Abstract
Ubiquitination regulates important cellular processes, including the DNA damage response (DDR) and DNA repair. The complexity of the ubiquitin-mediated signals is decoded by ubiquitin receptors, which contain protein modules named ubiquitin binding domains (UBDs). We previously identified a new ubiquitin ligase, RNF168, involved in DDR and endowed with two UBDs named MIU (motif interacting with ubiquitin). Here we have provided the identification of a novel UBD, the UMI (UIM- and MIU-related UBD), present in RNF168, and characterized the interaction surface with ubiquitin, centered on two Leu residues. We have demonstrated that integrity of the UMI, in addition to the MIUs, is necessary for the proper localization of RNF168 and for ubiquitination of nuclear proteins, including histone H2A. Finally, we have shown that simultaneous inactivation of UMI and MIUs prevents the recruitment to DDR foci of the crucial downstream mediator 53BP1.
Posttranslational modifications constitute one of the most versatile and effective intracellular signaling systems adopted by cells to rapidly counteract environmental changes. Among them, ubiquitination has a pivotal role as a master regulator of the most important cellular functions (4).
Ubiquitination is a multistep process involving a ubiquitin (Ub)-activating enzyme (E1) that activates the C terminus of free Ub, which in turn is passed to an E2-conjugating enzyme and finally, with the help of E3 Ub ligase, targets a lysine residue (Lys) of the substrate (21). Notably, since Ub comprises seven Lys (K) residues within its sequence, Ub itself can be a substrate of ubiquitination, resulting in the formation of different types of poly-Ub chains endowed with distinct signaling significance (28, 30). Indeed, while K48 polyubiquitination is involved in targeting proteins for proteasomal degradation, K63 chains are known to regulate a variety of cellular processes, including NF-κB signaling (3), DNA repair (2, 17), endocytosis, and vesicle trafficking (1). The functions of other types of Ub linkages have now being clarified (15).
The reversible covalent attachment of the bulky Ub moiety physically remodels the target protein, conferring new interaction interfaces that can be recognized by protein modules known as Ub binding domains (UBDs) (12). UBDs have the crucial role of decoding the Ub-mediated network, allowing ubiquitinated proteins and Ub receptors (i.e., proteins carrying UBD) to communicate and to translate the Ub signals into specific cellular functions. Recently, the key role of nonproteolytic ubiquitination and the UBD system in processes relevant to the surveillance and maintenance of genome integrity has been emerging (10). Indeed, upon formation of DNA lesions, a number of proteins were found to be ubiquitinated, including proliferating cell nuclear antigen (PCNA), proteins belonging to the Fanconi pathway, and histones H2A and H2AX (2, 17, 19). Of particular interest, ubiquitination assumes a prominent role in the signaling pathway triggered by formation of DNA double strand breaks (DSBs), known as the DNA damage response (DDR). It has been demonstrated that upon formation of DSBs, two Ub ligases, namely, RNF8 and RNF168, ubiquitinate histones H2A and H2AX at the site of damage (6, 11, 14, 16, 22, 26, 27). RNF8 and RNF168 act epistatically in the DNA damage signaling cascade, allowing relaxation of the chromatin structure and recruitment of important downstream effectors, such as 53BP1 and BRCA1. It is noteworthy that the localization of RNF8 to DDR foci is due to upstream phosphorylation events induced by the PI3K-related kinases (PIKKs), such as ATM, while subsequent recruitment of RNF168 follows the first ubiquitination events induced by RNF8 (6, 26). Two UBDs, named MIU1 and MIU2, are important for RNF168 localization and hence for its activity (6, 20, 22, 26). However, the inactivation of these domains dramatically reduces RNF168 recruitment without abolishing it, thus suggesting the involvement of additional mechanisms.
UBDs are structurally heterogeneous motifs and can recognize different types of Ub chains (5, 12). So far, six classes of UBDs have been involved in, although not restricted to, the regulation of DDR: UIM (Ub interacting motif), MIU (motif interacting with Ub), UBA (Ub associated), and UBM (Ub binding motif) present an α-helical structure; UEV (Ub-conjugating enzyme variant) displays the Ub-conjugating (UBC) domain; and UBZ (Ub binding zinc finger) is a zinc finger (10). Here we have identified and characterized in RNF168 a novel UBD that shows similarities to the previously described UIM (23) and MIU (20) domains, and therefore, we called it UMI (UIM- and MIU-related UBD). We found that the isolated UMI domain binds poly-Ub chains, with a preference for the K63 linkage, and we determined the amino acid residues within the UMI sequence crucial for Ub binding. We found that simultaneous inactivation of UMI, MIU1, and MIU2 almost completely abolishes RNF168-dependent formation of K63-specific polyubiquitinated proteins within the nucleus and the ubiquitination of histone H2A. Functionally, we demonstrated that this impairment has dramatic consequences in the signaling cascade activated by DSBs, resulting in the inadequate recruitment of 53BP1 at the DDR foci, which is a critical step for the proper activation of the downstream effectors of DDR.
MATERIALS AND METHODS
Cell culture and DNA transfection.
293T cells were grown in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (Gibco) and 2 mM l-glutamine (Sigma). U2OS cell lines expressing the RNF168-targeting short hairpin RNA (shRNA) in a doxycycline-inducible manner, kindly provided by J. Lukas, were cultured in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% fetal bovine serum (tetracycline free) (EuroClone), 1 μg/ml puromycin (Sigma), and 5 μg/ml blasticidin S (Sigma). Etoposide (Sigma) was used at a concentration of 5 μM for 1 h to induce DSBs. Depletion of endogenous RNF168 was obtained by treating a U2OS derivative cell line with 0.1 μg/ml doxycycline (Sigma) for 72 h (see Fig. S2 in the supplemental material). Cells were transfected either by the calcium phosphate method for biochemical analysis (293T cells) or with FuGENE (Roche) for immunofluorescence (IF) (U2OS cells).
Antibodies and construct design.
Antibodies used in this study included mouse monoclonal anti-Ub P4D1 (Santa Cruz) and FK2 (Stressgen Bioreagents), rabbit polyclonal anti-glutathione S-transferase (anti-GST) (made in-house), mouse monoclonal anti-FLAG (M2; Sigma), rabbit polyclonal anti-FLAG (Sigma), mouse monoclonal anti-ubiquityl-histone H2A (Upstate), and anti-phospho-histone H2AX (Ser139; Upstate). The linkage-specific antibody directed to K63 (Apu3.A8) was from Genentec. Mouse anti-53BP1 was a gift from T. Halazonetis. Anti-RNF168 polyclonal antibodies were raised against two different GST-RNF168 constructs, comprising the following regions: amino acid 1 to 191 (RF-MIU1) and 439 to 571 (MIU2-C-term); rabbit immunization was performed by Yorkshire Bioscience (United Kingdom), and the purified antibodies were tested as described in Fig. S2C and D in the supplemental material.
The cDNAs of human full-length RNF168 wild type and MIU mutants were obtained as described previously (22). The point mutations in the UMI domain were introduced by site-directed mutagenesis using the following nucleotides: forward, GAAGAATACATACAGAGGGCAGCAGCAGAGGAGGAAGAAGAG, and reverse, CTCTTCTTCCTCCTCTGCTGCTGCCCTCTGTATGTATTCTTC. The truncated forms of RNF168 constructs were generated by PCR amplification followed by cloning into the pGEX6P2 vector. The oligonucleotide sequences are available upon request. The constructs were made resistant to shRNA by site-directed mutagenesis using the following oligonucleotides: forward, AGACAGGCAGAAAAAAGAAGGCGAGCGATGGAAGAACAAC, and reverse, GTTGTTCTTCCATCGCTCGCCTTCTTTTTTCTGCCTGTCT. All the constructs were sequence verified.
GST pulldown assays.
Recombinant proteins were expressed in Escherichia coli strain BL21 pLys with a glutathione S-transferase (GST) tag and purified as previously described (14). Chromatin was extracted from 293T cells, treated or not with etoposide (20 μM), by resuspending cells in phosphate-buffered saline (PBS) buffer with inhibitors (1 mM phenylmethylsulfonylfluoride [PMSF], protease inhibitor cocktail [Sigma], 20 mM sodium pyruvate, 50 mM NaF, 1 mM Na3PO4, 20 μM N-ethylmalmeimide (NEM), 80 U/ml Benzonase), and clarified by centrifugation at 3,500 rpm for 5 min at 4°C. Pellet was lysed with lysis buffer (50 mM Tris-HCl [pH 8.1], 10 mM EDTA, 1% Triton, supplemented with inhibitors), incubated on ice for 10 min, and sonicated to shear DNA at 80% amplitude, two 10-s impulses, 20-s pauses, on ice. The shared chromatin was centrifuged at 14,000 rpm at 4°C for 30 min. For Ub chain pulldown, 1 μM GST fusion proteins, immobilized on glutathione (GSH) beads, were incubated with 0.5 μg of K48- or K63-linked poly-Ub2-7 chains for 1 h at 4°C in a buffer containing 50 mM HEPES (pH 7.5), 150 mM NaCl, 10% glycerol, and 0.5% Triton X-100. Proteins were separated by SDS-PAGE (15%) and transferred onto polyvinylidene difluoride (PVDF) membranes (Sigma). Membranes were denatured using a solution containing 6 M guanidine hydrochloride, 20 mM Tris-HCl (pH 7.4), 1 mM PMSF, and 5 μM β-mercaptoethanol for 30 min at 4°C. After extensive washing in Tris-buffered saline (TBS) buffer, membranes were blocked in TBS buffer containing bovine serum albumin (BSA) (5%) overnight and then incubated with anti-Ub P4D1 antibody (Santa Cruz) for 1 h. For chromatin binding, 10 μg of GST fusion proteins immobilized on GSH beads were incubated with 1 mg of the chromatin extracts for 2 h at 4°C. Bound proteins were resolved by SDS-PAGE (15%), transferred to nitrocellulose, and analyzed by immunoblotting as indicated.
In vivo ubiquitination assay.
Transfected 293T cells were collected in PBS containing protease inhibitor cocktail (Sigma), 1 mM PMSF, and 20 μM NEM. One-tenth of the samples were separately processed for protein normalization, while the remaining cell pellets were subjected to acid extraction of histones as previously described (22). Samples were resolved by SDS-PAGE, transferred onto PVDF membranes (Sigma), and detected with the indicated antibodies.
In vitro ubiquitination assay.
Five micrograms of purified GST-RNF168 constructs were incubated with 0.1 μg human recombinant E1 Ub-activating enzyme (Boston Biochem), 200 ng of purified Ubc13-Mms2 complex (provided by E. Maspero, IFOM, Milan, Italy), 2 μg of Ub (made in-house) in 25 mM Tris-HCl, pH 7.4, 5 mM MgCl2, 100 mM NaCl, 1 μM dithiothreitol (DTT), and 2 mM ATP at 30°C for 1.5 h. The reaction was stopped by boiling in Laemmli buffer. Ubiquitination was detected by anti-Ub (P4D1) immunoblotting.
Immunofluorescence analysis.
Twenty-four hours after transfection, U2OS cells were fixed in 4% paraformaldehyde, permeabilized with a solution of 0.5% Triton X-100 in 0.2% bovine serum albumin (BSA) for 5 min at room temperature, and blocked with PBG (PBS, 0.5% BSA, and 0.2% gelatin) for 1 h. Coverslips were incubated for 1 h with a primary antibody and, after extensive washing, incubated with the appropriate secondary antibody (Alexa Fluor 488 goat anti-mouse or anti-rabbit IgG, Alexa Fluor 546 goat anti-mouse IgG, or anti-rabbit IgG; Invitrogen) for 30 min at room temperature. Images were acquired by confocal scanning laser microscopy (Leica TCS2; Leica Lasertechnik, Heidelberg, Germany).
RESULTS
Identification of a novel Ub binding region in RNF168.
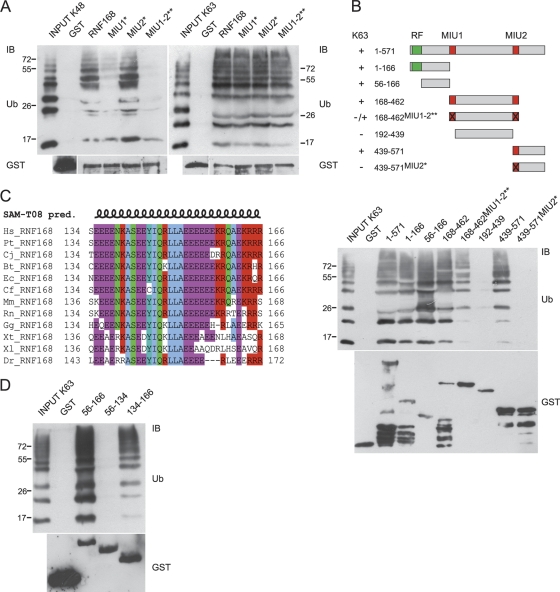
We previously found that RNF168 is endowed with two UBDs named MIU (motif interacting with Ub), and we demonstrated that isolated MIU1 (amino acids 168 to 191) and MIU2 (amino acids 439 to 462) are able to interact with both K48 and K63 Ub chains (20). The integrity of the MIU domains is required for the binding of full-length RNF168 to K48 Ub chains, for its appropriate localization to the site of genomic lesions, and for the correct assembly of the DDR foci (6, 22, 26). Hence, due to the importance of the MIU domains in RNF168 function, we further investigated its Ub binding ability. By in vitro pulldown experiments, we found that MIU1 plays a prominent role in RNF168 binding to K48 Ub chains. In fact, MIU1 inactivation by point mutation (A179G; MIU1*) strongly affected K48 Ub chain binding, while inactivation of MIU2 (A450G; MIU2*) resulted in only a slight reduction (Fig. 1A, left panel). This result is in accordance with our previous finding revealing that MIU1 is more efficient in binding K48 Ub chains than MIU2 (20). Consistently, the double mutation affecting integrity of the MIU domains (A179G A450G; MIU1-2**) almost completely abolished Ub binding.
FIG. 1.
Identification of a new Ub binding region in RNF168. (A) We performed an in vitro pulldown assay using the indicated GST-tagged RNF168 constructs. GST fusion proteins were incubated with synthetic K48-linked (left panel) or K63-linked (right panel) poly-Ub2-7 chains and separated by SDS-PAGE. Immunoblotting (IB) was performed with antibodies directed against Ub and GST, as described in Materials and Methods. (B) Schematic representation of RNF168 deletion constructs used in pulldown experiments (numbers refer to the amino acid positions within the sequence; RF, RING finger domain); their ability to bind K63 poly-Ub chains, resumed on the left, is shown in the anti-Ub immunoblot of the in vitro pulldown assay (lower panel). Normalization is visualized by anti-GST immunoblotting. (C) Multiple alignments of region 134 to 166 RNF168 homologues in vertebrates. Secondary structure prediction (pred.) was obtained using SAM-T08, a hidden Markov model (HMM)-based protein structure prediction software program (http://compbio.soe.ucsc.edu/SAM_T08/T08-query.html). (D) Mapping of the minimal sequence responsible for Ub binding. An in vitro pulldown assay was performed using the indicated GST-tagged deletion mutants of RNF168, incubated with K63 poly-Ub chains. IB was performed with anti-Ub and anti-GST antibodies.
K48 ubiquitination is generally considered a signal for proteasomal degradation, while other types of poly-Ub chains target proteins to different fates. In particular, since K63 ubiquitination is a signaling device largely used in DNA damage response and repair (9, 13, 25), we asked whether RNF168 shows the same specificity for binding to K48- and K63-linked Ub chains. Surprisingly, we found that the mutant MIU1-2** still interacts with K63 Ub chains, unveiling the existence of an additional Ub binding region within the protein, which shows preferential binding to the K63 linkage compared to the K48 one (Fig. 1A, right panel).
Therefore, to map the region involved in binding to K63 poly-Ub chains, we used a panel of deletion mutants encompassing the whole protein, in combination with point mutations addressing MIU1 and MIU2, summarized in Fig. 1B. We found that the sequence between amino acids 56 and 166, which does not contain any obvious UBD, interacts with K63 poly-Ub chains. Secondary structure prediction of this sequence highlighted the presence of a coiled-coil motif (amino acids 115 to 166). Within this sequence, a putative amphipathic α-helix is present (amino acids 134 to 166), highly conserved throughout evolution (Fig. 1C). Thus, we asked whether this short sequence was responsible for the binding to Ub chains. Indeed, we found that the region including amino acids 134 to 166 retained Ub binding ability, while the adjacent region (amino acids 56 to 134) did not (Fig. 1D). Interestingly, we observed that this isolated region also binds to K48 poly-Ub chains, although to a lesser extent (see Fig. S1 in the supplemental material), suggesting that Ub chain specificity might be influenced by the surrounding sequence.
Our bioinformatic analysis suggested an amino acid spatial distribution that is reminiscent of the amphipathic α-helix in the UIM/MIU motifs (Fig. 2A), with Y145, I146, L149, and L150 providing the core of the hydrophobic side, while E143, E144, Q147, and R148 make up the hydrophilic side (Fig. 2A). Consistently, a single substitution of each of the two leucine residues at position 149 and 150 almost abrogated the Ub binding of the domain (Fig. 2B). In contrast, mutation of alanine 151 (marked with an asterisk in Fig. 2A), crucial for the function of the MIU domain (20), did not exert a major effect on the interaction with Ub chains, as predicted by its positioning in the hydrophilic side of the helix in our model (Fig. 2B). Due to the similarity with the UIM and MIU domains, we named it UMI (UIM- and MIU-related Ub binding domain).
FIG. 2.
Mapping of the amino acid residues required for Ub binding. (A) Comparison of the UMI, UIM, and MIU motifs, where residues involved in the binding to Ub are in bold; underlined sequences were drawn as projected helices using the Helical Wheel Projections software tool (http://rzlab.ucr.edu/scripts/wheel/wheel.cgi). (B) The point mutants were generated by introduction of single (A151G, L149A, or L150A) or double (L149A and L150A) amino acid substitutions addressing the sequence 134 to 166 and tested by in vitro pulldown assay as in Fig. 1D. (C) The indicated RNF168 point mutations were inserted within the sequence encompassing amino acids 56 to 571 and analyzed by in vitro pulldown assay using K48 (left panel) and K63 (right panel) poly-Ub chains. Anti-Ub and anti-GST immunoblots are shown.
To better evaluate the Ub linkage specificity, we analyzed the effect of the point mutations addressing UMI and MIU domains on the binding to K63 and K48 poly-Ub chains. Since we observed that the N terminus of the protein displayed a residual Ub binding that is currently under investigation, we introduced the point mutations in a sequence deleted of this region (amino acids 56 to 571). As expected, the mutant that was simultaneously defective in UMI, MIU1, and MIU2 (UMI*MIU1-2**) was unable to interact with K48 and interacted poorly with K63 Ub chains, indicating that there are no additional UBDs in this region (Fig. 2C). Oddly, we found that the 56-to-571 mutant (MIU1-2**) still interacted with K48 Ub chains, in contrast to what was observed in the case of the full-length protein (Fig. 1A). This result, in agreement with other data obtained in the lab, suggests that the region encompassing the RING finger of RNF168 might influence the Ub binding specificity by limiting the type of Ub chain that can access the UMI domain.
The proper localization of RNF168 relies on the integrity of the UMI domain.
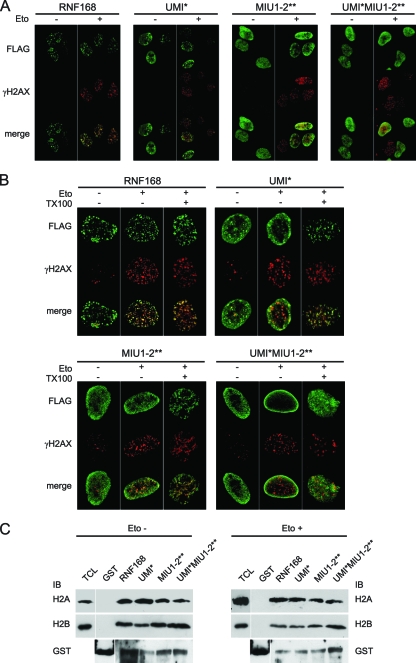
Localization of DDR proteins at the site of DNA damage is mandatory for the full activation of the cellular response aimed at counteracting the formation of harmful DNA DSBs (7). Proper subcellular localization of RNF168 largely depends on the integrity of the MIU domains, with a pivotal role of MIU2 (22, 26). In fact, inactivation of MIU2 by point mutation (A450G) highly impairs the accumulation of RNF168 at the DSBs upon etoposide treatment. Nevertheless, we previously showed that such impairment is not complete, since a small population of MIU1-2** still localizes at DDR foci (22), suggesting a possible role of other domains in RNF168 localization. To investigate the role of the UMI domain in RNF168 localization, we adopted experimental conditions where the expression of endogenous protein is abrogated, by using U2OS cells conditionally expressing RNF168-targeting shRNA (6) (see Fig. S2A in the supplemental material); here, we introduced the shRNA-resistant version of the different RNF168 constructs (see Fig. S2B in the supplemental material). We observed that inactivation of the UMI by Leu-to-Ala substitution at positions 149 and 150 (UMI*) partially altered the localization of RNF168, resulting in a more diffuse localization compared to the wild-type protein (Fig. 3A). A more dramatic effect is obtained by replacing the endogenous protein with the construct carrying simultaneously the UMI, MIU1, and MIU2 inactivating mutations (UMI*MIU1-2**). In this case, the protein displayed a diffuse nuclear pattern, and it failed to localize at the DSB foci even in the presence of DNA damage (Fig. 3A). We previously showed that a significant population of RNF168 still localized to DDR foci even when MIUs were inactivated (22), suggesting the existence of a MIU-independent mechanism for RNF168 recruitment to DDR foci. Thus, we adopted the same approach to clarify if the simultaneous inactivation of the three UBDs further affects localization of the protein at the sites of DNA lesion. Indeed, we found that the triple mutant (UMI*MIU1-2**) was unable to relocate to the DDR foci, although it still resided in detergent-resistant chromatin structures (Fig. 3B, right panels). This result is quite surprising, since we expected the mutant UMI*MIU1-2** to be soluble upon detergent treatment. To investigate this point, we performed a GST pulldown using chromatin extracts to evaluate the ability of the mutants to interact with chromatin (Fig. 3C). Consistent with the IF data, we found that all of RNF168 mutants, including UMI*MIU1-2**, retained the ability to bind histones at comparable levels, either in damaged or undamaged cells. These results indicate that in addition to the UBD-dependent binding of RNF168 to ubiquitinated histone H2A upon DNA lesions (26), there are additional sites on RNF168 for binding to chromatin.
FIG. 3.
Inactivation of the UMI domain affects localization of RNF168. (A) U2OS cells conditionally expressing RNF168-targeting shRNA were transfected with the indicated FLAG-tagged shRNA-resistant constructs. After 24 h, cells were treated with etoposide for 1 h or left untreated (Eto + and −, respectively). (B) The U2OS cells, treated as described for panel A, were either fixed (TX100 −) or pretreated with Triton X-100 before fixing (TX100 +). Immunostaining of both panels was performed using anti-FLAG and anti-phospho-H2AX (γH2AX) antibodies. (C) A Pulldown assay was performed using GST-RNF168 constructs and chromatin extracted from 293T cells treated or not with etoposide. H2A and H2B antibodies (upper and lower panels, respectively) revealed the presence of specific bands (of about 15 kDa) in all RNF168 constructs but not in the lane with GST alone.
RNF168-induced ubiquitination is impaired in cells expressing the UMI-defective mutant.
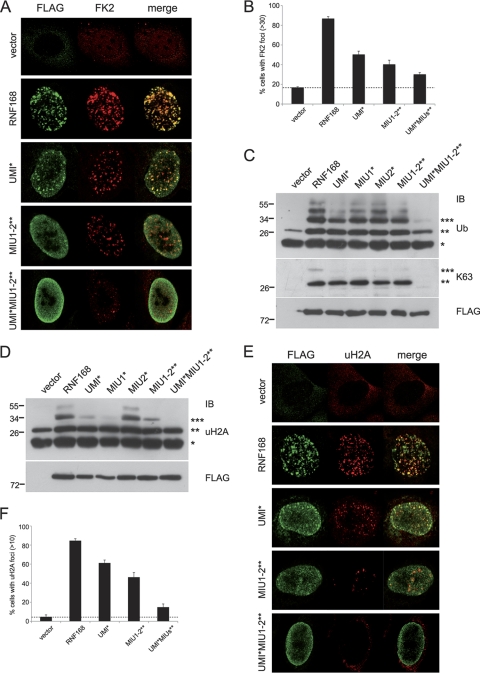
We and others previously demonstrated that RNF168 is a nuclear E3 Ub ligase which induces in vitro ubiquitination of histones H2A and H2AX in a RING finger-dependent manner (6, 22, 26). In addition, an important role in in vivo ubiquitination activity has also been ascribed to the two MIU domains. Indeed, inactivation of MIU1 and MIU2 caused a 70% reduction in the amount of ubiquitinated proteins at the DDR foci compared to that for the wild-type protein (6). Thus, we asked whether inactivation of the UMI domain could also impair the activity of RNF168 as a consequence of the altered localization. By using the U2OS RNF168-targeting shRNA cells complemented with the shRNA-resistant constructs of either wild-type RNF168 or the different mutants, we found that the sole inactivation of the UMI domain reduced the number of Ub-positive foci in the nucleus (UMI*), as revealed by the use of anti-Ub antibody (FK2) that recognized Ub only when conjugated in chains (Fig. 4A and B). This resembles what was observed with the MIU domain mutant (MIU1-2**). It is noteworthy that inactivation of the UMI domain in the context of a MIU-defective protein completely abolished the formation of Ub conjugates at the DSBs (UMI*MIU1-2**; Fig. 4A and B). The same results were recapitulated by the use of the K63-linkage-specific anti-Ub antibody (Apu3.A8) (18), as shown in Fig. S3 in the supplemental material. Furthermore, we addressed the ubiquitination status of chromatin-bound proteins by performing acidic extraction of nuclear components derived from cells ectopically expressing the wild type and different RNF168 mutants (Fig. 4C). In the presence of the wild-type protein, anti-Ub immunoblotting revealed the formation of a ladder of about five bands, which differ in molecular mass, reminiscent of the multiple ubiquitinated forms of histone H2A. Notably, inactivation of the three UBDs (UMI*MIU1-2**) prevented the formation of these ubiquitinated proteins, similar to what was observed in cells transfected with the empty vector. Apu3.A8 anti-Ub immunoblotting showed that expression of wild-type RNF168 induces formation of K63-ubiquitinated proteins with molecular masses of about 30 and 38 kDa (the latter with lower intensity), compatible with the di- and triubiquitinated forms of histone H2A. Inactivation of any single UBD (UMI*, MIU1*, and MIU2*) and of the double MIU mutant (MIU1-2**) still induced the formation of the 30-kDa band but not that of the higher-molecular mass band (Fig. 4C). Strikingly, the UMI*MIU1-2** mutant failed to form any K63-linked ubiquitinated proteins, strongly indicating that inactivation of the sole MIU domains is not enough to abolish RNF168 Ub ligase activity but that the concurrent inactivation of the UMI is strictly required. Interestingly, the decreased ubiquitinating activity of the mutants is not ascribable to a reduced intrinsic Ub ligase activity, since we performed an in vitro ubiquitination assay showing that the activity of the RNF168 mutants is comparable to that of the wild-type protein (see Fig. S4).
FIG. 4.
UMI, MIU1, and MIU2 are required for the ubiquitination events mediated by RNF168. (A) U2OS shRNF168 cells transfected either with the indicated FLAG-tagged shRNA-resistant constructs or with the vector alone were treated with etoposide for 1 h before fixing and immunostained using anti-FLAG and anti-Ub (FK2) antibodies. (B) Quantitation of RNF168-positive cells with more than 30 foci labeled with anti-FK2. (C) Acid extraction of histones from 293T cells transfected with the indicated FLAG-tagged RNF168 constructs. Ubiquitinated proteins were detected by immunoblotting using P4D1 (Ub; upper panel) and Apu3.A8 (K63; medium panel) antibodies. Cell extracts were analyzed for equal expression of the different constructs (FLAG; lower panel). (D) 293T cells were processed as described for panel C and immunodecorated with antibody directed to ubiquitinated forms of histone H2A (uH2A). Cell loading was normalized by FLAG immunoblotting. (E) Immunofluorescence analysis was performed with anti-FLAG and with anti-uH2A antibodies. (F) Quantitation of RNF168-positive cells with more than 10 foci labeled with anti-Ub.
Inactivation of UMI affects in vivo histone ubiquitination.
Histones H2A and H2AX are the sole RNF168 substrates identified at the moment. With the purpose of verifying the role of the new UBD in histone ubiquitination, we performed a biochemical analysis of chromatin histones using an antibody that specifically recognizes the ubiquitinated forms of histone H2A (uH2A) (Fig. 4D). Anti-uH2A immunoblotting revealed that inactivation of UMI and of MIU1, reduced the level of histone H2A ubiquitination induced by ectopical expression of RNF168, with a failure to form the signal corresponding to triubiquitinated histone H2A present in the wild-type protein and markedly reducing the diubiquitinated form. Simultaneous inactivation of the three UBDs (UMI*MIU1-2**) completely abolished both di- and triubiquitination of H2A, with a level of ubiquitination comparable to that of cells transfected with the empty vector alone (Fig. 4D). Interestingly, inactivation of the sole MIU2 did not have a significant effect despite its major role in the localization of RNF168 at DDR foci. We confirmed these results by immunofluorescence analysis, where we used anti-uH2A antibody to detect the amount of ubiquitinated histone H2A in U2OS (shRNF168) cells complemented with RNF168 mutants (Fig. 4E and F).
Overall, these results clearly indicated that the UMI domain has a pivotal role in ensuring proper localization of RNF168, which is instrumental for the execution of its full Ub ligase activity on physiological substrates, which are histones.
Integrity of the UMI domain is required for the proper recruitment of downstream signaling proteins.
The ubiquitination events driven by RNF8/RNF168 are strictly required for the proper execution of the DDR program. It has been demonstrated that the depletion of RNF168, either by naturally occurring mutations (26) or by using the U2OS RNF168-targeting shRNA (6), prevents the recruitment of 53BP1 to DDR foci, thereby affecting the downstream signaling events. Relevantly, such an effect may be reverted by reintroducing the wild-type form of RNF168.
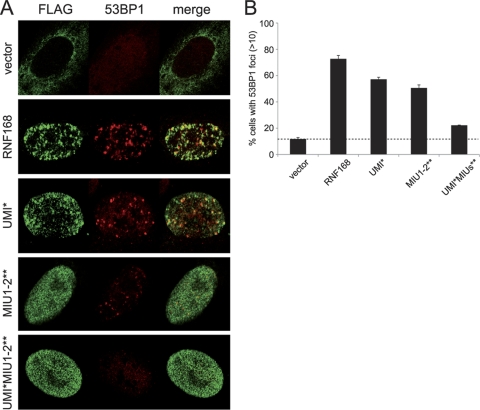
The results obtained, supporting the functional relevance of the UMI domain in the localization and activity of RNF168, prompted us to test whether this domain is also required for the recruitment of 53BP1 to DDR foci. To this purpose, we adopted the U2OS cell system, previously used in other experiments, to analyze the abilities of the different RNF168 mutants with the lack of endogenous protein complemented. In accordance with what has already been described in the literature (26), we found that upon DNA damage, the reintroduction of a construct carrying both MIU1 and MIU2 inactivating mutations (MIU1-2**) restores 53BP1 localization only partially (Fig. 5A and quantitation in Fig. 5B). Similar results were obtained upon expression of the UMI* mutant. Strikingly, the simultaneous inactivation of UMI and MIUs in RNF168 (UMI*MIU1-2**) generates a protein that is almost completely unable to complement the phenotype (Fig. 5A and B), with an effect comparable to that obtained in the absence of the protein.
FIG. 5.
Inactivation of RNF168 UBDs abolishes 53BP1 recruitment to DDR foci. (A) The U2OS cells conditionally expressing RNF168-targeting shRNA were transfected with the indicated FLAG-tagged shRNA-resistant RNF168 constructs. Twenty-four hours after transfection, cells were treated with etoposide for 1 h, fixed, and immunostained with anti-FLAG and anti-53BP1 antibodies. (B) Quantitation of the two independent experiments described for panel A, representing the percentage of transfected cells with 53BP1-positive foci.
DISCUSSION
UBDs have a crucial role in Ub-mediated events by determining localization, activity, and interaction partners of proteins. We previously identified and characterized a new family of UBDs, named MIU, present in a number of proteins with unrelated functions, such as the guanine nucleotide exchange factor Rabex 5, the epidermal growth factor (EGF)-induced phosphoprotein Ymer, Myosin 6, and the RING finger protein RNF168 (20). Further, we demonstrated that the two isolated MIUs of RNF168 are able to interact with Ub chains and that their inactivation by point mutation impedes interaction with K48-linked Ub chains.
Here we report the identification of a novel UBD in RNF168, called UMI, which manifests a preference for binding to K63-linked poly-Ub chains compared to K48-linked ones. At the sequence level, UMI shows similarity to the previously characterized UIM and MIU, being a short amphipathic α-helix with adjacent glutamate-rich regions at both the N- and C termini. Interestingly, the interaction surface of the UMI with Ub is centered on two Leu residues (L149 and L150), which are expected, for the predicted positioning of amino acid residues on the helix, to be exposed to the hydrophobic part of the helix. On the contrary, the Ala residue, crucial for UIM and MIU activity (20), is embedded in the hydrophilic side of the helix and does not participate in the Ub binding ability of UMI.
The MIU domains of RNF168 bind ubiquitinated histone H2A, and this event accounts for the proper localization of RNF168 at DDR foci and for the formation of RNF168-dependent ubiquitinated proteins, including histones, within the nucleus. In line with this, the MIU-defective mutant impairs 53BP1 recruitment at the site of lesions (26) without abolishing it. These data indicate the existence of other mechanisms responsible for RNF168 recruitment to DDR foci, in addition to the one mediated by the MIU domains.
Now we have shown that integrity of the new UMI domain is largely required for RNF168 function, since the simultaneous inactivation of the three UBDs generates a protein unable to localize at DDR foci (Fig. 3A and B), to induce K63-specific polyubiquitination of proteins (Fig. 4C; see also Fig. S3 in the supplemental material), including ubiquitination of histone H2A (Fig. 4D, E, and F), and to allow the recruitment of 53BP1 (Fig. 5).
The need for three different UBDs for the proper activity of RNF168 can be explained in different, not mutually exclusive, ways. The first invokes the “avidity effect” (12), with the three UBDs collaborating to ensure stable binding of RNF168 to chromatin at the site of DNA damage, thereby allowing sustained ubiquitination of histones. The second regards the functions of the three UBDs in regulating RNF168 activity that have to be considered separately. In this respect, we can envision the possibility that the UMI binds to a different, yet unidentified, substrate and that the contemporary binding of RNF168 to histones H2A and to this unknown substrate may be mandatory for recruitment/stability of the protein to DNA damage sites.
The third model relies on two different data: (i) inactivation of UMI and MIU1 affects in vivo ubiquitination of histone H2A (Fig. 4D), while MIU2 is mainly involved in the proper localization of RNF168 at DDR foci (22, 26); (ii) in cells, RNF168 self-associates to form dimers/oligomers (our unpublished data). In addition, it is well established that upon formation of DSBs, a vast area surrounding the damaged chromosome undergoes massive phosphorylation and ubiquitination (24). How this event occurs is still under investigation. Here, we hypothesize that RNF168 MIU domains, and MIU2 in particular, might help in keeping the dynamic status of RNF168-uH2A association, thereby allowing the protein to “walk” along the ubiquitinated histones. In such a way, RNF168 would be able to propagate the damage signal by stepping from one nucleosome to the other. In this scenario, due to the contiguity of UMI and MIU1 and their proximity with the RING finger domain, we hypothesize that UMI and MIU1 might facilitate the local polyubiquitination of histones by transiently sustaining the binding of RNF168 to the growing Ub chain appended to uH2A.
Finally, a speculative explanation takes into consideration the ubiquitination status of the protein. We observed that in cells, RNF168 is ubiquitinated (our unpublished data), although the meaning of such modification is still elusive. It has been described that a number of UBDs, including UIM and MIU, sustain the so-called “coupled monoubiquitination” (8, 29), a process in which ubiquitination of a target protein is driven by its own UBD through an unclear mechanism. It will be of interest to investigate the role of UMI, MIU1, and MIU2 in RNF168 ubiquitination to verify whether the UBDs are important not only for their intrinsic Ub binding ability but also because they affect RNF168 function by regulating its own ubiquitination status.
In conclusion, our results give new insights into the complex Ub-mediated regulation of RNF168 activation. Nevertheless, much more needs to be done to characterize in more detail how a single UBD regulates RNF168 localization and function. This will necessarily involve the identification of additional interacting partners, substrates, and regulators of RNF168.
Supplementary Material
Acknowledgments
We thank Fabrizio Condorelli for reading the manuscript and Grant Stewart for discussion. We are grateful to Jiri Lukas for kindly providing U2OS cells conditionally expressing RNF168-targeting shRNA and to Genentec for providing antibodies specific for K63 poly-Ub chains.
L.P. designed research, S.P. and M.G. performed research, C.S. contributed new reagents/analytical tools, S.C. performed sequence analysis, and S.P. and L.P. wrote the article.
This work was supported by grants from Associazione Italiana per la Ricerca sul Cancro and Regione Piemonte RSF. M.G. is supported by a Lagrange fellowship.
Footnotes
Published ahead of print on 1 November 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Acconcia, F., S. Sigismund, and S. Polo. 2009. Ubiquitin in trafficking: the network at work. Exp. Cell Res. 315:1610-1618. [DOI] [PubMed] [Google Scholar]
- 2.Bergink, S., and S. Jentsch. 2009. Principles of ubiquitin and SUMO modifications in DNA repair. Nature 458:461-467. [DOI] [PubMed] [Google Scholar]
- 3.Bhoj, V. G., and Z. J. Chen. 2009. Ubiquitylation in innate and adaptive immunity. Nature 458:430-437. [DOI] [PubMed] [Google Scholar]
- 4.Chen, Z. J., and L. J. Sun. 2009. Nonproteolytic functions of ubiquitin in cell signaling. Mol. Cell 33:275-286. [DOI] [PubMed] [Google Scholar]
- 5.Dikic, I., S. Wakatsuki, and K. J. Walters. 2009. Ubiquitin-binding domains—from structures to functions. Nat. Rev. Mol. Cell Biol. 10:659-671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doil, C., N. Mailand, S. Bekker-Jensen, P. Menard, D. H. Larsen, R. Pepperkok, J. Ellenberg, S. Panier, D. Durocher, J. Bartek, J. Lukas, and C. Lukas. 2009. RNF168 binds and amplifies ubiquitin conjugates on damaged chromosomes to allow accumulation of repair proteins. Cell 136:435-446. [DOI] [PubMed] [Google Scholar]
- 7.Harper, J. W., and S. J. Elledge. 2007. The DNA damage response: ten years after. Mol. Cell 28:739-745. [DOI] [PubMed] [Google Scholar]
- 8.Hicke, L., H. L. Schubert, and C. P. Hill. 2005. Ubiquitin-binding domains. Nat. Rev. Mol. Cell Biol. 6:610-621. [DOI] [PubMed] [Google Scholar]
- 9.Hoege, C., B. Pfander, G. L. Moldovan, G. Pyrowolakis, and S. Jentsch. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419:135-141. [DOI] [PubMed] [Google Scholar]
- 10.Hofmann, K. 2009. Ubiquitin-binding domains and their role in the DNA damage response. DNA Repair (Amsterdam) 8:544-556. [DOI] [PubMed] [Google Scholar]
- 11.Huen, M. S., R. Grant, I. Manke, K. Minn, X. Yu, M. B. Yaffe, and J. Chen. 2007. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 131:901-914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hurley, J. H., S. Lee, and G. Prag. 2006. Ubiquitin-binding domains. Biochem. J. 399:361-372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, H., J. Chen, and X. Yu. 2007. Ubiquitin-binding protein RAP80 mediates BRCA1-dependent DNA damage response. Science 316:1202-1205. [DOI] [PubMed] [Google Scholar]
- 14.Kolas, N. K., J. R. Chapman, S. Nakada, J. Ylanko, R. Chahwan, F. D. Sweeney, S. Panier, M. Mendez, J. Wildenhain, T. M. Thomson, L. Pelletier, S. P. Jackson, and D. Durocher. 2007. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 318:1637-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Komander, D. 2009. The emerging complexity of protein ubiquitination. Biochem. Soc. Trans. 37:937-953. [DOI] [PubMed] [Google Scholar]
- 16.Mailand, N., S. Bekker-Jensen, H. Faustrup, F. Melander, J. Bartek, C. Lukas, and J. Lukas. 2007. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 131:887-900. [DOI] [PubMed] [Google Scholar]
- 17.Messick, T. E., and R. A. Greenberg. 2009. The ubiquitin landscape at DNA double-strand breaks. J. Cell Biol. 187:319-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newton, K., M. L. Matsumoto, I. E. Wertz, D. S. Kirkpatrick, J. R. Lill, J. Tan, D. Dugger, N. Gordon, S. S. Sidhu, F. A. Fellouse, L. Komuves, D. M. French, R. E. Ferrando, C. Lam, D. Compaan, C. Yu, I. Bosanac, S. G. Hymowitz, R. F. Kelley, and V. M. Dixit. 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell 134:668-678. [DOI] [PubMed] [Google Scholar]
- 19.Panier, S., and D. Durocher. 2009. Regulatory ubiquitylation in response to DNA double-strand breaks. DNA Repair (Amsterdam) 8:436-443. [DOI] [PubMed] [Google Scholar]
- 20.Penengo, L., M. Mapelli, A. G. Murachelli, S. Confalonieri, L. Magri, A. Musacchio, P. P. Di Fiore, S. Polo, and T. R. Schneider. 2006. Crystal structure of the ubiquitin binding domains of rabex-5 reveals two modes of interaction with ubiquitin. Cell 124:1183-1195. [DOI] [PubMed] [Google Scholar]
- 21.Pickart, C. M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503-533. [DOI] [PubMed] [Google Scholar]
- 22.Pinato, S., C. Scandiuzzi, N. Arnaudo, E. Citterio, G. Gaudino, and L. Penengo. 2009. RNF168, a new RING finger, MIU-containing protein that modifies chromatin by ubiquitination of histones H2A and H2AX. BMC Mol. Biol. 10:55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polo, S., S. Sigismund, M. Faretta, M. Guidi, M. R. Capua, G. Bossi, H. Chen, P. De Camilli, and P. P. Di Fiore. 2002. A single motif responsible for ubiquitin recognition and monoubiquitination in endocytic proteins. Nature 416:451-455. [DOI] [PubMed] [Google Scholar]
- 24.Rogakou, E. P., C. Boon, C. Redon, and W. M. Bonner. 1999. Megabase chromatin domains involved in DNA double-strand breaks in vivo. J. Cell Biol. 146:905-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sobhian, B., G. Shao, D. R. Lilli, A. C. Culhane, L. A. Moreau, B. Xia, D. M. Livingston, and R. A. Greenberg. 2007. RAP80 targets BRCA1 to specific ubiquitin structures at DNA damage sites. Science 316:1198-1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stewart, G. S., S. Panier, K. Townsend, A. K. Al-Hakim, N. K. Kolas, E. S. Miller, S. Nakada, J. Ylanko, S. Olivarius, M. Mendez, C. Oldreive, J. Wildenhain, A. Tagliaferro, L. Pelletier, N. Taubenheim, A. Durandy, P. J. Byrd, T. Stankovic, A. M. Taylor, and D. Durocher. 2009. The RIDDLE syndrome protein mediates a ubiquitin-dependent signaling cascade at sites of DNA damage. Cell 136:420-434. [DOI] [PubMed] [Google Scholar]
- 27.Wang, B., and S. J. Elledge. 2007. Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc. Natl. Acad. Sci. U. S. A. 104:20759-20763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Winget, J. M., and T. Mayor. 2010. The diversity of ubiquitin recognition: hot spots and varied specificity. Mol. Cell 38:627-635. [DOI] [PubMed] [Google Scholar]
- 29.Woelk, T., B. Oldrini, E. Maspero, S. Confalonieri, E. Cavallaro, P. P. Di Fiore, and S. Polo. 2006. Molecular mechanisms of coupled monoubiquitination. Nat. Cell Biol. 8:1246-1254. [DOI] [PubMed] [Google Scholar]
- 30.Woelk, T., S. Sigismund, L. Penengo, and S. Polo. 2007. The ubiquitination code: a signalling problem. Cell Div. 2:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.