Abstract
The ibeA gene is located on a genomic island, GimA, which is involved in the pathogenesis of neonatal meningitis Escherichia coli (NMEC) and avian pathogenic E. coli (APEC). The prevalence of ibeA in the APEC collection in China was investigated, and 20 of 467 strains (4.3%) were positive. In addition, analysis of the association of the E. coli reference (ECOR) groups with positive strains revealed that ibeA was linked to group B2. The ibeA gene in DE205B was analyzed and compared to those of APEC and NMEC, which indicated that the specificity of ibeA was not consistent along pathotypes. The invasion of chicken embryo fibroblast DF-1 cells by APEC DE205B and RS218 was observed, which suggested that DF-1 cells could be a model to study the mechanism of APEC invasion. The inactivation of ibeA in APEC DE205B led to the reduced capacity to invade DF-1 cells, defective virulence in vivo, and decreased biofilm formation compared to the wild-type strain. In addition, strain AAEC189 expressing ibeA exhibited enhanced invasion capacity and biofilm formation. The results of the quantitative real-time reverse transcription-PCR (qRT-PCR) analysis and animal system infection experiments indicated that the loss of ibeA decreased the colonization and proliferation capacities of APEC in the brain during system infection.
Escherichia coli typically colonizes the mammalian and avian gastrointestinal tract and other mucosal surfaces. While many of these strains are commensal, certain pathogenic strains can cause severe diseases (33). Extraintestinal pathogenic E. coli (ExPEC) is a group of strains that have been implicated in a large range of infections in humans and animals, such as neonatal meningitis, urinary tract infections (UTIs), pneumonia, osteomyelitis, and septicemia (14, 16, 30, 35). Among these, typical infections caused by ExPEC in humans are UTIs and neonatal meningitis (5). Similarly, systemic infections caused by avian pathogenic E. coli (APEC) are economically devastating to poultry industries (14, 16). APEC enters and colonizes the avian respiratory tract by inhalation of fecal dust, leading to localized infections, such as airsacculitis and pneumonia. In certain cases, they spread into various internal organs, typically causing pericarditis, perihepatitis, peritonitis, salpingitis, and other extraintestinal diseases. Systemic infection of poultry is characterized in its acute form by septicemia, commonly resulting in sudden death (14, 16, 52).
Previous studies showed that certain subsets of ExPEC strains isolated from different host organisms are highly similar (17, 31, 44, 51), thus increasing the need to study their zoonotic potential. Virulence determinants common to uropathogenic E. coli (UPEC), APEC, neonatal meningitis Escherichia coli (NMEC), or septicemia-associated E. coli (SEPEC), such as the aerobactin iron transport system, the K1 capsule, and type 1 and P fimbriae, have been identified (2, 14, 19, 21, 43, 47, 59). Furthermore, the function of the K1 capsule was similar in virulence to those of APEC and NMEC. The K1 capsule mutant of NMEC strain E44 showed higher binding and internalization rates than the parent strain but decreased intracellular survival (34). A similar result was observed with the K1 mutant of the APEC strain (42).
Recently, several microbial determinants, such as Ibe proteins, OmpA, AslA, and the K1 capsule contributing to brain microvascular epithelial cells (BMEC) invasiveness, were identified and are common to both APEC and NMEC (8, 18, 20, 21, 23-26, 37, 48, 49, 63, 64, 66). Mutants of these genetic determinants were found to be significantly less invasive in BMEC monolayers in vitro and in a newborn rat model of hematogenous E. coli meningitis (36). The ibeA gene is located on the 20.3-kb island GimA, inserted between yjiD and yjiE and proposed to contain four operons, some of which are potentially involved in energy metabolism (24). ibeA encodes a protein which plays an important role in the invasion of ExPEC in BMEC. The inactivation of ibeA in the NMEC strain and APEC has been shown to result in defective invasion of BMEC both in vitro and in vivo (18, 26, 27). Moreover, IbeA was identified to interact with its receptor, an albumin-like protein, on the surface of both human (45-kDa) and bovine (55-kDa) BMEC. Furthermore, an IbeA-binding protein was purified from intestinal epithelial Caco-2 cells (28, 48, 67, 68). IbeA could therefore be involved in the receptor ligand-mediated invasion of BMEC. However, the invasion phenotypes of the double and triple knockouts for the ibe genes and ompA gene were similar to that of the single-gene deletion (37, 64), and the precise reason for these results was not clear.
Biofilm is composed of surface-bound or -sessile microbes enclosed in an amorphous extracellular matrix (15), often composed of exopolysaccharide, proteins, and nucleic acids (13). Residence in a biofilm community offers to bacteria certain advantages, such as the enhanced ability to cause disease, which is detrimental to animal and human public health. The associations between the potential for biofilm formation and some virulence-associated genes have been described previously (10, 32, 45, 54, 57, 60, 65). In addition, ibeA was observed to be more prevalent in biofilm-forming strains (41), but no studies have definitively shown that ibeA is involved in biofilm formation.
In the present study, the prevalence of ibeA in the APEC collection in China was investigated. We characterized the ibeA gene in APEC DE205B and compared it to those of APEC and NMEC. An ibeA mutant was also constructed, and the virulence, invasion capacity, biofilm formation, and expression of virulence factors associated with invasion and adhesion were evaluated, which helped us to understand the precise role of the ibeA gene in the pathogenicity of APEC.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The strains and plasmids used in this study are listed in Table 1. The APEC strain DE205B was isolated from the brain of a duck with septicemia and neurological symptoms. It was phylogenetically analyzed using multiplex PCR and was found to belong to the phylogenetic E. coli reference (ECOR) group B2. The strain harbors virulence-associated genes tsh, mat, fyuA, irp2, iucD, iutA, iss, vat, malX, fimC, ompA, ibeA, ibeB, yijp, gimB, and aslA but is negative for papC and hlyA by PCR analysis. DE205B was used to construct the mutants and served as a positive control in functional assays.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Characteristics | Source or reference |
|---|---|---|
| Strains | ||
| DE205B | O2:K1 Nalr | This study |
| DE205M | DE205B ΔibeA | This study |
| DE205P | DE205M with plasmid pUC18 | This study |
| DE205C | DE205M with plasmid pUC18-ibeAC | This study |
| AAEC189 | MG1655 Δfim | 4 |
| AAEC189P | AAEC189 with plasmid pUC18 | This study |
| AAEC189C | AAEC189 with plasmid pUC18-ibeAC | This study |
| DH5α | F− Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK+) phoA supE44 λ− | Tiangen |
| BL21(DE3) | F−ompT hsdS(rB− mB−) gal dcm (DE3) | Tiangen |
| Plasmids | ||
| pET28a(+) | Kan, F1 origin, His tag | Novagen |
| pET28a-ibeA | pET28a(+) carrying ibeA gene | This study |
| pMD 18-T vector | Amp, lacZ | Takara |
| pUC18 | Amp, lacZ | Takara |
| pUC18-ibeAC | pUC18 carrying ibeA ORF and its putative promoter | This study |
| pKD46 | Amp; expresses λ Red recombinase | 11 |
| pKD4 | kan gene, template plasmid | 11 |
| pCP20 | Cm, Amp, yeast Flp recombinase gene, FLP | 11 |
In addition, a total of 467 E. coli strains were used for studying the prevalence of ibeA, which was isolated from ducks with clinical signs of colibacillosis at different times and in different areas in the east of China. The E. coli strain DH5α was used for cloning procedures, BL21(DE3) was used for protein expression (12, 58), and the fim-negative E. coli strain AAEC189 (4) was used in biofilm formation and invasion experiments. All E. coli strains were grown in Luria-Bertani (LB) medium at 37°C with aeration. When necessary, LB medium was supplemented with an appropriate antibiotic: ampicillin (Amp; 100 μg ml−1), kanamycin (Kan; 50 μg ml−1), or nalidixic acid (Nal; 50 μg ml−1) unless otherwise specified.
DNA and genetic manipulations.
DNA manipulations and transformations were performed using standard methods. All restriction enzymes were purchased from Takara (Dalian, China). Plasmid DNA was isolated using the High Pure plasmid miniprep kit (Invitrogen, Shanghai, China). PCR products and DNA extractions from agarose gels were purified using the High Pure PCR product purification kit (Invitrogen, Shanghai, China) according to the manufacturer's guidelines. For sequencing and expression of ibeA, Takara PrimeSTAR HS DNA polymerase was used for PCR, whereas the 2× PCR premix (Tiangen, Beijing, China) was used in screening assays according to the manufacturer's instructions. The primers used in this study are shown in Table 2. The prevalence of ibeA was assessed by PCR primers ibeA-F and ibeA-R. Determination of the phylogenetic ECOR group of the E. coli collection was performed according to the method described by Clermont et al. (7). DNA and amino acid sequence analyses were performed using the DNASTAR Lasergene 7 software and online BLAST program of the National Center for Biotechnology Information (NCBI).
TABLE 2.
Primers used in this study
| Primer | Sequence (5′ to 3′)a | Target gene, plasmid, or region |
|---|---|---|
| ibeA-F | TGGAACCCGCTCGTAATATAC | ibeA |
| ibeA-R | CTGCCTGTTCAAGCATTGCA | ibeA |
| WSH30F | GCAGGATCC ATGGAATTTTATCTGGAA | ibeA |
| WSH31R | GCGAAGCTTTTAAAAGACTTTTACGCC | ibeA |
| WSH52F | TATAAGCGCGGGGGATTGTTTTACTCAATTATTGAATACGGAGATAAAGTGTGTAGGCTGGAGCTGCTTC | pKD4 |
| WSH53R | TGTAAGCGCGACATAAAAACTGGGTTTTTCTCTCATAACTTTATTCCCTGCATATGAATATCCTCCTTAG | pKD4 |
| WSH54F | GGTTATTAGCCAGGGAGACG | Upstream region of ibeA |
| WSH55R | TGGATTTCATCCCAGGTATA | Downstream region of ibeA |
| WSH56R | GCAGGATCCCAAATGTTGAGCATGCAG | ibeA |
| WSH57F | GCGAAGCTTGTGTACCTGCATAGCTCC | ibeA |
| k1 | CAGTCATAGCCGAATAGCCT | pKD4 |
| k2 | CGGTGCCCTGAATGAACTGC | pKD4 |
| dnaE RT-F | ATGTCGGAGGCGTAAGGCT | dnaE |
| dnaE RT-R | TCCAGGGCGTCAGTAAACAA | dnaE |
| ibeB RT-F | GTTAAATTACCGGCGGGCTT | ibeB |
| ibeB RT-R | GGTCAGGCTGATAGACGGGAA | ibeB |
| yijp RT-F | AGCGTTCTGTTTGTGATGTTCG | yijp |
| yijp RT-R | ATAGGCCAGCGCGATAAGCA | yijp |
| fimC RT-F | GCCGATGGTGTAAAGGATGG | fimC |
| fimC RT-R | AACTTTCCCGATCCTGTGGC | fimC |
| ompA RT-F | GCTGAGCCTGGGTGTTTCCT | ompA |
| ompA RT-R | TCCAGAGCAGCCTGACCTTC | ompA |
| aatA RT-F | CCGTACCCGTGTCGCTGTTAC | aatA |
| aatA RT-R | CAGCATTATCAGCATTGCCACT | aatA |
| tsh RT-F | GCACGAACTGGGAAGTATGGA | tsh |
| tsh RT-R | GGCATAGAAACCACCACCCC | tsh |
BamHI and HindIII restriction sites are underlined.
Expression and purification of the invasion protein IbeA.
The open reading frame (ORF) of ibeA was amplified by PCR with oligonucleotides WSH30F and WSH31R with added BamHI and HindIII recognition sites (underlined in sequences listed in Table 2). The obtained PCR fragment was subcloned into the pMD18T vector for sequencing. The fragment of ibeA was then digested with BamHI and HindIII and ligated into a BamHI/XhoI-digested pET28a(+) vector (Novagen). The resulting plasmid, pET28a-ibeA, was transformed into competent E. coli BL21(DE3) cells, and the IbeA protein was expressed by IPTG (isopropyl-β-d-thiogalactopyranoside) induction. The fusion protein was purified using a HisTrap HP column (GE Healthcare, Shanghai, China) according to the manufacturer's guidelines. Purified IbeA protein was dialyzed overnight at 4°C against dialysis buffer. The final protein concentration was determined by the Bradford method using SmartSpec3000 (Bio-Rad) (6). Expression and purity of the fusion protein were determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE).
Antibody production.
The polyclonal anti-IbeA antibody was produced in New Zealand White rabbits as follows: 300 μg highly purified fusion protein in phosphate-buffered saline (PBS) was mixed with an equal volume of adjuvant ISA 206 (Seppic, France) and subcutaneously injected into the back of the rabbits at seven different sites. Immunizations were repeated three times at 2-week intervals. Ten days after the final immunization, the blood was collected by cardiac puncture under terminal anesthesia, and serum samples were prepared and frozen at −20°C.
Immunoblotting.
For immunoblotting analysis, protein samples were loaded on 10% SDS gels and transferred onto a polyvinylidene fluoride membrane (Amersham Pharmacia Biotech) using a semidry blotting apparatus (TE77; Amersham Pharmacia Biotech) and a buffer containing 39 mM glycine, 48 mM Tris base, 20% methanol, and 0.037% SDS. Immune serum against purified IbeA (anti-IbeA) was used as the primary antibody, and horseradish peroxidase-conjugated antirabbit immunoglobulin as the secondary antibody. Diaminobenzidine (DAB) was used as the substrate.
Construction of an ibeA mutant.
The mutant with an in-frame deletion in the ibeA gene was generated using the lambda Red recombinase method essentially as described previously (11). The ibeA gene was replaced with a kanamycin resistance cassette, which was amplified from plasmid pKD4 using PCR with primers WSH52F and WSH53R (Table 2). The 5′ regions of the primers were homologous to the corresponding flanking region of ibeA. The PCR products were then transformed by electroporation into DE205B containing the lambda Red recombinase expression plasmid pKD46. After electroporation, samples were incubated at 37°C for 1 h in SOC broth (11) and plated on LB agar with kanamycin to select for ibeA mutants. Mutants were confirmed by PCR and sequencing using primers k1 and k2 in combination with primers WSH54F and WSH55R, which flanked ibeA. The kanamycin resistance cassette was cured by transforming the pCP20 plasmid into the mutant and selecting for a kanamycin-sensitive mutant strain, which was finally designated DE205M.
Complementation of the ibeA mutant and fimbria-negative E. coli strain AAEC189.
For complementation studies, the ibeA operon, including its putative promoters, was amplified and subcloned into the pMD18T vector using primer pairs WSH56F and WSH57R with restriction enzyme recognition sites BamHI and HindIII, and the resulting plasmid was transformed into E. coli DH5α. Positive colonies were selected and identified by PCR and sequencing. The resulting plasmid pMD18T-ibeAC and pUC18 were then digested with restriction enzymes BamHI and HindIII and ligated using T4 DNA ligase for 2 h at 16°C. Five microliters of the ligation mix was then transformed into E. coli DH5α and plated on LB agar containing ampicillin. Colonies were tested for the presence of ibeA. The wild-type strain DE205B and mutant strain DE205M were transformed with control vector pUC18 and the recombinant plasmid pUC18-ibeAC. For expression of the ibeA gene in fimbria-negative E. coli strain AAEC189, plasmids pUC18 and pUC18-ibeAC were transformed into AAEC189.
Biofilm formation assay.
Biofilm formation assays were performed using a previously described method in 96-well plates with some modifications (46, 53, 56). In brief, strains were grown to stationary phase in LB at 37°C and then diluted 1:100 in LB supplemented with 5 g liter−1 glucose. Aliquots of 200 μl for each dilution were dispensed per well into a microtiter plate. Negative-control wells contained uninoculated medium. Plates were cultured aerobically without shaking at 37°C for 24 h. The medium of the plates was then poured off, and the plates were washed twice with sterile PBS. Microplates were then stained with 200 μl of 1% (wt/vol) crystal violet for 30 min, washed four times with PBS to remove unbound crystal violet dye, and air dried for 1 h. After drying, adherent cells were resolubilized with 200 μl of an 80:20 solution of ethanol and acetone. The absorbance was measured at 595 nm in an enzyme-linked immunosorbent assay (ELISA) reader (Bio-Rad microtiter plate reader, model 550). All tests were carried out three times, and the results were averaged.
Invasion assays.
For invasion assays, chicken embryo fibroblast DF-1 cells were seeded at about 1 × 105 cells per well in 24-well tissue culture trays (TPP; Shanghai, China). Cells were grown in Dulbecco modified Eagle medium (DMEM) with 10% fetal bovine serum (FBS) at 37°C in a 5% CO2 humidified atmosphere and incubated for 36 h prior to invasion assays. Semiconfluent monolayers were washed and incubated with experimental medium (DMEM without FBS) containing bacteria with a multiplicity of infection (MOI) of 100. Infected monolayers were incubated for 2 h at 37°C under a 5% CO2 atmosphere to allow invasion into the cells. The extracellular bacteria were eliminated by incubation of the monolayers with experimental medium containing gentamicin (100 μg ml−1) prior to washing DF-1 cells using PBS, and the number of intracellular bacteria was determined by treatment with Triton X-100. Negative-control wells containing only DF-1 cells were used in all experiments. The percentage of invaded bacteria was determined by dividing the number of invaded bacteria by initial inoculation bacterial numbers. The assay was performed in triplicate.
Virulence testing.
To test the virulence of strains DE205B, DE205M, and DE205C, the 50% lethal dose (LD50) of each strain was determined. Strains were grown to exponential phase, collected, washed twice in PBS, and then adjusted to the appropriate doses. Ducks were inoculated intratracheally with 0.2 ml of each bacterial suspension containing different numbers of CFU. The numbers of bacterial CFU contained in the injected inoculum were confirmed by plating on LB agar. Negative controls were injected with PBS. Ten animals were used per dose. Mortality was monitored until 7 days postinfection. The experiment was repeated three times. The LD50 of each strain was also tested with a mouse model. The results were averaged and were calculated using the method by Reed and Muench (50).
Animal infection studies.
Animals in experiments were infected with wild-type strains and mutant and complementation strains to determine the colonization and proliferative abilities of strains during system infection. Briefly, groups of 15 7-day-old ducks each were infected intratracheally with a bacterial suspension containing 108 CFU (1). At 24 h postinfection, ducks were euthanized and dissected. Bacteria were isolated from the lungs and brains as follows: organ samples were weighed, suspended in PBS (1 ml/g), and homogenized, and then they were appropriately diluted and plated onto LB agar containing nalidixic acid to determine the number of bacteria colonizing the internal organs of the duck during systemic infection.
All the animal experimental protocols were approved by the Laboratory Animal Monitoring Committee of Jiangsu province.
qRT-PCR.
Overnight cultures of E. coli were diluted 1:100 into fresh LB. The bacteria were grown to the logarithmic phase, and RNA was isolated using the E.Z.N.A. bacterial RNA isolation kit (Omega, Beijing, China) according to the manufacturer's instructions. Contaminating DNA was removed from the samples with RNase-free DNase I (Takara, Dalian, China), and cDNA synthesis was performed using the PrimeScript RT reagent kit (Takara, Dalian, China) according to the manufacturer's instructions. Quantitative real-time reverse transcription-PCR (qRT-PCR) was performed to determine the transcription levels of the virulence factors using SYBR premix Ex Taq (Takara, Dalian, China) and gene-specific primers (Table 2), and the data were normalized to the housekeeping gene dnaE transcript. The relative fold change was calculated using the threshold cycle (ΔΔCT) method (40).
Statistical analyses.
Statistical analysis for in vitro and in vivo experiments was carried out using SPSS software (Statistical Package for the Social Sciences) and Graphpad Software package (GraphPad Software, La Jolla, CA). One-way analysis of variance (ANOVA) was used in the analysis of the invasion data in vitro and in biofilm formation, and two-way ANOVA was performed on the qRT-PCR results. For the analyses of the animal infection study, the nonparametric Mann-Whitney U test was carried out. The mean values are shown in the figures. Statistical significance was established at P values of <0.05.
RESULTS
Characterization and prevalence of the ibeA gene in APEC.
The ibeA gene from APEC DE205B was first sequenced and submitted to GenBank under accession no. FJ158545.1. The ibeA gene from DE205B is 1,371 bp long and 100% identical to that of the other two APEC strains, BEN2908 and APECO1, and one NMEC strain, IHE3034, whereas the ibeA gene of another NMEC strain, RS218, has 99.3% identity to that of DE205B. The differences resulted in six amino acid changes in the IbeA protein of DE205B compared to that of RS218 (Glu-104 to Ala, Leu-242 to Phe, Arg-243 to Ala, Glu-245 to Gly, Arg-246 to Lys, and Met-416 to Ile).
The prevalence of ibeA was assessed by PCR screening of APEC strains from our laboratory collections, and 20 strains were positive for ibeA, accounting for 4.3% of the 467 APEC strains. For all strains examined in this study, our results were compared with the phylogenetic ECOR group data. Of the isolates positive for ibeA, 8 isolates belonged to ECOR group A (3.1%), 5 to B1 (6.67%), and 7 to B2 (17.5%), but no positive strain belonged to ECOR group D. Thus, B2 seems to be the predominant group for isolates harboring ibeA.
Expression and purification of IbeA from DE205B.
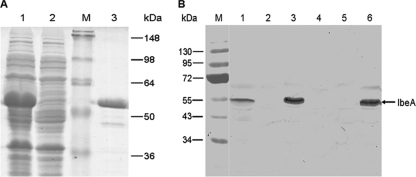
For expression, the entire ORF of ibeA was amplified by PCR from DE205B and cloned into the expression vector pET28a(+) under the control of the IPTG-inducible T7 promoter. The recombinant vector pET28a-ibeA led to the expression of a 53.2-kDa fusion protein in E. coli BL21(DE3) (Fig. 1 A). The refolded and purified IbeA proteins were dialyzed at 4°C against dialysis buffer containing diminishing concentrations of urea, and the final concentration of IbeA was 0.65 mg ml−1.
FIG. 1.
(A) Purification of IbeA expressed in E. coli BL21(DE3). The whole ORF of ibeA was cloned into pET28a(+), leading to the expression of the 53.2-kDa fusion protein IbeA. BL21(DE3) cells were incubated in LB at 37°C with (lane 1) or without (lane 2) addition of IPTG, which induces the expression of IbeA. Protein of the total extracts (lanes 1 and 2) and the elution of the purified IbeA (lane 3) were separated on an SDS-PAGE gel and stained with Coomassie blue. Lane M, protein marker. (B) Immunoblotting analysis of total cell lysates prepared from DE205B, DE205M, DE205C, AAEC189P, and AAEC189C using a serum directed against IbeA. Expression of IbeA was detected in wild-type strain DE205B and complementation strains DE205C and AAEC189C but not the isogenic ibeA mutants. Lane M, prestained protein marker; lane 1, DE205B; lane 2, DE205M; lane 3, DE205C; lane 4, DE205P; lane 5, AAEC189P; lane 6, AAEC189C.
Deletion of ibeA does not affect growth kinetics of APEC DE205B.
A clean ibeA deletion mutant of DE205B was created using the method described by Datsenko and Wanner (11). The deletion encompasses the entire ibeA gene. For genetic complementation, the ORF and putative promoter of ibeA were amplified from DE205B chromosomal DNA and cloned into plasmid pUC18. The resulting vector pUC18-ibeAC was transformed into the mutant strain DE205M, yielding the complementation strain. No significant differences in generation times and final optical densities were observed for all three strains during growth in LB medium (data not shown).
Analysis of ibeA expression.
The expressions of ibeA in the wild-type strain, mutant strain, and complementation strain were studied by immunoblotting using our anti-IbeA serum. As shown in Fig. 1B, the IbeA proteins in the lysates of only the wild-type and complementation strains, but not of DE205M and AAEC189P, were detected with the specific anti-IbeA antibody, indicating that the proteins identified by immunoblotting from strain DE205B were indeed IbeA.
The expression of ibeA contributes to biofilm formation.
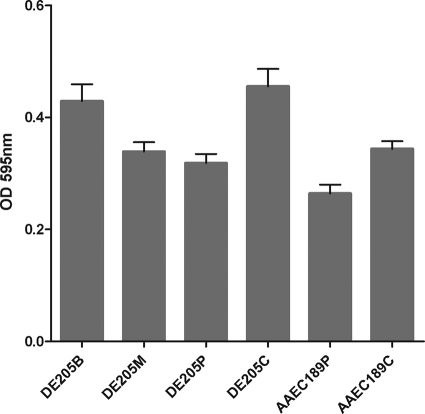
To examine whether the IbeA protein was involved in biofilm formation, strains DE205B, DE205M, DE205P, and DE205C were tested for their ability to form biofilm on polystyrene surfaces as previously described (53, 56). As shown in Fig. 2, a significant reduction of biofilm formation in the ibeA mutant DE205M, which was 0.79-fold lower than that of DE205B (P < 0.01), was observed. Moreover, the biofilm formation capacity was fully restored in the complementation strain. The complementation of ibeA in fimbria-negative E. coli strain AAEC189 was performed and resulted in significantly (P < 0.01) higher biofilm formation than that of control AAEC189P. Furthermore, the expression of ibeA was examined by immunoblotting, and the results indicated that the expression of ibeA contributed to the biofilm formation.
FIG. 2.
Biofilm formation by E. coli strains DE205B (ibeA+ strain), DE205M (ΔibeA mutant), DE205P (ΔibeA mutant), DE205C (ibeA+ strain), AAEC189P (ΔibeA mutant), and AAEC189C (ibeA+ strain). All strains were cultured in LB supplemented with 5 g liter−1 glucose. Biofilm formation was examined in polystyrene microtiter plates. All experiments were repeated at least three times. The columns represent the mean ± standard deviation of the data. The expression of ibeA led to significant increased biofilm formation (P < 0.01) by ibeA-positive strains DE205B and DE205C compared to the mutant strain DE205M. No significant difference was found between DE205B and DE205C. The biofilm formation capacity of AAEC189C was significantly (P < 0.01) higher than that of AAEC189P. Statistical significance analysis was performed using one-way ANOVA. OD 595 nm, optical density at 595 nm.
Since most virulence factors contributing to biofilm formation could induce the autoaggregation of cell to cell (38, 39, 54, 55, 65), the autoaggregation of AAEC189P and AAEC189C was detected, which indicated that the expression of ibeA did not lead to autoaggregation.
The ibeA deletion attenuates APEC DE205B virulence in vivo.
The virulence of the parent strain and mutant and complementation strains was evaluated with the duck and mouse models. Animals were infected with different strains intratracheally, and the mortality of animals was observed for 7 days after the challenge. The LD50 values were 4.0 × 106 CFU/duck with the DE205M strain and 1.0 × 106 CFU/duck with the wild-type strain DE205B, whereas the LD50 of the complementation strain was similar to that of the wild type (1.6 × 106 CFU/duck). Therefore, the virulence of the ibeA mutant was lower than that of the parent strain but could be restored in the complementation strain. Similar results were also observed with the mouse model (data not shown). Together, these results provide more evidence that ibeA is an important virulence factor in the pathogenesis of APEC infection.
Expression of ibeA leads to the enhanced abilities of APEC to invade DF-1 cells.
The ibeA gene has been shown to be involved in the invasion of human brain microvascular epithelial cells (HBMEC) by the NMEC strain RS218 and APEC BEN2908, and APEC has been demonstrated to be able to cross the blood-brain barrier (BBB) in vivo and to cause disease. However, since we did not have HBMEC on hand, the chicken embryo fibroblast cell line DF-1 was used instead in our invasion assays. We initially observed that the DF-1 cells could be invaded by DE205B and RS218 (data not shown), demonstrating that these cells could be used as a model to study the mechanism of APEC invasion.
Subsequently, DF-1 cells were infected with DE205B, DE205M, DE205P, and the complementation strain DE205C. The numbers of bacteria that invaded the DF-1 cells were estimated after a set of washing and plating of the bacteria for viable titers. Figure 3 shows a significant reduction of 35% in the invasion of mutant strain DE205M compared with that of the parent strain DE205B (P < 0.01), whereas the invasion ability was fully restored in the complementation strain DE205C (Fig. 3). On the other hand, ibeA was expressed under the control of its native promoter in E. coli strain AAEC189, and the invasion of AAEC189P (pUC18) and AAEC189C (pUC18-ibeAC) into DF-1 cells was evaluated. The results showed that AAEC189C acquired the invasion capacity due to the expression of ibeA, compared to the noninvasion strain AAEC189P. Thus, we can assume that ibeA is involved in the invasion of APEC into DF-1 cells.
FIG. 3.
IbeA plays a role in invasion of E. coli into DF-1 cells. Invasion of bacteria into DF-1 cells was examined. The values represent the average data of three independent experiments. The standard deviations are expressed as lines above the bars. Absence of a column indicates that no invasion was observed. Difference between DE205B and DE205M was statistically significant, with a P value of <0.01. No significant difference was found between DE205B and complementation strain DE205C. AAEC189C acquired the invasion capacity due to the expression of ibeA, compared to the noninvasion strain AAEC189P. One-way ANOVA was performed for the statistical significance analysis.
Effect of ibeA during systemic infection in vivo.
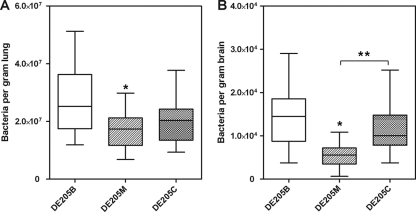
To determine the effect of ibeA during systemic infection in vivo, the ducks were infected intratracheally with strains DE205B, DE205M, and DE205C. Bacteria were recovered from the lungs and the brains of infected birds, which showed clinical signs of colibacillosis at 24 h postinoculation. As shown in Fig. 4, when ducks were infected with DE205M, a distinct reduction of recovered bacterial numbers in the lungs and brains were observed compared to those in ducks infected with DE205B (P < 0.05). The recovered complementation strain in the lungs and brains was restored by plasmid pUC18-ibeAC with intact ibeA on pUC18 so that the differences between DE205B and DE205C were not statistically significant (P > 0.05). The number of recovered bacteria of DE205C in brains was significantly higher than that of DE205M (P < 0.05). While the complementation strain DE205C showed increased numbers of colonies in the lungs, it was not significantly different from the mutant strain DE205M. The statistical analysis was performed using the nonparametric Mann-Whitney U test. These results indicated that the ibeA gene was involved in the process of system infection of APEC in vivo, which was consistent with the conclusion of Germon et al. (18).
FIG. 4.
In vivo infection study. Fifteen 7-day-old ducks were intratracheally infected with 108 CFU of bacteria. Bacterial reisolation of DE205B, DE205M, and DE205C from the lung (A) and brain (B) at 24 h postinoculation was determined by plate counting as described in Materials and Methods. The bars in the middle of columns indicate the average number of recovered bacteria from the organ for each group of animal. *, differences between DE205B and DE205M were statistically significant, with a P value of <0.05 with nonparametric Mann-Whitney U test; **, the numbers of recovered bacteria were significantly (P < 0.05) different between DE205M and DE205C. No significant difference was found between DE205B and DE205C.
Expression profiling of virulence genes.
Several microbial determinants, such as OmpA, Ibe proteins, and AslA, contribute to the invasion phenotype (18, 20, 23, 37, 48, 63, 64, 66). In addition, the inactivation of ibeA and ibeT leads to the decreased expression of type 1 fimbriae in BEN2908, which results in decreased adhesion to host cells (18). Thus, the expression profiling of virulence genes associated with invasion, adhesion, and other known virulence genes was analyzed by qRT-PCR with the various strains in vitro. The expression levels of the ibeB, yijp, fimC, ompA, aatA, and tsh genes were quantified by qRT-PCR. There were no significant differences in the expression levels of the ompA and yijp genes between DE205B and DE205M (P > 0.05). The other genes of DE205M were clearly degraded. As shown in Fig. 5, the expression levels of the virulence factors ibeB, fimC, aatA, and tsh were significantly decreased in the ibeA mutant DE205M by 0.17, 0.74, 0.29, and 0.74, respectively (P < 0.05). The expression levels of the virulence genes were restored in the ibeA complementation strain DE205C, except for gene fimC.
FIG. 5.
Quantification of virulence gene expression. Expression levels of ibeB, fimC, ompA, aatA, and tsh in strains DE205B, DE205M, and DE205C were measured by qRT-PCR, and data were normalized to the housekeeping gene dnaE. Results are shown as relative expression ratios compared to expression in the parent strain DE205B. Differences between DE205B and DE205M were statistically significant with a P value of <0.05 as determined by two-way ANOVA, except with gene ompA.
DISCUSSION
IbeA is involved in the pathogenesis of several ExPEC pathotypes, including NMEC and APEC. This virulence factor was observed to contribute to the invasion of ExPEC into host cells (18, 27). Furthermore, the IbeA receptor PSF was described for both human and bovine BMEC (28, 48, 67, 68), but the precise role of ibeA in the process of infection was still unclear. Thus, this study was conducted to investigate the role of ibeA in APEC.
The ibeA gene from APEC DE205B was identified and compared to those of APEC and NMEC strains collected in the public database. The sequences of ibeA in three APEC strains and one NMEC strain, IHE3034, shared 100% identity and showed 99.3% identity to that of another NMEC strain, RS218. It can be concluded that the differences in the ibeA genes between NMEC and APEC were not consistent along pathotypes, whereas the high identity of ibeA genes in ExPEC strains implies that APEC is a reservoir for the infection of neonatal meningitis. Thus, close monitoring of the possible contamination of persons by APEC should be reinforced.
The distribution of ibeA in the APEC collection was examined, and 4.3% of the E. coli collection was positive for ibeA, similar to the findings of other studies, in which only 3% or 4% of the strains are ibeA positive (29, 62). However, this distribution was very different from that observed with another APEC collection (26%) and NMEC strains (40 to 33%) by other researchers (18, 22, 29). A possible reason for this difference is the different sources and pathogenicities of E. coli collections. The E. coli strains in our lab were isolated from ducks with colibacillosis, which may be the cause of the different distribution of ibeA. It is well known that different strains of the species E. coli show a high degree of variability, despite a conserved core genome (3, 61). E. coli is divided into four main phylogenetic groups, namely, A, B1, B2, and D. Our analysis of the association between ibeA genes of the phylogenetic ECOR groups suggested that the phylogenetic ECOR group B2 is the predominant group for isolates harboring ibeA. Furthermore, there was no positive strain that belonged to phylogenetic ECOR group D. A parallel finding in which GimA-positive and GimA remnant strains occurred almost exclusively in ancestral group B2 was reported (22).
Since the invasion of HBMEC by NMEC RS218 and APEC BEN2908 was previously observed (18), in this study we found that chicken embryo fibroblast DF-1 cells could be invaded by both APEC DE205B and NMEC RS218 (data not shown).
A possible reason for the cross-interaction of ExPEC in human and avian cells is that the host cells share similar receptors for IbeA. An IbeA receptor, PSF, described for both human and bovine BMEC, has an N-terminal region that shares some homology with serum albumin (27, 47, 66, 67). Therefore, we predicted that PSF-like receptors for the binding of IbeA in avian cells would be identified in the future. There was a finding that the E. coli K1 invasion of endothelial cells is specific to BMEC, and no invasion characteristics were observed for non-brain-original endothelial cells (37). However, our invasion assays demonstrated that DF-1 cells could be invaded by ExPEC and could be used as another model to study the mechanism of APEC invasion due to the rarity of HBMEC.
The role of ibeA involved in the invasion of DF-1 cells by APEC was evaluated. The invasion ability of the ibeA mutant strain was significantly reduced compared to that of the wild-type strain. Moreover, the invasion ability was fully restored in the complementation strain, which indicated that ibeA mediated the invasion of APEC into DF-1 cells. While the ibeA mutant strain also maintained its ability to invade DF-1 cells, it might have been due to multiple factors, such as ibeB, yijp, and ompA interactions during the invasion process. On the other hand, the expression of ibeA in noninvasion strain AAEC189 leads to the acquirement of the invasion capacity. Thus, it could be concluded that ibeA was a virulence factor which was required for the invasion of E. coli into DF-1 cells.
The influence of ibeA on the virulence of APEC was also examined in this study. The LD50 of the ibeA mutant was significantly increased compared with that of the parent strain in the duck and mouse models, whereas the virulence was restored in the complementation strain, indicating that the deletion of ibeA resulted in the attenuated virulence of APEC DE205B.
The role of ibeA in the pathogenicity of DE205B was investigated in vivo by comparing its effects with those of the mutant strain. When the birds were infected with different strains, the colonization and proliferation capacities of bacteria in the lungs and brains were compared. The results indicated that the loss of ibeA resulted in significantly reduced numbers of recovered bacteria in the tissues compared to those of the wild-type strain. Similar results were also observed in another study, in which the inactivation of ibeA led to the reduction of bacteria in the liver and blood (18). This phenomenon might be due to the downregulation of other virulence factors, such as type 1 fimbriae and autotransporter adhesin (8, 9) and the invasion-associated gene ibeB (Fig. 5) in the mutant strain, which mediated the adhesion and invasion of APEC to host cells. The number of recovered bacteria in the brains was restored with the complementation strain DE205C in vivo and was significantly different than that for the mutant strain DE205M, which indicated that ibeA is involved in brain colonization and proliferation during system infection. Although the colonization capacity of bacteria in the lungs was restored for the complementation strain DE205C, it was not significantly different than that for mutant strain DE205M. This might be due to the fact that the expression levels of chromosome-encoded virulence genes were not fully restored in the ibeA complementation strain, which mediated the adhesion of APEC to host cells (8, 9). A parallel result was observed with another ibeA mutant strain, BEN2908, in which the expression level of type 1 fimbriae was not fully restored in the complementation strain as determined by an immunoblotting test (8), whereas the expression level of type 1 fimbriae was not restored in our experiment. The different methods for the detection of expression levels might be a reason for this phenomenon. Thus, the results of the qRT-PCR analysis and animal infection experiments demonstrated that the inactivation of ibeA decreased the expression level of virulence factors involved in the process of invasion and adhesion, which resulted in the reduced colonization and proliferation capacities of APEC.
Production of biofilm aids in the survival of bacteria and enhances their ability to cause disease, and the presence of ibeA and biofilm formation was observed, indicating that ibeA was found to be more prevalent in the biofilm-forming strains (41). To further determine the association of ibeA and biofilm, ibeA was expressed in an E. coli K-12 background strain, AAEC189, which promoted biofilm formation. Unlike other virulence factors which contributed to autoaggregation and biofilm, ibeA had no effect on the aggregation. The biofilm formation of the ibeA mutant was found to be attenuated compared to that of the parent strain, whereas it was restored when the ibeA gene was complemented in the mutant. However, the mutant strains still produce strong biofilm, which might be due to other surface factors contributing to biofilm formation, such as antigen 43 (39), since this single deletion in ibeA would not lead to the loss of ability in biofilm formation. Therefore, it is clear that ibeA is involved in biofilm formation, although the precise function of the IbeA protein in this process is still unclear and should be investigated in future studies.
In summary, the invasion protein IbeA from APEC DE205B was identified and compared with those of APEC and NMEC strains collected in the public database. We demonstrated that the DF-1 cells could be invaded by APEC DE205B and NMEC RS218, indicating that DF-1 cells could be used to further study the invasive properties of APEC. The deletion of the ibeA gene resulted in lower invasiveness in vitro and defective virulence in vivo. In addition, the results of system infection indicated that ibeA was involved in the colonization and proliferation of APEC in the brain. Furthermore, the results reported here represent the first demonstration of ibeA function in biofilm formation.
Acknowledgments
We thank Aijian Qin (Yangzhou University) and Aizhen Guo (Huazhong Agricultural University) for kindly providing chicken embryo fibroblast DF-1 cells and fimbria-negative E. coli strain AAEC189, respectively.
Editor: S. M. Payne
Footnotes
Published ahead of print on 25 October 2010.
REFERENCES
- 1.Antão, E. M., C. Ewers, D. Gurlebeck, R. Preisinger, T. Homeier, G. Li, and L. H. Wieler. 2009. Signature-tagged mutagenesis in a chicken infection model leads to the identification of a novel avian pathogenic Escherichia coli fimbrial adhesin. PLoS One 4:e7796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bahrani-Mougeot, F. K., E. L. Buckles, C. V. Lockatell, J. R. Hebel, D. E. Johnson, C. M. Tang, and M. S. Donnenberg. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079-1093. [DOI] [PubMed] [Google Scholar]
- 3.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 4.Blomfield, I. C., M. S. McClain, and B. I. Eisenstein. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439-1445. [DOI] [PubMed] [Google Scholar]
- 5.Blondeau, J. M. 2004. Current issues in the management of urinary tract infections: extended-release ciprofloxacin as a novel treatment option. Drugs 64:611-628. [DOI] [PubMed] [Google Scholar]
- 6.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 7.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cortes, M. A., J. Gibon, N. K. Chanteloup, M. Moulin-Schouleur, P. Gilot, and P. Germon. 2008. Inactivation of ibeA and ibeT results in decreased expression of type 1 fimbriae in extraintestinal pathogenic Escherichia coli strain BEN2908. Infect. Immun. 76:4129-4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dai, J., S. Wang, D. Guerlebeck, C. Laturnus, S. Guenther, Z. Shi, C. Lu, and C. Ewers. 2010. Suppression subtractive hybridization identifies an autotransporter adhesin gene of E. coli IMT5155 specifically associated with avian pathogenic Escherichia coli (APEC). BMC Microbiol. 10:236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Danese, P. N., L. A. Pratt, S. L. Dove, and R. Kolter. 2000. The outer membrane protein, antigen 43, mediates cell-to-cell interactions within Escherichia coli biofilms. Mol. Microbiol. 37:424-432. [DOI] [PubMed] [Google Scholar]
- 11.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davanloo, P., A. H. Rosenberg, J. J. Dunn, and F. W. Studier. 1984. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. U. S. A. 81:2035-2039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 15.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ewers, C., T. Janssen, and L. H. Wieler. 2003. Avian pathogenic Escherichia coli (APEC). Berl. Munch. Tierarztl. Wochenschr. 116:381-395. (In German.) [PubMed] [Google Scholar]
- 17.Ewers, C., G. Li, H. Wilking, S. Kiessling, K. Alt, E. M. Antao, C. Laturnus, I. Diehl, S. Glodde, T. Homeier, U. Bohnke, H. Steinruck, H. C. Philipp, and L. H. Wieler. 2007. Avian pathogenic, uropathogenic, and newborn meningitis-causing Escherichia coli: how closely related are they? Int. J. Med. Microbiol. 297:163-176. [DOI] [PubMed] [Google Scholar]
- 18.Germon, P., Y. H. Chen, L. He, J. E. Blanco, A. Bree, C. Schouler, S. H. Huang, and M. Moulin-Schouleur. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179-1186. [DOI] [PubMed] [Google Scholar]
- 19.Gunther, N. W. T., J. A. Snyder, V. Lockatell, I. Blomfield, D. E. Johnson, and H. L. Mobley. 2002. Assessment of virulence of uropathogenic Escherichia coli type 1 fimbrial mutants in which the invertible element is phase-locked on or off. Infect. Immun. 70:3344-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffman, J. A., J. L. Badger, Y. Zhang, S. H. Huang, and K. S. Kim. 2000. Escherichia coli K1 aslA contributes to invasion of brain microvascular endothelial cells in vitro and in vivo. Infect. Immun. 68:5062-5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman, J. A., C. Wass, M. F. Stins, and K. S. Kim. 1999. The capsule supports survival but not traversal of Escherichia coli K1 across the blood-brain barrier. Infect. Immun. 67:3566-3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Homeier, T., T. Semmler, L. H. Wieler, and C. Ewers. 2010. The GimA locus of extraintestinal pathogenic E. coli: does reductive evolution correlate with habitat and pathotype? PLoS One 5:e10877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, S. H., Y. H. Chen, Q. Fu, M. Stins, Y. Wang, C. Wass, and K. S. Kim. 1999. Identification and characterization of an Escherichia coli invasion gene locus, ibeB, required for penetration of brain microvascular endothelial cells. Infect. Immun. 67:2103-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Huang, S. H., Y. H. Chen, G. Kong, S. H. Chen, J. Besemer, M. Borodovsky, and A. Jong. 2001. A novel genetic island of meningitic Escherichia coli K1 containing the ibeA invasion gene (GimA): functional annotation and carbon-source-regulated invasion of human brain microvascular endothelial cells. Funct. Integr. Genomics 1:312-322. [DOI] [PubMed] [Google Scholar]
- 25.Huang, S. H., M. F. Stins, and K. S. Kim. 2000. Bacterial penetration across the blood-brain barrier during the development of neonatal meningitis. Microbes Infect. 2:1237-1244. [DOI] [PubMed] [Google Scholar]
- 26.Huang, S. H., Z. S. Wan, Y. H. Chen, A. Y. Jong, and K. S. Kim. 2001. Further characterization of Escherichia coli brain microvascular endothelial cell invasion gene ibeA by deletion, complementation, and protein expression. J. Infect. Dis. 183:1071-1078. [DOI] [PubMed] [Google Scholar]
- 27.Huang, S. H., C. Wass, Q. Fu, N. V. Prasadarao, M. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hui, C. Y., Y. Guo, J. Li, X. Y. Hao, H. Cao, and S. H. Huang. 2009. Purification of E. coli invasin IbeA-binding protein in intestinal epithelial cells. Nan Fang Yi Ke Da Xue Xue Bao 29:2375-2378. (In Chinese.) [PubMed] [Google Scholar]
- 29.Johnson, J. R., P. Delavari, M. Kuskowski, and A. L. Stell. 2001. Phylogenetic distribution of extraintestinal virulence-associated traits in Escherichia coli. J. Infect. Dis. 183:78-88. [DOI] [PubMed] [Google Scholar]
- 30.Johnson, J. R., and T. A. Russo. 2002. Uropathogenic Escherichia coli as agents of diverse non-urinary tract extraintestinal infections. J. Infect. Dis. 186:859-864. [DOI] [PubMed] [Google Scholar]
- 31.Johnson, T. J., Y. Wannemuehler, S. J. Johnson, A. L. Stell, C. Doetkott, J. R. Johnson, K. S. Kim, L. Spanjaard, and L. K. Nolan. 2008. Comparison of extraintestinal pathogenic Escherichia coli strains from human and avian sources reveals a mixed subset representing potential zoonotic pathogens. Appl. Environ. Microbiol. 74:7043-7050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanamaru, S., H. Kurazono, A. Terai, K. Monden, H. Kumon, Y. Mizunoe, O. Ogawa, and S. Yamamoto. 2006. Increased biofilm formation in Escherichia coli isolated from acute prostatitis. Int. J. Antimicrob. Agents 28(Suppl. 1):S21-S25. [DOI] [PubMed] [Google Scholar]
- 33.Kaper, J. B. 2005. Pathogenic Escherichia coli. Int. J. Med. Microbiol. 295:355-356. [DOI] [PubMed] [Google Scholar]
- 34.Kim, K. J., S. J. Elliott, F. Di Cello, M. F. Stins, and K. S. Kim. 2003. The K1 capsule modulates trafficking of E. coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell. Microbiol. 5:245-252. [DOI] [PubMed] [Google Scholar]
- 35.Kim, K. S. 2000. E. coli invasion of brain microvascular endothelial cells as a pathogenetic basis of meningitis. Subcell. Biochem. 33:47-59. [DOI] [PubMed] [Google Scholar]
- 36.Kim, K. S. 2001. Escherichia coli translocation at the blood-brain barrier. Infect. Immun. 69:5217-5222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim, K. S. 2002. Strategy of Escherichia coli for crossing the blood-brain barrier. J. Infect. Dis. 186(Suppl. 2):S220-S224. [DOI] [PubMed] [Google Scholar]
- 38.Kjaergaard, K., M. A. Schembri, H. Hasman, and P. Klemm. 2000. Antigen 43 from Escherichia coli induces inter- and intraspecies cell aggregation and changes in colony morphology of Pseudomonas fluorescens. J. Bacteriol. 182:4789-4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kjaergaard, K., M. A. Schembri, C. Ramos, S. Molin, and P. Klemm. 2000. Antigen 43 facilitates formation of multispecies biofilms. Environ. Microbiol. 2:695-702. [DOI] [PubMed] [Google Scholar]
- 40.Livak, K. J., and T. D. Schmittgen. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25:402-408. [DOI] [PubMed] [Google Scholar]
- 41.Martinez-Medina, M., P. Naves, J. Blanco, X. Aldeguer, J. E. Blanco, M. Blanco, C. Ponte, F. Soriano, A. Darfeuille-Michaud, and L. J. Garcia-Gil. 2009. Biofilm formation as a novel phenotypic feature of adherent-invasive Escherichia coli (AIEC). BMC Microbiol. 9:202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mellata, M., M. Dho-Moulin, C. M. Dozois, R. Curtiss III, B. Lehoux, and J. M. Fairbrother. 2003. Role of avian pathogenic Escherichia coli virulence factors in bacterial interaction with chicken heterophils and macrophages. Infect. Immun. 71:494-503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mobley, H. L., K. G. Jarvis, J. P. Elwood, D. I. Whittle, C. V. Lockatell, R. G. Russell, D. E. Johnson, M. S. Donnenberg, and J. W. Warren. 1993. Isogenic P-fimbrial deletion mutants of pyelonephritogenic Escherichia coli: the role of alpha Gal(1-4) beta Gal binding in virulence of a wild-type strain. Mol. Microbiol. 10:143-155. [DOI] [PubMed] [Google Scholar]
- 44.Moulin-Schouleur, M., C. Schouler, P. Tailliez, M. R. Kao, A. Bree, P. Germon, E. Oswald, J. Mainil, M. Blanco, and J. Blanco. 2006. Common virulence factors and genetic relationships between O18:K1:H7 Escherichia coli isolates of human and avian origin. J. Clin. Microbiol. 44:3484-3492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ong, C. L., G. C. Ulett, A. N. Mabbett, S. A. Beatson, R. I. Webb, W. Monaghan, G. R. Nimmo, D. F. Looke, A. G. McEwan, and M. A. Schembri. 2008. Identification of type 3 fimbriae in uropathogenic Escherichia coli reveals a role in biofilm formation. J. Bacteriol. 190:1054-1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 47.Pourbakhsh, S. A., M. Dho-Moulin, A. Bree, C. Desautels, B. Martineau-Doize, and J. M. Fairbrother. 1997. Localization of the in vivo expression of P and F1 fimbriae in chickens experimentally inoculated with pathogenic Escherichia coli. Microb. Pathog. 22:331-341. [DOI] [PubMed] [Google Scholar]
- 48.Prasadarao, N. V., C. A. Wass, S. H. Huang, and K. S. Kim. 1999. Identification and characterization of a novel Ibe10 binding protein that contributes to Escherichia coli invasion of brain microvascular endothelial cells. Infect. Immun. 67:1131-1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Prasadarao, N. V., C. A. Wass, J. N. Weiser, M. F. Stins, S. H. Huang, and K. S. Kim. 1996. Outer membrane protein A of Escherichia coli contributes to invasion of brain microvascular endothelial cells. Infect. Immun. 64:146-153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Reed, L. J., and H. Muench. 1938. A simple method of estimating fifty percent endpoints. Am. J. Hyg. (Lond.) 27:493-497. [Google Scholar]
- 51.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097-2110. [DOI] [PubMed] [Google Scholar]
- 52.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36:241-256. [DOI] [PubMed] [Google Scholar]
- 53.Schembri, M. A., and P. Klemm. 2001. Biofilm formation in a hydrodynamic environment by novel fimh variants and ramifications for virulence. Infect. Immun. 69:1322-1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sherlock, O., M. A. Schembri, A. Reisner, and P. Klemm. 2004. Novel roles for the AIDA adhesin from diarrheagenic Escherichia coli: cell aggregation and biofilm formation. J. Bacteriol. 186:8058-8065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sherlock, O., R. M. Vejborg, and P. Klemm. 2005. The TibA adhesin/invasin from enterotoxigenic Escherichia coli is self recognizing and induces bacterial aggregation and biofilm formation. Infect. Immun. 73:1954-1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Skyberg, J. A., K. E. Siek, C. Doetkott, and L. K. Nolan. 2007. Biofilm formation by avian Escherichia coli in relation to media, source and phylogeny. J. Appl. Microbiol. 102:548-554. [DOI] [PubMed] [Google Scholar]
- 57.Soto, S. M., A. Smithson, J. A. Martinez, J. P. Horcajada, J. Mensa, and J. Vila. 2007. Biofilm formation in uropathogenic Escherichia coli strains: relationship with prostatitis, urovirulence factors and antimicrobial resistance. J. Urol. 177:365-368. [DOI] [PubMed] [Google Scholar]
- 58.Studier, F. W., and B. A. Moffatt. 1986. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189:113-130. [DOI] [PubMed] [Google Scholar]
- 59.Torres, A. G., P. Redford, R. A. Welch, and S. M. Payne. 2001. TonB-dependent systems of uropathogenic Escherichia coli: aerobactin and heme transport and TonB are required for virulence in the mouse. Infect. Immun. 69:6179-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ulett, G. C., A. N. Mabbett, K. C. Fung, R. I. Webb, and M. A. Schembri. 2007. The role of F9 fimbriae of uropathogenic Escherichia coli in biofilm formation. Microbiology 153:2321-2331. [DOI] [PubMed] [Google Scholar]
- 61.van Passel, M. W., P. R. Marri, and H. Ochman. 2008. The emergence and fate of horizontally acquired genes in Escherichia coli. PLoS Comput. Biol. 4:e1000059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Vidotto, M. C., M. B. Queiroz, N. C. de Lima, and L. C. Gaziri. 2007. Prevalence of ibeA gene in avian pathogenic Escherichia coli (APEC). Vet. Microbiol. 119:88-89. [DOI] [PubMed] [Google Scholar]
- 63.Wang, Y., S. H. Huang, C. A. Wass, M. F. Stins, and K. S. Kim. 1999. The gene locus yijP contributes to Escherichia coli K1 invasion of brain microvascular endothelial cells. Infect. Immun. 67:4751-4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang, Y., and K. S. Kim. 2002. Role of OmpA and IbeB in Escherichia coli K1 invasion of brain microvascular endothelial cells in vitro and in vivo. Pediatr. Res. 51:559-563. [DOI] [PubMed] [Google Scholar]
- 65.Wells, T. J., O. Sherlock, L. Rivas, A. Mahajan, S. A. Beatson, M. Torpdahl, R. I. Webb, L. P. Allsopp, K. S. Gobius, D. L. Gally, and M. A. Schembri. 2008. EhaA is a novel autotransporter protein of enterohemorrhagic Escherichia coli O157:H7 that contributes to adhesion and biofilm formation. Environ. Microbiol. 10:589-604. [DOI] [PubMed] [Google Scholar]
- 66.Zou, Y., L. He, F. Chi, A. Jong, and S. H. Huang. 2008. Involvement of Escherichia coli K1 ibeT in bacterial adhesion that is associated with the entry into human brain microvascular endothelial cells. Med. Microbiol. Immunol. 197:337-344. [DOI] [PubMed] [Google Scholar]
- 67.Zou, Y., L. He, and S. H. Huang. 2006. Identification of a surface protein on human brain microvascular endothelial cells as vimentin interacting with Escherichia coli invasion protein IbeA. Biochem. Biophys. Res. Commun. 351:625-630. [DOI] [PubMed] [Google Scholar]
- 68.Zou, Y., L. He, C. H. Wu, H. Cao, Z. H. Xie, Y. Ouyang, Y. Wang, A. Jong, and S. H. Huang. 2007. PSF is an IbeA-binding protein contributing to meningitic Escherichia coli K1 invasion of human brain microvascular endothelial cells. Med. Microbiol. Immunol. 196:135-143. [DOI] [PubMed] [Google Scholar]