are adjacent to each other, as are rhlI and rhlR (9, 10, 35). The primary products of acyl-HSL synthases are the signals to which the cognate signal receptor responds at lowest concentrations, but the specificities of the synthases are not absolute. For example, the primary product of LasI is 3OC12-HSL, but it also produces smaller amounts of other acyl-HSLs (13, 22, 25).
In addition to LasR and RhlR, there is a third orphan (5) or solo (34) LasR-RhlR homolog, QscR, which does not have a cognate acyl-HSL synthase. QscR responds to a variety of acyl-HSLs, including 3OC12-HSL. QscR represses a number of LasR- and RhlR-activated genes and suppresses virulence in a Drosophila infection model (5). There are at least two promoters that serve as targets for QscR binding, the PA1897 and PA5351 promoters (15). Transcription of both genes is activated by QscR (16). PA1897 codes for a polypeptide of unknown function, and PA5351 codes for rubredoxin 1, an electron carrier protein that functions in an alkane hydroxylase system (17, 32, 33, 38). Direct binding to PA1897 and PA5351 was established by electrophoretic mobility shift assays (EMSAs) with purified His-tagged QscR (15). Transcriptomics have shown that there is a large QscR-controlled regulon that overlaps with the LasR- and RhlR-controlled regulons and that many genes controlled by QscR are likely regulated in an indirect fashion (16).
The TraR protein from Agrobacterium tumefaciens is the best-understood QscR homolog at a biochemical level. The structure of TraR bound to the cognate signal N-3-oxooctanoyl-homoserine lactone (3OC8-HSL) and to target DNA has been solved (40, 43). Synthesis of active TraR is thought to require 3OC8-HSL around which the nascent polypeptide must fold (45). 3OC8-HSL is bound very tightly to TraR such that it can be removed from the functional protein only by prolonged dialysis in the presence of 3% Tween 20 (44, 45). The tight ligand binding is consistent with the finding that 3OC8-HSL is fully embedded within the protein (40, 43) and with the idea that polypeptide folding requires 3OC8-HSL as a scaffold. LasR shows similar properties (3, 29).
QscR appears to be different from TraR and LasR in several respects. First, QscR purified as a His-tagged polypeptide was a monomer in solution, whereas TraR and LasR were dimers (24, 29). Because QscR binds the PA1897 promoter cooperatively and because the region in this promoter protected by QscR from DNase I is palindromic and its length is similar to the lengths of regions protected by dimeric QscR homologs (15), we believe that QscR exists as a dimer when bound to target promoters. Second, His-tagged QscR, which was produced by bacteria grown in the presence of 3OC12-HSL and purified using buffers without an acyl-HSL, required additional 3OC12-HSL for DNA binding activity (15). This suggests that QscR does not bind 3OC12-HSL as avidly as TraR and LasR bind their cognate acyl-HSLs. Third, in vivo experiments indicate that the acyl-HSL binding specificity of QscR is broader than that of LasR (15). Fourth, the expression of qscR is considerably lower than the expression of lasR (J.-H. Lee and E. P. Greenberg, unpublished data).
Here we report on the activity of purified native QscR. We show that the purified protein retains 3OC12-HSL, and we provide an explanation for why it nevertheless depends on exogenous addition of 3OC12-HSL for target DNA binding. We also show that purified QscR does have a broad signal binding capability. Our results provide some insight about possible roles for QscR in P. aeruginosa and generally about the biochemistry of members of the large family of acyl-HSL-responsive transcription factors represented by QscR.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
We used Escherichia coli strain DH5α (Invitrogen) for DNA manipulations and recombinant E. coli DH5α for 3OC12-HSL bioassays. Recombinant E. coli DH12S (Invitrogen) and BL21(DE3) pLysS (Novagen) were used for 3OC6-HSL bioassays and for overexpression of QscR, respectively. Routine growth of E. coli was in Luria-Bertani (LB) broth at 37°C with shaking. Growth was monitored as the optical density at 600 nm (OD600). Ampicillin (100 μg/ml), kanamycin (50 μg/ml), gentamicin (15 μg/ml), and chloramphenicol (34 μg/ml) were added to LB broth as appropriate.
For construction of the QscR expression vector, pET3a-qscR, we amplified qscR from P. aeruginosa PAO1 genomic DNA by PCR with the following primers: 5′-AAGCTCATATGCATGATGAGAG-3′ (the NdeI restriction site is underlined) and 5′-AACGGGATCCGGCCATTCGG-3′ (the BamHI restriction site is underlined). The PCR product was digested with NdeI and BamHI, and the resulting DNA fragment was ligated with NdeI-BamHI-digested pET3a (Novagen) to form pET3a-qscR. The size, orientation, and integrity of the construct were confirmed by restriction pattern analysis and DNA sequencing.
Overexpression and purification of native QscR.
For purification of QscR, E. coli BL21(DE3) pLysS carrying pET3a-qscR was grown in LB broth plus ampicillin, chloramphenicol, and 10 μM 3OC12-HSL unless otherwise indicated. The inoculum (1%) was from an overnight culture grown in LB broth containing ampicillin and chloramphenicol. When the cell density reached an OD600 of 0.4 to 0.6, cultures were shifted to 16°C with the addition of 0.5 mM isopropyl-β-d-thiogalactoside (IPTG) to induce qscR expression. After 16 to 18 h at 16°C, the cells were harvested by centrifugation at 10,000 × g for 20 min and stored at −80°C.
All purification steps were performed at 0 to 4°C in a buffer consisting of 25 mM Tris-HCl (pH 7.8), 1 mM EDTA, 1 mM dithiothreitol (DTT), and 10% glycerol (TEDG buffer). Cells from 4 liters of culture were thawed, suspended in 80 ml of TEDG buffer, and lysed by sonication. After insoluble material was removed by ultracentrifugation at 150,000 × g for 30 min, QscR in the cleared cell extract was precipitated by adding solid ammonium sulfate to 40% saturation. After an overnight incubation, the precipitate was pelleted by centrifugation at 14,000 × g for 30 min. The pellet was suspended in 32 ml of buffer and dialyzed against 2 liters of buffer. The dialysate was divided into two equal parts, each of which was subjected to HiTrap Q HP column (GE Healthcare) chromatography as follows. Material was applied to a series of three connected 5-ml columns and washed with 30 ml of TEDG buffer with 0.1 M NaCl. The bound proteins were eluted in a 150-ml linear 0.1 to 0.4 M NaCl gradient. We used sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) to identify QscR-containing fractions, which were pooled and diluted in TEDG buffer (2 parts of partially purified protein solution into 1 part of TEDG buffer). The resulting protein solution was further purified in four batches as follows. Material was applied to a HiPrep 16/10 heparin FF column (GE Healthcare) equilibrated with TEDG buffer. After the column was washed with 40 ml of buffer, QscR was eluted in a 200-ml linear 0 M to 0.6 M NaCl gradient. QscR-containing fractions were identified by SDS-PAGE, pooled, and stored at −80°C at a QscR concentration of 0.44 mg/ml (16 μM). N-3-oxohexanoyl (3OC6)-bound QscR was purified basically the same way as described above. A few modifications were made in the number or size of the columns used and in the number of times that chromatography was performed in each step depending on the amount of QscR obtained in the preceding step. When necessary, QscR was concentrated using Amicon Ultra centrifugal filter devices (nominal molecular weight limit of 5,000).
Gel filtration analysis.
Purified QscR (0.2 ml of 1.4, 7.1, 33, or 160 μM solutions as indicated) was applied to a Superdex 200 10/300 GL column (GE Healthcare) and eluted from the column in TEDG buffer containing 0.15 M NaCl and 5 μM 3OC12-HSL at a flow rate of 0.5 ml/min at 4°C. Protein in the eluate was monitored by UV absorption at 280 nm. The molecular mass of QscR was estimated from the elution profile relative to the following standard proteins (gel filtration calibration kit; GE Healthcare): aldolase (158 kDa), conalbumin (75 kDa), ovalbumin (43 kDa), carbonic anhydrase (29 kDa), and RNase A (13.7 kDa).
EMSAs.
Electrophoretic mobility shift assays were performed as described elsewhere (15) with a few modifications. Each EMSA reaction mixture contained both specific and nonspecific DNA probes. Specific DNA probes were prepared by PCR amplification of the PA1897 promoter region (from nucleotides 2068691 to 2069029 in the P. aeruginosa chromosome). The nonspecific probe (223 bp) was generated by PCR amplification of the mini-CTX-lacZ multiple cloning site (2). The PCR products were end labeled using [γ-32P]ATP and T4 polynucleotide kinase. Binding reaction mixtures contained 10 to 20 pM concentrations of specific and nonspecific DNA in 10 μl of DNA binding buffer (20 mM Tris-HCl [pH 7.5], 1 mM EDTA, 1 mM DTT, 10% glycerol, 50 mM KCl, and 0.1 mg/ml bovine serum albumin). Purified QscR and acyl-HSLs were added as indicated, and the reaction mixtures were incubated at room temperature for 20 min. The reaction mixtures were then loaded on native 5% Tris-glycine-EDTA polyacrylamide gels (29:1 acrylamide to bisacrylamide ratio) and separated at 100 V for 60 min at room temperature using a Mini-Protean tetra cell (Bio-Rad Laboratories). After electrophoresis, the gels were dried and used to expose a storage phosphor screen (GE Healthcare). The image on the screen was visualized by using a Storm 840 phosphorimager with ImageQuant software (GE Healthcare).
For measurements of affinities of QscR binding to acyl-HSLs and DNA, we used QscR purified from 3OC6-HSL-grown E. coli(pET3a-qscR), and the TEDG buffer for protein purification contained 10 μM 3OC6-HSL. Note that we concentrated QscR, which was originally 11 μM, to approximately 250 μM by ultrafiltration as described above. Ultrafiltration removed free 3OC6-HSL prior to EMSAs.
Measurements of acyl-HSLs retained with purified QscR.
We digested 0.5 nmol of QscR in 500 μl of buffer with 5 μg of proteinase K for 1 h at room temperature and then extracted acyl-HSLs with three equal volumes of ethyl acetate acidified with 0.01% glacial acetic acid. The ethyl acetate extracts were combined and evaporated to dryness under a stream of nitrogen gas, and the acyl-HSLs were dissolved in 500 μl of acidified ethyl acetate. To measure 3OC12-HSL, we used a bioassay with E. coli DH5α carrying pJN105L and pSC11 as described elsewhere (15), and synthetic 3OC12-HSL was used to prepare a standard curve. To measure 3OC6-HSL in ethyl acetate extracts, we used a recombinant E. coli system described by Antunes et al. (1). The E. coli reporter contained the LuxR expression vector pHV402 and a luxI promoter-gfp-[LVA] fusion. The reporter was grown in LB medium plus kanamycin and chloramphenicol overnight at 37°C and diluted to an OD600 of 0.1 in fresh LB plus kanamycin and chloramphenicol. Bioassays were in 2-ml plastic tubes to which ethyl acetate extracts had been added. The ethyl acetate evaporated spontaneously or under a stream of nitrogen gas. Five hundred microliters of diluted reporter culture (see above) was added to each tube, and fluorescence was measured after 4 h at 30°C with shaking by using a GENios Pro 96-well plate reader (TECAN). A standard curve with synthetic 3OC6-HSL was used to determine the amount of this molecule extracted from QscR.
Other analytical methods and reagents.
Protein concentrations in crude extracts and partially purified QscR solutions were determined by using a Bio-Rad protein assay (Bio-Rad Laboratories) with bovine serum albumin as the standard. The concentration of purified QscR was estimated from the absorbance at 280 nm in the presence of 5 to 6 M guanidine hydrochloride. We used a QscR molar extinction coefficient of 46,900 M−1 cm−1, which was calculated by the method of Gill and von Hippel (12). SDS-PAGE was performed in a 12% polyacrylamide (acrylamide-to-bisacrylamide ratio of 29:1) slab gel according to Laemmli (14). Gels were stained with Coomassie brilliant blue R-250. Precision Plus Protein standards (Bio-Rad Laboratories) were used as size markers. 3OC12-HSL was custom synthesized by RTI International, N-3-oxodecanoyl-homoserine lactone (3OC10-HSL) and 3OC6-HSL were purchased from Sigma-Aldrich, and N-dodecanoyl-homoserine lactone (C12-HSL) and N-decanoyl-homoserine lactone (C10-HSL) were purchased from Cayman Chemical Company.
RESULTS
Purification of QscR.
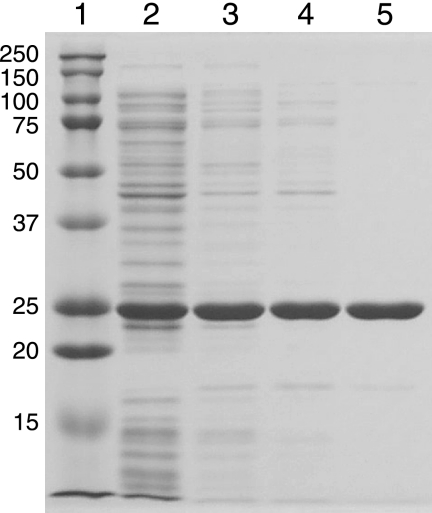
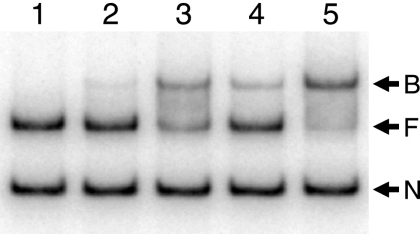
About 30 to 35% of the total QscR produced by E. coli BL21(DE3) pLysS carrying pET3a-qscR grown in the presence of 3OC12-HSL was found in the soluble cell extract with the rest pelleted by centrifugation and presumably in the form of inclusion bodies. As expected from the previous study of His-tagged QscR (15) and from studies of other QscR homologs (6, 29, 37, 45), very little soluble QscR was present in lysates of cells grown without added 3OC12-HSL (data not shown). Starting with the soluble protein fraction from cells grown with 3OC12-HSL, we purified QscR to >98% homogeneity in three steps (Fig. 1) with a recovery rate of about 40%. Activity of the purified material was assessed by using EMSA (Fig. 2). As was the case for purified His-tagged QscR, DNA binding activity of QscR was stimulated by the addition of 3OC12-HSL to the reaction buffer. However, there was a significant amount of DNA binding even in the absence of added 3OC12-HSL. This was particularly evident at the highest QscR concentration tested (Fig. 2). The 3OC12-HSL-independent target DNA-specific binding suggests that either acyl-HSL-free QscR retains some binding activity or that QscR retains some 3OC12-HSL through the purification process.
FIG. 1.
Purification of native QscR. Samples were analyzed by SDS-PAGE. Lane 1, molecular weight markers (the molecular mass of each marker in kilodaltons is indicated to the left of the gel); lane 2, cleared cell extract; lane 3, 40% ammonium sulfate precipitate; lane 4, pooled QscR-containing fractions from HiTrap Q HP column chromatography; lane 5, pooled QscR-containing fractions from HiPrep 16/10 heparin FF column chromatography.
FIG. 2.
An electrophoretic mobility shift assay for binding of purified QscR to a PA1897 promoter fragment. Lane 1, control with no QscR in the reaction mixture; lane 2, 1 nM QscR and no added 3OC12-HSL; lane 3, 5 nM QscR and no added 3OC12-HSL; lane 4, 1 nM QscR plus 5 μM 3OC12-HSL; lane 5, 5 nM QscR plus 5 μM 3OC12-HSL. The positions of nonspecific DNA (N), QscR-free target DNA (F), and QscR-bound DNA (B) are indicated by the arrows to the right of the gel. The promoter fragment is 339 bp, and it includes 301 bp upstream of PA1897.
Measurement of 3OC12-HSL retained with QscR.
To discriminate between whether purified QscR exhibited some 3OC12-HSL-independent DNA binding activity or whether it retained some bound 3OC12-HSL throughout purification, we measured 3OC12-HSL extracted from purified protein. We found about the same amount of 3OC12-HSL as a QscR monomer. Assuming an acyl-HSL-to-monomer stoichiometry of one-to-one, as is true for several QscR homologs (4, 20, 29, 42-44), this indicates that QscR completely retained 3OC12-HSL throughout the purification process. This was surprising, because the addition of 3OC12-HSL to the DNA binding buffer stimulated DNA binding activity significantly (Fig. 2). We believe 3OC12-HSL stimulation of DNA binding can be explained by considering the equilibrium of binding between 3OC12-HSL and QscR. Although it depends on temperature and other experimental conditions, the concentrations of each reaction component at equilibrium are determined by the following equation: [3OC12-HSL][QscR]/[3OC12-HSL-QscR] = Kd, where [3OC12-HSL], [QscR], [3OC12-HSL-QscR], and Kd are unbound 3OC12-HSL concentration, ligand-free QscR concentration, 3OC12-HSL-bound QscR concentration, and the dissociation constant, respectively. On the basis of this equation, one can predict that, at a high concentration, QscR in solution will contain an equimolar amount of (essentially bound) 3OC12-HSL, and if QscR is diluted, the equilibrium will shift toward the ligand-free form with release of 3OC12-HSL into the surrounding buffer. Our purified QscR is at a high concentration, but it is diluted into DNA binding reaction buffer to low concentrations (from 16 μM to 1 or 5 nM). We believe that QscR and 3OC12-HSL dissociate upon dilution in reaction buffer. This explanation is consistent with the fact that dependence of DNA binding on added 3OC12-HSL is reduced at the higher of the two QscR concentrations we used in our EMSA experiments (Fig. 2).
The QscR oligomeric state is concentration dependent.
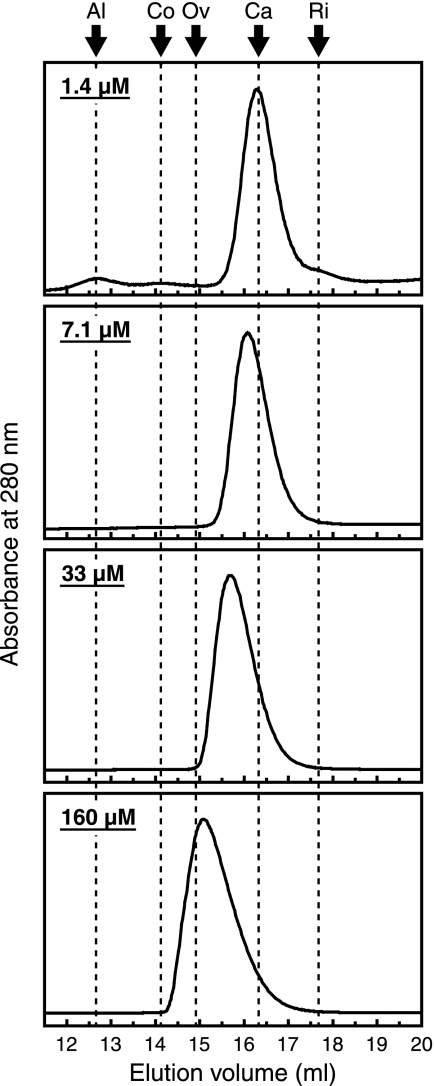
Unlike other well-studied QscR homologs (20, 21, 24, 29), which are homodimers in solution, purified His-tagged QscR was shown to exist as a monomer in solution (15). Because the oligomerization states of proteins often depend on the concentration of each component polypeptide, we assumed that QscR might form dimers at high concentrations. To test the hypothesis, we estimated the molecular mass of QscR by gel filtration at several different protein concentrations (Fig. 3). At the lowest concentration tested (1.4 μM), QscR was eluted in the position of a monomer (26 kDa; the predicted value is 27.3 kDa). However, as we increased the protein concentration, the elution peak shifted to positions corresponding to higher molecular masses (29, 35, and 47 kDa at 7.1, 33, and 160 μM, respectively). These results are consistent with the hypothesis that QscR dimerizes in a concentration-dependent fashion. We believe that the formation and dissociation of dimers from and into monomers is rapid compared to the time scale of the analysis. If so, existing monomers and dimers should continuously interchange during column chromatography except at extremely high or extremely low concentrations. It follows that the position of an elution peak does not simply indicate the size of the molecule but rather it indicates how long those molecules have existed as a monomer (or as a dimer) on average during the analysis. This can explain why we observed a single peak, and not two separate peaks, at any QscR concentration tested (note that observation of two separate peaks representing monomers and dimers requires the existence of two distinct populations of QscR that always exist as monomers and dimers, respectively, throughout the column chromatography).
FIG. 3.
Relationship between QscR concentration and its oligomeric state. Gel filtration chromatography was performed with purified QscR loaded at 1.4 μM, 7.1 μM, 33 μM, or 160 μM. The buffer contained 5 μM 3OC12-HSL. The elution of protein was monitored by UV absorption at 280 nm. The arrows indicate peaks of the following standards: aldolase (Al; 158 kDa), conalbumin (Co; 75 kDa), ovalbumin (Ov; 43 kDa), carbonic anhydrase (Ca; 29 kDa), and RNase A (Ri; 13.7 kDa).
Expression of soluble QscR in E. coli can be stimulated by acyl-HSLs other than 3OC12-HSL.
Previous work showed that QscR in recombinant E. coli activated transcription of a reporter in response to several acyl-HSLs, including N-octanoyl (C8)-, C10-, and C12-HSLs, but QscR did not respond to other acyl-HSLs, such as 3OC6-HSL at least at the concentrations tested (15). We asked whether growth of E. coli(pET3a-qscR) in the presence of acyl-HSLs other than 3OC12-HSL would stimulate accumulation of soluble QscR. Not surprisingly, the acyl-HSLs that served as coactivators of transcription together with QscR (15) also facilitated production of soluble QscR, but so did 3OC6-HSL (data not shown), which did not serve as a coactivator (15).
Because QscR was not active in E. coli grown in the presence of 3OC6-HSL (15), we thought purified preparations of this material would be useful to study interactions between QscR and various acyl-HSLs. When we subjected QscR from recombinant E. coli grown in the presence of 3OC6-HSL to the purification procedure described above, the product was about 95% pure. However, we recovered only about 2% of the starting soluble QscR. The bulk of the protein was lost as insoluble aggregated material especially during dialysis and right after elution from the chromatography columns (data not shown). Perhaps 3OC6-HSL-bound QscR is less stable than 3OC12-HSL-bound QscR, or perhaps QscR does not bind 3OC6-HSL as tightly as it binds 3OC12-HSL, and at the relatively high protein concentrations during purification, acyl-HSL-free QscR aggregates. Thus, we measured 3OC6-HSL retained in a purified QscR solution after removing aggregates and found 1.6 mol of 3OC6-HSL per mol of soluble QscR monomer. We also measured 3OC6-HSL in the aggregated insoluble QscR and found <0.1 3OC6-HSL per QscR monomer. The data support the conclusion that QscR does not bind 3OC6-HSL as tightly as it binds 3OC12-HSL and that ligand-free QscR is unstable and forms insoluble aggregates in TEDG buffer. The fact that the 3OC6-HSL is a little in excess of the predicted 1:1 ratio with QscR might be the result of several factors. The 3OC6-HSL released by the aggregated QscR will contribute to the final concentration in solution. The amount of aggregated QscR in the sample was 25% of the soluble QscR. There is also some error inherent in values based solely on bioassays.
Activity of 3OC6-HSL-bound QscR.
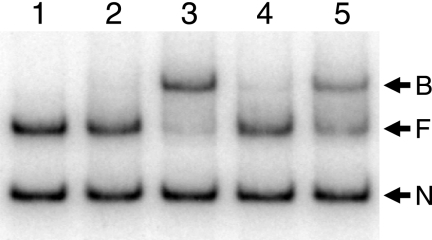
Several different acyl-HSLs could function together with QscR to activate expression of the P. aeruginosa PA1897 promoter in recombinant E. coli, but 3OC6-HSL did not serve as a coactivator (15). Nevertheless, we show that 3OC6-HSL can bind to QscR, and it can serve to keep QscR from forming insoluble aggregates. These disparate results raise the question of whether 3OC6-HSL can serve as an activator of QscR binding to target DNA in vitro. Thus, we performed an EMSA with purified 3OC6-HSL-bound QscR (Fig. 4). When we diluted 3OC6-HSL-bound QscR into DNA binding buffer containing 5 μM 3OC12-HSL, the target DNA was completely shifted to the QscR-bound state. When we diluted the QscR preparation into buffer containing 5 μM 3OC6-HSL, very little of the target DNA migrated in the QscR-bound position. However, when we included 500 μM 3OC6-HSL in the DNA binding buffer, about half of the target DNA appeared to be bound to QscR. These results indicate that compared with 3OC12-HSL, QscR has a very low affinity for 3OC6-HSL, but that sufficiently high concentrations of 3OC6-HSL are capable of affecting DNA binding by QscR. In fact, the highest concentration of acyl-HSLs tested in recombinant E. coli experiments published previously (15) was 200 nM. This is well below the 3OC6-HSL concentrations required for QscR binding to target DNA in our EMSA. We tested whether higher concentrations of 3OC6-HSL can facilitate activation of the PA1897 promoter by QscR in recombinant E. coli by using the reporter system described previously (15) except that we used very high concentrations of 3OC6-HSL (up to 600 μM). There was a slight activation of transcription at 1 μM 3OC6-HSL, and activation increased such that the response to 600 μM 3OC6-HSL was equivalent to the response to 200 nM 3OC12-HSL (data not shown).
FIG. 4.
An electrophoretic mobility shift assay with QscR purified from E. coli grown with 3OC6-HSL. Lane 1, control with no QscR in the reaction mixture; lane 2, 5 nM QscR and no added acyl-HSL; lane 3, 5 nM QscR plus 5 μM 3OC12-HSL; lane 4, 5 nM QscR plus 5 μM 3OC6-HSL; lane 5, 5 nM QscR plus 500 μM 3OC6-HSL. The positions of nonspecific DNA (N), QscR-free target DNA (F), and QscR-bound DNA (B) are indicated by the arrows to the right of the gel.
Affinity of QscR for acyl-HSLs and DNA.
Our evidence indicates that when we dilute 3OC6-HSL-bound QscR into buffer for DNA binding experiments, the 3OC6-HSL and QscR dissociate and the acyl-HSL-free QscR does not bind to DNA. Furthermore, if present in the reaction mixture, 3OC12-HSL can bind to the ligand-free QscR before it aggregates and triggers target DNA binding (Fig. 4). Thus, we can use purified 3OC6-HSL-QscR in experiments to estimate the affinity of QscR bound to different acyl-HSLs for target DNA and the affinity of QscR for different acyl-HSLs.
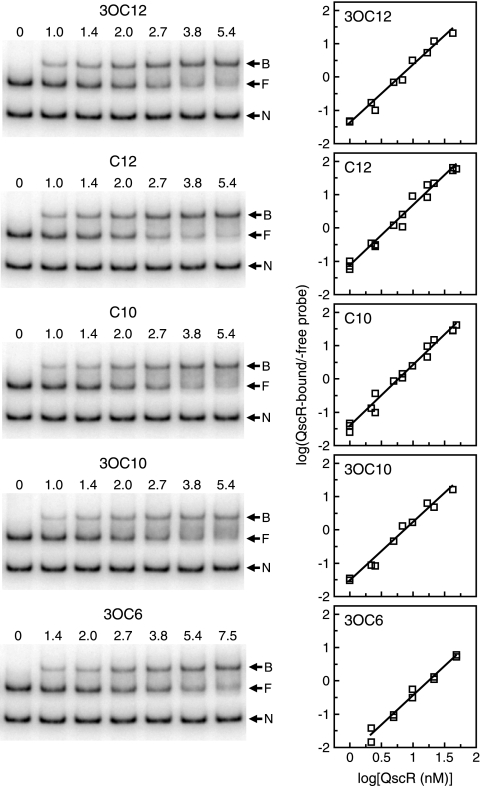
The apparent affinities of different acyl-HSL-bound QscR preparations for target DNA were measured by EMSAs with different concentrations of QscR in the presence of fixed concentrations of DNA and acyl-HSLs (5 μM for 3OC12-, C12-, C10-, and 3OC10-HSLs and 5 mM for 3OC6-HSL) (Fig. 5). Prior to this experiment, we performed a series of preliminary EMSAs and established that these concentrations of acyl-HSLs are sufficient to saturate QscR in the DNA binding reaction mixtures. The concentrations of QscR at which half of the added specific probe is shifted (K0.5s) were calculated to be 2.2 nM, 1.9 nM, 2.2 nM, 2.4 nM, and 3.5 nM in the presence of 3OC12-HSL, C12-HSL, C10-HSL, 3OC10-HSL, and 3OC6-HSL, respectively. The Hill coefficient was about 1.8, regardless of which acyl-HSL was included in the DNA binding buffer. The value indicates that binding of QscR to target DNA is cooperative.
FIG. 5.
Estimation of the binding affinities of different acyl-HSL-bound QscR preparations for target DNA. EMSAs were performed by using QscR purified from 3OC6-HSL-grown E. coli with the following acyl-HSLs added to the DNA binding buffer: 3OC12-, C12-, C10-, and 3OC10-HSLs (5 μM) or 3OC6-HSL (5 mM). Assays were performed two or three times for each acyl-HSL. (Left) A set of representative EMSA results. The numbers above the lanes are the QscR concentrations (in nanomolar concentrations). The positions of nonspecific DNA (N), QscR-free target DNA (F), and QscR-bound DNA (B) are indicated by the arrows to the right of the gels. (Right) Hill plots generated from the EMSA data. Two or three independent data sets are plotted together for each acyl-HSL. The amount of QscR-bound target DNA was calculated as the intensity of the free target DNA band in the control lane (no QscR) minus the intensity of the free DNA band in the presence of the indicated amount of QscR.
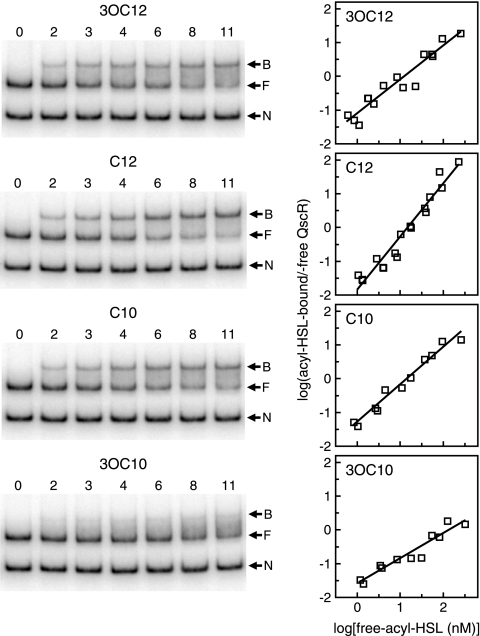
The apparent affinities of acyl-HSLs for QscR were measured by changing the acyl-HSL concentration in the presence of fixed amounts of QscR and DNA (Fig. 6). K0.5 values, the concentrations of each acyl-HSL required for half saturation of QscR were calculated to be 3.1 nM, 3.3 nM, 3.3 nM, and 8.7 nM for 3OC12-HSL, C12-HSL, C10-HSL, and 3OC10-HSL, respectively. These results indicate that QscR has about the same affinity for 3OC12-, C12-, and C10-HSLs, and the affinity for 3OC10-HSL is lower than for the other acyl-HSLs. The Hill coefficient with C12-HSL was unexpectedly high (1.6; the expected value is 1.0). We do not know whether this was caused by some technical error or whether it has physiological meaning.
FIG. 6.
Estimation of the binding affinities of QscR for different acyl-HSLs. EMSAs were performed with a fixed amount of 3OC6-HSL-QscR (5 nM). The different acyl-HSLs were added at different concentrations as indicated. Assays were performed two or three times for each acyl-HSL. (Left) Representative EMSA results. The numbers above the lanes are acyl-HSL concentrations (in nanomolar concentrations). The negative control (lanes 0) contained no QscR. The positions of nonspecific DNA (N), QscR-free target DNA (F), and QscR-bound DNA (B) are indicated by the arrows to the right of the gels. (Right) Hill plots derived from two or three independent data sets for each acyl-HSL. The amount of QscR-bound target DNA was calculated as the intensity of the free target DNA in the control lane (no QscR added) minus free target DNA in the lane of interest. The amount of acyl-HSL-bound QscR in each reaction mixture was then estimated from the amount of band shifting by using the fitted lines of the Hill plots in Fig. 5As standard curves. The numbers indicated on the x axis are logarithms of QscR-free acyl-HSL concentrations, which were calculated as the amount of total acyl-HSL added in each reaction mixture minus the estimated amount of signal-bound QscR.
DISCUSSION
Our analysis of purified native QscR provides answers to several questions about this orphan quorum-sensing signal receptor, and we also postulate a new general model for QscR homologs. The first question to be answered was why does purified QscR depend on an added acyl-HSL for activity in EMSAs? This is unlike the well-studied TraR (44). The obvious assumption is that QscR binds 3OC12-HSL less avidly than TraR binds 3OC8-HSL, and during purification, 3OC12-HSL is lost (15). To test this hypothesis, we measured 3OC12-HSL retained with QscR during purification, and in fact, 3OC12-HSL is not lost from the protein. Thus, the obvious assumption is not correct. Instead, our results (Fig. 2) support the view that when 3OC12-HSL-bound QscR is diluted into the buffer used for the DNA binding reaction, the concentrations of 3OC12-HSL and protein drop to a level at which the new equilibrium leaves QscR primarily in the acyl-HSL-free state. This predicts that there should be a correlation between protein dilution and dependence of DNA binding on acyl-HSL addition, just as we observed (Fig. 2). In fact, on the basis of our calculated K0.5 of 3.1 nM for 3OC12-HSL binding to QscR, when purified QscR containing the same amount of 3OC12-HSL is diluted to 1 nM, about 20% of the total protein should retain 3OC12-HSL. When diluted to 5 nM, nearly half of the QscR should retain 3OC12-HSL.
A similar dependence on 3OC12-HSL was observed for purified LasR. As assessed by EMSAs, there was a 3-fold difference in the apparent dissociation constant of LasR and a target DNA in the absence versus presence of 5 μM exogenously added 3OC12-HSL (29). Because we previously believed that 3OC12-HSL binding by LasR was virtually irreversible, the 3OC12-HSL dependence of DNA binding affinity was difficult to interpret. We now realize that the data are similar to those obtained with native QscR and that they can be interpreted the same way as described above for QscR. If this is so, it leads to the conclusion that 3OC12-HSL binding to QscR and LasR is fundamentally similar except that LasR has a greater affinity for 3OC12-HSL than does QscR (note that LasR was diluted to much higher extents in the published EMSA reactions than QscR in our experiments, and therefore a much higher affinity was required for LasR to retain 3OC12-HSL). We believe that our findings have important implications for quorum-sensing control of gene expression in P. aeruginosa. For example, if LasR signal binding is reversible, then one would predict that gene activation by 3OC12-HSL-LasR would not persist when the environmental signal concentrations decrease below a certain level. This simple ability to quickly stop transcription of quorum-sensing-controlled genes could be important for cells especially when they move from a high-population-density environment to a low-population-density environment.
Our investigations of QscR obtained from recombinant E. coli grown in the presence of 3OC6-HSL show that active purified protein retains 3OC6-HSL but that the majority of protein is lost to aggregation during purification. We found very little 3OC6-HSL associated with the aggregated material. We believe that ligand-free QscR is rather unstable and that acyl-HSL binding stabilizes the protein. This is consistent with the fact that some acyl-HSL is required to achieve significant levels of soluble active QscR (and other QscR homologs) during bacterial growth. It is also consistent with in vivo results showing that 3OC8-HSL-free Agrobacterium TraR is targeted for proteolysis in bacteria (44, 45). We speculate that most QscR homologs are unstable in their acyl-HSL-free state. If this is true for LasR, it can explain why functional ligand-free LasR cannot be obtained even after prolonged dialysis against ligand-free buffer (29). We believe that differences in the behavior of purified QscR homologs can be explained by considering the equilibrium of the protein-acyl-HSL binding reactions and stability of acyl-HSL-free proteins.
The previous report indicating that His-tagged QscR exists as a monomer in solution is in contrast to findings with several other QscR homologs (15). The oligomeric state of TraR has been shown to depend on protein concentration. Gel filtration experiments indicated that TraR existed as a dimer at a concentration of 300 nM but that a subpopulation existed in a monomeric state at concentrations of ≤75 nM. A concentration of 75 nM is equivalent to about 10 TraR monomers per cell. Thus, it was suggested that functional TraR exists in the dimeric form in vivo (45); however, one must interpret the data with caution, as it is clear that cytoplasmic conditions are distinct from the in vitro conditions. Our gel filtration chromatography of native QscR showed that it mainly exists as a monomer at concentrations as low as 1.4 μM but forms a dimer at higher concentrations (there is at least some dimer formation at around 7 μM; Fig. 3). We do not know the concentration of QscR in P. aeruginosa, but we reason it is quite low for the following reasons. Although we can detect LasR and RhlR by Western immunoblotting of P. aeruginosa cell extracts (26), we have not been able to detect QscR by this method (data not shown). Furthermore, qscR transcript levels, even at their maximum in late logarithmic and early stationary phase are much lower than either lasR or rhlR transcript levels (Lee and Greenberg, unpublished). Although one should be cautious in comparing in vivo and in vitro conditions, it is reasonable to believe that, in the absence of sufficient signal, QscR may exist primarily as a monomer in P. aeruginosa grown under standard laboratory conditions.
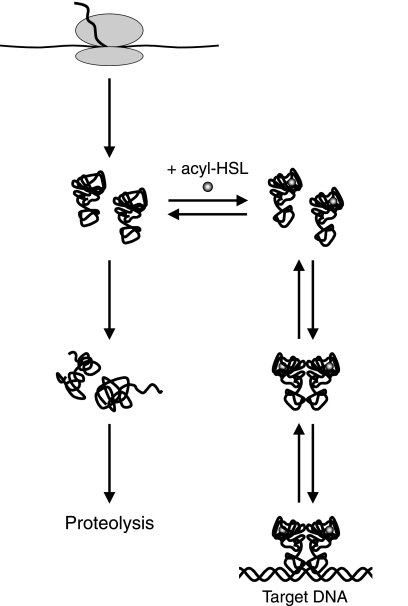
We suggest a model for QscR interactions where functional monomers can be synthesized by P. aeruginosa even in the absence of an acyl-HSL. This is a departure from the current view that for most QscR homologs, folding of the nascent polypeptide into a functional conformation requires folding around its cognate acyl-HSL (27, 45). According to the model, nascent QscR folds into a functional state without an acyl-HSL, but it is unstable and rapidly refolds into nonfunctional conformations, which are prone to aggregation and perhaps targeted for proteolysis. In the presence of an appropriate acyl-HSL, the functional folded protein binds the ligand and becomes more stable, therefore accumulating in cells to higher levels. QscR exists as a monomer at low concentrations, but as the monomer concentration increases, it starts to form dimers capable of high-affinity binding to target promoters. The formation of dimers may also increase the stability of QscR, as in the case of TraR (23). Although the model is speculative, we believe it may apply to the majority of QscR homologs studied thus far. Exceptions are EsaR from Pantoea stewartii and other members of the EsaR subfamily. Unlike other studied acyl-HSL-responsive transcription factors, these proteins function primarily as repressors in the absence of an acyl-HSL, and DNA binding is antagonized by the cognate acyl-HSLs (4, 8, 19, 20, 36). Evidence indicates that EsaR forms signal-free dimers (20), and signal may shift the equilibrium between dimers and monomers toward a monomeric state. We hypothesize that most QscR homologs (with EsaR family members as exceptions) are relatively unstable in the ligand-free state, but some may be more or less stable than others. In fact, EsaR family members may exhibit similar behavior except that we presume they are much more stable in the signal-free state than other QscR homologs. Our model is captured in the diagram shown in Fig. 7. Although we do not include a step that involves a required folding of nascent polypeptide around its acyl-HSL ligand, we cannot absolutely exclude such a possibility. However, we do not believe that the existing data necessitate such a step.
FIG. 7.
General model for acyl-HSL receptor states in vivo. Nascent polypeptides fold into a functional but relatively unstable state. In the absence of an appropriate acyl-HSL, the polypeptides refold into nonfunctional conformations, which aggregate or are targeted for proteolysis. The folded polypeptides exist as monomers at sufficiently low concentrations, but as the acyl-HSL-bound monomers accumulate, they form homodimers capable of high-affinity binding to target promoters. The concentration of monomers required for dimer formation varies among different QscR homologs.
We find it particularly interesting that QscR seems to have very little selectivity for the different long-chain acyl-HSLs we tested (>C6 acyl groups) (Fig. 6). This extends previous findings that in recombinant E. coli, C12-HSL, C10-HSL, and 3OC10-HSL were equivalent to or slightly better than 3OC12-HSL as QscR ligands for activation of PA1897 transcription (15). We also found that 3OC6-HSL can stimulate QscR to bind target DNA but only at very high and likely physiologically irrelevant concentrations relative to the long-acyl-chain HSLs. The broad signal specificity suggests that QscR, an orphan quorum-sensing regulator, might function as a receptor for signals produced by other bacteria in natural settings with mixed-species microbiota. QscR might activate gene expression when there is a sufficient population of P. aeruginosa generating 3OC12-HSL or when there is a low population of P. aeruginosa together with a high population of other Proteobacteria that produce long-chain fatty acyl-HSLs. The precedent for this idea can be drawn from studies of Salmonella enterica serovar Typhimurium and Escherichia coli, both of which contain a QscR homolog, SdiA. Neither of these bacteria produce any acyl-HSLs, but SdiA responds to acyl-HSLs produced by other bacterial species in specific ecological habitats (7, 11, 18, 30, 39).
Acknowledgments
This work was supported by U.S. Public Health Service grant GM-59026. K.-I.O. received a Postdoctoral Fellowship for Research Abroad from the Japan Society for the Promotion of Science.
Footnotes
Published ahead of print on 19 November 2010.
REFERENCES
- 1.Antunes, L. C., R. B. Ferreira, C. P. Lostroh, and E. P. Greenberg. 2008. A mutational analysis defines Vibrio fischeri LuxR binding sites. J. Bacteriol. 190:4392-4397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Becher, A., and H. P. Schweizer. 2000. Integration-proficient Pseudomonas aeruginosa vectors for isolation of single-copy chromosomal lacZ and lux gene fusions. Biotechniques 29:948-952. [DOI] [PubMed] [Google Scholar]
- 3.Bottomley, M. J., E. Muraglia, R. Bazzo, and A. Carfi. 2007. Molecular insights into quorum sensing in the human pathogen Pseudomonas aeruginosa from the structure of the virulence regulator LasR bound to its autoinducer. J. Biol. Chem. 282:13592-13600. [DOI] [PubMed] [Google Scholar]
- 4.Castang, S., S. Reverchon, P. Gouet, and W. Nasser. 2006. Direct evidence for the modulation of the activity of the Erwinia chrysanthemi quorum-sensing regulator ExpR by acylhomoserine lactone pheromone. J. Biol. Chem. 281:29972-29987. [DOI] [PubMed] [Google Scholar]
- 5.Chugani, S. A., et al. 2001. QscR, a modulator of quorum-sensing signal synthesis and virulence in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 98:2752-2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duerkop, B. A., R. L. Ulrich, and E. P. Greenberg. 2007. Octanoyl-homoserine lactone is the cognate signal for Burkholderia mallei BmaR1-BmaI1 quorum sensing. J. Bacteriol. 189:5034-5040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dyszel, J. L., et al. 2010. Salmonella enterica serovar Typhimurium can detect acyl homoserine lactone production by Yersinia enterocolitica in mice. J. Bacteriol. 192:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fineran, P. C., H. Slater, L. Everson, K. Hughes, and G. P. Salmond. 2005. Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol. Microbiol. 56:1495-1517. [DOI] [PubMed] [Google Scholar]
- 9.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 10.Fuqua, C., M. R. Parsek, and E. P. Greenberg. 2001. Regulation of gene expression by cell-to-cell communication: acyl-homoserine lactone quorum sensing. Annu. Rev. Genet. 35:439-468. [DOI] [PubMed] [Google Scholar]
- 11.Ghosh, D., et al. 2009. Acyl-homoserine lactones can induce virus production in lysogenic bacteria: an alternative paradigm for prophage induction. Appl. Environ. Microbiol. 75:7142-7152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 13.Gould, T. A., J. Herman, J. Krank, R. C. Murphy, and M. E. Churchill. 2006. Specificity of acyl-homoserine lactone synthases examined by mass spectrometry. J. Bacteriol. 188:773-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 15.Lee, J. H., Y. Lequette, and E. P. Greenberg. 2006. Activity of purified QscR, a Pseudomonas aeruginosa orphan quorum-sensing transcription factor. Mol. Microbiol. 59:602-609. [DOI] [PubMed] [Google Scholar]
- 16.Lequette, Y., J. H. Lee, F. Ledgham, A. Lazdunski, and E. P. Greenberg. 2006. A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit. J. Bacteriol. 188:3365-3370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marin, M. M., L. Yuste, and F. Rojo. 2003. Differential expression of the components of the two alkane hydroxylases from Pseudomonas aeruginosa. J. Bacteriol. 185:3232-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Michael, B., J. N. Smith, S. Swift, F. Heffron, and B. M. Ahmer. 2001. SdiA of Salmonella enterica is a LuxR homolog that detects mixed microbial communities. J. Bacteriol. 183:5733-5742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Minogue, T. D., A. L. Carlier, M. D. Koutsoudis, and S. B. von Bodman. 2005. The cell density-dependent expression of stewartan exopolysaccharide in Pantoea stewartii ssp. stewartii is a function of EsaR-mediated repression of the rcsA gene. Mol. Microbiol. 56:189-203. [DOI] [PubMed] [Google Scholar]
- 20.Minogue, T. D., M. Wehland-von Trebra, F. Bernhard, and S. B. von Bodman. 2002. The autoregulatory role of EsaR, a quorum-sensing regulator in Pantoea stewartii ssp. stewartii: evidence for a repressor function. Mol. Microbiol. 44:1625-1635. [DOI] [PubMed] [Google Scholar]
- 21.Nasser, W., M. L. Bouillant, G. Salmond, and S. Reverchon. 1998. Characterization of the Erwinia chrysanthemi expI-expR locus directing the synthesis of two N-acyl-homoserine lactone signal molecules. Mol. Microbiol. 29:1391-1405. [DOI] [PubMed] [Google Scholar]
- 22.Pearson, J. P., et al. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. U. S. A. 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pinto, U. M., and S. C. Winans. 2009. Dimerization of the quorum-sensing transcription factor TraR enhances resistance to cytoplasmic proteolysis. Mol. Microbiol. 73:32-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Qin, Y., et al. 2000. Quorum-sensing signal binding results in dimerization of TraR and its release from membranes into the cytoplasm. EMBO J. 19:5212-5221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaefer, A. L., D. L. Val, B. L. Hanzelka, J. E. Cronan, Jr., and E. P. Greenberg. 1996. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc. Natl. Acad. Sci. U. S. A. 93:9505-9509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schuster, M., and E. P. Greenberg. 2007. Early activation of quorum sensing in Pseudomonas aeruginosa reveals the architecture of a complex regulon. BMC Genomics 8:287-297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuster, M., and E. P. Greenberg. 2008. LuxR-type proteins in Pseudomonas aeruginosa quorum sensing: distinct mechanisms with global implications, p. 133-144. In S. C. Winans and B. L. Bassler (ed.), Chemical communication among bacteria. ASM Press, Washington, DC.
- 28.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schuster, M., M. L. Urbanowski, and E. P. Greenberg. 2004. Promoter specificity in Pseudomonas aeruginosa quorum sensing revealed by DNA binding of purified LasR. Proc. Natl. Acad. Sci. U. S. A. 101:15833-15839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Smith, J. N., et al. 2008. SdiA, an N-acylhomoserine lactone receptor, becomes active during the transit of Salmonella enterica through the gastrointestinal tract of turtles. PLoS One 3:e2826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith, R. S., and B. H. Iglewski. 2003. Pseudomonas aeruginosa quorum sensing as a potential antimicrobial target. J. Clin. Invest. 112:1460-1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Smits, T. H., S. B. Balada, B. Witholt, and J. B. van Beilen. 2002. Functional analysis of alkane hydroxylases from gram-negative and gram-positive bacteria. J. Bacteriol. 184:1733-1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smits, T. H., B. Witholt, and J. B. van Beilen. 2003. Functional characterization of genes involved in alkane oxidation by Pseudomonas aeruginosa. Antonie Van Leeuwenhoek 84:193-200. [DOI] [PubMed] [Google Scholar]
- 34.Subramoni, S., and V. Venturi. 2009. LuxR-family ‘solos’: bachelor sensors/regulators of signalling molecules. Microbiology 155:1377-1385. [DOI] [PubMed] [Google Scholar]
- 35.Swift, S., et al. 2001. Quorum sensing as a population-density-dependent determinant of bacterial physiology. Adv. Microb. Physiol. 45:199-270. [DOI] [PubMed] [Google Scholar]
- 36.Tsai, C. S., and S. C. Winans. 2010. LuxR-type quorum-sensing regulators that are detached from common scents. Mol. Microbiol. 77:1072-1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Urbanowski, M. L., C. P. Lostroh, and E. P. Greenberg. 2004. Reversible acyl-homoserine lactone binding to purified Vibrio fischeri LuxR protein. J. Bacteriol. 186:631-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Beilen, J. B., et al. 2002. Rubredoxins involved in alkane oxidation. J. Bacteriol. 184:1722-1732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Van Houdt, R., A. Aertsen, P. Moons, K. Vanoirbeek, and C. W. Michiels. 2006. N-Acyl-l-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol. Lett. 256:83-89. [DOI] [PubMed] [Google Scholar]
- 40.Vannini, A., et al. 2002. The crystal structure of the quorum sensing protein TraR bound to its autoinducer and target DNA. EMBO J. 21:4393-4401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Welch, M., et al. 2000. N-Acyl homoserine lactone binding to the CarR receptor determines quorum-sensing specificity in Erwinia. EMBO J. 19:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang, R. G., et al. 2002. Structure of a bacterial quorum-sensing transcription factor complexed with pheromone and DNA. Nature 417:971-974. [DOI] [PubMed] [Google Scholar]
- 44.Zhu, J., and S. C. Winans. 1999. Autoinducer binding by the quorum-sensing regulator TraR increases affinity for target promoters in vitro and decreases TraR turnover rates in whole cells. Proc. Natl. Acad. Sci. U. S. A. 96:4832-4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhu, J., and S. C. Winans. 2001. The quorum-sensing transcriptional regulator TraR requires its cognate signaling ligand for protein folding, protease resistance, and dimerization. Proc. Natl. Acad. Sci. U. S. A. 98:1507-1512. [DOI] [PMC free article] [PubMed] [Google Scholar]