Abstract
Protein phosphatase 2A (PP2A), a central Tor pathway phosphatase consisting of a catalytic subunit (Pph21 or Pph22), a scaffold subunit (Tpd3), and one of two regulatory subunits (Cdc55 or Rts1), has been repeatedly shown to play important roles in cytoplasmically localized signal transduction activities. In contrast, its involvement in intranuclear control of mRNA production has heretofore not been reported. Here, we demonstrate for the first time that binding of the nitrogen catabolite repression-responsive GATA transcription activators (Gln3 and Gat1) to the DAL5 promoter and DAL5 expression require Pph21/22-Tpd3-Cdc55/Rts1 in rapamycin-treated glutamine-grown cells. This conclusion is supported by the following observations. (i) Rapamycin-induced DAL5 expression along with Gln3 and Gat1 binding to the DAL5 promoter fails to occur in pph21Δ pph22Δ, tpd3Δ, and cdc55Δ rts1Δ mutants. (ii) The Pph21/22 requirement persists even when Gat1 and Gln3 are rendered constitutively nuclear, thus dissociating the intranuclear requirement of PP2A from its partial requirement for rapamycin-induced nuclear Gat1 localization. (iii) Pph21-Myc13 (Ppp21 tagged at the C terminus with 13 copies of the Myc epitope) weakly associates with the DAL5 promoter in a Gat1-dependent manner, whereas a similar Pph22-Myc13 association requires both Gln3 and Gat1. Finally, we demonstrate that a pph21Δ pph22Δ double mutant is epistatic to ure2Δ for nuclear Gat1 localization in untreated glutamine-grown cells, whereas for Gln3, just the opposite occurs: i.e., ure2Δ is epistatic to pph21Δ pph22Δ. This final observation adds additional support to our previous conclusion that the Gln3 and Gat1 GATA factor localizations are predominantly controlled by different regulatory pathways.
The centrality of mTor and its rapamycin family inhibitors in the treatment of diseases associated with cell proliferation has been a powerful impetus to investigate the mechanisms through which cellular processes are directly and/or indirectly regulated by the Tor signal transduction cascades (7, 17, 21, 29, 30, 38, 42). In the yeast Saccharomyces cerevisiae, there are two Tor protein kinases, Tor1 and Tor2, whose inhibition alters the expression level of over 500 genes and an ever growing list of cellular processes (4, 25, 43, 55). Through the work of many laboratories, relationships between the Tor cascade and its downstream targets have been identified and studied in detail. Among the best-studied Tor-responsive transcription factors are the GATA factors, Gln3 and Gat1/Nil1, which activate nitrogen catabolite repression (NCR)-sensitive gene expression (54).
Gln3 and Gat1 are largely quiescent in cells growing in a nitrogen-rich environment (e.g., glutamine) (8, 9, 28, 37). Available data are consistent with the view that their ability to mediate transcription is inhibited by their Ure2-dependent sequestration in the cytoplasm (1-3, 6, 15, 34). As their environmental nitrogen supply dwindles, or when only poor nitrogen sources (e.g., proline) are available, Gln3 and Gat1 increasingly accumulate in the nucleus, where they bind to DNA and activate transcription of NCR-sensitive genes, required to transport and degrade poor nitrogen sources (1, 2, 6, 11, 16, 24).
Regulatory relationships between Tor and the GATA factors are supported by several important correlations: treatment of cells growing in a nitrogen-rich environment with the Tor inhibitor rapamycin elicits Gln3 dephosphorylation, nuclear Gln3 and Gat1 localization, elevated Gln3 and Gat1 binding to NCR-sensitive promoters, and increased, GATA factor-dependent, NCR-sensitive gene expression (1, 2, 4-6, 18, 23-25, 41, 43; see the review by Zaman et al. [54] for a comprehensive list of contributions).
Initially, control by the Tor pathway was thought to explain how the signal for nitrogen sufficiency was sensed and a GATA transcription factor response to it was implemented (1). Multiple years of subsequent work, however, have substantially altered and expanded our view of how the cell responds to its nitrogen supply and the role played by the Tor pathway in this response. Overall, nitrogen supply-dependent GATA factor regulation is significantly more complex than first envisioned (12, 14, 19, 20, 31-33, 40, 52, 53). A pivotal discovery for the present work was our recent demonstration that two distinctly different regulatory pathways implement the cell's response to nitrogen availability by controlling GATA factor localization (24, 35, 47, 49). Gln3 localization predominantly responds to intracellular nitrogen levels, as evidenced by its greater sensitivity to NCR or by treatment of cells with the glutamine synthetase inhibitor methionine sulfoximine (Msx) and lower sensitivity to the effects of rapamycin treatment relative to parallel responses by Gat1 (47). Conversely, Gat1 localization predominantly responds to rapamycin-induced processes, as evidenced by its relative insensitivity to NCR, insensitivity to Msx treatment, and strong sensitivity to rapamycin (47).
Not only does GATA factor localization respond differently to the two regulatory pathways, but it exhibits different Tor pathway phosphatase requirements (10, 26, 27, 46-48). The phosphatases most often associated with Tor signaling are Sit4 and protein phosphatase 2A (PP2A) (19, 32, 33, 54). Sit4 is a type 2A-like phosphatase catalytic subunit which is active in a complex with the phosphoprotein, Tap42 (1, 19, 33, 54). PP2A phosphatase, on the other hand, is composed of a catalytic subunit, Pph21 or Pph22 complexed with a scaffold subunit, Tpd3, and one of two regulatory/specificity subunits, Cdc55 or Rts1 (32, 33, 54). Pph21 or Pph22 can also form complexes with Tap42 (33, 50, 52). Rapamycin-induced nuclear Gln3 localization in glutamine-grown cells exhibits absolute requirements for Sit4, Pph21/22, Tpd3, Cdc55, and Rts1 (10, 46-48). Gat1 localization, under this condition, exhibits only limited requirements for these proteins (46-48). On the other hand, nuclear Gat1 localization in response to growth on poor nitrogen sources, (i.e., NCR) exhibits absolute requirements for Sit4, Pph21/22, and Tpd3 and limited requirements for Cdc55 and Rts1 (24, 46-48). Nuclear Gln3 localization under these conditions requires only Sit4 in the TB123 genetic background (22, 46-48). Additionally, in glutamine-grown cells, Gat1 subcellular localization is much less sensitive to a URE2 deletion than that of Gln3 (24).
GATA factor- and Tor pathway component-specific differences have also been demonstrated to occur within the nucleus. For example, in the TB123 genetic background, rapamycin-induced Gln3 binding to its DAL5 promoter (PDAL5) target in glutamine-grown cells exhibits a strong requirement for Gat1, whereas Gat1 binding is largely Gln3 independent (24). With respect to the Tor pathway phosphatase Sit4, rapamycin-induced Gln3 binding to the DAL5 promoter is Sit4 dependent in ure2Δ cells, where Gln3 is constitutively nuclear (24). Analogous rapamycin-induced Gat1 binding to the DAL5 promoter exhibits only a partial requirement for Sit4. As a consequence of these GATA factor localization and binding characteristics, Sit4 is partially to largely dispensable for rapamycin-induced DAL5 expression (24).
The strikingly different phosphatase requirements for nuclear Gln3 and Gat1 localization in response to rapamycin treatment prompted us to query whether similarly distinguishable requirements could be observed with respect to GATA factors binding to their targets in the DAL5 promoter and to DAL5 transcription. Here, we show this to be the case, as follows. (i) PP2A, along with all three components of its Tpd3 complexes, Tpd3, Cdc55, and Rts1, is required for binding of rapamycin-induced Gat1-Myc13 (Gat1 tagged with 13 copies of the Myc epitope) to the DAL5 promoter and DAL5 transcription, whereas a demonstrable Sit4 requirement was previously shown to be limited in these assays. (ii) PP2A is also required for promoter binding of Gln3-Myc13 and DAL5 transcription in strains lacking Ure2, where Gln3 is highly nuclear, and for promoter binding of Gat1-NLSSV40-Myc13, which is also constitutively nuclear. Together, our data support the conclusion that Pph21/22 and its Tpd3-associated components have a heretofore unrecognized strong intranuclear function, at minimum being required for binding of the GATA transcription factors to their promoter targets.
MATERIALS AND METHODS
Strains and culture conditions.
The Saccharomyces cerevisiae strains used in this work appear in Table 1. The growth conditions were identical to those described earlier (24, 48). Cultures (50 ml) were grown to mid-log phase (A600 = 0.4 to 0.5) in YNB minimal medium (without ammonium sulfate or amino acids) containing 0.1% glutamine, and appropriate supplements (120 μg/ml leucine and 20 μg/ml each of uracil, histidine, and tryptophan) were added to the medium as necessary to cover auxotrophic requirements. Where indicated, cells were treated with 200 ng/ml (final concentration) rapamycin (Sigma and LC Laboratories) for 20 or 30 min.
TABLE 1.
Strains used in this work
| Strain | Pertinent genotype | Parent | Complete genotype | Reference |
|---|---|---|---|---|
| TB50 | WT | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLa | 1 | |
| TB123 | WT Gln3-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[KanMX] | 1 | |
| TB136-2a | sit4Δ Gln3-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[KanMX] sit4::kanMX | 1 | |
| TB138-1a | ure2Δ Gln3-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[KanMX] ure2::URA3 | 1 | |
| 03643c | sit4Δ pph22Δ Gln3-Myc13 | leu2-3,112 ura3-52 trp1 his3 rme1 GLN3-MYC13[KanMX] sit4::kanMX pph22::kanMX | 48 | |
| 03647a | sit4Δ pph22Δ | MATα leu2-3,112 ura3-52 trp1 his3 rme1 HMLα sit4::kanMX pph22::kanMX | 48 | |
| 03666c | sit4Δ pph21Δ Gln3-Myc13 | leu2-3,112 ura3-52 trp1 his3 rme1 GLN3-MYC13[KanMX] sit4::kanMX pph21::kanMX | 48 | |
| 03705d | pph21Δ pph22Δ Gln3-Myc13 | MATα leu2-3,112 ura3-52 trp1 his3 rme1 HMLα GLN3-MYC13[KanMX] pph21::kanMX pph22::kanMX | 48 | |
| 03879c | pph21 Δpph22Δ Gat1-Myc13 | MATα leu2-3,112 ura3-52 trp1 his3 rme1 HMLα GAT1-MYC13[HIS3] pph21::kanMX pph22::kanMX | 47 | |
| 06040d | sit4Δ Gat1-NLSSV40-Myc13 | 03647a × FV404 | leu2-3,112 ura3-52 trp1 his3 rme1 GAT1-NLSSV40-MYC13[HIS3] sit4::kanMX | This work |
| FV063 | WT Gat1-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] | 24 | |
| FV088 | ure2Δ Gat1-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] ure2::natMX | 24 | |
| FV103 | ure2Δ pph21Δ pph22Δ Gat1-Myc13 | 03879c | MATα leu2-3,112 ura3-52 trp1 his3 rme1 HMLα GAT1-MYC13[HIS3] pph21::kanMX pph22::kanMX ure2::natMX | This work |
| FV165 | ure2Δ pph21Δ pph22Δ Gln3-Myc13 | 03705d | MATα leu2-3,112 ura3-52 trp1 his3 rme1 HMLα GLN3-MYC13[KanMX] pph21::kanMX pph22::kanMX ure2::natMX | This work |
| FV177 | tpd3Δ Gln3-Myc13 | MATa leu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[KanMX] tpd3::natMX | 48 | |
| FV202 | WT Sit4-Myc13 | TB50 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaSIT4-MYC13[HIS3] | This work |
| FV203 | WT Pph21-Myc13 | TB50 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaPPH21-MYC13[HIS3] | This work |
| FV206 | cdc55Δ Gln3-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[KanMX] cdc55::natMX | 48 | |
| FV207 | cdc55Δ Gat1-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] cdc55::natMX | 47 | |
| FV208 | rts1Δ Gat1-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] rts1::natMX | 47 | |
| FV209 | rts1Δ Gln3-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[KanMX] rts1::natMX | 48 | |
| FV210 | tpd3Δ Gat1-Myc13 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] tpd3::natMX | 47 | |
| FV216 | WT Pph22-Myc13 | TB50 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaPPH22-MYC13[HIS3] | This work |
| FV224 | cdc55Δ ure2Δ Gat1-Myc13 | FV207 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] cdc55::natMX ure2::kanMX | This work |
| FV226 | cdc55Δ rts1Δ Gat1-Myc13 | FV208 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] rts1::natMX cdc55::kanMX | 47 |
| FV250 | WT Gln3-Myc13 | TB50 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] | This work |
| FV261 | rts1Δ Gln3-Myc13 | FV250 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] rts1::kanMX | This work |
| FV263 | cdc55Δ Gln3-Myc13 | FV250 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] cdc55::kanMX | This work |
| FV266 | cdc55Δ rts1Δ Gln3-Myc13 | FV263 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] cdc55::kanMX rts1::natMX | This work |
| FV272 | ure2Δ rts1Δ Gat1-Myc13 | FV208 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] rts1::natMX ure2::kanMX | This work |
| FV273 | ure2Δ rts1Δ Gln3-Myc13 | FV261 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] rts1::kanMX ure2::natMX | This work |
| FV274 | ure2Δ cdc55Δ Gln3-Myc13 | FV263 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] cdc55::kanMX ure2::natMX | This work |
| FV276 | tpd3Δ Gln3-Myc13 | FV250 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] tpd3::kanMX | This work |
| FV277 | ure2Δ tpd3Δ Gln3-Myc13 | FV276 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGLN3-MYC13[HIS3] tpd3::kanMX ure2::natMX | This work |
| FV278 | ure2Δ tpd3Δ Gat1-Myc14 | FV210 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-MYC13[HIS3] tpd3::natMX ure2::kanMX | This work |
| FV301 | gln3Δ Pph21-Myc13 | FV203 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaPPH21-MYC13[HIS3] gln3::kanMX | This work |
| FV303 | gat1Δ Pph21-Myc13 | FV203 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaPPH21-MYC13[HIS3] gat1::kanMX | This work |
| FV305 | gln3Δ Pph22-Myc13 | FV216 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaPPH22-MYC13[HIS3] gln3::kanMX | This work |
| FV307 | gat1Δ Pph22-Myc13 | FV216 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaPPH22-MYC13[HIS3] gat1::kanMX | This work |
| FV404 | WT Gat1-NLSSV40-Myc13 | TB50 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-NLSSV40-MYC13[HIS3] | This work |
| FV414 | pph22Δ Gat1-NLSSV40-Myc13 | FV404 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-NLSSV40-MYC13[HIS3] pph22::kanMX | This work |
| FV415 | pph21Δ pph22Δ Gat1-NLSSV40-Myc13 | FV414 | MATaleu2-3,112 ura3-52 trp1 his3 rme1 HMLaGAT1-NLSSV40-MYC13[HIS3] pph21::natMX pph22::kanMX | This work |
Strain construction.
Deletion strains involving insertion of kanMX or natMX cassettes were constructed as described earlier (24, 46, 48) by using the long flanking homology strategy of Wach (51) described by Georis et al. (24) and Tate et al. (48). Chromosomal GLN3, PPH21, PPH22, or SIT4 was tagged at the C terminus with 13 copies of the Myc epitope (Myc13), as described by Longtine et al. (36), using primers GLN3-F2 and GLN3-R1 for GLN3 (24), 5′-TGAACCTAGCGTCAGCAGAAAGACGCCAGATTACTTTTTACGGATCCCCGGGTTAATTAA-3′ and 5′-AATATATATCTATATAGATGCATATATGTATACATACTCAAGAATTCGAGCTCGTTTAAAC-3′ for PPH21, 5′-TGAACCAACCGTCACCAGGAAGACACCGGATTATTTCTTACGGATCCCCGGGTTAATTAA-3′ and 5′-AATAGCGTAGTAAGGATAAAGGTGTAATAGATATATATTAAGAATTCGAGCTCGTTTAAAC-3′ for PPH22, and 5′-CACGGCAAACCATAATAATCAAAGAGCCGGCTATTTCTTACGGATCCCCGGGTTAATTAA-3′ and 5′-TTTTATTCGTCGAGTTAGGGAGGGCATGCCGTCGTGTTAAGAATTCGAGCTCGTTTAAAC-3′ for SIT4. Chromosomal GAT1 was tagged at its C terminus with simian virus 40 nuclear localization signal (NLSSV40) and 13 copies of the Myc epitope (Myc13) as described by Longtine et al. (36) using primers OA483 (5′-CCAAATGGCAATCTGAGCCTGGATTG-GTTGAATCTGAATTTACCTCCAAAAAAGAAGAGAAAGGTCCGGATCCCCGGGTTAATTAA-3′) and GAT1-R1 (24).
Quantitative RT-PCR.
RNA isolation and cDNA synthesis were conducted as described in reference 23. Quantification of specific cDNA targets was measured by real-time PCR (RT-PCR) performed on a StepOnePlus device (Applied Biosystems), using primers described in Georis et al. (24). The mean represents at least two measurements from independent cultures.
ChIP.
For chromatin immunoprecipitation (ChIP), cell extracts were obtained and immunoprecipitations were conducted as described in references 23 and 24. Concentrations of specific DNA targets in immunoprecipitation (IP) and input (IN) samples were measured by real-time PCR performed on a StepOnePlus device (Applied Biosystems), using the primers described by Georis et al. (24). IP/IN values obtained for the unbound control (DAL5U) were subtracted from the corresponding IP/IN values obtained for the DAL5 promoter (DAL5P). The mean represents at least two replicate immunoprecipitations performed on a minimum of two independent cultures.
Western blot.
Total protein extracts were obtained from 10-ml cultures by trichloroacetic acid (TCA) precipitation as described in reference 43. The electrophoresis, blotting, and detection procedures have been described previously (23). Anti-Pgk1 antibodies were purchased from Invitrogen (catalogue no. 459250).
Intracellular Gln3-Myc13 and Gat1-GFP localization.
Cell collection and immunofluorescent staining procedures were performed as described earlier (13, 24, 44-49). Live and fixed cells were imaged at room temperature with a Zeiss Axioplan 2 imaging microscope with a 100× (Gln3-Myc13) or 63× (Gat1-green fluorescent protein [GFP]) 1.40 Plan-Apochromat oil objective, a Zeiss Axio camera, and AxioVision 3.0 (Zeiss) software. Images were processed for publication with Zeiss AxioVision 4.6.3, Adobe Photoshop CS3 and Illustrator CS3 software. Changes were made to the black and white set points exclusively to decrease background fluorescence. On those occasions when gamma settings were changed, it was done conservatively and only when necessary to avoid loss of detail relative to what was observed in the microscope; all changes were applied uniformly to the image presented and as similarly as possible from one image panel to another. In contrast, only unaltered .zvi file images were used for scoring intracellular Gln3 and Gat1 distributions.
To more representatively and completely describe images of Gln3-Myc13 localization appearing in the figures of this paper, we manually scored Gln3-Myc13 localization in 200 or more cells from the microscopic field (and additional randomly chosen fields on the same slide) from which each image presented was taken. Gat1-GFP localization was determined by counting all of the cells in six images from three culture samples (90 cells on average/sample). Cells were classified into one of three categories: cytoplasmic (fluorescence in the cytoplasm only), nuclear-cytoplasmic (fluorescence in the cytoplasm and nucleus), and nuclear (fluorescence in the nucleus only). Detailed examples of the three scoring categories for Gln3 and Gat1 as well as scoring precision (within and between experiments) and comparison of data from indirect immunofluorescence and GFP assays appear in references 47 to 49. Similar experiments were repeated two or more times with similar results.
RESULTS
Pph21/22 functions upstream of Ure2 for rapamycin-induced nuclear Gln3-Myc13 localization.
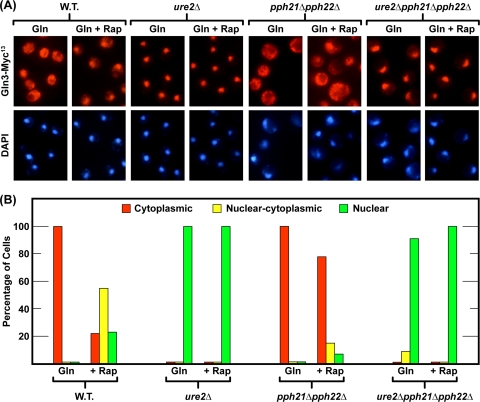
We recently showed that Pph21/22-Tpd3-Cdc55, Pph21/22-Tpd3-Rts1, and Sit4 are strongly required for rapamycin-induced nuclear Gln3-Myc13 localization in glutamine-grown cells (47): i.e., nuclear accumulation does not occur in the sit4Δ, pph21Δ pph22Δ, tpd3Δ, cdc55Δ, or rts1Δ mutants under these conditions (46-48). In contrast, sit4Δ and pph21Δ pph22Δ mutants exhibit opposite phenotypes for nuclear Gln3-Myc13 localization in response to growth with derepressive nitrogen sources such as proline. Sit4 is required in this situation, whereas Pph21/22, Tpd3, Cdc55, and Rts1 are not (46-48). In other words, the specific phosphatase requirements depend on the nature of the stimulus. To further investigate these requirements, we determined the epistatic relationship between the ure2Δ and pph21Δ pph22Δ mutations. Deletion of URE2 rendered Gln3-Myc13 constitutively nuclear, irrespective of Pph21/22 or rapamycin (Fig. 1A and B). These and previous reported results (24) indicated Pph21/22 and Sit4 functioned upstream of Ure2 for rapamycin-induced nuclear Gln3-Myc13 localization.
FIG. 1.
Pph21/22 is required upstream of Ure2 for rapamycin-induced nuclear Gln3-Myc13 localization in glutamine-grown cells. Wild-type (W.T.) (TB123) and ure2Δ (TB138-1a), pph21Δ pph22Δ (03705d), and ure2Δ pph21Δ pph22Δ (FV165) mutant cultures were grown in YNB-glutamine medium to an A600 of ∼0.5. Each culture was sampled for indirect immunofluorescence assay of Gln3-Myc13 localization, rapamycin (200 ng/ml) was added, and 20 min later, the cultures were sampled again. Indirect immunofluorescence assays were performed and images were obtained as described in Materials and Methods. For each histogram, a minimum of 200 cells were scored for intracellular Gln3-Myc13 localization, using the criteria described along with examples of each category in Fig. 2 from Tate et al. (48, 49). Images from which the histograms were derived are shown in pairs above each one. The upper member of each pair depicts Gln3-Myc13-derived fluorescence, and the lower one shows DAPI (4′,6-diamidino-2-phenylindole)-positive material fluorescence.
Pph21/22 is required downstream of Ure2 for rapamycin-induced DAL5 transcription.
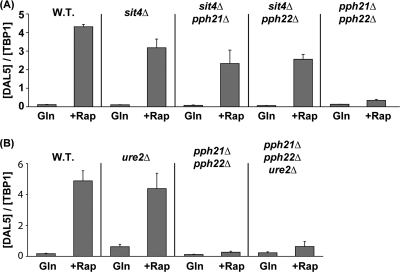
In contrast with Gln3, rapamycin-induced nuclear Gat1 localization in glutamine-grown cells exhibited only partial requirements for Sit4 (24) and Pph21/22 (47). Taken together, the known actions of these phosphatases (54) and their requirements for GATA factor localization raised the question of whether Sit4 and Pph21/22 functioned only outside of the nucleus in glutamine-grown cells. To begin investigating this question, we assayed overall DAL5 expression in glutamine-grown untreated or rapamycin-treated wild-type (WT) and mutant cells that either contained or were devoid of Ure2. As previously shown (24), the Sit4 requirement for DAL5 transcription coincided with that of rapamycin-induced Gat1 localization in glutamine-grown cells: i.e., Sit4 was largely dispensable (Fig. 2A). Deletion of either PPH21 or PPH22, in combination with SIT4, reduced DAL5 expression equivalently and also modestly more than sit4Δ alone, suggesting the likelihood of only limited nonredundant functions for Pph21 and Pph22 in the absence of Sit4 (Fig. 2A). Unexpectedly, however, DAL5 was not expressed in the pph21Δ pph22Δ double mutant (Fig. 2A), and, more important, this phenotype occurred irrespective of Ure2 (Fig. 2B). This indicated that, in addition to its requirement upstream of Ure2 to control Gln3 localization, Pph21/22 was required either downstream of Ure2 or in a separate regulatory pathway for rapamycin to elicit DAL5 transcription.
FIG. 2.
Phosphatase requirements for rapamycin-induced DAL5 transcription. Cells expressing GLN3-MYC13, which replaced the native GLN3 gene, were grown in YNB-glutamine medium and treated with rapamycin (Rap) (200 ng/ml) for 30 min. Control cells were similarly grown, but untreated (Gln). DAL5 mRNA levels were quantified by quantitative RT-PCR as described in Materials and Methods. DAL5 values were normalized with TBP1. The values represent the averages of at least two experiments from independent cultures, and the error bars indicate standard errors. (A) Pph21/22 is required for rapamycin-induced DAL5 expression. Total RNA was isolated from wild-type (TB123) or sit4Δ (TB136-2a), sit4Δ pph21Δ (03666c), sit4Δ pph22Δ (03643c), and pph21Δ pph22Δ (03705d) mutant cells. (B) Pph21/22 is required downstream of Ure2 for rapamycin-induced DAL5 expression. Total RNA was isolated from wild-type (TB123) or ure2Δ (TB138-1a), pph21Δ pph22Δ (03705d), and pph21Δ pph22Δ ure2Δ (FV165) cells.
Pph21/22 is required downstream of Ure2 or in a separate pathway to control Gat1-GFP localization.
Although the most straightforward interpretation of the above transcription results was that Pph21 and Pph22 functioned downstream of Ure2, they might alternatively derive from Pph21/22 controlling Gat1 localization in a regulatory pathway distinct from the one predominantly controlling Gln3 localization (24, 47). Three observations were consistent with the latter alternative. (i) Gat1 rather than Gln3 accounts for the majority of rapamycin-induced DAL5 expression in glutamine-grown cells (24). (ii) Gat1 and Gln3 localizations predominantly respond to different stimuli; i.e., Gat1 localization responds more strongly to induction by rapamycin than Gln3 and less strongly to NCR (47). (iii) Gat1 localization exhibits different phosphatase requirements and Ure2 sensitivity from those of Gln3 (24, 47). Therefore, did the above observations derive from distinct regulatory pathways for Gln3 and Gat1 and/or did Pph21/22 function both in the cytoplasm and nucleus to achieve DAL5 transcription? To begin distinguishing these possibilities, we determined the epistatic relationship of the ure2Δ and pph21Δ pph22Δ mutations with respect to Gat1-GFP localization.
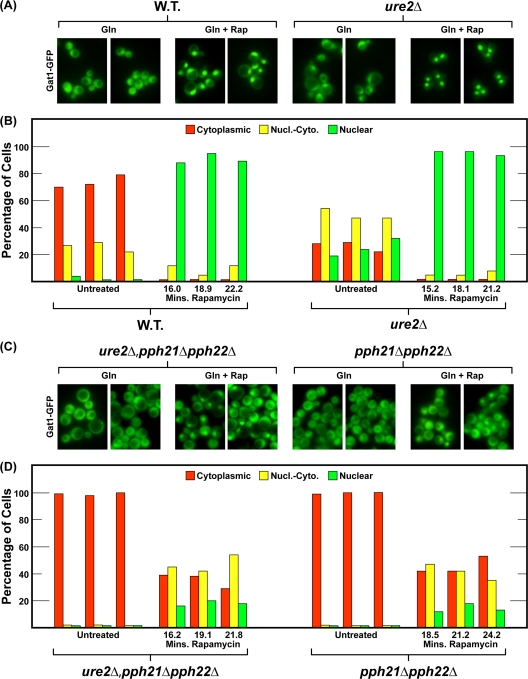
In untreated, glutamine-grown cells, deletion of URE2 increased nuclear Gat1-GFP localization: i.e., Gat1-GFP was nuclear-cytoplasmic or nuclear in a greater proportion of the cells compared to wild type (Fig. 3A and B). However, the nuclear shift in Gat1-GFP localization was not as great as that observed with Gln3 (Fig. 1A and B of this work; see Fig. 6A and 6B in reference 24). In sharp contrast, Gat1-GFP remained completely cytoplasmic in all glutamine-grown pph21Δ pph22Δ and ure2Δ pph21Δ pph22Δ cells (Fig. 3C and D). The pph21Δ pph22Δ mutations were epistatic to the ure2Δ mutation.
FIG. 3.
Pph21/22 is required downstream of Ure2 for rapamycin-induced nuclear Gat1-GFP localization. Wild-type (FV063) or ure2Δ (FV088), pph21Δ pph22Δ (03879c), and ure2Δ pph21Δ pph22Δ (FV103) cultures were transformed with plasmid RS416-Gat1-GFP. Fresh transformants were then grown in YNB-glutamine medium to an A600 of ∼0.5. Cells were then continuously sampled, and Gat1-GFP fluorescence images were acquired. Two microscopic images were acquired as quickly as possible from each sample; they were routinely completed within 90 to 100 and 130 to 160 s of sampling, respectively (47). After 12 images were collected, rapamycin (200 ng/ml) was added to the cultures and sampling continued for at least 30 additional min. The three bars of the histogram represent the averages derived from cells scored in the two images of each sample. Data from three successive samples are provided to assess scoring reproducibility (39). The times that appear on the abscissa are the averages of the two times at which the pairs of images from each sample were completed following addition of rapamycin to the culture. Microscopic images in panels A and C were derived from images used to determine intracellular Gat1-GFP localization in the histograms of panels B and D.
In rapamycin-treated, glutamine-grown cells, Gat1-GFP localization was highly nuclear in nearly all wild-type and ure2Δ cells (Fig. 3A and B); a similar result had been previously seen with Gat1-Myc13 (24). In contrast, rapamycin-induced nuclear Gat1-GFP localization substantially decreased in the pph21Δ pph22Δ mutant, relative to wild-type or ure2Δ cells: i.e., Gat1-GFP was now cytoplasmic in about half of the cells and completely nuclear in only about 20% of them (Fig. 3C and D). Gat1-GFP localization in the ure2Δ pph21Δ pph22Δ triple mutant was indistinguishable from that in pph21Δ pph22Δ cells (Fig. 3C and D). The most straightforward overall interpretation of these data is that the pph21Δ pph22Δ mutant is epistatic to a ure2Δ mutant. However, since Gat1-GFP localization in rapamycin-treated wild-type and ure2Δ cells exhibits the same phenotype, we cannot exclude the possibility that Pph21/22 interferes with Gat1 localization in a Ure2-independent way.
Pph21/22 is required for GATA factor binding to the DAL5 promoter.
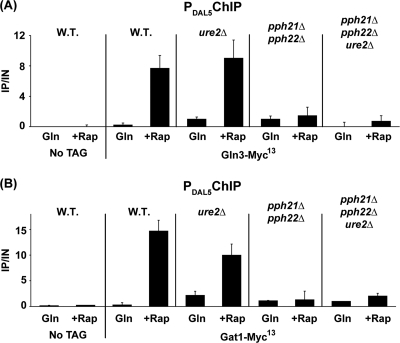
The above data cumulatively pointed to Pph21/22 functioning downstream of GATA factor accumulation in the nucleus to support DAL5 transcription. To identify the putative intranuclear event in which Pph21/22 participated, we analyzed Gln3-Myc13 and Gat1-Myc13 binding to the DAL5 promoter, a major event between nuclear localization of the GATA transcription factors and transcription itself. As previously reported (24), rapamycin strongly induced Gln3-Myc13 binding to the DAL5 promoter in both wild-type and ure2Δ mutant cells (Fig. 4A). In contrast, deletion of PPH21 and PPH22 largely abolished rapamycin-induced Gln3-Myc13 binding to the DAL5 promoter in either an otherwise wild-type or ure2Δ background (Fig. 4A). Thus, nuclear Gln3-Myc13 localization in rapamycin-treated ure2Δ pph21Δ pph22Δ cells was insufficient to permit its binding to the DAL5 promoter and hence participate in the activation of DAL5 transcription.
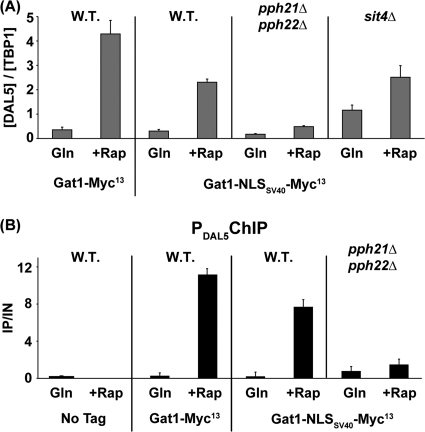
FIG. 4.
Pph21/22 is required for rapamycin-induced GATA factor binding to the DAL5 promoter (PDAL5). Cells were grown in YNB-glutamine medium and treated with rapamycin (Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). ChIP was performed using antibodies against c-myc as described in Materials and Methods. qPCR of IP and IN fractions was performed with primers specific for the DAL5 promoter (DAL5P) and for a region 2.5 kb upstream of the DAL5 open reading frame as a control (DAL5U). For each immunoprecipitation, IP/IN values were calculated as follows: [DAL5P]IP/[DAL5P]IN − [DAL5U]IP/[DAL5U]IN. Histograms represent the averages of two immunoprecipitations performed on at least two experiments from independent cultures. Error bars indicate standard errors. (A) Gln3-Myc13 does not bind PDAL5 in the absence of Pph21/22. Untagged wild-type (TB50), GLN3-MYC13 wild-type (TB123), GLN3-MYC13 ure2Δ (TB138-1a), GLN3-MYC13 pph21Δ pph22Δ (03705d), and GLN3-MYC13 pph21Δ pph22Δ ure2Δ (FV165) cells were subjected to ChIP analysis. (B) Gat1-Myc13 does not bind PDAL5 in the absence of Pph21/22 following rapamycin treatment. Untagged wild-type (TB50), GAT1-MYC13 wild-type (FV063), GAT1-MYC13 ure2Δ (FV088), GAT1-MYC13 pph21Δ pph22Δ (03879c), and GAT1-MYC13 pph21Δ pph22Δ ure2Δ (FV103) cells were subjected to ChIP analysis.
Although the above result could be construed as demonstrating an intranuclear function for PP2A, this conclusion carried the caveat that Gln3 binding to the DAL5 promoter has been shown to be Gat1 dependent in this strain background (24). Hence the effect, while striking, might be indirect. Therefore, we compared rapamycin-induced binding of Gat1-Myc13 to the DAL5 promoter in wild-type, ure2Δ, pph21Δ pph22Δ, and ure2Δ pph21Δ pph22Δ cells. Gat1-Myc13 binding to its DAL5 target increased in the untreated ure2Δ cells relative to the wild type, roughly paralleling Gat1-GFP localization (Fig. 4B). In rapamycin-treated wild-type and ure2Δ cells, nuclear Gat1-GFP localization and Gat1-Myc13 binding to DAL5 were similarly high (Fig. 4B) and commensurate with the rapamycin-induced DAL5 transcription observed in Fig. 2B. In sharp contrast, Gat1-Myc13 binding to the DAL5 promoter was nearly abolished in the pph21Δ pph22Δ and ure2Δ pph21Δ pph22Δ mutants (Fig. 4B). Here, as with Gat1-GFP localization, the pph21Δ pph22Δ phenotype was more prominent than that of a ure2Δ mutant.
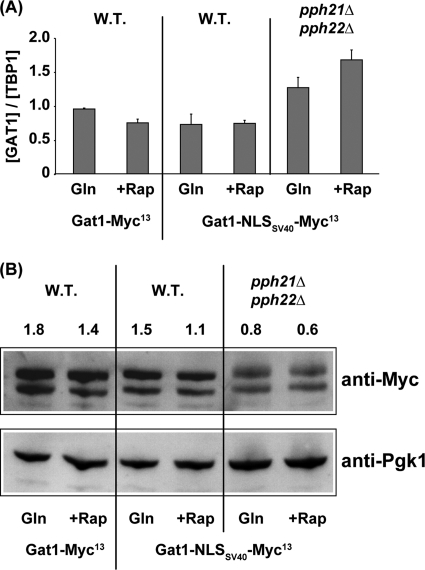
Together, the above data point to an intranuclear function for Pph21/22 phosphatase in the binding of GATA factors to the DAL5 promoter. This conclusion was not unequivocal, however, because our data did not unambiguously eliminate the possibility that loss of Gat1-Myc13 binding to DAL5 derived from insufficient intranuclear Gat1-Myc13 due to the partial Pph21/22 requirement for its rapamycin-induced nuclear localization. The most convincing way of eliminating this caveat was to determine the Pph21/22 requirement under conditions in which Gat1-Myc13 was completely nuclear. Therefore, it was necessary to artificially locate Gat1 to the nuclei of a pph21Δ pph22Δ mutant. This we accomplished, as previously described for Ure2, by fusing the SV40 nuclear localization sequence (NLSSV40) to the C terminus of Gat1 (34). Gat1-NLSSV40-Myc13 was completely nuclear in both glutamine-grown wild-type and pph21Δ pph22Δ cells (Fig. 5A and B), irrespective of the presence or absence of rapamycin. Yet there was only basal-level DAL5 transcription in the rapamycin-treated pph21Δ pph22Δ mutant (Fig. 6A). This requirement was specific to Pph21/22: i.e., there was no more of a Sit4 requirement for DAL5 transcription in the Gat1-NLSSV40-Myc13-containing strain, in which Gat1-NLSSV40-Myc13 was constitutively nuclear, than for one lacking the NLS sequence (Fig. 6A). Consistently, Pph21/22 was strongly required for Gat1-NLSSV40-Myc13 binding to the DAL5 promoter (Fig. 6B). Control experiments demonstrated that fusing the SV40 NLS to GAT1 did not alter its expression (Fig. 7A), Gat1 protein levels (Fig. 7B), or its function (Fig. 6A and B; WT rapamycin treated). Importantly, although Gat1-NLSSV40-Myc13 was constitutively nuclear, it could not bind to the DAL5 promoter in glutamine-grown cells (Fig. 6B).
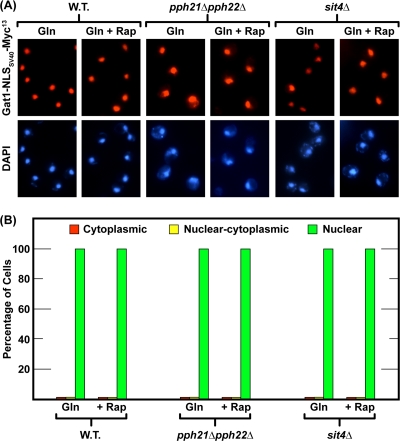
FIG. 5.
Gat1-NLSSV40-Myc13 is constitutively localized to the nucleus in untreated and rapamycin-treated, glutamine-grown wild-type, sit4Δ, and pph21Δ pph22Δ cells. The experimental format and data analyses were performed as described in the legend to Fig. 1; the strains employed were GAT1-NLSSV40-MYC13 wild type (FV404), GAT1-NLSSV40-MYC13 sit4Δ (06040d), and GAT1-NLSSV40-MYC13 pph21Δ pph22Δ (FV415).
FIG. 6.
Deletion of PPH21/22 affects DAL5 regulation in cells expressing constitutively nuclear Gat1. (A) Deletion of PPH21/22 adversely affects DAL5 expression in GAT1-NLSSV40-Myc13 cells. Total RNA was isolated from GAT1-MYC13 wild type (FV063), GAT1-NLSSV40-MYC13 wild type (FV404), GAT1-NLSSV40-MYC13 pph21Δ pph22Δ (FV415), and GAT1-NLSSV40-MYC13 sit4Δ (06040d) cells grown in YNB-glutamine medium and treated with rapamycin (+Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). DAL5 mRNA levels were quantified by quantitative RT-PCR, as described in the legend to Fig. 2. (B) Deletion of PPH21/22 adversely affects Gat1-NLSSV40-Myc13 binding to the DAL5 promoter. Untagged wild-type (TB50), GAT1-MYC13 wild type (FV063), GAT1-NLSSV40-MYC13 wild type (FV404), and GAT1-NLSSV40-MYC13 pph21Δ pph22Δ (FV415) cells were grown in YNB-glutamine medium and treated with rapamycin (+Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). ChIP was performed as described in the legend to Fig. 4.
FIG. 7.
The NLSSV40-Myc13 tag and deletion of PPH21/22 do not affect GAT1 expression. (A) Quantitative analysis of GAT1 mRNA levels. Total RNA was isolated from GAT1-MYC13 wild-type (FV063), GAT1-NLSSV40-MYC13 wild type (FV404), and GAT1-NLSSV40-MYC13 pph21Δ pph22Δ mutant (FV415) cells grown in YNB-glutamine medium and treated with rapamycin (+Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). GAT1 mRNA levels were quantified by quantitative RT-PCR as described in the legend to Fig. 2. (B) Western blot analysis of Gat1-Myc13 and Gat1-NLSSV40-Myc13 protein levels. GAT1-MYC13 wild type (FV063), GAT1-NLSSV40-MYC13 wild type (FV404), and GAT1-NLSSV40-MYC13 pph21Δ pph22Δ mutant (FV415) cells were grown in YNB-glutamine medium and treated with rapamycin (+Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). Total protein extracts were isolated and subjected to anti-Myc Western blot analysis. Pgk1 was used as the loading standard.
GATA factor-dependent Pph21/22 binding to the DAL5 promoter.
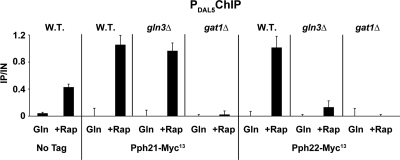
Since Pph21/22 appeared to perform a critical intranuclear function required for rapamycin to induce Gat1-Myc13 binding to the DAL5 promoter, it was worth asking whether Pph21 or Pph22 could be shown to be associated, like the GATA factors, with the DAL5 promoter. To this end, we used ChIP assays to assess the presence of Pph21-Myc13 and Pph22-Myc13 at the DAL5 promoter. The results of these assays demonstrated association of the Pph21/22 phosphatase catalytic subunits with DAL5 promoter DNA (Fig. 8), but did not unambiguously indicate that this occurred as result of a direct phosphatase subunit-Gat1 interaction. The occurrence of such an interaction was circumstantially supported, however, by the following findings: Pph21-Myc13 binding to DAL5 depended only on Gat1 (i.e., it was lost in a gat1Δ mutant but not in a gln3Δ mutant), whereas Pph22-Myc13 binding to DAL5 required both Gln3 and Gat1 (i.e., they were lost following deletion of either GATA factor gene).
FIG. 8.
Pph21 and Pph22 bind to the DAL5 promoter in a Gat1-dependent manner. Untagged wild-type (TB50), PPH21-MYC13 wild-type (FV203), PPH21-MYC13 gln3Δ (FV301), PPH21-MYC13 gat1Δ (FV303), PPH22-MYC13 wild-type (FV216), PPH22-MYC13 gln3Δ (FV305), and PPH22-MYC13 gat1Δ (FV307) cells were grown in YNB-glutamine medium and treated with rapamycin (+Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). ChIP was performed as described in the legend to Fig. 4.
We noted one unexplained anomaly in the ChIP data. The anti-myc ChIP signal in the untagged strain was markedly increased by the presence of rapamycin (Fig. 8). While we cannot explain this observation, it is important to recognize that the weak ChIP signals generated by Pph21-Myc13 and Pph22-Myc13 were such that the data were quite close to background levels, accentuating signals from the untagged strain.
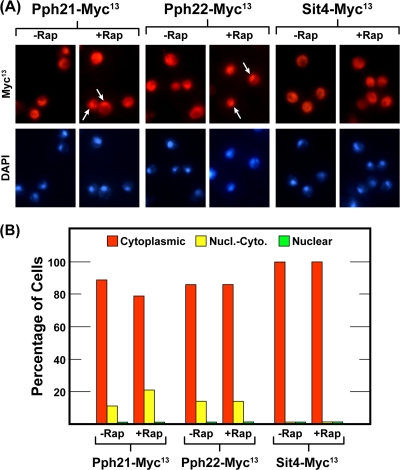
The above data supported the contention that PP2A functioned within the nucleus. To see if visual evidence of their intranuclear presence could be obtained, we determined the intracellular distributions of Sit4-Myc13, Pph21-Myc13, and Pph22-Myc13 in glutamine-grown untreated or rapamycin-treated wild-type cells (Fig. 9). Although we occasionally observed cells in which one of the phosphatase subunits exhibited nuclear-cytoplasmic localization (arrows in Fig. 9A), in a large majority of them, Pph21-Myc13 and Pph22-Myc13 were cytoplasmic (Fig. 9B). Consistent with the DAL5 transcription data, we were unable to find cells in which Sit4-Myc13 exhibited a demonstrable nuclear or nuclear-cytoplasmic distribution (Fig. 9B). Thus, we observed Pph21 and Pph22 proteins within some nuclei, but they were not preferentially accumulated in that cellular compartment.
FIG. 9.
Subcellular localization of the catalytic subunits of Sit4 and Pph21 and Pph22 phosphatase catalytic subunits. Cultures of strains FV202 (WT SIT4-MYC13), FV203 (WT PPH21-MYC13), and FV216 (WT PPH22-Myc13) were grown in YNB-glutamine medium in the absence or presence of rapamycin (200 ng/ml) and assayed as described in the legend to Fig. 1. Data are formatted as in Fig. 1.
When transcription factors are functioning within the nucleus, their preferential accumulation within the nucleus can usually be convincingly demonstrated as well. This, however, is an unlikely expectation for the PP2A catalytic subunits. Given their many well-documented cytoplasmic functions, it would be surprising indeed to see them localized in the nucleus to the extent that their intranuclear concentrations were two to four times greater than those in the surrounding cytoplasm. This is the concentration differential likely required for their localization to be scored nuclear-cytoplasmic by using the criteria described in Materials and Methods.
Both Tpd3 complexes of PP2A are required for rapamycin-induced DAL5 gene expression and GATA factor binding to the DAL5 promoter.
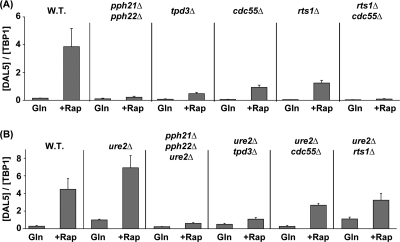
The Pph21 and Pph22 catalytic subunits of PP2A function in one of at least three different protein complexes, being associated with either Tap42, Tpd3-Cdc55, or Tpd3-Rts1 (32). Aware that the Tpd3 complexes participate heavily in rapamycin-induced regulation of Gln3 and Gat1 localization (47, 48), we queried whether they were similarly involved in the control of NCR-sensitive transcription and GATA factor binding to the promoter of an NCR-sensitive gene. To this end, we compared the expression of DAL5 in glutamine-grown untreated or rapamycin-treated wild-type, pph21Δ pph22Δ, tpd3Δ, cdc55Δ, rts1Δ, and cdc55Δ rts1Δ cells. DAL5 transcription was abolished in the pph21Δ pph22Δ and cdc55Δ rts1Δ double mutants and nearly so in the tpd3Δ mutant. There was a limited amount of DAL5 transcription in the cdc55Δ and rts1Δ mutants (Fig. 10A). Similar results were observed when each of the above mutations was introduced into a ure2Δ background (Fig. 10B), although the rapamycin-induced levels in the ure2Δ cdc55Δ and ure2Δ rts1Δ mutants were 2- to 3-fold higher than those in the corresponding cdc55Δ and rts1Δ single mutants.
FIG. 10.
Requirements of the PP2A regulatory subunits for rapamycin-induced DAL5 transcription. Cells expressing GLN3-MYC13, which replaced the native GLN3 gene, were grown in YNB-glutamine medium and treated with rapamycin (+Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). DAL5 mRNA levels were quantified by quantitative RT-PCR as described in the legend to Fig. 2. (A) Both Pph21/22-Tpd3-Cdc55 and Pph21/22-Tpd3-Rts1 complexes are required for DAL5 expression. Total RNA was isolated from wild-type (TB123), pph21Δ pph22Δ (03705d), tpd3Δ (FV177), cdc55Δ (FV206), rts1Δ (FV209), and rts1Δ cdc55Δ (FV266) cells. (B) PP2A regulatory subunits are required downstream of Ure2 for DAL5 expression. Total RNA was isolated from wild-type (TB123), ure2Δ (TB138-1a), ure2Δ pph21Δ pph22Δ (FV165), ure2Δ tpd3Δ (FV277), ure2Δ cdc55Δ (FV274), and ure2Δ rts1Δ (FV273) cells.
Thus, Pph21/22 and Tpd3 were strongly required for rapamycin-induced DAL5 transcription. Requirements for the Cdc55 and Rts1 subunits were more limited. However, the complete loss of DAL5 transcription in the cdc55Δ rts1Δ double mutant indicated the participation of both subunits and suggested that they might be performing partially redundant functions. Increasing the nuclear concentration of Gln3, as occurs in the ure2Δ background, increased overall transcription but did not alter its requirements.
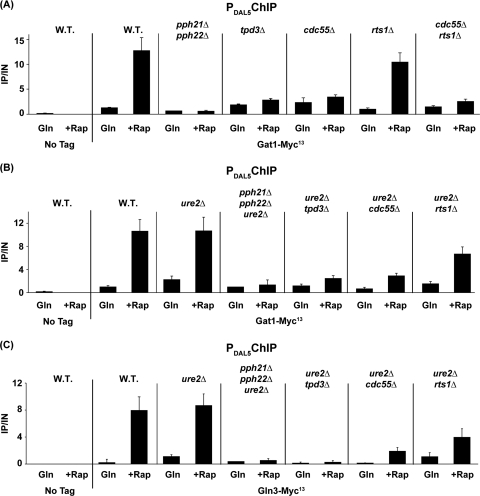
Given the requirements of the Pph21/22-Tpd3-Cdc55 and Pph21/22-Tpd3-Rts1 complexes for DAL5 transcription, we determined the extent of their participation in Gat1-Myc13 binding to the DAL5 promoter by using the mutants described above in Fig. 10. Pph21 and -22 were the most required subunits, followed closely by Tpd3 and Cdc55 (Fig. 11A). In contrast with transcription results, there was only a quite limited Rts1 requirement for Gat1-Myc13 binding to DAL5. Recall that it was also this subunit that was least required for rapamycin-induced nuclear Gat1-GFP localization (47).
FIG. 11.
Requirements of the PP2A regulatory subunits for rapamycin-induced GATA factor binding to the DAL5 promoter. Cells were grown in YNB-glutamine medium and treated with rapamycin (+Rap) (200 ng/ml) for 30 min. Control cells were similarly grown but untreated (Gln). ChIP was performed as described in the legend to Fig. 4. (A) Only the Pph21/22-Tpd3-Cdc55 complex is required to enable Gat1-Myc13 binding to PDAL5. Untagged wild-type (TB50), GAT1-MYC13 wild-type (FV063), GAT1-MYC13 pph21Δ pph22Δ (03879c), GAT1-MYC13 tpd3Δ (FV210), GAT1-MYC13 cdc55Δ (FV207), GAT1-MYC13 rts1Δ (FV208), and GAT1-MYC13 cdc55Δ rts1Δ (FV226) cells were subjected to ChIP analysis. (B) The Pph21/22-Tpd3-Cdc55 complex is required downstream of Ure2 to allow Gat1-Myc13 binding to PDAL5. Untagged wild-type (TB50), GAT1-MYC13 wild-type (FV063), GAT1-MYC13 ure2Δ (FV088), GAT1-MYC13 ure2Δ pph21Δ pph22Δ (FV103), GAT1-MYC13 ure2Δ tpd3Δ (FV278), GAT1-MYC13 ure2Δ cdc55Δ (FV224), and GAT1-MYC13 ure2Δ rts1Δ (FV272) cells were subjected to ChIP analysis. (C) The Pph21/22-Tpd3-Cdc55 complex is required downstream of Ure2 to allow Gln3-Myc13 binding to PDAL5. Untagged wild-type (TB50), GLN3-MYC13 wild-type (TB123), GLN3-MYC13 ure2Δ (TB138-1a), GLN3-MYC13 ure2Δ pph21Δ pph22Δ (FV165), GLN3-MYC13 ure2Δ tpd3Δ (FV277), GLN3-MYC13 ure2Δ cdc55Δ (FV274), and GLN3-MYC13 ure2Δ rts1Δ (FV273) cells were subjected to ChIP analysis.
Additionally, deletion of URE2 did not substantially alter the above requirements for Gat1-Myc13 binding (Fig. 11B). The PP2A requirements observed above for Gat1-Myc13 were exaggerated in the case of Gln3-Myc13 binding to the DAL5 promoter (Fig. 11C). This is an expected result since Gat1 bound to the DAL5 promoter positively influences Gln3-Myc13 binding (24).
DISCUSSION
Experiments presented in this work clearly demonstrate for the first time that in addition to its well-documented cytoplasmic functions, PP2A phosphatase is required within the nucleus for rapamycin-induced Gln3 and Gat1 binding to their target sequences in the DAL5 promoter and hence for DAL5 transcription in rapamycin-treated, glutamine-grown cells. These results confirm a similar conclusion derived from the correlations that Pph21/22-dependent Gln3 and Gat1 dephosphorylation is greatest under conditions in which Gln3 is nuclear and least under those where it localizes to the cytoplasm (47, 48). With respect to its cytoplasmic functions, our previous work has shown that PP2A is absolutely required for rapamycin-induced nuclear Gln3-Myc13 localization and partially so for Gat1-Myc13 (Fig. 1 and Fig. 3) (47, 48).
In support of these observations, epistasis analyses shown in Fig. 1 indicate that Pph21/22 functions upstream of Ure2 as far as Gln3-Myc13 localization is concerned in both glutamine-grown, untreated and rapamycin-treated cells. Importantly, the epistasis relationship observed in untreated cells when Gat1 localization was assayed is the reverse of that observed for Gln3, i.e., pph21Δ pph22Δ was epistatic to ure2Δ (Fig. 3). This striking difference in the epistasis relationships for the Ure2 and Pph21/22 requirements of Gat1 and Gln3 localizations in untreated cells supports our previous conclusion that Gln3 and Gat1 localizations are regulated by distinct pathways (47).
Several lines of evidence support an intranuclear function for Pph21/22. First, rapamycin-induced DAL5 expression and GATA factor binding to the DAL5 promoter were abolished in a pph21Δ pph22Δ mutant even in a ure2Δ background or in a GAT1-NLSSV40-Myc13 strain, where Gln3 or Gat1 is constitutively nuclear. Given the fact that rapamycin-induced Gln3 binding to the DAL5 promoter requires Gat1, we could not assess whether the lack of Gln3 binding in a triple ure2Δ pph21Δ pph22Δ mutant resulted from a direct PP2A requirement for Gln3 binding or is caused by the Gat1 binding impairment. Second, Pph21-Myc13 and Pph22-Myc13 were shown to be nuclear-cytoplasmic in some cells (∼15 to 20%) (Fig. 9) and furthermore to be associated with the DAL5 promoter in a GATA factor-dependent manner (Fig. 8). The GATA factor requirements of Pph21-Myc13 and Pph22-Myc13 binding—i.e., Gat1-dependent, Gln3-independent binding for Pph21-Myc13 and Gat1- as well as Gln3-dependent binding of Pph22-Myc13—increased our confidence in these measurements. PP2A catalytic subunit binding to the DAL5 promoter was about 10-fold lower than that of Gat1-Myc13 or Gln3-Myc13. This, however, is an expected result because GATA factors bind directly and stoichiometrically to the DNA, whereas Pph21-Myc13 and Pph22-Myc13 likely function catalytically and are probably not in direct contact with the DNA. Sit4-Myc13, in contrast, was not required for rapamycin-induced DAL5 transcription in glutamine-grown cells and could not be found in the nuclei of any cells irrespective of growth conditions (Fig. 9), leading us to conclude that Sit4 was not playing a demonstrable intranuclear role in DAL5 transcription.
Our experiments additionally identified the specific PP2A complexes participating in its two functions. As noted earlier, PP2A catalytic subunits Pph21 and Pph22 function in one of at least three distinct protein complexes: Pph21/22-Tap42, Pph21/22-Tpd3-Cdc55, or Pph21/22-Tpd3-Rts1 (32). The observations that the Tpd3 and combined Cdc55 and Rts1 requirements for DAL5 expression and GATA factor binding were similar to those of the PP2A catalytic subunits themselves suggested the Pph21/22-Tap42 complex did not play a major demonstrable role in these processes. If it had been performing a function that overlapped with the Pph21/22-Tpd3-Cdc55/Rts1 complexes, GATA factor DNA binding and DAL5 expression would have been greater in the tpd3Δ mutant than in the pph21Δ pph22Δ mutant. Pph21/22-Tpd3-Cdc55 participation was greater than that of Pph21/22-Tpd3-Rts1 in Gat1-Myc13 binding, although both complexes were similarly involved in DAL5 expression. However, the complete loss of DAL5 transcription in the cdc55Δ rts1Δ double mutant indicated that Cdc55 and Rts1 might be performing at least partially redundant functions.
The above experiments systematically analyzed the participation of Tor pathway phosphatase PP2A in rapamycin-induced transcription of the NCR-sensitive DAL5 gene, GATA factor binding to its promoter, and Gat1 localization. Comparison of the present results measuring the requirements for PP2A with previous observations investigating Sit4 involvement in these processes (24, 46, 48) revealed that the two phosphatases play quite different roles in DAL5 transcription. Sit4 was partially dispensable for rapamycin-induced DAL5 transcription in glutamine-grown cells. In contrast, deletion of both PPH21 and PPH22 severely impaired DAL5 expression under similar conditions. There was also diversity with respect to GATA factor-specific requirements of PP2A for rapamycin-induced nuclear localization in glutamine-grown cells. As with Sit4, PP2A was partially dispensable for nuclear Gat1 localization (Fig. 3; see Fig. 2 in reference 47). In contrast, both Sit4 and PP2A were strongly required for nuclear Gln3 localization under these conditions (Fig. 1; see Fig. 6 in reference 24 and Fig. 5 in reference 48). What remains unclear is whether the GATA factor-specific differences reflect quantitative differences in common functions associated with Gln3 and Gat1 nuclear localization or pathway-specific differences in the way nuclear localization is regulated.
In sum, the preponderance of our data point to an intranuclear function for Pph21/22-Tpd3-Cdc55/Rts1 phosphatase in the binding of GATA factors to their target sequences in the DAL5 promoter. Even though DAL5 has long been used as a model NCR-sensitive gene, it would be ill advised, in the absence of far broader data involving a variety of NCR-sensitive genes in more than one genetic background, to generalize the phosphatase requirements observed for GATA factor binding to DAL5 as a model of GATA factor binding per se to NCR-sensitive promoters. For example, since GAP1 expression occurs in the absence of either Gat1 or Gln3, and binding of Gln3-Myc13 to the GAP1 promoter does not require Gat1 (22), it would not be surprising for the profiles of GAP1 expression in the various PP2A subunit mutants to be different. Similar arguments are equally applicable to the other NCR-sensitive reporter genes, MEP2, GDH2, and PUT1. Detailed investigations of the cis- and trans-acting factors that function in a range of NCR-sensitive promoters will be required before such generalizations are justified. We have, however, made a solid beginning toward that goal, gaining significant new insights into GATA factor regulation of DAL5 gene expression and clearly demonstrating an intranuclear function for the PP2A phosphatase.
Acknowledgments
We thank Fabienne Vierendeels for excellent technical assistance and Rajendra Rai for suggestions to improve the manuscript.
Work by E.D. and I.G. was supported by the COCOF (Commission Communautaire Française). Work by J.J.T. and T.G.C. was supported by NIH grant GM-35642.
Footnotes
Published ahead of print on 25 October 2010.
REFERENCES
- 1.Beck, T., and M. N. Hall. 1999. The TOR signalling pathway controls nuclear localization of nutrient-regulated transcription factors. Nature 402:689-692. [DOI] [PubMed] [Google Scholar]
- 2.Bertram, P. G., J. H. Choi, J. Carvalho, W. Ai, C. Zeng, T. F. Chan, and X. F. Zheng. 2000. Tripartite regulation of Gln3p by TOR, Ure2p, and phosphatases. J. Biol. Chem. 275:35727-35733. [DOI] [PubMed] [Google Scholar]
- 3.Blinder, D., P. W. Coschigano, and B. Magasanik. 1996. Interaction of the GATA factor Gln3p with the nitrogen regulator Ure2p in Saccharomyces cerevisiae. J. Bacteriol. 178:4734-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cardenas, M. E., N. S. Cutler, M. C. Lorenz, C. J. Di Como, and J. Heitman. 1999. The TOR signaling cascade regulates gene expression in response to nutrients Genes Dev. 13:3271-3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carvalho, J., P. G. Bertram, S. R. Wente, and X. F. Zheng. 2001. Phosphorylation regulates the interaction between Gln3p and the nuclear import factor Srp1p. J. Biol. Chem. 276:25359-25365. [DOI] [PubMed] [Google Scholar]
- 6.Carvalho, J., and X. F. Zheng. 2003. Domains of Gln3p interacting with karyopherins, Ure2p, and the target of rapamycin protein. J. Biol. Chem. 278:16878-16886. [DOI] [PubMed] [Google Scholar]
- 7.Ciuffreda, L., C. Di Sanza, and M. Milella. 12 April 2010, posting date. The mTOR pathway: a new target in cancer therapy. Curr. Cancer Drug Targets. [Epub ahead of print.] [DOI] [PubMed]
- 8.Cooper, T. G. 1982. Nitrogen metabolism in Saccharomyces cerevisiae, p. 39-99. In J. N. Strathern, E. W. Jones, and J. R. Broach (ed.), Molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 9.Cooper, T. G. 2004. Integrated regulation of the nitrogen-carbon interface. Top. Curr. Genet. 7:225-257. [Google Scholar]
- 10.Cox, K. H., A. Kulkarni, J. J. Tate, and T. G. Cooper. 2004. Gln3 phosphorylation and intracellular localization in nutrient limitation and starvation differ from those generated by rapamycin inhibition of Tor1/2 in Saccharomyces cerevisiae. J. Biol. Chem. 279:10270-10278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cox, K. H., R. Rai, M. Distler, J. R. Daugherty, J. A. Coffman, and T. G. Cooper. 2000. Saccharomyces cerevisiae GATA sequences function as TATA elements during nitrogen catabolite repression and when Gln3p is excluded from the nucleus by overproduction of Ure2p. J. Biol. Chem. 275:17611-17618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox, K. H., J. J. Tate, and T. G. Cooper. 2002. Cytoplasmic compartmentation of Gln3 during nitrogen catabolite repression and the mechanism of its nuclear localization during carbon starvation in Saccharomyces cerevisiae. J. Biol. Chem. 277:37559-37566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, K. H., J. J. Tate, and T. G. Cooper. 2004. Actin cytoskeleton is required for nuclear accumulation of Gln3 in response to nitrogen limitation but not rapamycin treatment in Saccharomyces cerevisiae. J. Biol. Chem. 279:19294-19301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crespo, J. L., T. Powers, B. Fowler, and M. N. Hall. 2002. The TOR-controlled transcription activators GLN3, RTG1, and RTG3 are regulated in response to intracellular levels of glutamine. Proc. Natl. Acad. Sci. U. S. A. 99:6784-6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cunningham, T. S., R. Andhare, and T. G. Cooper. 2000. Nitrogen catabolite repression of DAL80 expression depends on the relative levels of Gat1p and Ure2p production in Saccharomyces cerevisiae. J. Biol. Chem. 275:14408-14414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cunningham, T. S., V. V. Svetlov, R. Rai, W. Smart, and T. G. Cooper. 1996. G1n3p is capable of binding to UASNTR elements and activating transcription in Saccharomyces cerevisiae. J. Bacteriol. 178:3470-3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dancey, J. E., R. Curiel, and J. Purvis. 2009. Evaluating temsirolimus activity in multiple tumors: a review of clinical trials. Semin. Oncol. 36(Suppl. 3):S46-S58. [DOI] [PubMed] [Google Scholar]
- 18.De Virgilio, C., and R. Loewith. 2006. Cell growth control: little eukaryotes make big contributions. Oncogene 25:6392-6415. [DOI] [PubMed] [Google Scholar]
- 19.Di Como, C. J., and K. T. Arndt. 1996. Nutrients, via the Tor proteins, stimulate the association of Tap42 with type 2A phosphatases. Genes Dev. 10:1904-1916. [DOI] [PubMed] [Google Scholar]
- 20.Düvel, K., A. Santhanam, S. Garrett, L. Schneper, and J. R. Broach. 2003. Multiple roles of Tap42 in mediating rapamycin-induced transcriptional changes in yeast. Mol. Cell 11:1467-1478. [DOI] [PubMed] [Google Scholar]
- 21.Fingar, D. C., and J. Blenis. 2004. Target of rapamycin (TOR): an integrator of nutrient and growth factor signals and coordinator of cell growth and cell cycle progression. Oncogene 23:3151-3171. [DOI] [PubMed] [Google Scholar]
- 22.Georis, I., A. Feller, J. J. Tate, T. G. Cooper, and E. Dubois. 2009. Nitrogen catabolite repression-sensitive transcription as a readout of Tor pathway regulation: the genetic background, reporter gene and GATA factor assayed determine the outcomes. Genetics 181:861-874. (Erratum, 182:927, 2010.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georis, I., A. Feller, F. Vierendeels, and E. Dubois. 2009. The yeast GATA factor Gat1 occupies a central position in nitrogen catabolite repression-sensitive gene activation. Mol. Cell. Biol. 29:3803-3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Georis, I., J. J. Tate, T. G. Cooper, and E. Dubois. 2008. Tor pathway control of the nitrogen-responsive DAL5 gene bifurcates at the level of Gln3 and Gat1 regulation in Saccharomyces cerevisiae. J. Biol. Chem. 283:8919-8929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardwick, J. S., F. G. Kuruvilla, J. K. Tong, A. F. Shamji, and S. L. Schreiber. 1999. Rapamycin-modulated transcription defines the subset of nutrient-sensitive signaling pathways directly controlled by the Tor proteins. Proc. Natl. Acad. Sci. U. S. A. 96:14866-14870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hirasaki, M., Y. Kaneko, and S. Harashima. 2008. Protein phosphatase Siw14 controls intracellular localization of Gln3 in cooperation with Npr1 kinase in Saccharomyces cerevisiae. Gene 409:34-43. [DOI] [PubMed] [Google Scholar]
- 27.Hirasaki, M., F. Nakamura, K. Yamagishi, M. Numamoto, Y. Shimada, K. Uehashi, S. Muta, M. Sugiyama, Y. Kaneko, S. Kuhara, and S. Harashima. 2010. Deciphering cellular functions of protein phosphatases by comparison of gene expression profiles in Saccharomyces cerevisiae. J. Biosci. Bioeng. 109:433-441. [DOI] [PubMed] [Google Scholar]
- 28.Hofman-Bang, J. 1999. Nitrogen catabolite repression in Saccharomyces cerevisiae. Mol. Biotechnol. 12:35-73. [DOI] [PubMed] [Google Scholar]
- 29.Huynh, H. 3 April 2010, posting date. Molecularly targeted therapy in hepatocellular carcinoma. Biochem. Pharmacol. [Epub ahead of print.] [DOI] [PubMed]
- 30.Inoki, K., and K. L. Guan. 2009. Tuberous sclerosis complex, implication from a rare genetic disease to common cancer treatment. Hum. Mol. Genet. 18:R94-R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jacinto, E., B. Guo, K. T. Arndt, T. Schmelzle, and M. N. Hall. 2001. TIP41 interacts with TAP42 and negatively regulates the TOR signaling pathway. Mol. Cell 8:1017-1026. [DOI] [PubMed] [Google Scholar]
- 32.Jiang, Y. 2006. Regulation of the cell cycle by protein phosphatase 2A in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:440-449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang, Y., and J. R. Broach. 1999. Tor proteins and protein phosphatase 2A reciprocally regulate Tap42 in controlling cell growth in yeast. EMBO J. 18:2782-2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kulkarni, A., A. T. Abul-Hamd, R. Rai, H. El Berry, and T. G. Cooper. 2001. Gln3p nuclear localization and interaction with Ure2p in Saccharomyces cerevisiae. J. Biol. Chem. 276:32136-32144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kulkarni, A., T. D. Buford, R. Rai, and T. G. Cooper. 2006. Differing responses of Gat1 and Gln3 phosphorylation and localization to rapamycin and methionine sulfoximine treatment in Saccharomyces cerevisiae. FEMS Yeast Res. 6:218-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Longtine, M. S., A. McKenzie III, D. J. Demarini, N. G. Shah, A. Wach, A. Brachat, P. Philippsen, and J. R. Pringle. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953-961. [DOI] [PubMed] [Google Scholar]
- 37.Magasanik, B., and C. A. Kaiser. 2002. Nitrogen regulation in Saccharomyces cerevisiae. Gene 290:1-18. [DOI] [PubMed] [Google Scholar]
- 38.Memmott, R. M., and P. A. Dennis. 7 January 2009, posting date. Akt-dependent and -independent mechanisms of mTOR regulation in cancer. Cell Signal. 21:656-664. [Epub ahead of print.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nature Cell Biology. 2009. How robust is your data? New rules for the presentation of statistics. Nature Cell Biol. 11:667. [Google Scholar]
- 40.Puria, R., S. A. Zurita-Martinez, and M. E. Cardenas. 2008. Nuclear translocation of Gln3 in response to nutrient signals requires Golgi-to-endosome trafficking in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U. S. A. 105:7194-7199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rohde, J. R., and M. E. Cardenas. 2004. Nutrient signaling through TOR kinases controls gene expression and cellular differentiation in fungi. Curr. Top. Microbiol. Immunol. 279:53-72. [DOI] [PubMed] [Google Scholar]
- 42.Sarbassov, D. D., S. M. Ali, and D. M. Sabatini. 2005. Growing roles for the mTOR pathway. Curr. Opin. Cell Biol. 17:596-603. [DOI] [PubMed] [Google Scholar]
- 43.Shamji, A. F., F. G. Kuruvilla, and S. L. Schreiber. 2000. Partitioning the transcriptional program induced by rapamycin among the effectors of the Tor proteins. Curr. Biol. 10:1574-1581. [DOI] [PubMed] [Google Scholar]
- 44.Tate, J. J., and T. G. Cooper. 2007. Stress-responsive Gln3 localization in Saccharomyces cerevisiae is separable from and can overwhelm nitrogen source regulation. J. Biol. Chem. 282:18467-18480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tate, J. J., and T. G. Cooper. 2008. Formalin can alter the intracellular localization of some transcription factors in Saccharomyces cerevisiae. FEMS Yeast Res. 8:1223-1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tate, J. J., A. Feller, E. Dubois, and T. G. Cooper. 2006. Saccharomyces cerevisiae Sit4 phosphatase is active irrespective of the nitrogen source provided, and Gln3 phosphorylation levels become nitrogen source-responsive in a sit4-deleted strain. J. Biol. Chem. 281:37980-37992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tate, J. J., I. Georis, E. Dubois, and T. G. Cooper. 2010. Distinct phosphatase requirements and GATA factor responses to nitrogen catabolite repression and rapamycin treatment in Saccharomyces cerevisiae. J. Biol. Chem. 285:17880-17895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tate, J. J., I. Georis, A. Feller, E. Dubois, and T. G. Cooper. 2009. Rapamycin-induced Gln3 dephosphorylation is insufficient for nuclear localization: Sit4 and PP2A phosphatases are regulated and function differently. J. Biol. Chem. 284:2522-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tate, J. J., R. Rai, and T. G. Cooper. 2005. Methionine sulfoximine treatment and carbon starvation elicit Snf1-independent phosphorylation of the transcription activator Gln3 in Saccharomyces cerevisiae. J. Biol. Chem. 280:27195-27204. (Erratum, 282:13139, 2007.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Van Hoof, C., E. Martens, S. Longin, J. Jordens, I. Stevens, V. Janssens, and J. Goris. 2005. Specific interactions of PP2A and PP2A-like phosphatases with the yeast PTPA homologues, Ypa1 and Ypa2. Biochem. J. 386:93-102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 52.Wang, H., X. Wang, and Y. Jiang. 2003. Interaction with Tap42 is required for the essential function of Sit4 and type 2A phosphatases. Mol. Biol. Cell 14:4342-4351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yan, G., X. Shen, and Y. Jiang. 2006. Rapamycin activates Tap42-associated phosphatase by abrogating their association with Tor complex 1. EMBO J. 25:3546-3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zaman, S., S. I. Lippman, X. Zhao, and J. R. Broach. 2009. How Saccharomyces responds to nutrients. Annu. Rev. Genet. 42:2.1-2.55. [DOI] [PubMed] [Google Scholar]
- 55.Zaragoza, D., A. Ghavidel, J. Heitman, and M. C. Schultz. 1998. Rapamycin induces the G0 program of transcriptional repression in yeast by interfering with the TOR signaling pathway. Mol. Cell. Biol. 18:4463-4470. [DOI] [PMC free article] [PubMed] [Google Scholar]