Abstract
Cwc22 was previously identified to associate with the pre-mRNA splicing factor Cef1/Ntc85, a component of the Prp19-associated complex (nineteen complex [NTC]) involved in spliceosome activation. We show here that Cwc22 is required for pre-mRNA splicing both in vivo and in vitro but is neither tightly associated with the NTC nor required for spliceosome activation. Cwc22 is associated with the spliceosome prior to catalytic steps and remains associated throughout the reaction. The stable association of Cwc22 with the spliceosome requires the presence of the NTC but is independent of Prp2. Although Cwc22 is not required for the recruitment of Prp2 to the spliceosome, it is essential for the function of Prp2 in promoting the release of the U2 components SF3a and SF3b. In the absence of Cwc22, Prp2 can bind to the spliceosome but is dissociated upon ATP hydrolysis without promoting the release of SF3a/b. Thus, Cwc22 represents a novel ATP-dependent step one factor besides Prp2 and Spp2 and has a distinct role from that of Spp2 in mediating the function of Prp2.
The splicing of precursor mRNAs (pre-mRNAs) requires five small nuclear RNAs (snRNAs), U1, U2, U4, U5, and U6, and numerous protein factors. These factors bind to the pre-mRNA in a sequential manner to form a large ribonucleoprotein complex, called the spliceosome, which catalyzes two consecutive steps of transesterification to excise the intron. After the binding of all five snRNAs, a major structural change occurs on the spliceosome, leading to the release of U1 and U4 and the formation of the active spliceosome, which is competent for catalyzing transesterification reactions (for a review, see references 45 and 47).
The spliceosome is a highly dynamic structure and undergoes repetitive remodeling throughout the assembly pathway to rearrange its structure at the expense of ATP (3, 23, 45). Eight DExD/H-box ATPases are required for the entire splicing process (36). Among them, Prp2 is required for the first catalytic step, and Prp16 is required for the second step. After the spliceosome is activated, the U2 components SF3a and SF3b, which bind to the branch site, are removed in a Prp2-dependent manner (21, 46). The binding of Prp2 to the spliceosome requires Spp2, originally identified as a multicopy suppressor of the prp2-1 mutation (32, 35). Cwc25 is then recruited to the spliceosome to promote the first transesterification reaction (12). The second transesterification reaction is promoted by Prp22, Prp18, and Slu7 but requires the prior action of Prp16 in an ATP-dependent manner. After the reaction is complete, mature mRNA is first released and the spliceosome is then disassembled. Both steps require ATP and the DExD/H-box proteins Prp22 and Prp43, respectively.
During the activation of the spliceosome, a protein complex associated with Prp19, known as the NTC (for nineteen complex), is added to the spliceosome after the release of U1 and U4 (37). The NTC plays a role in stabilizing the association of U5 and U6 by specifying base pair interactions between U6 and the 5′ splice site and between U5 and the exon sequence near the splice junction (4, 5). Seven proteins have been shown to associate with the NTC (7-9, 42) and to bind to the spliceosome concomitantly with Prp19, presumably as an integral complex. However, several of these components, Syf1/Ntc90, Syf2/Ntc31, and Isy1/Ntc30, are not required for NTC-mediated spliceosome activation but are required for the recruitment of the splicing factor Yju2 (6), which is involved in the first catalytic reaction after the activation of the spliceosome (22). Other proteins, including Prp45, Prp46, and Cwc2, have also been implicated as being putative NTC components (1, 29).
Proteomic analyses of proteins associated with the NTC component Cef1/Ntc85 and its orthologues have identified complexes homologous to the NTC in both human and the fission yeast Schizosaccharomyces pombe (28), suggesting the evolutionary conservation of the complex. At least 26 proteins are found to be associated with Cef1/Ntc85 (named CWC [for complexed with Cef1]) in Saccharomyces cerevisiae, many of which comprise previously uncharacterized proteins. However, not all of these components are true NTC components. Cwc25, for instance, was later shown not to be tightly associated with the NTC but to function in the first catalytic reaction after the Prp2 step (12, 46).
Here we show that another CWC component, Cwc22, is also involved in the first catalytic reaction after the spliceosome is activated. Cwc22 is neither tightly associated with known NTC components nor required for spliceosome activation, but like NTC, it is associated with the spliceosome from the precatalytic stage until the completion of the catalytic reactions. The binding of Cwc22 to the spliceosome requires the presence of the NTC yet is independent of Prp2. Prp2 can bind to the spliceosome independently of Cwc22 and, upon ATP hydrolysis, is dissociated from the spliceosome without promoting the release of SF3a/b. The productive function of Prp2 requires the presence of Cwc22. Thus, Cwc22 and Spp2 are both required for Prp2 to function in the first catalytic step but in distinct ways.
MATERIALS AND METHODS
Yeast strains.
The yeast strains used were BJ2168 (MATa prc1 prb1 pep4 leu2 trp1 ura3), YSCC1 (MATa prc1 prb1 pep4 leu2 trp1 ura3 PRP19HA), YSCC12 (MATa his3 his7 ade3 ura3 prp2-1 PRP19-HA), YSCC227 (MATa prc1 prb1 pep4 leu2 trp1 ura3 CWC22HA), and YSCC228 (MATa prc1 prb1 pep4 leu2 trp1 GAL-CWC22::URA3).
Oligonucleotides.
The following oligonucleotides were used: C22-1 (GGCCGGATCCGACCAACTGCGAATCTT), C22-2 (GGCCGACGTCGTATGGGTATCTATGCCGTTGCCTTC), C22-3 (GGCCGACGTCCCAGACTACGCTTGAAGAGCCCGGCCAAA), C22-4 (CCGGCTCGAGGACTTACGGTGTTGCTG), C22-5 (GGCCGGATCCGCATGTCTACCGCTACCAT), C22-6 (CCGTCAAGCTTGCTGGA), C22-7 (CAGCAGCTTCTGTCG), C22-8 (GGCCCGGCCGCCCAATGCGGTG), C22-9 (CCGGGGATCCGTAGCCTTTTTATTCC), C22-10 (GGCCGGATCCGGATGCTGTTTATATTGGACCC), C22-11 (CCGGGACGTCTCATCCTTCCAGCGGAAAC), C22-12 (GGCCACTAGTGACGCCGAACATATAAG), C22-13 (GCGGAGTCCTGGATCTG), C22-14 (CCGGGACGTCTCACTCTGCATCCTCAAC), and R13 (GAGTGACGATTCCTATAG).
Antibodies and reagents.
Antihemagglutinin (anti-HA) monoclonal antibody 8G5F was produced by immunizing mice with a keyhole limpet hemocyanin (KLH)-conjugated HA peptide (our unpublished data) and the anti-Cwc22 polyclonal antibody by immunizing rabbits with the full-length protein expressed in Escherichia coli cells. The anti-Cwc22 antibody was affinity purified on a Cwc22-conjugated Sepharose column for depletion experiments. Protein A-Sepharose was obtained from Amersham Inc., and streptavidin-Sepharose was obtained from Sigma-Aldrich.
Construction of the CWC22-HA and GAL-CWC22 strains.
The construction of the HA-tagged strain was described previously by Tsai et al. (40). For the construction of pRS406.CWC22-HA, DNA fragments A and B were generated by PCR using primers C22-1 and C22-2 and primers C22-3 and C22-4, respectively. Following digestion with BamHI and AatII and with AatII and XhoI, respectively, fragments A and B were ligated with BamHI- and XhoI-digested pRS406. Plasmid pRS406.CWC22C-HA was linearized with HindIII and transformed into yeast strain BJ2168 to displace the wild-type allele with the tagged allele by the pop-in and pop-out gene displacement method (49) to generate strain YSCC227. To construct the GAL-CWC22 strain, a 1.34-kb DNA fragment containing the 5′ end of the CWC22 open reading frame (ORF) was generated by PCR using primers C22-5 and C22-6, digested with BamHI and HindIII, and ligated with BamHI- and HindIII-digested pRS406.CWC22C-HA to yield plasmid pRS406.CWC22-HA, which contained the full-length ORF of CWC22. Plasmid pRS406.CWC22-HA was digested with BamHI and SpeI, and the 0.65-kb BamHI-SpeI fragment was cloned into BamHI-SpeI sites of pRS406.GAL to generate plasmid pRS406.GAL.CWC22N, which was linearized with EcoRI for transformation into strain BJ2168 to generate strain YSCC228.
Construction of deletion mutants.
A 0.6-kb DNA fragment upstream of the CWC22 ORF was generated by PCR using primers C22-8 and C22-9, digested with EagI and BamHI, and ligated with EagI- and BamHI-digested pRS406.CWC22-HA to yield plasmid pTY1. A 2.8-kb fragment was isolated by the digestion of pTY1 with EagI and XhoI and ligated with EagI- and XhoI-digested pRS414 to yield plasmid pTY2. For the generation of the 212-577 mutant, a 121-bp DNA fragment was generated by PCR using primers C22-7 and C22-10, digested with BamHI and NcoI, and ligated with BamHI- and NcoI-digested pTY2 to yield plasmid pTY3. For the generation of the 1-453 mutant, a 141-bp DNA fragment was generated by PCR using primers C22-1 and C22-11, digested with HpaI and AatII, and ligated with HpaI- and AatII-digested pTY2 to yield plasmid pTY4. For the generation of the Δ220-453 mutant, a 250-bp DNA fragment was generated by PCR using primers C22-12 and C22-13, digested with SpeI and BsaBI, and ligated with SpeI- and BsaBI-digested pTY2 to yield plasmid pTY5. For the generation of the 212-453 mutant, a 121-bp DNA fragment was generated by PCR using primers C22-7 and C22-10, digested with BamHI and NcoI, and ligated with BamHI- and NcoI-digested pTY4 to yield plasmid pTY6. For the generation of the 212-491 mutant, a 255-bp DNA fragment was generated by PCR using primers C22-1 and C22-14, digested with HpaI and AatII, and ligated with HpaI- and AatII-digested pTY6 to yield plasmid pTY7.
Splicing extracts, substrates, and assays.
Yeast whole-cell extracts were prepared according to a method described previously by Cheng et al. (11). Actin precursors were synthesized in vitro, using SP6 RNA polymerase according to a method described previously by Cheng and Abelson (10). Biotinylated pre-mRNA was synthesized according to a procedure described previously by Chan et al. (5). Splicing assays were carried out according to methods described previously by Cheng and Abelson (10). To assay for the stable association of U5 and U6, splicing was carried out in a volume of 300 μl, and the spliceosome was precipitated with 75 μl of streptavidin-Sepharose. After the washing off of unbound materials, the pellet was separated into three fractions: one was used for total precipitate, and 100 μl each of the splicing buffer with or without ATP was added to the other two fractions, followed by incubation at room temperature for 20 min. After removing the supernatant, the pellet was further washed, and RNAs in the pellet and supernatant fractions were analyzed by Northern blotting.
Immunodepletion, immunoprecipitation, and precipitation of the spliceosome by streptavidin-agarose.
The immunodepletion of the NTC was performed as described previously by Chan et al. (5). The immunodepletion of Yju2 and Cwc22 was performed by the incubation of 100 μl of splicing extracts with 100 μl of anti-Yju2 antiserum or 20 μl of 9 mg/ml affinity-purified anti-Cwc22 antibody coupled to 50 μl of protein A-Sepharose. Immunoprecipitation was performed as described by previously by Tarn et al. (38), using 25 μl of anti-HA for Cwc22-HA or 5 μl of anti-Ntc20 antibody for each 100-μl splicing reaction mixture. Precipitation of the spliceosome with streptavidin-agarose was carried out according to methods described previously by Chan et al. (5).
Purification of recombinant Cwc22.
The full-length CWC22 gene and the fragment of the CWC22 gene at residues 212 to 491 were cloned into a modified version of pET15b for the expression of His-tagged proteins in E. coli. Recombinant proteins were purified on an Ni affinity column (Novagen) according to the manufacturer's instructions.
Assay for release of Prp2 from the spliceosome.
Splicing reactions were carried out with V5-tagged Prp2 extracts under normal conditions. The reaction mixtures were incubated for 10 min following the addition of 2% glucose, and each 20 μl of the reaction mixture was precipitated with 1 μl of anti-V5 antibody conjugated to 10 μl of protein A-Sepharose. The precipitates were then incubated for 20 min with 20 μl splicing buffer containing 3 mM MgCl2 with or without 2 mM ATP, and the pellet and supernatant fractions were separated. In the assay for Prp2 function, the prp2S378L-V5 protein was added at 100 nM in the splicing reaction mixture, which used nontagged extracts. For the chase assay, 6 μl of micrococcal nuclease (MN)-treated extracts was added to 20 μl of the released spliceosome, followed by incubation for 20 min.
Complementation of the affinity-purified spliceosome.
Splicing reactions were carried out with Cwc22-depleted extracts under normal conditions. Each 20 μl of the reaction mixture was precipitated with 1 μl of anti-Ntc20 antibody conjugated to 10 μl of protein A-Sepharose. After the washing of unbound materials, 9 μl of micrococcal nuclease-treated extracts and 3 μl of Prp2 protein in 30 μl of splicing buffer with or without 2 mM ATP were added to the precipitates, followed by incubation for 20 min at 25°C.
RESULTS
Cwc22 is not tightly associated with the NTC.
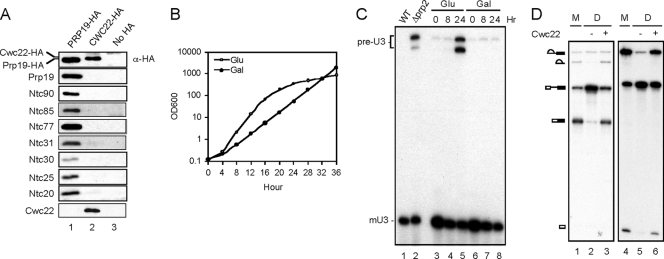
To examine whether Cwc22 is a component of the NTC, Cwc22 was tagged with the HA epitope at its carboxy terminus for immunoprecipitation analyses. Extracts prepared from PRP19-HA, CWC22-HA, or untagged strains were precipitated with anti-HA antibody, followed by Western blotting with antibodies against NTC components as probes (Fig. 1A). Cwc22-HA was not precipitated as efficiently as Prp19-HA with anti-HA antibody; the amount of Cwc22-HA precipitated was lower than that of Prp19-HA despite the fact that 10-times-more antibody was used for precipitation and 3-times-more precipitated materials were used for analysis, indicating that the HA epitope on Cwc22 may not be as accessible. Nevertheless, our results clearly showed that Cwc22 is not tightly associated with the NTC since none of the identified NTC components were significantly coprecipitated with Cwc22 (Fig. 1A, lane 2). Consistent with these results, Cwc22 was not detected to coprecipitate with Prp19 when probed with antibody raised against recombinant Cwc22 (lane 1). We conclude that Cwc22 is not an intrinsic component of the NTC.
FIG. 1.
Cwc22 is not a component of the NTC but is essential for pre-mRNA splicing both in vivo and in vitro. (A) Extracts prepared from PRP19-HA (lane 1), CWC22-HA (lane 2), and nontagged (lane 3) strains were immunoprecipitated with anti-HA antibody, followed by Western blotting using antibodies against the HA epitope and components of the NTC. (B) Growth curves of GAL-CWC22 cells in yeast extract-peptone-dextrose (YPD) and yeast extract-peptone galactose (YPG). Cells were grown in galactose-containing synthetic minimum medium to mid-log phase and then shifted to YPD or YPG medium. Cells were collected at 0, 4, 8, 12, 16, 20, 24, 28, 32, and 36 h after the shift for measurements of the optical density at 600 nm (OD600). (C) Total RNA extracted from collected cells was analyzed by primer extension using a U3 primer, R13. The prp2 mutant was grown at 37°C for 2 h before harvesting. Gal, galactose; Glu, glucose; WT, wild type. (D) Splicing was carried out in mock-treated (lanes 1 and 4) or Cwc22-depleted (lanes 2, 3, 5, and 6) extracts using wild-type (lanes 1 to 3) or ACAC (lanes 4 to 6) actin pre-mRNA with (lanes 3 and 6) or without (lanes 1, 2, 4, and 5) the addition of 100 ng recombinant Cwc22. M, mock; D, depletion.
CWC22 was previously reported to be essential for cellular viability by a large-scale survey of the yeast genome (13). To investigate whether CWC22 is essential for splicing, we constructed a yeast strain in which CWC22 was placed under the control of the inducible GAL1 promoter. The growth of such cells was retarded 20 h after transfer from galactose- to glucose-containing medium (Fig. 1B). RNA isolated from cells grown in either of the two media was probed for the accumulation of precursor RNA by primer extension analysis using primer R13 in the exon 2 region of the U3 gene (Fig. 1C). An accumulation of pre-U3 was seen in cells grown for 24 h in glucose-containing medium (Fig. 1C, lane 5) but not in galactose-containing medium (lane 8), indicating a splicing defect upon the repression of CWC22. This result shows that CWC22 is essential for splicing in vivo.
To see whether Cwc22 is required for the in vitro splicing reaction, we depleted Cwc22 from splicing extracts with antibodies against Cwc22 for splicing assays (Fig. 1D). Using either the wild type (Fig. 1D, lanes 1 to 3) or a 3′ splice site mutant, ACAC (44) (lanes 4 to 6), of actin pre-mRNA as a substrate, the splicing activity was nearly completely abolished upon Cwc22 depletion (lanes 2 and 5). The addition of recombinant Cwc22 restored the splicing activity (Fig. 1D, lanes 3 and 6), demonstrating that Cwc22 is required for splicing in vitro, either for the first catalytic reaction or for the prior assembly of the spliceosome. This result further confirms that Cwc22 is not tightly associated with the NTC since the depletion of Cwc22 did not codeplete other essential splicing factors to significant levels.
A segment of Cwc22 containing the MA-3 domain and the conserved downstream region is sufficient for the function of Cwc22.
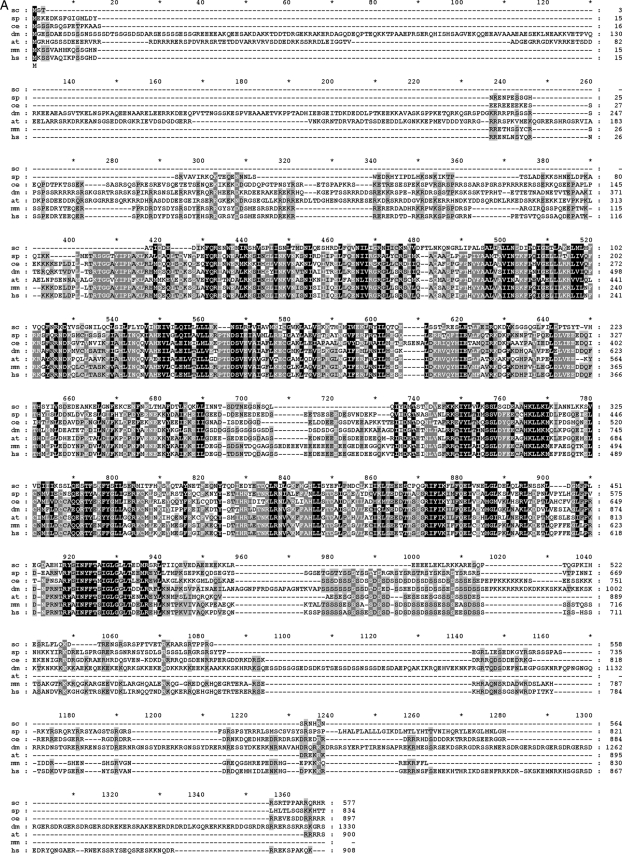
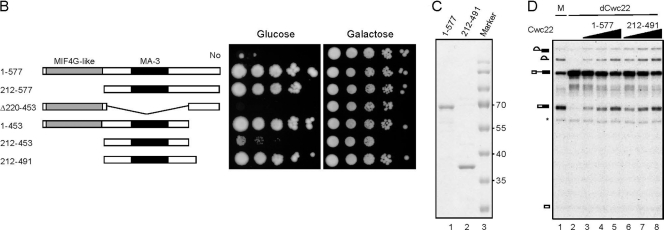
Cwc22 is a protein of 577 amino acid residues with an MIF4G-like domain in the N-terminal region and a MA-3 domain in the middle region. Sequence alignment of Cwc22 orthologues revealed that the budding yeast Cwc22 represents a short form of the protein containing only the core region (Fig. 2A). While proteins from most other species have a length of around 900 amino acid residues, the fly protein is extraordinarily long, with 1,330 amino acid residues, and contains extra domains in both the amino and carboxy termini of the protein. To determine the contribution of different domains to the function of Cwc22, we expressed Cwc22 with deletions in different regions of the protein in strain YSCC228 and grew cells in galactose- or glucose-based medium. Figure 2B shows that the deletion of the N-terminal MIF4G-like domain or the C-terminal region of the protein did not affect the growth of yeast cells, whereas the deletion of the middle region containing the MA-3 domain resulted in lethality. A Cwc22 segment encompassing amino acid residues 212 to 453 poorly supported cellular growth. However, if the segment was extended to amino acid residue 491 to include the entire conserved sequence downstream of the MA-3 domain, cells were viable, with a slightly reduced growth rate. To verify that this segment, which is sufficient for the cellular function of Cwc22, largely carries out the function of Cwc22 in pre-mRNA splicing, we purified the recombinant protein for in vitro complementation assays. The titration of the purified recombinant proteins (Fig. 2C) revealed that the complementation activity of the segment at residues 212 to 491 is around 30% of that of the full-length protein (Fig. 2D), validating that the segment at residues 212 to 491, although not in full activity, is sufficient for the function of Cwc22 in pre-mRNA splicing.
FIG. 2.
Deletion analysis of Cwc22. (A) Protein sequence alignment of Cwc22 orthologues using MAFFT and GeneDoc. Putative Cwc22 orthologues were identified by using the KEGG (Kyoto Encyclopedia of Genes and Genomes) database (17). Conserved residues are shaded in black for 100%, in gray for 80%, and in light gray for 60% conservation. Abbreviations: Sc, Saccharomyces cerevisiae; Sp, Schizosaccharomyces pombe; Dm, Drosophila melanogaster; At, Arabidopsis thaliana; Ce, Caenorhabditis elegans; Rn, Rattus norvegicus; Mm, Mus musculus; Hs, Homo sapiens. (B) CWC22 deletion clones in yeast strain YSCC228 were grown in galactose-containing synthetic minimum medium and spotted onto glucose- or galactose-based plates after serial dilutions. (C) An estimated 1.5 μg each of purified Cwc22 (lane 1) and the segment at residues 212 to 491 (lane 2) was analyzed on 10% SDS-PAGE gels stained by Coomassie blue. (D) Recombinant Cwc22 at 5, 10, or 20 ng (lanes 3 to 5, respectively) or the segment at residues 212 to 491 at 10, 20, or 50 ng (lanes 6 to 8, respectively) was added to Cwc22-depleted extracts (lane 2) for splicing assays. M, mock; dCwc22, Cwc22 depletion; *, nonspecific band.
Cwc22 is associated with the spliceosome after NTC-mediated spliceosome activation.
An immunoprecipitation analysis using CWC22-HA extracts was performed to examine whether Cwc22 is associated with the spliceosome (Fig. 3). Splicing was carried out with CWC22-HA extracts at various ATP concentrations, and the reaction mixture was precipitated with anti-HA or anti-Ntc20 antibody, respectively, for comparison. Although a large amount of the spliceosome containing pre-mRNA was coprecipitated with Ntc20 at 0.1 mM ATP (Fig. 3, lane 3), none was significantly coprecipitated with Cwc22 (lane 4). In contrast, at 0.5 mM or 2 mM ATP, both Ntc20 and Cwc22 were found to associate with the spliceosome at precatalytic and catalytic stages, as pre-mRNA, splicing intermediates, and the lariat intron were all coprecipitated (Fig. 3, lanes 7, 8, 11, and 12). The amount of the spliceosome coprecipitated with Cwc22 was approximately one-half of that coprecipitated with Ntc20, suggesting a difference in the efficiency of precipitation of Cwc22 and Ntc20 by 2-fold. Taken together, these results suggest that Cwc22 might be associated with the spliceosome in a step after the binding of the NTC but prior to the first catalytic reaction.
FIG. 3.
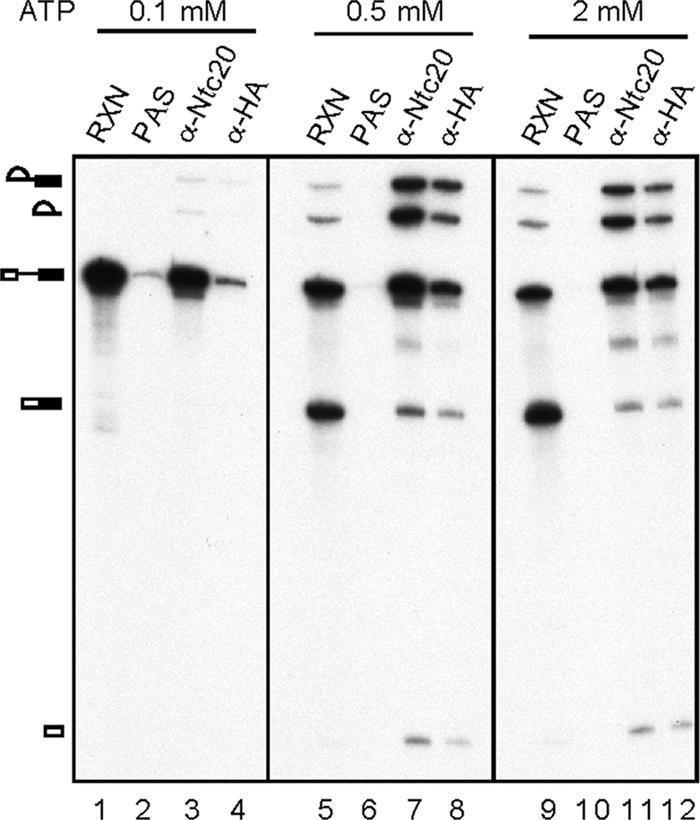
Cwc22 is associated with the spliceosome prior to the catalytic step and retained until completion of the reaction. Splicing was carried out with CWC22-HA extracts in the presence of 0.1 mM (lanes 1 to 4), 0.5 mM (lanes 5 to 8), or 2 mM (lanes 9 to 12) ATP, and the reaction mixtures were precipitated with anti-Ntc20 or anti-HA antibody. RXN, 1/10 of the reaction mixture used for immunoprecipitation; PAS, protein A-Sepharose.
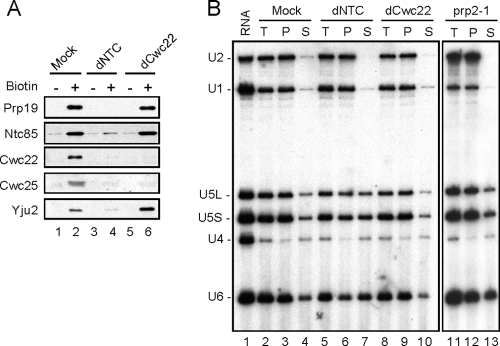
To confirm the binding order of Cwc22 and the NTC, we analyzed components of the spliceosome formed in Cwc22- and NTC-depleted extracts using biotinylated pre-mRNA for precipitation with streptavidin-Sepharose (Fig. 4A). The ACAC pre-mRNA was used in order to assemble and accumulate the spliceosome within one round of splicing, preventing the spliceosome from maturation and disassembly. Western blotting of the pulled-down spliceosome revealed that the depletion of the NTC from the extract prevented the binding of Cwc22 to the spliceosome (Fig. 4A, lane 4), whereas the depletion of Cwc22 did not affect the binding of the NTC components Prp19 and Ntc85 to the spliceosome (lane 6). The results therefore indicate that the stable association of Cwc22 with the spliceosome requires the presence of the NTC and are in agreement with data from the immunoprecipitation analysis of the spliceosome shown in Fig. 3. Two splicing factors, Yju2 and Cwc25, were recently reported to be required for the first catalytic reaction after the Prp2-mediated structural change of the spliceosome (12, 22). Yju2, although required for the first catalytic reaction, can bind to the spliceosome in an earlier step via its interaction with the NTC component Syf1/Ntc90 (6). Consistent with the previously reported observations, we found that the binding of Yju2 to the spliceosome did not require Cwc22 (Fig. 4A, lane 6). In contrast, the binding of Cwc25 to the spliceosome required the presence of Cwc22, suggesting that Cwc22 is required either for spliceosome activation after the binding of the NTC or for the first catalytic reaction.
FIG. 4.
Cwc22 is not required for NTC-mediated spliceosome activation. (A) The spliceosome formed with nonbiotinylated (lanes 1, 3, and 5) or biotinylated (lanes 2, 4, and 6) ACAC pre-mRNA in mock-treated (lanes 1 and 2), NTC-depleted (dNTC) (lanes 3 and 4), or Cwc22-depleted (lanes 5 and 6) extracts was isolated by precipitation with streptavidin-Sepharose, and the components were analyzed by Western blotting. (B) Splicing reactions were carried out with mock-depleted (lanes 2 to 4), NTC-depleted (lanes 5 to 7), Cwc22-depleted (lanes 8 to 10), or prp2-1 (lanes 11 to 13) extracts using biotinylated ACAC pre-mRNA as the substrate, and the spliceosome was precipitated with streptavidin-Sepharose. After washing off unbound materials, the pellet was separated into two fractions: one was used for total precipitate (lanes 2, 5, 8, and 11), and the other was added to splicing buffer and incubated for 20 min at room temperature. After separating the supernatant and pellet fractions, RNA was extracted and analyzed by Northern blotting. RNA, RNA from 2 μl of extracts; T, total precipitate; P, pellet; S, supernatant.
We have previously demonstrated that the NTC is required for spliceosome activation and that the depletion of the NTC results in the destabilization of U5 and U6 due to the lack of specific interactions between U6 and the intron and between U5 and the exon sequence near splice junctions (4, 5). We thus examined whether the stable association of U5 and U6 with the spliceosome required Cwc22 to determine a possible role of Cwc22 in spliceosome activation. Spliceosomes formed with biotinylated ACAC pre-mRNA in Cwc22- or NTC-depleted extracts were isolated by precipitation with streptavidin-Sepharose and then reincubated in splicing buffer at 25°C. The RNA retained on and released from the spliceosome was analyzed by Northern blotting using snRNAs as probes. Figure 4B shows that, similarly to mock-treated extracts (lanes 2 to 4), the majority of U5 and U6 remained associated with the spliceosome formed in Cwc22-depleted extracts (lanes 8 to 10), whereas around 50% of U5 and a higher percentage of U6 were dissociated from the spliceosome formed in NTC-depleted extracts (lanes 5 to 7). These results indicate that Cwc22 has a distinct function from that of the NTC, as Cwc22 is not required for the stabilization of U5 and U6 after the release of U1 and U4. When the experiment was performed with heat-treated prp2-1 mutant extracts (Fig. 4B, lanes 11 to 13), the majority of U5 and U6 was also retained on the spliceosome, as in mock-treated extracts, indicating that Prp2 is not required for the stable association of U5 and U6. If the spliceosome is defined as being activated when U5 and U6 become stably bound, then neither Prp2 nor Cwc22 is required for the activation of the spliceosome. The ATP-dependent action of Prp2 in mediating SF3a/b release may function in preparing the activated spliceosome for the first catalytic reaction, which requires Yju2, Cwc25, and HP-X (12, 22). Our data concur with the possibilities that Cwc22 is involved in the Prp2 step or the post-Prp2 step.
Cwc22 is involved in an ATP-dependent step of the splicing reaction.
To distinguish between these two steps, we examined whether the function of Cwc22 required ATP (Fig. 5A). Splicing was carried out with Cwc22-depleted extracts for 20 min using ACAC pre-mRNA, followed by the addition of glucose to the reaction mixtures to exhaust ATP (Fig. 5A, lanes 2 and 3) or of more ATP as a control (lanes 4 and 5). Recombinant Cwc22 was then added to the reaction mixtures for complementation. Figure 5A shows that recombinant Cwc22 was able to restore the splicing activity only in the presence of ATP (lane 5) although to a level slightly lower than that in the control of mock-treated extracts (lane 1) or in the control in which Cwc22 was readded prior to the splicing reaction (lane 7). This result indicates that Cwc22 is involved in the same step as Prp2 that requires ATP.
FIG. 5.
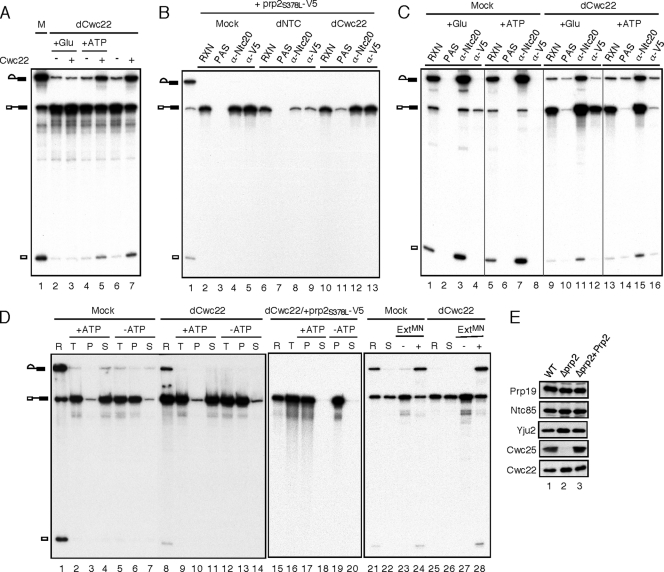
Cwc22 and Prp2 bind to the spliceosome independently of each other. Splicing was carried with ACAC pre-mRNA in these experiments. (A) Splicing was carried out with mock-treated (lane 1) or Cwc22-depleted (lanes 2 to 7) extracts without (lanes 2 to 6) or with (lane 7) the addition of recombinant Cwc22. Glucose (lanes 2 and 3) or ATP (lanes 4 and 5) was added to the reaction mixtures and incubated for 5 min. Cwc22 was then added, and the reaction mixture was incubated for 20 min. (B) Splicing was carried out with wild-type (lane 1), mock-treated (lanes 2 to 5), NTC-depleted (lanes 6 to 9), or Cwc22-depleted (lanes 10 to 13) extracts. After the addition of recombinant V5-tagged prp2S378L, the reaction mixtures were precipitated with anti-Ntc20 or anti-V5 antibody. RXN, 1/5 of the reaction mixture used for immunoprecipitation; PAS, protein A-Sepharose. (C) Splicing was carried out with mock-treated (lanes 1 to 8) or Cwc22-depleted (lanes 9 to 16) Prp2-V5 extracts. Following the addition of glucose (lanes 1 to 4 and 9 to 12) or ATP (lanes 5 to 8 and 13 to 16) and incubation for 5 min, the reaction mixtures were precipitated with anti-Ntc20 or anti-V5 antibody. RXN, 1/10 of the reaction mixture used for immunoprecipitation; PAS, protein A-Sepharose. (D) Splicing was carried out with mock-treated (lanes 1 to 7) or Cwc22-depleted (lanes 8 to 14) Prp2-V5 extracts or with Cwc22-depleted extracts with the addition of the prp2S378L-V5 protein (lanes 15 to 20). Following the addition of glucose and incubation for 5 min, the reaction mixtures were precipitated with anti-V5 antibody. The precipitates were reincubated in splicing buffer containing (lanes 2 to 4, 9 to 11, 17, and 18) or not containing (lanes 5 to 7, 12 to 14, 19, and 20) ATP, and supernatant and pellet fractions were separated. The released materials (lanes 22 and 26, as from lanes 4 and 11) were incubated for 20 min following the addition (lanes 24 and 28) or no addition (lanes 23 and 27) of micrococcal nuclease-treated extracts. R, 1/10 of the reaction mixture used for immunoprecipitation; T, total precipitates; P, pellet; S, supernatant; ExtMN, micrococcal nuclease-treated extracts. (E) The spliceosome formed with biotinylated ACAC pre-mRNA in wild-type (lane 1) or Δprp2 extracts without (lane 2) or with (lane 3) the addition of the Prp2 protein was isolated by precipitation with streptavidin-Sepharose, and the components were analyzed by Western blotting.
Independent binding of Cwc22 and Prp2 to the spliceosome.
Prp2 and Spp2 are required for the first catalytic step in an ATP-dependent manner. The interaction of Spp2 with Prp2 is required for the association of Prp2 with the spliceosome to execute its function in this step. To determine whether Cwc22 is also required for the function of Prp2, we first examined whether Cwc22 is required for the binding of Prp2 to the spliceosome. During the splicing reaction, Prp2 leaves the spliceosome immediately after its action and is barely detected on the spliceosome in the presence of ATP (19) (see below). To retain Prp2 on the spliceosome, we used a dominant negative mutant of PRP2, S378L, which carries a mutation in the SAT motif (24, 30). Splicing was carried out with Cwc22-depleted or NTC-depleted extracts, followed by the addition of V5-tagged recombinant prp2S378L. The reaction mixtures were then precipitated with anti-Ntc20 or anti-V5 antibody to examine the association of the NTC and Prp2 with the spliceosome (Fig. 5B). A higher background was observed for the immunoprecipitation of reactions carried out with Cwc22-depleted extracts (Fig. 5B, lane 11), possibly due to a small amount of anti-Cwc22 antibody released into the extract during its prior incubation with the antibody to deplete Cwc22 (data not shown). As expected, prp2S378L could bind to the spliceosome in mock-treated extracts (Fig. 5B, lane 5) but failed to bind when the NTC was depleted (lane 9). In Cwc22-depleted extracts, prp2S378L was also able to bind to the spliceosome (Fig. 5B, lane 13), indicating that the binding of prp2S378L is independent of Cwc22.
A complementary experiment was also performed to show that Cwc22 is not required for the binding of wild-type Prp2 (Fig. 5C). Splicing was carried out with mock-treated or Cwc22-depleted extracts prepared from a Prp2-V5 strain using ACAC pre-mRNA, followed by the addition of glucose to the reaction mixtures to exhaust ATP (Fig. 5C, lanes 1 to 4 and 9 to 12) or more ATP as controls (lanes 5 to 8 and 13 to 16). The depletion of ATP allows the retention of Prp2 on the spliceosome, which could be revealed by the precipitation of the spliceosome with anti-V5 antibody. Like prp2S378L, wild-type Prp2 was able to bind to the spliceosome containing pre-mRNA in the absence of ATP in Cwc22-depleted extracts (Fig. 5C, lane 12), as in mock-treated extracts (lane 4), further confirming that Cwc22 is not required for the binding of Prp2 to the spliceosome. In Cwc22-depleted extracts, small amounts of splicing intermediates were seen when precipitated with anti-V5 antibody (Fig. 5C, lanes 12 and 16) due to the presence of anti-Cwc22 antibody released from beads during the incubation for depletion, as similar amounts of intermediates were also present in no-antibody controls (lanes 10 and 14). It is interesting that Prp2 was not retained on the spliceosome in the presence of ATP when Cwc22 was depleted (Fig. 5C, lane 16). This finding suggests that Cwc22 is also not required for the dissociation of Prp2 from the spliceosome. To provide further evidence for this conclusion, the Prp2-associated spliceosome formed in Cwc22-depleted extracts was isolated by immunoprecipitation with anti-V5 antibody as described above and then reincubated in the presence or absence of ATP to see whether the spliceosome would be released from beads, which indicates the release of Prp2 from the spliceosome (Fig. 5D). Indeed, Prp2 was released from the spliceosome after incubation in the presence of ATP with (Fig. 5D, lane 11) or without (lane 4) the depletion of Cwc22. When the V5-tagged prp2S378L mutant protein was added to Cwc22-depleted extracts, the prp2S378L-associated spliceosome precipitated with anti-V5 antibody was retained on the spliceosome after incubation regardless of the presence of ATP (Fig. 5D, lanes 17 to 20), indicating that the ATP-dependent release required functional Prp2. Furthermore, the spliceosome released from Prp2-conjugated beads, regardless of the Cwc22 depletion, remained intact and functional as the addition of micrococcal nuclease (MN)-treated extracts allowed the first catalytic reaction to proceed (Fig. 5D, lanes 24 and 28). These results demonstrate that the Prp2 cycle on the spliceosome is independent of Cwc22 such that Prp2 can bind to the spliceosome, hydrolyze ATP, and then dissociate without requiring Cwc22.
Interestingly, the binding of Cwc22 to the spliceosome is also independent of Prp2 (Fig. 5E). When the spliceosome formed with biotinylated ACAC pre-mRNA in heat-treated prp2 mutant extracts was isolated for Western blotting, Cwc22 was found to be associated (Fig. 5E, lane 2), in contrast to Cwc25, whose binding required functional Prp2 (12). Taken together, Cwc22 and Prp2 bind to the spliceosome independently of each other.
The productive function of Prp2 requires Cwc22.
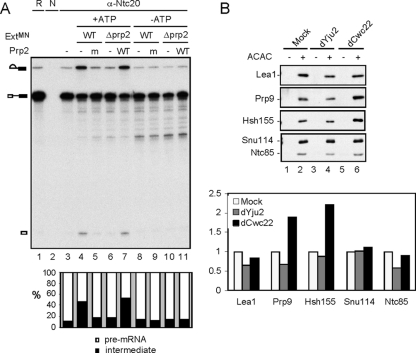
Since both Cwc22 and Prp2 require ATP for their function in the first catalytic step but can independently bind to the spliceosome, they act either in concert or in sequence to mediate the reaction. To determine the functional order of Cwc22 and Prp2, we isolated the spliceosome formed in Cwc22-depleted extracts for chase experiments (Fig. 6A). The spliceosome formed in Cwc22-depleted extracts with ACAC pre-mRNA was isolated by precipitation with anti-Ntc20 antibody. MN-treated extracts prepared from wild-type or Δprp2 extracts were then added in the presence or absence of ATP to chase the reaction. If the isolated spliceosome was at the post-Prp2 stage, either of the MN-treated wild-type or Δprp2 extracts would provide all the factors required for the first catalytic reaction. However, the result shows that only MN-treated wild-type but not Δprp2 extracts could promote the first catalytic reaction (Fig. 6A, lanes 4 and 6). The further addition of recombinant Prp2 to MN-treated Δprp2 extracts was also able to chase the reaction (Fig. 6A, lane 7). This result shows that despite the independent binding of these two proteins, Cwc22 is required for the productive function of Prp2 in the first catalytic step. In the absence of Cwc22, Prp2 binds to the spliceosome and dissociates upon ATP hydrolysis in a nonproductive manner.
FIG. 6.
Cwc22 is required for the function of Prp2. (A) Splicing was carried out with Cwc22-depleted extracts with ACAC pre-mRNA, and the reaction mixtures were precipitated with anti-Ntc20 antibody (lanes 3 to 11). Micrococcal nuclease-treated wild-type (lanes 4, 5, 8, and 9) or Δprp2 (lanes 6, 7, 10, and 11) extracts and wild-type (lanes 7 and 11), S378L (lanes 5 and 9), or no (lanes 4, 6, 8, and 10) Prp2 protein were added to the precipitated spliceosome and incubated in the presence (lanes 4 to 7) or absence (lanes 8 to 11) of ATP. The amounts of pre-mRNA and lariat intron-exon 2 were quantified by PhosphorImager analysis and, after conversion into molar amounts, plotted in a bar graph, with their sum as 100%. R, 1/10 of the reaction mixture; N, no antibody; WT, wild type; Δprp2, heat-treated prp2; m, mutant; ExtMN, micrococcal nuclease-treated extracts. (B) Splicing was carried out in the absence (lanes 1, 3, and 5) or presence (lanes 2, 4, and 6) of biotinylated ACAC pre-mRNA in mock-treated (lanes 1 and 2), Yju2-depleted (lanes 3 and 4), or Cwc22-depleted (lanes 5 and 6) Lea1-V5, Prp9-V5, or Hsh155-HA extracts. The spliceosomes were isolated by precipitation with streptavidin-Sepharose and analyzed by Western blotting using anti-V5, anti-HA, anti-Snu114, and anti-Ntc85 antibodies. Protein bands were quantified by using the UVP Biospectrum 600 imaging system and plotted in a bar graph, with each protein normalized to mock. dYju2, Yju2 depletion; dCwc22, Cwc22 depletion.
It was recently demonstrated that the U2 components SF3a and SF3b are dissociated from the spliceosome during the first catalytic step and that Prp2 plays a role in mediating the release of SF3a/b (21, 46). If Cwc22 was required for the function of Prp2 in the spliceosome pathway, the depletion of Cwc22 would prevent SF3a/b from dissociation from the spliceosome. We examined the role of Cwc22 in this connection by Western blotting of the spliceosome formed in Cwc22- and Yju2-depleted extracts. Yju2 is required for the first catalytic step after the action of Prp2 (22) and consequently is not required for the release of SF3a/b. In this experiment, Prp9 and Hsh155, as representative components of SF3a and SF3b, respectively, were tagged with V5 and HA. Another U2 component, Lea1, which remains associated with the spliceosome until the completion of splicing, was also tagged with V5 as a control. Spliceosomes formed with biotinylated ACAC pre-mRNA were formed in Yju2- or Cwc22-depleted extracts, using LEA1-V5, PRP9-V5, or HSH155-HA extracts, and isolated by precipitation with streptavidin-Sepharose followed by Western blotting probed with anti-HA or anti-V5 antibody and antibodies against Snu114 and Ntc85. As shown in Fig. 6B, the amounts of Prp9 or Hsh155 associated with the spliceosome were about twice as much in Cwc22-depleted extracts as those in Yju2- or mock-depleted extracts, whereas the amounts of Lea1, Snu114, and Ntc85 remained largely unchanged under all three conditions. These results indicate that the release of SF3a/b from the spliceosome also requires Cwc22, in agreement with the requirement of Cwc22 for the function of Prp2 in this step. A scheme illustrating how Cwc22 is involved in the first catalytic step is shown in Fig. 7.
FIG. 7.
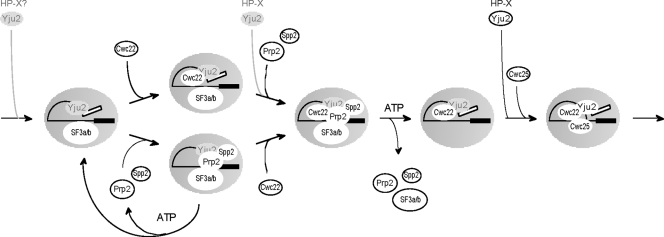
Schematic of the Cwc22 requirement to direct Prp2 into the productive pathway. After the activation of the spliceosome, Cwc22 and Prp2/Spp2 can bind to the spliceosome independently of each other. In the absence of Cwc22, Prp2 and Spp2 are dissociated from the spliceosome upon ATP hydrolysis with SF3a/b unreleased. In the presence of Cwc22, SF3a and SF3b are released with Prp2/Spp2 upon ATP hydrolysis. Cwc25 is then recruited to the spliceosome to promote the first catalytic reaction. Yju2 and HP-X are required for the first catalytic reaction after Prp2 action but can join the spliceosome at earlier steps, which are shown in gray to indicate their presence being functionally irrelevant. The question mark for HP-X indicates no experimental evidence for whether it can bind to the spliceosome prior to Cwc22 binding.
DISCUSSION
In this study, we show that Cwc22 is essential for pre-mRNA splicing both in vivo and in vitro. Although Cwc22 was previously reported to associate with the NTC component Cef1/Ntc85 (28), we found that the association of Cwc22 with the NTC was very weak, as only tiny amounts of known NTC components were coprecipitated with Cwc22. Our data thus suggest that Cwc22 is not an integral component of the NTC. Furthermore, we demonstrate that Cwc22 has a function different from that of the NTC. It is not required for NTC-mediated spliceosome activation but is required for the first catalytic step in promoting the Prp2-mediated release of SF3a/b.
The catalytic steps of splicing involve more protein factors than expected. Each one proceeds through an ATP-dependent reaction followed by an ATP-independent reaction, and each requires a DExD/H-box RNA helicase, Prp2 and Prp16, respectively, for the ATP-dependent function. The ATPase activity of Prp2 is required for the release of SF3a/b (21, 46), presumably to clear the branch site for the binding of other factors to promote the first transesterification, which does not require ATP but requires Yju2, Cwc25, and HP-X (12, 22). In the second step, Prp16 mediates an ATP-dependent structural change of the spliceosome, resulting in the protection of the 3′ splice site (33). Prp22, Prp18, and Slu7 are then required to promote the second transesterification (2, 15, 16, 34). Although the functions of Yju2 and HP-X are required only after the action of Prp2, they can be recruited to the spliceosome before the Prp2 step (22, 46). The findings that the binding of Cwc25 to the spliceosome depends on the function of Prp2 and Yju2 and that the catalytic reaction occurs upon its binding suggest that Cwc25 may play a critical role in the positioning of the 5′ splice site to the branch site. Prp2 requires a cofactor, Spp2, for its binding to the spliceosome and, consequently, for its function (32, 35), but how Spp2 regulates the function of Prp2 is not known.
In contrast, Cwc22 and Prp2 bind to the spliceosome independently of each other. Neither Cwc22 nor Prp2/Spp2 is associated with the spliceosome in the absence of the NTC, suggesting that these proteins bind only after the spliceosome is activated. Prp2 or Spp2 was not detected to interact with any of the known NTC components by two-hybrid assays (data not shown). Cwc22 is not tightly associated with the NTC but shows weak interactions with Syf3/Ntc77 and Isy1/Ntc30 in two-hybrid assays (data not shown). Whether such interactions are involved in the recruitment of Cwc22 to the spliceosome remains unknown. The stable association of Cwc22 with the spliceosome may involve interactions of Cwc22 with multiple spliceosomal components at a specific stage when the proper conformation of the spliceosome is achieved. Prp2 has been reported to interact with Brr2, which was shown previously to interact with splicing factors involved in various steps of the spliceosome pathway by two-hybrid assays (43). The interaction of Brr2 with Ntr2 is responsible for the recruitment of the NTR (for NTC-related) complex to the spliceosome to mediate its disassembly (41). Whether the recruitment of Prp2 is mediated by its interaction with Brr2 also remains to be investigated.
The productive action of Prp2 requires the presence of Cwc22. In the absence of Cwc22, Prp2 could bind to the spliceosome but is dissociated upon ATP hydrolysis, with SF3a/b still retained on the spliceosome. Only in the presence of Cwc22 could SF3a/b be released. Cwc22 thus prevents the spliceosome from entering a futile pathway. How Cwc22 acts in concert with Prp2 to promote the release of SF3a/b remains an open question. It is possible that the binding of Cwc22 may induce a conformational change in the spliceosome to allow the access of Prp2 to SF3a/b or that Cwc22 may interact with Prp2 directly or indirectly to reposition Prp2 from its docking site. Studies of Prp22-mediated mRNA release have revealed a mechanism that involves the repositioning of Prp22 (33). Prp22 has two roles in the splicing reaction, an ATP-independent role for exon ligation and an ATP-dependent role for mRNA release. Site-specific cross-linking studies have revealed an initial docking of Prp22 on the intron immediately upstream of the 3′ splice site (25, 26). Prp22 was found to cross-link to mRNA downstream of the splice junction after exon ligation, suggesting that the repositioning of Prp22 during a conformational change of the spliceosome accompanies the second transesterification reaction. Upon ATP hydrolysis, Prp22 moves in the 3′-to-5′ direction to disrupt U5-mRNA interactions in releasing mRNA (33). Like Prp22, Prp2 exhibits RNA-dependent NTPase activity (18); however, Prp2 has never been demonstrated in vitro to unwind RNA duplexes or to disrupt RNA-protein interactions. SF3b is known to bind to the branch site, and components of SF3b cross-link to the branch site (14, 31, 48). Prp2 has also been shown to directly interact with pre-mRNA by UV cross-linking, likely downstream of the branch site (39). Whether Prp2 acts by a similar mechanism to disrupt interactions of SF3a/b with the branch site awaits further study.
Unlike Prp2 and Spp2, Cwc22 remains associated with the spliceosome after SF3a and SF3b are released. It is worth noting that Cwc22 is the only factor known that is required for catalytic steps but remains associated after its action. This raises the question of whether Cwc22 has additional roles in subsequent steps of the spliceosome pathway. In the view that Cwc22 modulates the function of Prp2 in the first catalytic step, it will be interesting to know whether Cwc22 also regulates the function of other DExD/H-box ATPases, Prp16 and Prp22, in the second step and for mRNA release, respectively.
Cwc22 has an MIF4G domain at the amino terminus and a MA-3 domain in the middle region. The deletion of the MIF4G domain did not affect cellular growth, whereas the deletion of the middle region containing the MA-3 domain resulted in lethality. The segment of Cwc22 at residues 212 to 453, containing the entire MA-3 domain with extra 80- and 55-amino-acid residues flanking each side, hardly supported cellular growth. The extension of the carboxy-terminal flanking region (to amino acid residue 491) featuring a stretch of the evolutionarily conserved sequence recovered the growth phenotype. Thus, the MA-3 domain together with its flanking regions may constitute a structural domain for the function of Cwc22. Nevertheless, residues 453 to 491 become dispensable in the presence of the MIF4G domain, suggesting a functional redundancy of these two regions.
The MIF4G and MA-3 domains are present in the eukaryotic translational initiation factor eIF4G, in the middle and C-terminal regions, respectively. Each of these domains can interact with DExD/H-box RNA helicase eIF4A in forming the initiation complex (20, 27). It is tempting to think that Cwc22 may interact with Prp2 in a similar way. Two-hybrid analysis revealed Prp2 to weakly interact with the carboxy-terminal segment of Cwc22 (residues 212 to 577) but not the full-length protein (data not shown). This suggests that although the MIF4G domain is dispensable, its presence affects the interaction of Prp2 with Cwc22 and may also affect their interactions with other spliceosomal components. How such an interaction might affect the function of Prp2 remains to be investigated.
Acknowledgments
We thank Heiko Kuhn for English editing and members of the Cheng laboratory for helpful discussion.
This work was supported by a grant from Academia Sinica and the National Science Council (Taiwan), NSC98-2745-B-001-ASP.
Footnotes
Published ahead of print on 18 October 2010.
REFERENCES
- 1.Albers, M., A. Diment, M. Muraru, C. S. Russell, and J. D. Beggs. 2003. Identification and characterization of Prp45p and Prp46p, essential pre-mRNA splicing factors. RNA 9:138-150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ansari, A., and B. Schwer. 1995. SLU7 and a novel activity, SSF1, act during the PRP16-dependent step of yeast pre-mRNA splicing. EMBO J. 14:4001-4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bessonov, S., M. Anokhina, C. L. Will, H. Urlaub, and R. Lührmann. 2008. Isolation of an active step I spliceosome and composition of its RNP core. Nature 452:846-850. [DOI] [PubMed] [Google Scholar]
- 4.Chan, S.-P., and S.-C. Cheng. 2005. The Prp19-associated complex is required for specifying interactions of U5 and U6 with pre-mRNA during spliceosome activation. J. Biol. Chem. 280:31190-31199. [DOI] [PubMed] [Google Scholar]
- 5.Chan, S.-P., D.-I. Kao, W.-Y. Tsai, and S.-C. Cheng. 2003. The Prp19p-associated complex in spliceosome activation. Science 302:279-282. [DOI] [PubMed] [Google Scholar]
- 6.Chang, K.-J., H.-C. Chen, and S.-C. Cheng. 2009. Ntc90 is required for recruiting first step factor Yju2 but not for spliceosome activation. RNA 15:1729-1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C.-H., W.-Y. Tsai, H.-R. Chen, C.-H. Wang, and S.-C. Cheng. 2001. Identification and characterization of two novel components of the Prp19p-associated complex, Ntc30p and Ntc20p. J. Biol. Chem. 276:488-494. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C.-H., W.-C. Yu, T. Y. Tsao, L.-Y. Wang, H.-R. Chen, J.-Y. Lin, W.-Y. Tsai, and S.-C. Cheng. 2002. Functional and physical interactions between components of the Prp19p-associated complex. Nucleic Acids Res. 30:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen, H.-R., S.-P. Jan, T. Y. Tsao, Y.-J. Sheu, J. Banroques, and S.-C. Cheng. 1998. Snt309p, a component of the Prp19p-associated complex that interacts with Prp19p and associates with the spliceosome simultaneously with or immediately after dissociation of U4 in the same manner as Prp19p. Mol. Cell. Biol. 18:2196-2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cheng, S.-C., and J. Abelson. 1986. Fractionation and characterization of a yeast mRNA splicing extract. Proc. Natl. Acad. Sci. U. S. A. 83:2387-2391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cheng, S.-C., A. Newman, R.-J. Lin, G. D. McFarland, and J. N. Abelson. 1990. Preparation and fractionation of yeast splicing extract. Methods Enzymol. 181:89-96. [DOI] [PubMed] [Google Scholar]
- 12.Chiu, Y.-F., Y.-C. Liu, T.-W. Chiang, T.-C. Yeh, C.-K. Tseng, N. Y. Wu, and S.-C. Cheng. 2009. Cwc25 is a novel splicing factor required after Prp2 and Yju2 to facilitate the first catalytic reaction. Mol. Cell. Biol. 29:5671-5678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giaever, G., A. M. Chu, L. Ni, C. Connelly, L. Riles, S. Véronneau, S. Dow, A. Lucau-Danila, K. Anderson, B. André, A. P. Arkin, A. Astromoff, M. El Bakkoury, R. Bangham, R. Benito, S. Brachat, S. Campanaro, M. Curtiss, K. Davis, A. Deutschbauer, K.-D. Entian, P. Flaherty, F. Foury, D. J. Garfinkel, M. Gerstein, D. Gotte, U. Güldener, J. H. Hegemann, S. Hempel, Z. Herman, D. F. Jaramillo, D. E. Kelly, S. L. Kelly, P. Kötter, D. LaBonte, D. C. Lamb, N. Lan, H. Liang, H. Liao, L. Liu, C. Luo, M. Lussier, R. Mao, P. Menard, S. L. Ooi, J. L. Revuelta, C. J. Roberts, M. Rose, P. Ross-Macdonald, B. Scherens, G. Schimmack, B. Shafer, D. D. Shoemaker, S. Sookhai-Mahadeo, R. K. Storms, J. N. Strathern, G. Valle, M. Voet, G. Volckaert, C.-Y. Wang, T. R. Ward, J. Wilhelmy, E. A. Winzeler, Y. Yang, G. Yen, E. Youngman, K. Yu, H. Bussey, J. D. Boeke, M. Snyder, P. Philippsen, R. W. Davis, and M. Johnston. 2002. Functional profiling of the Saccharomyces cerevisiae genome. Nature 418:387-391. [DOI] [PubMed] [Google Scholar]
- 14.Gozani, O., J. Potashkin, and R. Reed. 1998. A potential role for U2AF SAP155 interactions in recruiting U2 snRNP to the branch site. Mol. Cell. Biol. 18:4752-4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horowitz, D. S., and J. Abelson. 1993. Stages in the second reaction of pre-mRNA splicing: the final step is ATP independent. Genes Dev. 7:320-329. [DOI] [PubMed] [Google Scholar]
- 16.James, S., W. Turner, and B. Schwer. 2002. How Slu7 and Prp18 cooperate in the second step of yeast pre-mRNA splicing. RNA 8:1068-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa, M., S. Goto, S. Kawashima, and A. Nakaya. 2002. The KEGG databases at GenomeNet. Nucleic Acids Res. 30:42-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim, S.-H., J. Smith, A. Claude, and R.-J. Lin. 1992. The purified yeast pre-mRNA splicing factor PRP2 is an RNA-dependent NTPase. EMBO J. 11:2319-2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King, D. S., and J. D. Beggs. 1990. Interactions of PRP2 protein with pre-mRNA splicing complexes in Saccharomyces cerevisiae. Nucleic Acids Res. 18:6559-6564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korneeva, N. L., B. J. Lamphear, F. L. C. Hennigan, W. C. Merrick, and R. E. Rhoads. 2001. Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem. 276:2872-2879. [DOI] [PubMed] [Google Scholar]
- 21.Lardelli, R. M., J. X. Thompson, J. R. Yates III, and S. W. Stevens. 2010. Release of SF3 from the intron branchpoint activates the first step of pre-mRNA splicing. RNA 16:516-528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y.-C., H.-C. Chen, N.-Y. Wu, and S.-C. Cheng. 2007. A novel splicing factor, Yju2, is associated with NTC and acts after Prp2 in promoting the first catalytic reaction of pre-mRNA splicing. Mol. Cell. Biol. 27:5403-5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makarov, E. M., O. V. Makarova, H. Urlaub, M. Gentzel, C. L. Will, M. Wilm, and R. Lührmann. 2002. Small nuclear ribonucleoprotein remodeling during catalytic activation of the spliceosome. Science 298:2205-2208. [DOI] [PubMed] [Google Scholar]
- 24.McGrail, J. C., and R. T. O'Keefe. 2008. The U1, U2 and U5 snRNAs crosslink to the 5′ exon during yeast pre-mRNA splicing. Nucleic Acids Res. 36:814-825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McPheeters, D. S., and P. Muhlenkamp. 2003. Spatial organization of protein-RNA interactions in the branch site-3′ splice site region during pre-mRNA splicing in yeast. Mol. Cell. Biol. 23:4174-4186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McPheeters, D. S., B. Schwer, and P. Muhlenkamp. 2000. Interaction of the yeast DExH-box RNA helicase Prp22p with the 3′ splice site during the second step of nuclear pre-mRNA splicing. Nucleic Acids Res. 28:1313-1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Morino, S., H. Imataka, Y. V. Svitkin, T. V. Pestova, and N. Sonenberg. 2000. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 20:468-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ohi, M. D., and K. L. Gould. 2002. Characterization of interactions among the Cef1p-Prp19p-associated splicing complex. RNA 8:798-815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ohi, M. D., C. W. V. Kooi, J. A. Rosenberg, L. Ren, J. P. Hirsch, W. J. Chazin, T. Walz, and K. Gould. 2005. Structural and functional analysis of essential pre-mRNA splicing factor Prp19p. Mol. Cell. Biol. 25:451-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Plumpton, M., M. McGarvey, and J. D. Beggs. 1994. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 13:879-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Query, C. C., and P. A. Sharp. 1996. Three recognition events at the branch-site adenine. EMBO J. 15:1392-1402. [PMC free article] [PubMed] [Google Scholar]
- 32.Roy, J., K. Kim, J. R. Maddock, J. G. Anthony, and J. L. Woolford. 1995. The final stages of spliceosome maturation require Spp2p that can interact with the DEAH box protein Prp2p and promote step 1 of splicing. RNA 1:375-390. [PMC free article] [PubMed] [Google Scholar]
- 33.Schwer, B. 2008. A conformational rearrangement in the spliceosome sets the stage for Prp22-dependent mRNA release. Mol. Cell 30:743-754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schwer, B., and C. H. Gross. 1998. Prp22, a DExH-box RNA helicase, plays two distinct roles in yeast pre-mRNA splicing. EMBO J. 17:2086-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silverman, E. J., A. Maeda, J. Wei, P. Smith, J. D. Beggs, and R.-J. Lin. 2004. Interaction between a G-patch protein and a spliceosome DEXD/H-box ATPase that is critical for splicing. Mol. Cell. Biol. 24:10101-10110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Staley, J. P., and C. Guthrie. 1998. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell 92:315-326. [DOI] [PubMed] [Google Scholar]
- 37.Tarn, W.-Y., C.-H. Hsu, K.-T. Huang, H.-R. Chen, H.-Y. Kao, K.-R. Lee, and S.-C. Cheng. 1994. Functional association of essential splicing factor(s) with PRP19 in a protein complex. EMBO J. 13:2421-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tarn, W.-Y., K.-R. Lee, and S.-C. Cheng. 1993. The yeast PRP19 protein is not tightly associated with small nuclear RNAs, but appears to associate with the spliceosome after binding of U2 to the pre-mRNA and prior to formation of the functional spliceosome. Mol. Cell. Biol. 13:1883-1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Teigelkamp, S., M. McGarvey, M. Plumpton, and J. D. Beggs. 1994. The splicing factor PRP2, a putative RNA helicase, interacts directly with pre-mRNA. EMBO J. 13:888-897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsai, R.-T., R.-H. Fu, F.-L. Yeh, C.-K. Tseng, Y.-C. Lin, Y.-H. Huang, and S.-C. Cheng. 2005. Spliceosome disassembly catalyzed by Prp43 and its associated components Ntr1 and Ntr2. Genes Dev. 19:2991-3003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsai, R.-T., C.-K. Tseng, P.-J. Lee, H.-C. Chen, R.-H. Fu, K.-J. Chang, F.-L. Yeh, and S.-C. Cheng. 2007. Dynamic interactions of Ntr1-Ntr2 with Prp43 and with U5 govern the recruitment of Prp43 to mediate spliceosome disassembly. Mol. Cell. Biol. 27:8027-8037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai, W.-Y., Y.-T. Chow, H.-R. Chen, K.-T. Huang, R.-I. Hong, S.-P. Jan, N.-Y. Kuo, T. Y. Tsao, C.-H. Chen, and S.-C. Cheng. 1999. Cef1p is a component of the Prp19p-asociated complex and essential for pre-mRNA splicing. J. Biol. Chem. 274:9455-9462. [DOI] [PubMed] [Google Scholar]
- 43.van Nues, R. W., and J. D. Beggs. 2001. Functional contacts with a range of splicing proteins suggest a central role for Brr2p in the dynamic control of the order of events in spliceosomes of Saccharomyces cerevisiae. Genetics 157:1451-1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vijayraghavan, U., R. Parker, J. Tamm, Y. Iimura, J. Rossi, J. Abelson, and C. Gurthrie. 1986. Mutations in conserved intron sequences affect multiple steps in the yeast splicing pathway, particularly assembly of the spliceosome. EMBO J. 5:1683-1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wahl, M. C., C. L. Will, and R. L. Lührmann. 2009. The spliceosome: design principles of a dynamic RNP machine. Cell 136:701-718. [DOI] [PubMed] [Google Scholar]
- 46.Warkocki, Z., P. Odenwälder, J. Schmitzová, F. Platzmann, H. Stark, H. Urlaub, R. Ficner, P. Fabrizio, and R. Lührmann. 2009. Reconstitution of both steps of Saccharomyces cerevisiae splicing with purified spliceosomal components. Nat. Struct. Mol. Biol. 16:1237-1243. [DOI] [PubMed] [Google Scholar]
- 47.Will, C. L., and R. Lührmann. 2006. Spliceosome structure and function, p. 369-400. In R. F. Gesteland, T. R. Cech, and J. F. Atkins (ed.), The RNA world, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 48.Will, C. L., C. Schneider, A. M. MacMillan, N. F. Katopodis, G. Neubauer, M. Wilm, R. Lührmann, and C. C. Query. 2001. A novel U2 and U11/U12 snRNP protein that associates with the pre-mRNA branch site. EMBO J. 20:4536-4546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winston, F., F. Chumley, and G. R. Fink. 1983. Eviction and transplacement of mutant genes in yeast. Methods Enzymol. 101:211-228. [DOI] [PubMed] [Google Scholar]