Abstract
Chlamydia trachomatis contains a conserved ∼7.5-kb plasmid. Loss of the plasmid results in reduced glycogen accumulation, failure to activate TLR2, and reduced infectivity. We hypothesized that reduced infectivity functions as a means of selection for plasmid maintenance. We directly examined the biological significance of the reduced infectivity associated with plasmid deficiency by determining the relative fitness of plasmid-deficient CM972 versus that of wild-type C. muridarum Nigg in mixed inocula in vitro and in vivo. C. muridarum Nigg rapidly out-competed its plasmid-cured derivative CM972 in vitro but was not competitive with CM3.1, a derivative of CM972 that has reverted to a normal infectivity phenotype. C. muridarum Nigg also effectively competed with CM972 during lower and upper genital tract infection in the mouse, demonstrating that strong selective pressure for plasmid maintenance occurs during infection. The severity of oviduct inflammation and dilatation resulting from these mixed infections correlated directly with the amount of C. muridarum Nigg in the initial inoculum, confirming the role of the plasmid in virulence. Genetic characterization of CM972 and CM3.1 revealed no additional mutations (other than loss of the plasmid) to account for the reduced infectivity of CM972 and detected a single base substitution in TC_0236 in CM3.1 that may be responsible for its restored infectivity. These data demonstrate that a chlamydial strain that differs genetically from its wild-type parent only with respect to the lack of the chlamydial plasmid is unable to compete in vitro and in vivo, likely explaining the rarity of plasmid-deficient isolates in nature.
Chlamydia trachomatis is an obligate intracellular Gram-negative bacterium. It is the world's leading cause of preventable blindness and the most common sexually transmitted pathogen. In females, C. trachomatis infects epithelial cells of the cervix and ascends canalicularly to infect the oviduct mucosa, where it elicits inflammation and scarring.
The developmental cycle of C. trachomatis is unusual, as represented by two distinct forms: the elementary body (EB), which is infectious but metabolically inert, and the reticulate body (RB), which replicates intracellularly within an intracytoplasmic vesicle or inclusion. Attachment and entry of EBs into epithelial cells is essential for establishment of infection. Multiple candidate host cell receptors (7, 8, 21, 27, 28) and chlamydial adhesins (9, 23, 31, 34) have been previously identified, and there is evidence that several mechanisms of entry may be utilized for uptake (see a review by A. Dautry-Varsat et al. in reference 6).
C. trachomatis carries a highly conserved plasmid of approximately 7.5 kb. Naturally occurring plasmid-deficient clinical isolates are very rare; only three such strains have been previously described and characterized (10, 20, 29). Loss of the plasmid has been associated with several chlamydial phenotypes, including (i) failure to accumulate glycogen within intracellular inclusions (14, 19); (ii) inability to activate innate immune signaling via TLR2 (18), a critical step in the development of immunopathology (5); and (iii) reduced ability to establish infection both in vitro (19) and in vivo (18). The strong selection serving to maintain the plasmid in strains infecting humans suggests that at least one of these phenotypes confers a survival advantage to the organism.
Vaginal inoculation of mice with the plasmid-cured derivative of C. muridarum Nigg, strain CM972, resulted in an infection of normal duration, but reduced bacterial burden (∼10-fold), and an absence of oviduct pathology (18). In vivo, the impact of reduced infectivity seemed minor compared to the ∼100-fold reduction observed in cell culture. The reduced bacterial load did not reflect a replication defect, because this strain's growth properties were indistinguishable from those of the Nigg parent once inside the cell (19). The absence of pathology was attributed to failure of the organism to activate TLR2, since a derivative of CM972 called CM3.1 fails to elicit oviduct pathology although it has reverted to normal infectivity without reestablishing TLR2-dependent signaling (18).
We hypothesized that reduced infectivity functions as a selection mechanism for plasmid maintenance. We directly examined the biological significance of the reduced infectivity that we have found to be associated with plasmid deficiency by determining the relative fitness of C. muridarum Nigg versus that of CM972 in mixed inocula, where plasmid-deficient chlamydiae must compete with wild-type microorganisms to establish infection. In vitro experiments revealed that C. muridarum Nigg rapidly out-competed its plasmid-cured derivative CM972 but was not competitive with CM3.1. In addition, C. muridarum Nigg effectively competed with CM972 during lower and upper genital tract infection in the mouse, demonstrating strong selective pressure for plasmid maintenance during infection. The severity of oviduct inflammation and dilatation resulting from these mixed infections correlated directly with the amount of plasmid-containing C. muridarum Nigg in the initial inoculum, confirming the role of the plasmid in virulence. Genetic characterization of both CM972 and CM3.1 revealed no additional mutations (other than loss of the plasmid) to account for the reduced infectivity of CM972 and detected a single base substitution in TC_0236 in CM3.1 that may be responsible for the restored infectivity observed for this strain.
MATERIALS AND METHODS
Animals.
Female C57BL/6J female mice were purchased from The Jackson Laboratory (Bar Harbor, ME) and were provided food and water ad libitum in an environmentally controlled room with a cycle of 12 h of light and 12 h of darkness. Age-matched mice (7 to 12 weeks old) were used for all of the experiments. All experimental protocols were approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh.
Strains, cell lines, and culture conditions.
C. muridarum strains Nigg (17), CM972 (19), and CM3.1 (18) were cultured in L929 fibroblast cells. Properties of these strains are described in Table 1. Cells were infected at an approximate multiplicity of infection (MOI) of 0.5 to 1 before being centrifuged for 1 h at 37°C. The cell culture medium was then removed and replaced with 1× Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (FBS), gentamicin (20 μg ml−1), and 0.1 μg of cycloheximide ml−1. Infected cells were harvested and transferred into sucrose phosphate glutamate (SPG) buffer at 40 h postinfection, sonicated, and maintained at −80°C. Bacteria were subsequently titrated as inclusion-forming units (IFU) (12) by the use of a genus-specific fluorescently tagged anti-chlamydial lipopolysaccharide (LPS) monoclonal antibody (Bio-Rad, Hercules, CA).
TABLE 1.
Characteristics of C. muridarum strains used in this study
| C. muridarum strain | Plasmid | Glycogen | Infectivity | LPS staining | TCA08 staining |
|---|---|---|---|---|---|
| Nigg | + | + | Wild type | + | + |
| CM972 | − | − | Reduced | + | − |
| CM3.1 | − | − | Wild type | + | − |
In vitro competition.
Suspensions comprising C. muridarum Nigg and CM972 or CM3.1 were inoculated onto L929 cell monolayers at fixed ratios ranging from 1:10 to 1:106 under culture conditions favoring entry of plasmid-containing chlamydiae (i.e., without centrifugation) and harvested after 40 h of incubation, a time at which we have determined bacterial yield is at the steady state (17), by lysing the infected cells in 1 ml of SPG buffer. A 0.1-ml volume of the resulting chlamydial suspension was then applied to a fresh L929 cell monolayer for a total of 5 sequential passages. The remaining suspension was frozen at −80°C for subsequent analysis using iodine staining (26) to detect glycogen within chlamydial inclusions and to distinguish inclusions formed by plasmid-deficient chlamydiae from the wild type.
In vivo competition.
Groups of 5 mice were subcutaneously treated with 2.5 mg of medroxyprogesterone acetate (Depo-Provera; Upjohn, Kalamazoo, MI) 7 days prior to vaginal inoculation. The progesterone was given to synchronize all mice in an anestrous state. Groups of anesthetized mice were intravaginally inoculated with 30 μl of SPG buffer containing a total of 1 × 105 IFU of chlamydiae in mixed amounts of Nigg and CM972 (Table 2). Infection was monitored by swabbing the vaginal vault and cervix on alternate days from day 3 through day 9 postinfection. Swab eluates were titrated for IFUs by the use of a mouse monoclonal antibody directed against chlamydial LPS (Santa Cruz Biotechnologies, Santa Cruz, CA) or a custom rabbit polyclonal antibody raised against recombinant TCA08, a protein encoded by the C. muridarum plasmid. Groups of mice were sacrificed on either day 10 or day 14 of infection, and oviducts were harvested and analyzed for chlamydial infectious burden via plaque assay (day 10) as previously described (18) or for pathology (day 14). Individual plaques were immunofluorescently stained using the rabbit polyclonal antibody recognizing TCA08 (expressed by C. muridarum Nigg but not by CM972).
TABLE 2.
Inocula for in vivo competition analysis
| Nigg/CM972 ratio | Mouse inoculum (IFU/mouse) |
|
|---|---|---|
| Nigg | CM972 | |
| 1:0 | 1 × 105 | 0 |
| 0:1 | 0 | 1 × 105 |
| 1:1 | 5 × 104 | 5 × 104 |
| 1:10 | 1 × 104 | 1 × 105 |
| 1:100 | 1 × 103 | 1 × 105 |
Genome analysis.
Total genomic DNA from gradient-purified, DNase-treated C. muridarum Nigg, CM972, and CM3.1 EBs was isolated using a Masterpure complete DNA isolation kit (Epicenter Biotechnologies, Madison, WI), according to the manufacturer's protocol. Comparative genome hybridization was performed using a C. muridarum-specific DNA microarray (NimbleGen Systems, Inc., Madison, WI), the design of which was derived from the published and annotated sequence (24). CM972 was subjected to a single pass, while CM3.1 was analyzed twice and hybridized with C. muridarum Nigg genomic DNA in each case. Genomic DNA purified from the C. muridarum Nigg strain passaged in the laboratory of Harlan Caldwell was generously provided by him for use as the hybridization control for the analysis of CM972 and for one of the two analyses of CM3.1. Putative single nucleotide polymorphisms (SNPs) identified by hybridization were PCR amplified and sequenced on both strands at the University of Pittsburgh Genomics and Proteomics Core Facility to confirm their presence or absence in the genome. Sequence differences were ultimately confirmed by manual examination of the trace data.
Histopathology.
Genital tract tissues from each mouse were removed and fixed in 10% buffered formalin. Longitudinal 4-μm sections were cut and stained with hematoxylin and eosin. Each anatomic site (ectocervix, endocervix, uterine horn, and oviduct) was independently assessed by a pathologist in a blinded experimental design for the presence of neutrophilic inflammation, lymphocytic-monocytic inflammation, plasma cell erosion, and fibrosis. A four-tiered semiquantitative scoring system was used to evaluate the extent of inflammation and fibrosis as follows: 0, normal; 1+, rare foci (minimal presence of parameter); 2+, scattered (1 to 4) aggregates or mild diffuse increase in parameter; 3+, numerous aggregates (more than four) or moderate diffuse or confluent areas of parameter; and 4+, severe diffuse infiltration or confluence of parameters. In addition, luminal distention of the uterine horns and dilatation of the oviducts was graded from 1 to 4, with grade 4 representing the greatest severity. Right and left uterine horns and right and left oviducts were evaluated individually.
Statistics.
Statistical comparisons between the groups of mice for levels of infection determined from endocervical swabs were made by two-way repeated-measure (RM; days and infection group) analysis of variance (ANOVA) with a post hoc Tukey test as a multiple-comparison procedure. Differences in oviduct bacterial burden among the groups were determined by ANOVA. The Kruskal-Wallis one-way ANOVA rank test was used to determine significant differences in the pathological data between groups. A Spearman rank order correlation coefficient was used to measure the strength of association between inoculation doses and oviduct inflammatory scores. Linear regression analysis was used to determine best-fit values and for comparisons of slopes for in vitro competition data using a method equivalent to analysis of covariance (16), while a nonlinear regression method was used to evaluate in vivo competition in order to accommodate logarithmic replication of infecting bacteria. Both analyses were performed using GraphPad Prism 5 (GraphPad Software Inc., San Diego, CA). SigmaStat software was utilized for all other statistical analysis (SPSS). P values < 0.05 were considered significant.
RESULTS
C. muridarum Nigg successfully competes with CM972 but not with CM3.1 in vitro.
Attempts to evaluate the significance of infectivity defects associated with plasmid-deficient chlamydiae have been challenging. Carlson et al. did not detect any reduction in the infectivity of strain 25667R, a plasmid-deficient clinical isolate, during in vitro tissue culture experiments, although the strain was unable to establish infection in the mouse genital tract, unlike L2/434/Bu, a plasmid-containing strain of the same serovar (4). We devised an in vitro competitive assay that could be used to examine the role of the plasmid in infectivity.
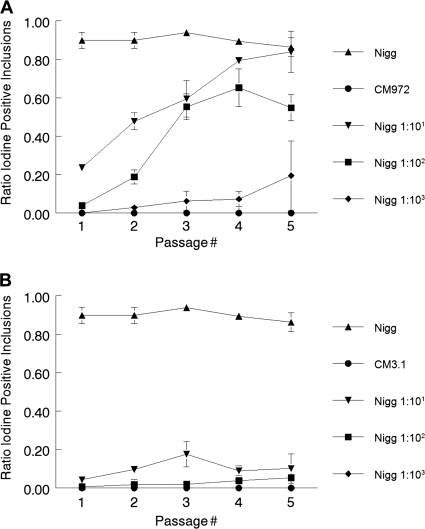
Suspensions comprising C. muridarum Nigg and CM972 or CM3.1 were sequentially passaged 5 times by inoculation onto L929 cell monolayers at fixed ratios ranging from 1:10 to 1:106 under culture conditions favoring the entry of plasmid-containing chlamydiae (i.e., without centrifugation). Samples from each passage were cultured in L929 cells under conditions that equally favored infection by both strains at an MOI of ∼0.3, under which conditions fewer than 40% of the cells were infected and the likelihood of an individual cell becoming infected with two or more bacteria was significantly reduced. The cells were fixed and stained with iodine to detect Nigg [iodine(+)] and CM972 [iodine(−)] inclusions. The ratio of iodine positivity {[iodine(+) inclusions]/[iodine(−) + iodine(+)]} for each sample was determined and plotted against passage number (Fig. 1A). C. muridarum Nigg and CM972 passaged alone were used as controls. Regardless of passage number, ≥86% of Nigg inclusions in monoculture stained positively with iodine, while inclusions formed by CM972 always stained negatively. When mixed inocula comprising Nigg and CM972 at ratios of 1:10, 1:100, or 1:103 were passaged, the population of iodine(+) inclusions in each harvested passage increased by ∼1.4-fold to 1.8-fold and, in the case of infections where Nigg was inoculated at 1:10 and 1:100, it became the predominant strain within 3 to 5 passages. When inoculated at a ratio of 1:103, iodine(+) inclusions expanded from being undetectable to representing ∼20% of the population by the fifth passage. In mixed infections where Nigg was present at ratios ≥ 1:104, iodine(+) inclusions were never detected (data not shown). Linear regression analysis revealed that the slope of the line best fitting the data obtained from Nigg in monoculture experiments did not differ significantly from zero (P = 0.31). The slopes of the lines fitted to the 1:10 (0.15 ± 0.02) and 1:100 (0.14 ± 0.04) infections were almost identical, indicating that the rate of change observed was likely associated with Nigg's infectivity advantage and was not dynamic with respect to the size of the population over time.
FIG. 1.
C. muridarum Nigg successfully competes with CM972 in vitro but not with CM3.1. (A) Competition with CM972. L929 fibroblasts were infected without centrifugation with C. muridarum Nigg, with CM972, or with a mixed inoculum and were incubated for 40 h before harvest. This process was repeated 5 times, using 1/10 of the prior passage volume as the inoculum each time. Samples from each passage were subsequently infected into L929 fibroblasts with centrifugation and incubated for 40 h before being fixed and stained with iodine to detect Nigg (iodine-positive) and CM972 (iodine-negative) inclusions. (B) Competition with CM3.1. L929 fibroblasts were infected (without centrifugation) with C. muridarum Nigg or CM3.1 or with a mixed inoculum for passage and analysis as described above. Data presented were obtained from the simultaneous counting and iodine scoring of 150 to 200 inclusions/well, with two replicates per passage. Each experiment was performed twice.
In contrast, parallel experiments performed identically using CM3.1 instead of CM972 (Fig. 1B) revealed that Nigg was unable to expand within the overall chlamydial population regardless of input ratio or passage number, indicating that Nigg had no advantage in the face of a plasmid-deficient strain with normal infectivity. In all cases, linear regression analysis yielded best-fit lines with slopes which were not significantly different (P = 0.24) and a pooled slope estimate of 0.003. Significantly, we also noted that the slope of the line fitted to data from the Nigg-CM3.1 mixed infection at a ratio of 1:10 did not deviate significantly from zero (P = 0.42), suggesting that additional passaging of mixed inocula would not likely result in expansion of Nigg numbers.
Genomic characterization of C. muridarum plasmid-deficient derivatives CM972 and CM3.1.
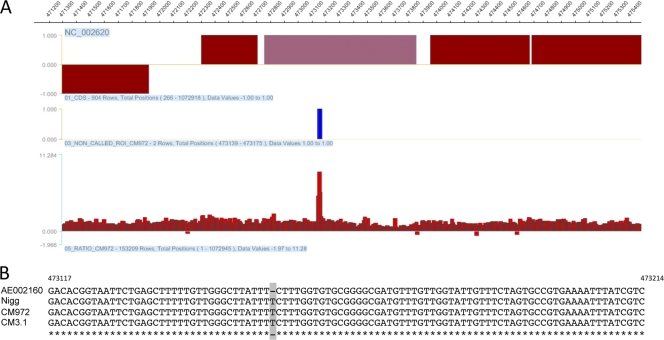
To exclude the possibility that an unrecognized chromosomal mutation was responsible for the infectivity defect in CM972 and to characterize the mutation predicted in CM3.1, we employed comparative genomic hybridization to investigate the genetic relatedness of Nigg to its plasmid-deficient derivatives. This is a powerful technique that can be used to detect copy number differences and the presence of genomic aberrations (33) and to identify mutations in bacterial genomes for evolutionary, epidemiological, or virulence studies (1, 4). Using the service provided by Nimblegen Corp. Inc. (Madison, WI), we analyzed genomic DNA obtained from CM972. A single mutation was detected in CM972 within TC_0412: specifically, an additional T at 473155 (Fig. 2A) that prematurely terminates the open reading frame. This gene is predicted to encode a protein of unknown function that is 365 amino acids in length. The presence of the additional base within the coding sequence of TC_0412 is predicted to result in a truncated protein of 149 amino acids. Significantly, we determined that this mutation was also present in our parental Nigg strain (Fig. 2B), indicating that the mutation in CM972 was present prior to novobiocin treatment. No additional differences between the strains were detected to the limit of resolution provided by this technique, indicating that the phenotypic changes observed can be attributed solely to the absence of the plasmid.
FIG. 2.
Comparative genome hybridization detects an SNP present in C. muridarum Nigg and its derivatives CM972 and CM3.1. (A) Graphic representation of data obtained from tiled-microarray analysis of the C. muridarum strain Nigg genome. A single nucleotide polymorphism was identified at 473155. (B) PCR products encompassing TC_0412 were generated and sequenced. The aligned sequence obtained from this analysis, representing the genomes from bp 473117 to bp 473214 and with the additional T insertion indicated, is presented.
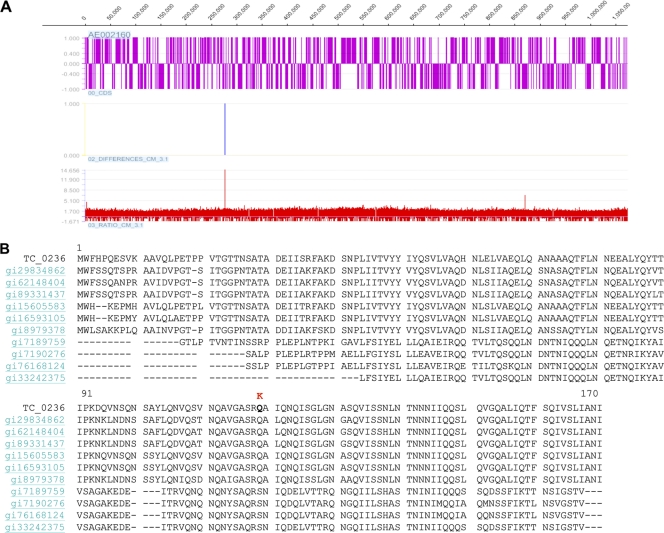
A similar analysis of CM3.1 was performed using the same technique, and two independent passes performed using the CM3.1 genome detected only the previously identified mutation in TC_0412 and a G→T single base substitution in TC_0236 (Fig. 3A), a highly conserved open reading frame (ORF) of unknown function which encodes a member of the Pfam DUF 720 family. This family represents a group of 22 highly conserved proteins of unknown function present in Chlamydia and Chlamydophila species (11). The result of this mutation was a Q119K nonsynonymous amino acid substitution at the C-terminal end of the protein encoded by this gene (Fig. 3B). The substitution was not anticipated to have polar effects on coding sequences located downstream, because the TC_0236 coding sequence remained in frame.
FIG. 3.
Comparative genomic hybridization detects a novel SNP in C. muridarum CM3.1. (A) Graphic representation of data obtained from a tiled microarray analysis of the C. muridarum CM3.1 genome. An SNP is identified at bp 276796. (The TC_0412 SNP was not detected in this analysis, because the C. muridarum Nigg isolate also carrying this mutation was used as the hybridization control). (B) Sequence comparison of TC_0236 with the most closely related members of pfam DUF 720 family. The nonconservative amino acid substitution Q119K is indicated.
Plasmid-containing C. muridarum Nigg competes effectively with CM972 to establish itself and predominate during infection of the lower genital tract of mice.
Our previous characterization of C. muridarum CM972 infection in the lower genital tract of C3H/HeOuJ mice indicated that, although the course was similar to that of Nigg, peaking in the lower genital tract on day 5, a consistent reduction of ∼10-fold in CM972 shedding was observed during the first week of infection compared to Nigg results (18). However, the CM972 infection became indistinguishable from Nigg infection by day 10 and mice ultimately cleared infection by day 42, suggesting that the plasmid-associated infectivity defect of CM972 was not as profound as it appeared to be in vitro and might be important only during early infection, prior to the development of an adaptive immune response specific for chlamydia.
We investigated the competitive advantage associated with plasmid carriage in vivo by inoculating groups of five female C57BL/6J mice with C. muridarum Nigg alone, with CM972 alone, or with mixed inocula as indicated in Table 2. The intensity and duration of infection in each group were determined by quantitative culture of endocervical swabs obtained on alternate days from day 3 to day 9 after infection. To distinguish between C. muridarum Nigg and CM972, we used a rabbit polyclonal antibody raised against recombinant TCA08, a protein encoded by the C. muridarum plasmid. Immunofluorescent staining with anti-TCA08 successfully discriminated between plasmid-containing Nigg and the plasmid-cured CM972 that did not stain. We used a mouse monoclonal antibody specific for chlamydial LPS that stained both strains to enumerate the total bacterial burden. We did not perform costaining with both anti-LPS and anti-TCA08, because this would have necessitated the use of confocal imaging to discriminate between inclusions staining positively with both antibodies (Nigg) and those staining positively for LPS alone (CM972) in titrations of all vaginal swabs collected from the infected mice over the course of the experiment.
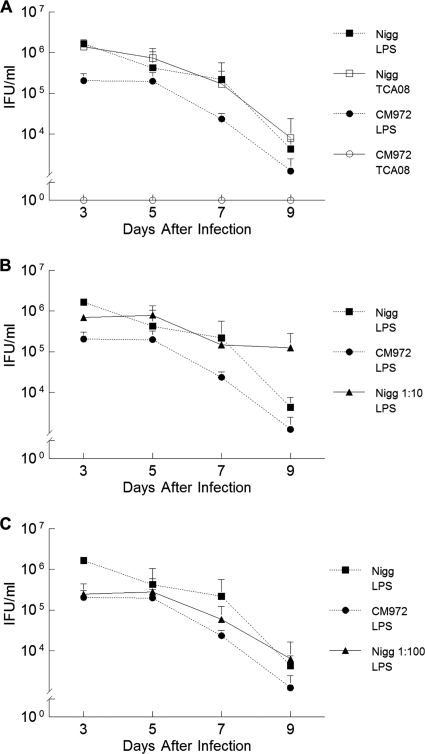
In C57BL/6J mice infected with either Nigg or CM972, peak infection was observed on day 3, the first day of sampling (Fig. 4A). Thereafter, infection began to clear and CM972 maintained a statistically significant reduction in chlamydial shedding of ∼1 log (P = 0.03; two-way RM ANOVA) over the remaining days, a finding consistent with our previous studies. As expected, anti-TCA08 staining, which detects only plasmid-containing chlamydiae, revealed an infection course no different than that observed via anti-LPS staining of the infection course in mice infected solely with Nigg (P = 0.84) (Fig. 4A).
FIG. 4.
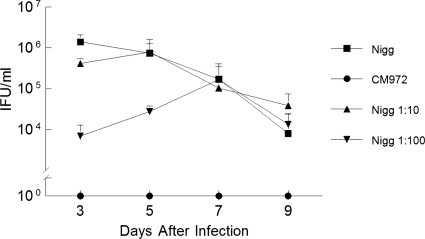
Cervicovaginal shedding of chlamydiae by mice infected with C. muridarum Nigg and CM972. (A) Groups of mice were infected with either Nigg or CM972, and cervicovaginal shedding over subsequent days was assessed by immunofluorescent staining using a mouse monoclonal antibody directed against chlamydial LPS (Nigg plus CM972) or a rabbit polyclonal antibody directed against pMoPn-encoded TCA08. Immunofluorescent staining with an anti-TCA08 antibody successfully discriminates between C. muridarum Nigg (open squares) and plasmid-deficient CM972 (open circles at 100) shed by infected mice. CM972 infection indicated by anti-LPS staining (closed circles) was less infectious than Nigg (closed squares) (P = 0.03 by two-way RM ANOVA). (B) Cervicovaginal shedding of chlamydiae by mice infected with C. muridarum Nigg and CM972 at ratio of 1:10. The total bacterial burden was detected by immunofluorescent staining with anti-LPS antibody. Results of shedding by parallel groups infected with Nigg or CM972 alone are indicated as controls. By day 3 of infection, mice infected with the mixed inoculum were shedding more chlamydiae than mice infected solely with CM972 (P = 0.04; two-way RM ANOVA with multiple comparisons), indicating that Nigg had quickly become predominant. (C) Cervicovaginal shedding of chlamydiae by mice infected with C. muridarum Nigg and CM972 at ratio of 1:100. The total bacterial burden was detected by immunofluorescent staining with anti-LPS antibody. Results of shedding by parallel groups infected with Nigg or CM972 alone are indicated as controls. Mice infected with 103 Nigg plus 105 CM972 shed chlamydiae in amounts similar to those seen with mice infected with wild-type Nigg by day 5 of infection, and levels were equivalent to those seen with mice infected with Nigg alone by day 9. Data represent the means ± standard deviations of the results for each group of 5 mice, and the experiment was performed twice.
Cervicovaginal shedding of chlamydiae by mice infected with C. muridarum Nigg and CM972 at a ratio of 1:10.
Once again, the total bacterial burden was detected by immunofluorescent staining with anti-LPS antibody. Shedding by parallel groups infected with Nigg or CM972 alone was determined to serve as a control. Mice infected with 1 × 104 Nigg and 1 × 105 CM972 (at a ratio of 1:10) rapidly shed chlamydiae in amounts similar to those seen with mice infected with 105 wild-type Nigg. By day 5, mice infected with mixed inocula were shedding more bacteria than mice infected with CM972 (Fig. 4B). The overall trend of infection at a 1:10 ratio of Nigg to CM972 seemed to follow that of an infection due solely to Nigg. When the early infection course for mice infected with Nigg and CM972 at a 1:10 ratio was compared with that for mice infected with CM972 only, a statistically significant difference was determined on each day by the Holm-Sidak method of multiple comparisons. Although the groups were statistically significantly different by two-way RM ANOVA, a significant interaction was noted between the factors of groups and days. Thus, Nigg was the predominant strain in the mixed infection, despite CM972 being present in a 10-fold-higher proportion in the inoculum. The detection of increased amounts of bacteria on day 9 in this group may have been due to a delay in the adaptive immune response, although no similar increase on day 9 was observed when a 1:100 ratio of Nigg to CM972 was used for infection (Fig. 4B and C).
Mice infected with 103 Nigg and 105 CM972 (at a ratio of 1:100) shed plasmid-containing chlamydiae in amounts similar to those seen with mice infected with 105 wild-type Nigg by 7 days postinfection.
At day 3, the total bacterial burden indicated by anti-LPS staining was equivalent to that seen with mice infected solely with CM972 (Fig. 4C). No statistically significant difference was seen between the monitored course of the mixed infection (P = 0.585) and that of mice infected solely with CM972. However, the bacterial burden began approaching that of mice infected solely with Nigg on day 7, and by day 9, the numbers of bacteria cultured were approximately equivalent to those of mice infected solely with Nigg.
Mixed infections reach the infection levels of mice infected with Nigg alone over time.
Use of the anti-TC08 antibody to identify plasmid-carrying chlamydiae allowed us to evaluate the ability of Nigg to out-compete CM972 over time (Fig. 5). Infection with 1 × 104 Nigg and 1 × 105 CM972 (1:10) revealed an infection course that approximated that of mice infected solely with Nigg (P = 0.848). This again verified that Nigg rapidly became the predominant bacteria present and that it established itself early when inoculated at 10-fold-smaller amount than the CM972 inoculum. Nonlinear regression was applied to the data obtained from the experiments performed with the mice infected with Nigg and CM972 at a ratio of 1:100 in order to quantify the rate of Nigg expansion in vivo. The slope of the line best fitting the data obtained by sampling on days 3 to 7 was 0.43 ± 0.56, suggesting that Nigg expanded within the population of chlamydiae infecting the lower genital tract by ∼2.7-fold per day during the first week of infection.
FIG. 5.
Detection of C. muridarum Nigg expansion in vivo using anti-TCA08 antibody. Mixed-inoculum infections were compared with wild-type Nigg and CM972 infections using anti-TCA08 antibody to detect plasmid-containing bacteria. The results of infection with Nigg plus CM972 (1:10) were similar to the results of infection with Nigg alone (squares) on all days examined (P = 0.85). Plasmid-positive chlamydiae detected in mice infected with Nigg plus CM972 (1:100) became equal in number to those detected in mice infected with Nigg alone by day 7. Data represent the means ± standard deviations of the results for each group of 5 mice, and the experiment was performed twice.
Oviduct inflammation directly correlates with the inoculum dose of Nigg.
We previously demonstrated that CM972 ascended to the oviducts of mice but failed to cause chronic oviduct pathology. We attributed the lack of oviduct pathology to the combination of the inability of CM972 to activate TLR2 and the infectivity defect associated with this strain. The inability of CM972 to compete with Nigg in vitro and in the lower genital tract in vivo led us to hypothesize that Nigg would out-compete CM972 during ascension in vivo, resulting in an absence of CM972 in the oviducts. Groups of mice were sacrificed on day 10 for titration of the oviduct burden or on day 14 for evaluation of pathology. Oviducts were titrated by plaque assay. The monolayers were stained with neutral red to reveal the total number of plaques and were subsequently fixed and stained with anti-TCA08 antibody to distinguish plaques formed by Nigg. Plaque titration of oviduct tissue is sensitive and also facilitates simultaneous quantitation of bacterial burden and detection of plasmid carriage without a need for confocal imaging.
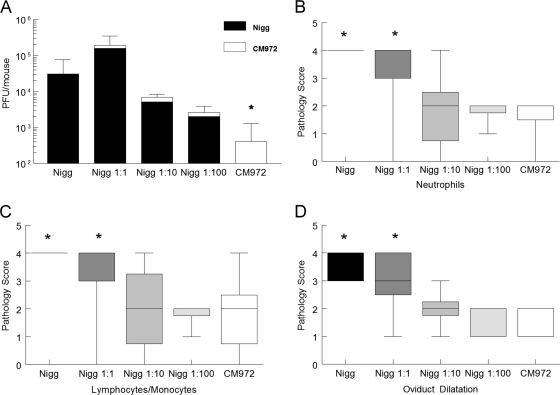
Anti-TCA08 staining indicated that Nigg predominated in the oviducts regardless of its initial proportion in the inoculums (Fig. 6A), confirming the competitive advantage of Nigg. In every mixed infection group, CM972 contributed minimally to oviduct infection. Regardless of the ratio of Nigg to CM972, in all groups infected with Nigg, the mean oviduct bacterial burden was not statistically significantly different from those of mice infected with 105 Nigg alone but was statistically significantly greater than the mean burden of mice infected solely with 105 CM972. These data demonstrate further the significant survival advantage of Nigg compared to CM972 (Fig. 6A).
FIG. 6.
Nigg is the predominant strain in the oviduct. (A) Oviducts were harvested from mice sacrificed on day 10 of infection. The bacterial load associated with the oviduct homogenates from each mouse was determined by a plaque assay. Plaques formed by Nigg were identified via immunofluorescent staining using anti-TCAO8 antibody. *, P < 0.05 for CM972 versus each group infected with Nigg (one-way ANOVA with multiple comparisons). The Nigg groups were not statistically significantly different from each other. (B to D) Genital tracts from mice in each group sacrificed on day 14 were scored for histopathology. Median pathology scores for acute inflammatory infiltrates (neutrophils) (B), chronic inflammatory infiltrates (lymphocytes and monocytes) (C), and oviduct dilatation (D) were significantly increased in mice infected with either 105 Nigg or 5 × 104 Nigg (Nigg ratio of 1:1) compared to the results obtained with mice in each of the other three groups. *, P < 0.05 by ANOVA on ranks with multiple-comparison test. Results for mice infected with 1:10 and 1:100 ratios of Nigg were not different from those seen with mice infected with CM972. Data represent the means ± standard deviations of the results obtained for each group of 5 mice in a single experiment. Boxes extend from the 25th to 75th percentiles, and whiskers indicate the 5th to 95th percentiles.
Despite the detection of similar bacterial loads in the oviducts of mice infected with Nigg alone and in those in which the inoculum combined Nigg with CM972, median pathology scores for levels of neutrophils (acute inflammation) (Fig. 6B), lymphocytes-monocytes (chronic inflammation) (Fig. 6C), erosion, and dilatation (Fig. 6D) were each significantly increased in the oviducts of mice infected solely with Nigg (105) and in mice infected with 5 × 104 Nigg plus 5 × 104 CM972 (1:1 Nigg) compared to the results seen with groups infected with Nigg inocula at ≤104 or with the group solely infected with CM972. In contrast, no statistical difference was determined for median oviduct pathology scores for mice infected with CM972 alone determined to those calculated for mice infected with Nigg inocula at ≤104. Median pathology scores determined for the oviducts of mice infected with a 1:1 ratio of Nigg to CM972 were similar to those determined for mice infected solely with Nigg.
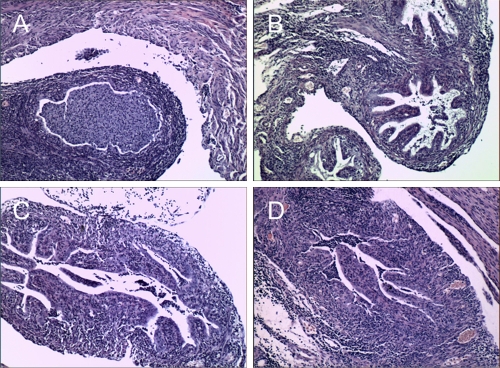
A significant positive correlation was determined for the level of neutrophils in the oviducts and the dose of Nigg used for infection (r = 0.975; P = 0.02). Although not statistically significant, trends for positive correlations were also noted for levels of lymphocytes and monocytes (r = 0.87; P = 0.08) and oviduct dilatation (r = 0.89; P = 0.08). Thus, the degrees of inflammation and pathology detected at 14 days postinfection were directly proportional to the amount of Nigg in the inoculum despite the presence of 105 CM972 in each mixed inoculum group (Fig. 7 ). The increase in oviduct pathology observed on day 14 postinfection with increasing doses of Nigg may have been due to a combined effect of the increasing rapidity of effect resulting from increased doses and the enhanced oviduct burden with plasmid-carrying chlamydiae resulting in a more rapid induction of inflammation via TLR2, a plasmid-associated property not expressed by CM972.
FIG. 7.
Severity of oviduct pathology is proportional to the amount of Nigg in the inoculum. The figure panels show representative oviduct histologic sections from a mouse sacrificed for histopathology on day 14 after infection with the following inocula: (A) 105 Nigg; (B) 104 Nigg plus 105 CM972 (Nigg ratio, 1:10); (C) 103 Nigg plus 105 CM972 (Nigg ratio, 1:100); (D) 105 CM972. Each section is shown at ×100 magnification. Data represent the means ± standard deviations of the results obtained for the individually scored oviducts from each group of 5 mice in a single experiment.
DISCUSSION
Plasmid-deficient strains infecting humans are extremely rare despite the high prevalence of chlamydial infection. Furthermore, they do not appear to spread within the human population, despite their potential for avoidance of detection by the plasmid-targeted diagnostic tests currently in use. The recent emergence of a new variant of C. trachomatis in Sweden that was undetectable because it carried a 377-bp deletion in the plasmid that overlapped the diagnostic detection sequence of several commercial PCR-based tests (25) demonstrates that lack of the plasmid could be advantageous in the face of standard clinical screening and treatment programs, provided that there is no “fitness” compromise that leads to reduced transmission. Although multiplication delays arising from mutations in genes central to DNA replication or protein biosynthesis have previously been demonstrated to impair chlamydial fitness (2, 3), plasmid loss has not been associated with replication defects in vitro (4, 18). In this paper, we demonstrate that the plasmid-associated infectivity defect that we have identified in C. muridarum CM972 significantly impairs its competitiveness during in vivo infection and that this likely serves as a primary mechanism of selection for plasmid maintenance in chlamydiae infecting the genital tract.
In vitro competition assays have recently been employed to compare the relative levels of fitness of related chlamydial strains (32) and may model in vivo processes more effectively than a tissue culture plaque assay, which requires relatively prolonged incubation, or than assays in which developmental cycle asynchrony over multiple rounds of reinfection may amplify minor differences that are of limited in vivo relevance. We observed that C. muridarum Nigg competed effectively with plasmid-deficient CM972 over 5 serial passages, almost doubling its share of the total bacterial population with each passage. In contrast, the Nigg population was unable to expand when cocultured with CM3.1, a plasmid-deficient derivative of CM972 which carries a secondary mutation restoring infectivity to wild-type levels. Use of CM3.1, the normally infectious strain, allowed us to control for additional plasmid-associated factors that may have contributed to infection, including the absence of glycogen, which has been proposed to enhance chlamydial infection in the mouse genital tract infection model, possibly by promoting enhanced adherence (4). CM3.1 remained predominant over Nigg through all passages, suggesting that failure to accumulate glycogen as a consequence of plasmid loss does not impair competitive fitness in vitro. Linear regression analysis revealed that the rate at which Nigg competed with CM972 was constant regardless of input ratio, indicating that the infectivity defect associated with the absence of the plasmid is fixed and is not additionally influenced by population dynamics. Strikingly, the rate at which Nigg expanded within the chlamydial population in vivo versus in vitro appeared somewhat higher (∼5.4-fold/44 h versus ∼1.6-fold/44 h) but was considerably lower than the ∼100-fold infectivity difference estimated via plaquing efficiency. Thus, we conclude that the in vitro competition assay more accurately reflects differences in infectivity in vivo than the assay examining plaquing efficiency. Nevertheless, plaquing efficiency remains useful for the selection and characterization of mutants that have reverted to wild-type infectivity and provides quantitative comparisons rapidly and easily.
In vivo, C. muridarum Nigg competed very effectively with CM972 in the lower genital tract, ultimately becoming the predominant strain. In all groups of mice receiving mixed inocula, Nigg levels exceeded CM972 levels by day 9 postinfection. The impact of reduced infectivity on chlamydial burden was most pronounced during the first week of infection, a time point at which mice infected with Nigg and CM972 at a ratio of 1:100 shed fewer chlamydiae than mice infected at ratios of 1:1 and 1:10. The impact of host immune responses on chlamydial infection by both strains was shown by a decrease in chlamydial shedding in all groups (including controls) by day 5 that continued to the conclusion of the experiment. Our previous study revealed no differences in the quality of T cell-mediated immunity induced by C. muridarum Nigg or its plasmid-deficient derivatives, and these strains appear equally subject to host clearance mechanisms, with no differences in the duration of infection (18). Consequently, it is unlikely that differential host responses to Nigg or CM972 influenced the proportion of each strain detected in cervicovaginal swabs. Together, these findings suggest that when individual plasmid-deficient chlamydiae attempt to arise spontaneously during infection, they struggle to expand within the population of chlamydiae infecting the lower genital tract and thus would be noncompetitive with the cotransmitted parental population if transferred to a new host. One caveat with respect to our study is that we used a model of female genital tract infection, and the male genital tract may impose different selective pressures. All three of the characterized plasmid-deficient clinical isolates were recovered from infected males (10, 20, 29), a surprising observation, considering the high rates of genital tract infection and chlamydial screening in women. It is possible that males might represent a more permissive environment for carriage and transmission of plasmid-deficient strains than females.
Although we have yet to determine the mechanism by which the chlamydial plasmid controls chlamydial infectivity, comparative genome hybridization clarified the genetic lineage of Nigg and its plasmid-deficient derivatives and confirmed that CM972 differs genetically from its parent, C. muridarum Nigg, only by the absence of the plasmid. Interestingly, the single G→T nucleotide polymorphism detected in CM3.1 results in a nonconservative Q-K substitution in the protein encoded by the TC_0236 gene and implicates TC_0236 in the restoration of infectivity observed in this strain.
The derivation of both CM972 and CM3.1 from the parental Nigg strain is apparent from our detection of a unique single base deletion in the TC_0412 coding sequence that is common to all. In a recent study, Ramsey et al. reported the sequencing of strain “Nigg2,” the C. muridarum isolate maintained in the laboratory of Roger Rank (22). They noted a total of 16 SNPs or indels in this strain compared with the published sequence (24). The C. muridarum parent of CM972 is a plaque-purified isolate obtained from Roger Rank's stock, and yet we failed to detect any of the identified mutations by comparative hybridization whereas we identified a deletion in TC_0412 at bp 473155 that was not observed in their analysis. This may reflect reduced sensitivity of comparative genome hybridization compared with direct sequencing. We therefore PCR amplified and sequenced the regions of the genome that contained some of the mutations that they noted (specifically, TC_0107 [bp 126406 to 126475] and TC_0341-342 [bp 403623 to 404884]) and detected their presence (7/7) in our Nigg isolate, indicating, as expected, that our strain is closely related to the Nigg2 strain, although the sequence of our isolate matches the published Nigg sequence at bp 473705 (TC_0412) rather than the base deletion that they observed. Nevertheless, hybridization was sensitive enough to consistently detect the mutation in TC_0412 associated with our Nigg lineage when CM972 and CM3.1 were hybridized with genomic DNA from a C. muridarum Nigg isolate provided by Harlan Caldwell that is homologous to the published Nigg strain at this position and did not detect this mutation when hybridized with genomic DNA from our parental Nigg isolate (both strains mutated at this position). A significant difference is not registered in the analysis when both the control and test strains diverge from the sequence used to generate the oligonucleotide probes assembled on the array, explaining why the sequence differences identified by Ramsey et al. were not identified. Consequently, we agree that the commonly passaged stock of C. muridarum Nigg is heterogeneous (22) and conclude that the C. muridarum Nigg strain that we have used in our experiments represents a single clone closely related but not identical to the Nigg2 strain. That the two derivatives of the Rank stock contain different SNPs in TC_0412 suggests that this gene may be a mutagenic “hot spot” for chlamydiae. Interestingly, TC_0412 is the C. muridarum homolog of the CT135 locus that has been recently associated with enhancement of C. trachomatis D/UW-3/Cx genital infection in mice when inactivated by frameshift mutations (30). Our studies indicate that inactivation of TC_0412 does not impair the pathogenicity of our parental Nigg strain and that the presence of this mutation in its plasmid-deficient derivatives does not contribute to the infectivity defect under investigation. We previously selected novobiocin for use as a curing agent in the derivation of CM972, because it is not genotoxic but acts by inhibiting the GyrB subunit of DNA gyrase to alter supercoiling and consequently disproportionately impacts replication of plasmid molecules, ultimately leading to daughter cells with plasmids that are reduced in copy numbers or absent with respect to the genome equivalent (13). However, our findings do not preclude the possibility of the presence of other, still undetected, mutations in these strains. Studies directed toward identifying the chlamydial effectors of plasmid-associated infectivity are ongoing via the derivation and characterization of additional “CM3.1-like” mutants by direct genome sequencing and proteomic analysis.
The results of examination of the extent of oviduct infection and pathological outcome after infection with mixed inocula were limited but nevertheless revealing. Nigg became the predominant strain in the oviducts in all groups, reflecting the findings determined in experiments investigating the lower genital tract. Furthermore, the degree of oviduct inflammation and pathology correlated directly with the amount of Nigg inoculated into the lower genital tract. The CM972 burden detected in the oviducts of mice receiving mixed inocula did not differ from that of mice inoculated with CM972 alone, suggesting that its ability to ascend was neither enhanced nor inhibited by coinfection with the Nigg strain. Interestingly, oviduct inflammation and pathology results determined on day 14 postinfection correlated directly with the amount of Nigg inoculated into the lower genital tract, despite detection of similar levels of Nigg bacteria in the oviducts on day 10. These data indicate that higher doses of TLR2-activating chlamydiae facilitate rapid ascension to the oviduct, resulting in enhanced induction of inflammation. These data are important because they demonstrate that chlamydial factors that facilitate the establishment of ascending infections contribute to virulence directly and that host responses that prevent or limit upper tract infection are likely to be protective against immunopathology. It is possible that inflammation may increase at later time points in mice infected with lower inocula of Nigg (15). However, we speculate that coinfection with non-TLR2-activating CM972 at the level of the cervix would result in the induction of protective T cell responses, leading to normal clearance of chlamydiae from the oviducts of mice infected with lower inoculum doses, and would consequently limit pathology associated with TLR2-activation.
In summary, using a plasmid-cured strain of C. muridarum that differs genetically from its parent only with respect to lack of the plasmid, we have demonstrated that the plasmid confers a significant survival advantage in vitro and in vivo. During mixed infections in vivo, Nigg numbers steadily increased as a percentage of the lower genital tract bacterial burden and predominated in the oviducts even when the strain was inoculated at a ratio of 1:100 with respect to CM972, and the amount of Nigg in the original inoculum correlated directly with degree of oviduct pathology. These results clearly demonstrate a competitive advantage associated with normal infectivity that may serve as a mechanism of selective pressure for plasmid maintenance sufficient to account for the predominance of plasmid-containing strains in clinical infection.
Acknowledgments
We greatly appreciate the support and assistance provided by Harlan Caldwell and Lazlo Kari.
Editor: S. R. Blanke
Footnotes
Published ahead of print on 25 October 2010.
REFERENCES
- 1.Albert, T. J., D. Dailidiene, G. Dailide, J. E. Norton, A. Kalia, T. A. Richmond, M. Molla, J. Singh, R. D. Green, and D. E. Berg. 2005. Mutation discovery in bacterial genomes: metronidazole resistance in Helicobacter pylori. Nat. Methods 2:951-953. [DOI] [PubMed] [Google Scholar]
- 2.Binet, R., and A. T. Maurelli. 2007. Frequency of development and associated physiological cost of azithromycin resistance in Chlamydia psittaci 6BC and C. trachomatis L2. Antimicrob. Agents Chemother. 51:4267-4275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binet, R., and A. T. Maurelli. 2005. Fitness cost due to mutations in the 16S rRNA associated with spectinomycin resistance in Chlamydia psittaci 6BC. Antimicrob. Agents Chemother. 49:4455-4464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Carlson, J. H., W. M. Whitmire, D. D. Crane, L. Wicke, K. Virtaneva, D. E. Sturdevant, J. J. Kupko III, S. F. Porcella, N. Martinez-Orengo, R. A. Heinzen, L. Kari, and H. D. Caldwell. 2008. The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 76:2273-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Darville, T., J. M. O'Neill, C. W. Andrews, Jr., U. M. Nagarajan, L. Stahl, and D. M. Ojcius. 2003. Toll-like receptor-2, but not toll-like receptor-4, is essential for development of oviduct pathology in chlamydial genital tract infection. J. Immunol. 171:6187-6197. [DOI] [PubMed] [Google Scholar]
- 6.Dautry-Varsat, A., A. Subtil, and T. Hackstadt. 2005. Recent insights into the mechanisms of Chlamydia entry. Cell Microbiol. 7:1714-1722. [DOI] [PubMed] [Google Scholar]
- 7.Davis, C. H., J. E. Raulston, and P. B. Wyrick. 2002. Protein disulfide isomerase, a component of the estrogen receptor complex, is associated with Chlamydia trachomatis serovar E attached to human endometrial epithelial cells. Infect. Immun. 70:3413-3418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elwell, C. A., A. Ceesay, J. H. Kim, D. Kalman, and J. N. Engel. 2008. RNA interference screen identifies Abl kinase and PDGFR signaling in Chlamydia trachomatis entry. PLoS Pathog. 4:e1000021. doi: 10.1371/journal.ppat.1000021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fadel, S., and A. Eley. 2007. Chlamydia trachomatis OmcB protein is a surface-exposed glycosaminoglycan-dependent adhesin. J. Med. Microbiol. 56:15-22. [DOI] [PubMed] [Google Scholar]
- 10.Farencena, A., M. Comanducci, M. Donati, G. Ratti, and R. Cevenini. 1997. Characterization of a new isolate of Chlamydia trachomatis which lacks the common plasmid and has properties of biovar trachoma. Infect. Immun. 65:2965-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Finn, R. D., J. Tate, J. Mistry, P. C. Coggill, S. J. Sammut, H. R. Hotz, G. Ceric, K. Forslund, S. R. Eddy, E. L. L. Sonnhammer, and A. Bateman. 2008. The Pfam protein families database. Nucleic Acids Res. 36:D281-D288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kelly, K. A., E. A. Robinson, and R. G. Rank. 1996. Initial route of antigen administration alters the T-cell cytokine profile produced in response to the mouse pneumonitis biovar of Chlamydia trachomatis following genital infection. Infect. Immun. 64:4976-4983. (Erratum, 65:2508.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luttinger, A. 1995. The twisted ‘life’ of DNA in the cell: bacterial topoisomerases. Mol. Microbiol. 15:601-606. [DOI] [PubMed] [Google Scholar]
- 14.Matsumoto, A., H. Izutsu, N. Miyashita, and M. Ohuchi. 1998. Plaque formation by and plaque cloning of Chlamydia trachomatis biovar trachoma. J. Clin. Microbiol. 36:3013-3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Maxion, H. K., W. Liu, M. H. Chang, and K. A. Kelly. 2004. The infecting dose of Chlamydia muridarum modulates the innate immune response and ascending infection. Infect. Immun. 72:6330-6340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Motulsky, H., and A. Christopoulos. 2004. Fitting models to biological data using linear and non-linear regression: a practical guide to curve fitting. Oxford University Press Inc., New York, NY.
- 17.Nigg, C. 1942. An unidentified virus which produces pneumonia and systemic infection in mice. Science 95:49-50. [DOI] [PubMed] [Google Scholar]
- 18.O'Connell, C. M., R. R. Ingalls, C. W. Andrews, Jr., A. M. Skurlock, and T. Darville. 2007. Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 179:4027-4034. [DOI] [PubMed] [Google Scholar]
- 19.O'Connell, C. M., and K. M. Nicks. 2006. A plasmid-cured Chlamydia muridarum strain displays altered plaque morphology and reduced infectivity in cell culture. Microbiology 152:1601-1607. [DOI] [PubMed] [Google Scholar]
- 20.Peterson, E. M., B. A. Markoff, J. Schachter, and L. M. De La Maza. 1990. The 7.5-kb plasmid present in Chlamydia trachomatis is not essential for the growth of this microorganism. Plasmid 23:144-148. [DOI] [PubMed] [Google Scholar]
- 21.Puolakkainen, M., C. C. Kuo, and L. A. Campbell. 2005. Chlamydia pneumoniae uses the mannose 6-phosphate/insulin-like growth factor 2 receptor for infection of endothelial cells. Infect. Immun. 73:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ramsey, K. H., I. M. Sigar, J. H. Schripsema, C. J. Denman, A. K. Bowlin, G. A. Myers, and R. G. Rank. 2009. Strain and virulence diversity in the mouse pathogen Chlamydia muridarum. Infect. Immun. 77:3284-3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Raulston, J. E., C. H. Davis, T. R. Paul, J. D. Hobbs, and P. B. Wyrick. 2002. Surface accessibility of the 70-kilodalton Chlamydia trachomatis heat shock protein following reduction of outer membrane protein disulfide bonds. Infect. Immun. 70:535-543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ripa, T., and P. Nilsson. 2006. A variant of Chlamydia trachomatis with deletion in cryptic plasmid: implications for use of PCR diagnostic tests. Euro. Surveill. 11:E061109. [DOI] [PubMed] [Google Scholar]
- 26.Schachter, J., and C. R. Dawson. 1978. Human chlamydial infections, p. 83-88. PSG Publishing Company, Littleton, MA.
- 27.Shimazaki, K., A. M. Chan, R. J. Moniz, M. Wadehra, A. Nagy, C. P. Coulam, S. Mareninov, E. M. Lepin, A. M. Wu, K. A. Kelly, J. Braun, and L. K. Gordon. 2009. Blockade of epithelial membrane protein 2 (EMP2) abrogates infection of Chlamydia muridarum murine genital infection model. FEMS Immunol. Med. Microbiol. 55:240-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shimazaki, K., M. Wadehra, A. Forbes, A. M. Chan, L. Goodglick, K. A. Kelly, J. Braun, and L. K. Gordon. 2007. Epithelial membrane protein 2 modulates infectivity of Chlamydia muridarum (MoPn). Microbes Infect. 9:1003-1010. [DOI] [PubMed] [Google Scholar]
- 29.Stothard, D. R., J. A. Williams, B. Van Der Pol, and R. B. Jones. 1998. Identification of a Chlamydia trachomatis serovar E urogenital isolate which lacks the cryptic plasmid. Infect. Immun. 66:6010-6013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sturdevant, G. L., L. Kari, D. J. Gardner, N. Olivares-Zavaleta, L. B. Randall, W. M. Whitmire, J. H. Carlson, M. M. Goheen, E. M. Selleck, C. Martens, and H. D. Caldwell. 2010. Frameshift mutations in a single novel virulence factor alter the in vivo pathogenicity of Chlamydia trachomatis for the female murine genital tract. Infect. Immun. 78:3660-3668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Su, H., L. Raymond, D. D. Rockey, E. Fischer, T. Hackstadt, and H. D. Caldwell. 1996. A recombinant Chlamydia trachomatis major outer membrane protein binds to heparan sulfate receptors on epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 93:11143-11148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Unemo, M., H. M. Seth-Smith, L. T. Cutcliffe, R. J. Skilton, D. Barlow, D. Goulding, K. Persson, S. R. Harris, A. Kelly, C. Bjartling, H. Fredlund, P. Olcen, N. R. Thomson, and I. N. Clarke. 2010. The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology 156:1394-1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walsh, T., J. M. McClellan, S. E. McCarthy, A. M. Addington, S. B. Pierce, G. M. Cooper, A. S. Nord, M. Kusenda, D. Malhotra, A. Bhandari, S. M. Stray, C. F. Rippey, P. Roccanova, V. Makarov, B. Lakshmi, R. L. Findling, L. Sikich, T. Stromberg, B. Merriman, N. Gogtay, P. Butler, K. Eckstrand, L. Noory, P. Gochman, R. Long, Z. Chen, S. Davis, C. Baker, E. E. Eichler, P. S. Meltzer, S. F. Nelson, A. B. Singleton, M. K. Lee, J. L. Rapoport, M. C. King, and J. Sebat. 2008. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320:539-543. [DOI] [PubMed] [Google Scholar]
- 34.Zhang, J. P., and R. S. Stephens. 1992. Mechanism of C. trachomatis attachment to eukaryotic host cells. Cell 69:861-869. [DOI] [PubMed] [Google Scholar]