Abstract
Porphyromonas gingivalis is a Gram-negative obligate anaerobe that has been implicated in the etiology of adult periodontitis. We recently introduced a Drosophila melanogaster killing model for examination of P. gingivalis-host interactions. In the current study, the Drosophila killing model was used to characterize the host response to P. gingivalis infection by identifying host components that play a role during infection. Drosophila immune response gene mutants were screened for altered susceptibility to killing by P. gingivalis. The Imd signaling pathway was shown to be important for the survival of Drosophila infected by nonencapsulated P. gingivalis strains but was dispensable for the survival of Drosophila infected by encapsulated P. gingivalis strains. The P. gingivalis capsule was shown to mediate resistance to killing by Drosophila antimicrobial peptides (Imd pathway-regulated cecropinA and drosocin) and human beta-defensin 3. Drosophila thiol-ester protein II (Tep II) and Tep IV and the tumor necrosis factor (TNF) homolog Eiger were also involved in the immune response against P. gingivalis infection, while the scavenger receptors Eater and Croquemort played no roles in the response to P. gingivalis infection. This study demonstrates that the Drosophila killing model is a useful high-throughput model for characterizing the host response to P. gingivalis infection and uncovering novel interactions between the bacterium and the host.
Porphyromonas gingivalis is a Gram-negative, obligate anaerobe that has been strongly implicated as a pathogen in adult (chronic) periodontitis (23, 29), a polymicrobial inflammatory disease that affects the gingiva and other tooth-supporting structures. In order to characterize P. gingivalis-host interactions a number of animal infection models have been developed, the most common of which are rodent models (6, 20, 25, 28, 40, 44). Rodent models have been used to identify P. gingivalis components that are involved in pathogenesis (26, 32, 43, 46, 48, 52, 56, 57, 67, 73) and to characterize the host response to P. gingivalis infection (3, 6, 7, 13, 22, 31, 34, 35, 41, 74).
The use of the fruit fly Drosophila melanogaster has been well established for examining host-pathogen interactions (5, 16, 19, 21, 53, 55, 65, 66). Numerous studies have demonstrated the high degree of conservation between the Drosophila immune system and the mammalian innate immune system (reviewed in reference 49). Like the mammalian innate immune system, the Drosophila immune system detects the presence of invading microbes by using pattern recognition receptors (PRRs), which recognize pathogen-associated molecular patterns (PAMPs) and trigger an immune response that is specific for the class of invading microbe. Other mammalian immune response features that are conserved in Drosophila include signaling pathways (e.g., Toll/interleukin-1 receptor [IL1R], tumor necrosis factor receptor [TNFR]), antimicrobial peptides (AMPs), macrophage-like blood cells, complement C3/α2-macroglobulin (C3/α2M) superfamily proteins, cytokines, reactive oxygen and nitrogen species, and iron-sequestering proteins. The use of Drosophila as a model host offers several important advantages. The absence of an adaptive immune response makes the model useful for studying pathogen interactions with the host's innate immune response in isolation. The Drosophila genome sequence is known, and the Berkeley Drosophila Genome Project has successfully inactivated 40% of the currently annotated Drosophila genes (8), with ongoing efforts to eventually inactivate all genes. Drosophila is genetically amenable, and well-developed genetic technologies are available that facilitate the identification of host factors that promote or fight infection (4, 12, 53, 70). Additionally, their short generation time, ease of use, and affordability allow for the use of sample sizes that are large enough to permit statistical analysis of the data.
We have recently developed a Drosophila killing model for examining P. gingivalis-host interactions (37). We observed that P. gingivalis is pathogenic in Drosophila and that differences in the virulence of P. gingivalis strains can be observed with the Drosophila killing model. Multiple P. gingivalis components are involved in the killing of Drosophila, and they are also involved in virulence in mammals. Additionally, our data suggest that P. gingivalis killing of Drosophila involves mechanisms that are host mediated. The objective of the current study was to use the Drosophila killing model to characterize the host response to P. gingivalis infection. Specifically, Drosophila immune response gene mutants were screened to identify host factors that play a role during infection. The Drosophila Imd pathway was found to be important for the immune response against infection by unencapsulated P. gingivalis, and in a novel finding the P. gingivalis capsule was shown to be involved in mediating resistance to antimicrobial peptides. Drosophila thiol-ester proteins II and IV and cytokine Eiger (TNF homolog, Janus kinase [JNK] pathway ligand) were also involved in the response against P. gingivalis infection.
MATERIALS AND METHODS
Bacterial and Drosophila strains and growth conditions.
Bacterial and Drosophila strains used in this study are described in Table 1. P. gingivalis strains were grown on brucella blood agar (BBA; Anaerobe Systems) at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). Drosophila stocks were maintained and propagated at 26°C in standard culture vials containing corn flour-molasses medium. Only 3- to 5-day-old female flies were used in experiments. Transheterozygous eater null progeny [Df(3R)D605/Df(3R)TI-l e1] were generated as previously described (45) by crossing the deficiency lines B32 [Df(3R)D605/TM3 Ser,GFP] and B34 [Df(3R)Tl-I e1/TM3 Ser,GFP].
TABLE 1.
Bacterial and Drosophila strains used in this study
| Strain or stock | Description or genotype | Source |
|---|---|---|
| P. gingivalis strains | ||
| W83 | Lab strain | Margaret Duncan |
| 381 | Lab strain | Joseph Zambon |
| W50 | Lab strain; renamed W50UK for these studies; wt for GPC | Mike Curtis (1) |
| GPC | Capsule mutant | Mike Curtis (1) |
| E. coli DH5α | Invitrogen | |
| D. melanogaster | ||
| Canton S | Wild type | |
| key1 | Kenny null; yw DD1;cn bw key1 | Neal Silverman (60) |
| DIG 672 | ywdpt-lacZ Drs-GFP | Bruno Lemaitre |
| 15936 | Tep IV null; y1w67c23;P{EPgy2}TepIVEY04656 | Bloomington Stock Center (8) |
| 20939 | Croquemort null; y1w67c23;P{EPgy2}crqEY14489 | Bloomington Stock Center (8) |
| f02756 | Tep II null; PBac{WH}TepIIf02756 | Exelexis Stock Center (8) |
| eiger3 | w[1118]egr[3] | Masayuki Miura (36) |
| w1118 | w[1118]; wt for eiger3 | Masayuki Miura (36) |
| BOS 32 | Df(3R)D605/TM3 Ser,GFP | Christine Kocks (45) |
| BOS 34 | Df(3R)Tl-I e1/TM3 Ser,GFP | Christine Kocks (45) |
| Eater | Eater null; Df(3R)D605/Df(3R)TI-l e1 | This study, and as previously described (45) |
Infection of adult female Drosophila.
Bacterial strains were grown in 40 ml of Trypticase soy broth (TSB) for 24 h at 37°C. Escherichia coli was grown aerobically with shaking, while P. gingivalis was supplemented with hemin (5 μg/ml) and vitamin K (1 μg/ml) and incubated anaerobically. The bacteria were harvested at 3,150 × g for 8 min and diluted in TSB to an optical density at 600 nm (OD600) of 2.0 (1.09 × 1011 CFU ml−1 of P. gingivalis; 1 × 1011 CFU ml−1 of E. coli DH5α). The bacteria were introduced into the hemocoel (body cavity) of CO2-anesthetized Drosophila through the thorax, using a 30-gauge (G) needle dipped into 500 μl of bacterial culture or sterile TSB for mock infections (vector controls [VC]). The Drosophila flies were returned to the original culture vials, and the number of surviving animals at time 0 h was recorded. The animals were incubated at 30°C, and the number of dead animals was recorded every 12 h for 7 days. All experiments were repeated.
β-Galactosidase assay and microscopy.
Drosophila were killed 4 h postinfection by immersion in 95% ethanol then rinsed once in 1× phosphate-buffered saline (PBS, pH 7.4), and holes were made in the cuticle of the dead animals by using a 30-G needle to facilitate penetration of the fixative (10). Fixing the carcasses and assaying for β-galactosidase activity were performed as previously described (9), with modifications. Drosophila carcasses were fixed in 5% paraformaldehyde in 1× PBS for 10 min, rinsed three times (10 min, 10 min, and 20 min) in 1× PBS, and incubated at 37°C for 24 h in staining solution (1.8 mM magnesium chloride, 0.9× PBS [pH 7.3], 5 mM potassium ferrocyanide, 5 mM potassium ferricyanide, 1.2 mg/ml 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside [X-Gal] in dimethylformamide). After staining, the carcasses were washed in 1× PBS for 2 h and immediately viewed under a microscope. Bright-field images were taken of representative carcasses by using a Nikon Eclipse E600 microscope equipped with a Nikon DXM 1200 digital camera.
AMP susceptibility assay.
P. gingivalis strains W50UK and GPC were grown in 40 ml of TSB with hemin (5 μg/ml), vitamin K (1 μg/ml), and anaerobic incubation until mid-log phase (13 to 17 h) and then harvested by centrifugation at 3,150 × g for 11 min. The bacteria were washed once in 2 volumes of 1× PBS and diluted to approximately 4 × 106 CFU/ml in 1× PBS with 1% TSB. Fifty microliters of this suspension was mixed in Eppendorf tubes with various concentrations of cecropin A from Hyalophora cecropia (Bachem, Torrance CA), drosocin from Drosophila melanogaster (Biosynthesis, Lewisville, TX), human beta-defensin-3 (hβd3; Aaron Weinberg, Case Western Reserve University), or 1× PBS for controls, in a final volume of 100 μl. The mixtures were incubated anaerobically at 37°C for 3 h, after which serial dilutions were spread on BBA plates and incubated anaerobically at 37°C for 6 days. Colony counts were determined, and the results are expressed as percentages of the colony counts of bacteria not exposed to AMPs. The experiments were performed with duplicate samples (triplicates for controls) on four independent occasions.
Statistical methods. (i) Sample size.
Power calculations based on pilot data estimated that a sample size of 136 Drosophila animals per group would be sufficient to detect a relative risk of mortality (RR) of at least 2.0 at an α-level of 0.05 with 90% power when comparing different infections. A sample size of 150 animals per group was used in all experiments. Depending on the number of experimental groups involved, each experiment was divided into four or five parts for feasibility. The experiments were repeated for a total sample size of 300 Drosophila animals per group.
(ii) Data analysis.
Survival data were analyzed using the SAS statistical software package (SAS Institute, Cary, NC). A Cox proportional hazards (P-H) model was fitted to the survival data. Likelihood ratio tests were performed, and RR values were obtained from the fitted Cox P-H model and adjusted for the individual “experiments” and “parts.” RR values with P values of <0.05 were considered significant.
The results of the antimicrobial peptide susceptibility assays are presented as means ± standard deviations of four independent experiments. Statistical analyses were performed via an unpaired, two-tailed t test using JMP software (SAS, Cary, NC). A P value of <0.05 was considered significant.
RESULTS
The rationale used to select Drosophila immune response components to test for a role in the response to P. gingivalis infection was as follows: Lemaitre et al. compiled a list of 370 Drosophila genes (50) that are induced in response to septic injury (17), known to function in immunity, or encode proteins with homology to immune response components of other organisms. From this list, we identified genes that encode proteins with homology to mammalian proteins for which we could predict a role in the interaction with P. gingivalis and for which Drosophila mutants are readily available. Using this rationale we selected six Drosophila immune response components (Tables 1 and 2) to test for a role in the response to P. gingivalis infection. Although the selected components play important and varied roles in the Drosophila immune response, they represent only a small fraction of the Drosophila immune response components. The Drosophila immune response gene mutants have all been previously characterized. Groups of wild-type (wt) and immune response-defective Drosophila animals were infected with P. gingivalis strain W83, strain 381, or mock infected, and the survival of the animals was compared. Based on results presented in our other report also appearing in this issue (37), between 5.10 × 103 CFU and 3.17 × 104 CFU of P. gingivalis were inoculated into the animals. RR values for pairwise comparisons of the survival of immune response-deficient Drosophila versus wt Drosophila animals are shown in Table 2. An RR value greater than 1 indicates that the Drosophila immune response gene mutant was more likely than the wt Drosophila animals to die from the P. gingivalis infection.
TABLE 2.
Drosophila immune response components tested for a role in the immune response to infection with various P. gingivalis strains
| Drosophila immune response component (comparison strains) | P. gingivalis infection strain | RR (comparison strain vs wt) | P valuea | Component involved? |
|---|---|---|---|---|
| Imd signaling pathway (key1 vs wt) | W83 (encapsulated) | 1 | 0.8 | No |
| 381 (unencapsulated) | 1.4 | 0.006 | Yes | |
| W50UK (encapsulated) | 1 | 0.7 | No | |
| GPC (W50UK mutant, unencapsulated) | 2.2 | <0.0001 | Yes | |
| Tep II (tepII−/− vs wt) | W83 | 1.68 | <0.0001 | Yes |
| 381 | 2.45 | <0.0001 | Yes | |
| Tep IV (tepIV−/− vs wt) | W83 | 1.5 | <0.0001 | Yes |
| 381 | 1.5 | 0.0006 | Yes | |
| Eiger (egr−/− vs wt) | W83 | 1.32 | 0.009 | Yes |
| 381 | 1.47 | 0.007 | Yes | |
| Croquemort (crq−/− vs wt) | W83 | 1.1 | 0.4 | No |
| 381 | 0.9 | 0.4 | No | |
| Eater (eater−/− vs wt)b | W83 | >0.9999 | No | |
| 381 | >0.9957 | No |
RR values for which P was <0.05 are shown in bold.
Because eater−/− animals are naturally more susceptible to death by injury than wt animals, the reported P values are for comparisons of the RR of eater−/− versus wt flies infected with W83 or 381 to the RR of mock-infected eater−/− versus wt flies.
Imd signaling pathway.
The Imd signaling pathway is homologous to the mammalian TNF receptor signaling pathway (39, 49), although they differ at the level of activation/detection. The Imd pathway is a major regulator of Drosophila immune response genes (18), most notably AMP genes. In fat body cells (the functional equivalent of the liver), the detection of Gram-negative-type peptidoglycan (PPG) by peptidoglycan recognition protein LC (PGRP-LC) activates the pathway. Activation of the Imd pathway triggers a signal transduction cascade that culminates in the activation of the NF-κB family protein Relish, which then activates the transcription of AMPs, e.g., diptericin, and other immune response genes. In key1 Drosophila the kenny gene, which encodes a subunit of the IκB kinase complex, is inactivated, which inactivates the Imd pathway (60). key1 Drosophila flies therefore tend to be highly susceptible to infections with Gram-negative bacteria, e.g., E. coli (27, 60).
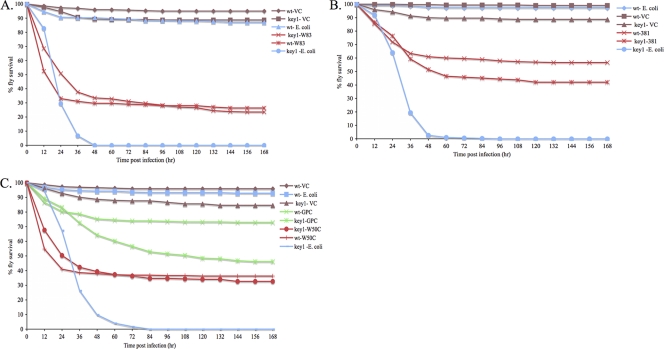
Survival curves of infected and mock-infected wt and key1 flies are shown in Fig. 1. key1 Drosophila were significantly more likely to die than wt Drosophila from an Escherichia coli DH5α infection, as previously reported (60). No difference was observed in the survival of wt and key1 Drosophila animals infected with P. gingivalis strain W83 (Fig. 1A; Table 2). To determine whether the capsule that surrounds strain W83 plays a role in the nullification of the Imd pathway, the survival rates of wt and key1 animals infected with the acapsular P. gingivalis strain 381 were compared (Fig. 1B). key1 Drosophila flies were more likely to die than wt Drosophila flies from an infection with strain 381 (Fig. 1B; Table 2). To further examine the role of the P. gingivalis capsule in the nullification of the Imd pathway, Drosophila flies were infected with P. gingivalis strain W50UK or its isogenic capsule mutant, GPC (1), and the survival rates of the animals were compared (Fig. 1C). Strain W50UK is highly similar to strain W83 (51). No difference was observed in the survival of wt and key1 Drosophila flies infected with the encapsulated strain W50UK; however, key1 Drosophila flies were significantly more likely to die than wt Drosophila flies from the GPC (acapsular mutant) infection (Fig. 1C; Table 2).
FIG. 1.
Survival rates of P. gingivalis-, E. coli DH5α-, and mock-infected wt and key1 Drosophila animals. wt and key1 Drosophila flies were infected with the indicated bacteria. The legend labels are of the form Drosophila strain-infection agent. (A) Survival of W83-, E. coli DH5α-, and mock-infected Drosophila animals. (B) Survival of 381-, E. coli DH5α-, and mock-infected Drosophila animals. (C) Survival of W50UK-, GPC (capsule mutant)-, E. coli DH5α-, and mock-infected Drosophila animals.
To determine whether the P. gingivalis capsule can hide the bacterium's peptidoglycan, thereby preventing activation of the Imd pathway, Drosophila animals that express β-galactosidase under the control of the diptericin promoter (dipt-LacZ) were used to monitor activation of the Imd pathway in response to W50UK, GPC, and mock infection. Images of representative animals are shown in Fig. 2. Animals infected with strain W50UK (encapsulated) as well as animals infected with strain GPC (acapsular mutant) displayed robust β-galactosidase activity at 4 h postinfection, while mock-infected animals (VC) displayed little or no β-galactosidase activity. The Imd pathway was activated by both the encapsulated and capsule mutant strains of W50UK.
FIG. 2.
Imd signaling pathway activation by P. gingivalis infection, as measured using a dipt-LacZ reporter. dipt-LacZ animals were infected with strain W50UK, strain GPC, or mock (VC) infected and killed 4 h postinfection. Representative animals from each of the infection groups and the tubes in which the assays were performed are shown.
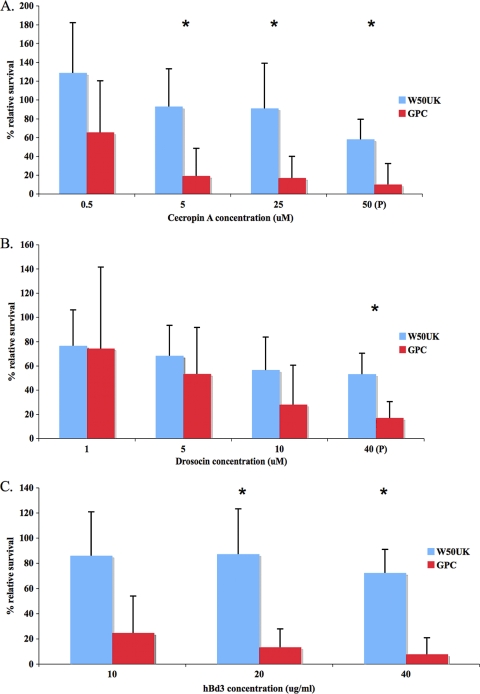
To determine whether the P. gingivalis capsule can provide resistance to Imd pathway-regulated AMPs, strains W50UK and GPC were exposed to a range of concentrations of cecropin A (from Hyalophora cecropia) and drosocin (from Drosophila melanogaster) up to their reported physiological concentrations (49, 61), and the survival rates of the bacteria were compared. Cecropin A from Hyalophora cecropia was used because it has an identical activity spectrum as cecropin A from Drosophila (61) and is readily available. The relative survival rates of W50UK versus GPC bacteria after exposure to cecropin A and drosocin are shown in Fig. 3A and B, respectively. Strain GPC was significantly (P < 0.05) more sensitive than strain W50UK to killing by cecropinA and drosocin, although only at the physiological concentration for drosocin. Strain GPC was also more sensitive than W50UK to killing by hβd3 (Fig. 4C) (P < 0.05).
FIG. 3.
Survival of P. gingivalis strains W50UK and GPC after exposure to insect and human antimicrobial peptides. P, physiological concentration (49, 60). Survival rates of bacteria after exposure to different concentrations of cecropin A (A), drosocin (B), or hβd3 (C) are shown. Each point represents the mean and standard deviation of eight samples from four independent experiments, and statistically significant (P < 0.05) survival differences between W50UK and GPC are indicated by asterisks.
FIG. 4.
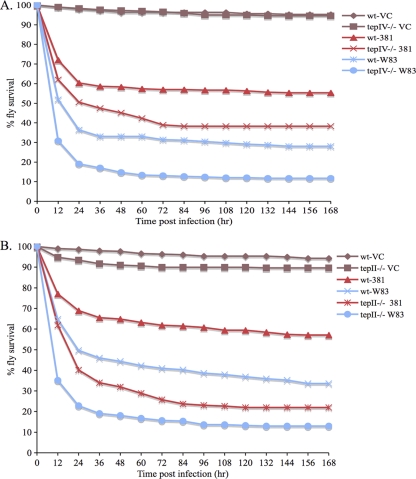
Survival curves of wt, tep II−/− and tep IV−/− Drosophila flies infected with P. gingivalis strains W83 or 381. wt and tep−/− Drosophila flies were infected with strain W83, strain 381, or mock infected (VC). Labels are of the form Drosophila strain-infection agent. (A) Survival of wt and tep II−/− Drosophila flies; (B) survival of wt and tep IV−/− Drosophila flies.
Teps II and IV.
Teps I to IV are members of the complement C3/α2M superfamily of proteins (2, 11, 47, 49). Teps are expressed by Drosophila fat body cells and plasmatocytes (47) in response to Upd3 (cytokine) signaling via the JAK/STAT pathway. Proteins in this family are involved in the opsonization of pathogens for phagocytosis (complement) and the inhibition of a broad spectrum of proteases (α2M). Three Teps (I, II, and IV) are strongly upregulated in response to bacterial challenge (47).
Tep II and Tep IV were tested for a role in the Drosophila immune response to P. gingivalis infection. Survival curves of infected and mock-infected wt and tep−/− animals are shown in Fig. 4. tepII−/− Drosophila flies were more likely to die than wt Drosophila flies from infections with P. gingivalis strains W83 and 381 (Fig. 4A; Table 2). Similarly, tepIV−/− Drosophila flies were more likely to die than wt Drosophila flies from infections with P. gingivalis strains W83 and 381 (Fig. 4B; Table 2). The data suggest a role for Tep II and Tep IV in the Drosophila immune response to P. gingivalis infection.
Eiger.
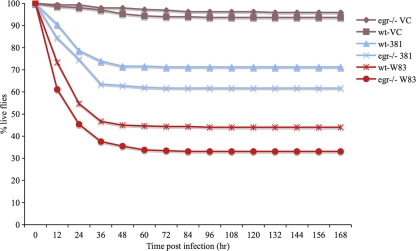
Eiger is the sole TNF superfamily homolog in Drosophila (42, 54), which upon binding to its receptor Wengen activates the JNK signaling pathway (36, 42, 54). Eiger was tested for a role in the Drosophila immune response to P. gingivalis infection. Survival curves of infected and mock-infected wt and egr−/− animals are shown in Fig. 5. egr−/− Drosophila animals were more likely to die than wt Drosophila animals from infections with P. gingivalis strains W83 and 381 (Fig. 5; Table 2), demonstrating a role for Eiger in the Drosophila immune response to P. gingivalis infection.
FIG. 5.
Survival curves for wt and egr−/− Drosophila flies infected with P. gingivalis strain W83 or 381. wt and egr−/− Drosophila flies were infected with strain W83, strain 381, or mock infected (VC). Labels are of the form Drosophila strain-infection agent.
Croquemort.
Croquemort is a CD36 superfamily protein that is expressed on Drosophila plasmatocytes (24). It is a scavenger receptor that has been implicated in the phagocytosis of S. aureus (71). No differences were observed in the survival of crq−/− versus wt Drosophila animals infected with P. gingivalis strains W83 and 381 (Table 2), suggesting that Croquemort plays no role in the Drosophila immune response to P. gingivalis infection.
Eater.
Eater is an epidermal growth factor (EGF) domain-containing scavenger receptor that is expressed on Drosophila plasmatocytes and is involved in the phagocytosis of Gram-positive (Staphylococcus aureus) and Gram-negative (E. coli) bacteria (45). The most similar mammalian protein to Eater is SREC-I (scavenger receptor expressed by endothelial cells I). Mock-infected eater−/− Drosophila animals were more likely to die than mock-infected wt Drosophila animals (data not shown), suggesting that the eater−/− animals are defective in wound healing. The differences in the survival of eater−/− versus wt Drosophila animals infected with P. gingivalis strains W83 and 381 were similar to the differences in survival of mock-infected eater−/− versus wt animals (Table 2), suggesting no role for Eater in the Drosophila immune response to P. gingivalis infection.
DISCUSSION
We recently developed a Drosophila melanogaster killing model for examining P. gingivalis-host interactions (37), and in the current study the killing model was used to characterize the host response to P. gingivalis infection. The Drosophila model has been widely used to examine the host responses to many other bacterial pathogens. For example, Brandt et al. observed that the Drosophila cytokine Eiger (TNF homolog) contributes to Salmonella enterica serovar Typhimurium-induced pathology (12), and Mansfield et al. observed that the Toll pathway is important for the Drosophila immune response against Listeria monocytogenes infection (53).
In this study the Drosophila Imd signaling pathway was shown to be important for the immune response against unencapsulated strains of P. gingivalis but ineffective for the immune response against encapsulated strains of the bacterium. key1 Drosophila flies infected with the encapsulated P. gingivalis strain W83 survived as well as wt Drosophila flies with the same infection, indicating that the Imd signaling pathway is dispensable for the immune response against W83 infection. This is unlike E. coli DH5α infection in Drosophila, for which a functional Imd pathway is necessary and sufficient to control the infection (27, 60). We hypothesized that the capsule, which is present on strain W83 and absent from E. coli DH5α, may be involved in nullification of the Imd pathway by the bacterium. To test this hypothesis, the survival of wt and key1 Drosophila flies infected with P. gingivalis strain W50UK (highly similar if not identical to strain W83) versus its isogenic capsule mutant (GPC) were compared. Similarly to the W83 infection, infection with W50UK did not result in significant differences in the survival of key1 and wt Drosophila animals; however, key1 Drosophila animals were significantly more susceptible than wt Drosophila animals to killing by the capsule mutant GPC. To summarize, when the capsule is removed from P. gingivalis strain W50UK the Imd signaling pathway becomes relevant for the Drosophila immune response against the infection. Thus, the capsule plays a role in the nullification of the Imd pathway by encapsulated P. gingivalis strains (e.g., W83 and W50UK). The presence of a capsule on P. gingivalis could shield the bacterium's PPG from detection by PGRP-LC, thus preventing activation of the pathway. The use of a dipt-LacZ reporter to monitor Imd pathway activation in response to W50UK and GPC (capsule mutant) infections demonstrated that the pathway was activated in response to both infections. Thus, the P. gingivalis capsule does not shield the bacterium's PPG from detection by PGRP-LC. It is also possible that the P. gingivalis capsule protects the bacterium from the actions of Imd pathway-regulated immune effectors. For example, the capsule could act as a physical barrier to Drosophila AMPs, limiting their interaction with the P. gingivalis cell membrane and thereby protecting the bacteria from AMP-induced killing, while the absence of a capsule could render P. gingivalis more sensitive to killing by AMPs. The sensitivities of strains W50UK and GPC to killing by the Imd-regulated AMPs cecropin A and drosocin were therefore tested. Cecropins kill bacteria by permeabilizing the cell membrane, which causes lysis (69). Drosocin functions intracellularly, entering bacterial cells via lipopolysaccharide-mediated entry and kills them by interfering with the actions of DnaK (58). Strain W50UK was more resistant than its capsule mutant GPC to killing by the antimicrobial peptides, demonstrating that the P. gingivalis capsule is involved in the resistance to killing by Drosophila Imd-regulated AMPs. These results are physiologically relevant, as the AMP concentrations that were tested are within the range reported to be present in Drosophila hemolymph (49, 61). The findings are also relevant to humans, as strain W50UK was also more resistant than GPC to killing by hβd3, which has the same mode of action as cecropin A. The involvement of the P. gingivalis capsule in mediating resistance to host AMPs is a novel finding; however, similar protective effects of bacterial capsules against host AMPs have been observed with Klebsiella pneumoniae (14) and Neisseria meningitidis (68). It is important to note that in addition to AMPs the Imd pathway activates multiple immune response genes and processes (an estimated 30% of Drosophila immune response genes are regulated by the pathway). Therefore, P. gingivalis capsule interaction with and nullification of Imd pathway-regulated responses are likely to be multifaceted. The Imd pathway is a major regulator of Drosophila immune response genes (18), and although it is activated in response to infection by encapsulated P. gingivalis, it is dispensable for the survival of the animals. Therefore, Imd activation by encapsulated P. gingivalis could be directly toxic and/or energetically wasteful to Drosophila and could contribute to the pathology induced by the bacterium in this model. Schneider et al. suggested that Eiger signaling induced upon infection by some intracellular bacteria, which does not help fight infection, results in lethality via mechanisms that may be directly toxic or energetically wasteful (66). As heat-killed P. gingivalis kills Drosophila as readily as live P. gingivalis (37), nullification of the Imd pathway by the P. gingivalis capsule likely involves mechanisms that do not require bacterial viability.
This study demonstrated that the C3/α2M superfamily proteins Tep II and Tep IV are involved in the Drosophila immune response against P. gingivalis infection. Tep II appears to play a bigger role in the response to infection with strain 381 versus strain W83 (Fig. 4A). It has been suggested that Tep II functions as an α2M and that alternative splicing (38) serves to increase the diversity of proteases that can be inhibited by this protein (11). At least one Tep II isoform and Tep IV have been demonstrated to function as opsonins in the phagocytosis of E. coli and S. aureus, respectively (70). It is likely that Tep IV functions as an opsonin in the response to P. gingivalis infection, while Tep II may function as both an opsonin and a protease inhibitor by virtue of its multiple isoforms. It has been demonstrated that purified human α2M can inhibit the activity of P. gingivalis arginine-specific proteases in vitro (30, 59) and that C3 can bind to (15, 63) and promote the phagocytosis of (64) P. gingivalis in vitro. The results of this study provide strong evidence of an in vivo role for C3/α2M family proteins in the host immune response against P. gingivalis infection.
This study demonstrated that Eiger (TNF homolog) is involved in the Drosophila immune response against P. gingivalis infection. Schneider et al. showed that Eiger fights infection by extracellular pathogens (66) and suggested that Eiger signaling increases the potency of Drosophila phagocytes against these microbes. Eiger also contributes to pathology during infections by some facultative intracellular pathogens (12, 66).
There are a number of transgenic mouse strains with defined immune response gene mutations that have been used to study P. gingivalis-host interactions (3, 7, 13, 33, 34, 62, 72, 74). However, the powerful genetics of Drosophila has made it more feasible to generate a large group of animals with defined mutations in immune response genes and other genes. The availability of a large number of Drosophila immune response mutants will facilitate large-scale screening to identify host components that play a role during P. gingivalis infection. Also, as the Drosophila immune response continues to be characterized, additional components and pathways that could potentially interact with the bacterium will be identified. Additionally, well-developed Drosophila genetic tools, e.g., microarray and RNA interference libraries are available for genome-wide analysis of P. gingivalis-Drosophila interactions. As with any animal model, the Drosophila killing model has its limitations. Drosophila is not a natural host for P. gingivalis, and due to the chronic, polymicrobial, multifactorial nature of adult periodontitis, the Drosophila killing model does not mimic the natural disease process. P. gingivalis interactions with the host's adaptive immune system cannot be studied in Drosophila, which lacks this system. While the Drosophila model cannot be used to identify all host components that are involved in the immune response to P. gingivalis infection, the results of this study clearly show that important host factors that are involved in the immune response to P. gingivalis infection can be identified and novel findings about the interaction between the bacterium and the host can be made using this model.
The results of this study demonstrate that Drosophila melanogaster is a powerful model system for characterizing the host response to P. gingivalis infection. We have identified several Drosophila immune system components that are important in the response to P. gingivalis infection. The interaction between P. gingivalis and Drosophila is clearly multifaceted, and there are likely additional host factors involved. Future studies involving genome-wide examination of the Drosophila response to P. gingivalis infection should provide new insights into the interaction between the bacterium and the host.
Acknowledgments
We thank Amanda Simcox (Ohio State University) for helpful advice, discussions, demonstrations, and for the kindly supplied Drosophila husbandry materials. We thank Aaron Weinberg and Zhinmin Feng (Case Western Reserve University) for providing hβd3 and for helpful discussions. We thank Gregory Young (Ohio State University) for help with statistical analyses.
This study was supported by grant DE10467 from the National Institutes of Health.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Aduse-Opoku, J., J. M. Slaney, A. Hashim, A. Gallagher, R. P. Gallagher, M. Rangarajan, K. Boutaga, M. L. Laine, A. J. Van Winkelhoff, and M. A. Curtis. 2006. Identification and characterization of the capsular polysaccharide (K-antigen) locus of Porphyromonas gingivalis. Infect. Immun. 74:449-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Agaisse, H., and N. Perrimon. 2004. The roles of JAK/STAT signaling in Drosophila immune responses. Immunol. Rev. 198:72-82. [DOI] [PubMed] [Google Scholar]
- 3.Alayan, J., S. Ivanovski, E. Gemmell, P. Ford, S. Hamlet, and C. S. Farah. 2006. Deficiency of iNOS contributes to Porphyromonas gingivalis-induced tissue damage. Oral Microbiol. Immunol. 21:360-365. [DOI] [PubMed] [Google Scholar]
- 4.Apidianakis, Y., M. N. Mindrinos, W. Xiao, G. W. Lau, R. L. Baldini, R. W. Davis, and L. G. Rahme. 2005. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc. Natl. Acad. Sci. U. S. A. 102:2573-2578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Apidianakis, Y., L. G. Rahme, J. Heitman, F. M. Ausubel, S. B. Calderwood, and E. Mylonakis. 2004. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot. Cell 3:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baker, P. J., R. T. Evans, and D. C. Roopenian. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035-1040. [DOI] [PubMed] [Google Scholar]
- 7.Baker, P. J., L. Howe, J. Garneau, and D. C. Roopenian. 2002. T cell knockout mice have diminished alveolar bone loss after oral infection with Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 34:45-50. [DOI] [PubMed] [Google Scholar]
- 8.Bellen, H. J., R. W. Levis, G. Liao, Y. He, J. W. Carlson, G. Tsang, M. Evans-Holm, P. R. Hiesinger, K. L. Schulze, G. M. Rubin, R. A. Hoskins, and A. C. Spradling. 2004. The BDGP gene disruption project: single transposon insertions associated with 40% of Drosophila genes. Genetics 167:761-781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bellen, H. J., C. J. O'Kane, C. Wilson, U. Grossniklaus, R. K. Pearson, and W. J. Gehring. 1989. P-element-mediated enhancer detection: a versatile method to study development in Drosophila. Genes Dev. 3:1288-1300. [DOI] [PubMed] [Google Scholar]
- 10.Bello, B., D. Resendez-Perez, and W. J. Gehring. 1998. Spatial and temporal targeting of gene expression in Drosophila by means of a tetracycline-dependent transactivator system. Development 125:2193-2202. [DOI] [PubMed] [Google Scholar]
- 11.Blandin, S., and E. A. Levashina. 2004. Thioester-containing proteins and insect immunity. Mol. Immunol. 40:903-908. [DOI] [PubMed] [Google Scholar]
- 12.Brandt, S. M., M. S. Dionne, R. S. Khush, L. N. Pham, T. J. Vigdal, and D. S. Schneider. 2004. Secreted bacterial effectors and host-produced Eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol. 2:e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burns, E., G. Bachrach, L. Shapira, and G. Nussbaum. 2006. Cutting edge. TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J. Immunol. 177:8296-8300. [DOI] [PubMed] [Google Scholar]
- 14.Campos, M. A., M. A. Vargas, V. Regueiro, C. M. Llompart, S. Alberti, and J. A. Bengoechea. 2004. Capsule polysaccharide mediates bacterial resistance to antimicrobial peptides. Infect. Immun. 72:7107-7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cutler, C. W., R. R. Arnold, and H. A. Schenkein. 1993. Inhibition of C3 and IgG proteolysis enhances phagocytosis of Porphyromonas gingivalis. J. Immunol. 151:7016-7029. [PubMed] [Google Scholar]
- 16.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Gregorio, E., P. T. Spellman, G. M. Rubin, and B. Lemaitre. 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U. S. A. 98:12590-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Gregorio, E., P. T. Spellman, P. Tzou, G. M. Rubin, and B. Lemaitre. 2002. The Toll and Imd pathways are the major regulators of the immune response in Drosophila. EMBO J. 21:2568-2579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dionne, M. S., N. Ghori, and D. S. Schneider. 2003. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71:3540-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eke, P. I., V. O. Rotimi, and B. E. Laughon. 1996. Experimental model for Porphyromonas gingivalis infection in animals. Afr. J. Med. Med. Sci. 25:31-39. [PubMed] [Google Scholar]
- 21.Elwell, C., and J. N. Engel. 2005. Drosophila melanogaster S2 cells: a model system to study Chlamydia interaction with host cells. Cell. Microbiol. 7:725-739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evans, R. T., B. Klausen, N. S. Ramamurthy, L. M. Golub, C. Sfintescu, and R. J. Genco. 1992. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch. Oral Biol. 37:813-819. [DOI] [PubMed] [Google Scholar]
- 23.Ezzo, P. J., and C. W. Cutler. 2003. Microorganisms as risk indicators for periodontal disease. Periodontol. 2000 32:24-35. [DOI] [PubMed] [Google Scholar]
- 24.Franc, N. C., J. L. Dimarcq, M. Lagueux, J. Hoffmann, and R. A. Ezekowitz. 1996. Croquemort, a novel Drosophila hemocyte/macrophage receptor that recognizes apoptotic cells. Immunity 4:431-443. [DOI] [PubMed] [Google Scholar]
- 25.Genco, C. A., C. W. Cutler, D. Kapczynski, K. Maloney, and R. R. Arnold. 1991. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gibson, F. C., III, and C. A. Genco. 2001. Prevention of Porphyromonas gingivalis-induced oral bone loss following immunization with gingipain R1. Infect. Immun. 69:7959-7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gottar, M., V. Gobert, T. Michel, M. Belvin, G. Duyk, J. A. Hoffmann, D. Ferrandon, and J. Royet. 2002. The Drosophila immune response against Gram-negative bacteria is mediated by a peptidoglycan recognition protein. Nature 416:640-644. [DOI] [PubMed] [Google Scholar]
- 28.Grenier, D., and D. Mayrand. 1987. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J. Clin. Microbiol. 25:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffen, A. L., M. R. Becker, S. R. Lyons, M. L. Moeschberger, and E. J. Leys. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 36:3239-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gron, H., R. Pike, J. Potempa, J. Travis, I. B. Thogersen, J. J. Enghild, and S. V. Pizzo. 1997. The potential role of alpha 2-macroglobulin in the control of cysteine proteinases (gingipains) from Porphyromonas gingivalis. J. Periodontal Res. 32:61-68. [DOI] [PubMed] [Google Scholar]
- 31.Gyurko, R., G. Boustany, P. L. Huang, A. Kantarci, T. E. Van Dyke, C. A. Genco, and F. C. Gibson III. 2003. Mice lacking inducible nitric oxide synthase demonstrate impaired killing of Porphyromonas gingivalis. Infect. Immun. 71:4917-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamada, N., K. Watanabe, T. Tahara, K. Nakazawa, I. Ishida, Y. Shibata, T. Kobayashi, H. Yoshie, Y. Abiko, and T. Umemoto. 2007. The r40-kDa outer membrane protein human monoclonal antibody protects against Porphyromonas gingivalis-induced bone loss in rats. J. Periodontol. 78:933-939. [DOI] [PubMed] [Google Scholar]
- 33.Holzhausen, M., L. C. Spolidorio, R. P. Ellen, M. C. Jobin, M. Steinhoff, P. Andrade-Gordon, and N. Vergnolle. 2006. Protease-activated receptor-2 activation: a major role in the pathogenesis of Porphyromonas gingivalis infection. Am. J. Pathol. 168:1189-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Houri-Haddad, Y., W. A. Soskolne, E. Shai, A. Palmon, and L. Shapira. 2002. Interferon-gamma deficiency attenuates local P. gingivalis-induced inflammation. J. Dent. Res. 81:395-398. [DOI] [PubMed] [Google Scholar]
- 35.Huang, J. H., Y. Y. Lin, Y. Y. Lai, and S. W. Hu. 2006. Lethal outcome caused by Porphyromonas gingivalis A7436 in a mouse chamber model is associated with elevated titers of host serum interferon-gamma. Oral Microbiol. Immunol. 21:100-106. [DOI] [PubMed] [Google Scholar]
- 36.Igaki, T., H. Kanda, Y. Yamamoto-Goto, H. Kanuka, E. Kuranaga, T. Aigaki, and M. Miura. 2002. Eiger, a TNF superfamily ligand that triggers the Drosophila JNK pathway. EMBO J. 21:3009-3018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Igboin, C. O., M. L. Moeschberger, A. L. Griffen, and E. J. Leys. 2011. Porphyromonas gingivalis virulence in a Drosophila melanogaster model. Infect. Immun. 79:439-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiggins, F. M., and K. W. Kim. 2006. Contrasting evolutionary patterns in Drosophila immune receptors. J. Mol. Evol. 63:769-780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaneko, T., and N. Silverman. 2005. Bacterial recognition and signalling by the Drosophila IMD pathway. Cell. Microbiol. 7:461-469. [DOI] [PubMed] [Google Scholar]
- 40.Kastelein, P., T. J. van Steenbergen, J. M. Bras, and J. de Graaff. 1981. An experimentally induced phlegmonous abscess by a strain of Bacteroides gingivalis in guinea pigs and mice. Antonie Van Leeuwenhoek 47:1-9. [DOI] [PubMed] [Google Scholar]
- 41.Katz, J., D. C. Ward, and S. M. Michalek. 1996. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:309-318. [DOI] [PubMed] [Google Scholar]
- 42.Kauppila, S., W. S. Maaty, P. Chen, R. S. Tomar, M. T. Eby, J. Chapo, S. Chew, N. Rathore, S. Zachariah, S. K. Sinha, J. M. Abrams, and P. M. Chaudhary. 2003. Eiger and its receptor, Wengen, comprise a TNF-like system in Drosophila. Oncogene 22:4860-4867. [DOI] [PubMed] [Google Scholar]
- 43.Kesavalu, L., S. C. Holt, and J. L. Ebersole. 1996. Trypsin-like protease activity of Porphyromonas gingivalis as a potential virulence factor in a murine lesion model. Microb. Pathog. 20:1-10. [DOI] [PubMed] [Google Scholar]
- 44.Klausen, B. 1991. Microbiological and immunological aspects of experimental periodontal disease in rats: a review article. J. Periodontol. 62:59-73. [DOI] [PubMed] [Google Scholar]
- 45.Kocks, C., J. H. Cho, N. Nehme, J. Ulvila, A. M. Pearson, M. Meister, C. Strom, S. L. Conto, C. Hetru, L. M. Stuart, T. Stehle, J. A. Hoffmann, J. M. Reichhart, D. Ferrandon, M. Ramet, and R. A. Ezekowitz. 2005. Eater, a transmembrane protein mediating phagocytosis of bacterial pathogens in Drosophila. Cell 123:335-346. [DOI] [PubMed] [Google Scholar]
- 46.Kumagai, Y., K. Konishi, T. Gomi, H. Yagishita, A. Yajima, and M. Yoshikawa. 2000. Enzymatic properties of dipeptidyl aminopeptidase IV produced by the periodontal pathogen Porphyromonas gingivalis and its participation in virulence. Infect. Immun. 68:716-724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lagueux, M., E. Perrodou, E. A. Levashina, M. Capovilla, and J. A. Hoffmann. 2000. Constitutive expression of a complement-like protein in toll and JAK gain-of-function mutants of Drosophila. Proc. Natl. Acad. Sci. U. S. A. 97:11427-11432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Laine, M. L., and A. J. van Winkelhoff. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 13:322-325. [DOI] [PubMed] [Google Scholar]
- 49.Lemaitre, B., and J. Hoffmann. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697-743. [DOI] [PubMed] [Google Scholar]
- 50.Lemaitre, L. 2007. Drosophila genes potentially involved in responses to microbial infection. http://www.cgn.cnrs-gif.fr/immunity/drosophila_immunity_genes.html.
- 51.Leys, E. J., J. H. Smith, S. R. Lyons, and A. L. Griffen. 1999. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J. Clin. Microbiol. 37:3906-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Malek, R., J. G. Fisher, A. Caleca, M. Stinson, C. J. van Oss, J. Y. Lee, M. I. Cho, R. J. Genco, R. T. Evans, and D. W. Dyer. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mansfield, B. E., M. S. Dionne, D. S. Schneider, and N. E. Freitag. 2003. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell. Microbiol. 5:901-911. [DOI] [PubMed] [Google Scholar]
- 54.Moreno, E., M. Yan, and K. Basler. 2002. Evolution of TNF signaling mechanisms: JNK-dependent apoptosis triggered by Eiger, the Drosophila homolog of the TNF superfamily. Curr. Biol. 12:1263-1268. [DOI] [PubMed] [Google Scholar]
- 55.Nehme, N. T., S. Liegeois, B. Kele, P. Giammarinaro, E. Pradel, J. A. Hoffmann, J. J. Ewbank, and D. Ferrandon. 2007. A model of bacterial intestinal infections in Drosophila melanogaster. PLoS Pathog. 3:e173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.O'Brien-Simpson, N. M., R. D. Pathirana, R. A. Paolini, Y. Y. Chen, P. D. Veith, V. Tam, N. Ally, R. N. Pike, and E. C. Reynolds. 2005. An immune response directed to proteinase and adhesin functional epitopes protects against Porphyromonas gingivalis-induced periodontal bone loss. J. Immunol. 175:3980-3989. [DOI] [PubMed] [Google Scholar]
- 58.Otvos, L., Jr., I. O., M. E. Rogers, P. J. Consolvo, B. A. Condie, S. Lovas, P. Bulet, and M. Blaszczyk-Thurin. 2000. Interaction between heat shock proteins and antimicrobial peptides. Biochemistry 39:14150-14159. [DOI] [PubMed] [Google Scholar]
- 59.Rangarajan, M., M. A. Scragg, and M. A. Curtis. 2000. Bait region cleavage and complex formation of human α2M with a Porphyromonas gingivalis W50 protease is not accompanied by enzyme inhibition. Biol. Chem. 381:57-65. [DOI] [PubMed] [Google Scholar]
- 60.Rutschmann, S., A. C. Jung, R. Zhou, N. Silverman, J. A. Hoffmann, and D. Ferrandon. 2000. Role of Drosophila IKK gamma in a toll-independent antibacterial immune response. Nat. Immunol. 1:342-347. [DOI] [PubMed] [Google Scholar]
- 61.Samakovlis, C., D. A. Kimbrell, P. Kylsten, A. Engstrom, and D. Hultmark. 1990. The immune response in Drosophila: pattern of cecropin expression and biological activity. EMBO J. 9:2969-2976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sasaki, H., Y. Okamatsu, T. Kawai, R. Kent, M. Taubman, and P. Stashenko. 2004. The interleukin-10 knockout mouse is highly susceptible to Porphyromonas gingivalis-induced alveolar bone loss. J. Periodontal Res. 39:432-441. [DOI] [PubMed] [Google Scholar]
- 63.Schenkein, H. A. 1989. Failure of Bacteroides gingivalis W83 to accumulate bound C3 following opsonization with serum. J. Periodontal Res. 24:20-27. [DOI] [PubMed] [Google Scholar]
- 64.Schenkein, H. A., H. M. Fletcher, M. Bodnar, and F. L. Macrina. 1995. Increased opsonization of a prtH-defective mutant of Porphyromonas gingivalis W83 is caused by reduced degradation of complement-derived opsonins. J. Immunol. 154:5331-5337. [PubMed] [Google Scholar]
- 65.Schneider, D., and M. Shahabuddin. 2000. Malaria parasite development in a Drosophila model. Science 288:2376-2379. [DOI] [PubMed] [Google Scholar]
- 66.Schneider, D. S., J. S. Ayres, S. M. Brandt, A. Costa, M. S. Dionne, M. D. Gordon, E. M. Mabery, M. G. Moule, L. N. Pham, and M. M. Shirasu-Hiza. 2007. Drosophila Eiger mutants are sensitive to extracellular pathogens. PLoS Pathog. 3:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shi, X., S. A. Hanley, M. C. Faray-Kele, S. C. Fawell, J. Aduse-Opoku, R. A. Whiley, M. A. Curtis, and L. M. Hall. 2007. The rag locus of Porphyromonas gingivalis contributes to virulence in a murine model of soft tissue destruction. Infect. Immun. 75:2071-2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Spinosa, M. R., C. Progida, A. Tala, L. Cogli, P. Alifano, and C. Bucci. 2007. The Neisseria meningitidis capsule is important for intracellular survival in human cells. Infect. Immun. 75:3594-3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Steiner, H., D. Andreu, and R. B. Merrifield. 1988. Binding and action of cecropin and cecropin analogues: antibacterial peptides from insects. Biochim. Biophys. Acta 939:260-266. [DOI] [PubMed] [Google Scholar]
- 70.Stroschein-Stevenson, S. L., E. Foley, P. H. O'Farrell, and A. D. Johnson. 2006. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 4:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Stuart, L. M., J. Deng, J. M. Silver, K. Takahashi, A. A. Tseng, E. J. Hennessy, R. A. Ezekowitz, and K. J. Moore. 2005. Response to Staphylococcus aureus requires CD36-mediated phagocytosis triggered by the COOH-terminal cytoplasmic domain. J. Cell Biol. 170:477-485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsukuba, T., S. Yamamoto, M. Yanagawa, K. Okamoto, Y. Okamoto, K. I. Nakayama, T. Kadowaki, and K. Yamamoto. 2006. Cathepsin E-deficient mice show increased susceptibility to bacterial infection associated with the decreased expression of multiple cell surface Toll-like receptors. J. Biochem. 140:57-66. [DOI] [PubMed] [Google Scholar]
- 73.Yoshimura, M., Y. Nakano, Y. Yamashita, T. Oho, T. Saito, and T. Koga. 2000. Formation of methyl mercaptan from L-methionine by Porphyromonas gingivalis. Infect. Immun. 68:6912-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu, J. J., M. J. Ruddy, G. C. Wong, C. Sfintescu, P. J. Baker, J. B. Smith, R. T. Evans, and S. L. Gaffen. 2007. An essential role for IL-17 in preventing pathogen-initiated bone destruction: recruitment of neutrophils to inflamed bone requires IL-17 receptor-dependent signals. Blood 109:3794-3802. [DOI] [PMC free article] [PubMed] [Google Scholar]