Abstract
Clostridium difficile causes chronic intestinal disease, yet little is understood about how the bacterium interacts with and survives in the host. To colonize the intestine and cause persistent disease, the bacterium must circumvent killing by host innate immune factors, such as cationic antimicrobial peptides (CAMPs). In this study, we investigated the effect of model CAMPs on growth and found that C. difficile is not only sensitive to these compounds but also responds to low levels of CAMPs by expressing genes that lead to CAMP resistance. By plating the bacterium on medium containing the CAMP nisin, we isolated a mutant capable of growing in three times the inhibitory concentration of CAMPs. This mutant also showed increased resistance to the CAMPs gallidermin and polymyxin B, demonstrating tolerance to different types of antimicrobial peptides. We identified the mutated gene responsible for the resistance phenotype as CD1352. This gene encodes a putative orphan histidine kinase that lies adjacent to a predicted ABC transporter operon (CD1349 to CD1351). Transcriptional analysis of the ABC transporter genes revealed that this operon was upregulated in the presence of nisin in wild-type cells and was more highly expressed in the CD1352 mutant. The insertional disruption of the CD1349 gene resulted in significant decreases in resistance to the CAMPs nisin and gallidermin but not polymyxin B. Because of their role in cationic antimicrobial peptide resistance, we propose the designation cprABC for genes CD1349 to CD1351 and cprK for the CD1352 gene. These results provide the first evidence of a C. difficile gene associated with antimicrobial peptide resistance.
Clostridium difficile is a major nosocomial pathogen that causes chronic intestinal disease that is both difficult and costly to treat (12, 53). A number of risk factors have been identified for infection with C. difficile, namely, antibiotic exposure, the disturbance of the normal colonic flora, advanced age, and hospitalization (11, 28, 38, 66). Though C. difficile is a strict anaerobe, it can survive outside the host intestinal environment as a dormant spore, which allows the bacterium to spread to other hosts via the fecal-oral route of infection (67). The progression of C. difficile disease is dependent on the ability of the bacteria to germinate from the spore form, multiply, reside, and produce toxins within the intestinal tract (37, 53, 69).
Aside from the principal toxins A and B, a few additional factors have been implicated in the colonization or virulence of C. difficile. These include the S-layer proteins (6), the Cwp84 protease (29), Cwp66 adhesin (68), the Fbp68 fibronectin-binding protein (21), CDT binary toxin (3, 57), and flagellar proteins (64). While these factors may play a role in the initial adherence and colonization of the bacteria, little is known about how C. difficile is able to survive the host immune response to cause persistent infections. There is evidence that host immune factors, such as alpha-defensins, can inhibit C. difficile toxin B activity, and they may play an important role in susceptibility to C. difficile disease (13). Diseases such as ulcerative colitis and Crohn's disease, which appear to be immune related, are linked to an increased risk of C. difficile infection, although the reason for this association is unknown (1, 25). Experiments in animal models of C. difficile disease have demonstrated that defects in the host innate immune response lead to greater colonization and disease progression (33). These studies underscore that the host immune response plays a key role in the clearance of C. difficile infection, but details of the host-pathogen interactions are not understood.
The intestinal tract is a dynamic organ that relies on many factors to prevent being overrun by pathogenic bacteria. The innate immune response in particular provides a constant and immediate line of host defenses that protect the intestines from invasion. The production of cationic antimicrobial peptides (CAMPs) represents a critical component of host defense that bacteria must overcome to cause disease (4, 16). CAMPs are small, positively charged peptides that are made by bacteria, fungi, plants, and animals, and they have microbicidal activities (15). Humans produce a variety of CAMPs, including defensins, cathelicidins, and thrombocidins, that accumulate in areas of the body that routinely encounter microorganisms, such as the intestines (42, 50). These compounds are able to kill bacteria directly, and many can stimulate the host to produce additional immune effectors that contribute to microbial death (17).
Resistance to antimicrobial peptides is essential for bacteria to establish persistent infections, and consequently many human bacterial pathogens are resistant to the killing effects of CAMPs (16, 43, 50). Because C. difficile causes chronic intestinal disease, we hypothesized that it has evolved mechanisms for defense against host antimicrobial peptides that allow it to colonize and persist in the intestinal environment. In the current work, we investigated the response of C. difficile to CAMPs in vitro. We found that the bacteria can adapt to the presence of these compounds and identified a broad-range CAMP resistance mechanism and a component of its regulatory system. This is the first identified CAMP resistance mechanism in C. difficile and appears to be related to lantibiotic self-tolerance systems.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Clostridium difficile strains were grown in BHIS medium (61) supplemented with 0.1% L-cysteine. Media for the growth of C. difficile were supplemented with 250 μg D-cycloserine ml−1, 50 μg kanamycin ml−1, 10 to 20 μg thiamphenicol ml−1, 5 μg erythromycin ml−1, or 5 μg erythromycin ml−1 and 50 μg kanamycin ml−1 as needed. C. difficile strains were maintained at 37°C in an anaerobic chamber (Coy Laboratory Products) with an atmosphere of 10% H2, 5% CO2, and 85% N2. Bacillus subtilis strains were routinely grown at 37°C in L broth (34) or BHIS medium supplemented with 1 μg erythromycin ml−1 when needed. For the propagation of B. subtilis in the anaerobic chamber, the growth medium was supplemented with 5 mM KNO3. Escherichia coli strains were grown at 37°C in L or BHIS medium supplemented with 20 μg chloramphenicol ml−1 or 100 μg ampicillin ml−1 as needed.
TABLE 1.
Bacterial strains and plasmids
| Plasmid or strain | Relevant genotype or features | Source or reference |
|---|---|---|
| Strains | ||
| E. coli | ||
| HB101 | F−mcrB mrr hsdS20(rB− mB−)recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 | B. Dupuy |
| MC135 | HB101 containing pRK24 and pMC123 | This study |
| MC138 | HB101 containing pRK24 and pMC125 | This study |
| MC161 | HB101 containing pRK24 and pMC147 | This study |
| C. difficile | ||
| 630 | Clinical isolate | 71 |
| JIR8094 | Erms derivative of strain 630 | 44 |
| CCUG37769 | Serotype A4; ToxA+, ToxB+ | Lars Burman |
| MC119 | JIR8094 cprK1 | This study |
| MC137 | JIR8094 pMC123 | This study |
| MC141 | JIR8094 cprA::ermB | This study |
| MC146 | MC141::Tn916 (cprABC); Ermr | This study |
| MC162 | JIR8094 pMC147 | This study |
| MC163 | MC119 pMC147 | This study |
| MC164 | MC119 pMC123 | This study |
| B. subtilis | ||
| BS49 | CU2189::Tn916 | P. Mullany |
| MC144 | BS49 Tn916::pMC130 | This study |
| Plasmids | ||
| pRK24 | Tra+, Mob+; bla, tet | 65 |
| pCR2.1 | bla, kan | Invitrogen |
| pUC19 | Cloning vector; bla | 72 |
| pCE240 | C. difficile TargeTron construct based on pJIR750ai (group II intron, ermB::RAM, ltrA); catP | C. Ellermeier |
| pSMB47 | Tn916 integrational vector; Cmr, Ermr | 36 |
| pJIR1456 | E. coli-C. perfringens shuttle vector; catP | 35 |
| pMC122 | pCE240 retargeted to cprA | This study |
| pMC123 | E. coli-C. difficile shuttle vector; bla, catP | This study |
| pBL58 | pMC123 with BsrGI and two HindIII sites removed | This study |
| pMC125 | 5.48 kb SfoI/SmaI fragment from pMC122 cloned in SmaI site of pMC123 | This study |
| pMC130 | 2,945-bp DNA sequence corresponding to cprABC plus 500-bp upstream sequence cloned as BamHI/SphI fragment in pSMB47 | This study |
| pMC147 | pBL58 with 1,798 bp of CD1352 and upstream promoter region (PcprK) | This study |
Bacterial strain and plasmid construction.
Oligonucleotides used in this study are listed in Table 2 . C. difficile strain 630 DNA, GenBank accession number AM180355 (58), was used as a template for PCR amplification unless otherwise noted. The sequencing of cloned DNA fragments was performed by the Tufts University Core Facility using an ABI 3130XL DNA sequencer. A null mutation in CD1349 (cprA) was created in several steps. First, plasmid pMC122 was created by the retargeting of the group II intron from pCE240 (kindly supplied by Craig Ellermeier, University of Iowa) using primers oMC123, oMC124, and oMC125 and the EBS Universal primer as outlined in the TargeTron users manual (Sigma-Aldrich), followed by the initial cloning of the retargeted fragment in pCE240 digested with BsrGI/HindIII. Plasmid pCE240 is a derivative of pJIR750ai (Sigma-Aldrich) that is similar to pMTL007 (19). pMC123 was constructed by the stepwise addition of three DNA fragments to pUC19. A 390-bp oriT fragment was amplified from pJIR1456 (35) using primers oMC15 and oMC16 and cloned as an EcoO1091/AatII fragment in pUC19. A 1,043-bp fragment containing catP from pJIR1456, amplified using primers oMC13 and oMC14, then was added as a PciI/SapI fragment. Lastly, a 3.1-kb plasmid origin of replication (ori69) amplified from C. difficile strain CCUG37769 using primers oMC122 and oMC123 was inserted at the SfoI site. The intron-containing part of pMC122 was excised as a 5.48-kb SfoI/SmaI fragment and cloned in the SmaI site of pMC123, creating pMC125, which was introduced by transformation into E. coli strain HB101(pRK24), resulting in strain MC138. pRK24 is a derivative of the broad-host-range plasmid RP4 that mobilizes IncP oriT plasmids. MC138 then was mated with C. difficile strain JIR8094, resulting in the transfer of pMC125 by conjugation as previously described (8), except that transconjugants were selected on BHIS plates supplemented with D-cycloserine, kanamycin, and thiamphenicol. The inactivation of CD1349 (cprA) was selected for by screening transconjugants for erythromycin resistance and thiamphenicol sensitivity, resulting in strain MC141. The insertional disruption of cprA was confirmed using primers oMC130 and oMC97, which are located outside the region of insertion in the cpr locus.
TABLE 2.
Oligonucleotides
| Primer | Sequence (5′→3′) | Use/location |
|---|---|---|
| oMC13 | 5′-GCACATGTCCTTGGTTGTGTTGCTTTTCG-3′ | catP gene PCR |
| oMC14 | 5′-GCTCTTCTAGCGCCTACGGGGAATT-3′ | catP gene PCR |
| oMC15 | 5′-GCAGGCCCTCGGATCTTTTCCGCTGCA-3′ | oriT PCR |
| oMC16 | 5′-GCGACGTCCTTATCGGCCAGCCTCG-3′ | oriT PCR |
| oMC44 | 5′-CTAGCTGCTCCTATGTCTCACATC-3′ | rpoC qPCR |
| oMC45 | 5′-CCAGTCTCTCCTGGATCAACTA-3′ | rpoC qPCR |
| oMC55 | 5′-GGTTTTCTAAATGGGAAGGTAAA-3′ | cprK sequencing |
| oMC56 | 5′-CCAGATAAGTCATTAATTGCTGCG-3′ | cprK sequencing |
| oMC57 | 5′-GCTCTGAATAACAGTTCTCTATCTA-3′ | cprK sequencing |
| oMC96 | 5′-CGTTCAGGTCAATTCTCTCTAGGC-3′ | cprA qPCR/sequencing |
| oMC97 | 5′-GGTCAAGACCATTTGTAGGCTC-3′ | cprA qPCR/sequencing |
| oMC121 | 5′-CGTATTGGCGCCAACCAGGAATATAGTGTATGCA-3′ | ori69 PCR |
| oMC122 | 5′-CGTATTGGCGCCCTAGAGAACCAAACGACGG-3′ | ori69 PCR |
| oMC123a | 5′-AAAAGCTTTTGCAACCCACGTCGATCGTGAAGAAAGGATTTATGTGCGCCCAGATAGGGTG-3′ | cprA intron retargeting |
| oMC124a | 5′-CAGATTGTACAAATGTGGTGATAACAGATAAGTCATTTATCATAACTTACCTTTCTTTGT-3′ | cprA intron retargeting |
| oMC125a | 5′-CGCAAGTTTCTAATTTCGGTTCTTTCTCGATAGAGGAAAGTGTCT-3′ | cprA intron retargeting |
| EBS universal | 5′-CGAAATTAGAAACTTGCGTTCAGTAAAC-3′ | Sigma-Aldrich |
| oMC126 | 5′-GCTACTACTTTATTGAGTACGGCA-3′ | CD1348 qPCR |
| oMC127 | 5′-CTTCATCCTTCTTTACAACTGCTG-3′ | CD1348 qPCR |
| oMC128 | 5′-GCGTATTACACAGGAGTTTGAACC-3′ | ori69 sequencing |
| oMC129 | 5′-GCATTGTATGTATCTTTTATTCCTAGC-3′ | ori69 sequencing |
| oMC130 | 5′-GGAGCAGATGGCAGTTGATAAC-3′ | cprA PCR |
| oMC137 | 5′-GCTGCTTTCAACTGGTGGTA-3′ | cprB qPCR |
| oMC138 | 5′-CCAGTCCAAACGTCTTTCATTTC-3′ | cprB qPCR |
| oMC139 | 5′-GCACATTTGCATTGCTTTAATGGG-3′ | cprC qPCR |
| oMC140 | 5′-CATTGAACACACAACACCTGAC-3′ | cprC qPCR |
| oMC141 | 5′-GAAGAAAGGCATGCATATTCAGA-3′ | cprK qPCR |
| oMC142 | 5′-TCATACCCAATGTCTCTGGT-3 | cprK qPCR |
| oMC144 | 5′-GCGGATCCGGTTCGGAAAAGGAACATC-3 | PcprK cloning |
| oMC145 | 5′- GCAAGCTTGTTACCATAGCACCACCAG-3′ | PcprK cloning |
| oMC146 | 5′-GCGGATCCGCTACTACTTTATTGAGTACGG C-3′ | PcprABC cloning |
| oMC147 | 5′-GCGCATGCCAAGCATTTGTACCCTGTCCTC-3′ | PcprABC cloning |
| BC1ab | 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGACGCTCTTCCGATCTAACCT-3′ | DNA barcoding |
| BC1bb,c | 5′-*GGTTAGATCGGAAGAGCGGTTCAGCAGGAATGC CGAGACCGATCTCGTATGCCGTCTTCTGCTTG-3′ | DNA barcoding |
| Olj139 | 5′-AATGATACGGCGACCACCGAGATCTACACTCTTTCCCTACACGA-3′ | DNA barcoding |
| Olj140 | 5′-CAAGCAGAAGACGGCATACGAGATCGGTCTCGGCATTCCTGCTGAAC-3′ | DNA barcoding |
Underlined bases denote retargeted sequences.
Underlined bases define the barcode.
Asterisk denotes the position of 5′ phosphorylation.
To complement the cprA disruption, a 2,945-bp fragment containing the cprABC operon and its upstream region was amplified using oMC146 and oMC147 as primers and was cloned between the BamHI and SphI sites of pSMB47, generating pMC130. MC144 was created by the integration of pMC130 into the chromosomal Tn916 locus of B. subtilis strain BS49. MC146 is a transconjugant from the mating of B. subtilis strain MC144 and C. difficile strain MC141, resulting in the integration of the plasmid::Tn916 fusions of the donor strains into the C. difficile chromosome, as previously described (18).
To investigate the impact of the overexpression of cprK, a plasmid-based copy of the cprK gene and its upstream region (PcprK) was introduced into strains of C. difficile as follows. A 1,798-bp fragment corresponding to cprK and the entire cprC-cprK intergenic region was amplified using primers oMC144 and oMC145 and cloned between the BamHI and HindIII sites of pBL58 to create pMC147. pBL58 is a derivative of plasmid pMC123 in which one HindIII site (outside the multiple cloning site [MCS]) and all BsrGI sites were removed (L. Bouillaut, personal communication). pMC147 then was introduced into E. coli strain HB101(pRK24) by transformation, resulting in strain MC161. MC161 then was mated with C. difficile strains JIR8094 and MC119 to transfer pMC147 by conjugation, resulting in strains MC162 and MC163, respectively. As a control, E. coli strain HB101(pRK24) was transformed with pMC123, yielding strain MC135. pMC123 then was transferred to C. difficile strains JIR8094 and MC119 by conjugation with MC135, creating strains MC137 and MC164, respectively.
MIC determination.
Susceptibility tests were performed anaerobically in BHIS broth as follows. C. difficile strain JIR8094 and derivatives were grown to an optical density at 600 nm (OD600) of 0.5 (exponential growth phase) and then diluted 1:100 into 1 ml of fresh medium to give an inoculum of approximately 5 × 105 CFU/ml. The medium was supplemented with a range of concentrations of nisin (MP Biomedicals), polymyxin B (≥6,000 USP U/mg; Sigma-Aldrich), gallidermin (Alexis Biochemicals), LL-37 (17-29)/FK-13 (cathelicidin) (Phoenix Pharmaceuticals), or Magainin II (Phoenix Pharmaceuticals), and cultures were incubated for 18 h at 37°C. The MIC was defined as the lowest concentration of antimicrobial peptide that prevented visible turbidity after 18 h.
qRT-PCR.
Exponential-phase cultures of C. difficile grown in BHIS medium or BHIS supplemented with nisin, gallidermin, or polymyxin B were diluted 1:50 in BHIS or BHIS supplemented with the same antimicrobial peptides and incubated anaerobically at 37°C. Samples for RNA isolation were taken when cultures reached an OD600 of 0.4 (active growth), diluted with an equal volume of cold 1:1 ethanol-acetone, and stored at −80°C. RNA was extracted and treated to remove contaminating DNA as previously described (8). SuperScript II reverse transcriptase (Invitrogen) was used to generate randomly primed cDNA pools from 1 μg of RNA per sample, as instructed by the manufacturer. To control for chromosomal DNA contamination, mock cDNA synthesis reaction mixtures containing no reverse transcriptase were used as negative controls in subsequent amplifications. cDNA samples were diluted 4-fold and used as templates for quantitative, real-time PCR (qRT-PCR) of rpoC (primers oMC44/oMC45), cprA (oMC96/oMC97), cprB (oMC137/oMC138), cprC (oMC139/oMC140), cprK (oMC141/oMC142), and CD1348 (oMC126/oMC127), using Qiagen SYBR green PCR mix and an MXP3005 thermocycler (Stratagene/Agilent Technologies). Reactions were performed in a final volume of 25 μl using 4 μl of diluted cDNA and 1 μM each primer. Reactions were performed in triplicate using cDNA extracted from each of a minimum of two biological replicates, and results are presented as the means and standard deviations of these experiments. Amplification included 40 cycles of the following steps: 30 s at 95°C, 60 s at 50°C, and 30 s at 72°C. Results were calculated using the comparative cycle threshold method (56), in which the amount of target mRNA is normalized relative to an internal control transcript (rpoC). The two-tailed Student's t test was used to analyze the data.
Identification of a chromosomal mutation through SNP analysis.
Purified genomic DNA of strain MC119 was sheared, barcoded, and amplified for high-throughput DNA sequencing as previously described (7a). Deep sequencing was performed using an Illumina Genome Analyzer II by the Tufts University Core Facility. Sequenced genomic DNA reads were aligned to the C. difficile 630 genome (NCBI accession number NC_009089) using MAQ 0.6.6 (http://maq.sourceforge.net). Reads with more than two mismatches from their aligned positions were removed from the assembly. A consensus sequence was called from the assembled reads, and single-nucleotide polymorphisms (SNPs) and short insertions and deletions were identified. The list was filtered using MAQ with the following parameters: a minimum read depth of 3, a minimum consensus quality of 20, and a minimum adjacent consensus quality of 20.
RESULTS
Adaptation of C. difficile to nisin.
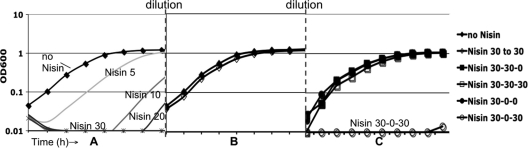
To understand the effects of CAMPs on the colonization and virulence of C. difficile, we first determined the sensitivity of C. difficile to CAMPs and its ability to adapt to the presence of these molecules. As a model, we first tested the CAMP nisin, a commercially available CAMP produced by Lactococcus lactis that is routinely used to study the effects of CAMPs on bacteria (5, 7, 10, 31). The MIC of nisin for strain JIR8094 was approximately 90 μg/ml (Table 3). Based on the MIC, we then assessed the growth effects of a range of sublethal nisin concentrations. As shown in Fig. 1 A, increasing the concentration of nisin in the medium resulted in an increasing delay in logarithmic growth.
TABLE 3.
MICs
| Drug | MIC (μg/ml) for strain: |
|||
|---|---|---|---|---|
| 630 | MC119 | MC141 | MC146 | |
| Nisin | 90 | 350 | 25 | 120 |
| Polymyxin B | 300 | 500 | 300 | ≥300 |
| Gallidermin | 0.5 | ≥1.3 | ≥0.3 | 0.6 |
FIG. 1.
Growth and adaptation of C. difficile to nisin. Cells were grown in BHIS medium with or without nisin as indicated (μg/ml). (A) C. difficile growth is inhibited by increasing concentrations of nisin. (B) C. difficile cells preconditioned in nisin at 30 μg/ml are able to grow at normal rates when resuspended in medium without nisin (30-0) or with nisin at 30 μg/ml (30-30). (C) Cells preconditioned in nisin maintain the ability to grow in nisin at the same concentration (30-30-30), but cells preconditioned in nisin (30) and then grown without nisin (30-0) lose the ability to grow in nisin without significant lag (30-0-30).
Many bacterial species have been shown to adapt to low concentrations of CAMPs by inducing the expression of CAMP resistance mechanisms. The induction of these resistance mechanisms allows the bacteria to grow in previously inhibitory concentrations (14, 47, 54). We found that preconditioning C. difficile to inhibitory, but not lethal, levels of nisin (30 μg/ml) enabled the bacteria to grow in otherwise-inhibitory concentrations (Fig. 1B). Cells that were preconditioned in nisin at 30 μg/ml and then diluted into fresh medium containing nisin at the same concentration were able to grow without a lag and at the same rate as wild-type cells grown without nisin. When preconditioned cells were subcultured without nisin and then reexposed to nisin, however, growth again was inhibited (Fig. 1C). These results demonstrate that preconditioning allows cells to grow in nisin due to transient adaptation and not to mutation.
Isolation and identification of a nisin-resistant mutant.
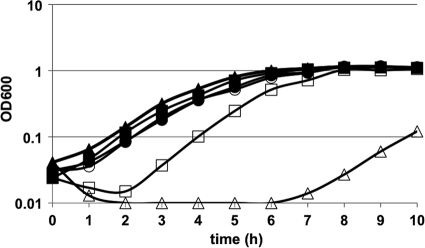
The ability of C. difficile to adapt to CAMPs most likely is accomplished by the induction of genes encoding resistance mechanisms. To identify such genes, we isolated spontaneous CAMP-resistant C. difficile mutants by plating wild-type cells on BHIS agar supplemented with nisin. Nisin-resistant colonies that appeared on plates containing 225 μg nisin per ml were tested for their ability to grow in BHIS broth with increasing concentrations of nisin. One such mutant, MC119, was able to grow in approximately four times the nisin MIC for the wild-type without preconditioning (Table 3). In addition, this mutant grew at wild-type rates in sublethal concentrations, a condition that delays the growth of wild-type bacteria (Fig. 2 ). MC119 also showed increased resistance to the CAMPs gallidermin and polymyxin B, demonstrating tolerance to a variety of antimicrobial peptides (Table 3).
FIG. 2.
Growth of cpr mutants in nisin. C. difficile wild-type (JIR8094, squares), MC119 (cprK W235C, circles), and MC141 (cprA::intron::ermB, triangles) cells grown in BHIS medium (filled shapes) or BHIS medium supplemented with 10 μg/ml nisin (open shapes).
To map the CAMP resistance mutation in MC119, we resequenced the entire genome (41), the only method currently available to map spontaneous mutations in C. difficile. Using this approach, we identified four single-nucleotide polymorphisms that were represented at greater-than 85% frequency in the MC119 genome compared to that of the parent strain, JIR8094. These mutations were located in the coding sequences of genes CD1352 (100%), CD2125 (100%), CD2667 (85%), and CD3089 (95%). Five additional independently isolated mutants were obtained that also were resistant to high levels of nisin. Because the phenotypes of these mutants were identical to that of MC119, we sequenced the CD1352 gene of each and found that all had a mutation within CD1352. Because there was a 100% correlation between the high-nisin-resistance phenotype and mutations in the CD1352 gene, we further investigated this correlation. The mutation in MC119 lies within gene CD1352 and resulted in a tryptophan-to-cysteine alteration at amino acid position 235 (W235C), whereas the other five mutants had serine-to-tyrosine substitutions at amino acid 230 (S230Y). Based on the genome annotation (58), CD1352 is predicted to encode a lantibiotic sensor histidine kinase. Concordantly, by protein prediction modeling (SOSUI) (23, 39, 40), the CD1352 protein product appears to have all of the characteristics of a sensor histidine kinase (HK) member of a two-component regulatory system (TCS). The mutated amino acids at positions 230 and 235 are predicted to be located on the cytoplasmic side of the molecule, which includes the kinase-phosphatase and protein interaction regions, near but not within the catalytic active site (24). TCSs typically contain a sensor histidine kinase that receives an extracellular stimulus and transmits a signal to a partner protein, the response regulator (RR). The activated RR acts by binding DNA to regulate gene transcription (24). In most cases, the cognate HK and RR genes are located adjacently to each other on the chromosome, but there is no RR encoded near CD1352. If CD1352 is in fact an HK involved in regulating the transcription of CAMP resistance genes through a cognate RR, then this RR must be encoded elsewhere on the chromosome.
Effects of the CD1352 mutation on transcription of CD1348 to CD1352.
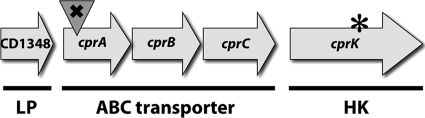
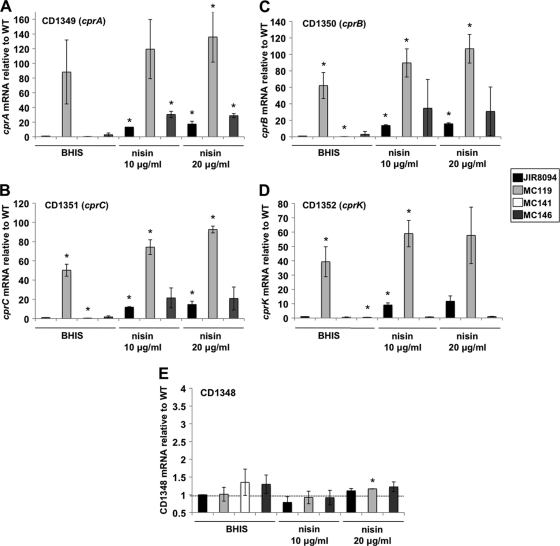
As CD1352 appears to be a regulatory gene, we next sought to identify the potential target(s) of its regulation. CD1352 lies immediately downstream of a gene cluster that putatively encodes an ABC transport system (Fig. 3). This putative transporter and the CD1352 histidine kinase both share homology to lantibiotic immunity mechanisms, which are found in bacteria that produce such antimicrobial peptides (e.g., nisin) and are highly specific for individual peptides (55, 70). To test whether the CD1348-CD1351 cluster is regulated by the CD1352 gene product, we performed real-time quantitative PCR analysis of the CD1349 to CD1351 transcripts in the wild-type and MC119 mutant strains grown in the presence or absence of CAMPs. As shown in Fig. 4, levels of CD1349, CD1350, and CD1351 mRNA increased when wild-type cells were grown in the presence of nisin or gallidermin at sublethal concentrations. Moreover, the level of expression of these genes was higher in cells grown with nisin at 20 μg/ml than in cells grown at 10 μg/ml. Increased expression also was seen when cells were grown in sublethal concentrations of gallidermin (Fig. 5 D to F). As predicted, the ABC transporter genes were very highly expressed in the MC119 mutant, even in the absence of CAMPs (Fig. 4). The expression of the CD1352 histidine kinase also increased in the wild-type strain during growth in nisin (Fig. 4D) or gallidermin (Fig. 5F) and was highly expressed in the presence or absence of CAMPs in the MC119 mutant, indicating that CD1352 also is regulated in response to CAMPs. These data demonstrated that the CD1349-CD1352 gene cluster is induced in response to CAMPs in wild-type C. difficile and raised the possibility that the overexpression of these genes contributes to the high CAMP resistance of the MC119 mutant. We therefore propose the designation cprABC for genes CD1349 to CD1351 and cprK for the CD1352 gene (cpr is a mnemonic for cationic peptide resistance).
FIG. 3.
Putative ABC transporter system and surrounding genes (CD1348 to CD1352) in JIR8094. CD1348 lipoprotein (LP), the CD1349 to CD1351 ABC transporter ABC-binding cassettes and permeases (cprABC), and orphan histidine kinase (HK) and CD1352 (cprK) are shown. × indicates the insertional disruption of cprA; * denotes a W235C spontaneous mutation in cprK.
FIG. 4.
qRT-PCR analysis of cprA, cprB, cprC, cprK, and CD1348 expression during growth in nisin. C. difficile wild-type (JIR8094), MC119 (cprK W235C), MC141 (cprA::intron::ermB), and MC146 (cprA::intron::ermB, PcprABC) strains were grown in BHIS supplemented with 0, 10, or 20 μg/ml nisin to an OD600 of 0.4 as indicated. RNA was harvested, cDNA synthesized, and qPCR performed using gene specific primers for cprA (A), cprB (B), cprC (C), cprK (D), and CD1348 (E). Results were normalized to an internal control gene (rpoC) and are presented as the ratio of each transcript level relative to wild-type and no-nisin controls. The means and standard deviations of biological replicates are shown (*, P ≤ 0.05 by Student's t test).
FIG. 5.
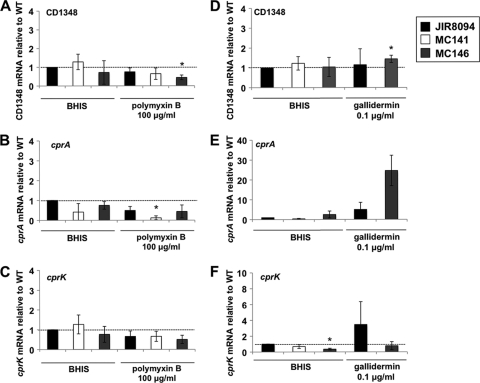
qRT-PCR analysis of CD1348, cprA, and cprK expression during growth in polymyxin B or gallidermin. C. difficile wild-type (JIR8094), MC141 (cprA::intron::ermB), and MC146 (cprA::intron::ermB, PcprABC) strains were grown in BHIS alone or BHIS supplemented with 100 μg/ml polymyxin B or 0.1 μg/ml gallidermin to an OD600 of 0.4 as indicated. RNA was harvested, cDNA synthesized, and qPCR performed using gene-specific primers for CD1348 (A and D), cprA (B and E), or cprK (C and F). Results were normalized to an internal control gene (rpoC) and graphed as the ratio of each transcript level relative to wild-type and no-nisin controls. The means and standard deviations of biological replicates are shown (*, P ≤ 0.05 by Student's t test).
ABC transporter systems often work in association with a lipoprotein carrier; auspiciously, a predicted lipoprotein is encoded by the upstream gene, CD1348 (Fig. 3). We tested the level of the transcription of CD1348 in the presence and absence of nisin, gallidermin, and polymyxin B, and we found no change in gene expression in the wild-type or MC119 (Fig. 4 and 5), indicating that this gene is not induced in response to these CAMPs and is not regulated by the cprK histidine kinase.
Effects of disruption of the cprABC operon on CAMP resistance.
To test whether the cprABC genes play a role in CAMP resistance, we introduced an insertion mutation in the first gene of the operon, cprA, thereby creating a polar disruption of the cprABC operon. To do so, we used a TargeTron-based group II intron (27), which we retargeted for integration into cprA (see Materials and Methods). The resulting strain, MC141, then was tested for its ability to grow in the presence of nisin, polymyxin B, or gallidermin. The MICs of nisin and gallidermin, but not for polymyxin B, for MC141 exhibited significant decreases (Table 3). We also tested the resistance of the wild-type, MC119, and MC141 strains against two CAMPs of animal origin, LL-37 and Magainin II, and we found that all of the strains tested were resistant to the highest concentrations of these compounds tested (greater than 100 μM and 100 μg/ml, respectively); as a result, the animal CAMPs were not used in further analyses.
Figure 2 illustrates the considerable growth delay seen for MC141 exposed to nisin (10 μg/ml) compared to the growth of the wild type and the MC119 mutant. A similar delay in growth was observed when MC141 was grown in gallidermin (data not shown). Although MC141 had a pronounced lag when grown in CAMPs, it did eventually grow at wild-type rates at sublethal concentrations of nisin and gallidermin (less than 20 and 0.1 μg/ml, respectively). This result implies that other CAMP resistance mechanisms can be induced and provide some compensation for the absence of cpr expression.
As expected, the transcription of the cprA, cprB, and cprC genes was markedly decreased in the MC141 mutant compared to levels for the wild type (Fig. 4), suggesting that these genes are cotranscribed as part of an operon. The expression of cprK in MC141, however, was at wild-type levels in the absence of CAMPs, indicating that cprK expression is not disrupted by the polar effects of the insertion in the cprA gene.
To confirm that the CAMP resistance defect of MC141 was due to the disruption of the putative ABC transporter system, we complemented MC141 with the native version of the CD1349-CD1351 region, including a 500-bp upstream region predicted to contain the promoter for these genes. The cprABC genes and the upstream region were integrated into the Tn916 locus of B. subtilis strain BS49 (MC144) and transferred via conjugation to C. difficile strain MC141, creating a cprABC-complemented strain, MC146 (see Materials and Methods). The MICs for MC146 and growth in CAMPs were similar to those of the wild type (Table 3). In the wild type and in MC146, the expression of cprABC was induced during growth in nisin and gallidermin, but cprK transcription did not increase for MC146 (Fig. 4D and 5F). The disruption of the cprABC genes does not disrupt cprK expression, although both the cprABC mutant and cprABC-trans-complemented strains no longer induce the expression of cprK in CAMPs, suggesting that cprK is transcribed from its own promoter as well as from an upstream promoter for cprABC, although this has not been verified. In addition, the B. subtilis strain MC144, which contains the cprABC locus within Tn916, also had increased resistance to nisin (data not shown). These data imply that the cpr locus is directly involved in resistance to multiple CAMPs.
Unlike the case for nisin and gallidermin, the growth of the cprABC mutant, MC141, in polymyxin B was similar to that of the wild type (Table 3). In contrast, the polymyxin B MIC for the cprK mutant, MC119, was higher than that for the wild-type parent (500 versus 300 μg/ml). The expression analysis of the cprABC and cprK genes during growth in a sublethal concentration of polymyxin B (100 μg/ml) demonstrated that neither the cprABC nor cprK gene are induced under these conditions (Fig. 5A to C). In fact, all of the strains, including MC119 (data not shown), appeared to have the decreased expression of cprABC in the presence of polymyxin B. Thus, the cprABC-encoded resistance mechanism does not contribute to resistance to polymyxin B, but cprK may regulate the transcription of an additional, as-yet unknown CAMP resistance mechanism.
Impact of wild-type cprK overexpression on transcription of cpr genes.
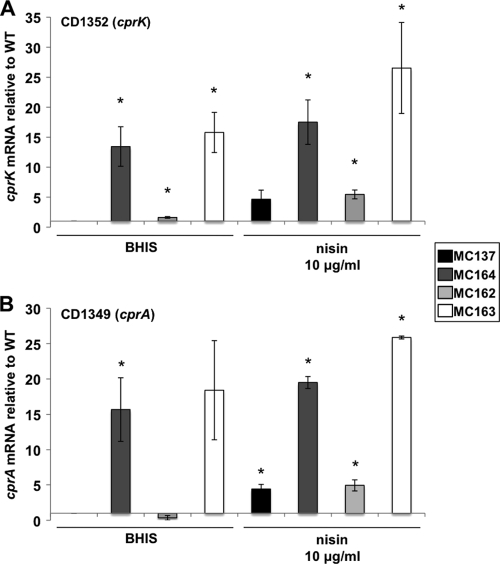
To further investigate the role of cprK in the regulation of the cpr genes, an extrachromosally replicating, plasmid-based copy of the cprK gene and its upstream region (pMC147) was introduced into the wild-type and MC119 strains of C. difficile. The parent vector, pMC123, also was introduced into the wild-type and MC119 strains to control for variability due to plasmid replication and growth in the presence of the selective antibiotic thiamphenicol. Wild-type and MC119 strains carrying pMC123 (MC137 and MC164) or pMC147 (MC162 and MC163) then were grown in BHIS medium supplemented with 10 μg/ml thiamphenicol, with or without the addition of 10 μg/ml nisin. All of these strains grew at the same rate in the absence of nisin, and their growth in nisin paralleled the growth observed for the parent strains, with the exception of MC162 (JIR8094 pMC147), which had a more-than 2-h delay compared to the rate of the vector control strain (MC137) (data not shown). As anticipated, the introduction of a plasmid-borne copy of cprK resulted in modestly higher expression of cprK in MC162 than in MC137 in the absence of nisin (Fig. 6 A). The expression of cprA was reduced in the MC162 strain in the absence of nisin, suggesting that an excess in cprK in a wild-type background results in the more tightly controlled transcription of the cpr genes, although the statistical significance of this result is uncertain (Fig. 6B). As suggested by the growth observed in nisin, the expression of the cprA transcript was unchanged in the MC119 mutant containing the plasmid copy of PcprK (MC163) compared to that of MC119 with vector alone (MC164). These data indicate that the wild-type cprK allele, even in multicopy, is recessive to the mutant version of cprK in MC119, leading us to conclude that the cprK mutation results in a gain-of-function defect. Because the phenotype of the MC119 gain-of-function mutation leads to the greater expression of the cpr operon, we can conclude that CprK functions as a positive regulator of cprABC expression in the presence of nisin.
FIG. 6.
qRT-PCR analysis of cprA and cprK expression with the addition of plasmid-encoded cprK. C. difficile wild-type and MC119 strains carrying control vector pMC123 (MC137 and MC164, respectively) or pMC147 (MC162 and MC163) were grown in BHIS medium supplemented with 10 μg/ml thiamphenicol with or without the addition of 10 μg/ml nisin to an OD600 of 0.4. RNA was harvested, cDNA synthesized, and qPCR performed using gene-specific primers for cprK (A) or cprA (B). Results were normalized to an internal control gene (rpoC) and graphed as the ratio of each transcript level to that of rpoC and then normalized to the wild type grown without nisin. The means and standard deviations of biological replicates are shown (*, P ≤ 0.05 by Student's t test).
DISCUSSION
CAMPs are critical components of the innate immune defense against bacterial pathogens. Accordingly, resistance to these peptides is a demonstrated virulence factor for many bacterial pathogens (16, 43, 48, 50). In this work, we show that C. difficile, like some other bacterial pathogens (14, 47, 54), is able to adapt to antimicrobial peptides by increasing the expression of CAMP resistance genes.
The principles of CAMP resistance are similar among different bacterial species, but the mechanisms for achieving resistance vary considerably. Mechanisms of CAMP resistance identified in bacteria include increasing the net positive charge of the cell wall or membrane (repulsion) (10, 49), the proteolytic cleavage of CAMPs (degradation) (60), sequestration by proteins that bind CAMPs (trapping) (26, 32), and efflux pumping (export) (59). By selecting a mutant resistant to nisin, we isolated a strain that can grow in the presence of elevated concentrations of multiple CAMPs (nisin, gallidermin, and polymyxin B) due to a mutation in a predicted histidine kinase (cprK). We also identified a nearby putative ABC transporter operon, designated cprABC, that is directly involved in resistance to nisin and gallidermin (but not polymyxin B) and is regulated by CprK. The cprABC transporter operon and the putative histidine kinase encoded by cprK have sequence similarity to lantibiotic immunity systems found in bacteria that make lantibiotics (e.g., nisin), suggesting that resistance to CAMPs is accomplished through the export of the peptides by the ABC transporter. The fact that the cprABC mechanism can provide resistance to both nisin and gallidermin and, to a lesser extent, polymyxin B, is highly unusual, as lantibiotic immunity mechanisms are highly specific for individual peptides (55, 70) and cross-immunity is very rare and of limited range (2, 9, 20). Limited cross-immunity has been demonstrated for the immunity proteins of some bacteria that produce structurally similar lantibiotics (2, 20, 22). Low-level immunity to lacticin 3147 can be conferred by overexpressing transporter proteins from Bacillus licheniformis and Enterococcus faecium in an L. lactis background, although the resistance effects of these genes in their natural hosts are not clear (9). The native immunity mechanisms for the CAMPs tested (nisin, gallidermin, and polymyxin B) do not exhibit cross-immunity (22, 46). In contrast, the C. difficile cprABC and cprK CAMP resistance genes do not appear to be associated with a lantibiotic synthesis gene cluster, can protect against multiple CAMPs, and provide significant resistance to these compounds.
The immunity genes for lantibiotics and other bacteriocins are diverse but often are composed of an ABC transporter system with or without an additional distinct immunity gene (55). The immunity gene product may provide protection independently of the ABC transporter or function synergistically with the transporter to generate resistance (22, 45, 51). For example, immunity to the lantibiotic subtilin is conferred by an ABC transporter system (SpaFEG) in conjunction with the lipoprotein immunity factor SpaI, encoded in the gene order spaIFEG on the B. subtilis chromosome (30, 63). Separately, SpaI and SpaFEG confer partial immunity; both are required for full immunity to subtilin. A predicted lipoprotein, CD1348, also is encoded upstream of the cprABC genes. Unlike the case for spaI-like genes that typically are increased during lantibiotic production (52, 55, 62), no change in CD1348 expression was detected in the wild type or MC119, indicating that this gene is not induced in response to these CAMPs. Our results, however, do not rule out the possibility that CD1348 provides or is part of an independent, constitutively expressed mechanism of resistance to CAMPs.
The most likely scenario is that the extracellular domain of CprK senses CAMPs, and as a result either the kinase or phosphatase activity of the cytoplasmic domain becomes activated. A response regulator that controls the cprABC operon and other genes whose identities are not known then becomes altered in activity, resulting in the induction of cprABC and other CAMP resistance genes. It is evident that cprK is a positive regulator of cprABC and expression in response to CAMPs and also is autoregulated. We have attempted to create an insertional disruption in cprK to further define its role as a positive or negative regulator of the cpr genes, but these attempts thus far have been unsuccessful. As there are no apparent RR genes near cprK on the chromosome, the presumed RR is encoded at another location. The C. difficile genome encodes 51 putative response regulators, most of which are uncharacterized (58).
A BLAST homology search for the cprABC operon revealed homologs of these genes in all of the other C. difficile isolates sequenced to date, suggesting that this operon encodes a universal mechanism of CAMP resistance for the species. The fact that a cprABC-disrupted mutant (MC141) can eventually adapt, without additional mutations, to sub-MIC levels of nisin indicates that other, unidentified CAMP resistance mechanisms also are present in C. difficile. The implications of CAMP resistance in C. difficile may include resistance to CAMPs produced by the host innate immune system, resistance to bacteriocins and lantibiotics produced by indigenous bacteria, and resistance to food preservation processes involving bacteriocins. Considering the importance of CAMPs in defending against infection and the public health threat posed by C. difficile, the resistance mechanisms of these bacteria may present a target for the design of anti-infective therapeutics.
Acknowledgments
We thank J. Sorg, L. Bouillaut, B. Belitsky, R. Tamayo, A. Camilli, and M. Malamy for helpful suggestions and discussions during the course of this work and for criticism of the manuscript; C. Ellermeier (University of Iowa) for pCE240; L. Bouillaut for pBL58; J.P. van Pijkeren and the Britton laboratory at Michigan State University for the group II intron algorithm; and K. Bodi for help with Illumina analysis.
This work was supported by a research grant (AI057637 to A.L.S.) and a National Research Service Award (DK082156 to S.M.M.) from the U.S. National Institutes of Health, a Natalie V. Zucker Research grant to S.M.M., and a core facility grant (NS047243) to the Tufts University Center for Neuroscience Research.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 25 October 2010.
REFERENCES
- 1.Ananthakrishnan, A. N., E. L. McGinley, and D. G. Binion. 2008. Excess hospitalisation burden associated with Clostridium difficile in patients with inflammatory bowel disease. Gut 57:205-210. [DOI] [PubMed] [Google Scholar]
- 2.Aso, Y., K. Okuda, J. Nagao, Y. Kanemasa, N. Thi Bich Phuong, H. Koga, K. Shioya, T. Sashihara, J. Nakayama, and K. Sonomoto. 2005. A novel type of immunity protein, NukH, for the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. Biosci. Biotechnol. Biochem. 69:1403-1410. [DOI] [PubMed] [Google Scholar]
- 3.Barbut, F., D. Decre, V. Lalande, B. Burghoffer, L. Noussair, A. Gigandon, F. Espinasse, L. Raskine, J. Robert, A. Mangeol, C. Branger, and J. C. Petit. 2005. Clinical features of Clostridium difficile-associated diarrhoea due to binary toxin (actin-specific ADP-ribosyltransferase)-producing strains. J. Med. Microbiol. 54:181-185. [DOI] [PubMed] [Google Scholar]
- 4.Boman, H. G. 1991. Antibacterial peptides: key components needed in immunity. Cell 65:205-207. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet, M., M. M. Rafi, M. L. Chikindas, and T. J. Montville. 2006. Bioenergetic mechanism for nisin resistance, induced by the acid tolerance response of Listeria monocytogenes. Appl. Environ. Microbiol. 72:2556-2563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Calabi, E., F. Calabi, A. D. Phillips, and N. F. Fairweather. 2002. Binding of Clostridium difficile surface layer proteins to gastrointestinal tissues. Infect. Immun. 70:5770-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cao, M., and J. D. Helmann. 2004. The Bacillus subtilis extracytoplasmic-function sigmaX factor regulates modification of the cell envelope and resistance to cationic antimicrobial peptides. J. Bacteriol. 186:1136-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7a.Dineen, S. S., S. M. McBride, and A. L. Sonenshein. 2010. Integration of metabolism and virulence by Clostridium difficile CodY. J. Bacteriol. 192:5350-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dineen, S. S., A. C. Villapakkam, J. T. Nordman, and A. L. Sonenshein. 2007. Repression of Clostridium difficile toxin gene expression by CodY. Mol. Microbiol. 66:206-219. [DOI] [PubMed] [Google Scholar]
- 9.Draper, L. A., K. Grainger, L. H. Deegan, P. D. Cotter, C. Hill, and R. P. Ross. 2009. Cross-immunity and immune mimicry as mechanisms of resistance to the lantibiotic lacticin 3147. Mol. Microbiol. 71:1043-1054. [DOI] [PubMed] [Google Scholar]
- 10.Fabretti, F., C. Theilacker, L. Baldassarri, Z. Kaczynski, A. Kropec, O. Holst, and J. Huebner. 2006. Alanine esters of enterococcal lipoteichoic acid play a role in biofilm formation and resistance to antimicrobial peptides. Infect. Immun. 74:4164-4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fekety, R., and A. B. Shah. 1993. Diagnosis and treatment of Clostridium difficile colitis. JAMA 269:71-75. [PubMed] [Google Scholar]
- 12.Ghantoji, S. S., K. Sail, D. R. Lairson, H. L. DuPont, and K. W. Garey. 2010. Economic healthcare costs of Clostridium difficile infection: a systematic review. J. Hosp. Infect. 74:309-318. [DOI] [PubMed] [Google Scholar]
- 13.Giesemann, T., G. Guttenberg, and K. Aktories. 2008. Human alpha-defensins inhibit Clostridium difficile toxin B. Gastroenterology 134:2049-2058. [DOI] [PubMed] [Google Scholar]
- 14.Guo, L., K. B. Lim, C. M. Poduje, M. Daniel, J. S. Gunn, M. Hackett, and S. I. Miller. 1998. Lipid A acylation and bacterial resistance against vertebrate antimicrobial peptides. Cell 95:189-198. [DOI] [PubMed] [Google Scholar]
- 15.Hancock, R. E., and D. S. Chapple. 1999. Peptide antibiotics. Antimicrob. Agents Chemother. 43:1317-1323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hancock, R. E., and G. Diamond. 2000. The role of cationic antimicrobial peptides in innate host defences. Trends Microbiol. 8:402-410. [DOI] [PubMed] [Google Scholar]
- 17.Hancock, R. E., and M. G. Scott. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. U. S. A. 97:8856-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haraldsen, J. D., and A. L. Sonenshein. 2003. Efficient sporulation in Clostridium difficile requires disruption of the sigmaK gene. Mol. Microbiol. 48:811-821. [DOI] [PubMed] [Google Scholar]
- 19.Heap, J. T., O. J. Pennington, S. T. Cartman, G. P. Carter, and N. P. Minton. 2007. The ClosTron: a universal gene knock-out system for the genus Clostridium. J. Microbiol. Methods 70:452-464. [DOI] [PubMed] [Google Scholar]
- 20.Heidrich, C., U. Pag, M. Josten, J. Metzger, R. W. Jack, G. Bierbaum, G. Jung, and H. G. Sahl. 1998. Isolation, characterization, and heterologous expression of the novel lantibiotic epicidin 280 and analysis of its biosynthetic gene cluster. Appl. Environ. Microbiol. 64:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hennequin, C., C. Janoir, M. C. Barc, A. Collignon, and T. Karjalainen. 2003. Identification and characterization of a fibronectin-binding protein from Clostridium difficile. Microbiology 149:2779-2787. [DOI] [PubMed] [Google Scholar]
- 22.Hille, M., S. Kies, F. Gotz, and A. Peschel. 2001. Dual role of GdmH in producer immunity and secretion of the staphylococcal lantibiotics gallidermin and epidermin. Appl. Environ. Microbiol. 67:1380-1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hirokawa, T., S. Boon-Chieng, and S. Mitaku. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14:378-379. [DOI] [PubMed] [Google Scholar]
- 24.Hoch, J. A. 2000. Two-component and phosphorelay signal transduction. Curr. Opin. Microbiol. 3:165-170. [DOI] [PubMed] [Google Scholar]
- 25.Issa, M., A. N. Ananthakrishnan, and D. G. Binion. 2008. Clostridium difficile and inflammatory bowel disease. Inflamm. Bowel Dis. 14:1432-1442. [DOI] [PubMed] [Google Scholar]
- 26.Jin, T., M. Bokarewa, T. Foster, J. Mitchell, J. Higgins, and A. Tarkowski. 2004. Staphylococcus aureus resists human defensins by production of staphylokinase, a novel bacterial evasion mechanism. J. Immunol. 172:1169-1176. [DOI] [PubMed] [Google Scholar]
- 27.Karberg, M., H. Guo, J. Zhong, R. Coon, J. Perutka, and A. M. Lambowitz. 2001. Group II introns as controllable gene targeting vectors for genetic manipulation of bacteria. Nat. Biotechnol. 19:1162-1167. [DOI] [PubMed] [Google Scholar]
- 28.Karlstrom, O., B. Fryklund, K. Tullus, and L. G. Burman. 1998. A prospective nationwide study of Clostridium difficile-associated diarrhea in Sweden. The Swedish C. difficile Study Group. Clin. Infect. Dis. 26:141-145. [DOI] [PubMed] [Google Scholar]
- 29.Kirby, J. M., H. Ahern, A. K. Roberts, V. Kumar, Z. Freeman, K. R. Acharya, and C. C. Shone. 2009. Cwp84, a surface-associated cysteine protease, plays a role in the maturation of the surface layer of Clostridium difficile. J. Biol. Chem. 284:34666-34673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klein, C., and K. D. Entian. 1994. Genes involved in self-protection against the lantibiotic subtilin produced by Bacillus subtilis ATCC 6633. Appl. Environ. Microbiol. 60:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovács, M., A. Halfmann, I. Fedtke, M. Heintz, A. Peschel, W. Vollmer, R. Hakenbeck, and R. Bruckner. 2006. A functional dlt operon, encoding proteins required for incorporation of d-alanine in teichoic acids in gram-positive bacteria, confers resistance to cationic antimicrobial peptides in Streptococcus pneumoniae. J. Bacteriol. 188:5797-5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lauth, X., C. W. McNamara, S. Myskowski, E. Igwe, B. Beall, P. Ghosh, R. L. Gallo, and V. Nizet. 2004. A new virulence role for group A streptococcal M1 protein is protection against cathelicidin antimicrobial peptides, abstr. E-84. Abstr. 104th Gen. Meet. Am. Soc. Microbiol. American Society for Microbiology, Washington, DC.
- 33.Lawley, T. D., S. Clare, A. W. Walker, D. Goulding, R. A. Stabler, N. Croucher, P. Mastroeni, P. Scott, C. Raisen, L. Mottram, N. F. Fairweather, B. W. Wren, J. Parkhill, and G. Dougan. 2009. Antibiotic treatment of Clostridium difficile carrier mice triggers a supershedder state, spore-mediated transmission, and severe disease in immunocompromised hosts. Infect. Immun. 77:3661-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luria, S. E., and J. W. Burrous. 1957. Hybridization between Escherichia coli and Shigella. J. Bacteriol. 74:461-476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lyras, D., and J. I. Rood. 1998. Conjugative transfer of RP4-oriT shuttle vectors from Escherichia coli to Clostridium perfringens. Plasmid 39:160-164. [DOI] [PubMed] [Google Scholar]
- 36.Manganelli, R., R. Provvedi, C. Berneri, M. R. Oggioni, and G. Pozzi. 1998. Insertion vectors for construction of recombinant conjugative transposons in Bacillus subtilis and Enterococcus faecalis. FEMS Microbiol. Lett. 168:259-268. [DOI] [PubMed] [Google Scholar]
- 37.McDonald, L. C., G. E. Killgore, A. Thompson, R. C. Owens, Jr., S. V. Kazakova, S. P. Sambol, S. Johnson, and D. N. Gerding. 2005. An epidemic, toxin gene-variant strain of Clostridium difficile. N. Engl. J. Med. 353:2433-2441. [DOI] [PubMed] [Google Scholar]
- 38.McFarland, L. V., M. E. Mulligan, R. Y. Kwok, and W. E. Stamm. 1989. Nosocomial acquisition of Clostridium difficile infection. N. Engl. J. Med. 320:204-210. [DOI] [PubMed] [Google Scholar]
- 39.Mitaku, S., and T. Hirokawa. 1999. Physicochemical factors for discriminating between soluble and membrane proteins: hydrophobicity of helical segments and protein length. Protein Eng. 12:953-957. [DOI] [PubMed] [Google Scholar]
- 40.Mitaku, S., T. Hirokawa, and T. Tsuji. 2002. Amphiphilicity index of polar amino acids as an aid in the characterization of amino acid preference at membrane-water interfaces. Bioinformatics 18:608-616. [DOI] [PubMed] [Google Scholar]
- 41.Morozova, O., and M. A. Marra. 2008. Applications of next-generation sequencing technologies in functional genomics. Genomics 92:255-264. [DOI] [PubMed] [Google Scholar]
- 42.Müller, C. A., I. B. Autenrieth, and A. Peschel. 2005. Innate defenses of the intestinal epithelial barrier. Cell Mol. Life Sci. 62:1297-1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nizet, V. 2006. Antimicrobial peptide resistance mechanisms of human bacterial pathogens. Curr. Issues Mol. Biol. 8:11-26. [PubMed] [Google Scholar]
- 44.O'Connor, J. R., D. Lyras, K. A. Farrow, V. Adams, D. R. Powell, J. Hinds, J. K. Cheung, and J. I. Rood. 2006. Construction and analysis of chromosomal Clostridium difficile mutants. Mol. Microbiol. 61:1335-1351. [DOI] [PubMed] [Google Scholar]
- 45.Okuda, K., Y. Aso, J. Nakayama, and K. Sonomoto. 2008. Cooperative transport between NukFEG and NukH in immunity against the lantibiotic nukacin ISK-1 produced by Staphylococcus warneri ISK-1. J. Bacteriol. 190:356-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Otto, M., A. Peschel, and F. Gotz. 1998. Producer self-protection against the lantibiotic epidermin by the ABC transporter EpiFEG of Staphylococcus epidermidis Tu3298. FEMS Microbiol. Lett. 166:203-211. [DOI] [PubMed] [Google Scholar]
- 47.Pamp, S. J., M. Gjermansen, H. K. Johansen, and T. Tolker-Nielsen. 2008. Tolerance to the antimicrobial peptide colistin in Pseudomonas aeruginosa biofilms is linked to metabolically active cells, and depends on the pmr and mexAB-oprM genes. Mol. Microbiol. 68:223-240. [DOI] [PubMed] [Google Scholar]
- 48.Peschel, A. 2002. How do bacteria resist human antimicrobial peptides? Trends Microbiol. 10:179-186. [DOI] [PubMed] [Google Scholar]
- 49.Peschel, A., R. W. Jack, M. Otto, L. V. Collins, P. Staubitz, G. Nicholson, H. Kalbacher, W. F. Nieuwenhuizen, G. Jung, A. Tarkowski, K. P. van Kessel, and J. A. van Strijp. 2001. Staphylococcus aureus resistance to human defensins and evasion of neutrophil killing via the novel virulence factor MprF is based on modification of membrane lipids with l-lysine. J. Exp. Med. 193:1067-1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peschel, A., and H. G. Sahl. 2006. The co-evolution of host cationic antimicrobial peptides and microbial resistance. Nat. Rev. Microbiol. 4:529-536. [DOI] [PubMed] [Google Scholar]
- 51.Qiao, M., T. Immonen, O. Koponen, and P. E. Saris. 1995. The cellular location and effect on nisin immunity of the NisI protein from Lactococcus lactis N8 expressed in Escherichia coli and L. lactis. FEMS Microbiol. Lett. 131:75-80. [DOI] [PubMed] [Google Scholar]
- 52.Ra, R., M. M. Beerthuyzen, W. M. de Vos, P. E. Saris, and O. P. Kuipers. 1999. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145:1227-1233. [DOI] [PubMed] [Google Scholar]
- 53.Redelings, M. D., F. Sorvillo, and L. Mascola. 2007. Increase in Clostridium difficile-related mortality rates, United States, 1999-2004. Emerg. Infect. Dis. 13:1417-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sallum, U. W., and T. T. Chen. 2008. Inducible resistance of fish bacterial pathogens to the antimicrobial peptide cecropin B. Antimicrob. Agents Chemother. 52:3006-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saris, P. E., T. Immonen, M. Reis, and H. G. Sahl. 1996. Immunity to lantibiotics. Antonie Van Leeuwenhoek 69:151-159. [DOI] [PubMed] [Google Scholar]
- 56.Schmittgen, T. D., and K. J. Livak. 2008. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 3:1101-1108. [DOI] [PubMed] [Google Scholar]
- 57.Schwan, C., B. Stecher, T. Tzivelekidis, M. van Ham, M. Rohde, W. D. Hardt, J. Wehland, and K. Aktories. 2009. Clostridium difficile toxin CDT induces formation of microtubule-based protrusions and increases adherence of bacteria. PLoS Pathog. 5:e1000626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sebaihia, M., B. W. Wren, P. Mullany, N. F. Fairweather, N. Minton, R. Stabler, N. R. Thomson, A. P. Roberts, A. M. Cerdeno-Tarraga, H. Wang, M. T. Holden, A. Wright, C. Churcher, M. A. Quail, S. Baker, N. Bason, K. Brooks, T. Chillingworth, A. Cronin, P. Davis, L. Dowd, A. Fraser, T. Feltwell, Z. Hance, S. Holroyd, K. Jagels, S. Moule, K. Mungall, C. Price, E. Rabbinowitsch, S. Sharp, M. Simmonds, K. Stevens, L. Unwin, S. Whithead, B. Dupuy, G. Dougan, B. Barrell, and J. Parkhill. 2006. The multidrug-resistant human pathogen Clostridium difficile has a highly mobile, mosaic genome. Nat. Genet. 38:779-786. [DOI] [PubMed] [Google Scholar]
- 59.Shafer, W. M., X. Qu, A. J. Waring, and R. I. Lehrer. 1998. Modulation of Neisseria gonorrhoeae susceptibility to vertebrate antibacterial peptides due to a member of the resistance/nodulation/division efflux pump family. Proc. Natl. Acad. Sci. U. S. A. 95:1829-1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sieprawska-Lupa, M., P. Mydel, K. Krawczyk, K. Wojcik, M. Puklo, B. Lupa, P. Suder, J. Silberring, M. Reed, J. Pohl, W. Shafer, F. McAleese, T. Foster, J. Travis, and J. Potempa. 2004. Degradation of human antimicrobial peptide LL-37 by Staphylococcus aureus-derived proteinases. Antimicrob. Agents Chemother. 48:4673-4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Smith, C. J., S. M. Markowitz, and F. L. Macrina. 1981. Transferable tetracycline resistance in Clostridium difficile. Antimicrob. Agents Chemother. 19:997-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stein, T., S. Borchert, P. Kiesau, S. Heinzmann, S. Kloss, C. Klein, M. Helfrich, and K. D. Entian. 2002. Dual control of subtilin biosynthesis and immunity in Bacillus subtilis. Mol. Microbiol. 44:403-416. [DOI] [PubMed] [Google Scholar]
- 63.Stein, T., S. Heinzmann, S. Dusterhus, S. Borchert, and K. D. Entian. 2005. Expression and functional analysis of the subtilin immunity genes spaIFEG in the subtilin-sensitive host Bacillus subtilis MO1099. J. Bacteriol. 187:822-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tasteyre, A., M. C. Barc, A. Collignon, H. Boureau, and T. Karjalainen. 2001. Role of FliC and FliD flagellar proteins of Clostridium difficile in adherence and gut colonization. Infect. Immun. 69:7937-7940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Thomas, C. M., and C. A. Smith. 1987. Incompatibility group P plasmids: genetics, evolution, and use in genetic manipulation. Annu. Rev. Microbiol. 41:77-101. [DOI] [PubMed] [Google Scholar]
- 66.Viscidi, R., S. Willey, and J. G. Bartlett. 1981. Isolation rates and toxigenic potential of Clostridium difficile isolates from various patient populations. Gastroenterology 81:5-9. [PubMed] [Google Scholar]
- 67.Vonberg, R. P., E. J. Kuijper, M. H. Wilcox, F. Barbut, P. Tull, P. Gastmeier, P. J. van den Broek, A. Colville, B. Coignard, T. Daha, S. Debast, B. I. Duerden, S. van den Hof, T. van der Kooi, H. J. Maarleveld, E. Nagy, D. W. Notermans, J. O'Driscoll, B. Patel, S. Stone, and C. Wiuff. 2008. Infection control measures to limit the spread of Clostridium difficile. Clin. Microbiol. Infect. 14(Suppl. 5):2-20. [DOI] [PubMed] [Google Scholar]
- 68.Waligora, A. J., C. Hennequin, P. Mullany, P. Bourlioux, A. Collignon, and T. Karjalainen. 2001. Characterization of a cell surface protein of Clostridium difficile with adhesive properties. Infect. Immun. 69:2144-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Warny, M., J. Pepin, A. Fang, G. Killgore, A. Thompson, J. Brazier, E. Frost, and L. C. McDonald. 2005. Toxin production by an emerging strain of Clostridium difficile associated with outbreaks of severe disease in North America and Europe. Lancet 366:1079-1084. [DOI] [PubMed] [Google Scholar]
- 70.Willey, J. M., and W. A. van der Donk. 2007. Lantibiotics: peptides of diverse structure and function. Annu. Rev. Microbiol. 61:477-501. [DOI] [PubMed] [Google Scholar]
- 71.Wüst, J., and U. Hardegger. 1983. Transferable resistance to clindamycin, erythromycin, and tetracycline in Clostridium difficile. Antimicrob. Agents Chemother. 23:784-786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yanisch-Perron, C., J. Vieira, and J. Messing. 1985. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene 33:103-119. [DOI] [PubMed] [Google Scholar]