Abstract
Nontypeable Haemophilus influenzae (NTHI) is a leading cause of otitis media infections, which are often chronic and/or recurrent in nature. NTHI and other bacterial species persist in vivo within biofilms during otitis media and other persistent infections. These biofilms have a significant host component that includes neutrophil extracellular traps (NETs). These NETs do not mediate clearance of NTHI, which survives within NET structures by means of specific subpopulations of lipooligosaccharides on the bacterial surface that are determinants of biofilm formation in vitro. In this study, the ability of NTHI and NTHI components to initiate NET formation was examined using an in vitro model system. Both viable and nonviable NTHI strains were shown to promote NET formation, as did preparations of bacterial DNA, outer membrane proteins, and lipooligosaccharide (endotoxin). However, only endotoxin from a parental strain of NTHI exhibited equivalent potency in NET formation to that of NTHI. Additional studies showed that NTHI entrapped within NET structures is resistant to both extracellular killing within NETs and phagocytic killing by incoming neutrophils, due to oligosaccharide moieties within the lipooligosaccharides. Thus, we concluded that NTHI elicits NET formation by means of multiple pathogen-associated molecular patterns (most notably endotoxin) and is highly resistant to killing within NET structures. These data support the conclusion that, for NTHI, formation of NET structures may be a persistence determinant by providing a niche within the middle-ear chamber.
Nontypeable Haemophilus influenzae (NTHI) is a common commensal of the human nasopharynx, in which setting carriage is usually asymptomatic and without adverse effect. When host mucociliary clearance is impaired, NTHI can cause localized opportunistic infections within the airway (14). For example, NTHI is a leading cause of otitis media (OM), which is among the most common and costly pediatric infections and can be a persistent and/or recurrent infection (30). Persistent populations of NTHI in vivo during chronic and recurrent otitis media are found within biofilm communities within the middle-ear chamber (19, 40). Other factors that provide an environment for NTHI to cause infections include age, genetic predisposition, atopy, and immune system impairment (44), as well as Eustachian tube dysfunction and pressure dysregulation caused by virus-induced congestion (11, 12, 33). Biofilms have long been thought to promote microbial resistance to pharmaceutical or immune clearance during persistent infections (17, 20). Bacterial factors important to NTHI biofilms include specific subsets of lipooligosaccharides (LOS) containing sialic acid and/or phosphorylcholine (23, 24, 45), pili (28), and double-stranded DNA (25, 27). Our recent work demonstrated that NTHI biofilms also have a significant host component, including a double-stranded DNA lattice decorated with histones and elastase, that fits the defining traits of a neutrophil extracellular trap (NET) (22). NETs have microbicidal activities that are distinct from phagocytosis and are due to entrapment within the DNA scaffold and killing by granular components (7, 8). Notably, viable NTHI biofilms have been observed throughout these NET structures, and the presence of a biofilm/NET was positively correlated with higher bacterial loads during experimental otitis media infections (22). Based on these data, we concluded that NTHI biofilms survive within NET structures in vivo.
The objective of the present study was to define determinants of NET formation and bacterial survival within NETs following addition of NTHI and bacterial components to polymorphonuclear leukocytes (PMNs). The data show that NTHI elicits NET formation due to host stimulation by endotoxin and, to a lesser extent, other NTHI components, such as outer membrane proteins (OMP) and DNA.
In an attempt to elucidate a mechanism for the induction of NETs, we investigated the potential role of cellular Toll-like receptor (TLR) signaling. TLR signaling has been shown to be critical in immune defense during NTHI infections. In one example, a murine lung model was utilized, and TLR4 knockout mice showed attenuation in early inflammation that correlated with decreased bacterial clearance; this effect was MyD88 dependent and was not observed as dramatically in TLR2 knockout mice (50). In this study, the data suggest that TLR4 signaling through MyD88 may be important for the ability of NTHI to induce NET formation as well.
Moreover, NTHI survived within NET structures and resisted both extracellular killing and phagocytic killing by additional incoming neutrophils. These data support the conclusion that NET formation during otitis media is a determinant of bacterial persistence rather than clearance.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Nontypeable H. influenzae 2019 is a well-described clinical isolate (10, 34). NTHI 2019 rfaD and NTHI 2019 htrB have defects relating to biosynthesis and assembly of the lipooligosaccharides (32, 37), and both are attenuated in the chinchilla infection model of otitis media (13). NTHI 86-028NP is a pediatric OM isolate (4). NTHI 1479 and NTHI 7502 are sputum isolates which have also been described extensively (10, 34). Bacteria were cultured on brain heart infusion agar (Difco) supplemented with hemin (10 μg/ml; ICN Biochemicals) and NAD (10 μg/ml; Sigma). Prior to inoculation, bacteria were suspended at an optical density at 600 nm (OD600) of 0.15 (∼108 CFU/ml) in RPMI 1640 without phenol red (Gibco), and bacterial numbers were confirmed by plate counts. For preparation of nonviable NTHI, bacterial suspensions were heated to 65°C for 30 min and plated to ensure bacterial killing.
Bacterial outer membrane purification.
Outer membrane protein fractions were prepared from NTHI as described previously (35). Briefly, NTHI cells were harvested from overnight plate cultures and suspended in 0.01 M HEPES buffer (pH 7.4) before sonication. Intact cells and debris were removed by centrifugation in a Beckman 78-70 M ultracentrifuge (SW41 rotor) at 180 × g for 30 min at room temperature. Cell envelopes were collected from supernatants by centrifugation at 110,000 × g for 1 h at 4°C. The pellet was resuspended in equal volumes of 0.01 M HEPES buffer and 2% sarcosyl (Sigma) in 0.01 M HEPES and then incubated at room temperature for 30 min. Centrifugation at 110,000 × g was repeated for 1 h at 4°C. After removal of the supernatant, the pellet was rinsed in double-distilled H2O (ddH2O) before resuspension in ddH2O. After determination of protein content using a bicinchoninic acid (BCA) assay (Thermo Scientific), the samples were stored at 4°C until use.
NTHI DNA isolation.
Genomic DNA was purified from overnight cultures of NTHI by use of a Wizard genomic DNA isolation kit (Promega). The DNA preparations were resuspended in ddH2O, quantified by absorbance using a NanoDrop system, and stored at 4°C until use.
LOS extraction.
Lipooligosaccharides from NTHI 2019 or NTHI 2019 htrB were isolated as described previously (1, 26). Briefly, NTHI was harvested from overnight plate cultures and suspended in cold phosphate-buffered saline (PBS). The suspension was centrifuged for 10 min at 8,000 × g to pellet bacteria; pellets were resuspended in ddH2O and incubated at 65°C for 30 min. One volume of 65°C phenol was added, vortexed, and placed on ice for 1 h. The suspensions were then centrifuged at 3,600 × g for 45 min, the aqueous phase was removed and placed in a fresh tube, and back extraction of the phenol was performed on the initial sample by reheating it to 65°C and adding 1 volume of 65°C ddH2O. The sample was mixed and placed on ice for 1 h before centrifugation at 3,600 × g for 45 min and combination of the aqueous phases. LOS was precipitated by adding a 1/10 volume of 3 M sodium acetate and 2 volumes of 100% ethanol, with incubation at −80°C overnight. LOS was pelleted by centrifugation at 32,500 × g for 1 h and was resuspended in buffer containing 60 mM Tris, 10 mM EDTA, and 2% SDS (pH 6.8). Samples were boiled for 5 min, and after they returned to room temperature, 100 μg of proteinase K (Sigma) was added. Samples were then incubated overnight at 37°C and precipitated as described above, by centrifugation at 32,500 × g followed by dialysis against ddH2O. LOS was lyophilized and, after recording of the dry mass, diluted to 1 mg/ml in water. Samples were then stored at 4°C until diluted to the indicated final concentration during PMN treatment. Biologically relevant concentrations were calculated by multiplying the number of live or heat-killed bacteria used in other assays by ∼8 fg (LOS accounts for roughly 4% of the dry weight of NTHI), the average mass contribution of LOS (15).
In vitro NET induction assays.
Neutrophils were isolated from peripheral blood obtained from healthy donors according to a standard method, using Isolymph density gradient centrifugation, and viability was assessed by trypan blue exclusion. Analysis of the cellular populations recovered from the density gradients by Cytospin centrifugation and hematoxylin and eosin staining confirmed the presence of over 97% polymorphonuclear cells (i.e., neutrophils). The blood donations were obtained by informed consent under protocols reviewed and approved by the WFUHS Institutional Review Board. MyD88 studies utilized bone marrow-derived PMNs, isolated by standard Isolymph density gradient centrifugation, from the tibias and femurs of male wild-type, MyD88−/−, and TLR2−/− C57BL/6 mice of 6 to 9 weeks of age (kindly provided by S. Akira, Osaka, Japan). TLR4 studies utilized bone marrow-derived PMNs from male C3H/HeN and C3H/HeJ mice of 7 to 10 weeks of age (NCI), isolated as described above. Neutrophils were suspended in RPMI culture medium and seeded into wells of a chamber coverslip system (Nunc Nalge International) after treatment with 0.01% poly-l-lysine (Sigma). In some experiments, neutrophils were pretreated with 10 μM diphenyliodonium (DPI) (MP Biomedicals) 30 min prior to treatment, as indicated in the text. DPI is a nonspecific inhibitor of the NAD(P)H oxidase complex. Following adherence, cells were treated with NTHI 2019 (multiplicity of infection [MOI], ∼0.01), an equivalent amount of heat-killed NTHI 2019, purified NTHI LOS (0.032 ng/μl in each treatment well), purified NTHI DNA (0.032 ng/μl in each treatment well), purified NTHI outer membrane proteins (5 μg OMP in each treatment well), or purified NTHI 2019 htrB LOS (0.032 ng/μl in each treatment well). In some experiments, LOS was pretreated with 50 μg/ml polymyxin B (Sigma) for 20 min prior to treatment of PMNs, as indicated in the text. Amounts of bacterial components used in these treatments were extrapolated to be consistent with those present within the numbers of NTHI cells used in the infections. Samples were incubated for 5 h in 5% CO2 at 37°C. Following incubation, samples were fixed in 4% paraformaldehyde-PBS and blocked in 1% bovine serum albumin (BSA; Sigma)-PBS. Next, samples were processed for immunofluorescence by use of primary antibodies directed against histone protein (panreactive monoclonal antibody [MAb] MAB3422; Chemicon) or neutrophil elastase (MAb NP57; Thermo Scientific), followed with an appropriate fluorescent secondary antibody conjugate (Alexa Fluor 488-goat anti-mouse IgG; Molecular Probes). Murine studies utilized a primary antibody directed against murine histone proteins (MAb H5300-10; U.S. Biologicals) and the same secondary antibody as that described above. Five 5-min washes were performed after each antibody treatment with 1% BSA-PBS. Samples were also stained with 0.3 mM propidium iodide (PI; Molecular Probes) to visualize DNA. Microscopy was performed using a Zeiss LSM 510 confocal laser scanning microscope. Immunofluorescent images were selected to show the size and structural characteristics of NETs, and although some panels show fewer or more PMNs than others, the average number of PI-stained nuclei remained constant among conditions. In bacterial fractionation and murine assays, fields of view positive for NET formation were quantified. Secondary antibody-only controls were used in each experiment to confirm the absence of significant nonspecific background staining.
NET-associated NTHI killing assay.
Primary human neutrophils were isolated as described above and seeded into wells of a 24-well tissue culture dish (Corning) at a concentration of 106 cells/well. Following adherence, PMNs were treated with approximately 104 heat-killed NTHI 2019 cells for 5 h at 37°C and 5% CO2. Following the induction of NETs, cytochalasin D (Sigma) was added to the wells, to a final concentration of 10 μg/ml. After 15 min of cytochalasin D treatment, approximately 104 NTHI 2019 or NTHI 2019 rfaD cells were added to the wells and allowed to incubate for 24 h at 37°C and 5% CO2. Following the incubation, supernatants were removed, and freshly isolated neutrophils (106/well) were added to the wells and incubated with the overnight surface-attached communities for 30 min at 37°C and 5% CO2. A subset of the wells received neutrophils that were pretreated with 10 μg/ml cytochalasin D for 15 min. Wells were then thoroughly scraped prior to serial dilutions of the suspensions. Plate counts were used to assess bacterial viability.
Statistics.
Data were analyzed using paired and nonparametric t tests and two-way analysis of variance (ANOVA) with post hoc tests of significance. Data groups with P values of ≤0.05 were deemed statistically significant. Analyses were performed using GraphPad Prism, version 5 (GraphPad Software, San Diego, CA).
RESULTS
NTHI induces the formation of NETs in vitro.
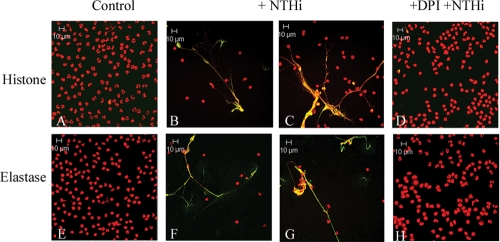
We hypothesized that exposure of neutrophils to NTHI would elicit formation of NETs in vitro. Polymorphonuclear leukocytes were obtained from healthy human donors, treated with live NTHI 2019, and stained for analysis of NET formation. As shown in the immunofluorescent micrographs in Fig. 1, it is clear that NTHI elicited NET formation, as evidenced by the web-like structures showing colocalization of DNA with histone protein (Fig. 1B and C) or with neutrophil elastase (Fig. 1F and G). Evidence of NET formation was seen at earlier time points as well (1 h and 3.5 h) (data not shown). Untreated controls (Fig. 1A and E) presented normal nuclear morphology, and no evidence of NET formation was observed. Previous work has shown a requirement of the neutrophil oxidative burst for NET formation, as neutrophils from individuals suffering from chronic granulomatous disease or neutrophils treated with the NADPH oxidase inhibitor DPI are unable to undergo this process (16). Our studies are consistent with these findings, as neutrophils treated with NTHI 2019 following pretreatment with DPI (Fig. 1D and H) resembled untreated controls after 5 h of incubation.
FIG. 1.
NTHI induces the formation of NETs in vitro. Neutrophils (106/well) were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells remained untreated (A and E) or were treated with NTHI 2019 at an MOI of 0.01 bacterium/neutrophil. Neutrophils in panels D and H were pretreated with 10 μM DPI for 30 min. Samples were incubated for 5 h and then stained with antibodies against histone protein (A to D) (green) or neutrophil elastase (E to H) (green) and with secondary antibody-Alexa 488 conjugate. All samples were also stained with propidium iodide (red), and yellow indicates areas of colocalization between DNA and either histone protein (A to D) or neutrophil elastase (E to H). Merged images of 1-μm optical slices are shown. Magnification, ×63.
Nonviable NTHI induces the formation of NETs in vitro.
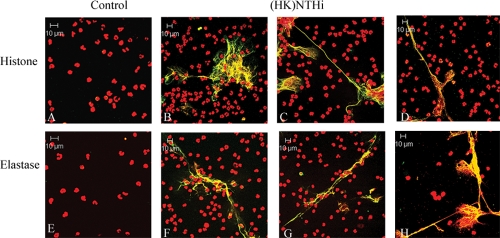
In order to determine if NET induction by NTHI is dependent upon some characteristic of viable bacteria, we used heat-killed NTHI in the in vitro NET induction assay. An equivalent amount of NTHI 2019 to that used in previous experiments was heated to 65°C for 30 min, and killing was confirmed by plating the inocula. After incubation, the samples were processed for immunofluorescence as described above. Just as with viable NTHI, heat-killed bacteria elicited formation of NETs in vitro, as colocalization of DNA with either histone protein (Fig. 2 B to D) or neutrophil elastase (Fig. 2F to H) was observed. As in previous experiments, some signs of neutrophil death prior to fixation and concentrated nuclear colocalization were observed in untreated wells after 5 h (data not shown), while the characteristic pattern of NET staining remained absent. Equivalent amounts of heat-killed NTHI strains 86-028, 1479, and 7502 induced the formation of NETs as well (data not shown).
FIG. 2.
Nonviable NTHI induces the formation of NETs in vitro. Neutrophils (106/well) were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells remained untreated (A and E) or were treated with ∼104 heat-killed (HK) NTHI 2019 cells at 65°C. Samples were incubated for 5 h and then stained with antibodies against histone protein (A to D) (green) or neutrophil elastase (E to H) (green) and with secondary antibody-Alexa 488 conjugate. All samples were also stained with propidium iodide (red), and yellow indicates areas of colocalization between DNA and either histone protein (A to D) or neutrophil elastase (E to H). Merged images of 1-μm optical slices are shown. Magnification, ×63.
Purified NTHI lipooligosaccharides induce the formation of NETs in vitro.
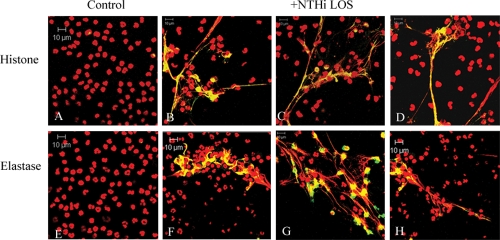
Next, we asked which component(s) of NTHI was involved in initiation of NET formation. Using purified NTHI LOS, we performed NET induction assays to observe the effect of this portion of the NTHI outer membrane on neutrophils. After determining the approximate amount of LOS corresponding to the number of live or heat-killed NTHI cells used in previous experiments, neutrophils were treated with 0.032 ng/μl of purified LOS or left untreated. Following incubation, the samples were processed for immunofluorescence as described above. As seen with both live NTHI and heat-killed bacteria, purified LOS was able to induce the formation of NETs in vitro. Colocalization of DNA with histone protein (Fig. 3 B to D) or with neutrophil elastase (Fig. 3F to H) was observed in treated wells and was not seen in untreated controls (Fig. 3A and E). In addition, quantitative analyses enumerating fields of view positive for NET formation showed no difference among wells treated with either live NTHI, heat-killed NTHI, or purified NTHI LOS; also, no differences were seen when a 10-fold greater amount of purified LOS was used in this assay (data not shown).
FIG. 3.
Purified NTHI LOS induces the formation of NETs in vitro. Neutrophils (106/well) were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells remained untreated (A and E) or were treated with 0.032 ng/μl LOS purified from NTHI 2019, a dose that was consistent with the lowest MOI that yielded NET formation. Samples were incubated for 5 h and then stained with antibodies against histone protein (A to D) (green) or neutrophil elastase (E to H) (green) as in the preceding figures. All samples were also stained with propidium iodide (red), and yellow indicates areas of colocalization between DNA and either histone protein (A to D) or neutrophil elastase (E to H). Merged images of 1-μm optical slices are shown. Magnification, ×63.
Purified NTHI LOS induces NET formation to a higher degree than does purified NTHI DNA or OMP in vitro.
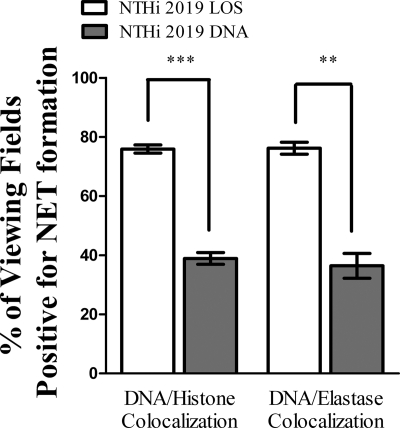
Since the above experiments showed that purified NTHI LOS induced NET formation to the levels seen with whole bacteria, we compared the abilities of other NTHI components to induce NETs. Neutrophils were left untreated or were treated with either purified NTHI LOS or DNA. After incubation, the samples were processed for immunofluorescence as described above. Colocalization of DNA with either neutrophil histone or elastase showed that both treatment conditions resulted in some level of NET formation. However, in enumerating the number of viewing fields positive for NET formation (Fig. 4), we observed that approximately 80% of the LOS-treated fields of view showed DNA-histone colocalization (left white bar), and 76% showed DNA-neutrophil elastase colocalization (right white bar). In contrast, the percentages fell to approximately 39% (left gray bar) and 36% (right gray bar), respectively, when neutrophils were instead treated with DNA. Also, 10-fold higher concentrations of NTHI DNA did not produce NETs at the level observed for LOS induction (data not shown). In additional experiments, the amount of NET formation in response to bacterial DNA was shown to be unaffected by treatment with DNase (data not shown), suggesting that trace endotoxin contamination may have been responsible for the lesser degree of NET formation observed in these fractions.
FIG. 4.
NTHI LOS is a more potent inducer of NET formation than is NTHI DNA in vitro. Neutrophils (106/well) were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells remained untreated or were treated with 0.032 ng/μl NTHI LOS or DNA. After 5 h of incubation, samples were processed for immunofluorescence as described in the text and viewed using immunofluorescence confocal microscopy. At least 65 randomly selected fields of view from four wells per treatment condition and staining condition were observed to quantify the percentage of viewing fields positive for NET formation, based on colocalization of DNA with either histone (left) or neutrophil elastase (right). White bars, LOS treatment; gray bars, DNA treatment. Data were obtained from three separate experiments, and statistical significance was determined by two-way ANOVA with a post hoc test of significance. **, P < 0.005; ***, P < 0.0005.
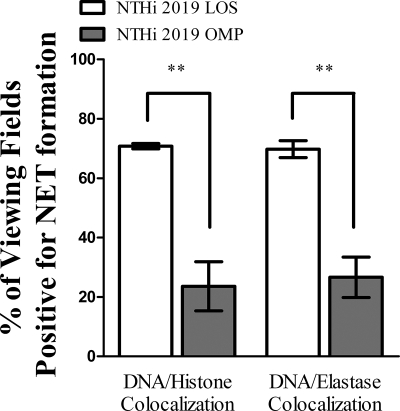
Similar experiments were performed in order to compare the ability of purified NTHI OMP to induce NET formation to that of LOS. Again, neutrophils remained untreated or were treated with either NTHI LOS or a high concentration (5 μg) of OMP. Immunofluorescence assay and quantification of these samples (Fig. 5) showed that approximately 71% of the LOS-treated fields of view showed DNA-histone colocalization (left white bar), and 70% showed DNA-neutrophil elastase colocalization (right white bar). In contrast, the percentages fell to approximately 24% (left gray bar) and 27% (right gray bar), respectively, when neutrophils were instead treated with OMP. Also, when either a higher (10 μg) or lower (1 μg) concentration of OMP was used for induction, no significant differences were seen in the level of NET formation (data not shown). Treatment of protein fractions with proteinase K had no effect on the degree of NET formation (data not shown), suggesting that trace contamination with another component(s) may be involved.
FIG. 5.
NTHI LOS is a more potent inducer of NET formation than is NTHI OMP in vitro. Neutrophils (106/well) were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells remained untreated or were treated with either 0.032 ng/μl NTHI LOS or 5 μg NTHI OMP. After 5 h of incubation, samples were processed for immunofluorescence as described in the text and viewed using immunofluorescence confocal microscopy. At least 65 randomly selected fields of view from four wells per treatment condition and staining condition were observed to quantify the percentage of viewing fields positive for NET formation, based on colocalization of DNA with either histone (left) or neutrophil elastase (right). White bars, LOS treatment; gray bars, OMP treatment. Data were obtained from three separate experiments, and statistical significance was determined by two-way ANOVA with a post hoc test of significance. **, P < 0.005.
Full-length NTHI 2019 LOS optimally induces NET formation.
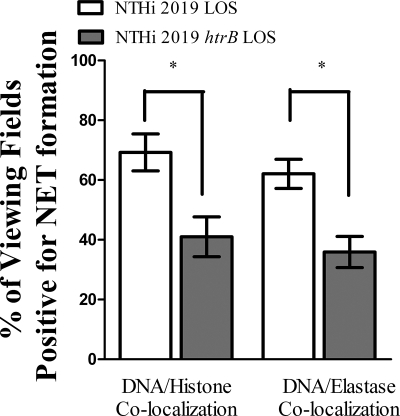
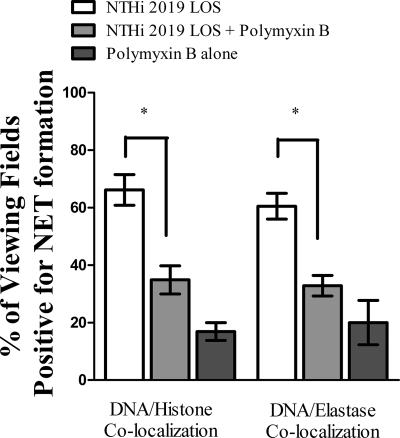
Observing that NTHI LOS is a potent inducer of NET formation, we modified the induction assays to compare the ability of LOS isolated from NTHI 2019 htrB to that of LOS isolated from the parental strain. In infection studies, the htrB strain required a 3-log greater dose than the parental strain to induce otitis media in the chinchilla middle-ear model (18). Neutrophils again remained untreated or were treated with equivalent amounts of LOS from either the htrB mutant or the parental strain. Immunofluorescence assay and quantification of these samples (Fig. 6) showed that approximately 69% of the fields of view observed were positive for DNA-histone colocalization (left white bar), and 62% showed DNA-neutrophil elastase colocalization (right white bar), when PMNs were treated with LOS from the parental strain. The percentages dropped to approximately 41% (left gray bar) and 40% (right gray bar), respectively, when PMNs were treated with LOS isolated from the htrB mutant strain. In addition, to bind and inactivate endotoxin, purified LOS was pretreated with polymyxin B. Neutrophils then received either untreated 2019 LOS, polymyxin B-treated 2019 LOS, or polymyxin B alone. Immunofluorescence assay and quantification of these samples (Fig. 7) showed that approximately 66% of the fields of view observed were positive for DNA-histone colocalization (left white bar), and 65% showed DNA-neutrophil elastase colocalization (right white bar), when PMNs were treated with untreated LOS. The percentages dropped to approximately 35% (left light gray bar) and 33% (right light gray bar), respectively, when PMNs received polymyxin B-treated LOS. Treatment with polymyxin B alone did actually result in low levels of NET formation, although levels of NET formation with polymyxin B-treated LOS were not significantly above these levels (dark gray bars).
FIG. 6.
Full-length NTHI LOS optimally induces NET formation in vitro. Neutrophils (106/well) were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells remained untreated or were treated with either 0.032 ng/μl NTHI LOS or 0.032 ng/μl NTHI htrB LOS. After 5 h of incubation, samples were processed for immunofluorescence as described in the text and viewed using immunofluorescence confocal microscopy. At least 65 randomly selected fields of view from four wells per treatment condition and staining condition were observed to quantify the percentage of viewing fields positive for NET formation, based on colocalization of DNA with either histone (left) or neutrophil elastase (right). White bars, parental strain LOS treatment; gray bars, htrB mutant strain LOS treatment. Data were obtained from three separate experiments, and statistical significance was determined by a nonparametric t test, *, P < 0.05.
FIG. 7.
Polymyxin B-treated LOS induces NET formation to lower levels than those induced by untreated LOS. Neutrophils (106/well) were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells remained untreated or were treated with either 0.032 ng/μl NTHI LOS, 0.032 ng/μl NTHI LOS pretreated with 50 μg/ml polymyxin B for 20 min, or an equivalent amount of polymyxin B alone (∼0.53 μg/ml). After 5 h of incubation, samples were processed for immunofluorescence as described in the text and viewed using immunofluorescence confocal microscopy. At least 65 randomly selected fields of view from four wells per treatment condition and staining condition were observed to quantify the percentage of viewing fields positive for NET formation, based on colocalization of DNA with either histone (left) or neutrophil elastase (right). White bars, untreated LOS; light gray bars, polymyxin B-treated LOS; dark gray bars, polymyxin B alone. Data were obtained from three separate experiments, and statistical significance was determined by a nonparametric t test. *, P < 0.05.
Murine neutrophils require MyD88 to undergo optimal NET production.
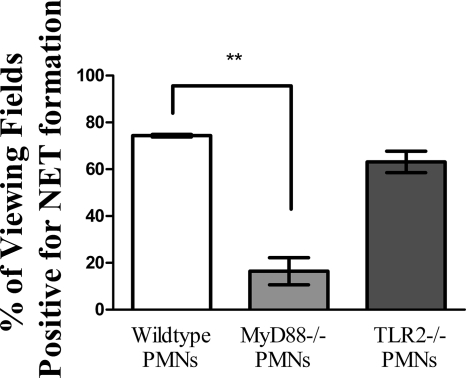
Immunofluorescence assays confirmed that neutrophils isolated from the bone marrow of C57BL/6 mice formed NETs in response to NTHI LOS (data not shown). We next wanted to determine possible cellular signaling mediators that may be important for a neutrophil's ability to undergo NET formation in response to NTHI. Bone marrow-derived neutrophils from wild-type, MyD88-deficient, and TLR2-deficient C57BL/6 mice were isolated, treated with heat-killed NTHI, and processed for immunofluorescence to determine levels of NET formation by the presence of characteristic web-like structures containing DNA and histone colocalization. In these assays, the percentage of viewing fields positive for NET formation was significantly higher for samples containing PMNs from wild-type animals than for those from MyD88-deficient animals, at approximately 74.3% and 16.4%, respectively; the ability of PMNs deficient in TLR2 to undergo NET formation was not significantly lower than that seen under wild-type conditions (Fig. 8).
FIG. 8.
Murine PMNs lacking MyD88 have decreased levels of NET formation compared to wild-type PMNs in vitro. Neutrophils (∼5 × 105) isolated from the bone marrow of wild-type (white bar), MyD88-deficient (light gray bar), and TLR2-deficient (dark gray bar) C57BL/6 mice were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells then received heat-killed NTHI at an MOI of 0.01 for 5 h. Following incubation, wells were fixed, processed for immunofluorescence, and viewed by confocal microscopy. At least 65 randomly selected fields of view from four wells per treatment condition were observed. Bars indicate the percentages of viewing fields that contained NET structures positive for DNA and histone protein colocalization. Data were obtained from three separate experiments, and statistical significance was determined by a paired t test. **, P < 0.01.
Murine neutrophils deficient in Toll-like receptor 4 show decreased levels of NET formation.
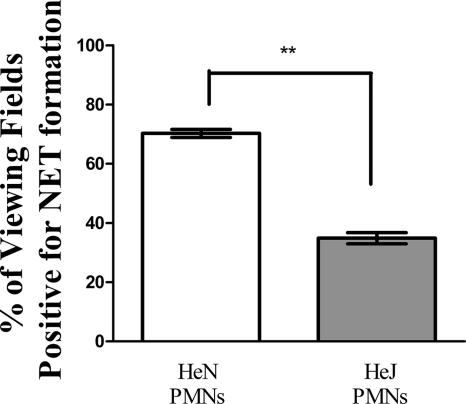
Human neutrophils express mRNAs for all Toll-like receptors except for TLR3 (21), and Toll-like receptors 1, 2, 4, 5, and 6 are expressed on the neutrophil surface (46). Recognizing that LOS is a major determinant for NET formation by NTHI and following up on the observation that MyD88-deficient PMNs have a decreased ability to undergo this program, we tested the ability of TLR4-deficient neutrophils to form NETs. In a similar set of experiments, PMNs were isolated from the bone marrow of either C3H/HeN (TLR4+) or C3H/HeJ (TLR4−) mice and treated with heat-killed NTHI. Immunofluorescence staining consistently showed higher levels of NET formation from PMNs expressing TLR4 than from those that did not express TLR4; averages of 70.3% and 34.9% of viewing fields positive for NET formation were observed for C3H/HeN and C3H/HeJ neutrophils, respectively (Fig. 9).
FIG. 9.
Absence of TLR4 on murine PMNs results in decreased levels of NET formation. Neutrophils (∼5 × 105) isolated from the bone marrow of C3H/HeN (white bar) or C3H/HeJ (gray bar) mice were seeded into wells of a chamber glass coverslip and allowed to adhere. Wells then received heat-killed NTHI at an MOI of 0.01 for 5 h. Following incubation, wells were fixed, processed for immunofluorescence, and viewed by confocal microscopy. At least 65 randomly selected fields of view from four wells per treatment condition were observed. Bars indicate the percentages of viewing fields that contained NET structures positive for DNA and histone protein colocalization. Data were obtained from three separate experiments, and statistical significance was determined by a paired t test. **, P < 0.01.
Primary human neutrophils fail to kill NTHI associated with NETs in vitro.
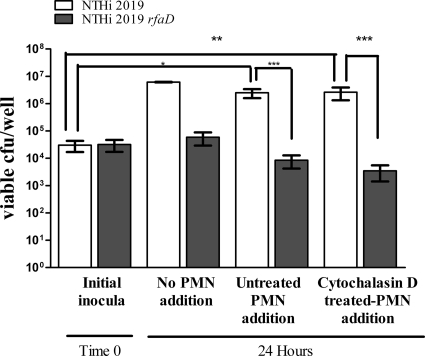
Survival of NTHI within macroscopically visible masses taken from the chinchilla middle ear for up to 2 weeks following infection is observed, even in the face of a large PMN infiltrate evident in toluidine blue-stained sections (22). Using an in vitro approach, we observed the effects of freshly isolated, intact neutrophils on NTHI communities associated with preformed, bacterium-induced NETs. Our previous data showed that phorbol myristate acetate (PMA)-treated neutrophils do not efficiently kill NTHI by either the phagocytic or extracellular route, except in the case of biofilm-deficient mutants with altered LOS composition (22). Consistently, neutrophils treated with heat-killed NTHI to induce NETs prior to phagocytosis inhibition were unable to kill NTHI 2019 (data not shown). To follow up on this observation, and in an effort to mimic infiltrating neutrophils, NETs were formed by treatment of PMNs with heat-killed NTHI 2019 for 5 h. The potential for phagocytic uptake was then eliminated by treatment with cytochalasin D, and wells in which NETs had been induced were inoculated with NTHI 2019 or NTHI 2019 rfaD for 24 h. The rfaD mutant has core/lipid A assembly defects and has been shown to be attenuated in the chinchilla model of otitis media (13). Our previous work also showed susceptibility of this mutant to NET-mediated killing in vitro, with the percentage of extracellular bacterial killing approaching the level of total (NET-mediated and phagocytic) bacterial killing (22). After incubation, the wells received newly isolated PMNs, either left untreated or pretreated with cytochalasin D. Wells were incubated for an additional 30 min before their contents were plated for bacterial enumeration. The contents of replicate wells that did not receive freshly isolated PMNs after the 24-h incubation were also plated for bacterial enumeration. Statistically higher counts were observed in wells containing NTHI 2019 (an approximately 2-log increase) than in those containing NTHI 2019 rfaD or inoculum only, regardless of the phagocytic ability of the added neutrophils; also, numbers of NTHI 2019 in replicate wells that did not receive freshly isolated PMNs were not above those taken from wells that did receive neutrophils. Only a slight but insignificant decrease was observed for NTHI 2019 rfaD under these conditions (Fig. 10). Additionally, large numbers of parental NTHI cells were not recovered from wells that were inoculated with bacteria in the absence of NETs for the 24-h incubation before exposure to intact neutrophils (data not shown).
FIG. 10.
NET-associated NTHI evades killing by neutrophils. Neutrophils (106/well) were seeded into wells of a 24-well plate and allowed to adhere. NETs were induced by treatment with approximately 104 heat-killed NTHI cells for 5 h before cytochalasin D was added to the wells, at a concentration of 10 μg/ml. Approximately 104 live NTHI 2019 (white bars) or NTHI 2019 rfaD (gray bars) cells were added to wells (inocula, time zero). Sample wells were incubated for 24 h and then were assayed for bacterial viability (no PMN addition) or received 106 freshly isolated neutrophils that were left untreated (untreated PMN addition) or treated with 10 μg/ml cytochalasin D for 15 min prior to addition (cytochalasin D treated-PMN addition). These remaining sample wells were incubated for 30 min and then thoroughly scraped, and serial dilutions of well contents were plated to assess viable CFU per well. Statistical significance was determined by two-way ANOVA with a post hoc test of significance. *, P < 0.05; **, P < 0.005; ***, P < 0.0005.
DISCUSSION
The presence of biofilms during otitis media has been recognized for some time (19, 40-42). For NTHI, determinants of biofilm formation include lipooligosaccharides (18, 23, 24, 29, 36, 45), pili (27, 28), and extracellular DNA (27). In most cases, these factors have been shown to be required for persistent infections in animal models (2, 3, 23, 24, 29, 38, 39, 45).
Based on recent work from our group, it is now clear that biofilms formed during experimental otitis media include a significant host component (22, 43). Biofilms recovered from experimental otitis media infections contain a double-stranded DNA lattice (27) to which bactericidal factors such as histone and elastase are attached (22). Thus, these structures fit the definition of NET structures. For some microbes, these NET structures can mediate extracellular microbial killing and possibly facilitate phagocytic killing by additional infiltrating phagocytes (7, 8, 47). Bacterial pathogens causing persistent infections can be highly resistant to killing within NETs, often by means of LOS moieties or modifications to the bacterial surface (5, 6, 9, 22, 31, 49). For NTHI, the available evidence indicates that NETs do not mediate significant bacterial clearance; in fact, they are positively correlated with higher bacterial loads within middle-ear fluids and surface-attached communities in the chinchilla infection model (22). Notably, our recent work shows striking parallels between lipooligosaccharide modifications that promote NTHI survival within NETs (22) and those that promote NTHI biofilm formation (18, 23, 24, 29, 45).
The results of the present study clearly establish that NTHI LOS mediates NET formation by neutrophils following exposure to NTHI. While NET formation was induced by NTHI DNA and protein fractions, this activity was unaffected by DNase or proteinase K and thus could be conferred by trace cross-contamination with other bacterial components. Consistent with this interpretation, Limulus assays of these fractions did show trace levels of endotoxin (data not shown). Interestingly, a viability assay in which NTHI 2019 was exposed to NETs induced by either LOS, OMP, or DNA showed equivalent ineffectiveness in killing the bacteria, consistent with previous in vitro NET killing assays (22); however, the rfaD mutant, previously shown to be more susceptible than the parental strain to NET-mediated killing, exhibited a lower percentage of death for exposure to OMP- or DNA-induced NETs than for exposure to LOS-induced NETs (see Fig. S1 in the supplemental material). This is consistent with the observation that LOS optimally induces the formation of NETs. Also, compared to full-length LOS molecules from NTHI 2019, a detoxified form from the htrB mutant elicited significantly less induction of NETs. Additionally, binding of LOS through use of polymyxin B significantly reduced its ability to induce NET formation; in fact, no significant differences in levels of NET formation were seen between polymyxin B-treated LOS and the agent polymyxin B alone. It is likely that there are various programs of NET formation, as well as various triggers in their initiation. We have shown that for NTHI infections, potential signaling through the mediator MyD88 may be required for optimal NET formation, as murine PMNs deficient in this molecule have drastically reduced abilities to form NETs. Also, an NTHI LOS-host PMN TLR4 interaction appears to represent one mechanism of NET induction, since neutrophils from a TLR4-deficient line of mice showed decreased levels of NET formation.
Notably, we also observed NTHI survival within NETs for a prolonged period in vitro and no significant killing of NTHI within NETs by additional neutrophils. Both of these phenotypes were conferred at least in part by LOS moieties on the bacterial surface, as what appears to be a balance between survival, phagocytic killing, and NET-mediated killing was observed for a deep-rough rfaD mutant. It is also important that mast cells form mast cell extracellular traps (MCETs), which may have more potent extracellular killing activity than NETs (48). Since mast cells are found in a significant proportion of effusions from otitis media infections, this will be an important topic for future study.
Taken together, these results support the conclusion that the formation of neutrophil extracellular traps is not a significant determinant of NTHI clearance during chronic and recurrent otitis media. Rather, NTHI communities with biofilm phenotypes appear to survive within the NET structure, which appears to promote rather than inhibit bacterial persistence. The continual influx and death of neutrophils within the middle-ear chamber likely promote the periodically severe inflammation that is observed during the various clinical presentations of otitis media. It is also certainly plausible that the significant increase in biomass associated with the NET structure could present a passive diffusion barrier that may enhance resistance to some types of antibiotics (17, 20). On the other hand, it is also important that formation of a NET structure around a bacterial community could provide a means for the immune system to contain a persistent infection. Because otitis media is among the most common pediatric infections worldwide, a better in vivo definition of determinants of bacterial persistence within biofilm communities during these infections is of great importance.
Supplementary Material
Acknowledgments
We acknowledge outstanding technical assistance by Gayle Foster and members of the WFUHS microscopy core facility and helpful comments and critiques from colleagues in the WFUHS Department of Microbiology and Immunology.
This work was supported by grants from the U.S. Public Health Service (DC007444 and DC10051), awarded to W.E.S. Kristin E. D. Weimer and Chelsie E. Armbruster were supported by T32 training grant AI007401.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 18 October 2010.
Supplemental material for this article may be found at http://iai.asm.org/.
REFERENCES
- 1.Apicella, M. A., J. M. Griffiss, and H. Schneider. 1994. Isolation and characterization of lipopolysaccharides, lipooligosaccharides and lipid A. Methods Enzymol. 235:242-252. [DOI] [PubMed] [Google Scholar]
- 2.Armbruster, C., M. Byrd, C. Love, R. Juneau, N. D. Kock, and W. E. Swords. 2009. LuxS promotes biofilm maturation and persistence of nontypeable Haemophilus influenzae in experimental otitis media by modulation of lipooligosaccharide composition. Infect. Immun. 77:4081-4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Armbruster, C. E., W. Hong, B. Pang, K. E. Weimer, R. A. Juneau, J. Turner, and W. E. Swords. 2010. Indirect pathogenicity of Haemophilus influenzae and Moraxella catarrhalis in polymicrobial otitis media occurs via interspecies quorum signaling. MBio 1:e00102-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakaletz, L. O., B. M. Tallan, T. Hoepf, T. F. DeMaria, H. G. Birck, and D. J. Lim. 1988. Frequency of fimbriation of nontypable Haemophilus influenzae and its ability to adhere to chinchilla and human respiratory epithelium. Infect. Immun. 56:331-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beiter, K., F. Wartha, B. Albiger, S. Normark, A. Zychlinsky, and B. Henriques-Normark. 2006. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 16:401-407. [DOI] [PubMed] [Google Scholar]
- 6.Berends, E. T., A. R. Horswill, N. M. Haste, M. Monestier, V. Nizet, and M. von Kockritz-Blickwede. 2010. Nuclease expression by Staphylococcus aureus facilitates escape from neutrophil extracellular traps. J. Innate Immun. 2:576-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brinkmann, V., U. Reichard, C. Goosmann, B. Fauler, Y. Uhlemann, D. S. Weiss, Y. Weinrauch, and A. Zychlinsky. 2004. Neutrophil extracellular traps kill bacteria. Science 303:1532-1535. [DOI] [PubMed] [Google Scholar]
- 8.Brinkmann, V., and A. Zychlinsky. 2007. Beneficial suicide: why neutrophils die to make NETs. Nat. Rev. Microbiol. 5:577-582. [DOI] [PubMed] [Google Scholar]
- 9.Buchanan, J. T., A. J. Simpson, R. K. Aziz, G. Y. Liu, S. A. Kristian, M. Kotb, J. Feramisco, and V. Nizet. 2006. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 16:396-400. [DOI] [PubMed] [Google Scholar]
- 10.Campagnari, A. A., M. R. Gupta, K. C. Dudas, T. F. Murphy, and M. A. Apicella. 1987. Antigenic diversity of lipooligosaccharides of nontypable Haemophilus influenzae. Infect. Immun. 55:882-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chonmaitree, T., K. Revai, J. J. Grady, A. Clos, J. A. Patel, S. Nair, J. Fan, and K. J. Henrickson. 2008. Viral upper respiratory tract infection and otitis media complication in young children. Clin. Infect. Dis. 46:815-823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Daly, K. A., L. L. Hunter, and G. S. Giebink. 1999. Chronic otitis media with effusion. Pediatr. Rev. 20:85-93. [DOI] [PubMed] [Google Scholar]
- 13.DeMaria, T. F., M. A. Apicella, W. A. Nichols, and E. R. Leake. 1997. Evaluation of the virulence of nontypeable Haemophilus influenzae lipopolysaccharide htrB and rfaD mutants in the chinchilla model of otitis media. Infect. Immun. 65:4431-4435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Erwin, A. L., and A. L. Smith. 2007. Nontypeable Haemophilus influenzae: understanding virulence and commensal behavior. Trends Microbiol. 15:355-362. [DOI] [PubMed] [Google Scholar]
- 15.Flesher, A. R., and R. A. Insel. 1978. Characterization of lipopolysaccharide of Haemophilus influenzae. J. Infect. Dis. 138:719-730. [DOI] [PubMed] [Google Scholar]
- 16.Fuchs, T. A., U. Abed, C. Goosmann, R. Hurwitz, I. Schulze, V. Wahn, Y. Weinrauch, V. Brinkmann, and A. Zychlinsky. 2007. Novel cell death program leads to neutrophil extracellular traps. J. Cell Biol. 176:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fux, C. A., J. W. Costerton, P. S. Stewart, and P. Stoodley. 2005. Survival strategies of infectious biofilms. Trends Microbiol. 13:34-40. [DOI] [PubMed] [Google Scholar]
- 18.Greiner, L., H. Watanabe, N. J. Phillips, J. Shao, A. Morgan, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2004. Nontypeable Haemophilus influenzae strain 2019 produces a biofilm containing N-acetylneuraminic acid that may mimic sialylated O-linked glycans. Infect. Immun. 72:4249-4260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall-Stoodley, L., F. Z. Hu, A. Gieseke, L. Nistico, D. Nguyen, J. D. Hayes, M. Forbes, D. P. Greenberg, B. Dice, A. Burrows, P. Wackym, P. Stoodley, J. C. Post, G. D. Ehrlich, and J. E. Kerschner. 2006. Direct detection of bacterial biofilms on the middle ear mucosa of children with chronic otitis media. JAMA 296:202-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hall-Stoodley, L., and P. Stoodley. 2009. Evolving concepts in biofilm infections. Cell. Microbiol. 11:1034-1043. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi, F., T. K. Means, and A. D. Luster. 2003. Toll-like receptors stimulate human neutrophil function. Blood 102:2660-2669. [DOI] [PubMed] [Google Scholar]
- 22.Hong, W., R. Juneau, B. Pang, and W. E. Swords. 2009. Survival of bacterial biofilms within neutrophil extracellular traps promotes nontypeable Haemophilus influenzae persistence in the chinchilla model for otitis media. J. Innate Immun. 1:215-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong, W., K. Mason, J. A. Jurcisek, L. A. Novotny, L. O. Bakaletz, and W. E. Swords. 2007. Phosphorylcholine decreases early inflammation and promotes the establishment of stable biofilm communities of nontypeable Haemophilus influenzae strain 86-028NP in a chinchilla model of otitis media. Infect. Immun. 75:958-965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hong, W., B. Pang, S. West-Barnette, and W. E. Swords. 2007. Phosphorylcholine expression by nontypeable Haemophilus influenzae correlates with maturation of biofilm communities in vitro and in vivo. J. Bacteriol. 189:8300-8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Izano, E. A., S. M. Shah, and J. B. Kaplan. 2009. Intercellular adhesion and biocide resistance in nontypeable Haemophilus influenzae biofilms. Microb. Pathog. 46:207-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones, P. A., N. A. Samuels, N. J. Phillips, R. S. Munson, J. A. Bozue, J. A. Arseneau, W. A. Nichols, A. Zaleski, B. W. Gibson, and M. A. Apicella. 2002. Haemophilus influenzae type B strain A2 has multiple sialyltransferases involved in lipooligosaccharide sialylation. J. Biol. Chem. 277:14598-14611. [DOI] [PubMed] [Google Scholar]
- 27.Jurcisek, J. A., and L. O. Bakaletz. 2007. Biofilms formed by nontypeable Haemophilus influenzae in vivo contain both double-stranded DNA and type IV pilin protein. J. Bacteriol. 189:3868-3875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jurcisek, J. A., J. Bookwalter, B. Baker, S. Fernandez, L. A. Novotny, R. S. Munson, Jr., and L. O. Bakaletz. 2007. The PilA protein of nontypeable Haemophilus influenzae plays a role in biofilm formation, adherence to epithelial cells and colonization of the mammalian upper respiratory tract. Mol. Microbiol. 65:1288-1299. [DOI] [PubMed] [Google Scholar]
- 29.Jurcisek, J. A., L. Greiner, H. Watanabe, A. Zaleski, M. A. Apicella, and L. O. Bakaletz. 2005. Role of sialic acid and complex carbohydrate biosynthesis in biofilm formation by nontypeable Haemophilus influenzae in the chinchilla middle ear. Infect. Immun. 73:3210-3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein, J. O. 2000. The burden of otitis media. Vaccine 19:S2-S8. [DOI] [PubMed] [Google Scholar]
- 31.Lauth, X., M. von Kockritz-Blickwede, C. W. McNamara, S. Myskowski, A. S. Zinkernagel, B. Beall, P. Ghosh, R. L. Gallo, and V. Nizet. 2009. M1 protein allows group A streptococcal survival in phagocyte extracellular traps through cathelicidin inhibition. J. Innate Immun. 1:202-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee, N.-G., M. G. Sunshine, J. J. Engstrom, B. W. Gibson, and M. A. Apicella. 1995. Mutation of the htrB locus of Haemophilus influenzae nontypable strain 2019 is associated with modifications of lipid A and phosphorylation of the lipo-oligosaccharide. J. Biol. Chem. 270:27151-27159. [PubMed] [Google Scholar]
- 33.Mandel, E. M., W. J. Doyle, B. Winther, and C. M. Alper. 2008. The incidence, prevalence and burden of OM in unselected children aged 1-8 years followed by weekly otoscopy through the “common cold” season. Int. J. Pediatr. Otorhinolaryngol. 72:491-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy, T. F., L. C. Bartos, A. A. Campagnari, M. B. Nelson, and M. A. Apicella. 1986. Antigenic characterization of the P6 protein of nontypable Haemophilus influenzae. Infect. Immun. 54:774-779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Murphy, T. F., K. C. Didas, J. M. Mylotte, and M. A. Apicella. 1983. A subtyping system for nontypable Haemophilus influenzae based on outer-membrane proteins. J. Infect. Dis. 147:838-846. [DOI] [PubMed] [Google Scholar]
- 36.Murphy, T. F., and C. Kirkham. 2002. Biofilm formation by nontypeable Haemophilus influenzae: strain variability, outer membrane antigen expression and role of pili. BMC Microbiol. 2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nichols, W. A., B. W. Gibson, W. Melaugh, N. G. Lee, M. Sunshine, and M. A. Apicella. 1997. Identification of the ADP-l-glycero-d-manno-heptose-6-epimerase (rfaD) and heptosyltransferase II (rfaF) biosynthesis genes from nontypeable Haemophilus influenzae 2019. Infect. Immun. 65:1377-1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang, B., W. Hong, S. L. West-Barnette, N. D. Kock, and W. E. Swords. 2008. Diminished ICAM-1 expression and impaired pulmonary clearance of nontypeable Haemophilus influenzae in a mouse model for COPD/emphysema. Infect. Immun. 76:4959-4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pang, B., D. Winn, R. Johnson, W. Hong, S. West-Barnette, N. Kock, and W. E. Swords. 2008. Lipooligosaccharides containing phosphorylcholine delay pulmonary clearance of nontypeable Haemophilus influenzae. Infect. Immun. 76:2037-2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Post, J. C. 2001. Direct evidence of bacterial biofilms in otitis media. Laryngoscope 111:2083-2094. [DOI] [PubMed] [Google Scholar]
- 41.Post, J. C., R. A. Preston, J. J. Aul, M. Larkins-Pettigrew, J. Rydquist-White, K. W. Anderson, R. M. Wadowsky, D. R. Reagan, E. S. Walker, L. A. Kingsley, A. E. Magit, and G. D. Ehrlich. 1995. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA 273:1598-1604. [PubMed] [Google Scholar]
- 42.Rayner, M. G., Y. Zhang, M. C. Gorry, Y. Chen, J. C. Post, and G. D. Ehrlich. 1998. Evidence of bacterial metabolic activity in culture-negative otitis media with effusion. JAMA 279:296-299. [DOI] [PubMed] [Google Scholar]
- 43.Reid, S. D., W. Hong, K. E. Dew, D. R. Winn, B. Pang, J. Watt, D. T. Glover, S. K. Hollingshead, and W. E. Swords. 2009. Streptococcus pneumoniae forms surface-attached communities in the middle ear of experimentally infected chinchillas. J. Infect. Dis. 199:786-794. [DOI] [PubMed] [Google Scholar]
- 44.Rovers, M. M., A. G. Schilder, G. A. Zielhuis, and R. M. Rosenfeld. 2004. Otitis media. Lancet 363:465-473. [DOI] [PubMed] [Google Scholar]
- 45.Swords, W. E., M. L. Moore, L. Godzicki, G. Bukofzer, M. J. Mitten, and J. VonCannon. 2004. Sialylation of lipooligosaccharides promotes biofilm formation by nontypeable Haemophilus influenzae. Infect. Immun. 72:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Takeda, K., and S. Akira. 2004. Microbial recognition by Toll-like receptors. J. Dermatol. Sci. 34:73-82. [DOI] [PubMed] [Google Scholar]
- 47.Urban, C. F., S. Lourido, and A. Zychlinsky. 2006. How do microbes evade neutrophil killing? Cell. Microbiol. 8:1687-1696. [DOI] [PubMed] [Google Scholar]
- 48.von Kockritz-Blickwede, M., O. Goldmann, P. Thulin, K. Heinemann, A. Norrby-Teglund, M. Rohde, and E. Medina. 2008. Phagocytosis-independent antimicrobial activity of mast cells by means of extracellular trap formation. Blood 111:3070-3080. [DOI] [PubMed] [Google Scholar]
- 49.Wartha, F., K. Beiter, B. Albiger, J. Fernebro, A. Zychlinsky, S. Normark, and B. Henriques-Normark. 2007. Capsule and d-alanylated lipoteichoic acids protect Streptococcus pneumoniae against neutrophil extracellular traps. Cell. Microbiol. 9:1162-1171. [DOI] [PubMed] [Google Scholar]
- 50.Wieland, C. W., S. Florquin, N. A. Maris, K. Hoebe, B. Beutler, K. Takeda, S. Akira, and T. van der Poll. 2005. The MyD88-dependent, but not the MyD88-independent, pathway of TLR4 signaling is important in clearing nontypeable Haemophilus influenzae from the mouse lung. J. Immunol. 175:6042-6049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.