Abstract
Staphylococcus aureus is a prominent human pathogen and a leading cause of community- and hospital-acquired bacterial infections worldwide. Herein, we describe the identification and characterization of the S. aureus 67.6-kDa hypothetical protein, named for the surface factor promoting resistance to oxidative killing (SOK) in this study. Sequence analysis showed that the SOK gene is conserved in all sequenced S. aureus strains and homologous to the myosin cross-reactive antigen of Streptococcus pyogenes. Immunoblotting and immunofluorescence analysis showed that SOK was copurified with membrane fractions and was exposed on the surface of S. aureus Newman and RN4220. Comparative analysis of wild-type S. aureus and an isogenic deletion strain indicated that SOK contributes to both resistance to killing by human neutrophils and to oxidative stress. In addition, the S. aureus sok deletion strain showed dramatically reduced aortic valve vegetation and bacterial cell number in a rabbit endocarditis model. These results, plus the suspected role of the streptococcal homologue in certain diseases such as acute rheumatic fever, suggest that SOK plays an important role in cardiovascular and other staphylococcal infections.
Staphylococcus aureus is a commensal that often colonizes skin and mucosal membranes (11, 28). This species is usually benign in healthy individuals, but it is a high-risk pathogen for immunocompromised individuals. As a consequence of its numerous virulence factors and its adaptability, S. aureus is one of the most significant human pathogens for both nosocomial- and community-associated infections (20). Moreover, an increasing resistance to antibacterial agents and the adaptation and emergence of methicillin- and vancomycin-resistant S. aureus (MRSA and VRSA, respectively) strains is alarming (2, 13).
S. aureus is the causative agent of diverse human and animal maladies, including, but not limited to, abscesses, food poisoning, toxic shock syndrome, septicemia, and endocarditis (3, 46, 49). This cadre of diseases results from S. aureus strain heterogeneity. Although numerous, most S. aureus virulence factors are categorized into one of the following groups according to their functions: (i) surface proteins that promote adhesion, internalization, and colonization; (ii) toxins and enzymes that promote tissue damage, inflammation, and invasion and dissemination; (iii) surface factors that affect phagocytosis by leukocytes; (iv) factors that enhance survival in phagocytes; or (v) superantigens and other molecules that modulate the immune system by altering the function of lymphocytes and antigen-presenting cells (1, 12, 44).
Our bioinformatics analysis of 13 S. aureus genomic sequences in search of potential virulence factors for staphylococcus-induced cardiovascular diseases revealed a conserved open reading frame ([ORF] 96 to 100% identity among all S. aureus sequences). The predicted translation products from these ORFs share 59% identity with the 67-kDa myosin cross-reactive antigen (MCRA) of Streptococcus pyogenes (19). The S. aureus homologue (ORF SA0102) was reported initially by Kuroda et al. (25) in reference to the N315 strain genome sequence. They described SA0102 as one of two major histocompatibility complex class II (MHC-II) β-chain homologues in the N315 genome. The 67-kDa S. pyogenes protein and the SA0102 predicted translation product share 62% and 34% similarity (19% and 21.2% identity), respectively, to the murine β1 domain of the mouse I-Au chain (19; also our unpublished results).
The 67-kDa streptococcal homologue is a putative virulence factor, and hybridization studies suggested that related proteins exist in streptococcal groups A, C, and G (19). This protein is a member of extensive MCRA protein family. It reacts with sera from patients with acute rheumatic fever (ARF), acute glomerulonephritis, and active streptococcal infections (19). It also reacts with anti-myosin antibody in sera of patients with ARF. Recently, this streptococcal protein was described as fatty acid double bond hydratase (47). Although members of the MCRA protein family are widely distributed among bacteria, only three proteins from this family have been biochemically characterized (6, 47), and the exact role of the vast majority of proteins and homologues belonging to this family remains unknown. The objective of this study is to characterize the 67-kDa myosin cross-reactive homologue of S. aureus. To address this goal, we constructed deletion mutants in two well-characterized S. aureus strains, RN4220 and Newman, compared the properties of the parental and mutant strains, and investigated the molecular relatedness of the structural gene in a number of clinical isolates. The data suggest that this protein is ubiquitous among S. aureus clinical isolates and could contribute to infectious diseases such as endocarditis by promoting survival in phagocytes and resistance to oxidative killing. Due to the latter property, we tentatively designated the S. aureus protein SOK, a surface factor promoting resistance to oxidative killing.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids used are described in Table 1. Escherichia coli was grown using Luria-Bertani (Difco Laboratories, Detroit, MI) medium that was supplemented with ampicillin (Am; 100 μg/ml) or chloramphenicol (Cm; 34 μg/ml) when necessary. Except where indicated, S. aureus was propagated using tryptic soy (TS) medium (Difco Laboratories, Detroit, MI). Plasmid selection for S. aureus was conducted with erythromycin (Em), Cm, or tetracycline (Tc) (5, 10, or 10 μg/ml, respectively).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Function or relevant properties | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| DH5α | General cloning applications | Life Technologies |
| BL21(DE3) pLysS | Recombinant protein expression | Stratagene |
| S. aureus | ||
| Newman | Clinical isolate | ATCC 25904 |
| NM10 | Newman sok::Tc; Tcr | This study |
| NM20 | NM10 complemented with the pMIN164-sok plasmid; Tcr Emr | This study |
| DU5875 | Isogenic mutant of 8325-4; spa::Tc; Tcr | 33 |
| RN4220 | Restriction negative derivative of 8325-4 | 24 |
| NM1 | RN4220 sok::Tc; Tcr | This study |
| NM2 | NM1 complemented with the pMIN164-sok plasmid; Tcr Emr | This study |
| Plasmids | ||
| pDG1515 | Source of Tcr cassette; Amr Tcr | 14 |
| pSOK1 | Plasmid pdg1515 with sok upstream and downstream fragments; Amr Tcr | This study |
| pCL10 | Temp-sensitive Amr Cmr Tcr | 42 |
| pSOK2 | Vector pcl10 with sok upstream, downstream, and Tcr cassette; Amr Cmr Tcr | This study |
| pMIN164 | E. coli-S. aureus shuttle vector; Amr Emr | 18 |
| pSOK3 | Plasmid pMIN164 with sok ORF; Amr Emr | This study |
| pGEX-5T | E. coli GST/His-tag vector; Amr | 5 |
Bioinformatics analysis.
The search for homologues was performed using BLAST on the NCBI web server (http://www.ncbi.nlm.nih.gov/), and sequences were aligned by the ClustalW program (http://www.ebi.ac.uk/Tools/clustalw/index.html). The following tools on the ExPASy Proteomic Server were used to analyze various properties of the SOK protein: Compute pI/Mw was used to calculate isoelectric point and molecular weight, SignalP, version 3.0, was used to calculate the signal peptide cleavage site by artificial neural networks (NN) and hidden Markov models (HMM), ProtScale was used to determine hydrophobicity, and PSIpred was used to determine transmembrane topology (using MEMSAT on the PSIpred Protein Structure Prediction Server). The BPROM program (Softberry, Inc., Mount Kisco, NY) was used to predict bacterial promoter.
DNA isolation.
Genomic DNA was isolated by using Genomic DNA Prep Plus kits (A&A Biotechnology, Gdynia, Poland) facilitated by a method to promote S. aureus lysis (41). Plasmids were isolated with a MiniPrep isolation kit (Qiagen GmbH, Hilden, Germany) using the manufacturer's protocol. S. aureus cells were treated with lysostaphin in the kit resuspension buffer. DNA was quantified using a NanoDrop ND-1000 instrument (Nanodrop Technologies, Wilmington, DE).
RFLP analysis.
A 1,789-bp fragment containing predicted sok coding region plus flanking promoter and ribosome binding sites was amplified using primers 12631 and 12632 (Table 2). PCR products were purified, quantified (see above), and digested with Csp6I, Hin6I, or TaqI. These enzymes were selected based on two criteria: (i) that at least three cleavage sites exist within the PCR-amplified sok gene and (ii) that predicted digestion patterns are readily discernible by agarose gel electrophoresis. Fragments were separated by electrophoresis in 2% agarose gels to compare sok genotypes according to digestion profiles. At least a one-band difference was used as the criterion for designating unique restriction fragment length polymorphism (RFLP) profiles. Concordance between spa typing results and PCR-RFLP was calculated as described previously (30).
TABLE 2.
Primers used in this study
| Name | Sequencea | Purposeb |
|---|---|---|
| Tet1F | 5′-TCAGAATCCAAATCTAGACGAGTGATAAAATTT-3′ | Upstream fragment for mutation (XbaI) |
| Tet1R | 5′-ATTTTAATATCTTCGGGATCCACATCGTATTCA-3′ | Upstream fragment for mutation (BamHI) |
| Tet2F | 5′-ATTACAATCAATGATTGAATTCGGCAAATGAGTTT-3′ | Downstream fragment for mutation (EcoRI) |
| Tet2R | 5′-ATTTCTGTCCCGGTACCATGATTTGAAAT-3′ | Downstream fragment for mutation (KpnI) |
| 12631 | 5′-CTTGGTGGATATGTATTACAGT-3′ | PCR-RFLP |
| 12632 | 5′-TCGTTATAACAATTTGTGTTCTTTT-3′ | PCR-RFLP |
| RT-Fsok | 5′-AGCGCCACCAACTGACGA-3′ | RT-PCR sok gene |
| RT-Rsok | 5′-CCTGCAAGTGGGTCACGTTTA-3′ | RT-PCR sok gene |
| RT-Fdn | 5′-TATTAGGTGTTATTGCAGGTATCGTTG-3′ | RT-PCR downstream gene |
| RT-Rdn | 5′-AAATTGGCATTGCATATTCGC-3′ | RT-PCR downstream gene |
| SR4 | 5′-GGCCTTTGCAGGGCTGGCAAGCCACG-3′ | Sequencing of the pGEX-5T insert |
| SR5 | 5′-GCTGCATGTGTCAGAGGTTTTCACCG-3′ | Sequencing of the pGEX-5T insert |
| MExpF | 5′-AAAGGACTTGGGATCCATGTATTACAGT-3′ | Complementation of sok deletion strains (BamHI) |
| MExpR | 5′-ATAAAAATCTATTAATGGGTCGACTTATAACAAT-3′ | Complementation of sok deletion strains (SalI) |
Enzyme restriction sites are underlined.
Restriction enzymes used to digest PCR products are in parentheses.
RNA isolation and RT-PCR.
RNA was isolated with an RNeasy Mini Kit (Qiagen Science, Germantown, MD) according to the manufacturer's protocol. Bacteria were disrupted using a FastPrep FP120 homogenizer (ThermoSavant, Holbrook, NY) (45 s at 6.0 m/sec). DNA was removed using RNase-free DNase I (Ambion, Austin, TX), and RNA was further purified by treatment again with the same RNA isolation protocol. RNA samples with a ratio of the optical density at 260 nm (OD260)/OD280 of ≥2.0 were used. First-strand cDNA was synthesized from 1 μg of RNA using Superscript Reverse Transcriptase (Invitrogen, Carlsbad, CA), and 5 μl of a 100-fold diluted aliquot was used as the template (final sample volume, 25 μl; MicroAmp Optical 96-Well Reaction Plate; Applied Biosystems). Real-time PCR (RT-PCR) was performed in an ABI Prism 7500 system using SYBR green mix as recommended (Applied Biosystems, Foster City, CA).
Cloning and purification of SOK.
Recombinant SOK (rSOK) was expressed in Escherichia coli BL21(DE3) pLysS using pGEX-5T, which expresses proteins with an N-terminal His tag and a glutathione S-transferase (GST) label (5). The predicted SOK ORF was amplified with Pfu DNA polymerase (Fermentas, Lithuania); the 1,827-bp product was digested with BamHI and XhoI. The resulting 1,776-bp fragment was cloned into pGEX-5T and transformed into E. coli DH5α cells (Life Technologies, Inc., Gaithersburg, MD). After the appropriate construct was confirmed by PCR and sequencing, the plasmid was purified and transformed into E. coli BL21(DE3) pLysS (Stratagene). Expression was induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) when the OD600 of the culture reached 0.9. Following growth at 20°C for an additional 10 h, cells were collected by centrifugation, washed with phosphate-buffered saline ([PBS] 140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH 7.3), and disrupted using a French press (Thermo Fisher Scientific, Inc., Waltham, MA). rSOK was purified on the glutathione-Sepharose 4B resin, according to the manufacturer's recommendation (Amersham, Piscataway, NJ), and GST label was cleaved with thrombin (Sigma-Aldrich, St. Louis, MO). The recombinant protein was dialyzed against 10 mM morpholinepropanesulfonic acid (MOPS) buffer (pH 7.0) containing 10 mM NaCl and 1 mM EDTA. The apparently pure rSOK, as assessed by SDS-PAGE (26), was stored in dialysis buffer containing 10% glycerol.
Antibody production.
Sprague-Dawley rats (6 to 8 weeks old; Simonsen Laboratories, Inc., San Diego, CA) were given biweekly subcutaneous injections of rSOK (100 μg) in Freund's incomplete adjuvant (Gibco, Grand Island, NY). One week after the fourth boost, sera were harvested and pooled. The anti-rSOK titer, 15,000, was determined by immunoblotting (9), and diluted (1:10,000) antiserum was typically used for experiments.
Immunofluorescence analysis of S. aureus.
Cell samples were prepared by the method of Hiraga et al. (17). Slides were visualized using a Zeiss LSM 5 Pascal instrument and software (version 4.0, service pack 2; Carl Zeiss MicroImaging GmbH, Heidelberg, Germany) following treatment with anti-rSOK rat serum (see above) and Alexa Fluor 488-conjugated goat anti-rat IgG(H+L) (2 mg/ml) secondary antibodies (Molecular Probes, Inc., Eugene, OR).
SOK subcellular localization.
Cells and supernatants from overnight (o.n.) cultures (25 ml) were separated by centrifugation (at 8,000 × g for 10 min). Culture supernatant proteins were precipitated (2 h at −20°C) with 9 volumes of trichloroacetic acid-acetone (1:8, vol/vol), pelleted, and resuspended in distilled water (1 ml). The cell pellet was washed with deionized water and treated (18,000 lb/in2) in a French press (7), followed by the addition of protease inhibitor cocktail and nuclease mix (Amersham Bioscience Corp., Piscataway, NJ) and centrifugation (75,000 × g for 30 min). The obtained supernatant containing soluble cytoplasmic proteins was stored at −80°C until needed. The resulting pellet, containing crude membrane and wall fractions, was resuspended in rehydration buffer (7 M urea, 2 M thiourea, 2% amidosulfobetaine-14 [ASB-14], 0.5% Triton X-100, 2 mM tributylphosphine, 1% bromophenol blue), and incubated at room temperature for 2 h. The sample was clarified by centrifugation (75,000 × g for 20 min), and supernatant fluids were recovered for analysis of integral membrane proteins. Cell wall proteins in the pellet were released by incubation for 3 h in TE buffer (50 mM Tris-HCl and 10 mM EDTA, pH 8.0) with lysostaphin (40 μg/ml). Proteins were analyzed by SDS-PAGE and immunoblotting (see above).
SOK deletion mutagenesis.
DNA fragments, 918 (5′) and 698 (3′) bp in length, were amplified by PCR from within sok through external flanking regions. The 5′ fragment included nucleotides (nt) −154 through 764; the 3′ fragment encompassed nt 1138 through an additional 59 nt after the predicted stop codon (nucleotide numbering is relative to the predicted ATG initiation codon) (see Fig. S1A in the supplemental material). PCR products were digested with appropriate enzymes (Table 2) and ligated on opposite ends of the Tcr cassette in pDG1515 (50), resulting in pSOK1 vector. This construct was propagated in E. coli DH5α and digested with BamHI and KpnI. The Tcr cassette plus flanking sok fragments were cloned into the pCL10 shuttle vector, resulting in pSOK2. The plasmid was then electroporated into S. aureus RN4220 and Newman as recommended (ECM 600 electroporator; BTX Molecular Delivery Systems). Transformed cells were grown in TS broth (TSB) with Tc (24 h at 43°C) and plated on TS agar (TSA) containing Tc to select colonies with the first recombination. One colony was transferred to TSB and grown (at 30°C for 5 days) with daily transfer to fresh TSB. After 5 days, an aliquot was grown on TSA with Tc. Colonies were screened for a Tcr Cms phenotype indicating that the second recombination resulted in deletion of a 404-nt sok sequence and insertion of the Tc cassette. To complement the mutation, a fragment representing the sok ORF with 260 nt upstream was amplified using MExpF and MExpR primers and ligated into the BamHI and SalI sites of pMIN164 (18), resulting in pSOK3. This plasmid was electroporated into the S. aureus RN4220 and Newman strains (selection was accomplished on TSA with Tc and Em).
PMN killing assay.
Human polymorphonuclear leukocytes (PMNs) were isolated from heparinized venous blood of four different donors in accordance with a human subject protocol approved by the University of Idaho Institutional Review Board for Human Subjects (approval number 05-056). Donors were informed of the procedure risks and provided a written consent prior to participation. Killing of bacteria by human PMNs was determined as described previously (22), with the following modifications. PMNs (106) were combined with opsonized bacteria (107) in 96-well plates which were centrifuged at 400 × g for 5 min, followed by incubation at 37°C for 15 min. PMNs were treated with 400 μM gentamicin (Sigma-Aldrich Co.) for 10 min to remove any remaining extracellular bacteria (time [T] zero). Cultures were further maintained for time points up to 180 min. At the times indicated in Fig. 3A, gentamicin was removed by aspiration, and cells were gently washed with PBS and lysed in sterile water, and the bacteria were plated on TSA. CFU were enumerated following overnight incubation, and the percentage of bacteria killed was calculated using the following equation: (CFUPMN+/CFUT = 0) × 100, where CFUT = 0 indicates number of enumerated colonies for time point zero and CFUPMN+ is the number of CFU for each analyzed time point. The assay measures the percentage of the total number of viable ingested bacteria compared to the number at time point zero.
ROS analysis.
Human PMNs (see above) were mixed with 10 mM 2,7-dichlorodihydrofluorescein diacetate (DCF) (Molecular Probes Inc., Eugene, OR) and incubated for 30 min at room temperature in the dark (8). Opsonized bacteria (above) were mixed with the PMNs at a 10:1 ratio, and transferred to precoated wells of a 96-well plate. The plate was centrifuged (for 5 min at 700 × g at 4°C) to synchronize phagocytosis. Reactive oxygen species (ROS) production was monitored during incubation (at 37°C) using a SpectraMax M2 plate reader (Molecular Devices, Sunnyvale, CA) with 485-nm excitation and 538-nm emission wavelengths. Data were analyzed by SoftMax Pro software, version 5.0.1 (Molecular Devices), and are presented as the rate of change (Vmax) in fluorescence over time.
H2O2 and 1O2 susceptibility assays.
S. aureus cells (mid-exponential growth phase) were harvested by centrifugation, washed once with PBS, and adjusted to 1 × 108 and 2 × 109 cells/ml, respectively, for H2O2 and 1O2 assays (27). To assess the effect of H2O2 on S. aureus viability, the bacteria were incubated with various concentrations of H2O2 (Sigma-Aldrich Co., St. Louis, MO) in glass tubes for 1 h at 37°C, followed by addition of Micrococcus lysodeikticus catalase (1,000 U/ml) (Sigma-Aldrich Co.) to quench the remaining H2O2. The percentage of surviving bacteria was calculated by using the following equation: (CFUH2O2+/CFUT = 0) × 100, where CFUT = 0 indicates number of enumerated colonies for time point zero and CFUH2O2+ is number of CFU after incubation with H2O2 for 1 h. For 1O2 susceptibility, bacteria were incubated in 24-well plates (for 30 or 60 min at 37°C) with various concentrations (0.25 to 6.0 μg/ml) of methylene blue (Sigma-Aldrich Co.). The plates were placed 10 cm from a 100-W incandescent light bulb, and survival was measured by plate counting.
Endocarditis model.
New Zealand White rabbits (2 to 3 kg) were anesthetized with xylazine and ketamine (20 mg/kg each) and subjected to transaortic catheterization for 2 h to damage the aortic valves (43). After the catheters were removed and animals were closed, a washed suspension of S. aureus (2 ml; 1.0 × 109/ml for RN4220 strains or 5.0 × 108/ml for Newman strains in PBS) was administered to each rabbit in the marginal ear vein. Animals were observed daily. Except for rabbits infected with the parental Newman strain, which succumbed on days 2 and 3, animals were sacrificed on days 4 and 5 to assess vegetation formation. Aseptically harvested vegetations from all animals were weighed, and bacteria were enumerated by plate counts.
Statistical analysis.
A Student t test was carried out using GraphPad Prism software, version 4.02 (GraphPad Software, Inc., San Diego, CA).
RESULTS
Bioinformatics analysis results.
The initial analysis of published S. aureus genome sequences revealed that the sok gene is ubiquitous and that the sequence is highly conserved in these strains (96 to 100%). In all 13 strains, a 591-residue protein is predicted to be translated from a 1,776-bp gene (see Fig. S1A in the supplemental material). The theoretical isoelectric point (pI) and molecular mass of the S. aureus Newman sok translation product are 5.0 and 67,648 Da, respectively. A potential signal peptide cleavage site predicted using the neural networks (NN) algorithm is located between positions 36 and 37 (SLA-AA). Subsequent to the removal of the potential signal peptide, the deduced molecular size of the putative mature SOK protein in the Newman strain is 63,663 Da. One potential transmembrane-spanning region contains an internal helix cap (residues 25 to 28), a central transmembrane helix (residues 29 to 38), and an external helix cap (residues 39 to 42). This is consistent with the hydrophobicity profile of SOK, which predicts a highly hydrophobic region between residues 25 and 42 (results not shown). In the S. aureus Newman sequence (accession number AP009351) (4), sok is located between ORFs for two hypothetical proteins (NWMN0049 and NWMN0051), which by BLAST sequence comparison resemble a Na/P cotransporter and metabolic/drug transporter, respectively. sok homologues are found in a variety of genome sequences including those of Gram-positive and Gram-negative bacteria (see Fig. S1B in the supplemental material, created using MEGA software [45]). S. aureus SOK sequences share 89.9 to 91.2% homology with homologues in other staphylococcal species and >33.8% homology with those of other bacterial species (see Fig. S1B).
RFLP analysis.
The molecular genetic variability of sok was evaluated using an RFLP technique to analyze a 59-member culture collection (National Institute of Public Health, Warsaw, Poland) comprised predominantly of methicillin-resistant and methicillin-sensitive S. aureus human clinical isolates. The isolates are from a variety of human infections, predominantly from Poland but also from other European countries collected from 1992 through 2001. Initial characterization of these isolates by pulsed-field gel electrophoresis (PFGE), PCR typing, multilocus sequence typing (MLST), and spa typing was previously reported (30). RFLP analysis of the sok locus reveals that the 59 isolates segregate into eight unique RFLP types, and this segregation correlates closely with the clustering of spa types (Table 3). RFLP analysis of the sok gene reveals 97% concordance with spa clusters deduced previously by Malachowa et al. (30). The largest RFLP group correlates with the S2 spa cluster that includes 44% of all isolates. This particular spa cluster contains spa types comprised of the identical or similar repeat profiles (23, 30). Nearly all sok RFLP patterns correlate with a specific spa cluster with two exceptions: isolates 794 and 3502 belong to the S3 cluster, yet they fell into different groups by RFLP analysis of the sok locus. Only the BN4 isolate was not classified to any cluster type, either by spa typing or sok PCR-RFLP.
TABLE 3.
Comparison of sok PCR-RFLP results with spa typinga
| spa cluster |
sok PCR-RFLP pattern |
Isolate no. or name (country/yr)b | |||
|---|---|---|---|---|---|
| Type | Csp6I | Hin6I | TaqI | ||
| S1 | 1 | A | A | A | 2233 (PL/97), 2234b (PL/98), 2255 (PL/98), 2258 (PL/98), 2577 (PL/98), 303 (PL/00), 2260 (PL/98), 3028 (PL/96), 3498 (RU/98), 3521 (LT/98), NCTC8325c |
| S2 | 2 | B | B | B | N104A (PL/95), 1791 (PL/97), 1794 (PL/97), 1807 (PL/97), 3254 (TR/96), MR1003 (PL/92), 2689 (PL/98), N98 (PL/95), MR11 (PL/92), MR1064 (PL/92), MR89 (PL/92), B098 (PL/94), A005b (PL/94), 3497 (BG/98), MR47 (PL/92), MR76 (PL/92), J405 (PL/94), 771 (PL/97), 3301 (SL/98), 2690 (PL/98), 3121 (PL/96), H390 (PL/94), 3248 (CZ/96), EMRSA-16 (UK/92), 2956 (PL/01), 2684 (PL/98)c |
| S2 | 3 | B | D | B | C115 (PL/94), 2688 (PL/98), A005a (PL/98) |
| S3 | 4 | C | C | E | 1899 (PL/96), 2838 (PL/96), MR1010A (PL/92) |
| 5 | B | C | E | 3502 (BG/98) | |
| 6 | B | B | C | 794 (PL/97)c | |
| S4 | 7 | C | C | D | 3483 (SL/98), MR44 (PL/92), MR63 (PL/92), MR84 (PL/92), MR5 (PL/92), MR24 (PL/92), MR52 (PL/92), 2700 (PL/98), MR31 (PL/92), MR29 (PL/92), MR27 (PL/92), MR80 (PL/92), N39 (PL/95) |
| t159d | 8 | B | C | B | BN4 (PL/96) |
spa typing results used in this table were previously published by Malachowa et al. (30).
Countries are abbreviated as follows: BG, Bulgaria; CZ, Czech Republic; LT, Lithuania; PL, Poland; RU, Russia; SL, Slovenia; TR, Turkey; UK, United Kingdom. Years are abbreviated by the last two digits (e.g., 97 is 1997 and 00 is 2000).
Methicillin-susceptible S. aureus strain.
The BN4 strain does not belong to any spa cluster; the spa type is t159.
SOK is surface exposed and copurifies with membrane fractions.
The sok gene in S. aureus strain Newman was disrupted with a Tcr cassette introduced by allelic replacement, as described previously, to generate strain NM10 (Δsok). Strain NM10 was transformed with a plasmid harboring the promoter region plus the coding region of sok to produce the complemented strain designated NM20. RT-PCR confirmed the lack and restoration of detectable sok expression in the mutant and complemented strains, respectively, plus the absence of any detectable effect on expression of the adjacent downstream gene (results not shown).
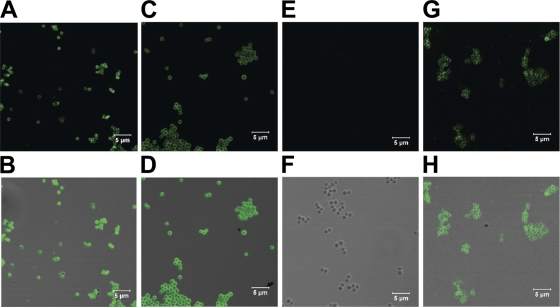
Gene expression studies were consistent with immunofluorescence microscopy using rat antiserum prepared against rSOK. The Newman, NM10, and NM20 strains and a strain lacking protein A (DU5875) were examined for SOK by immunofluorescence microscopy (Fig. 1). The S. aureus parental and complemented strains expressed detectable levels of SOK on the cell surface. SOK was also detectable on the surface of the strain lacking protein A. In contrast, SOK was not detected on the surface of the sok deletion strain (Fig. 1).
FIG. 1.
Surface localization of SOK. S. aureus bacterial strains DU5875 (A and B), Newman (C and D), NM10 (E and F), and NM20 (G and H) (magnification of ×227) were observed by immunofluorescence using SOK-specific rat antiserum, followed by Alexa Fluor 488-conjugated goat anti-rat Ig(H+L) secondary antibody (A, C, E, and G) or combined fluorescence with differential interference contrast (B, D, F, and H).
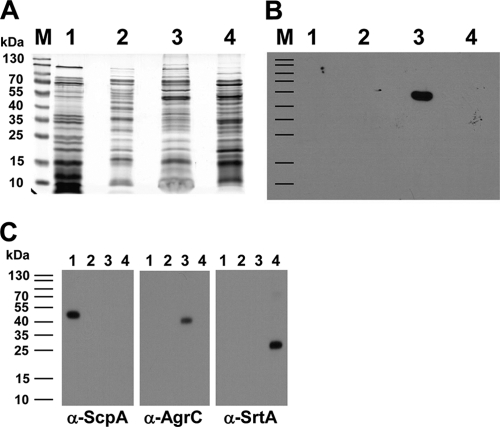
Proteins in culture supernatants and subcellular fractions were resolved by SDS-PAGE (Fig. 2 A) and analyzed by immunoblotting (Fig. 2B). The SOK protein was detected in integral membrane fractions but not in fractions containing cytoplasmic proteins or proteins covalently bound to the cell wall (Fig. 2). The membrane fraction contained an immunoreactive band with a migration consistent with a ∼55-kDa protein, which is smaller than predicted (67.6 kDa) based on bioinformatic analysis (see above). Internal controls for an extracellular protein (ScpA, 44 kDa), an integral membrane protein (AgrC, 42 kDa), and a covalently bound membrane protein (SrtA, 24 kDa) were appropriately detected from the corresponding protein fractions (Fig. 2C).
FIG. 2.
Subcellular localization of SOK in S. aureus Newman using SDS-PAGE (A), immunoblotting with anti-rSOK rat antiserum (B), and immunoblotting with control antibodies (for extracellular protein, anti-ScpA; for integral membrane protein, anti-AgrC; and for covalently bound protein, anti-SrtA) (C). Subcellular localization of SOK was assessed using various bacterial cellular proteins. Lanes M, PageRuler Prestained Protein Ladder (Fermentas); lanes 1, extracellular proteins; lanes 2, cytoplasmic fraction; lanes 3, integral membrane fraction; lanes 4, covalently bound proteins. α, anti.
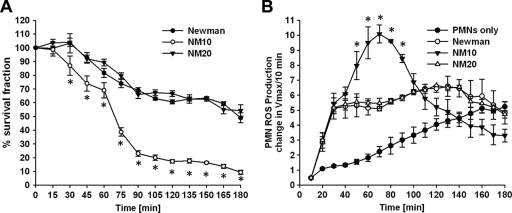
SOK affects S. aureus survival and ROS production in PMNs.
Human PMNs were cultured under conditions to promote synchronized phagocytosis of opsonized S. aureus. After 15 min, extracellular bacteria were killed with gentamicin, and the percentages of viable intracellular bacteria were quantified after incubation for additional time periods. The parental strain displayed nearly linear killing kinetics throughout the 3-h incubation period following addition of gentamicin, after which 43.7% of intracellular bacteria remained viable (Fig. 3 A). The isogenic sok deletion strain (NM10) was more sensitive to PMN killing: 49.5% of bacteria were killed after 65 min, and after 3 h, only 12.7% remained viable. The complemented strain (NM20) showed killing kinetics similar to that of the parental strain.
FIG. 3.
sok affects interactions of S. aureus with PMNs. To assess the effect of sok on innate immunity, we measured bacterial survival in PMNs and ROS production in PMNs. (A) For the survival assay, PMNs were incubated with opsonized S. aureus for 15 min to allow engulfment of bacteria and for an additional 10 min at 37°C with gentamicin prior to commencement of the assay (T = 0). The PMNs were then incubated for up to 180 min. Data shown are the means of four experiments which were performed in triplicate (n = 12). Statistical significance (P < 0.01) between the Newman and NM10 strains is indicated by asterisks. (B) The ROS production is shown as a second-order kinetics plot of the maximum increase in fluorescence (Vmax) over 10-min intervals up to 180 min. Data shown are the means of four experiments which were performed in triplicate (n = 12). For each experiment PMNs and sera were isolated from four different donors. Statistical significance (P < 0.01) between Newman and NM10 is indicated by asterisks.
Disruption of the sok gene affected the PMN ROS production following phagocytosis of S. aureus Newman. Phagocytosis of the parental Newman isolate rapidly induced PMN ROS production compared to resting PMNs (Fig. 3B). Interestingly, ROS levels in cultures harboring phagocytosed S. aureus bacteria lacking SOK were dramatically different from levels in cultures containing the parental strain (Fig. 3B). Despite nearly identical levels of ROS for the first 40 min, levels rose dramatically for the next 30 min until they eventually declined to levels at or below those of PMN cultures harboring the phagocytosed Newman parental strain. The pattern of ROS production was restored to the parental strain by SOK complementation.
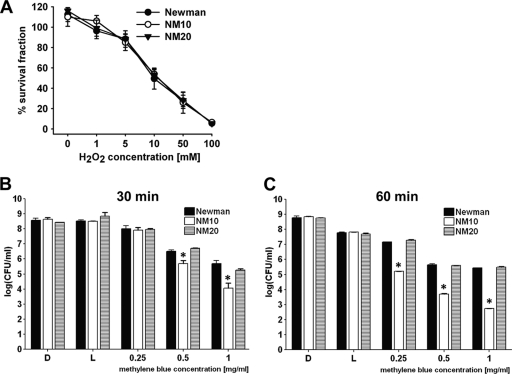
SOK affects S. aureus sensitivity to 1O2 killing but not H2O2.
Since SOK expression influenced ROS levels and survival in PMNs harboring intracellular staphylococci, we assessed whether SOK expression affects susceptibility to the ROS molecules H2O2 or 1O2. Analysis of the bacterial cell viability of the Newman and NM10 strains after 1 h of incubation with various concentrations of H2O2 indicated that SOK expression did not affect the viability of S. aureus cells when they were exposed to H2O2. All strains showed dose-dependent killing effects with respect to increasing concentrations of H2O2. Following exposure to 10 mM H2O2, all strains showed approximately 50% survival (49.5% ± 10.3%, 53.8% ± 4.3%, and 53.1% ± 6.1% for Newman, NM10, and NM20, respectively). Following exposure to 100 mM H2O2, all strains showed approximately 5 to 6% survival (5.6% ± 0.68%, 6.6% ± 1.2%, and 5.5% ± 0.95% for Newman, NM10, and NM20, respectively) (Fig. 4 A).
FIG. 4.
sok affects susceptibility to 1O2 but not H2O2. (A) The susceptibility of various strains (Newman, NM10, and NM20) to treatment with various concentrations of H2O2 was measured by enumeration of CFU after incubation with H2O2. (B and C) The susceptibility of various strains (Newman, NM10, and NM20) to methylene blue-producing 1O2 by photoactivation was measured by enumeration of CFU after 30 min or 60 min. Two controls (L, incubation under a direct light source without methylene blue; D, incubation in darkness without methylene blue) were included for each experiment. Data shown are the means of four experiments which were performed in triplicate (n = 12). Statistical significance (P < 0.01) between the Newman and NM10 strains is indicated by asterisks.
Although sok did not affect survival of bacteria during incubation with H2O2, additional experiments indicated that the sok deletion strain was more sensitive to 1O2 than the parental strain (Fig. 4B and C). Methylene blue releases singlet oxygen (1O2) species when exposed to light and was therefore used to measure the susceptibility of S. aureus strains to 1O2. In the presence of 0.5 and 1 μg/ml methylene blue for a 60-min incubation time, the survival rates of both strains dropped significantly, compared to the same concentration of methylene blue for a 30-min incubation. The strain lacking SOK (NM10) was 4 and 24 times more sensitive to 1O2 than the parental strain after incubation for 30 min with 0.5 and 1 μg of methylene blue, respectively. The highest level (498 times) of difference in susceptibilities to 1O2 between mutant and wild-type strains was observed when bacteria were incubated with 1 μg of methylene blue for 60 min. Longer incubation times (2 and 3 h) as well as higher concentrations of methylene blue (3 and 6 μg/ml) were tested for the Newman and NM10 strains, but no bacteria were able to survive under such conditions (results not shown).
SOK enhances virulence in a model of staphylococcal endocarditis.
To investigate the effect of sok, and thus SOK, on virulence of S. aureus, an infective endocarditis model was used. New Zealand White rabbits with aortic valve leaflets previously damaged by cardiac catheterization were challenged with the parental S. aureus strain RN4220 or Newman or with sok deletion derivatives of the parent strains (NM1 or NM10, respectively). Animals were also challenged with sok deletion strains complemented with a plasmid encoding sok (NM2 or NM20). Rabbits were challenged intravenously with 1 × 109 CFU of Newman or Newman derivatives. RN4220 and its derivatives were used to infect rabbits at a dose of 2 × 109 CFU per animal. Hearts were harvested immediately from animals that died and from survivors that were euthanized after infection. Heart tissues were examined, and vegetations on aortic valves were removed, weighed, and homogenized to enumerate the bacteria contained in the vegetations. If vegetations were not observed, the aortic valves were removed from hearts and homogenized to enumerate bacteria adhering to host tissue.
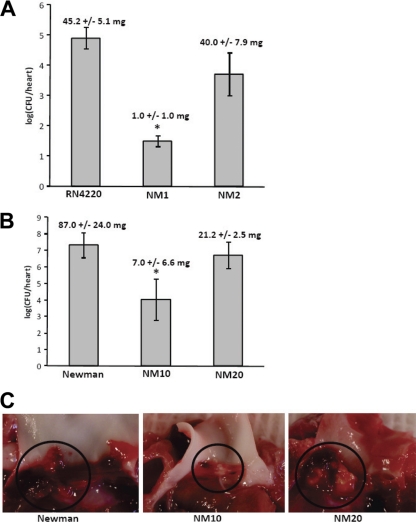
Infection with parent S. aureus RN4220 caused vegetations in all animals (45.2 ± 5.1 mg). In contrast, vegetations were observed in only one of six rabbits infected with NM1 (a sok deletion strain) (6.0 mg) (Fig. 5 A). Animals infected with the parental RN4220 strain showed greater weight loss than animals infected with NM1 and had diarrhea and mottled faces (Fig. 5B). Vegetations were also observed in animals infected with the complemented strain (NM2) (40.0 ± 7.9 mg). Statistical analysis of the vegetation sizes by an unpaired t test showed that the parental RN4220 strain and NM2 strain produced vegetation sizes that were significantly larger (P < 0.0001 and P = 0.0003, respectively) than the vegetations produced by the sok deletion mutant but not significantly different (P = 0.58) from each other. Consistent with vegetation sizes, S. aureus RN4220 and NM2 also produced vegetations with larger bacterial loads (log10 CFU of 4.90 ± 0.35 and 3.71 ± 0.71/heart, respectively) than the sok mutant (log10 CFU of 1.49 ± 0.18/heart). As measured by an unpaired t test, the bacterial loads of vegetations produced by RN4220 and NM2 were significantly different from the bacterial loads of vegetations produced by the sok deletion mutant NM1 (P < 0.00001 and P < 0.007, respectively).
FIG. 5.
SOK expression contributes to endocarditis vegetation formation. The rabbit model of endocarditis was used to examine the ability of RN4220 and RN4220-derived strains (NM1 and NM2) (A) or Newman and Newman-derived strains (NM10 and NM20) (B) to form vegetations on heart valves. Bacteria in vegetations or adhering to heart valves were enumerated, and vegetations were weighed. The mean weight of vegetations produced by each strain is indicated at the top of the bar for each strain. Statistical significance (P < 0.05) between the wild type and mutant is indicated by asterisks. (C) Representative vegetations from infections with strains Newman, NM10, and NM20 were photographed. Vegetations adhering to heart valves are indicated with circles. Vegetations from animals infected with Newman or NM20 were larger than vegetations from animals infected with NM10.
Infection with parent strain Newman and the sok deletion mutant NM10 produced vegetations in all infected animals, but vegetations produced by the Newman strain were significantly larger than vegetations produced by NM10 as measured by a t test (87.0 ± 24.0 mg and 7.0 ± 6.6 mg, respectively; P < 0.01). Statistical analysis by an unpaired t test showed that the bacterial load of vegetations produced by the Newman strain (log10 CFU of 7.31 ± 0.74/heart) was significantly higher than the bacterial load of vegetations produced by strain NM10 (log10 CFU of 4.02 ± 1.24/heart; P < 0.05). Infection with the complemented strain, NM20, caused vegetations with bacterial loads statistically similar to those of the parental Newman strain (log10 CFU of 6.71 ± 0.80/heart; P < 0.6).
DISCUSSION
This study characterized the molecular and biological properties of an S. aureus protein tentatively designated SOK. The protein was designated SOK to highlight its cell surface location and enhancement of resistance to oxidative killing. In silico sequence analysis for potential staphylococcal virulence factors involved in systemic or cardiovascular infections identified SOK, partly because of its relatedness to the S. pyogenes 67.7-kDa myosin cross-reactive protein. SOK has not been extensively studied but was previously noted during the report of the S. aureus N315 genome sequence (25). The N315 SOK is encoded by ORF SA0102 and was described as an MHC-II β-chain homologue. SOK and its S. aureus homologues share 20 to 21% identity and 34% similarity to murine HLA class II β-chain homologues (RefSeq NP_996988) on the almost 300-residue-long fragment (unpublished data).
SOK has homologues in other bacteria and shares 49.8 to 62.3% sequence homology with proteins in Bacillus, Bradyrhizobium, Bifidobacterium, Clostridium, Enterococcus, Lactococcus, Lactobacillus, Leuconostoc, Streptococcus, and Rhodopseudomonas genera and less, but significant, relatedness to numerous homologues in other Gram-positive and Gram-negative bacteria species (see Fig. S1B in the supplemental material). SOK and its relatives have several conserved sequences which, in SOK, encompass residues 75 to 102, 174 to 211, and 495 to 539, showing >90% homologies. Previously, the most well-characterized protein in this family was the S. pyogenes 67-kDa myosin cross-reactive antigen (19, 47), which reacts with sera from patients with streptococcal infections and poststreptococcal sequelae (19). Considering the numerous SOK homologues and various degrees of relatedness, plus the unique association of S. pyogenes with immunopathologies, the ability to cross-react with myosin antibodies may be unique to the streptococcal homologue. Studies to determine whether other SOK homologues possess this property are ongoing in our laboratories.
This high level of sequence conservation is also reflected in PCR-RFLP results, which generated only eight different pattern types among a collection of 59 clinical isolates digested with three different restriction endonucleases. The PCR-RFLP results for sok were compared with spa typing results, which were previously reported for these same isolates (30). The spa typing method was chosen here as one of the most suitable techniques for evaluation of long-term evolutionary changes (15, 40). The differences between RFLP patterns reflect the variability of the sok gene among the 13 sequenced S. aureus genomes analyzed, where this variability is caused by point mutations in sok (data not shown). Nearly 97% concordance between deduced spa typing clusters and sok PCR-RFLP groups illustrates the significance of the sok gene for staphylococcal epidemiology. It is possible that the high concordance between spa typing and sok PCR-RFLP is the result of the close genetic linkage of these two genes. In the majority of sequenced S. aureus genomes, these two genes are separated by less than 7.5 kb, including four ORFs, and in some genomes, such as those of the MW2 and MSSA476 strain, these two genes are separated by less than 3 kb, which include only two ORFs (4, 25).
Consistent with immunoblotting results showing colocalization of SOK in the membrane fractions, immunofluorescence experiments indicated that SOK is exposed on the S. aureus cell surface. However, sequence analysis did not reveal the presence of a typical LPXTG cell wall-anchoring motif although SOK has one predicted N-terminal transmembrane domain encompassing residues 25 through 42 within a potential signal peptide sequence (10). Repeated attempts to obtain a pure or partially purified mature form of SOK for the purpose of N-terminal sequencing to help explain the size discrepancy between bioinformatic analysis (∼67.6 kDa) and immunoblotting results (∼55 kDa) were unsuccessful. Thus, whether SOK has a signal peptide which is removed during maturation and localization is still to be resolved. It has been shown that staphylococcal proteases such as metalloprotease and metallocysteine protease are involved in the maturation process for staphylococcal virulence factors such as lipase and serine protease (35, 38). It is possible that SOK might be processed by staphylococcal protease in the maturation and localization process. This possibility is currently under investigation.
One striking effect of SOK is its ability to confer resistance to killing by PMNs following phagocytosis. The mechanism by which SOK could contribute to virulence is not completely understood but likely is due at least partly to its role in promoting survival in phagocytes, mediated by the resistance to PMN oxidative killing. The human innate immune response is an essential first line of defense against bacterial pathogens (16, 21, 39, 44). PMNs are recruited early to sites of infection and typically efficiently recognize and ingest invading S. aureus. The ability of PMNs to kill S. aureus is based on the bactericidal activity of ROS produced by the NADPH-dependent oxidase and antimicrobial compounds contained within granules. The importance of ROS in protection against S. aureus is exemplified by the increased susceptibility of NADPH oxidase-deficient chronic granulomatous disease patients to severe staphylococcal infections. To test the involvement of SOK in resistance to the innate immune response, PMN bactericidal activity was assessed following phagocytosis of a serum-opsonized S. aureus Newman wild-type strain and an isogenic sok mutant derivative. Our data indicate that survival of the S. aureus sok mutant strain in human PMNs was decreased compared to survival of the isogenic parent strain at all times tested. In addition, PMN ROS production was increased following interactions with the S. aureus sok deletion strain compared to that induced by the wild-type strain. Further examination of the role of SOK in ROS resistance indicates that SOK is involved in resistance to singlet oxygen and not hydrogen peroxide. Inasmuch as our study demonstrates that SOK is localized to the cell surface of S. aureus and that the mutant strain elicits altered PMN ROS production, it is possible that SOK participates in pathogen recognition by neutrophils. However, efficient PMN ROS production and killing in response to wild-type strains of S. aureus have been demonstrated previously (36, 48). It is also possible that the apparent increase in the PMN oxidative burst following interactions with the sok mutant strain is due to alterations in metabolic pathways controlling the S. aureus oxidative stress response. S. aureus has been shown to produce both catalase and superoxide dismutase in response to oxidative stress (29). Increased production of either enzyme following ingestion by PMNs would result in a decrease in oxidation of the DCF substrate in the in vitro ROS assay. Alternatively, the differential effects in ROS levels in cells following the parental versus deletion mutant could be the result of more efficient quenching of singlet oxygen by the parental strain. There are a number of additional reactive oxygen species that are present in the phagosome of polymorphonuclear phagocytes. Activation of the NADPH oxidase generates superoxide, a relatively unstable molecule that rapidly dismutates itself to form hydrogen peroxide and oxygen (or is catalyzed by superoxide dismutase). The diversity of ROS present in the phagosome, in addition to relative instability, confounds simulation of this environment through use of independent compounds. In addition, the direct role of ROS in microbicidal activity has been intensely debated (34). At the very least, PMN microbicidal activity is likely the result of integrated activities of both oxygen-dependent and -independent molecules. In our study we tested directly the ability of the S. aureus Δsok strain to survive following neutrophil phagocytosis through in vitro killing assays. It is unclear whether the ability of the S. aureus Δsok strain to alter PMN ROS production kinetics and the increased sensitivity to singlet oxygen/PMN killing are linked.
With use of deletion mutagenesis, we demonstrated that SOK affects in vivo staphylococcal virulence. Under the conditions tested, the lack of the SOK protein severely debilitated the virulence of two S. aureus strains tested in the rabbit endocarditis model. Vegetation formation was completely abrogated or reduced to minimal levels, and the number of culturable bacteria in the hearts was reduced by several logs. We hypothesize that the inability of the SOK-deficient strains to cause vegetations as significant as those formed by the parent and complemented strains is due to their increased elimination before vegetations can form. This is consistent with our prior studies with Enterococcus faecalis, related to the surface protein enterococcal aggregation substance, where it has been shown that the aggregation substance also interferes with phagocyte killing (37) and contributes significantly to vegetation formation (43). Our prior studies also suggest that once vegetations begin to form in E. faecalis endocarditis, the enterococci become trapped within host- and bacterium-derived matrices that significantly reduce phagocytic removal (32). Interestingly, McAleese et al. (31) demonstrated that sok expression was elevated 2.6-fold in isolates from a heart valve compared to bloodstream isolates of the same patient. The reduced ability of sok deletion strains to form vegetations suggests that sok expression may be important as S. aureus interacts with host cells in heart tissues.
Collectively, this study resulted in partial biochemical and biological characterization of the SOK protein. Certainly, to get a full characterization of the SOK protein and its complete role in S. aureus infections further investigation is required. SOK protects bacteria during the infection process from neutrophil killing, as well as promoting vegetation formation during heart infections. The presence of sok homologues in a diverse group of organisms that includes both pathogens and environmental bacteria suggests that SOK and its homologues may function to protect bacteria from ROS encountered both in animal hosts and in the environment.
Supplementary Material
Acknowledgments
These studies were supported by the Idaho Agricultural Experiment Station in addition to the following: Committee for Scientific Research grant PBZ-KBN-101/T09/2003/14 (J.M.) and Public Health Service grants P20-RR016454 (G.A.B.), P20-RR15587 (G.A.B.), and AI074283 (P.M.S.); U.S. Department of Agriculture NIFA AFRI grant 2008-35204-04582 (K.S.S. and G.A.B.); and National Centers for Research Resources, Centers of Biomedical Research Excellence, grant P20RR015587 (S.K.). P.M.S. also acknowledges membership in and support (U54 AI57153) from the Region V Great Lakes Regional Center of Excellence in Biodefense and Emerging Infectious Diseases Consortium.
We are grateful to Waleria Hryniewicz (National Medicines Institute, Warsaw, Poland) for providing the culture collection of human clinical isolates and to Ann Norton for technical assistance.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 11 October 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Alexander, E. H., and M. C. Hudson. 2001. Factors influencing the internalization of Staphylococcus aureus and impacts on the course of infections in humans. Appl. Microbiol. Biotechnol. 56:361-366. [DOI] [PubMed] [Google Scholar]
- 2.Appelbaum, P. C. 2007. Microbiology of antibiotic resistance in Staphylococcus aureus. Clin. Infect. Dis. 45:S165-S170. [DOI] [PubMed] [Google Scholar]
- 3.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 4.Baba, T., T. Bae, O. Schneewind, F. Takeuchi, and K. Hiramatsu. 2008. Genome sequence of Staphylococcus aureus strain Newman and comparative analysis of staphylococcal genomes: polymorphism and evolution of two major pathogenicity islands. J. Bacteriol. 190:300-310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berthold, H., B. Frorath, M. Scanarini, C. C. Abney, B. Ernst, and W. Northemann. 1992. Plasmid pGEX-5T: an alternative system for expression and purification of recombinant proteins. Biotechnol. Lett. 14:245-250. [Google Scholar]
- 6.Bevers, L. E., M. W. H. Pinkse, P. D. E. M. Verhaert, and W. R. Hagen. 2009. Oleate hydratase catalyzes the hydration of a nonactivated carbon-carbon bond. J. Bacteriol. 191:5010-5012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chevallet, M., V. Santoni, A. Poinas, D. Rouquié, A. Fuchs, S. Kieffer, M. Rossignol, J. Lunardi, J. Garin, and T. Rabilloud. 1998. New zwitterionic detergents improve the analysis of membrane proteins by two-dimensional electrophoresis. Electrophoresis 19:1901-1909. [DOI] [PubMed] [Google Scholar]
- 8.DeLeo, F. R., L. A. Allen, M. Apicella, and W. M. Nauseef. 1999. NADPH oxidase activation and assembly during phagocytosis. J. Immunol. 163:6732-6740. [PubMed] [Google Scholar]
- 9.Dziewanowska, K., A. R. Carson, J. M. Patti, C. F. Deobald, K. W. Bayles, and G. A. Bohach. 2000. Staphylococcal fibronectin binding protein interacts with heat shock protein 60 and integrins: role in internalization by epithelial cells. Infect. Immun. 68:6321-6328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fekkes, P., and A. J. Driessen. 1999. Protein targeting to the bacterial cytoplasmic membrane. Microbiol. Mol. Biol. Rev. 63:161-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischetti, V. A., R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. J. Rood. 2000. Gram-positive pathogens. ASM Press, Washington, DC.
- 12.Foster, T. J. 2004. The Staphylococcus aureus “superbug.” J. Clin. Invest. 114:1693-1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox, P. M., R. J. Lampen, K. S. Stumpf, G. L. Archer, and M. W. Climo. 2006. Successful therapy of experimental endocarditis caused by vancomycin-resistant Staphylococcus aureus with a combination of vancomycin and beta-lactam antibiotics. Antimicrob. Agents Chemother. 50:2951-2956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 15.Hallin, M., A. Deplano, O. Denis, R. De Mendonça, R. De Ryck, and M. J. Struelens. 2007. Validation of pulsed-field gel electrophoresis and spa typing for long-term, nationwide epidemiological surveillance studies of Staphylococcus aureus infections. J. Clin. Microbiol. 45:127-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hermann, M., M. E. Jaconi, C. Dahlgren, F. A. Waldvogel, O. Stendahl, and D. P. Lew. 1990. Neutrophil bactericidal activity against Staphylococcus aureus adherent on biological surfaces. Surface-bound extracellular matrix proteins activate intracellular killing by oxygen-dependent and -independent mechanisms. J. Clin. Invest. 86:942-951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hiraga, S., C. Ichinose, H. Niki, and M. Yamazoe. 1998. Cell cycle-dependent duplication and bidirectional migration of SeqA-associated DNA-protein complexes in E. coli. Mol. Cell 1:381-387. [DOI] [PubMed] [Google Scholar]
- 18.Hovde, C. J., S. P. Hackett, and G. A. Bohach. 1990. Nucleotide sequence of the staphylococcal enterotoxin C3 gene: sequence comparison of all three type C staphylococcal enterotoxins. Mol. Gen. Genet. 220:329-333. [DOI] [PubMed] [Google Scholar]
- 19.Kil, K. S., M. W. Cunningham, and L. A. Barnett. 1994. Cloning and sequence analysis of a gene encoding a 67-kilodalton myosin-cross-reactive antigen of Streptococcus pyogenes reveals its similarity with class II major histocompatibility antigens. Infect. Immun. 62:2440-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klevens, R. M., M. A. Morrison, J. Nadle, S. Petit, K. Gershman, S. Ray, L. H. Harrison, R. Lynfield, G. Dumyati, J. M. Townes, A. S. Craig, E. R. Zell, E. Fosheim, L. K. McDougal, R. B. Carey, S. K. Fridkin, and Active Bacterial Core Surveillance MRSA Investigators. 2007. Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298:1763-1771. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi, S. D., J. M. Voyich, C. Burlak, and F. R. DeLeo. 2005. Neutrophils in the innate immune response. Arch. Immunol. Ther. Exp. (Warsz.) 53:505-517. [PubMed] [Google Scholar]
- 22.Kobayashi, S. D., K. R. Braughton, A. R. Whitney, J. M. Voyich, T. G. Schwan, J. M. Musser, and F. R. DeLeo. 2003. Bacterial pathogens modulate an apoptosis differentiation program in human neutrophils. Proc. Natl. Acad. Sci. U. S. A. 100:10948-10953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koreen, L., S. V. Ramaswamy, E. A. Graviss, S. Naidich, J. M. Musser, and B. N. Kreiswirth. 2004. spa typing method for discriminating among Staphylococcus aureus isolates: implications for use of a single marker to detect genetic micro- and macrovariation. J. Clin. Microbiol. 42:792-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kreiswirth, B. N., S. Löfdahl, M. J. Betley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:709-712. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda, M., T. Ohta, I. Uchiyama, T. Baba, H. Yuzawa, I. Kobayashi, L. Cui, A. Oguchi, K. Aoki, Y. Nagai, J. Lian, T. Ito, M. Kanamori, H. Matsumaru, A. Maruyama, H. Murakami, A. Hosoyama, Y. Mizutani-Ui, N. K. Takahashi, T. Sawano, R. Inoue, C. Kaito, K. Sekimizu, H. Hirakawa, S. Kuhara, S. Goto, J. Yabuzaki, M. Kanehisa, A. Yamashita, K. Oshima, K. Furuya, C. Yoshino, T. Shiba, M. Hattori, N. Ogasawara, H. Hayashi, and K. Hiramatsu. 2001. Whole genome sequencing of methicillin-resistant Staphylococcus aureus. Lancet 357:1225-1240. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 27.Liu, G. Y., A. Essex, J. T. Buchanan, V. Datta, H. M. Hoffman, J. F. Bastian, J. Fierer, and V. Nizet. 2005. Staphylococcus aureus golden pigment impairs neutrophil killing and promotes virulence through its antioxidant activity. J. Exp. Med. 202:209-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lowy, F. D. 1998. Staphylococcus aureus infections. N. Engl. J. Med. 339:520-532. [DOI] [PubMed] [Google Scholar]
- 29.Maalej, S., I. Dammak, and S. Dukan. 2006. The impairment of superoxide dismutase coordinates the derepression of the PerR regulon in the response of Staphylococcus aureus to HOCL stress. Microbiol. 152:855-861. [DOI] [PubMed] [Google Scholar]
- 30.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyszton-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multiple-locus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McAleese, F., S. W. Wu, K. Sieradzki, P. Dunman, E. Murphy, S. Projan, and A. Tomasz. 2006. Overexpression of genes of the cell wall stimulon in clinical isolates of Staphylococcus aureus exhibiting vancomycin-intermediate-S. aureus-type resistance to vancomycin. J. Bacteriol. 188:1120-1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.McCormick, J. K., T. J. Tripp, G. M. Dunny, and P. M. Schlievert. 2002. Formation of vegetations during infective endocarditis excludes binding of bacterial-specific host antibodies to Enterococcus faecalis. J. Infect. Dis. 185:994-997. [DOI] [PubMed] [Google Scholar]
- 33.McDevitt, D., P. Francois, P. Vaudaux, and T. J. Foster. 1994. Molecular characterization of the clumping factor (fibrinogen receptor) of Staphylococcus aureus. Mol. Microbiol. 11:237-248. [DOI] [PubMed] [Google Scholar]
- 34.Nauseef, W. M. 2007. How human neutrophils kill and degrade microbes: an integrated view. Immunol. Rev. 219:88-102. [DOI] [PubMed] [Google Scholar]
- 35.Nickerson, N. N., L. Prasad, L. Jacob, L. T. Delbaere, and M. J. McGavin. 2007. Activation of the SspA serine protease zymogen of Staphylococcus aureus proceeds through unique variations of a trypsinogen-like mechanism and is dependent on both autocatalytic and metalloprotease-specific processing. J. Biol. Chem. 282:34129-34138. [DOI] [PubMed] [Google Scholar]
- 36.Palazzolo-Ballance, A. M., M. L. Reniere, K. R. Braughton, D. E. Sturdevant, M. Otto, B. N. Kreiswirth, E. P. Skaar, and F. R. DeLeo. 2008. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180:500-509. [DOI] [PubMed] [Google Scholar]
- 37.Rakita, R. M., N. N. Vanek, K. Jacques-Palaz, M. Mee, M. M. Mariscalco, G. M. Dunny, M. Snuggs, W. B. Van Winkle, and S. I. Simon. 1999. Enterococcus faecalis bearing aggregation substance is resistant to killing by human neutrophils despite phagocytosis and neutrophil activation. Infect. Immun. 67:6067-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rollof, J., and S. Normark. 1992. In vivo processing of Staphylococcus aureus lipase. J. Bacteriol. 174:1844-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rooijakkers, S. H. M., K. P. M. van Kessel, and J. A. G. van Strijp. 2005. Staphylococcal innate immune evasion. Trends Microbiol. 13:596-601. [DOI] [PubMed] [Google Scholar]
- 40.Ruppitsch, W., A. Indra, A. Stöger, B. Mayer, S. Stadlbauer, G. Wewalka, and F. Allerberger. 2006. Classifying spa types in complexes improves interpretation of typing results for methicillin-resistant Staphylococcus aureus. J. Clin. Microbiol. 44:2442-2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sau, S., J. Sun, and C. Y. Lee. 1997. Molecular characterization and transcriptional analysis of type 8 capsule genes in Staphylococcus aureus. J. Bacteriol. 179:1614-1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlievert, P. M., P. J. Gahr, A. P. Assimacopoulos, M. M. Dinges, J. A. Stoehr, J. W. Harmala, H. Hirt, and G. M. Dunny. 1998. Aggregation and binding substances enhance pathogenicity in rabbit models of Enterococcus faecalis endocarditis. Infect. Immun. 66:218-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sibbald, M. J. J. B., A. K. Ziebandt, S. Engelmann, M. Hecker, A. de Jong, H. J. M. Harmsen, G. C. Raangs, I. Stokroos, J. P. Arends, J. Y. F. Dubois, and J. M. van Dijl. 2006. Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70:755-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tamura, K., J. Dudley, M. Nei, and S. Kumar. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596-1599. [DOI] [PubMed] [Google Scholar]
- 46.van Belkum, A. 2006. Staphylococcal colonization and infection: homeostasis versus disbalance of human (innate) immunity and bacterial virulence. Curr. Opin. Infect. Dis. 19:339-344. [DOI] [PubMed] [Google Scholar]
- 47.Volkov, A., A. Liavonchanka, O. Kamneva, T. Fiedler, C. Goebel, B. Kreikemeyer, and I. Feussner. 2010. Myosin cross-reactive antigen of Streptococcus pyogenes M49 encodes a fatty acid double bond hydratase that plays a role in oleic acid detoxification and bacterial virulence. J. Biol. Chem. 285:10353-10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Voyich, J. M., K. R. Braughton, D. E. Sturdevant, A. R. Whitney, B. Saïd-Salim, S. F. Porcella, R. D. Long, D. W. Dorward, D. J. Gardner, B. N. Kreiswirth, J. M. Musser, and F. R. DeLeo. 2005. Insights into mechanisms used by Staphylococcus aureus to avoid destruction by human neutrophils. J. Immunol. 175:3907-3919. [DOI] [PubMed] [Google Scholar]
- 49.Weems, J. J. 2001. The many faces of Staphylococcus aureus infection. Recognizing and managing its life-threatening manifestations. Postgrad. Med. 110:24-26, 29-31, 35-36. [DOI] [PubMed] [Google Scholar]
- 50.Yang, S. J., K. C. Rice, R. J. Brown, T. G. Patton, L. E. Liou, Y. H. Park, and K. W. Bayles. 2005. A LysR-type regulator, CidR, is required for induction of the Staphylococcus aureus cidABC operon. J. Bacteriol. 187:5893-5900. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.