Abstract
The interaction of the minor fimbrial antigen (Mfa) with streptococcal antigen I/II (e.g., SspB) facilitates colonization of the dental biofilm by Porphyromonas gingivalis. We previously showed that a 27-mer peptide derived from SspB (designated BAR) resembles the nuclear receptor (NR) box protein-protein interacting domain and potently inhibits this interaction in vitro. Here, we show that the EXXP motif upstream of the NR core α-helix contributes to the Mfa-SspB interaction and that BAR reduces P. gingivalis colonization and alveolar bone loss in vivo in a murine model of periodontitis. Substitution of Gln for Pro1171 or Glu1168 increased the α-helicity of BAR and reduced its inhibitory activity in vitro by 10-fold and 2-fold, respectively. To determine if BAR prevents P. gingivalis infection in vivo, mice were first infected with Streptococcus gordonii and then challenged with P. gingivalis in the absence and presence of BAR. Animals that were infected with either 109 CFU of S. gordonii DL-1 or 107 CFU of P. gingivalis 33277 did not show a statistically significant increase in alveolar bone resorption over sham-infected controls. However, infection with 109 CFU of S. gordonii followed by 107 CFU of P. gingivalis induced significantly greater bone loss (P < 0.01) than sham infection or infection of mice with either organism alone. S. gordonii-infected mice that were subsequently challenged with 107 CFU of P. gingivalis in the presence of BAR exhibited levels of bone resorption similar to those of sham-infected animals. Together, these results indicate that both EXXP and the NR box are important for the Mfa-SspB interaction and that BAR peptide represents a potential therapeutic that may limit colonization of the oral cavity by P. gingivalis.
Periodontal disease is an oral inflammatory disorder that is initiated by a microbial biofilm that forms in the subgingival pocket. In addition to the oral manifestation of periodontitis, periodontal pathogens such as Porphyromonas gingivalis are associated with a variety of other systemic disorders such as atherosclerosis, pneumonia, rheumatoid arthritis, and nephritis (7, 10, 12, 23, 24, 26, 28, 29, 34). However, colonization of the subgingival pocket by P. gingivalis can occur only after the organism first becomes established in the supragingival biofilm or on tissue surfaces. Colonization of the supragingival biofilm occurs through interspecies interactions of P. gingivalis with specific species of oral streptococci (e.g., Streptococcus oralis and Streptococcus gordonii but not the mutans streptococci) (11, 20), and these initial interactions thus represent viable targets for therapeutic intervention to limit colonization of the oral cavity by P. gingivalis.
The selective adherence of P. gingivalis to specific oral streptococcal species is driven by a protein-protein interaction that occurs between the minor fimbrial antigen (Mfa) of P. gingivalis and the streptococcal antigen I/II, and inactivation of mfa completely prevents P. gingivalis adherence and formation of biofilms with streptococci (6, 11, 27). Brooks et al. (6) showed that a region encompassing amino acids 1167 to 1250 of the S. gordonii antigen I/II protein SspB was essential for adherence of P. gingivalis. Subsequently, Demuth et al. (11) compared this region of SspB with the corresponding region of the Streptococcus mutans antigen I/II and showed that a protein determinant comprising amino acids 1167 to 1193 was sufficient to promote P. gingivalis adherence. This study also demonstrated that sequence variability in this region between the S. gordonii and S. mutans antigen I/II proteins accounted for binding selectivity of P. gingivalis and that substitution of S. mutans residues for Asn1182 and Val1185 in SspB by site-specific mutagenesis rendered the protein inactive. More recently, Daep et al. (8) showed that a peptide comprising residues 1167 to 1193, designated BAR, potently inhibited the interaction of P. gingivalis with S. gordonii interaction (50% inhibitory concentration [IC50] of 1.3 μM) using a dual-species open-flow biofilm culture model. Two structural motifs in BAR, VXXLL and NITVK (amino acids 1171 to 1180 and 1182 to 1186, respectively, in the full-length antigen I/II protein of S. gordonii), were shown to be essential for the interaction of BAR peptide with Mfa (9). However, an analog of BAR that possessed VXXLL and NITVK but lacked amino acids upstream of VXXLL exhibited lower specific activity than BAR peptide itself, suggesting that additional structural motifs in BAR may contribute to its interaction with Mfa.
In this report, we show that substitutions of Gln for Glu1168 and Pro1171 affect the structure and activity of BAR, suggesting that these residues also contribute to Mfa interaction. In addition, using an in vivo mouse model of periodontitis, we show that S. gordonii promotes P. gingivalis oral colonization of mice and P. gingivalis-induced alveolar bone resorption. However, infection of mice in the presence of BAR peptide reduced bone loss in P. gingivalis-S. gordonii-infected animals to the level of sham-infected controls. Finally, we show that BAR peptide exhibits little cytotoxicity against primary human gingival epithelial cells or human cell lines, suggesting that BAR may be developed as a potential therapeutic agent to limit P. gingivalis colonization of the oral cavity.
MATERIALS AND METHODS
Bacterial culture.
S. gordonii ATCC strain DL-1 was cultured in brain heart infusion (Difco) broth supplemented with 1% (wt/vol) yeast extract (BHIYE) at 37°C for 24 h. P. gingivalis ATCC 33277 was grown in TSBYE medium, which consists of 30 g/liter Trypticase soy broth (Difco) supplemented with 2% (wt/vol) yeast extract, 1 μg/ml hemin (final concentration), and 5 μg/ml menadione (final concentration) under anaerobic conditions (10% CO2, 10% H2, and 80% N2) at 37°C for 48 h.
Peptide synthesis.
The synthetic peptides used in this study are listed and described in Table 1. All of the peptides were derived from the sequence of the BAR peptide comprising residues 1167 to 1193 of the antigen I/II (SspB) protein sequence of S. gordonii (6). Peptides were synthesized at ≥85% purity by Biosynthesis (Lewisville, TX) and were suspended in nuclease/protease-free water (Fisher Scientific Co., Fairlawn, NJ) at the desired concentration immediately before use.
TABLE 1.
Peptides used in this study
| Peptide | Description | Peptide sequence |
|---|---|---|
| BAR | NH2-LEAAPKKVQDLLKKANITVKGAFQLFS-OH | |
| BAR-15 | Gln/Pro1171-BAR | NH2-LEAAQKKVQDLLKKANITVKGAFQLFS-OH |
| BAR-16 | Gln/Glu1168-BAR | NH2-LQAAPKKVQDLLKKANITVKGAFQLFS-OH |
| BAR-17 | Cyclic BAR peptide | NH2-CEAAPKKVQDLLKKANITVKGAFQCFS-OHa |
The underlined portion is conformationally constrained by a disulfide bond between Cys1 and Cys24.
CD spectroscopy.
Circular dichroism (CD) experiments were carried out at 25°C using a Jasco J-810 spectropolarimeter (Jasco, Easton, MD). A 40-μl sample of the appropriate peptide at a concentration of 1.4 mM in filter-sterilized 0.1 M phosphate buffer (pH = 7.2) was analyzed in a 0.01-cm quartz cuvette (Starna Cells, Inc., Atascadero, CA) using the following parameters: sensitivity of 100 millidegrees (mdeg), start wavelength of 340 nm, end wavelength of 180 nm, continuous scanning mode at 200 nm/minute, response of 1 s, and bandwidth of 1 nm. Nitrogen was flushed into the system at a rate of 31.8 to 42.4 cubic feet/min during each experiment. A total of 10 scans were accumulated and averaged for each peptide sample with the appropriate blanks subtracted from the spectra. The resulting spectra were expressed in molar ellipticity (mdeg). Normalized CD data were analyzed using K2D software (http://www.embl-heidelberg.de/∼andrade/k2d.html) to estimate α-helical, β-sheet, and random coil content (2).
Biofilm formation of P. gingivalis and S. gordonii.
The formation of dual species P. gingivalis-S. gordonii biofilms was carried out essentially as previously described by Daep et al. (8). Cultures were grown in a BST FC 71 flow cell (Biosurface Technologies Corp., Bozeman, MT) attached to a Manostat Carter 4/8 cassette peristaltic pump (Fisher Scientific, Suwanee, GA) using 0.89-mm platinum-cured silicone tubing (Fisher Scientific, Suwanee, GA). A single surface of a 15- by 40-mm cover glass (Fisher Scientific, Suwanee, GA) was coated with 0.22-μm-pore-size-filter-sterilized saliva and incubated at 37°C for 30 min. The saliva-coated cover glass was then washed with sterile 1× phosphate-buffered saline (PBS) at a flow rate of 6 ml per hour for 30 min.
S. gordonii DL-1 cells were harvested by centrifugation at 7,000 × g at 4°C, suspended in 25 ml of sterile 1× PBS, and labeled with 20 μl of hexidium iodide (1.6 mg/ml; Molecular Probes, Eugene, OR) at 25°C for 30 min in the dark. After the cells were washed with PBS, adherence of streptococci to the saliva-coated cover glass was carried out by delivering S. gordonii cells (1010 CFU per ml) to the flow chamber at a rate of 6 ml per hour for approximately 2 h. Following inoculation with S. gordonii, the flow cell was washed with sterile 1× PBS for 30 min at 6 ml per hour to remove nonadherent bacteria from the cover glass.
P. gingivalis was harvested by centrifugation, suspended in 25 ml of sterile 1× PBS at 2 × 109 CFU per ml and introduced into the flow cell at a flow rate of 6 ml per hour for 2 h to allow P. gingivalis to adhere and accumulate on the streptococcal substrate. Flow cells were subsequently washed with sterile 1× PBS to remove nonadherent P. gingivalis cells. To visualize adherent P. gingivalis, rabbit anti-P. gingivalis 33277 polyclonal antibody diluted 1:5,000 in 5 ml of sterile 1× PBS was flowed into the cell at a rate of 6 ml per hour. The flow cells were washed with sterile 1× PBS for 1 h, reacted with anti-rabbit IgG-fluorescein isothiocyanate (FITC) conjugate (Sigma, St. Louis, MO) in sterile 1× PBS (1:5,000) for 1 h at 6 ml per hour, and washed again with sterile 1× PBS as above. For some experiments, adherent P. gingivalis was labeled with rabbit anti-P. gingivalis 33277 IgG conjugated with Alexa Fluor 488 diluted 1:5,000 in sterile PBS. Under the conditions described above, streptococci bound to the saliva-coated cover glass in a nonconfluent layer comprising mostly single cells and small aggregates. P. gingivalis subsequently formed distinct microcolonies on the immobilized streptococci. The biofilms that were formed were visualized and quantified by confocal microscopy as described below.
To assess the inhibitory activity of BAR peptide or BAR analogs, P. gingivalis (2 × 109 CFU per ml) was pretreated with various concentrations of peptide (0 to 16.8 μM) for 30 min at room temperature and then inoculated into the flow cell containing an established S. gordonii biofilm. The resulting P. gingivalis-S. gordonii biofilms were observed using Olympus FluoView 500 confocal laser scanning microscope (Olympus, Pittsburgh, PA) under a total magnification of ×600. Fluorescein isothiocyanate-labeled P. gingivalis was visualized using an argon laser while the hexidium iodide-labeled S. gordonii bacteria were observed using a helium-neon green (HeNe-G) laser. Columnar microcolonies (defined as P. gingivalis-S. gordonii coaggregates of ≥ 5μm in depth) were identified from 30 to 70 randomly chosen frames. Microcolony depth was determined via z-plane scans between 0 to 60 μm from the cover glass surface using the provided FluoView software (version 5). Statistical analyses were carried out using GraphPad InStat, version 3 (GraphPad Software Co.). A pairwise, nonparametric analysis of variance using Dunn's multiple comparison test was used to determine the statistical difference between the microcolony number and height between the experimental and control samples. A P value of less than 0.05 was considered significant.
In vivo model of periodontitis.
Specific-pathogen-free BALB/cByJ mice were obtained from the Jackson Laboratory (Bar Harbor, ME) at 10 weeks old and housed in the University of Louisville Research Resource Center animal facility. The mice were fed with Lab Diet 5001 meal (Purina Mills, LLC, Gray Summit, MO) during the entire experiment. The protocols used for the study were approved by the Institutional Animal Care and Use Committee as described in the federal guidelines for the care and use of laboratory animals.
Oral infection of mice was performed essentially as previously described by Baker et al. (3) but modified to first establish S. gordonii in the oral cavity prior to infection with P. gingivalis. A total of 12 mice per group were used per experiment. Animals were initially treated with sulfamethoxazole (MP Biomedical, Solon, OH) at a final concentration of 800 μg/ml and trimethoprim (Sigma, St. Louis, MO) at a final concentration of 400 μg/ml ad libitum for 10 days at 2-day intervals. Four days after the last antibiotic treatment, the mice were orally infected with 109 CFU of S. gordonii cells suspended in 1 ml of 2% carboxymethylcellulose (CMC; MP Biomedical, Solon, OH) in sterile PBS using a 2.25-mm feeding needle (Popper and Sons, Inc., New Hyde Park, NY). Following confirmation of S. gordonii colonization by PCR (see below), animals were infected five times with 107 CFU of P. gingivalis at 2-day intervals over a 10-day period. The animals were subsequently rested for 47 days and then euthanized via CO2 asphyxiation. Thus, the total duration of the experiment was 80 days.
To confirm S. gordonii colonization, oral samples were collected along the gingiva of the upper molars using a 15-cm sterile polyester-tipped applicator (Puritan Medical Products Co., Guilford, ME) on noninfection days and weekly for 4 weeks after the last oral infection. Samples were transported in BHIYE medium, then gently vortexed for 10 s, plated on Mitis-Salivarius agar (Difco, Sparks, MD), and incubated at 37°C for 48 h under aerobic conditions to select for streptococcal species. The resulting colonies were scraped from the plates and suspended in sterile nuclease-free water, and S. gordonii was identified by PCR using S. gordonii-specific primers that target the glucosyltransferase (gftG) gene, as described by Hoshino et al. (19) (Table 2). The PCR mixture contained the following: 1 μl of the total bacterial suspension, 11.25 μl of Platinum PCR Supermix (Invitrogen), 50 ng of forward and reverse S. gordonii-specific gftG primers, and 12.5 μl of nuclease-free water. The PCR was initially heated at 95°C for 10 min to lyse the bacteria, and the reaction was performed using the following conditions: 95°C for 30 s, 55°C for 30 s, and 72°C for 30 s for a total of 35 cycles. The products were visualized after electrophoresis in 2% agarose gels.
TABLE 2.
Primers used in the study
| Primer name | Sequence | Product size (bp) | Reference or source |
|---|---|---|---|
| gftG upstream | 5′-CTATGCGGATGATGCTAATCAAG-3′ | 440 | 19 |
| gftG downstream | 5′-GGAGTCGCTATAATCTTGTCAGA-3′ | ||
| Pg16SF | 5′-AGGCAGCTTGCCATACTGCG-3′ | 404 | 22 |
| Pg16SD | 5′-ACTGTTAGCAACTACCGATGT-3′ | ||
| Pg16SNF | 5′-CCCACCTGGGGAGTACGCCGGC-3′ | 231 | This study |
| Pg16SND | 5′-TGCGCTCGTTATGGCACTTAAGC-3′ |
Confirmation of P. gingivalis colonization was performed by nested PCR following the initial PCR using P. gingivalis-specific primers (22). Bacterial samples were collected from the mouse oral cavity as described above and stored in 300 μl of lysis solution (Promega, Madison WI). Total genomic DNA was purified using a Wizard Genomic DNA Purification Kit (Promega) as described by the manufacturer. The isolated DNA was then suspended in 20 μl of nuclease-free water. For the primary PCR, ∼50 ng of the isolated bacterial DNA was added to 11.25 μl of Platinum PCR Supermix (Invitrogen), and 50 ng of the forward and reverse P. gingivalis-specific 16S rRNA primers (Table 2, Pg16SF and Pg16SD), and the balance consisted of nuclease-free water. The reaction mixture was initially heated at 95°C for 10 min, followed by 35 cycles of the following conditions: 95°C for 30 s, 68°C for 45 s, and 72°C for 45 s. Approximately 1 μl of the resulting PCR product was then used as a template for the second-round nested PCR using primers Pg16SNF and Pg16SND (Table 2). The PCR was carried out essentially as previously described for the first-round PCR, and the resulting products were analyzed in a 2% agarose gel as described above.
Determination of alveolar bone loss in mouse maxilla.
Mouse skulls were defleshed by autoclaving for 15 min; they were then immersed in 3% hydrogen peroxide overnight at room temperature to remove any remaining musculature and washed with deionized water. Skulls were then soaked in 1% bleach solution for 30 s, sonicated at 14 V for 1 min, and then washed with water. To remove any remaining bacteria and tissues, the skulls were brushed with toothpaste and sonicated in 1% bleach solution for an additional 30 s at ∼14 V. The cleaned skulls were stained with 1% methylene blue for 15 s and rinsed with deionized water to remove excess dye. The stained skulls were allowed to air dry prior to measurement for alveolar bone loss.
Bone loss was assessed by measuring the distance between the alveolar bone crest (ABC) and the cemento-enamel junction (CEJ) at seven sites on the buccal side of the right and left maxillary molars, for a total of 14 measurements. This was accomplished using a dissecting microscope fitted with a video imaging marker measurement system (Sony model VIA-170K; Fryer) at a total magnification of ×40. Measurements were taken in millimeters. The average of the total bone loss for each mouse group was assessed and subtracted from the baseline bone loss observed in sham-infected mice.
Statistical differences in bone loss were analyzed using GraphPad Instat (La Jolla, CA). For pairwise parametric analysis of variance, a Tukey multiple comparison test was used to determine the statistical difference among the mouse groups. A P value of less than 0.05 was considered to be statistically significant.
Tissue culture.
Primary human gingival epithelial cells (HGEC) (kindly provided by Denis Kinane, University of Pennsylvania) were grown on 24-well collagen-coated plates (Becton Dickinson, Bedford, MA) and cultured using keratinocyte serum-free medium (KSFM; Gibco, Grand Island, NY) supplemented with penicillin/streptomycin (100 U/ml final concentration; St. Louis, MO), 10 μM β-mercaptoethanol (Sigma), 10 nM sodium selenite (Sigma), 10 μM l-aminoethanol (Sigma), insulin (10 μg/ml final concentration; Sigma), transferrin (5 μg/ml final concentration; Sigma), amphotericin B (50 ng/ml final concentration; Sigma), and bovine pituitary extract (50 mg/ml final concentration; Sigma). The epithelial cells were incubated at 37°C in the presence of 5% CO2 for 5 days until they reached ∼95% confluence.
Determination of BAR cytotoxicity.
The cytotoxicity of BAR peptide was determined using several approaches, including assessing hemolytic activity against sheep and human erythrocytes, assessing the effect of BAR on the viability of the human OBA-9 oral epithelial cell line and primary human gingival epithelial cells, and determining if BAR induces apoptosis of HGEC. The hemolytic activity of the BAR peptide was assessed as previously described by Gorr et al. (16). A total of 250 μl of 1% sheep or human erythrocytes was suspended in sterile phosphate-buffered saline containing 5% fetal bovine serum (FBS buffer). BAR peptide suspended in FBS (approximately 10 μg/ml and 100 μg/ml) was added to the erythrocyte suspension and incubated at 37°C for 0, 90, and 180 min. These peptide concentrations represent 1 and 10 times the amount of BAR required to inhibit P. gingivalis adherence to SspB by 100%. Cell lysis and hemoglobin release were analyzed by spectrophotometry at a wavelength of 541 nm.
Cell viability was assessed by quantifying total ATP levels in cell culture samples using CellTiter Glo reagent (Promega, Madison WI), which was reconstituted as described by the manufacturer. OBA-9 cells were incubated with the BAR peptide (34.0 μM final concentration) for 16 h at 37°C in 5% CO2. The supernatant was collected and assayed for lactate dehydrogenase content (see below). Cells were then washed three times with sterile PBS and lysed with 500 μl of 0.1% Triton X-100 for 30 min at 37°C. The lysates were collected and centrifuged at 1,000 × g for 10 min at 4°C, and 50 μl of supernatant was mixed with 50 μl of CellTiter Glo reagent and incubated at ambient temperature for 10 min in a black 96-well plate in the dark. Total light production was determined using a Vector 3 luminometer. As positive and negative controls, cells were incubated with 1 ng of staurosporine or medium only, respectively.
Cell lysis and/or membrane leakage was measured by the release of lactate dehydrogenase (LDH). LDH levels in the cell supernatant were measured using a Cytox96 nonradioactive cytotoxicity assay (Promega) as described by the manufacturer. Fifty microliters of supernatant from BAR-treated cells (see above) was added to LDH substrate and incubated at room temperature for 30 min; then reactions were terminated by the addition of 50 μl of stop solution. LDH activity was determined by measuring the optical density of the solution at a wavelength of 490 nm. Staurosporine-treated cells were used as positive controls while medium-only-treated cells served as a negative control.
Induction of apoptosis in HGEC following treatment with the BAR peptide was assessed through a direct terminal deoxynucleotidyltransferase-mediated dUTP-fluorescein nick end labeling (TUNEL) assay using a fluorescein in situ cell death detection kit (Roche, Indianapolis, IN). HGEC were grown to confluence prior to treatment with 34.0 μM (final concentration) BAR. The cells were incubated at 37°C under 5% CO2 for 16 h prior to labeling. TUNEL assays were performed essentially as described by the manufacturer. The cells were washed with sterile PBS three times, fixed with 4% paraformaldehyde (pH 7.4) at ambient temperature for 30 min, and washed two additional times with PBS. Fixed cells were then permeabilized with 0.1% Triton X-100 (Sigma, St. Louis, MO) and washed with PBS two times. Fifty microliters of TUNEL reaction mixture was added to the cells and incubated at 37°C for 60 min; then labeled cells were washed three times with PBS. An actin-specific counterstain of phalloidin was used to visualize whole cells. Analysis of the samples was performed by confocal microscopy using an Olympus FluoView 500 microscope (Olympus, Center Valley, PA). Samples were scanned using an argon laser (excitation wavelength, 488 nm) and helium-neon green laser (excitation wavelength, 543 nm). Cells positive for apoptosis are indicated by DNA fragmentation and thus the FITC staining of the nucleus.
RESULTS
The EXXP motif contributes to Mfa/SspB (BAR) interaction.
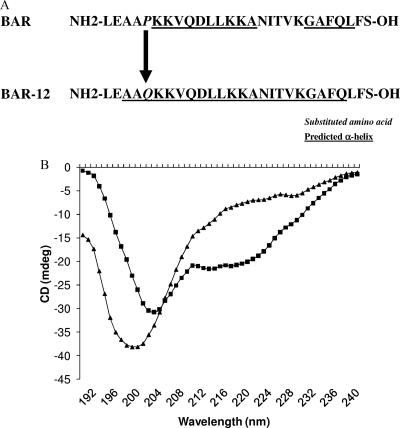
Our previous work identified two structural motifs in the BAR peptide that are essential for its interaction with Mfa (6, 8, 9). However, a 20-mer peptide containing these two motifs exhibited lower specific inhibitory activity than the parent peptide (27-mer), suggesting that additional amino acids in BAR may be involved in binding to Mfa. In addition, Duan et al. (13) previously showed that SspB is a calcium binding protein and that the BAR region was necessary for calcium binding activity. Interestingly, BAR possesses an EXXP motif upstream from VXXLL, and EXXP is known to contribute to calcium binding in some calcium transport channels. To determine if EXXP is important in the interaction of BAR with Mfa, peptides in which Glu1168 or Pro1171 was replaced with Gln were assessed as competitive inhibitors of P. gingivalis biofilm formation on streptococci, as described in Materials and Methods. As shown in Table 3, BAR-15 (containing a substitution of Gln for Pro1171) was a poor competitive inhibitor of P. gingivalis biofilm formation on S. gordonii relative to the native BAR peptide. At a concentration of 1.7 μM, BAR-15 inhibited biofilm formation by approximately 14%, whereas BAR inhibition was 70% (P < 0.001). At a concentration of 3.4 μM, BAR-15 inhibited P. gingivalis adherence by 35%, whereas BAR exhibited 82% inhibition (P < 0.01). In silico secondary structure analyses of the BAR and BAR-15 peptides predicted that the Gln/Pro1171 substitution conferred greater α-helical content of BAR-15 (Fig. 1A). Consistent with these predictions, circular dichroism spectroscopy showed that BAR-15 exhibited increased negative ellipticity at 222 nm (Fig. 1B), indicative of increased the α-helical character. Analysis of the CD spectrum using K2D software (2) indicated that the α-helical content of BAR-15 was 30%, whereas BAR was approximately 10% α-helix. Together, these results suggest that Pro1171 contributes to the interaction of BAR with Mfa and also contributes to the overall secondary structure of BAR.
TABLE 3.
Comparison of the specific inhibitory activity of BAR versus BAR analogues
| Peptide | Concn (μM) | No. of microcolonies per frame (mean ± SEM) | % Inhibitiona |
|---|---|---|---|
| No peptide | 0 | 6.02 ± 0.88 | 0 |
| BAR | 1.7 | 1.8 ± 0.47 | 70 |
| 3.4 | 1.08 ± 0.28 | 82 | |
| BAR-15 | 1.7 | 5.2 ± 0.87 | 14 |
| 3.4 | 3.9 ± 0.78 | 35 | |
| BAR-16 | 1.7 | 3.52 ± 0.72 | 42 |
Percent inhibition was determined relative to the no-peptide (0 μM) control using the following equation: (average number of microcolonies per frame at peptide concentration x/average number microcolonies per frame in the no-peptide control) × 100.
FIG. 1.
Secondary structure of BAR and BAR-15. (A) Secondary structure predictions of BAR and BAR-15 indicate that substitution of Gln for Pro1171 increases the α-helical content of BAR-15. (B) Circular dichroism spectra of BAR (triangles) and BAR-15 (squares) show increased negative ellipticity for BAR-15 at the λ of 222 nm, consistent with increased α-helical structure.
BAR-16, containing Gln for Glu1168, was a slightly less potent competitive inhibitor of P. gingivalis biofilm formation than BAR, inhibiting by 42% at 1.7 μM compared to 70% by the parent BAR peptide (Table 3). Thus, these structural and functional analyses showed that EXXP is a third important structural motif that contributes to the interaction of BAR with P. gingivalis (Mfa), but altering Pro1171 had a more profound effect on the interaction of BAR with P. gingivalis than substitution for Glu1168.
Constraining structural flexibility of BAR increases bioactivity.
The circular dichroism spectra analyzed in the experiments above indicated that BAR exhibits significant structural flexibility and contains approximately 50% random coil structure in solution, consistent with our previous observations (9). Since our previous studies indicated that the secondary structure of BAR was important for interaction with Mfa (8, 9), we hypothesized that limiting the structural flexibility of BAR in solution may improve its biofilm-inhibitory activity by locking the peptide into an “active” conformation. To address this, a conformationally constrained analog of BAR was synthesized by introducing cysteine residues for Leu1167 and Leu1191 to generate a cyclic disulfide bridged peptide (BAR-17) (Fig. 1A) that constrains peptide bond rotation in each of the important functional regions of BAR (i.e., EXXP, VXXLL, and NITVK). As shown in Table 4, BAR-17 was a more potent competitive inhibitor of P. gingivalis biofilm formation than the linear parent peptide. Indeed, BAR-17 inhibited P. gingivalis biofilm formation on streptococci by 47% and 59% at concentrations of 0.338 μM and 0.845 μM, respectively, whereas BAR exhibited only 10% and 34% inhibition at these peptide concentrations, suggesting that BAR-17 may assume a conformation that more closely resembles its structure in the intact SspB protein.
TABLE 4.
Inhibitory activity of BAR and BAR-17
| Peptide | Concn (μM) | No. of colonies per frame (mean ± SEM)a | % Inhibitionb |
|---|---|---|---|
| No peptide | 0 | 6.34 ± 0.71 | 0 |
| BAR | 0.17 | 7.16 ± 0.85 | −13.1 |
| 0.34 | 5.70 ± 0.60 | 9.9 | |
| 0.85 | 4.17 ± 0.64 | 34.0 | |
| 1.70 | 1.00 ± 0.25 | 84.2 | |
| BAR-17 | 0.17 | 4.95 ± 0.80 | 21.9 |
| 0.34 | 3.36 ± 0.66 | 46.9 | |
| 0.85 | 2.60 ± 0.54 | 58.9 | |
| 1.70 | 1.16 ± 0.31 | 81.7 |
The number of colonies was determined from a total of 30 to 51 frames.
After normalizing the average number of colonies/frame for each tested peptide concentration versus the no-peptide (0 μM) control, percent inhibition was determined using the following equation: (normalized average number of colonies per frame/average number of colonies in the no-peptide control per frame) × 100. An IC50of 0.52 μM was observed for BAR-17, which is significantly lower than the IC50of 1.25 μM observed for the original BAR peptide.
BAR inhibits P. gingivalis virulence and colonization of the oral cavity in vivo.
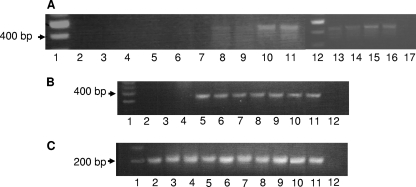
Baker et al. (3, 4) previously showed that P. gingivalis can successfully colonize the murine oral cavity and induce the resorption of alveolar bone, which is the clinical outcome of P. gingivalis oral infections in humans. To determine if BAR can inhibit P. gingivalis colonization of the oral cavity in vivo, we adapted the model of Baker et al. by first infecting mice with S. gordonii and subsequently challenging the animals with P. gingivalis in the presence and absence of BAR. Mice were initially infected with 1 × 109 CFU of S. gordonii DL-1 every other day for a period of 10 days. Samples of the oral flora were obtained and assessed for S. gordonii by PCR using primers specific for gtfG. As shown in Fig. 2A, the expected amplicon was detected in each animal that received S. gordonii inocula but not in the sham-infected animals, indicating that the mice were successfully colonized with streptococci. S. gordonii persisted in the oral cavity throughout the duration of the experiment (i.e., 7 weeks) (data not shown). As a control, other groups of animals were similarly infected with S. gordonii but in the presence of BAR. As shown in Fig. 2B, S. gordonii colonization of the murine oral cavity was not affected by treatment with BAR peptide. All plaque samples contained bacterial DNA and produced the expected amplicon from PCRs using primers specific for the 5S rRNA gene (Fig. 2C).
FIG. 2.
(A) S. gordonii colonization of the murine oral cavity. Successful infection and establishment of S. gordonii DL-1 was assessed by PCR using gtfG-specific primers. Microbial samples obtained from each of the sham-infected animals 1 week after the last round of infection (lanes 2 to 7) did not generate the 440-bp gtfG amplicon, whereas samples obtained at the same time point from S. gordonii-infected mice (lanes 8 to 11 and 13 to 16) each produced the gtfG amplicon. Lanes 1 and 12 contain DNA size markers, and lane 17 represents a no-template negative-control reaction. (B) BAR does not affect S. gordonii colonization of the mouse oral cavity. Murine oral plaque samples were analyzed by PCR for the presence of S. gordonii as described above. Lane 1, DNA size markers; lanes 2 to 4, sham-infected mice; lanes 5 to 7, mice infected with S. gordonii; lanes 8 to 10, mice infected with S. gordonii in the presence of 3.4 μM BAR peptide; lane 11, S. gordonii genomic DNA positive control; and lane 12, no-template negative control. (C) PCRs using universal primers for the 5S rRNA gene indicated that all the plaque samples contained bacterial DNA. Lane 1, DNA size markers, lanes 2 to 4, sham-infected mice; lanes 5 to 7, mice infected with S. gordonii; lanes 8 to 10, mice infected with S. gordonii in the presence of 3.4 μM BAR peptide; lane 11, S. gordonii genomic DNA positive control; and lane 12, no-template negative control.
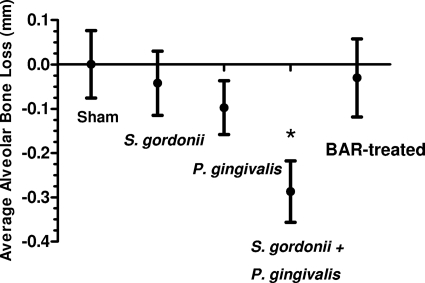
Mice that were infected with S. gordonii were subsequently infected with 107 CFU of P. gingivalis in the presence or absence of 3.4 μM BAR peptide, and the extent of alveolar bone resorption was determined 47 days after the final P. gingivalis inoculum. As shown in Fig. 3, animals that received a monospecies infection with 109 CFU of S. gordonii did not exhibit significant bone loss over the sham-infected controls. Monospecies infection with 107 CFU of P. gingivalis induced greater bone loss than sham infection of animals but this increase was not statistically significant (P = 0.057). Dual-species infection with both S. gordonii and P. gingivalis induced significantly greater bone loss (P = 0.01) than that in sham-infected or single-species-infected animals, suggesting that P. gingivalis colonization of the murine oral cavity occurs more efficiently in the presence of S. gordonii. However, S. gordonii-infected mice that were subsequently challenged with 107 CFU of P. gingivalis in the presence of BAR exhibited levels of bone loss that were similar to the level observed in the sham-infected controls, suggesting that BAR peptide potently inhibited the interaction of P. gingivalis with the established S. gordonii biofilm in the murine oral cavity, resulting in reduced bone loss.
FIG. 3.
BAR treatment reduces alveolar bone loss in P. gingivalis-infected mice. Animals that were infected with either 109 CFU of S. gordonii DL-1 or 107 CFU of P. gingivalis 33277 did not show a statistically significant increase in alveolar bone resorption over sham-infected controls. However, infection with 109 CFU of S. gordonii followed by 107 CFU of P. gingivalis induced significantly greater bone loss (*, P < 0.01) than sham-infected animals or mice infected with either organism alone. S. gordonii-infected mice that were subsequently challenged with 107 CFU of P. gingivalis in the presence of BAR exhibited levels of bone resorption similar to levels in sham-infected animals, suggesting that BAR inhibited adherence of P. gingivalis to streptococci in the oral cavity.
Cytotoxicity of BAR peptide.
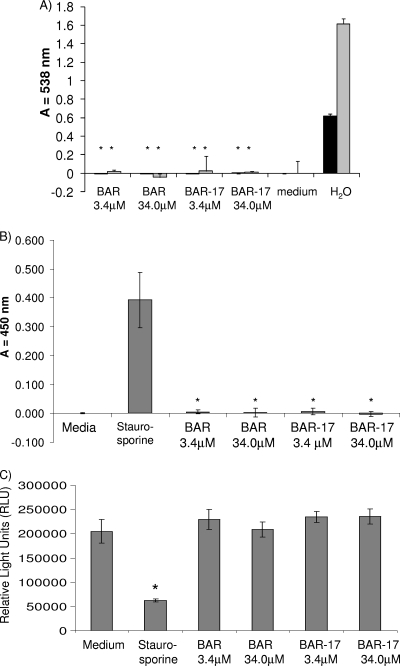
To begin to assess the potential therapeutic viability of BAR peptide, a series of toxicity tests was carried out to determine the effect of peptide exposure on primary human gingival epithelial cells and other cell types. To assess hemolytic activity of BAR, sheep and human erythrocytes were incubated with 3.4 μM or 34.0 μM BAR or BAR-17 (10-fold greater than required to completely block P. gingivalis adherence to streptococci in vitro or inhibit P. gingivalis virulence in vivo). As shown in Fig. 4A, the peptide did not possess detectable hemolytic activity at either concentration.
FIG. 4.
BAR is not toxic to human cells. (A) Hemolytic activity of BAR and BAR-17 was assessed by incubating sheep (gray bars) and human (black bars) erythrocytes with 3.4 μM and 34.0 μM BAR peptide for 180 min at 37°C. As a positive control, RBCs were lysed by incubation in distilled H2O. As a negative control, RBCs were incubated in FBS buffer without BAR peptide. No significant hemoglobin release was observed from peptide-treated cells relative to H2O-incubated samples (*, P < 0.001). (B) Peptide-induced necrosis was assessed by incubating OBA-9 oral epithelial cells with 3.4 μM or 34.0 μM BAR or BAR-17 as described in Materials and Methods. Compared with the staurosporine-treated positive-control reaction, no significant release of lactate dehydrogenase (*, P < 0.001) was measured from the peptide-treated or medium-treated cells. (C) Determination of total ATP content in BAR- or BAR-17-treated OBA-9 cells. ATP levels in peptide-treated cells did not differ significantly from levels in cells treated with medium alone, whereas ATP levels in the staurosporine-treated control cells was significantly lower (*, P < 0.001), suggesting that neither peptide significantly affects cell viability.
Since some amphipathic peptides possess membranolytic activity, we next sought to determine if BAR affected membrane structure or permeability. To accomplish this, human OBA-9 oral epithelial cells or HGEC were incubated with 34.0 μM BAR or BAR-17. As controls, cells were incubated overnight in medium containing 500 ng/ml staurosporine in medium containing 0.1% Triton X-100 or in sterile medium alone. The level of lactate dehydrogenase released from the cells into the culture supernatant was assayed and normalized against the medium-only samples. As shown in Fig. 4B, no release of LDH occurred from BAR-treated OBA-9 cells. In contrast, staurosporine (40% lysis; P < 0.001) exhibited significant release of LDH. To confirm that OBA-9 cells were viable following BAR treatment, total ATP was extracted and quantitated (Fig. 4C). There were no significant differences between the peptide-treated and medium-only samples. In contrast, staurosporine-treated OBA-9 cells had significantly lower ATP levels (P < 0.001). Together, these data show that BAR peptide does not induce cell necrosis or significantly affect OBA-9 cell viability. Similar results were obtained when HGEC were treated with BAR peptide (not shown).
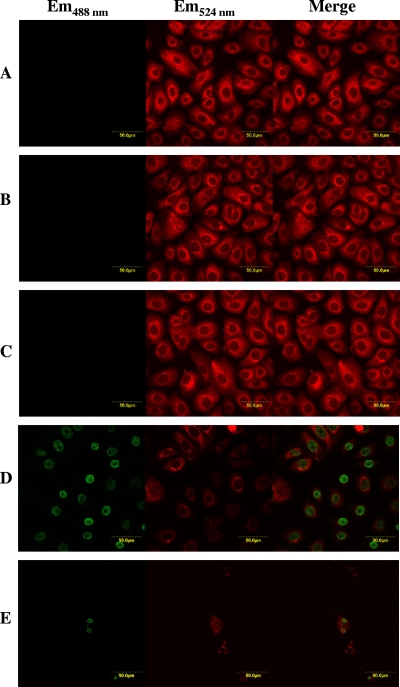
We next examined the ability of BAR to induce apoptosis in HGEC. Cells were treated with 34.0 μM BAR or BAR-17, 500 ng/ml staurosporine, or medium only, and apoptosis was determined using a TUNEL assay. As additional controls, terminal transferase-free labeling solution and DNase treatment were used as negative and positive controls, respectively. No FITC labeling occurred in cells treated with BAR or in terminal transferase-free labeling solution and medium-only negative-control cell groups (Fig. 5A to C). In contrast, the DNase-treated and staurosporine-treated cells exhibited significant labeling in the nuclei (Fig. 5D and E). Thus, BAR peptide does not induce apoptosis of HGEC. Finally, 34 μM BAR peptide did not inhibit planktonic growth of P. gingivalis or Aggregatibacter actinomycetemcomitans, suggesting that its inhibitory activity does not arise from an inherent antimicrobial activity (data not shown).
FIG. 5.
BAR does not induce apoptosis in HGEC. Apoptosis was determined using a TUNEL assay. Incubation of HGEC with BAR (A) did not induce apoptosis, and peptide-treated cells exhibited staining similar to cells incubated with medium only (B) or transferase-free buffer negative controls (C). DNase-treated (D) or staurosporine-treated (E) cells were TUNEL positive, as indicated by the green staining of the nuclei, suggesting that DNA cleavage and apoptosis occurred. All cell samples were counterstained with phalloidin (red). Em, emission wavelength.
DISCUSSION
P. gingivalis is a member of the “red group” of Gram-negative organisms that is strongly associated with adult periodontal disease (10, 18, 32). Although the main niche for P. gingivalis in the oral cavity is the subgingival pocket, it first colonizes the supragingival biofilm by interacting with specific bacteria such as Fusobacterium nucleatum and members of the commensal streptococci (e.g., Streptococcus sanguinis, S. oralis, and S. gordonii) (5, 21, 25, 27, 33). Thus, these initial interspecies interactions represent targets for therapeutic intervention to control periodontitis.
Our previous studies showed that adherence of P. gingivalis to streptococci is mediated by a protein-protein interaction between Mfa and antigen I/II (6, 27), and we identified a discrete structural region in antigen I/II that confers the adherence phenotype (11). Within this region, two essential functional motifs, VXXLL and NITVK, were identified. In this study, we show that the EXXP motif located upstream of VXXLL in BAR also contributes to the interaction of Mfa with BAR since peptides containing substitutions of Gln for either Glu1168 or Pro1171 were less potent competitive inhibitors of P. gingivalis adherence to streptococci. Interestingly, EXXP motifs have been suggested to contribute to calcium binding and transport (1, 15, 35, 36). In these proteins, the Pro residue confers a kink within an α-helical region, and Sansom et al. (30, 31) suggested that an acidic residue at position −3 from Pro may function in cation binding. Consistent with this, Duan et al. (13) showed that the S. gordonii antigen I/II protein, SspB, binds calcium and that the BAR region of antigen I/II was necessary for cation binding activity. However, while a recently published crystal structure of a portion of the C-terminal domain of SspB showed that calcium was bound by this domain, Glu1168 was not one of the residues that functioned to coordinate calcium in the reported structure (14). Interestingly, the crystal structure showed that Pro1171 constrained the amphipathic VXXLL α-helix such that its hydrophilic face was solvent exposed. Our results showing that replacement of Pro1171 with Gln significantly reduced inhibitory activity indicate that Pro1171 is important for the interaction of BAR with Mfa. Thus, our functional data of BAR peptide in solution, together with the crystal structure of Forsgren et al. (14), suggest that Pro1171 of SspB may play a role in maintaining the structural integrity of the core amphipathic α-helix of the nuclear receptor (NR) box in BAR rather than directly contributing to interaction with Mfa as a contact residue.
Our previous studies also suggested that the secondary structural characteristics of BAR were important for its interaction with Mfa (9, 11). The current results are consistent with this since CD spectroscopy showed that replacing Pro with Gln increased the α-helical content of the peptide and reduced the ability of the altered peptide to inhibit P. gingivalis biofilm formation. Furthermore, a cyclic structurally constrained peptide in which bond rotation along the peptide backbone was limited by a disulfide bond between Cys residues in place of Leu1167 and Leu1191 functioned as a better competitive inhibitor of P. gingivalis biofilm formation than the linear BAR peptide. This suggests that the EXXP, VXXLL, and NITVK motifs in the structurally constrained peptide may assume a conformation that more closely resembles the crystal structure of the BAR region determined by Forsgren et al. (14).
Previous studies have examined BAR-mediated inhibition of P. gingivalis adherence and biofilm development using in vitro binding and biofilm culture approaches. However, there is little in vivo evidence showing that P. gingivalis colonization and virulence are facilitated by commensal streptococci such as S. gordonii. To examine the in vivo effectiveness of BAR and to develop an in vivo approach to assess the activity of BAR structural analogs, we adapted the mouse model of periodontitis of Baker et al. (3) by first establishing S. gordonii in the oral cavity and then infecting animals with P. gingivalis. S. gordonii colonization of the murine oral cavity was readily established, was unaffected by BAR peptide, and persisted throughout the 70-day duration of the experiment. Single-species infection with S. gordonii DL-1 resulted in no significant resorption of alveolar bone even though streptococci were shown by PCR to persist in the oral cavity. Similarly, single-species infection using 107 CFU of P. gingivalis caused a small but not statistically significant increase in bone loss over sham-infected animals. Overall, these results are consistent with previous results from Baker et al. (3) and our prior studies (17, 37) that showed that infection using 109 CFU of P. gingivalis was required to induce significant levels of bone resorption. Interestingly, dual-species infection with 109 CFU of S. gordonii followed by 107 CFU of P. gingivalis induced significant bone loss, similar to that observed in the prior single infection experiments using 109 CFU of P. gingivalis (17, 37). This suggests that S. gordonii facilitates P. gingivalis colonization of the murine oral cavity. Animals that were infected with both species in the presence of 3.4 μM BAR peptide showed no increase in bone loss relative to sham-infected controls, suggesting that the BAR peptide inhibited colonization of the murine oral cavity by P. gingivalis by preventing its adherence to the streptococci. P. gingivalis could not be detected by PCR in animals that were infected with bacteria in the presence of BAR (data not shown). Thus, our in vivo results are consistent with prior in vitro results showing that 3.4 μM BAR completely inhibits P. gingivalis adherence to S. gordonii and its subsequent biofilm formation (8). These results, together with our initial observations that BAR peptide is nontoxic to P. gingivalis, human red blood cells (RBCs), and epithelial cells, suggest that the BAR peptide may represent a viable potential therapy to limit or reduce P. gingivalis colonization of the oral biofilm.
Acknowledgments
We thank J. B. Chaires and Nicola Garbett of the Biophysics Core Facility at the James Graham Brown Cancer Center at the University of Louisville for their assistance with circular dichroism spectroscopy. We also thank Denis Kinane (University of Pennsylvania School of Dental Medicine) for providing primary human gingival epithelial cells and Johnah Galicia for technical assistance.
This study was supported by PHS grant RO1DE12505 from the National Institute of Dental and Craniofacial Research.
Editor: A. Camilli
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Andersen, J. P., B. Vilsen, E. Leberer, and D. H. MacLennan. 1989. Functional consequences of mutations in the beta-strand sector of the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 264:21018-21023. [PubMed] [Google Scholar]
- 2.Andrade, M. A., P. Chacon, J. J. Merelo, and F. Moran. 1993. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 6:383-390. [DOI] [PubMed] [Google Scholar]
- 3.Baker, P. J., M. Dixon, and D. C. Roopenian. 2000. Genetic control of susceptibility to Porphyromonas gingivalis-induced alveolar bone loss in mice. Infect. Immun. 68:5864-5868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, P. J., L. Howe, J. Garneau, and D. C. Roopenian. 2002. T cell knockout mice have diminished alveolar bone loss after oral infection with Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 34:45-50. [DOI] [PubMed] [Google Scholar]
- 5.Bradshaw, D. J., P. D. Marsh, G. K. Watson, and C. Allison. 1998. Role of Fusobacterium nucleatum and coaggregation in anaerobe survival in planktonic and biofilm oral microbial communities during aeration. Infect. Immun. 66:4729-4732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brooks, W., D. R. Demuth, S. Gil, and R. J. Lamont. 1997. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infect. Immun. 65:3753-3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cengiz, M. I., S. Bal, S. Gokcay, and K. Cengiz. 2007. Does periodontal disease reflect atherosclerosis in continuous ambulatory peritoneal dialysis patients? J. Periodontol. 78:1926-1934. [DOI] [PubMed] [Google Scholar]
- 8.Daep, C. A., D. M. James, R. J. Lamont, and D. R. Demuth. 2006. Structural characterization of peptide-mediated inhibition of Porphyromonas gingivalis biofilm formation. Infect. Immun. 74:5756-5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daep, C. A., R. J. Lamont, and D. R. Demuth. 2008. Interaction of Porphyromonas gingivalis with oral streptococci requires a motif that resembles the eukaryotic nuclear receptor box protein-protein interaction domain. Infect. Immun. 76:3273-3280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Darveau, R. P., A. Tanner, and R. C. Page. 1997. The microbial challenge in periodontitis. Periodontol. 2000 14:12-32. [DOI] [PubMed] [Google Scholar]
- 11.Demuth, D. R., D. C. Irvine, J. W. Costerton, G. S. Cook, and R. J. Lamont. 2001. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infect. Immun. 69:5736-5741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dietrich, T., and R. I. Garcia. 2005. Associations between periodontal disease and systemic disease: evaluating the strength of the evidence. J. Periodontol. 76:2175-2184. [DOI] [PubMed] [Google Scholar]
- 13.Duan, Y., E. Fisher, D. Malamud, E. Golub, and D. R. Demuth. 1994. Calcium-binding properties of SSP-5, the Streptococcus gordonii M5 receptor for salivary agglutinin. Infect. Immun. 62:5220-5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Forsgren, N., R. J. Lamont, and K. Persson. 2010. Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. J. Mol. Biol. 397:740-751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fox, R. O., Jr., and F. M. Richards. 1982. A voltage-gated ion channel model inferred from the crystal structure of alamethicin at 1.5-Å resolution. Nature 300:325-330. [DOI] [PubMed] [Google Scholar]
- 16.Gorr, S. U., J. B. Sotsky, A. P. Shelar, and D. R. Demuth. 2008. Design of bacteria-agglutinating peptides derived from parotid secretory protein, a member of the bactericidal/permeability increasing-like protein family. Peptides 29:2118-2127. [DOI] [PubMed] [Google Scholar]
- 17.Hajishengallis, G., M. Wang, S. Liang, M. A. Shakhatreh, D. James, S. Nishiyama, F. Yoshimura, and D. R. Demuth. 2008. Subversion of innate immunity by periodontopathic bacteria via exploitation of complement receptor-3. Adv. Exp. Med. Biol. 632:203-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Holt, S. C., and J. L. Ebersole. 2005. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex,” a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38:72-122. [DOI] [PubMed] [Google Scholar]
- 19.Hoshino, T., M. Kawaguchi, N. Shimizu, N. Hoshino, T. Ooshima, and T. Fujiwara. 2004. PCR detection and identification of oral streptococci in saliva samples using gtf genes. Diagn. Microbiol. Infect. Dis. 48:195-199. [DOI] [PubMed] [Google Scholar]
- 20.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 21.Maeda, K., H. Nagata, Y. Yamamoto, M. Tanaka, J. Tanaka, N. Minamino, and S. Shizukuishi. 2004. Glyceraldehyde-3-phosphate dehydrogenase of Streptococcus oralis functions as a coadhesin for Porphyromonas gingivalis major fimbriae. Infect. Immun. 72:1341-1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matto, J., M. Saarela, S. Alaluusua, V. Oja, H. Jousimies-Somer, and S. Asikainen. 1998. Detection of Porphyromonas gingivalis from saliva by PCR by using a simple sample-processing method. J. Clin. Microbiol. 36:157-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nibali, L., F. D'Aiuto, G. Griffiths, K. Patel, J. Suvan, and M. S. Tonetti. 2007. Severe periodontitis is associated with systemic inflammation and a dysmetabolic status: a case-control study. J. Clin. Periodontol. 34:931-937. [DOI] [PubMed] [Google Scholar]
- 24.Okuda, K., R. Kimizuka, S. Abe, T. Kato, and K. Ishihara. 2005. Involvement of periodontopathic anaerobes in aspiration pneumonia. J. Periodontol. 76:2154-2160. [DOI] [PubMed] [Google Scholar]
- 25.Onagawa, M., K. Ishihara, and K. Okuda. 1994. Coaggregation between Porphyromonas gingivalis and Treponema denticola. Bull. Tokyo Dent. Coll. 35:171-181. [PubMed] [Google Scholar]
- 26.Paju, S., and F. A. Scannapieco. 2007. Oral biofilms, periodontitis, and pulmonary infections. Oral Dis. 13:508-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park, Y., M. R. Simionato, K. Sekiya, Y. Murakami, D. James, W. Chen, M. Hackett, F. Yoshimura, D. R. Demuth, and R. J. Lamont. 2005. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infect. Immun. 73:3983-3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ridker, P. M., and J. D. Silvertown. 2008. Inflammation, C-reactive protein, and atherothrombosis. J. Periodontol. 79:1544-1551. [DOI] [PubMed] [Google Scholar]
- 29.Robert, R., G. Grollier, J. P. Frat, C. Godet, M. Adoun, J. L. Fauchere, and P. Dore. 2003. Colonization of lower respiratory tract with anaerobic bacteria in mechanically ventilated patients. Intensive Care Med. 29:1062-1068. [DOI] [PubMed] [Google Scholar]
- 30.Sansom, M. S. 1991. The biophysics of peptide models of ion channels. Prog. Biophys. Mol. Biol. 55:139-235. [DOI] [PubMed] [Google Scholar]
- 31.Sansom, M. S., I. D. Kerr, and I. R. Mellor. 1991. Ion channels formed by amphipathic helical peptides. A molecular modelling study. Eur. Biophys. J. 20:229-240. [DOI] [PubMed] [Google Scholar]
- 32.Socransky, S. S., A. D. Haffajee, M. A. Cugini, C. Smith, and R. L. Kent, Jr. 1998. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 25:134-144. [DOI] [PubMed] [Google Scholar]
- 33.Stinson, M. W., K. Safulko, and M. J. Levine. 1991. Adherence of Porphyromonas (Bacteroides) gingivalis to Streptococcus sanguis in vitro. Infect. Immun. 59:102-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thorman, R., M. Neovius, and B. Hylander. 2009. Clinical findings in oral health during progression of chronic kidney disease to end-stage renal disease in a Swedish population. Scand. J. Urol. Nephrol. 43:154-159. [DOI] [PubMed] [Google Scholar]
- 35.Vilsen, B., J. P. Andersen, D. M. Clarke, and D. H. MacLennan. 1989. Functional consequences of proline mutations in the cytoplasmic and transmembrane sectors of the Ca2+-ATPase of sarcoplasmic reticulum. J. Biol. Chem. 264:21024-21030. [PubMed] [Google Scholar]
- 36.Vogel, H., and F. Jahnig. 1986. The structure of melittin in membranes. Biophys. J. 50:573-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang, M., M. A. Shakhatreh, D. James, S. Liang, S. Nishiyama, F. Yoshimura, D. R. Demuth, and G. Hajishengallis. 2007. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J. Immunol. 179:2349-2358. [DOI] [PubMed] [Google Scholar]