Abstract
The nuclear hormone receptor, estrogen receptor α (ERα), and mitogen-activated protein kinases (MAPKs) play key roles in hormone-dependent cancers, and yet their interplay and the integration of their signaling inputs remain poorly understood. In these studies, we document that estrogen-occupied ERα activates and interacts with extracellular signal-regulated kinase 2 (ERK2), a downstream effector in the MAPK pathway, resulting in ERK2 and ERα colocalization at chromatin binding sites across the genome of breast cancer cells. This genomic colocalization, predominantly at conserved distal enhancer sites, requires the activation of both ERα and ERK2 and enables ERK2 modulation of estrogen-dependent gene expression and proliferation programs. The ERK2 substrate CREB1 was also activated and recruited to ERK2-bound chromatin following estrogen treatment and found to cooperate with ERα/ERK2 in regulating gene transcription and cell cycle progression. Our study reveals a novel paradigm with convergence of ERK2 and ERα at the chromatin level that positions this kinase to support nuclear receptor activities in crucial and direct ways, a mode of collaboration likely to underlie MAPK regulation of gene expression by other nuclear receptors as well.
Estrogen receptor α (ERα), a member of the large superfamily of nuclear receptors, exerts profound effects on the gene expression, cellular response programs, and phenotypic properties of estrogen target cells, including over 70% of breast cancers. This hormone receptor also plays a central role in breast cancer development and progression. Because of these broad and important actions, ERα is usually considered the single most crucial predictor of breast cancer prognosis and is the key target of endocrine therapies. Blocking the activity of this receptor protein by use of selective estrogen receptor modulators (SERMs) or aromatase inhibitors, which reduce estrogen production, has proven highly effective in targeted treatment of hormone-responsive breast cancers (23, 24, 37) and also in the prevention of breast cancer in women at high risk for the disease (42).
Increased activity of the mitogen-activated protein kinase (MAPK) pathway is one of the hallmarks of more aggressive cancers and of endocrine resistance, in which ERα-positive tumors become refractory to endocrine therapies and relapse. It is believed that the balance of control of cellular physiology switches from ERα nuclear-initiated pathways to increased involvement of extranuclear-activated protein kinase pathways in these breast cancers (5, 20, 23, 32, 38, 39). However, the interplay and integration of the signaling inputs of this nuclear hormone receptor and MAPKs are poorly understood and were therefore aspects we examined here.
The MAPK family comprises well-conserved proteins that function as downstream effectors of a multitier signaling cascade, including a MAPK kinase (MAPKK, MEK) and a MAPKK kinase (MAPKKK), with MAPKs phosphorylating serine/threonine residues on target proteins to control a variety of cellular activities. Of the MAPKs, ERK1 and ERK2 are activated by mitogenic stimuli and are distributed throughout the cell, with more than half of ERK1 and ERK2 associating with microtubules in the cytoplasm (3, 27). However, overexpression or activation of MAPKs drives ERK1 and ERK2 into the nucleus (36), where these kinases phosphorylate downstream target transcription factors, including p53, Sp-1, c-Myc and c-Fos, and other kinases (e.g., Msk-1 and Rsk-2), which phosphorylate histone tails, providing a permissive environment for gene transcription (1, 3, 12, 26, 27, 36).
Recent studies from our laboratory and others have provided evidence that estrogens exert their effects by eliciting both direct nuclear actions and extranuclear-initiated actions that are integrated to regulate the diverse activities of the estrogen receptor (16, 17, 28, 31). Some of the most important actions of estrogens include stimulation of protein kinase signaling pathways. Not only are steroid receptors substrates for MAPK phosphorylation, but in our prior studies we found that estradiol (E2) regulation of gene expression in ERα-positive breast cancer cells required both ERα and active MAPK (31).
Therefore, in this report, we have explored the molecular basis of this collaboration between ERα and MAPK. Using genome-wide analysis of ERα and ERK2 chromatin binding sites and gene regulation, we have investigated interrelationships between ERα and ERK2 and show that their actions converge at the level of chromatin, where they colocalize at ERα distal enhancer binding sites across the genome from which ERK2 is well positioned to collaborate with ERα in exerting direct genomic actions. Our findings document extensive linkages and collaboration between this protein kinase and nuclear receptor that underlie the regulation of hormone-dependent gene activities to alter the proliferative program of breast cancer cells.
MATERIALS AND METHODS
Chemicals and treatments.
17β-Estradiol was from Sigma (St. Louis, MO). For MEK inhibitor experiments, cells were pretreated with 10 μM U0126 (Calbiochem) for 1 h and then treated with 0.1% control ethanol vehicle or 10 nM E2 in the presence of inhibitor for the indicated times.
GeneChip microarrays, statistical analysis, and functional categorization of target genes.
MCF-7 human breast cancer cells were transfected with 20 nM siGENOME ctrl (siGL3), ERK1, or ERK2 (Thermo Scientific) using Dharmafect1 according to the manufacturer's instructions. After 60 h of transfection, the cells were treated with 0.1% control ethanol vehicle or 10 nM E2 for 4 or 24 h in three separate experiments, and the total RNA was prepared from each sample, further purified, and used to generate cRNA, which was labeled with biotin. cRNAs were then hybridized on Affymetrix human Hu-133A2 GeneChips, which contain oligonucleotide probe sets representing approximately 56,000 human genes and expressed sequence tags. After washing, the chips were scanned and data analyzed as described previously (31). Briefly, data were analyzed using GeneChip operating software (Affymetrix, Santa Clara, CA). CEL files were then analyzed by using “affy” and “gcrma” package protocols in R/Bioconductor. Probesets with consistently low expression values were discarded, and then statistical multivariate analysis was done by the “limma” package. Probesets were also filtered based on best overall significance by the F-test statistic (8, 31). The criteria for genes regulated by E2 were set so that they have a false discovery rate of 1% and a fold change of ≥1.5 compared to vehicle-treated samples. Web-based Panther and ClueGO software were used for functional classification of the genes.
ChIP assays.
Chromatin immunoprecipitation (ChIP) assays were performed as described previously (31). MCF-7 cells were treated with 0.1% ethanol (Veh) or 10 nM E2 for the indicated times. The antibodies used, most obtained from Santa Cruz Biotechnology unless indicated otherwise, were ERα (HC-20), ERα Ab-10 mouse monoclonal antibody (Fischer), ERK1(K-23), ERK2 (D-2), and cyclic AMP response element binding protein 1 (CREB1; C-21). Controls using IgG were routinely done in all ChIP assays, as indicated in the figure legends. We also used a non-ER binding region of the pS2/TFF1 gene as an additional control for the specificity of the ChIP analyses. Quantitative real-time PCR was used to calculate recruitment to the regions studied, as described before (31). ChIP-reChIP experiments were performed as described previously (40).
ChIP-microarray analysis (ChIP-chip).
MCF-7 cells were grown for 6 days in estrogen-free conditions in phenol red-free medium supplemented with 5% charcoal-dextran-treated calf serum (CD-CS). Cells were then treated with 0.1% ethanol (Veh) or 10 nM E2 and cross-linked using 1% formaldehyde. Samples were sonicated (Fisher Sonic Dismembrator, model 500) and immunoprecipitated as previously described (6, 31, 40), using for ERα a 1:1 antibody mixture of ERα antibody F-10 (Santa Cruz) and ERα Ab-10 (Fischer) and, for ERK2, antibody D-2 (Santa Cruz). Purified samples were labeled as previously described (6). The microarray chips used were Affymetrix GeneChip Human Tiling 2.0R Array Sets. Genome-wide ChIP-on-Chip analysis was conducted using the model-based analysis of tiling arrays program (MAT) (22).
Cell culture, RNA extraction, and real-time PCR analysis of gene expression.
MCF-7 were maintained in culture as previously described (31, 40). At 6 days before E2 treatment, cells were switched to phenol red-free medium containing CD-CS. Medium was changed on days 2 and 4 of culture, and cells were then transfected with 20 nM siGENOME Ctrl, ERα, ERK1, or ERK2 using Dharmafect. After 48 h of transfection, cells were treated for 4 and 24 h with the indicated compounds. After treatments, total RNA was isolated, reverse transcribed, and analyzed by real-time PCR as described previously (31).
Immunoprecipitation and Western blot analysis.
MCF-7 cells were grown and then transfected with 20 nM siGENOME Ctrl, ERα, ERK1, or ERK2 using Dharmafect as described above. After 60 h of transfection, the cells were treated for 8 h with the indicated compounds. Total protein was extracted using radioimmunoprecipitation assay (RIPA) buffer, and 20 μg of protein was resolved by SDS-10% PAGE. For coimmunoprecipitation experiments, the cells were harvested in nondenaturing Cell Signaling lysis buffer, sonicated, and precleared by using agarose beads conjugated to normal IgG from the proper species (Santa Cruz). ERα-ERK2 complexes were precipitated either with ERK2 or ERα antibody (Santa Cruz) overnight at 4°C. Immunoprecipitated complexes were harvested by using protein A/G beads (Santa Cruz). Proteins were extracted by boiling beads in SDS loading buffer, and the samples were resolved on 4 to 20% SDS-PAGE gels. The proteins were transferred to nitrocellulose membrane. Antibodies against total MAPK, CCND1, E2F1, CREB1, and phosphoCREB1 were from Cell Signaling, and ERα (F10) and ERK2 (D2) monoclonal antibodies were from Santa Cruz Biotechnology. All antibodies were used at 1:500 dilution except the ERα antibody, which was used at a 1:1,000 dilution. The secondary antibodies were obtained from Odyssey and used at a 1:10,000 dilution. The membranes were scanned and analyzed by using Odyssey LI-COR infrared imaging device and software. Equal loading of the samples was assessed by blotting the membranes for β-actin (Sigma-Aldrich).
Data set accession number.
The entire microarray data set is available through the NCBI Gene Expression Omnibus (GEO) database under no. GSE 24592.
RESULTS
Interrelationships between ERα and MAPK signaling: phosphorylation of ERK1 and ERK2 and interaction of ERKs and ERα after hormone treatment of cells.
To characterize the interrelationships between ERα and MAPK signaling, we first examined the phosphorylation of ERK1 and ERK2, downstream effector kinases in the MAPK pathway, and observed robust phosphorylation after estradiol (E2) treatment of cells, which peaked by 15 min (∼6-fold increase) and still remained quite elevated at 45 min (Fig. 1A). We also observed in coimmunoprecipitation experiments that ERK1 and ERK2 interacted with ERα upon hormone treatment and that the interaction of ERK2 and ERα appeared to be stronger (Fig. 1B). We observed the same interaction in samples with extensive DNase treatment before coimmunoprecipitation (data not shown), implying that ERα and ERK2 are coming down via a protein-protein interaction and are not coimmunoprecipitating because they both might be binding to DNA.
FIG. 1.
cDNA microarray gene expression analysis after ERK1 or ERK2 knockdown in MCF-7 cells and effects of kinase depletion on estradiol (E2)-mediated gene regulation. (A) Time course of MAPK activation by E2. MCF-7 cells were treated with 10 nM E2 for the indicated times. Protein was harvested in RIPA buffer and subjected to SDS-PAGE analysis. pMAPK, ERK2, and ERα antibodies were used for Western blot analysis. (B) ERα and ERK2 interact upon E2 treatment of cells and can be immunoprecipitated from MCF-7 cells. Cells were treated with 10 nM E2 for the indicated times and then harvested with RIPA buffer. ERK2- or ERK1-containing complexes were immunoprecipitated from whole-cell extracts and immunoprecipitated proteins were subjected to SDS-PAGE and Western blot analysis for ERα and total MAPK. (C) Validation of selective ERK1 or ERK2 knockdowns in MCF-7 cells. Cells were transfected with siCtrl or single siRNA from each siGENOME or with siGENOME pool reagents for 60 h. ERK2, ERK1, and ERα protein levels after knockdowns were verified by Western blotting. (D) Cluster diagram of genes impacted by ERK2 or ERK1 knockdown. MCF-7 cells were treated with siCtrl, siERK2, or siERK1 for 60 h prior to treatment with 0.1% ethanol vehicle or 10 nM E2 for 4 or 24 h. Affymetrix gene expression microarrays were analyzed by LIMMA and Tightcluster software. The cluster map is visualized using Treeview Java. Fold expression is indicated below. Vertical red bar with star at right indicates gene cluster associated with cell proliferation. (E) Venn diagram depicting numbers of E2 regulated genes in each cell background (siCtrl, siERK2, and siERK1) at 4 and 24 h.
Gene expression microarray analysis in breast cancer cells depleted of ERK1 or ERK2 and examination of the impact of ERK knockdown on estrogen-stimulated cell proliferation and cell cycle-associated genes.
Based on our finding that estrogen enhanced the interaction of ERK1 and ERK2 with ERα, we sought to determine what effect these protein kinases might have on the pattern of ERα-mediated gene regulation. Using small interfering RNA (siRNA), we specifically depleted MCF-7 breast cancer cells of each kinase, using siGENOME reagents that contain a pool of four siRNAs targeting the gene of interest (siGENOME pool) or the individual siRNAs present in the pool. As shown in Fig. 1C, we observed very specific knockdown of each kinase. The knockdown of ERK1 or ERK2 did not affect ERα levels in MCF-7 cells, and likewise knockdown of ERα did not alter ERK1 or ERK2 levels (Fig. 1C). These data indicate that we can obtain a very efficient and selective knockdown of either kinase without cross-regulation or compensation by changes in the levels of ERα, ERK1, or ERK2 proteins.
To examine the effects of ERK1 and ERK2 on E2-regulated gene expression, MCF-7 cells were transfected with a control siRNA or with the pool of four siRNAs (siGENOME) targeting ERK1 or ERK2 for 60 h and were then treated with vehicle or E2 for 4 or 24 h. After RNA isolation and processing, we utilized Affymetrix Hu-133A2 Genechips to evaluate global gene expression profiles (Fig. 1D and E). Estrogen treatment resulted in the regulation of over 400 and 1,400 genes in control cells, at 4 and 24 h, respectively, as reported previously (7, 8, 13, 14), as well as in cells with knockdown of ERK1 or ERK2, but the genes regulated in the three cases showed some notable differences (Fig. 1E). Of the estrogen-regulated genes, approximately 160 (at 4 h) and 690 (at 24 h) were regulated only in cells with reduced ERK2, and approximately 60 (at 4 h) or 290 (at 24 h) were regulated selectively in cells with reduced ERK1, indicating differential gene regulation by E2 that is determined by the level of each kinase.
Using web-based Panther and ClueGO software, we analyzed the groups of genes whose estrogen regulation were most impacted by ERK2 or ERK1 knockdown, and we found that knockdown of each kinase affected different gene categories. ERK2 knockdown predominantly affected mitosis, DNA repair, and DNA metabolism-related genes (see Fig. S1 at www.life.illinois.edu/bkatzlab/supplementalfigure1.eps), whereas ERK1 knockdown had less of an effect on these groups of genes. In exploring this aspect further, cell proliferation assays showed that ERK2 knockdown fully blocked the E2-mediated increase in cell number (Fig. 2A). The depletion of ERK1 had a smaller impact on cell proliferation.
FIG. 2.
ERK2 controls E2-regulated cell proliferation and the expression of proliferation associated genes. (A) ERK2 is critical for E2-stimulated cell proliferation. MCF-7 cells were plated at 1,000 cells/well in 96-well plates. Cells were transfected with 20 nM siGENOME for Ctrl, ERK1, or ERK2 and the following day (day 0) were treated with 0.1% ethanol vehicle (Veh) or 10 nM E2. Treatment was repeated on day 2 and cell numbers were examined using the MTS assay at day 4. ***, P < 0.001; ###, P < 0.01 (versus vehicle). (B) ERK2 is essential for E2 stimulation of proliferation group genes. MCF-7 cells were transfected with 20 nM siGENOME reagent for Ctrl or ERK2 for 60 h and were then treated with 0.1% ethanol vehicle or 10 nM E2 for 24 h. Total RNA was isolated and reverse transcribed, and expression of the proliferation group genes from the 21 gene signature, which predicts tamoxifen responsiveness of ERα positive breast tumors, was examined by using quantitative PCR (Q-PCR). (C) ERK2 is essential for E2 stimulation of S-phase genes. MCF-7 cells were transfected with 20 nM siGENOME reagent for Ctrl, ERK1, or ERK2 for 60 h and then treated with control vehicle or 10 nM E2 for 24 h. Total RNA was isolated and reverse transcribed, and the expression of DNA synthesis-associated genes was examined by using Q-PCR. (D) CCND1 (cyclin D1) and E2F1 expression and E2 stimulation are affected by ERK2. MCF-7 cells were transfected with 20 nM siGENOME for Ctrl, ERK1, or ERK2 for 60 h and were then treated with control 0.1% ethanol vehicle or 10 nM E2 for 8 h. CCND1 and E2F1 protein levels were assessed by Western blotting. (E) ERK2 is recruited to the ERα binding site at the 3′ enhancer of CCND1. MCF-7 cells were treated with vehicle or 10 nM E2 for 45 min after exposure to siERα for 60 h or 10 μM MEK1 inhibitor U0126 for 1 h. Chromatin was cross-linked and sonicated. ERK2-DNA or background IgG complexes were immunoprecipitated using ERK2 antibody or normal mouse IgG antibody overnight. Recovered DNA was subjected to Q-PCR analysis. Values are expressed as the percent input and are means ± the standard errors of the mean (SEM) from four independent experiments.
We also verified E2-stimulated expression of several M-phase genes identified in our microarray analysis in siCtrl cells, including Ki-67, CCNB1, MYBL2, AURKB, and Survivin (BIRC5), which are part of a 21-gene signature used to predict the risk of breast cancer recurrence in patients on tamoxifen therapy (35). These genes are upregulated by E2, and knockdown of ERK2, but not ERK1 (data not shown), completely blocked their stimulation by E2, further supporting a major role for ERK2 in estrogen enhancement of cell proliferation (Fig. 2B). When we assessed the expression of S-phase genes required for DNA synthesis, we also observed reduced expression and loss of E2 regulation with ERK2 depletion (Fig. 2C), and two key mediators of cell cycle progression, CCND1 and E2F1, lost most or all of their estrogen stimulation when cells were depleted of ERK2 but not ERK1 (Fig. 2D).
To examine the possibility of a direct nuclear role for ERK2 in estrogen-stimulated expression of CCND1, we monitored recruitment of ERK2 to the ERα binding site at the 3′ enhancer of the CCND1 gene. We observed a hormone-stimulated recruitment, which was abrogated by inhibition of MAPK activation by the MEK inhibitor U0126 and also by ERα knock-down (Fig. 2E). Thus, our data show that ERK2 is a major regulator of proliferation and of the expression and E2 stimulation of genes promoting cell cycle progression. Further, estrogen stimulated the recruitment of ERK2 to the ERα binding site in the estrogen-regulated gene CCND1, and this recruitment required ERα and active MAPK, indicating a convergence of ERK2 and ERα at the level of chromatin. We explored this aspect further in genome-wide analyses described below.
Genome-wide analysis of ERα and ERK2 binding sites: overlapping binding sites and conservancy of the binding sites.
The marked effects of ERK2 depletion on the ability of hormone to regulate gene expression and cell proliferation, and the observed estrogen-stimulated interaction between ERK2 and ERα, suggested the possibility that ERK2 might in fact be recruited by the nuclear receptor to ER binding sites in chromatin. To investigate this, we first performed a limited ChIP-qPCR analysis in MCF-7 cells after E2 treatment at regions of a number of genes that we previously identified as being ERα binding sites. Notably, we detected estrogen-stimulated recruitment of ERK2 and ERK1 to the ER binding sites of all of these estrogen-regulated genes (see Fig. S2 at www.life.illinois.edu/bkatzlab/supplementalfigure2.eps). Since we obtained a stronger recruitment with ERK2 and observed more major effects of this kinase on estrogen-regulated gene expression and cell proliferation, we undertook ChIP-chip analyses to examine genome-wide ERα and ERK2 binding sites (Fig. 3).
FIG. 3.
ChIP-on-Chip analysis of genome-wide ERα and ERK2 binding sites. (A) UCSC Genome Browser view of ERα and ERK2 binding sites identified by our ChIP-chip studies. MCF-7 cells were treated with vehicle or 10 nM E2 for 45 min. After formaldehyde cross-linking and sonication, ERα and ERK2 containing complexes were immunoprecipitated. After amplification of the immunoprecipitated or input DNA, microarray analysis was performed using whole-genome Affymetrix GeneChip Human Tiling 2.0R Array sets. (B) Localization of binding sites relative to annotated genes. The location of binding sites was determined relative to the nearest gene in both upstream and downstream directions on both strands, within a 300-kb window. Distributions shown are percentage values. If the binding region is within a gene, CEAS software indicates whether it is in a 5′ untranslated region (5′UTR), a 3′UTR, a coding exon, or an intron. Proximal promoter is defined as 1 kb upstream from RefSeq 5′ start and immediate downstream is 1 kb downstream from RefSeq 3′ end. If a binding site is more than 1 kb away from the RefSeq TSS, it is considered an enhancer. (C) Conservancy of binding sites. Conservancy plots of binding sites were generated by using CEAS software.
MCF-7 cells were treated with vehicle or 10 nM E2 for 45 min and cross-linked with formaldehyde, and ERα and ERK2 containing chromatin complexes were immunoprecipitated with ERα or ERK2 specific antibodies. After DNA amplification, genome-wide microarray analysis was performed using Affymetrix GeneChip Human Tiling 2.0R arrays. We identified only background levels of ERα and ERK2 binding sites (less than 50) in cells in the absence of E2 treatment, but upon E2 treatment we obtained a mean number of 4547 ERα binding sites, similar to the number of ERα binding sites previously reported (6), and 1,303 ERK2 binding sites. Many genes, such as the well-known ERα target genes pS2 (also known as TFF1) and LRRC54 (also known as TSKU), harbored overlapping ERα and ERK2 binding sites (Fig. 3A).
Of note, 63% of ERK2 binding sites overlapped with ERα binding sites, suggesting that ERα might be the major transcription factor tethering ERK2 to chromatin after treatment of cells with E2. We then used the cis-regulatory element annotation system (CEAS) to further analyze the binding sites (21). The location of ERα and ERK2 binding sites were mapped to the nearest gene in both upstream and downstream directions on both strands, within 300 kb (Fig. 3B). About 50% of ERα binding sites and ERK2 binding sites were localized to distal enhancers (intergenic regions), followed by intronic regions (ca. 30%), proximal promoters (20% for ERK2 versus only 5% for ERα), and exon regions (3 to 7%). Interestingly, when we analyzed overlapping ERα and ERK2 binding sites relative to annotated genes, their distribution very much resembled the distribution of ERα binding sites. Further, using Oncomine concept maps, genes associated with ERK2 binding sites (see Fig. S3A at www.life.illinois.edu/bkatzlab/supplementalfigure3.eps) and with ERK2 and ERα overlapping binding sites after estrogen (see Fig. S3B at the same URL) were found to be those showing a strong positive correlation with ERα expression in breast tumors. These data imply that ERα is likely a major determinant of ERK2 binding to chromatin in estrogen-treated cells. Of interest, ERα and ERK2 cobound regions were biased two times more toward E2-regulated genes compared to ERα but not ERK2 regions. In addition, when we compared chromatin interaction analysis by paired-end tag sequencing (ChIA-PET) data (15) to our ERα and ERK2 overlapping binding sites, we observed that 85% of these sites mapped to ChIA-PETs, implying their association with chromatin looped regions.
We also performed a conservancy analysis of the different groups of binding sites, which compares the conservation of the binding site area across different species from zebrafish to human (and includes human, chimp, mouse, rat, dog, chicken, fugu, and zebrafish). This analysis (Fig. 3C) revealed that the three groups of binding sites (i.e., ERα, ERK2, and overlapping ERα and ERK2) showed high conservation across species, but, of note, the overlapping ERα and ERK2 binding sites showed the highest conservation. This might suggest an evolutionarily conserved function for these cooccupied genomic locations of ERα and ERK2.
Characterization of ERK2 recruitment to ERα binding sites.
To characterize the kinetics of ERK2 recruitment to ERα binding sites, we performed an E2 treatment time course. ERK2 recruitment, monitored at the ER binding sites of two estrogen-stimulated genes, increased by 5 min of E2 treatment, reached maximum levels at 30 to 45 min, and decreased somewhat by 1 h (Fig. 4A). Interestingly, this temporal profile of ERK2 recruitment after E2 is virtually identical to that which we have observed for ERα recruitment after hormone (40).
FIG. 4.
Characterization of ERK2 recruitment to ERα binding sites upon E2 treatment. (A) Time course of ERK2 recruitment to ERα binding sites in the estrogen-responsive LRRC54 and pS2 genes. MCF-7 cells were treated with 10 nM E2 for the indicated times. Chromatin was cross-linked and sonicated. ERK2-DNA or background IgG complexes were immunoprecipitated using ERK2 antibody or normal mouse IgG antibody overnight. Precipitated DNA was subjected to Q-PCR analysis. Values are the means ± the SEM from three independent experiments. (B) ERα is required for ERK2 recruitment to chromatin. MCF-7 cells were transfected with 20 nM siCtrl or siGENOME ERα for 60 h and were then treated with vehicle or 10 nM E2 for 45 min. ERK2-DNA or background IgG complexes were immunoprecipitated using ERK2 (D-2; Santa Cruz) antibody or normal mouse IgG antibody overnight. Precipitated DNA was subjected to Q-PCR analysis. Values are the means ± the SEM from four independent experiments. (C) ERα and ERK2 are present together at the ER binding sites. ChIP/reChIP experiments were performed using antibodies for ERK2 (D-2, sc-1647) and ERα (HC-20, sc-543). Recovered DNA was analyzed by Q-PCR. Values are the means ± the SEM from four independent experiments. (D) ERK2 is not required for ERα recruitment. MCF-7 cells were transfected with 20 nM siCtrl or siGENOME ERK2 for 60 h and then treated with vehicle or 10 nM E2 for 45 min. ERα-DNA or background IgG complexes were immunoprecipitated using ERα (F-10) or normal mouse IgG (Santa Cruz) antibodies overnight. Precipitated DNA was subjected to Q-PCR analysis. Values are the means ± the SEM from four independent experiments. (E) ERK2 activation by MEK1 is required for ERK2 but not ERα recruitment to ER binding sites. MCF-7 cells were pretreated with vehicle or 10 μM U0126 for 1 h and then treated with vehicle or 10 nM E2 for 45 min in the presence or absence of inhibitor. ERK2-DNA, ERα-DNA, or background IgG complexes were immunoprecipitated and precipitated DNA was subjected to Q-PCR analysis. Values are the means ± the SEM from four independent experiments.
To investigate the ERα dependency of ERK2 recruitment to genomic binding sites, we utilized siRNA-mediated knockdown of ERα. Knockdown of ERα (Fig. 4B) or treatment with the ER antagonist ICI182,780 (data not shown) completely abolished E2 stimulated recruitment of ERK2 to ER binding sites, indicating that functionally active ERα is required for recruitment of ERK2. Moreover, ChIP-reChIP experiments confirmed that ERα and ERK2 were present together at these binding sites (Fig. 4C). In contrast, depletion of ERK2 with siRNA did not impact ERα recruitment to binding sites of these estrogen-regulated genes (Fig. 4D).
Next, we queried the importance of ERK2 activation by MEK1 for the observed recruitment to chromatin of ERK2 and ERα. The MEK inhibitor U0126 nearly completely prevented ERK2 recruitment to ER binding sites, implying that activated ERK2 is required for recruitment (Fig. 4E). In contrast, U0126 did not affect recruitment of ERα to the regions studied (Fig. 4E). Hence, ERα is recruited upon E2 treatment to ER binding sites independent of ERK2, whereas ERK2 recruitment requires ERα and active ERK2 is required for its own chromatin localization.
Identification of CREB1 as a transcription factor regulating ERK2 chromatin binding and hormone-stimulated cell proliferation.
Based on our genome-wide mapping of ERα and ERK2 binding sites, we performed bioinformatic analysis to identify enriched transcription factor binding motifs in these binding sites. For this purpose, we used two programs: CEAS, which analyzes the full length of the binding site, and SeqPos, which analyzes enrichment around the center of the binding site. Both approaches revealed the response element for CREB1 to be highly enriched. We further assessed involvement of CREB1 with ERK2 and ERα actions, because CREB1 is a known MAPK target and is also highly expressed in these breast cancer cells.
As shown in Fig. 5A, ERK2 and CREB1 showed increased recruitment to overlapping binding sites for ERα and ERK2 in regulated genes after E2 treatment of cells. E2 also elicited a rapid increase in phosphoCREB1 (Fig. 5B). We confirmed by ChIP an E2-stimulated rapid recruitment of CREB1 to overlapping ERα and ERK2 binding sites in the E2-regulated genes LRRC54/TSKU and pS2/TFF1 (Fig. 5C), with the time course paralleling the recruitment of ERK2 and ERα after E2. The co-presence of CREB1 with ERK2 was also observed by ChIP-reChIP experiments (Fig. 5D). To establish whether this transcription factor is a putative tethering factor for ERK2, we examined the effect of knockdown of CREB1 on the recruitment of ERK2 to the estrogen-stimulated genes. As shown in Fig. 5E, knockdown of CREB1 with siRNA reduced the estrogen-stimulated recruitment of ERK2 to these estrogen-regulated genes while having no impact on recruitment of ERα. Furthermore, knockdown of CREB1 markedly reduced cell proliferation and prevented estrogen stimulation of proliferation (Fig. 5F) and the estrogen-stimulated expression of S-phase and proliferation-associated genes (Fig. 5G and H). Thus, the findings in Fig. 4 and 5 indicate that ERα and CREB1 are involved in ERK2 recruitment to chromatin upon estrogen treatment and that ERK2 and CREB1 greatly impact hormone-stimulated cell proliferation.
FIG. 5.
CREB1 is a cooperating transcription factor in ERK2 recruitment to chromatin binding sites and in E2 regulation of cell proliferation. (A) Box plots showing that E2 treatment stimulates recruitment of ERK2 and CREB1 to ERα binding sites (n = 7 genes evaluated). MCF-7 cells were treated with vehicle or 10 nM E2 for 45 min. ChIP was performed using specific antibodies for ERK2 or CREB1, and DNA was analyzed by Q-PCR. Values are the means ± the SEM of at least three independent experiments. (B) Time course of CREB1 phosphorylation after E2 treatment. Total CREB1 is also shown as a control for loading. (C) Time course of CREB1 recruitment. MCF-7 cells were treated with 10 nM E2 for the indicated times. Chromatin was cross-linked and sonicated, and CREB1-DNA or background IgG complexes were immunoprecipitated overnight using antibody for CREB1 or IgG as control. Precipitated DNA was subjected to Q-PCR analysis. Values are the means ± the SD of two experiments. (D) ChIP-reChIP for CREB1 and ERK2 shows that they are present together at ERα binding sites of the LRRC54 and pS2 genes. (E) CREB1 is required for full ERK2 recruitment to binding sites in the LRRC54 and pS2 genes. The left panel shows a Western blot for CREB1, ERα, and ERK2 after control siGL3 or siCREB1 transfection into MCF-7 cells for 72 h prior to vehicle or E2 treatment. ChIP was performed with specific antibodies as described in panel A. (F) MCF-7 cells were transfected with 20 nM siCtrl or siCREB1 and the following day (day 0) were treated with 0.1% ethanol vehicle or 10 nM E2. The treatment was repeated on day 2, and cell numbers were examined using the MTS assay at day 4. (G) CREB1 is required for the E2 stimulation of S-phase genes. MCF-7 cells were transfected with 20 nM siCtrl or siCREB1 for 60 h and then treated with vehicle or 10 nM E2 for 24 h. Total RNA was isolated and reverse transcribed, and expression of DNA synthesis-associated genes was examined by using Q-PCR. (H) CREB1 is required for the E2 stimulation of proliferation group genes. MCF-7 cells were transfected with 20 nM siCtrl or siCREB1 for 60 h and then treated with vehicle or 10 nM E2 for 24 h. Total RNA was isolated and reverse transcribed, and expression of proliferation group genes was examined by using Q-PCR.
DISCUSSION
This study reveals a novel paradigm for integration of MAPK and ERα actions in which ERK2 not only acts as a signaling protein but becomes colocalized with ERα at many chromatin binding sites across the genome, where it is positioned to collaborate with this receptor in its nuclear functions. This multilevel interplay between ERK2 and ERα is schematized in Fig. 6. Our whole-genome mapping of ERα and ERK2 chromatin binding showed that after estrogen treatment, two-thirds of ERK2 chromatin localization was at ERα binding sites. Hence, our findings bring to light previously unknown nuclear colocalization and functions of ERK2 in the hormone-dependent activity of the estrogen receptor. This intertwining of ERK2 and ERα at the level of chromatin enables this protein kinase to collaborate in proximate and crucial ways with ERα to support the actions of this nuclear hormone receptor in regulating gene expression and the proliferation of breast cancer cells.
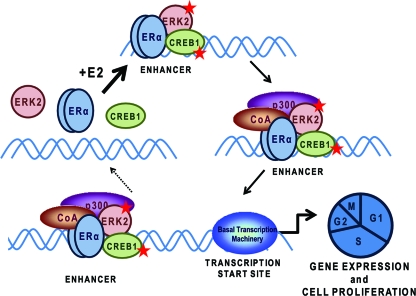
FIG. 6.
Model depicting the interrelationships elucidated in the present study between ERα, ERK2, and CREB1 in the hormonal regulation of gene expression and cell proliferation. Our findings reveal rapid activation of ERK2 and CREB1 (red stars) in response to estrogen and their colocalization with ERα at enhancer binding sites. ERK2 and CREB1 collaborate with ERα in regulating hormone stimulation of proliferation and of cell cycle-related genes. The findings indicate that ERK2 has not only a signaling function but also a nuclear role at chromatin in integrating with and supporting the actions of this nuclear hormone receptor. See the text for details. CoA, coactivator.
Estrogen stimulation of ERK2 and ERα chromatin localization: a convergence point of protein kinase and nuclear receptor signaling pathways.
By mapping ERα and ERK2 binding sites across the genome after hormone exposure, we have found that there is a convergence of the nuclear receptor and the protein kinase at chromatin sites associated with estrogen-regulated genes that suggest important outcomes relevant to the biology of estrogen target cells. There are a number of features of the ERα and ERK2 binding sites (cistromes) that indicate that ERα is the major transcription factor responsible for ERK2 chromatin localization after hormone treatment. First, two-thirds of the ERK2 sites overlap with ERα sites and, in the absence of hormone, we find only a very low background level of both ERK2 and ERα chromatin binding. Also, ERK2 recruitment to the ERα binding sites is abolished by ERα knockdown, and the distribution of ERK2 and ERα binding sites is similar, showing a predominance at distal enhancer regions where ERα works (6, 15, 30, 43). All of these observations are consistent with ERα being the major factor tethering ERK2 to chromatin after hormone treatment. Further, it is of note that although ERα and ERK2 binding sites show high conservancy across species, the ERα and ERK2 overlapping sites show the highest conservation, suggesting important functions of these binding sites.
Once localized at ERα chromatin binding sites, ERK2 is positioned to affect the state of phosphorylation and activity of other important components of ER transcription regulatory complexes, such as coregulators or mediator components. In this regard, a recent report has shown that estrogen stimulates ERK phosphorylation of MED1/TRAP220/DRIP205, a step required for its association with the mediator complex and its nuclear receptor coactivator activity (4). Likewise, estrogen-regulated MAPK phosphorylation of the coactivator SRC3 regulates its association with ERα and thereby receptor transcriptional activity (2). In other systems, RIP140 (18) and p300 (9) were also shown to be substrates for MAP kinases. Moreover, we show that CREB1, a known ERK2 substrate, is phosphorylated upon estrogen treatment and that this activation is prevented by the MEK inhibitor U0126. The presence of kinases in transcription complexes of regulated genes has been reported in a few prior studies (26, 27, 33, 41), but these examined kinase involvement at only a few genes and localization only in the proximal promoter region. Our studies, which have examined genome-wide ERα and ERK2 localization, have revealed their widespread colocalization, primarily at distal enhancer binding sites after hormone activated cell stimulation, thus providing physical and functional convergence points for the integration of kinase and nuclear receptor activities in the control of gene expression and cell proliferation.
Involvement of CREB in ERK2 and ERα colocalization and functional collaboration.
The response element for the transcription factor CREB was highly enriched in the overlapping ERα and ERK2 binding site group, and our studies indicated that CREB1 was recruited to these sites with estrogen treatment, as schematically shown in Fig. 6. ChIP-reChIP experiments showed colocalization of CREB and ERK2, and CREB depletion selectively reduced ERK2 but not ERα recruitment, indicating involvement of CREB in the estrogen-dependent recruitment of ERK2. Moreover, depletion of CREB1 reduced cell proliferation and prevented hormone-stimulated proliferation.
Although two-thirds of ERK2 binding sites colocalized with ERα, a portion (ca. one-third) of ERK2 binding sites did not colocalize with ERα. Although ERα cannot function as a tethering factor for ERK2 at these ERK2 only sites, ERK2 recruitment to these sites was still dependent on ERα, because we observed that ERα knockdown or treatment with anti-estrogen prevented recruitment to these sites, as well as to the sites in common with ERα. Because our studies show that activated ERK2 is required for ERK2 chromatin binding, we assume that the requirement of ERα for ERK2 localization to ERK2-only sites might also result from MAPK activation by the estrogen-occupied ERα. Notably, the ERK2-only sites had a distribution with a larger fraction being promoter proximal (20%) than that of ERK2 sites in common with ERα sites (only 5% promoter proximal).
A recent study showed that ERK2 is able to bind directly to DNA in vitro using protein microarrays and libraries of DNA sequences (19). The authors of that study identified a putative motif G/CAAAG/C for ERK2 direct DNA binding, and they performed studies in HeLa cells to support the hypothesis that ERK2 binding to this motif was important in repression of interferon-regulated genes. Although the possibility of direct ERK2-DNA binding is intriguing, we feel it is unlikely to be very important in regulating estrogen action. We found very few chromatin binding sites for ERK2 in breast cancer cells in the absence of estrogen, and many after estrogen treatment; so, we are dealing with a hormonally regulated process. In addition, we found that this motif is not enriched in our common ERα and ERK2 binding sites or in our ERK2-only binding sites.
Impact of ERK2 on ERα-mediated gene and cell proliferation programs.
ERα is a master regulator of gene expression in its target cells, with estrogen regulating the expression of over 1,000 genes in MCF-7 breast cancer cells, as found in the present study and studies by us and others previously (7, 8, 13, 14, 29, 34). It is known that MAPK activation by mitogenic hormonal signals, including estrogen and some growth factors, can impact ERα gene regulation by changing phosphorylation of ERα and its coregulators (12). Our findings provide evidence that the merging of signaling inputs from the ERα and MAPK pathways occurs in a very direct manner, through the hormone-dependent colocalization of ERK2 and ERα at chromatin binding sites across the genome, positioning ERK2 to work directly with ERα in providing an integrated outcome of steroid hormone receptor and protein kinase actions.
In conclusion, extensive interrelationships between ERα and MAPK pathways that converge on chromatin are evident from our findings. Estrogen exposure increases cellular MAPK activity and stimulates the association of ERα and ERK2, resulting in colocalization of both proteins at many chromatin binding sites. This convergence of ERα and ERK2 at the chromatin level is critical in ERK2 support of estrogen and ERα activities that regulate gene expression programs and cell cycle progression. That the MAPK pathway has been shown to also regulate gene expression by androgen receptor in prostate cancer cells (1) and by progesterone receptor in breast cancer cells (10, 11, 25, 41) suggests that the paradigm of convergence and functional collaboration of these two proteins that we have defined at the genome level may be more universal and likely will be observed for other members of the large nuclear receptor superfamily of proteins.
Acknowledgments
We thank Karen Kieser and Rosa Ventrella for assistance in these studies.
This study was supported by grants from the Breast Cancer Research Foundation (to B.S.K.), the National Institutes of Health (R01 CA18119 and P01AG024387 to B.S.K.; T32ES007326 to Z.M.E.; R01DK074967, P01CA8011105, and DF/HCC Breast Cancer SPORE Grant P50C89393 to M.B.), and U.S. Department of Defense Breast Cancer Research Program Award (W81XWH-08-1-0214 to M.L.).
We declare no conflict of interest.
Footnotes
Published ahead of print on 18 October 2010.
REFERENCES
- 1.Agoulnik, I. U., W. E. Bingman III, M. Nakka, W. Li, Q. Wang, X. S. Liu, M. Brown, and N. L. Weigel. 2008. Target gene-specific regulation of androgen receptor activity by p42/p44 mitogen-activated protein kinase. Mol. Endocrinol. 22:2420-2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amazit, L., L. Pasini, A. T. Szafran, V. Berno, R. C. Wu, M. Mielke, E. D. Jones, M. G. Mancini, C. A. Hinojos, B. W. O'Malley, and M. A. Mancini. 2007. Regulation of SRC-3 intercompartmental dynamics by estrogen receptor and phosphorylation. Mol. Cell. Biol. 27:6913-6932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Avruch, J. 2007. MAP kinase pathways: the first twenty years. Biochim. Biophys. Acta 1773:1150-1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belakavadi, M., P. K. Pandey, R. Vijayvargia, and J. D. Fondell. 2008. MED1 phosphorylation promotes its association with mediator: implications for nuclear receptor signaling. Mol. Cell. Biol. 28:3932-3942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Britton, D. J., I. R. Hutcheson, J. M. Knowlden, D. Barrow, M. Giles, R. A. McClelland, J. M. Gee, and R. I. Nicholson. 2006. Bidirectional cross talk between ERα and EGFR signaling pathways regulates tamoxifen-resistant growth. Breast Cancer Res. Treat 96:131-146. [DOI] [PubMed] [Google Scholar]
- 6.Carroll, J. S., X. S. Liu, A. S. Brodsky, W. Li, C. A. Meyer, A. J. Szary, J. Eeckhoute, W. Shao, E. V. Hestermann, T. R. Geistlinger, E. A. Fox, P. A. Silver, and M. Brown. 2005. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell 122:33-43. [DOI] [PubMed] [Google Scholar]
- 7.Chang, E. C., T. H. Charn, S. H. Park, W. G. Helferich, B. Komm, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2008. Estrogen Receptors alpha and beta as determinants of gene expression: influence of ligand, dose, and chromatin binding. Mol. Endocrinol. 22:1032-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chang, E. C., J. Frasor, B. Komm, and B. S. Katzenellenbogen. 2006. Impact of estrogen receptor beta on gene networks regulated by estrogen receptor alpha in breast cancer cells. Endocrinology 147:4831-4842. [DOI] [PubMed] [Google Scholar]
- 9.Chen, Y. J., Y. N. Wang, and W. C. Chang. 2007. ERK2-mediated C-terminal serine phosphorylation of p300 is vital to the regulation of epidermal growth factor-induced keratin 16 gene expression. J. Biol. Chem. 282:27215-27228. [DOI] [PubMed] [Google Scholar]
- 10.Edwards, D. P. 2005. Regulation of signal transduction pathways by estrogen and progesterone. Annu. Rev. Physiol. 67:335-376. [DOI] [PubMed] [Google Scholar]
- 11.Faivre, E. J., and C. A. Lange. 2007. Progesterone receptors upregulate Wnt-1 to induce epidermal growth factor receptor transactivation and c-Src-dependent sustained activation of Erk1/2 mitogen-activated protein kinase in breast cancer cells. Mol. Cell. Biol. 27:466-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Font de Mora, J., and M. Brown. 2000. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol. Cell. Biol. 20:5041-5047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frasor, J., E. C. Chang, B. Komm, C. Y. Lin, V. B. Vega, E. T. Liu, L. D. Miller, J. Smeds, J. Bergh, and B. S. Katzenellenbogen. 2006. Gene expression preferentially regulated by tamoxifen in breast cancer cells and correlations with clinical outcome. Cancer Res. 66:7334-7340. [DOI] [PubMed] [Google Scholar]
- 14.Frasor, J., J. M. Danes, B. Komm, K. C. Chang, C. R. Lyttle, and B. S. Katzenellenbogen. 2003. Profiling of estrogen up- and down-regulated gene expression in human breast cancer cells: insights into gene networks and pathways underlying estrogenic control of proliferation and cell phenotype. Endocrinology 144:4562-4574. [DOI] [PubMed] [Google Scholar]
- 15.Fullwood, M. J., M. H. Liu, Y. F. Pan, J. Liu, H. Xu, Y. B. Mohamed, Y. L. Orlov, S. Velkov, A. Ho, P. H. Mei, E. G. Chew, P. Y. Huang, W. J. Welboren, Y. Han, H. S. Ooi, P. N. Ariyaratne, V. B. Vega, Y. Luo, P. Y. Tan, P. Y. Choy, K. D. Wansa, B. Zhao, K. S. Lim, S. C. Leow, J. S. Yow, R. Joseph, H. Li, K. V. Desai, J. S. Thomsen, Y. K. Lee, R. K. Karuturi, T. Herve, G. Bourque, H. G. Stunnenberg, X. Ruan, V. Cacheux-Rataboul, W. K. Sung, E. T. Liu, C. L. Wei, E. Cheung, and Y. Ruan. 2009. An estrogen-receptor-alpha-bound human chromatin interactome. Nature 462:58-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hammes, S. R., and E. R. Levin. 2007. Extranuclear steroid receptors: nature and actions. Endocrinol. Rev. 28:726-741. [DOI] [PubMed] [Google Scholar]
- 17.Harrington, W. R., S. H. Kim, C. C. Funk, Z. Madak-Erdogan, R. Schiff, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2006. Estrogen dendrimer conjugates that preferentially activate extranuclear, nongenomic versus genomic pathways of estrogen action. Mol. Endocrinol. 20:491-502. [DOI] [PubMed] [Google Scholar]
- 18.Ho, P. C., P. Gupta, Y. C. Tsui, S. G. Ha, M. Huq, and L. N. Wei. 2008. Modulation of lysine acetylation-stimulated repressive activity by Erk2-mediated phosphorylation of RIP140 in adipocyte differentiation. Cell Signal. 20:1911-1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hu, S., Z. Xie, A. Onishi, X. Yu, L. Jiang, J. Lin, H. S. Rho, C. Woodard, H. Wang, J. S. Jeong, S. Long, X. He, H. Wade, S. Blackshaw, J. Qian, and H. Zhu. 2009. Profiling the human protein-DNA interactome reveals ERK2 as a transcriptional repressor of interferon signaling. Cell 139:610-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hutcheson, I. R., J. M. Knowlden, T. A. Madden, D. Barrow, J. M. Gee, A. E. Wakeling, and R. I. Nicholson. 2003. Oestrogen receptor-mediated modulation of the EGFR/MAPK pathway in tamoxifen-resistant MCF-7 cells. Breast Cancer Res. Treat. 81:81-93. [DOI] [PubMed] [Google Scholar]
- 21.Ji, X., W. Li, J. Song, L. Wei, and X. S. Liu. 2006. CEAS: cis-regulatory element annotation system. Nucleic Acids Res. 34:W551-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson, W. E., W. Li, C. A. Meyer, R. Gottardo, J. S. Carroll, M. Brown, and X. S. Liu. 2006. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl. Acad. Sci. U. S. A. 103:12457-12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jordan, V. C., and B. W. O'Malley. 2007. Selective estrogen-receptor modulators and antihormonal resistance in breast cancer. J. Clin. Oncol. 25:5815-5824. [DOI] [PubMed] [Google Scholar]
- 24.Katzenellenbogen, B. S., and J. Frasor. 2004. Therapeutic targeting in the estrogen receptor hormonal pathway. Semin. Oncol. 31:28-38. [DOI] [PubMed] [Google Scholar]
- 25.Lange, C. A. 2008. Integration of progesterone receptor action with rapid signaling events in breast cancer models. J. Steroid Biochem. Mol. Biol. 108:203-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lawrence, M., C. Shao, L. Duan, K. McGlynn, and M. H. Cobb. 2008. The protein kinases ERK1/2 and their roles in pancreatic beta cells. Acta Physiol. 192:11-17. [DOI] [PubMed] [Google Scholar]
- 27.Lawrence, M. C., K. McGlynn, C. Shao, L. Duan, B. Naziruddin, M. F. Levy, and M. H. Cobb. 2008. Chromatin-bound mitogen-activated protein kinases transmit dynamic signals in transcription complexes in beta-cells. Proc. Natl. Acad. Sci. U. S. A. 105:13315-13320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Levin, E. R., and R. J. Pietras. 2008. Estrogen receptors outside the nucleus in breast cancer. Breast Cancer Res. Treat. 108:351-361. [DOI] [PubMed] [Google Scholar]
- 29.Lin, C. Y., A. Strom, V. B. Vega, S. L. Kong, A. L. Yeo, J. S. Thomsen, W. C. Chan, B. Doray, D. K. Bangarusamy, A. Ramasamy, L. A. Vergara, S. Tang, A. Chong, V. B. Bajic, L. D. Miller, J. A. Gustafsson, and E. T. Liu. 2004. Discovery of estrogen receptor alpha target genes and response elements in breast tumor cells. Genome Biol. 5:R66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lin, C. Y., V. B. Vega, J. S. Thomsen, T. Zhang, S. L. Kong, M. Xie, K. P. Chiu, L. Lipovich, D. H. Barnett, F. Stossi, A. Yeo, J. George, V. A. Kuznetsov, Y. K. Lee, T. H. Charn, N. Palanisamy, L. D. Miller, E. Cheung, B. S. Katzenellenbogen, Y. Ruan, G. Bourque, C. L. Wei, and E. T. Liu. 2007. Whole-genome cartography of estrogen receptor alpha binding sites. PLoS Genet. 3:e87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Madak-Erdogan, Z., K. J. Kieser, S. H. Kim, B. Komm, J. A. Katzenellenbogen, and B. S. Katzenellenbogen. 2008. Nuclear and extranuclear pathway inputs in the regulation of global gene expression by estrogen receptors. Mol. Endocrinol. 22:2116-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McGlynn, L. M., T. Kirkegaard, J. Edwards, S. Tovey, D. Cameron, C. Twelves, J. M. Bartlett, and T. G. Cooke. 2009. Ras/Raf-1/MAPK pathway mediates response to tamoxifen but not chemotherapy in breast cancer patients. Clin. Cancer Res. 15:1487-1495. [DOI] [PubMed] [Google Scholar]
- 33.Narayanan, R., A. A. Adigun, D. P. Edwards, and N. L. Weigel. 2005. Cyclin-dependent kinase activity is required for progesterone receptor function: novel role for cyclin A/Cdk2 as a progesterone receptor coactivator. Mol. Cell. Biol. 25:264-277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ochsner, S. A., D. L. Steffen, S. G. Hilsenbeck, E. S. Chen, C. Watkins, and N. J. McKenna. 2009. GEMS (Gene Expr. MetaSignatures), a Web resource for querying meta-analysis of expression microarray datasets: 17beta-estradiol in MCF-7 cells. Cancer Res. 69:23-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paik, S., S. Shak, G. Tang, C. Kim, J. Baker, M. Cronin, F. L. Baehner, M. G. Walker, D. Watson, T. Park, W. Hiller, E. R. Fisher, D. L. Wickerham, J. Bryant, and N. Wolmark. 2004. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N. Engl. J. Med. 351:2817-2826. [DOI] [PubMed] [Google Scholar]
- 36.Raman, M., W. Chen, and M. H. Cobb. 2007. Differential regulation and properties of MAPKs. Oncogene 26:3100-3112. [DOI] [PubMed] [Google Scholar]
- 37.Santen, R. J., H. Brodie, E. R. Simpson, P. K. Siiteri, and A. Brodie. 2009. History of aromatase: saga of an important biological mediator and therapeutic target. Endocrinol. Rev. 30:343-375. [DOI] [PubMed] [Google Scholar]
- 38.Santen, R. J., R. X. Song, R. McPherson, R. Kumar, L. Adam, M. H. Jeng, and W. Yue. 2002. The role of mitogen-activated protein (MAP) kinase in breast cancer. J. Steroid Biochem. Mol. Biol. 80:239-256. [DOI] [PubMed] [Google Scholar]
- 39.Sivaraman, V. S., H. Wang, G. J. Nuovo, and C. C. Malbon. 1997. Hyperexpression of mitogen-activated protein kinase in human breast cancer. J. Clin. Invest. 99:1478-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stossi, F., Z. Madak-Erdogan, and B. S. Katzenellenbogen. 2009. Estrogen receptor alpha represses transcription of early target genes via p300 and CtBP1. Mol. Cell. Biol. 29:1749-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vicent, G. P., C. Ballare, A. S. Nacht, J. Clausell, A. Subtil-Rodriguez, I. Quiles, A. Jordan, and M. Beato. 2006. Induction of progesterone target genes requires activation of Erk and Msk kinases and phosphorylation of histone H3. Mol. Cell 24:367-381. [DOI] [PubMed] [Google Scholar]
- 42.Vogel, V. G. 2009. The NSABP Study of Tamoxifen and Raloxifene (STAR) trial. Expert Rev. Anticancer Ther. 9:51-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Welboren, W. J., M. A. van Driel, E. M. Janssen-Megens, S. J. van Heeringen, F. C. Sweep, P. N. Span, and H. G. Stunnenberg. 2009. ChIP-Seq of ERα and RNA polymerase II defines genes differentially responding to ligands. EMBO J. 28:1418-1428. [DOI] [PMC free article] [PubMed] [Google Scholar]