Abstract
The human symbiont Bacteroides thetaiotaomicron promotes intestinal function and health, whereas the phylogenetically related pathogen Porphyromonas gingivalis is associated with the chronic oral inflammatory disease periodontitis. Although both B. thetaiotaomicron and P. gingivalis synthesize lipopolysaccharides (LPS) consisting of penta-acylated, monophosphorylated lipid A in addition to immunologically silent, nonphosphorylated lipid A, they elicit strikingly distinct Toll-like receptor 4 (TLR4) responses. We show that the phosphate position of penta-acylated, monophosphorylated lipid A is a key feature for determining the differential TLR4 responses elicited by these evolutionarily related bacteria. B. thetaiotaomicron produces TLR4-stimulatory lipid A bearing a 1-phosphate, in contrast to P. gingivalis, which produces TLR4-evasive lipid A bearing a 4′-phosphate. Confirming these observations, recombinant Escherichia coli LPS containing penta-acylated, 1-phosphorylated lipid A is more TLR4 stimulatory than LPS containing 4′-phosphorylated lipid A. The specific capacity of a Gram-negative bacterium to alert or evade the host innate immune defense system through TLR4-dependent signaling is currently recognized as a critical aspect defining the relationship between the host and the bacterium. We propose that the distinct lipid A phosphate positions observed for the B. thetaiotaomicron and P. gingivalis LPS contributes to the manifestation of these bacteria as commensal or pathogen within the human host.
Bacteroides is the most numerous microbial genus found in the human ileum and large intestine, outnumbering Escherichia coli 100 to 1 (10, 24). The genus consists of anaerobic Gram-negative rods, including the important human symbiont Bacteroides thetaiotaomicron. B. thetaiotaomicron benefits its human host by metabolizing indigestible plant polysaccharides, promoting proper enterocyte fucosylation patterns (which assists colonization by other symbionts), and inducing intestinal angiogenesis (7, 11, 27). The phylogenetically related bacterium Porphyromonas gingivalis was formerly assigned to the genus Bacteroides (23). In contrast to B. thetaiotaomicron, P. gingivalis is considered a human pathogen capable of expressing a number of distinct virulence factors thought to play a major role in the development and progression of the chronic inflammatory disease periodontitis (8, 12, 13, 25).
Lipopolysaccharide (LPS) is responsible for septic shock resulting from Gram-negative bacterial infection, and it is also a major structural component in the bacterial outer membrane. LPS, specifically the lipid A portion, is known to play a role in the maintenance of homeostasis between the host and beneficial bacteria, as well as stimulation of innate immune defenses that are necessary for the effective clearance of Gram-negative bacterial pathogens (20, 21). The lipid A produced by E. coli is a prototypical ligand for stimulating Toll-like receptor 4 (TLR4)-dependent innate immune responses. It is among the most immunologically potent lipid A species known, consisting of a hexa-acylated, 1,4′-bis-phosphorylated glucosamine disaccharide (9). Modifications of this evolutionarily conserved structure typically result in dampened innate immune responses. For example, replacing the C12 fatty acid (laurate) normally found in E. coli lipid A with a C16 fatty acid (palmitate) yields a less potent LPS (2). Notably, it has been shown that synthetic bis-phosphorylated lipid A is significantly more potent than monophosphorylated lipid A bearing a 1-phosphate, which is in turn more potent than monophosphorylated lipid A bearing a 4′-phosphate in stimulating the host innate immune system (16).
Natural variations observed in lipid A structures often include differences in fatty acid or phosphate content. Both B. thetaiotaomicron and P. gingivalis produce LPS that contains penta-acylated, monophosphorylated lipid A (3, 14, 30). Paradoxically, we observed that despite the high degree of lipid A structural similarity, the LPS of B. thetaiotaomicron is significantly more potent than that of P. gingivalis in stimulating TLR4-dependent innate immune responses (3). We also recently reported that P. gingivalis can modulate its lipid A structure in response to the hemin concentration in the growth medium and that this is likely due to differential activities of lipid A 1- and 4′-phosphatases (1, 5). These and other data indicate that P. gingivalis grown under low-hemin conditions expresses both penta-acylated, monophosphorylated lipid A bearing a 4′-phosphate and tetra-acylated, nonphosphorylated lipid A (5, 22). This contrasts with structural studies of Bacteroides fragilis lipid A, which demonstrated the presence of penta-acylated, monophosphosphorylated lipid A bearing a 1-phosphate (30). These observations suggest that the phosphate position of P. gingivalis penta-acylated lipid A, possibly due to the presence of tetra-acylated, nonphosphorylated lipid A, determines the relatively weak immunostimulatory potency of its LPS compared to B. thetaiotaomicron LPS.
In the present study, we examined the molecular basis for the reduced immunostimulatory potency of P. gingivalis LPS compared to that of B. thetaiotaomicron LPS. We show that the magnitude of the host TLR4-dependent response is significantly influenced by the position of the phosphate occurring on monophosphorylated lipid A. The observed differences in TLR4 recognition between the two bacterial species involve the production of the more immunostimulatory 1-phosphorylated lipid A by B. thetaiotaomicron, as opposed to the production of the relatively nonimmunostimulatory 4′-phosphorylated lipid A by P. gingivalis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
P. gingivalis ATCC 33277 was obtained from our stock collection. The isogenic mutant strain P. gingivalis 1626KO exhibits a lipid A phenotype that is characteristic of P. gingivalis grown in hemin (1 μg/ml), which represents low-hemin-concentration growth conditions as described previously (1, 3, 5). The isogenic mutant strain of P. gingivalis designated P. gingivalis 1587KO displays penta-acylated, 4′-phosphorylated lipid A and lacks nonphosphorylated lipid A, as described previously (5). B. thetaiotaomicron VPI-5482 was also obtained from our stock collection. All B. thetaiotaomicron and P. gingivalis strains of bacteria were grown in TYHK medium consisting of Trypticase soy broth (30 g/liter) (Becton Dickinson, Sparks, MD), yeast extract (5 g/liter) (Becton Dickinson, Sparks, MD), and vitamin K3 (menadione) (Sigma-Aldrich, St. Louis, MO). The basal TYHK medium was sterilized by autoclaving, followed by the addition of filter-sterilized hemin (Sigma-Aldrich, St. Louis, MO) to a final concentration of 1 μg/ml. Cultures were grown in an anaerobic growth chamber (5% H2, 5% CO2, 90% N2) and maintained at 37°C on TYHK-agar plates (3, 5).
Construction of E. coli strains expressing recombinant LpxE and LpxF.
The E. coli msbB strain contains an inactivating mutation in the genetic locus that encodes the lipid A secondary myristoyl transferase, msbB. As a consequence, these bacteria produce penta-acylated, bis-phosphorylated lipid A lacking the secondary myristate at the 3′ position of the glucosamine disaccharide backbone (26). Therefore, this lipid A resembles the structure of the penta-acylated lipid A observed in P. gingivalis and B. thetaiotaomicron, with the exception that it contains C12 and C14 fatty acyl groups rather than C15, C16, and C17 fatty acyl groups. The coding sequences for Francisella novicida LpxE and LpxF lipid A phosphatases were previously cloned into the expression vector pWSK29, yielding the plpxE and plpxF constructs, respectively (28, 29). (The constructs were kindly provided by Christian Raetz.) To create the recombinant E. coli msbB strains used in this study, the vectors pWSK29, plpxE, and plpxF were transformed into the E. coli msbB strain by electroporation. The resulting recombinant strains, designated E. coli msbB WSK, E. coli msbB LpxE, and E. coli msbB LpxF, respectively, were grown in Luria broth (LB) supplemented with 100 μg/ml of ampicillin.
Isolation of LPS and lipid A.
All bacterial strains used in this study were derived from B. thetaiotaomicron, P. gingivalis, and E. coli msbB and were cultured as described above. LPS was isolated using a modified version of the Tri-reagent protocol for LPS isolation, as previously described (1, 5). To generate lipid A, dried LPS samples were resuspended in 10 mM sodium acetate (pH 4.5) containing 1% (wt/vol) sodium dodecyl sulfate. The solution was heated at 100°C for 1 h, followed by lyophilization overnight. The resulting lipid A pellets were washed once in ice-cold 95% ethanol containing 0.02 N HCl and three times in 95% ethanol. The lipid A samples (except for those derived from E. coli) were then subjected to a final Bligh-Dyer extraction, which consisted of 1,160 μl of a chloroform-methanol-water mixture (1:1:0.9 [vol/vol/vol]) to remove residual carbohydrate contaminants.
MALDI-TOF MS analyses.
For matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS) analyses, lipid A samples were dissolved in 10 μl of a mixture of 5-chloro-2-mercaptobenzothiazole (20 mg/ml) in 1:1 (vol/vol) chloroform-methanol, and 0.5 μl of each sample was analyzed in both positive- and negative-ion modes on an AutoFlex Analyzer (Bruker Daltonics). Data were acquired with a 50-Hz repletion rate, and up to 3,000 shots were accumulated for each spectrum. Instrument calibration and all other tuning parameters were optimized using HP Calmix (Sigma-Aldrich, St. Louis, MO). Data were acquired and processed using flexAnalysis software (Bruker Daltonics).
MALDI-TOF/TOF tandem MS analyses.
For MALDI-TOF/TOF tandem MS analyses, lipid A was analyzed by a MALDI-TOF mass spectrometer in the positive and negative-ion modes on a 4700 Proteomics Analyzer (Applied Biosystems, Framingham, MA). Samples were dissolved in 10 μl of a mixture of 5-chloro-2-mercaptobenzothiazole (20 mg/ml) in chloroform-methanol-water (4:4:1 [vol/vol/vol]), and 0.5 μl of sample was analyzed by MALDI-TOF MS and MALDI-TOF/TOF tandem MS. Both MALDI-TOF MS and MALDI-TOF/TOF tandem MS data were acquired in the reflectron mode with a neodymium-doped yttrium aluminum garnet (Nd:YAG) laser with a 200-Hz repetition rate, and up to 3,750 shots were accumulated for each spectrum. The precursor isolation window was set to ±5 Da. MALDI-TOF/TOF tandem mass spectra were acquired with collision energies of 1 keV, and air was used as the collision gas. Instrument calibration and all other tuning parameters were optimized using HP Calmix (Sigma-Aldrich, St. Louis, MO). Data were acquired and processed using Data Explorer (Applied Biosystems, Framingham, MA).
HEK293 cell TLR4 activation assays.
HEK293 cell TLR4 activation assays were performed essentially as described previously (5). Briefly, HEK293 cells were plated in 96-well plates at a density of 4 × 104 cells per well and transfected the following day with plasmids bearing firefly luciferase, Renilla luciferase, and human TLR4 and MD-2 by standard calcium phosphate precipitation. The test wells were stimulated in triplicate for 4 h at 37°C with the indicated doses of LPS isolates or intact bacteria in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum. For stimulations involving intact B. thetaiotaomicron and P. gingivalis bacteria, the bacteria were cultured in the appropriate growth medium as described above. The cultures were then centrifuged, the supernatant was removed, and the pellets were resuspended in stimulation medium. Following stimulation, the transfected HEK293 cells were rinsed with phosphate-buffered saline and lysed with 50 μl of passive lysis buffer (Promega, Madison, WI). Luciferase activity was measured using the Dual Luciferase Assay Reporter System (Promega, Madison, WI). Data are expressed as fold increase of NF-κB activity, which represents the ratio of NF-κB-dependent firefly luciferase activity to β-actin promoter-dependent Renilla luciferase activity.
Statistical analyses.
Data were analyzed by two-tailed unpaired Student's t tests (GraphPad Prism). A P value of <0.05 was considered indicative of statistical significance.
RESULTS
B. thetaiotaomicron LPS and P. gingivalis 1626KO LPS contain penta-acylated lipid A structures bearing a phosphate at different positions.
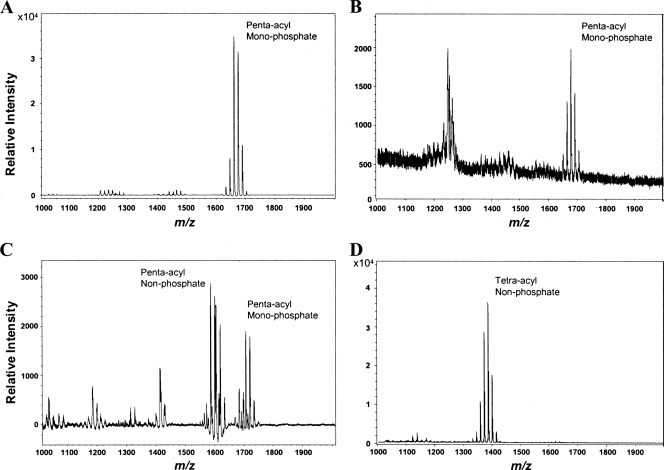
Negative-ion mode MALDI-TOF MS analyses were performed in order to compare the structural profiles of lipid A isolates derived from either B. thetaiotaomicron or an isogenic mutant strain of wild-type (WT) P. gingivalis ATCC 33277 that has been designated P. gingivalis 1626KO. P. gingivalis 1626KO displays a lipid A profile that is identical to the profile of P. gingivalis that has been grown in relatively low concentrations of the micronutrient hemin (1, 3). This mutant strain was chosen for this study because it maintains a consistent lipid A structure independent of the effects of the hemin concentrations in the growth media. As shown previously (3), both B. thetaiotaomicron-derived and P. gingivalis 1626KO-derived lipid A moieties display similar penta-acylated, monophosphorylated structures that resolve into a cluster of mass ion peaks centered around m/z 1,688 (Fig. 1A and B). However, B. thetaiotaomicron LPS and lipid A are significantly more potent than P. gingivalis 1626KO LPS and its derivative lipid A with regard to the ability to elicit TLR4-dependent NF-κB activation (3). This suggests that a functionally relevant chemical distinction exists between the penta-acylated B. thetaiotaomicron and P. gingivalis 1626KO lipid A molecules. A positive-ion mode MALDI-TOF MS analysis of the lipid A isolates derived from B. thetaiotaomicron LPS and P. gingivalis 1626KO LPS was employed to identify lipid A structures that might escape negative-ion mode detection, since the method has recently been validated by examinations of P. gingivalis lipid A (5). Penta-acylated, nonphosphorylated lipid A, as well as penta-acylated, monophosphorylated lipid A, was detected in the B. thetaiotaomicron LPS isolate (Fig. 1C). Also, P. gingivalis 1626KO LPS displays a major tetra-acylated, nonphosporylated lipid A structure (Fig. 1D).
FIG. 1.
B. thetaiotaomicron LPS and P. gingivalis LPS contain penta-acylated, monophosphorylated lipid A structures, as well as nonphosphorylated lipid A structures. (A) Negative-ion mode MALDI-TOF MS analysis of lipid A derived from B. thetaiotaomicron LPS. (B) Negative-ion mode MALDI-TOF MS analysis of lipid A derived from P. gingivalis 1626KO LPS. (C) Positive-ion mode MALDI-TOF MS analysis of lipid A derived from B. thetaiotaomicron LPS. (D) Positive-ion mode MALDI-TOF MS analysis of lipid A derived from P. gingivalis 1626KO LPS.
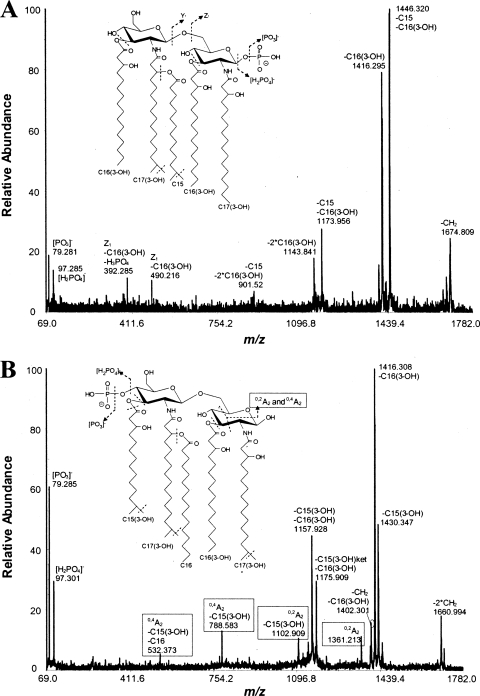
Further resolution of the penta-acylated, monophosphorylated lipid A structures (centered around m/z 1,688) that are present in B. thetaiotaomicron LPS and P. gingivalis 1626KO LPS was obtained by MALDI-TOF/TOF tandem MS analyses (Fig. 2A and B). These analyses indicate that the penta-acylated lipid A derived from B. thetaiotaomicron LPS contains a 1-phosphate rather than a 4′-phosphate, as indicated by the absence of A2-type ion fragments (Fig. 2A). Thus, the structure for B. thetaiotaomicron lipid A as determined by MALDI-TOF/TOF tandem MS analyses agrees with the previously reported structure for B. fragilis lipid A (30). However, the penta-acylated, monophosphorylated lipid A contained in P. gingivalis 1626KO LPS bears a 4′-phosphate, as indicated by the presence of diagnostic A2-type ion fragments (Fig. 2B). This interpretation contradicts the originally proposed penta-acylated, 1-phosphorylated lipid A, which was inferred from a tetra-acylated 1-phosphate lipid A structure (14). However, the lipid A structure reported here is consistent with the recently determined structure of the penta-acylated, 4′-phosphorylated P. gingivalis lipid A grown in relatively low concentrations of hemin (5). The combined MALDI-TOF MS and MALDI-TOF/TOF tandem MS data indicated that B. thetaiotaomicron LPS contains penta-acylated, 1-phosporylated lipid A and penta-acylated, nonphosporylated lipid A structures, whereas P. gingivalis 1626KO LPS (similar to P. gingivalis LPS) consists of penta-acylated, 4′-phosphorylated and tetra-acylated, nonphosphorylated lipid A structures. These data strongly suggest that the lipid A phosphate position, in addition to the presence of nonphosphorylated lipid A, determines the different abilities of B. thetaiotaomicron LPS and P. gingivalis 1626KO LPS to stimulate host TLR4 activation.
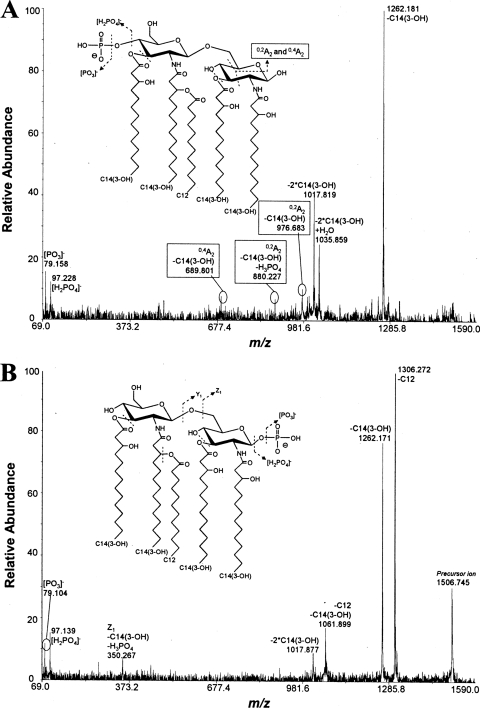
FIG. 2.
B. thetaiotaomicron and P. gingivalis produce differentially phosphorylated penta-acylated lipid A, as determined by negative-ion mode MALDI-TOF/TOF tandem MS spectra. (A and B) Analyses of a penta-acylated, 1-phosphorylated B. thetaiotaomicron lipid A (m/z 1,688) (A) and a penta-acylated, 4′-phosphorylated P. gingivalis 1626KO lipid A (m/z 1,688) (B). The inset cartoon structures illustrate the differentially located phosphate positions on the respective lipid A backbones, as determined by the presence or absence of A2-type ion fragments.
The lipid A phosphate position determines the relative abilities of B. thetaiotaomicron and P. gingivalis to stimulate host TLR4 responses.
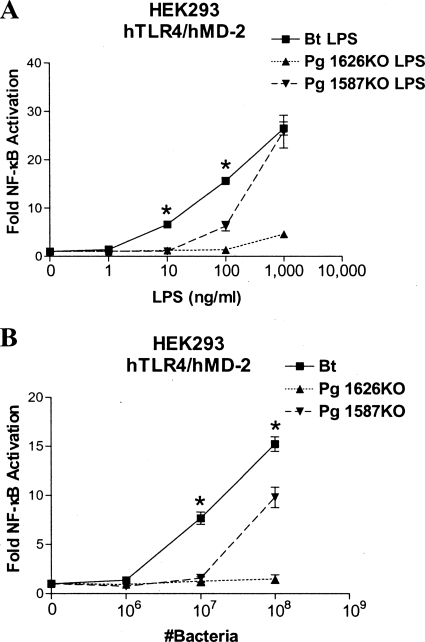
As indicated above, both B. thetaiotaomicron LPS and P. gingivalis 1626KO LPS contain novel nonphosphorylated lipid A structures in addition to the previously described penta-acylated, monophosphorylated lipid A structures (Fig. 1) (3). Previous observations by our laboratory and the Curtis laboratory have indicated that the unique, nonphosphorylated lipid A species are “silent” (i.e., neither agonistic nor antagonistic to TLR4) and that they do not significantly contribute to the observed immunostimulatory activity associated with LPS, presumably due to their inability to productively engage the TLR4 coreceptor, MD-2 (5, 22). To assist in further elucidating the relative contributions of lipid A nonphosphorylation or 4′-phosphorylation to TLR4-dependent stimulation, we employed a mutant strain of WT P. gingivalis, designated P. gingivalis 1587KO, that fails to produce nonphosphorylated lipid A but continues to produce the penta-acylated, 4′-phosphorylated lipid A structure (5). We then compared the relative immunostimulatory potencies of LPSs isolated from B. thetaiotaomicron, P. gingivalis 1626KO, and P. gingivalis 1587KO by a TLR4 activation assay in HEK293 cells (Fig. 3A). B. thetaiotaomicron LPS is significantly more potent than either P. gingivalis 1587KO LPS or P. gingivalis 1626KO LPS in stimulating TLR4-dependent NF-κB activation in HEK293 cells. These data indicate that LPS consisting of penta-acylated, 1-phosphorylated lipid A is inherently more immunostimulatory than LPS containing penta-acylated, 4′-phosphorylated lipid A, regardless of the nonphosphorylated lipid A content. These data also suggest that the appreciable accumulation of nonphosphorylated lipid A in P. gingivalis 1626KO LPS contributes to an overall reduced TLR4 response, since it is significantly less potent than P. gingivalis 1587KO LPS.
FIG. 3.
The different abilities of B. thetaiotaomicron (Bt) and P. gingivalis (Pg) to elicit robust innate immune recognition are determined by the positions of the phosphate present on penta-acylated, monophosphorylated lipid A. (A) B. thetaiotaomicron LPS is more potent than either P. gingivalis 1626KO LPS or P. gingivalis 1587KO LPS in stimulating TLR4-dependent NF-κB activation in HEK cells. (B) Intact B. thetaiotaomicron bacteria are more potent than either intact P. gingivalis 1626KO bacteria or P. gingivalis 1587KO bacteria in stimulating TLR4-dependent NF-κB activation in HEK cells. NF-κB activation was determined by measurement of firefly luciferase activity, and the results were plotted as the mean fold induction (±standard deviation [SD]) of triplicate determinations relative to the unstimulated control. The asterisks represent significant differences between either B. thetaiotaomicron LPS and P. gingivalis 1587KO LPS or B. thetaiotaomicron and P. gingivalis 1587KO bacteria (P < 0.05; unpaired Student t tests). hTLR4, human TLR4; hMD-2, human MD-2.
Importantly, the use of intact B. thetaiotaomicron, P. gingivalis 1587KO, and P. gingivalis 1626KO bacteria in the TLR4 activation assay (Fig. 3B) produced results that were similar to those obtained using LPS isolates. Altogether, the data in Fig. 3 provide compelling evidence that the penta-acylated, 1-phosphorylated lipid A produced by B. thetaiotaomicron is significantly more TLR4 immunostimulatory than the penta-acylated, 4′-phosphorylated lipid A produced by either P. gingivalis 1587KO or P. gingivalis 1626KO.
Penta-acylated E. coli lipid A bearing a 1-phosphate is more immunostimulatory than penta-acylated E. coli lipid A bearing a 4′-phosphate.
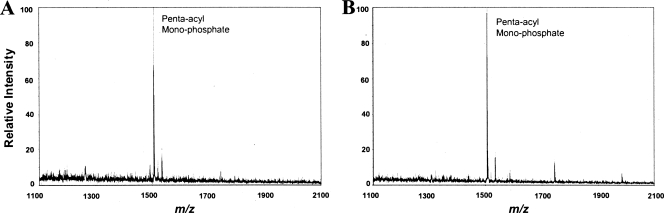
In order to obtain independent confirmation of the importance of the phosphate position in determining bacterially induced TLR4 activation, we examined the immunostimulatory ability of LPS isolated from a mutant strain of E. coli msbB, which expresses a penta-acylated, bis-phosphorylated version of the canonical hexa-acylated E. coli lipid A structure due to mutation of the msbB gene (26). The penta-acylated structure of E. coli msbB lipid A resembles those of B. thetaiotaomicron and P. gingivalis 1626KO lipid A with the important exception that it contains shorter fatty acids (C12 and C14) instead of longer fatty acids (C15 to C17) and is bis-phosphorylated. We were able to create E. coli msbB derivative strains expressing recombinant penta-acylated, monophosphorylated lipid A that bears either a 1- or 4′-phosphate by transforming plasmids encoding either F. novicida LpxE (lipid A 1-phosphatase) or LpxF (lipid A 4′-phosphatase) into E. coli msbB to yield strains E. coli msbB LpxE and E. coli msbB LpxF, respectively (28, 29). The empty expression vector pWSK29 was utilized to create the control strain, E. coli msbB WSK. Consequently, E. coli msbB WSK bacteria produce the expected penta-acylated, bis-phosphorylated lipid A (m/z 1,586) observed in the E. coli msbB parent strain (data not shown), whereas E. coli msbB LpxE bacteria and E. coli msbB LpxF bacteria both produce a major penta-acylated, monophosphorylated lipid A (m/z 1,506) (Fig. 4A and B) indicative of exogenous lipid A phosphatase activity. Further examination of the monophosphorylated lipid A structures by MALDI-TOF/TOF tandem MS analyses confirmed that the E. coli msbB LpxE strain produces lipid A bearing a 4′-phosphate (Fig. 5A) and that the E. coli msbB LpxF strain produces lipid A bearing a 1-phosphate (Fig. 5B). As shown in Fig. 6, E. coli msbB LpxF LPS is significantly more potent than either E. coli msbB LpxE LPS or E. coli msbB WSK LPS with respect to the ability to stimulate TLR4-dependent NF-κB activation in HEK293 cells. These data support the hypothesis that the lipid A phosphate position determines the TLR4-dependent immunostimulatory potency of bacteria producing penta-acylated lipid A structures bearing either short-chain fatty acids (Fig. 6) or long-chain fatty acids (Fig. 3A).
FIG. 4.
E. coli msbB bacteria transformed with plasmids expressing F. novicida lipid A phosphatase (LpxE or LpxF) produce penta-acylated, monophosphorylated lipid A structures. (A) Negative-ion mode MALDI-TOF MS analysis of lipid A derived from E. coli msbB bacteria expressing LpxE. (B) Negative-ion mode MALDI-TOF MS analysis of lipid A derived from E. coli msbB bacteria expressing LpxF.
FIG. 5.
E. coli msbB bacteria expressing either LpxE or LpxF produce differentially phosphorylated penta-acylated lipid A, as determined by negative-ion mode MALDI-TOF/TOF tandem MS spectra. (A and B) Analyses of a penta-acylated, 4′-phosphorylated E. coli msbB LpxE lipid A (m/z 1,506) (A) and a penta-acylated, 1-phosphorylated E. coli msbB LpxF lipid A (m/z 1,506) (B). The inset cartoon structures illustrate the differentially located phosphate positions on the respective lipid A backbones, as determined by the presence or absence of A2-type ion fragments.
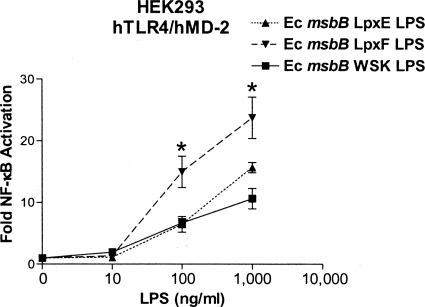
FIG. 6.
E. coli (Ec) msbB LPS consisting of either penta-acylated, 1-phosphorylated or 4′-phosphorylated lipid A structures differentially activate the innate immune system. E. coli msbB LpxF LPS containing 1-phosphorylated lipid A is more potent than E. coli msbB LpxE LPS bearing 4′-phosphorylated lipid A in stimulating TLR4-dependent NF-κB activation in HEK cells. NF-κB activation was determined by measurement of firefly luciferase activity, and the results were plotted as the mean fold induction (±SD) of triplicate determinations relative to the unstimulated control. The asterisks represent significant differences between E. coli msbB LpxE LPS and E. coli msbB LpxF LPS (P < 0.05; unpaired Student t tests).
DISCUSSION
We previously observed that despite bearing penta-acylated, monophosphorylated lipid A moieties that exhibit a high degree of structural similarity, LPS isolated from B. thetaiotaomicron was significantly more immunostimulatory than LPS isolated from P. gingivalis (3). We have recently shown that the LPS of P. gingivalis grown in relatively low levels of hemin in the culture medium produces penta-acylated lipid A that contains a 4′-phosphate rather than a 1-phosphate. In addition, the bacterium produces a tetra-acylated, nonphosphorylated lipid A which has been postulated to influence the low endotoxic potential that is typically associated with P. gingivalis LPS (5). In the present study, we discovered that the lipid A 1-phosphate of B. thetaiotaomicron LPS, compared to the lipid A 4′-phosphate of P. gingivalis LPS, plays a critical role in determining the different potencies of B. thetaiotaomicron and P. gingivalis LPS to activate TLR4-dependent innate immune responses. The relative abilities of E. coli msbB LPS bearing either penta-acylated, 1-phosphorylated or 4′-phosphorylated lipid A to elicit TLR4-dependent NF-κB activation provide strong independent support for this conclusion. In addition, other investigators have provided evidence that synthetic hexa-acylated, monophosphorylated lipid A (bearing a 4′-phosphate) is significantly less potent than the corresponding synthetic hexa-acylated, bis-phosphorylated lipid A (bearing both a 4′- and a 1-phosphate) in stimulating TLR4-dependent NF-κB activation (4). In light of this evidence, we propose that the lipid A structures of both B. thetaiotaomicron and P. gingivalis are under the control of lipid A phosphatases, which in turn are regulated by their respective local microenvironmental conditions. The genes encoding putative lipid A 1- and 4′-phosphatases in P. gingivalis have been analyzed and exhibit genetic functions that are consistent with this idea (5). In addition, the B. thetaiotaomicron genome encodes sequences that are homologous to those of the P. gingivalis lipid A phosphatases (unpublished data). This hypothetical paradigm provides a rational genetic mechanism to explain the distinct abilities of phylogenetically related bacteria that occupy distinct niches to differentially engage host TLR4-dependent innate immune responses depending upon the position of the phosphate in the lipid A of their LPS. The molecular basis for the postulated regulation of lipid A phosphatases requires further investigation.
Taking human health and disease into consideration, the structure-function relationships that exist between lipid A and the human innate immune defense system are currently under intense investigation and are at the forefront of medical research. For example, hexa-acylated, monophosphorylated lipid A bearing a 4′-phosphate selectively activates the TLR4-associated TRIF pathway and is a relatively weak activator of MyD88-dependent TLR4 responses, suggesting that this lipid A possesses unique adjuvant properties that render it valuable for promoting robust adaptive immune responses (17). The present study provides novel evidence that the genetic regulation of the phosphate position on penta-acylated lipid A from medically relevant and phylogenetically related bacteria is likely an important naturally occurring mechanism that contributes to the resolution of bacterial-host innate immune system interactions into either symbiotic or pathogenic relationships.
The present study also illuminates a novel aspect of lipid A structure-TLR4 function. Previous structural studies of crystal complexes between TLR4-MD-2 and lipid A revealed that the specific stereochemical interaction between lipid A and the TLR4-MD-2 complex can vary depending upon the number of fatty acyl groups on the lipid A. The specific binding conformation that a particular lipid A displays with MD-2 and TLR4 is thought to play a role in determining whether LPS functions as an agonist or an antagonist (18, 19). We observed that the penta-acylated, bis-phosphorylated lipid A from E. coli msbB WSK, which bears short-chain fatty acids (C12 and C14), is a weak TLR4-agonist compared to the 1-phosphorylated E. coli msbB LpxF lipid A (Fig. 6). Notably, the bis-phosphorylated E. coli msbB LPS is recognized as a TLR4 antagonist (6). In contrast, the penta-acylated, bis-phosphorylated version of P. gingivalis lipid A contains long-chain fatty acids (C15, C16, and C17), and it appears to be a relatively potent agonist for TLR4 activation (5, 15). These data suggest that the lipid A fatty acid chain length, as well as the lipid A fatty acid chain number and lipid A phosphate content, can significantly influence the ability of penta-acylated lipid A to function as a TLR4 agonist or antagonist. It remains to be determined how the fatty acid chain length and phosphate position of penta-acylated lipid A structures, which are present on the LPSs of multiple clinically important Gram-negative bacteria, dictate the specific molecular interactions and contact points occurring between TLR4-MD-2 and lipid A to guide the functional oligomerization of the TLR4-receptor complex.
Acknowledgments
This work was supported by National Institutes of Health grants DE012768 and DE018274.
Editor: B. A. McCormick
Footnotes
Published ahead of print on 25 October 2010.
REFERENCES
- 1.Al-Qutub, M. N., P. H. Braham, L. M. Karimi-Naser, X. Liu, C. A. Genco, and R. P. Darveau. 2006. Hemin-dependent modulation of the lipid A structure of Porphyromonas gingivalis lipopolysaccharide. Infect. Immun. 74:4474-4485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bainbridge, B. W., S. R. Coats, T. T. Pham, R. A. Reife, and R. P. Darveau. 2006. Expression of a Porphyromonas gingivalis lipid A palmitylacyltransferase in Escherichia coli yields a chimeric lipid A with altered ability to stimulate interleukin-8 secretion. Cell Microbiol. 8:120-129. [DOI] [PubMed] [Google Scholar]
- 3.Berezow, A. B., R. K. Ernst, S. R. Coats, P. H. Braham, L. M. Karimi-Naser, and R. P. Darveau. 2009. The structurally similar, penta-acylated lipopolysaccharides of Porphyromonas gingivalis and Bacteroides elicit strikingly different innate immune responses. Microb. Pathog. 47:68-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cekic, C., C. R. Casella, C. A. Eaves, A. Matsuzawa, H. Ichijo, and T. C. Mitchell. 2009. Selective activation of the p38 MAPK pathway by synthetic monophosphoryl lipid A. J. Biol. Chem. 284:31982-31991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Coats, S. R., J. W. Jones, C. T. Do, P. H. Braham, B. W. Bainbridge, T. T. To, D. R. Goodlett, R. K. Ernst, and R. P. Darveau. 2009. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4′-phosphatase activities. Cell Microbiol. 11:1587-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coats, S. R., T. T. Pham, B. W. Bainbridge, R. A. Reife, and R. P. Darveau. 2005. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 175:4490-4498. [DOI] [PubMed] [Google Scholar]
- 7.Comstock, L. E., and M. J. Coyne. 2003. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. Bioessays 25:926-929. [DOI] [PubMed] [Google Scholar]
- 8.Darveau, R. P. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat. Rev. Microbiol. 8:481-490. [DOI] [PubMed] [Google Scholar]
- 9.Dixon, D. R. 2005. Lipid A heterogeneity within Porphyromonas gingivalis and other oral bacteria: effect of lipid A content on hTLR4 utilization and E-selectin expression. Ph. D. Thesis. University of Washington, Seattle, WA.
- 10.Erridge, C., E. Bennett-Guerrero, and I. R. Poxton. 2002. Structure and function of lipopolysaccharides. Microbes Infect. 4:837-851. [DOI] [PubMed] [Google Scholar]
- 11.Falk, P. G., L. V. Hooper, T. Midtvedt, and J. I. Gordon. 1998. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiol. Mol. Biol. Rev. 62:1157-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hajishengallis, G. 2009. Porphyromonas gingivalis-host interactions: open war or intelligent guerilla tactics? Microbes Infect. 11:637-645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jain, S., and R. P. Darveau. 2010. Contribution of Porphyromonas gingivalis lipopolysaccharide to periodontitis. Periodontol. 2000 54:53-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kumada, H., Y. Haishima, T. Umemoto, and K. Tanamoto. 1995. Structural study on the free lipid A isolated from lipopolysaccharide of Porphyromonas gingivalis. J. Bacteriol. 177:2098-2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumada, H., Y. Haishima, K. Watanabe, C. Hasegawa, T. Tsuchiya, K. Tanamoto, and T. Umemoto. 2008. Biological properties of the native and synthetic lipid A of Porphyromonas gingivalis lipopolysaccharide. Oral Microbiol. Immunol. 23:60-69. [DOI] [PubMed] [Google Scholar]
- 16.Loppnow, H., H. Brade, I. Durrbaum, C. A. Dinarello, S. Kusumoto, E. T. Rietschel, and H. D. Flad. 1989. IL-1 induction-capacity of defined lipopolysaccharide partial structures. J. Immunol. 142:3229-3238. [PubMed] [Google Scholar]
- 17.Mata-Haro, V., C. Cekic, M. Martin, P. M. Chilton, C. R. Casella, and T. C. Mitchell. 2007. The vaccine adjuvant monophosphoryl lipid A as a TRIF-biased agonist of TLR4. Science 316:1628-1632. [DOI] [PubMed] [Google Scholar]
- 18.Ohto, U., K. Fukase, K. Miyake, and Y. Satow. 2007. Crystal structures of human MD-2 and its complex with antiendotoxic lipid IVa. Science 316:1632-1634. [DOI] [PubMed] [Google Scholar]
- 19.Park, B. S., D. H. Song, H. M. Kim, B. S. Choi, H. Lee, and J. O. Lee. 2009. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature 458:1191-1195. [DOI] [PubMed] [Google Scholar]
- 20.Raetz, C. R., C. M. Reynolds, M. S. Trent, and R. E. Bishop. 2007. Lipid A modification systems in gram-negative bacteria. Annu. Rev. Biochem. 76:295-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Raetz, C. R., and C. Whitfield. 2002. Lipopolysaccharide endotoxins. Annu. Rev. Biochem. 71:635-700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rangarajan, M., J. Aduse-Opoku, N. Paramonov, A. Hashim, N. Bostanci, O. P. Fraser, E. Tarelli, and M. A. Curtis. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190:2920-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shah, H. N., and M. D. Collins. 1988. Proposal for reclassification of Bacteroides asaccharolyticus, Bacteroides gingivalis, and Bacteroides endodontalis in a new genus, Porphyromonas. Int. J. Syst. Bacteriol. 38:128-131. [Google Scholar]
- 24.Shelburne, C. E., W. A. Coulter, D. Olguin, M. S. Lantz, and D. E. Lopatin. 2005. Induction of {beta}-defensin resistance in the oral anaerobe Porphyromonas gingivalis. Antimicrob. Agents Chemother. 49:183-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Socransky, S. S., and A. D. Haffajee. 2005. Periodontal microbial ecology. Periodontol. 2000 38:135-187. [DOI] [PubMed] [Google Scholar]
- 26.Somerville, J. E., Jr., L. Cassiano, B. Bainbridge, M. D. Cunningham, and R. P. Darveau. 1996. A novel Escherichia coli lipid A mutant that produces an antiinflammatory lipopolysaccharide. J. Clin. Invest. 97:359-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stappenbeck, T. S., L. V. Hooper, and J. I. Gordon. 2002. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. U. S. A. 99:15451-15455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang, X., M. J. Karbarz, S. C. McGrath, R. J. Cotter, and C. R. Raetz. 2004. MsbA transporter-dependent lipid A 1-dephosphorylation on the periplasmic surface of the inner membrane: topography of Francisella novicida LpxE expressed in Escherichia coli. J. Biol. Chem. 279:49470-49478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang, X., S. C. McGrath, R. J. Cotter, and C. R. Raetz. 2006. Expression cloning and periplasmic orientation of the Francisella novicida lipid A 4′-phosphatase LpxF. J. Biol. Chem. 281:9321-9330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weintraub, A., U. Zahringer, H. W. Wollenweber, U. Seydel, and E. T. Rietschel. 1989. Structural characterization of the lipid A component of Bacteroides fragilis strain NCTC 9343 lipopolysaccharide. Eur. J. Biochem. 183:425-431. [DOI] [PubMed] [Google Scholar]