Abstract
In Drosophila melanogaster, achaete (ac) and m8 are model basic helix-loop-helix activator (bHLH A) and repressor genes, respectively, that have the opposite cell expression pattern in proneural clusters during Notch signaling. Previous studies have shown that activation of m8 transcription in specific cells within proneural clusters by Notch signaling is programmed by a “combinatorial” and “architectural” DNA transcription code containing binding sites for the Su(H) and proneural bHLH A proteins. Here we show the novel result that the ac promoter contains a similar combinatorial code of Su(H) and bHLH A binding sites but contains a different Su(H) site architectural code that does not mediate activation during Notch signaling, thus programming a cell expression pattern opposite that of m8 in proneural clusters.
In Drosophila melanogaster neurogenesis, the proneural basic helix-loop-helix activator (bHLH A) genes are initially expressed in clusters of adjacent cells called “proneural clusters” (Fig. 1A). Although each cell within the proneural cluster has the potential to adopt a neural cell fate, only one cell or a few cells within the cluster become a neural precursor cell (NPC). Subsequently, the expression of both the proneural bHLH A genes and several putative downstream “panneural” target genes are strongly upregulated in the NPC. In contrast, the expression of proneural and panneural gene is not upregulated in the non-NPCs.
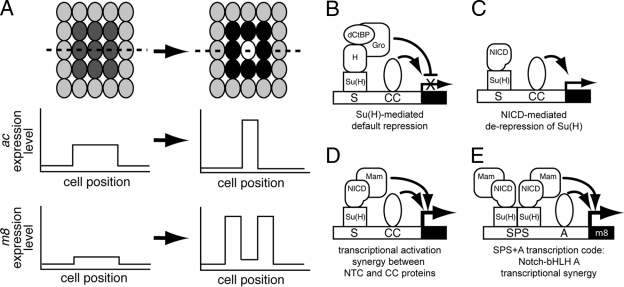
FIG. 1.
Proneural ac and E(spl)-C m8 gene expression patterns during Notch signaling mediated lateral inhibition in Drosophila proneural clusters. (A) In the early proneural cluster, both the model proneural and E(spl)-C genes, ac and m8, respectively, are expressed uniformly at low levels. In the late proneural cluster, after Notch signaling is activated, m8 is transcribed at high levels in the nonneural precursor cells (non-NPCs [black cells]). In contrast, at these later stages, ac is strongly expressed in the neural precursor cell (NPC [white cell]). The relative m8 and ac gene expression levels along the broken line bisecting the proneural cluster are shown below the proneural clusters. (B to E) Current models for Su(H)-regulated transcription. (B) In the absence of active Notch signaling and NICD, expression of target genes is blocked by Su(H)-mediated default repression. In this situation, corepressor proteins, such as Hairless (H), Groucho (Gro), and dCtBP, bind Su(H) and repress gene transcription. (C) When Notch signaling is activated, the cleaved Notch intracellular domain (NICD) translocates to the nucleus and displaces corepressor protein complexes bound to Su(H). Formation of the Su(H)/NICD binary complex and displacement of corepressors results in NICD-mediated derepression of Su(H). (D) The binary complex can recruit coactivators, such as Mastermind (Mam), and the resulting ternary complex can synergistically interact with other transcription factors (combinatorial cofactors [CC]) also bound nearby on the DNA. (E) On the model E(spl)-C promoter, m8, the SPS+A transcription code mediates synergistic interactions between Notch transcription complexes (NTC) and bHLH A combinatorial cofactors that strongly upregulate m8 expression in the non-NPCs.
Notch signaling-mediated lateral inhibition is critical for repression of proneural bHLH A gene expression in the non-NPCs. Several effector genes for the lateral inhibition pathway in proneural clusters are in the Enhancer of split Complex [E(spl)-C]. The E(spl)-C bHLH repressor (bHLH R) genes (m3, m5, m7, m8, mγ, and mδ) are well-characterized effector genes for Notch signaling (4), and the E(spl)-C m4 and mα Bearded-like (Brd-like) genes have also been proposed to mediate lateral inhibition (2). The bHLH R proteins can repress proneural gene expression by binding to R sites in proneural gene regulatory regions (33, 34, 40) as well as physically interacting with the proneural proteins and blocking proneural autoactivation (15, 16). The Brd-like proteins physically interact with the Neuralized panneural protein and modulate intracellular processing of the Notch signaling ligand Delta (2).
Activation of E(spl)-C gene transcription in proneural clusters is initially inhibited by a “default repression” mechanism that is mediated by the bifunctional protein Suppressor of Hairless [Su(H); also called CSL], which binds to S DNA binding sites (3, 5, 21, 28). In the absence of Notch signaling, Su(H) mediates repression of these genes by recruiting specific corepressors, including Hairless (H), Groucho (Gro), and dCtBP (Fig. 1B) (3, 30). However, once the NPC is established in proneural clusters, the Notch receptor becomes selectively activated in the non-NPCs, and Su(H)-mediated repression of the E(spl)-C genes in the non-NPCs is relieved. This derepression is due to the cleaved Notch intracellular domain (NICD) binding to Su(H) and displacing the corepressor proteins (Fig. 1C). The Su(H)/NICD binary complex then recruits additional coactivators, such as Mastermind (Mam) (5, 21, 24). The resulting ternary complex can also synergistically interact with other transcription factors bound nearby on the DNA (Fig. 1D). For example, synergistic interactions between Notch transcription complexes and bHLH A proteins is critical for strong expression of m8 and several neural E(spl)-C genes in non-NPCs (5, 7, 9).
Several E(spl)-C bHLH R and Brd-like genes have cell-specific expression patterns in proneural clusters that are the opposite of the proneural bHLH A genes during Notch signaling (Fig. 1A). These opposing expression patterns are programmed by “DNA transcription codes” embedded in regulatory DNA sequences. Transcription codes are the specific combinations and “architectures” (that is, the order, orientation, and spacing) of transcription factor binding sites clustered in small promoter or enhancer regions that program a specific component of the overall expression pattern (26). For example, the m8 model bHLH R gene contains an “SPS+A” transcription code that mediates synergistic interactions between Su(H)/NICD complexes and bHLH A protein complexes (Fig. 1E) (7). The SPS+A code contains an SPS element [Su(H) paired site] and at least one A site. The SPS element has a specific, inverted repeat architecture of S sites that is critical for programming Notch-proneural transcriptional synergy on the m8 promoter. The SPS element architecture is also present in vertebrate Notch pathway target genes (1, 20, 32) and can also mediate strong transcriptional synergy with vertebrate homologues to Drosophila proneural bHLH A proteins (7, 25). The SPS+A transcription code drives the upregulation of m8 only in non-NPCs during lateral inhibition (Fig. 1A). SPS+A modules are also present in several other E(spl)-C gene promoters, including m7, mγ, mδ, and m4, where they also are predicted to program Notch-proneural synergy and to mediate upregulation in non-NPCs (7). However, Notch-proneural synergy is not exclusively mediated by the SPS+A transcription code, since S sites in the E(spl)-C mα gene promoter, which lack an SPS architecture, can also mediate synergistic interactions between Notch signaling and proneural bHLH A proteins (5).
A key question in the Notch signaling field is how Notch can selectively activate distinct sets of target genes in different developmental pathways and yet always function through a single class of DNA site, the S site. On the basis of our previous demonstration that the SPS architecture is necessary for the cell-specific expression of m8 in Drosophila proneural clusters, we proposed that distinct S binding site “subcodes” function to program selective activation of Notch target genes (7). The different architectures of S sites within the subcodes program synergistic interactions between Notch complexes bound to the S sites and specific combinatorial cofactors bound to nearby DNA sites, thus allowing regulation of multiple developmental pathways.
In this study, we expand our understanding of the role that distinct S-site architectures play in mediating differential transcription responses during Notch signaling. Although canonical models of Notch signaling indicate that S sites always mediate activation of target genes during Notch signaling, we identify an S site in the promoter of the achaete (ac) proneural bHLH A gene that mediates repression, not activation, during Notch signaling. Mutation of this S site derepresses the native ac promoter in both cultured cells and transgenic flies. We show that the opposite transcriptional responses to NICD for ac and m8 can be interconverted simply by exchanging the S-site architectures of those promoters. NICD is shown to associate with the ac promoter after Notch signaling is activated, suggesting that Su(H)/NICD complexes do form on the ac promoter, but they are not sufficient to activate gene transcription. These findings challenge the current models of Notch signaling which would predict that recruitment of NICD to the ac promoter by Su(H) should mediate either activation or derepression of gene transcription. Together, these results show that distinct Su(H) binding site architectures in the ac and m8 promoters are critical for programming their differential responses to Notch signaling.
MATERIALS AND METHODS
Transcription assays.
Drosophila melanogaster S2 cells (from Invitrogen) were maintained according to standard protocols (8). Protocols for transfection and transcription assays with S2 cells have been described elsewhere (7). For both expression and reporter plasmids, 1 μg of each plasmid was transfected into cultures maintained in 24-well plates. The expression plasmids for Achaete protein (Ac), Daughterless protein (Da), and NICD as well as the wild-type and mutant m8 reporter plasmids have been previously described (5). Construction of the reporter plasmid for the wild-type 0.9-kb ac promoter has been described previously (34), and the Su(H) mutant of this promoter was generated by PCR. For all transcription assay experiments, the mean relative reporter gene expression levels are shown with error bars representing the standard deviations of the means. Statistical significance of pair-wise comparisons of transcription assay results was determined using the Student t test.
Transgenic fly analysis.
The construction of the transgenic expression plasmid for the wild-type ac transgene has been described elsewhere (34), and the transgene containing either mutated S site was generated by shuttling the mutant promoter from the luciferase reporter plasmid into the ac transgene expression plasmid. All transgenic lines were generated in w1118 flies. The bristle phenotypes of transgenic lines (n = 50 per line) were compared to those of 5 randomly picked samples (n = 50) of w1118 flies.
Chromatin immunoprecipitation (ChIP) assays with Drosophila embryos.
Embryos from w1118 flies from overnight collections were dechorinated and then fixed with 1.8% formaldehyde. The fixed embryos were stored at 4°C, and subsequent overnight collections were fixed and stored with previous collections. Embryo collections were pooled for a week before progressing with the immunoprecipitation protocol. Pooled embryo collections were homogenized and then sonicated. Cellular debris was removed by centrifugation, and the lysate was precleared with protein A/G-Sepharose beads (Santa Cruz Biotechnology). Antibodies (5 to 10 μg) to either Hairless (H) or β-actin (dC-19 and H300, respectively; Santa Cruz) were added to the lysate and incubated at 4°C overnight. Bovine serum albumin (BSA) and herring sperm DNA (Sigma) were also added (0.75 mg/ml and 0.15 mg/ml, respectively) to minimize nonspecific interactions. The antibody-protein-DNA complexes were precipitated with protein A/G Sepharose beads. Following several washes, the precipitated DNA was isolated by an overnight incubation in 0.1 M sodium bicarbonate and 1% SDS at 65°C. The isolated DNA was purified using the QIAquick PCR purification kit (Qiagen). The following primers were used for PCRs to test whether the ac, mα, and m8 regulatory regions were immunoprecipitated: 5′-GGGCCAGGTTTTCGTTTGGGGACGACAGGC-3′ and 5′-GGGCCTAGGGATCCCACCTGCGTGACTACC-3′ for the ac promoter; 5′-CTGGGGATTCGAAACTCAGAAACGGTCCCC-3′ and 5′-TATTCAAGTGCTGCGTGAAATCCCCAGAGG-3′ for the mα promoter; and 5′-GCGACAGCTGCAAAAATGTGCCCTGATCCTT-3′ and 5′-CACCCTCTGATACGCACCTTTCCTGCCCTC-3′ for the m8 promoter.
ChIP assays with DmD8 cells.
DmD8 cells were obtained from the Drosophila Genome Research Center and maintained according to recommended protocols (https://dgrc.cgb.indiana.edu/cells/support/protocols.html). Induction of Notch signaling by EDTA treatment and ChIP assays were performed using previously reported protocols (23). The ChIP studies used antibodies generated against Su(H) (dC-20; Santa Cruz Biotechology) and NICD (C17.9C6-c; Developmental Studies Hybridoma Bank). Quantitative real-time PCR was performed using Sybr green master mix (Applied Biosystems) and an Applied Biosystems 7500 reverse transcription-PCR (RT-PCR) system with the following primer sets: 5′-CGTTTGGGGACGACAGGCAG-3′ and 5′-GTAGTAATATTATCTCTCGTTCTCTCTG-3′ for the ac promoter; 5′-ACTGAAGATGAGGACATCCTCGAC-3′ and 5′-GGTTTGATGTGTGTTATGGTTGGG-3′ for the ac open reading frame (ORF); and 5′-CAACAGAGTGCGTCGCCGCTTC-3′ and 5′-ACCTCCAGCTCGCGCACGTTGT-3′ for rp49. Statistical significance of pair-wise comparisons was determined using the Student t test.
RESULTS AND DISCUSSION
Repression of the model proneural ac promoter by an S site.
S sites are essential for the non-NPC specific activation of certain neural E(spl)-C genes. Proximal promoter fragments from several of these genes that are sufficient to mediate non-NPC specific proneural cluster expression contain multiple S sites (Fig. 2A). Since the proneural bHLH A genes are repressed in the non-NPCs and not activated like the neural E(spl)-C genes, the proximal promoters of the proneural genes were expected to be devoid of functional S sites. However, with the exception of l'sc and ato, S sites were found in the 2-kb proximal promoter regions of all proneural genes, as well as some panneural genes (Fig. 2B). Moreover, there are S sites in many of the fragments derived from the proximal promoters of proneural and panneural genes that drive reporter genes specifically in NPCs (Fig. 2B). The potential regulatory role of S sites in proneural gene transcription is unexpected, since proneural genes are repressed by Notch signaling, and neither Su(H) or NICD has been reported, or predicted, to act directly on proneural promoters. In addition, NICD is believed to always mediate derepression and coactivation of Su(H), and not repression.
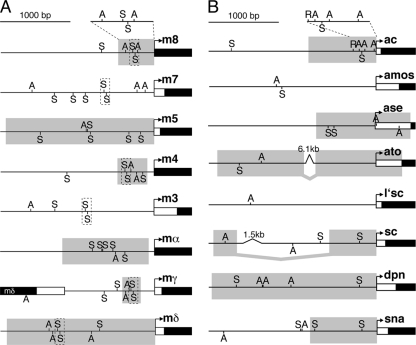
FIG. 2.
Su(H) and bHLH A protein binding sites in the proximal promoter regions of the neural E(spl)-C, proneural bHLH A, and certain panneural genes. (A) Neural E(spl)-C genes. bHLH R (m8, m7, m5, m3, mγ, and mδ) and Brd-like (m4 and mα) genes are shown. (B) Proneural bHLH A genes (ac, amos, ase, ato, l'sc, and sc) as well as certain panneural genes (dpn and sna). Expansions of the proximal promoters for ac and m8 are shown, since they are model promoter regions for the proneural and E(spl)-C genes, respectively. The binding sites for the Su(H) and bHLH A proteins are indicated by “S” and “A,” respectively. Boxes with dashed lines indicate SPS elements, which are an inverted repeat of high-affinity S sites separated by 15 to 17 bp that are present in several E(spl)-C promoters. Promoter regions that have been used to drive reporter gene expression in proneural clusters in vivo are indicated by the gray boxes: m4 and mγ (32), m5 and m8 (22), mα (5), mδ (9), ac (41), ase (19), dpn (13), and sna (18). Reporter gene constructs with ato (38) and sc (11) used noncontiguous regulatory regions.
The ac 0.9-kb promoter was selected as a model to study whether S sites in proneural gene proximal promoters are functional because it is a short, contiguous promoter region that is sufficient to mediate proper ac expression (27, 34, 40, 41). In cultured Drosophila S2 cells, the wild-type 0.9-kb ac promoter (ac-WT) mediated robust activation of reporter gene expression when Achaete and Daughterless (Ac/Da) proteins were coexpressed (Fig. 3A, compare experiment 1 with experiment 2). However, when the S site was mutated (ac-Sm), reporter gene expression was modestly, but significantly (P = 0.0008), increased relative to ac-WT (Fig. 3A, compare experiment 2 with experiment 4).
FIG. 3.
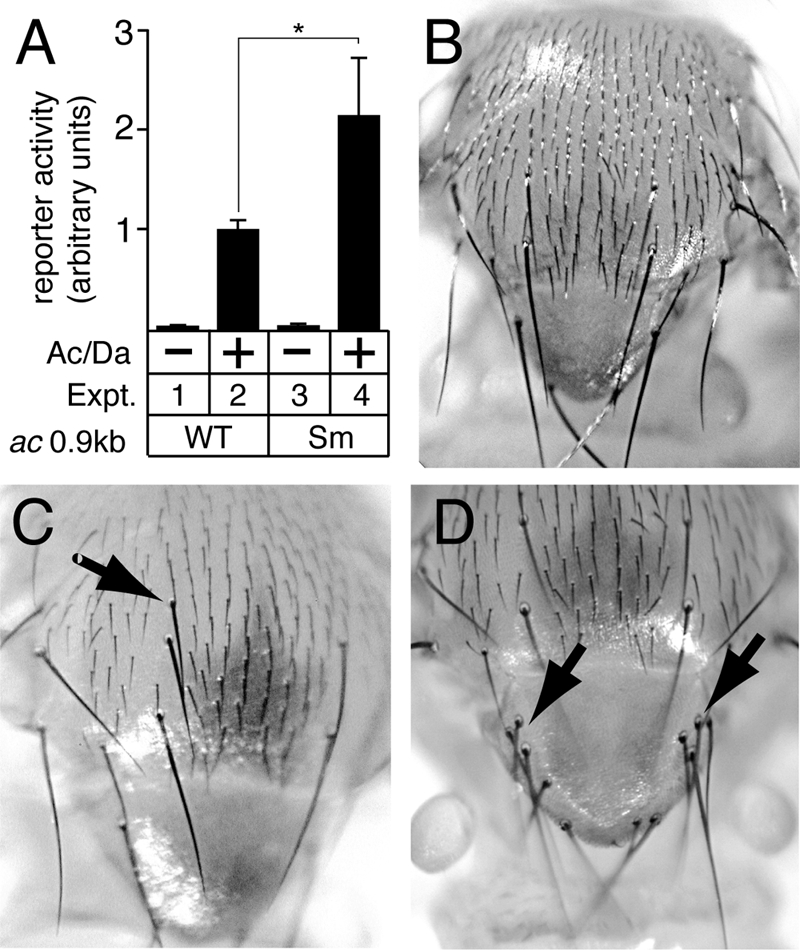
Functional analysis of S-site mutation in the 0.9-kb ac proximal promoter. (A) Transcription assays in S2 cells revealed that mutation of the ac S site (Sm) significantly increased reporter gene expression (P = 0.0008; indicated by asterisk), compared to the wild-type (WT) promoter, when Achaete and Daughterless proteins (Ac/Da) were coexpressed. (B) Bristle phenotype of the wild-type adult thorax. (C and D) Transgenic flies containing an ac transgene with the 0.9-kb S-site mutant promoter had extra dorsocentral macrochaete (arrow in panel C), and several transgenic lines of flies had clusters of extra macrochaete in the scutellum (arrows in panel D). Together, these results suggest that the ac promoter is repressed via the S site.
To test whether elimination of the S site within the ac 0.9-kb promoter could disrupt ac regulation in vivo, we constructed ac minigenes containing either the ac-WT or ac-Sm 0.9-kb promoters. The ac gene is essential for the development of the small, and some of the large, mechanosensory bristles on the adult notum (microchaete and macrochaete, respectively) (29). Both microchaete and macrochaete are bristle cells that are direct descendants of NPCs that have differentiated within the ectodermal epithelium. As a result, these bristles are highly sensitive markers for NPC generation under the present experimental conditions. Although the w1118 strain used to generate the transgenic lines had a high frequency of single extra scutellar macrochaete (i.e., duplications), neither multiple scutellar nor extra dorsocentral macrochaete bristles were common in this line (Tables 1 and 2). Control transgenic fly lines containing an extra copy of the wild-type ac minigene did not have any significant increase in either scutellar or dorsocentral bristles (n = 15 lines) (Tables 1 and 2), which was in agreement with previous studies (34, 40). In contrast, all transgenic lines containing the same ac minigene, but with the S site mutated (ac-Sm), had significant increases in multiple scutellar and extra dorsocentral macrochaete (n = 9 lines) (Fig. 3C and D and Tables 1 and 2). Together, these results indicate that the S site in the ac 0.9-kb promoter mediates repression of ac transcription, both in cultured cells and in vivo.
TABLE 1.
Percentage of flies containing extra scutellar and dorsocentral bristles in w1118 flies, ac-WT flies, and ac-Sm transgenic flies
| Line | % of flies (n = 50) containing an extra SC or DC bristle(s)a |
|||
|---|---|---|---|---|
| At least 1 extra SC bristle | Multiple (≥2) SC bristles | At least 1 extra DC bristle | Multiple (≥2) DC bristles | |
| w1118-1b | 46 | 16 | 2 | 0 |
| w1118-2b | 54 | 18 | 4 | 0 |
| w1118-3b | 44 | 14 | 6 | 2 |
| w1118-4b | 38 | 10 | 4 | 0 |
| w1118-5b | 42 | 14 | 6 | 0 |
| ac-WT-1 | 42 | 8 | 8 | 0 |
| ac-WT-2 | 44 | 12 | 12 | 0 |
| ac-WT-3 | 64 | 26 | 12 | 0 |
| ac-WT-4 | 50 | 12 | 2 | 0 |
| ac-WT-5 | 40 | 24 | 14 | 2 |
| ac-WT-6 | 64 | 40 | 6 | 0 |
| ac-WT-7 | 36 | 20 | 4 | 0 |
| ac-WT-8 | 35 | 10 | 6 | 0 |
| ac-WT-9 | 24 | 16 | 16 | 2 |
| ac-WT-10 | 46 | 20 | 8 | 2 |
| ac-WT-11 | 54 | 18 | 18 | 2 |
| ac-WT-12 | 40 | 8 | 4 | 0 |
| ac-WT-13 | 48 | 22 | 10 | 2 |
| ac-WT-14 | 38 | 6 | 4 | 0 |
| ac-WT-15 | 44 | 18 | 6 | 0 |
| ac-Sm-1 | 86 | 44 | 46 | 22 |
| ac-Sm-2 | 82 | 40 | 22 | 6 |
| ac-Sm-3 | 90 | 76 | 44 | 24 |
| ac-Sm-4 | 78 | 60 | 34 | 8 |
| ac-Sm-5 | 70 | 58 | 44 | 16 |
| ac-Sm-6 | 80 | 62 | 38 | 8 |
| ac-Sm-7 | 80 | 46 | 50 | 12 |
| ac-Sm-8 | 84 | 44 | 50 | 6 |
| ac-Sm-9 | 82 | 68 | 24 | 8 |
SC, scutellar; DC, dorsocentral.
w1118 flies were analyzed using 5 randomly sampled groups (n = 50).
TABLE 2.
Combined analysis of extra scutellar and dorsocentral macrochaete bristle phenotype in w1118 and ac-WT flies and ac-Sm transgenic flies
| Line | % of flies containing an extra SC or DC bristle(s) |
|||
|---|---|---|---|---|
| At least 1 extra SC bristle | ≥2 extra SC bristles | At least 1 extra DC bristle | ≥2 extra DC bristles | |
| w1118 | 50 ± 14 | 14 ± 3 | 4 ± 2 | 0 ± 1 |
| ac-WT | 44 ± 11 | 17 ± 9 | 9 ± 5 | 1 ± 1 |
| ac-Sm | 81 ± 6a | 55 ± 12a | 37 ± 10a | 12 ± 7a |
These values were significantly different (P < 0.005) from those for the w1118 and ac-WT lines.
Hairless and NICD recruitment to the ac promoter.
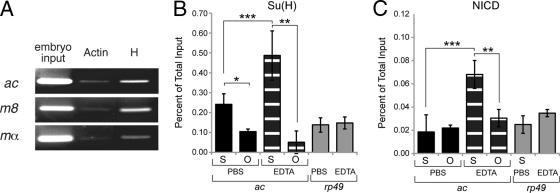
The increase of both reporter gene expression in transcription assays and the number of bristles in transgenic flies indicated that mutation of the S site disrupted Su(H)-mediated repression. Since Hairless (H) is a corepressor for Su(H) on E(spl)-C genes (3, 5, 28), we tested whether H was recruited to the ac promoter in vivo by performing chromatin immunoprecipitation (ChIP) experiments with whole-embryo lysate. As shown in Fig. 4A, the ac promoter was immunoprecipitated using anti-H antibodies, suggesting that ac is repressed by Su(H)/H complexes in vivo. For controls, we also observed that the mα and m8 promoters could be immunoprecipitated with anti-H antibodies, which was consistent with previous studies that reported that these promoters are also repressed in vivo by Su(H)/H complexes (3, 5). These results indicate that both proneural bHLH A and neural E(spl)-C genes may be regulated by the same or similar default repression mechanism(s) that include Su(H)/H complexes.
FIG. 4.
Recruitment of Su(H), Hairless, and NICD to the ac promoter. (A) ChIP assays with whole-embryo lysate reveal that antibodies to Hairless (H) immunoprecipitate the ac, m8, and mα promoter regions containing S binding sites. Parallel immunoprecipitation reactions using antibodies to β-actin were used to control for nonspecific interactions. (B and C) Quantitative PCR analysis of ac genomic DNA regions immunoprecipitated with antibodies to Su(H) and NICD, respectively, in DmD8 cells treated either with EDTA to activate Notch signaling or phosphate-buffered saline (PBS) as a control. In the absence of activated Notch signaling (PBS treated), the ac promoter region containing the S site (indicated by “S”) was preferentially bound by Su(H) compared to a region of the ORF that lacks S sites (indicated by “O”) (P = 0.04; indicated by a single asterisk). In contrast, NICD binding to either the ac promoter or ORF in PBS-treated cells was not above negative-control levels (rp49). In the EDTA-treated cells, both Su(H) and NICD preferentially bound the ac promoter compared to the ORF (P = 0.02 and P = 0.01, respectively; indicated by two asterisks). The binding of Su(H) and NICD to the ac promoter was also significantly greater in cells treated with EDTA compared to cells treated with PBS (P = 0.04 and P = 0.01, respectively; indicated by three asterisks). Together, these findings indicate that Su(H), but not NICD, is bound to the ac promoter in the absence of activated Notch signaling. When Notch signaling is activated, both Su(H) and NICD occupy the ac promoter, suggesting that an Su(H)/NICD complex is formed on the ac promoter S site.
A recent study showed that Notch signaling is activated in cultured Drosophila DmD8 cells by treatment with EDTA (23). This study also showed that both Su(H) and NICD occupancy on a subset of E(spl)-C gene promoters increased following EDTA treatment and Notch activation. Consistent with this previous study, our ChIP assays with DmD8 cells showed that Su(H) and NICD occupancy of the ac promoter also increased when Notch signaling was activated (Fig. 4B and C). Together, these results are also in agreement with the results of previous in vitro studies that showed that mammalian Su(H)/NICD complexes can assemble on promoters with either single S sites or SPS elements that are analogous to those in the ac or m8 promoter, respectively (31, 35).
Although Su(H) and NICD binding to the ac promoter increased when Notch signaling was activated in the DmD8 cells, coexpression of NICD with Ac/Da did not activate the wild-type ac promoter in S2 cells to levels above those with expression of Ac/Da alone (Fig. 5, compare experiments 2 and 4). In contrast to current models of Notch signaling, these results indicate that formation of the Su(H)/NICD protein complex on the ac promoter is not sufficient to increase gene expression levels under the conditions studied. Rather, the ac promoter was derepressed only when the S site was mutated (Fig. 3). These results suggest that NICD recruitment does not functionally displace all corepressor proteins recruited by Su(H) on the ac promoter.
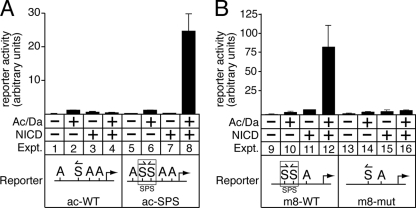
FIG. 5.
Interconversion of ac and m8 promoter responses to NICD upon mutation of their S-site architectures. (A) The single endogenous S site in the wild-type ac promoter (ac-WT) did not mediate depression or activation when NICD was coexpressed with the Ac/Da bHLH A proteins (experiments 1 to 4). In contrast, when a second S site is added to the ac promoter so that an SPS element is created (ac-SPS), coexpression of NICD and Ac/Da synergistically activated reporter gene transcription very strongly (experiments 5 to 8). (B) The wild-type m8 promoter, which contains an SPS element, mediates synergistic activation of reporter gene expression when NICD and Ac/Da are coexpressed (experiments 9 to 12). In contrast, NICD and Ac/Da proteins do not synergistically activate a modified m8 promoter (m8-mut) with one of the S sites in the SPS element mutated so that the remaining single S site is in the same orientation as the wild-type ac promoter (experiments 13 to 16). Together, these findings with ac and m8 demonstrate the functional importance of S-site architecture in mediating differential transcriptional responses to Notch signaling. The arrows indicate the S binding site orientation.
In addition to Su(H)/H complexes, the bHLH R proteins also directly bind the ac promoter and repress transcription (33, 34, 40). The bHLH R proteins have WRPW motifs that recruit the Groucho corepressor (12, 14, 36), and Groucho is also known to interact with Hairless and dCtBP to mediate repression through Su(H) proteins (3, 28). Therefore, interactions between multiple transcription factors may stabilize and prevent complete displacement of Hairless or other corepressors that are initially assembled on Su(H), even in the presence of NICD. Moreover, the bHLH R proteins also can physically interact with bHLH A proteins bound to the A sites to mediate repression (15, 16), and those interactions may also stabilize the non-DNA binding corepressors on the ac promoter, even if NICD is bound to Su(H).
An alternative mechanism that prevents Notch signaling from activating ac expression is that recruitment of coactivators, such as Mam, to the ac promoter by the Su(H)/NICD binary complex is blocked. Such blockage could result from either the failure to completely displace corepressors from Su(H) or if there are proteins specifically bound to the ac promoter that prevent formation of the Su(H)/NICD/Mam ternary complex. The latter mechanism is similar to a corepression mechanism recently described for the cell-specific regulation of the Notch target gene Pitx2 in Xenopus (37). We previously showed that coactivation by Mam is promoter specific and that Mam does not coactivate the ac promoter, even if an SPS element is present (6). These previous studies suggested that the ac promoter lacks a DNA binding site that is present in the m8 promoter to which an unknown coactivator binds. On the basis of the current and previous studies, we predict that Mam is unlikely to be playing a role in regulation of the ac promoter or to be associated with the Su(H)/NICD complex on the ac promoter.
Differential regulation of gene transcription by distinct S-site architectures.
To test the prediction that changing the S-site architecture could cause the ac promoter to have an m8-like response to NICD, we modified the ac 0.9-kb promoter by adding a second S site to create an SPS element. In striking contrast to the wild-type ac promoter, the ac-SPS promoter was strongly and synergistically activated by coexpression of NICD and Ac/Da proteins in S2 cells (Fig. 5A, compare experiment 4 with experiment 8). Conversely, a mutated m8 promoter containing only a single S site with S-site architecture similar to that in the ac promoter did not mediate Notch-proneural transcriptional synergy (Fig. 5B, compare experiment 12 with experiment 16). Thus, simply interchanging the S-site architectures of the ac and m8 promoters resulted in the functional interconversion of their response to Notch signaling. This clearly demonstrates the importance of S-site architecture in programming target gene transcription activation in response to Notch signaling.
Our results contrast with the results of a previous study that suggested an S+A “logic” or transcription code is the most accurate and most general description of the cis regulatory code that mediates activation of E(spl)-C genes in non-NPCs (5). This previous study showed that E(spl)-C mα expression was synergistically upregulated in non-NPC cells by a promoter that contains both S and A sites, but no SPS elements. These previous findings indicate that at least one additional S-site architecture, other than the SPS element, is able to mediate Notch-proneural transcriptional synergy. However, since the ac, m8, and mα promoters each contain functional S and A sites, our current results demonstrate that a purely combinatorial S+A transcription code is not sufficient for predicting or explaining whether expression of a gene is activated or repressed in non-NPCs during Notch signaling.
Although the differences in S-site architectures appears to underlie the differential responses of m8 and ac to Notch signaling in non-NPCs, the mechanisms that underlie the differential responses between mα and ac in non-NPCs are unclear. Both of these promoters have a combination of A sites and S sites, but only mα is strongly activated in response to Notch signaling in the non-NPCs. The mα promoter has multiple “unpaired” S sites (i.e., non-SPS) that may contain an alternative S-site architecture capable of mediating synergistic interactions between Notch transcription complexes and proneural bHLH A proteins. However, many different architectural variations of unpaired S sites have been tested for mediating Notch-bHLH synergy, and essentially none of the non-SPS architectures mediated strong synergy with HLH proteins bound to nearby A sites (7). Therefore, a more likely mechanism may be that there are additional, unknown DNA-binding cofactors specifically bound to the mα promoter that synergistically interact with both Notch transcription complexes and proneural bHLH A proteins.
Given that the S site in the ac promoter is functional and mediates repression in non-NPCs, a remaining question is how ac upregulation in the NPCs overcomes repression via this S site. This derepression mechanism cannot involve Notch signaling, since Notch is not activated in the NPCs. One potential mechanism is that the presence of multiple A sites in the ac promoter allows autoactivation to overcome Su(H)-mediated repression, without specific derepression or activation of Su(H) in the NPCs. Alternatively, or in addition, Su(H) bound to the ac promoter may be functionally derepressed by epidermal growth factor receptor (EGFR) signaling, which is known to counteract Su(H)-mediated repression in the developing eye (39) and to promote proneural gene expression in both mechanosensory and chordontonal NPCs (10, 17, 42).
Implications for transcriptional regulation of Notch target genes.
Our results provide important new insights into the DNA transcription codes that program cell-specific gene expression in response to Notch signaling. We have shown that the ac promoter contains an S-site architecture that mediates repression, not activation, during Notch signaling in proneural clusters. Given that there are unpaired S sites in the promoters of many other proneural and panneural genes (Fig. 2B), we predict that some, or potentially all, of these S sites could mediate repression in cells where Notch is activated. This differential activation versus repression of gene transcription programmed by distinct S-site architectures greatly expands the potential regulatory complexity of pathways mediated by Notch signaling. Our previous studies suggested that specific S-site architectures (S-site “subcodes”) programmed specific interactions between Notch complexes on S sites and specific combinatorial coactivator proteins bound to nearby DNA sites (7). Together with these previous findings, our current study provides an important and novel understanding of the role that S-site architecture plays in mediating differential transcriptional responses to Notch signaling. Given that at least some aspects of the S-site architectural codes are functionally conserved in mammals, it will be interesting and important to test whether the same differential regulation mechanisms are conserved in mammals.
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Bailey, A. M., and J. W. Posakony. 1995. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 9:2609-2622. [DOI] [PubMed] [Google Scholar]
- 2.Bardin, A. J., and F. Schweisguth. 2006. Bearded family members inhibit Neuralized-mediated endocytosis and signaling activity of Delta in Drosophila. Dev. Cell 10:245-255. [DOI] [PubMed] [Google Scholar]
- 3.Barolo, S., T. Stone, A. G. Bang, and J. W. Posakony. 2002. Default repression and Notch signaling: Hairless acts as an adaptor to recruit the corepressors Groucho and dCtBP to Suppressor of Hairless. Genes Dev. 16:1964-1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bray, S. J. 2006. Notch signalling: a simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7:678-689. [DOI] [PubMed] [Google Scholar]
- 5.Castro, B., S. Barolo, A. M. Bailey, and J. W. Posakony. 2005. Lateral inhibition in proneural clusters: cis-regulatory logic and default repression by Suppressor of Hairless. Development 132:3333-3344. [DOI] [PubMed] [Google Scholar]
- 6.Cave, J. W., and M. A. Caudy. 2008. Promoter-specific co-activation by Drosophila mastermind. Biochem. Biophys. Res. Commun. 377:658-661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cave, J. W., F. Loh, J. W. Surpris, X. Li, and M. A. Caudy. 2005. A DNA transcription code for cell-specific gene activation by Notch signaling. Curr. Biol. 15:94-104. [DOI] [PubMed] [Google Scholar]
- 8.Cherbas, L., and P. Cherbas. 2000. Drosophila cell culture and transformation, p. 373-387. In W. Sullivan, M. Ashburner, and R. S. Hawley (ed.), Drosophila protocols. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 9.Cooper, M. T., D. M. Tyler, M. Furriols, A. Chalkiadaki, C. Delidakis, and S. Bray. 2000. Spatially restricted factors cooperate with notch in the regulation of Enhancer of split genes. Dev. Biol. 221:390-403. [DOI] [PubMed] [Google Scholar]
- 10.Culi, J., E. Martin-Blanco, and J. Modolell. 2001. The EGF receptor and N signalling pathways act antagonistically in Drosophila mesothorax bristle patterning. Development 128:299-308. [DOI] [PubMed] [Google Scholar]
- 11.Culi, J., and J. Modolell. 1998. Proneural gene self-stimulation in neural precursors: an essential mechanism for sense organ development that is regulated by Notch signaling. Genes Dev. 12:2036-2047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson, S. R., D. L. Turner, H. Weintraub, and S. M. Parkhurst. 1995. Specificity for the hairy/enhancer of split basic helix-loop-helix (bHLH) proteins maps outside the bHLH domain and suggests two separable modes of transcriptional repression. Mol. Cell. Biol. 15:6923-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emery, J. F., and E. Bier. 1995. Specificity of CNS and PNS regulatory subelements comprising pan-neural enhancers of the deadpan and scratch genes is achieved by repression. Development 121:3549-3560. [DOI] [PubMed] [Google Scholar]
- 14.Fisher, A. L., S. Ohsako, and M. Caudy. 1996. The WRPW motif of the hairy-related basic helix-loop-helix repressor proteins acts as a 4-amino-acid transcription repression and protein-protein interaction domain. Mol. Cell. Biol. 16:2670-2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giagtzoglou, N., P. Alifragis, K. A. Koumbanakis, and C. Delidakis. 2003. Two modes of recruitment of E(spl) repressors onto target genes. Development 130:259-270. [DOI] [PubMed] [Google Scholar]
- 16.Giagtzoglou, N., K. A. Koumbanakis, J. Fullard, I. Zarifi, and C. Delidakis. 2005. Role of the Sc C terminus in transcriptional activation and E(spl) repressor recruitment. J. Biol. Chem. 280:1299-1305. [DOI] [PubMed] [Google Scholar]
- 17.Hasson, P., N. Egoz, C. Winkler, G. Volohonsky, S. Jia, T. Dinur, T. Volk, A. J. Courey, and Z. Paroush. 2005. EGFR signaling attenuates Groucho-dependent repression to antagonize Notch transcriptional output. Nat. Genet. 37:101-105. [DOI] [PubMed] [Google Scholar]
- 18.Ip, Y. T., M. Levine, and E. Bier. 1994. Neurogenic expression of snail is controlled by separable CNS and PNS promoter elements. Development 120:199-207. [DOI] [PubMed] [Google Scholar]
- 19.Jarman, A. P., Y. Grau, L. Y. Jan, and Y. N. Jan. 1993. atonal is a proneural gene that directs chordotonal organ formation in the Drosophila peripheral nervous system. Cell 73:1307-1321. [DOI] [PubMed] [Google Scholar]
- 20.Jarriault, S., C. Brou, F. Logeat, E. H. Schroeter, R. Kopan, and A. Israel. 1995. Signalling downstream of activated mammalian Notch. Nature 377:355-358. [DOI] [PubMed] [Google Scholar]
- 21.Koelzer, S., and T. Klein. 2003. A Notch-independent function of Suppressor of Hairless during the development of the bristle sensory organ precursor cell of Drosophila. Development 130:1973-1988. [DOI] [PubMed] [Google Scholar]
- 22.Kramatschek, B., and J. A. Campos-Ortega. 1994. Neuroectodermal transcription of the Drosophila neurogenic genes E(spl) and HLH-m5 is regulated by proneural genes. Development 120:815-826. [DOI] [PubMed] [Google Scholar]
- 23.Krejci, A., and S. Bray. 2007. Notch activation stimulates transient and selective binding of Su(H)/CSL to target enhancers. Genes Dev. 21:1322-1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, E. C. 2002. Keeping a good pathway down: transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 3:840-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lamar, E., and C. Kintner. 2005. The Notch targets Esr1 and Esr10 are differentially regulated in Xenopus neural precursors. Development 132:3619-3630. [DOI] [PubMed] [Google Scholar]
- 26.Levine, M., and R. Tjian. 2003. Transcription regulation and animal diversity. Nature 424:147-151. [DOI] [PubMed] [Google Scholar]
- 27.Martinez, C., J. Modolell, and J. Garrell. 1993. Regulation of the proneural gene achaete by helix-loop-helix proteins. Mol. Cell. Biol. 13:3514-3521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Morel, V., M. Lecourtois, O. Massiani, D. Maier, A. Preiss, and F. Schweisguth. 2001. Transcriptional repression by suppressor of hairless involves the binding of a hairless-dCtBP complex in Drosophila. Curr. Biol. 11:789-792. [DOI] [PubMed] [Google Scholar]
- 29.Moscoso del Prado, J., and A. Garcia-Bellido. 1984. Genetic regulation of the Acheate-scute complex of Drosophila melanogaster. Roux's Arch. Dev. Biol. 193:242-245. [DOI] [PubMed] [Google Scholar]
- 30.Nagel, A. C., A. Krejci, G. Tenin, A. Bravo-Patino, S. Bray, D. Maier, and A. Preiss. 2005. Hairless-mediated repression of notch target genes requires the combined activity of Groucho and CtBP corepressors. Mol. Cell. Biol. 25:10433-10441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nam, Y., P. Sliz, W. S. Pear, J. C. Aster, and S. C. Blacklow. 2007. Cooperative assembly of higher-order Notch complexes functions as a switch to induce transcription. Proc. Natl. Acad. Sci. U. S. A. 104:2103-2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nellesen, D. T., E. C. Lai, and J. W. Posakony. 1999. Discrete enhancer elements mediate selective responsiveness of enhancer of split complex genes to common transcriptional activators. Dev. Biol. 213:33-53. [DOI] [PubMed] [Google Scholar]
- 33.Oellers, N., M. Dehio, and E. Knust. 1994. bHLH proteins encoded by the Enhancer of split complex of Drosophila negatively interfere with transcriptional activation mediated by proneural genes. Mol. Gen. Genet. 244:465-473. [DOI] [PubMed] [Google Scholar]
- 34.Ohsako, S., J. Hyer, G. Panganiban, I. Oliver, and M. Caudy. 1994. Hairy function as a DNA-binding helix-loop-helix repressor of Drosophila sensory organ formation. Genes Dev. 8:2743-2755. [DOI] [PubMed] [Google Scholar]
- 35.Ong, C. T., H. T. Cheng, L. W. Chang, T. Ohtsuka, R. Kageyama, G. D. Stormo, and R. Kopan. 2006. Target selectivity of vertebrate notch proteins. Collaboration between discrete domains and CSL-binding site architecture determines activation probability. J. Biol. Chem. 281:5106-5119. [DOI] [PubMed] [Google Scholar]
- 36.Paroush, Z., R. L. Finley, Jr., T. Kidd, S. M. Wainwright, P. W. Ingham, R. Brent, and D. Ish-Horowicz. 1994. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell 79:805-815. [DOI] [PubMed] [Google Scholar]
- 37.Sakano, D., A. Kato, N. Parikh, K. McKnight, D. Terry, B. Stefanovic, and Y. Kato. 2010. BCL6 canalizes Notch-dependent transcription, excluding Mastermind-like1 from selected target genes during left-right patterning. Dev. Cell 18:450-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sun, Y., L. Y. Jan, and Y. N. Jan. 1998. Transcriptional regulation of atonal during development of the Drosophila peripheral nervous system. Development 125:3731-3740. [DOI] [PubMed] [Google Scholar]
- 39.Tsuda, L., R. Nagaraj, S. L. Zipursky, and U. Banerjee. 2002. An EGFR/Ebi/Sno pathway promotes delta expression by inactivating Su(H)/SMRTER repression during inductive notch signaling. Cell 110:625-637. [DOI] [PubMed] [Google Scholar]
- 40.Van Doren, M., A. M. Bailey, J. Esnayra, K. Ede, and J. W. Posakony. 1994. Negative regulation of proneural gene activity: hairy is a direct transcriptional repressor of achaete. Genes Dev. 8:2729-2742. [DOI] [PubMed] [Google Scholar]
- 41.Van Doren, M., P. A. Powell, D. Pasternak, A. Singson, and J. W. Posakony. 1992. Spatial regulation of proneural gene activity: auto- and cross-activation of achaete is antagonized by extramacrochaetae. Genes Dev. 6:2592-2605. [DOI] [PubMed] [Google Scholar]
- 42.zur Lage, P. I., L. M. Powell, D. R. Prentice, P. McLaughlin, and A. P. Jarman. 2004. EGF receptor signaling triggers recruitment of Drosophila sense organ precursors by stimulating proneural gene autoregulation. Dev. Cell 7:687-696. [DOI] [PubMed] [Google Scholar]