Abstract
Porphyromonas gingivalis has been implicated in the etiology of adult periodontitis. In this study, we examined the viability of Drosophila melanogaster as a new model for examining P. gingivalis-host interactions. P. gingivalis (W83) infection of Drosophila resulted in a systemic infection that killed in a dose-dependent manner. Differences in the virulence of several clinically prevalent P. gingivalis strains were observed in the Drosophila killing model, and the results correlated well with studies in mammalian infection models and human epidemiologic studies. P. gingivalis pathobiology in Drosophila did not result from uncontrolled growth of the bacterium in the Drosophila hemolymph (blood) or overt damage to Drosophila tissues. P. gingivalis killing of Drosophila was multifactorial, involving several bacterial factors that are also involved in virulence in mammals. The results from this study suggest that many aspects of P. gingivalis pathogenesis in mammals are conserved in Drosophila, and thus the Drosophila killing model should be useful for characterizing P. gingivalis-host interactions and, potentially, polymicrobe-host interactions.
Porphyromonas gingivalis is a Gram-negative, obligate anaerobe that has been strongly implicated in the etiology of adult (chronic) periodontitis (21, 29), a destructive disease that affects the gums and supporting structures of the teeth. P. gingivalis-host interactions have previously been studied using several animal models, the most common of which are murine models (5, 7, 18, 20, 28), including an abscess model (39), a subcutaneous chamber model (24), and a periodontal bone loss model (41). Studies performed using murine models have demonstrated that P. gingivalis strains vary in their ability to cause periodontal bone loss (20, 40) and soft tissue destruction and death (28, 43, 56) and that strain W83 is highly virulent relative to many other strains of P. gingivalis (28, 43, 56). Murine models have also been used to identify P. gingivalis components that are important for pathogenesis (25, 43, 51, 59) and to characterize the host response to P. gingivalis infection (6, 11, 31, 35).
The fruit fly, Drosophila melanogaster, has been well established as a nonmammalian model for studying host-pathogen interactions (1, 17, 52, 55, 63). Drosophila relies solely on an innate immune response to combat invading microbes, and this immune response strongly parallels the mammalian innate immune response (47, 48). Like the mammalian innate immune response, Drosophila uses pattern recognition receptors to detect conserved microbial motifs on invading microbes, and the flies activate an immune response that is specific for the type of invading microbe. The absence of an adaptive immune response makes Drosophila useful for studying the interactions between microbes and the host innate immune response, in isolation. Numerous tools exist for the genetic manipulation of Drosophila, and these have been used to generate thousands of transgenic and mutant lines, including lines useful for identifying host factors that promote or fight infection (9, 52). The Drosophila genome has been sequenced, which has facilitated the development of microarray, proteomic, and RNA interference (RNAi) technologies for genome-wide analysis of Drosophila processes (14, 23, 65, 66). Additionally, the relative affordability, short generation time (10 to 14 days), and ease of use allow sample sizes that are large enough to permit statistical analysis of the data and make Drosophila an attractive complement to mammalian models.
The Drosophila killing model has been successfully used to characterize host-microbe interactions of bacterial (9, 13, 15, 17, 22, 52, 55), fungal (1, 3), and parasitic (62) pathogens, and the results of these studies suggest that there is good correlation between pathogenesis in mammals and in nonmammalian animals such as Drosophila. In these studies the microbe of interest is either fed to Drosophila or introduced directly into the hemocoel (body cavity) through the thorax by using a needle, and the survival of the infected animals is monitored over time.
P. gingivalis grows optimally at 37°C, and in contrast to other bacteria studied using the Drosophila model, it (and other oral pathogens) is an obligate anaerobe. It was therefore unclear whether this model would be viable for studying P. gingivalis-host interactions. The objective of the current study was to determine the viability of the Drosophila killing model for studying P. gingivalis-host interactions. We demonstrate that P. gingivalis is pathogenic in Drosophila, killing the animals in a dose-dependent manner. P. gingivalis killing of Drosophila was not due to uncontrolled bacterial growth or overt damage to Drosophila tissues. Heat-killed and live P. gingivalis microorganisms were equally pathogenic in Drosophila, which suggests that P. gingivalis cell surface components and Drosophila immune responses play important roles in causing pathology in this model. Differences in the virulence of several clinically prevalent P. gingivalis strains were observed using the Drosophila killing model and correlated well with studies in mammalian infection models (24, 28, 43, 56) and human epidemiologic studies (30). Additionally, multiple P. gingivalis components that are involved in virulence in mammals were found to be involved in Drosophila killing.
MATERIALS AND METHODS
Bacterial and Drosophila strains and growth conditions.
Bacterial and Drosophila strains used in this study are described in Table 1. P. gingivalis strains were grown on brucella blood agar (BBA; Anaerobe Systems), at 37°C in an anaerobic chamber (85% N2, 10% H2, 5% CO2). Drosophila stocks were maintained and propagated at 26°C in standard culture vials containing corn flour-molasses medium. Only 3- to 5-day-old female flies were used in experiments.
TABLE 1.
Bacterial and Drosophila strains used in this study
| Strain or stock | Description or genotype | Source |
|---|---|---|
| P. gingivalis strains | ||
| W83 | Lab strain, originally isolated from an oral infection | Margaret Duncan |
| ATCC 49417 | Lab strain, originally isolated from an oral infection | Joseph Zambon |
| 381 | Lab strain, originally isolated from an oral infection | Joseph Zambon |
| ATCC 33277 | Lab strain, originally isolated from an oral infection | Joseph Zambon |
| A7A1 | Lab strain, originally isolated from an oral infection | Joseph Zambon |
| 23A4 | Lab strain, originally isolated from an oral infection | Mike Curtis |
| W50 | Lab strain; renamed W50UK for these studies; wt for E8 and GPC | Mike Curtis |
| E8 | rgpA rgpB double mutant (arginine-specific protease) | Mike Curtis |
| GPC | Capsule mutant | Mike Curtis |
| W83 2741 | Lab strain, renamed W83VA for these studies; wt for V2577 | Janina Lewis |
| V2577 | kgp mutant (lysine-specific protease) | Janina Lewis |
| ATCC 33277 | Lab strain, renamed 33277FL for these studies; wt for YPF1 and SMF1 | Richard Lamont |
| YPF1 | fimA mutant (major fimbriae) | Richard Lamont |
| SMF1 | mfa1 mutant (minor fimbriae) | Richard Lamont |
| W83 | Lab strain, renamed W83JP for these studies; wt for M1217 | Koji Nakayama |
| M1217 | mgl mutant (l-methionine-α-deamino-γ-mercaptomethane-lyase) | Koji Nakayama |
| P. aeruginosa PA01 | Lab strain | Neil Baker |
| E. coli DH5α | Invitrogen | |
| D. melanogaster Canton S | Wild type | Amanda Simcox |
Infection of adult female Drosophila.
Bacterial strains were grown in 40 ml of Trypticase soy broth (TSB) for 24 h at 37°C. Pseudomonas aeruginosa and Escherichia coli were grown aerobically with shaking. P. gingivalis was supplemented with hemin (5 μg/ml) and vitamin K (1 μg/ml) and incubated anaerobically. The bacteria were harvested by centrifugation at 4,000 rpm for 8 min and diluted in TSB to the following optical densities at 600 nm (OD600; unless otherwise noted): P. aeruginosa, 0.7; E. coli DH5α, 2.0 (1 × 1011 CFU ml−1); P. gingivalis, 2.0 (1.09 × 1011 CFU ml−1). For heat killing, P. gingivalis was incubated at 60°C in a water bath for 1 h, and an aliquot of the preparation was plated on BBA to confirm loss of viability. The bacteria were introduced into the hemocoel (body cavity) of CO2-anesthetized Drosophila through the thorax, using a 30-gauge needle dipped into 500 μl of bacterial culture, or sterile TSB was used for mock infections (vector controls [VC]). Drosophila flies were returned to culture vials, and the number of surviving animals at time point 0 h was recorded. Infected Drosophila flies were incubated at 30°C, and the number of dead flies was recorded every 12 h for 7 days (unless otherwise noted). All experiments were repeated.
FITC labeling of P. gingivalis and microscopy.
P. gingivalis strain W83 was grown in TSB and harvested as described above. The bacteria were washed once in phosphate-buffered saline (PBS, pH 7.4) and diluted in PBS to an OD of 2.0. A 0.2-ml volume of fluorescein isothiocyanate (FITC) solution (2 mg/ml) was added to 0.4 ml of the resuspended bacteria, and the mixture was incubated at room temperature in the dark with occasional vortexing for 30 min. After incubation with FITC, the bacteria were pelleted, washed two times with PBS to remove unbound FITC, and resuspended in 0.4 ml of PBS. Adult female Drosophila flies were infected as described above. Infected animals were killed 2 h postinfection (exposed to −20°C for 5 min) and immediately visualized using a Nikon Eclipse E600 microscope equipped with a FITC/Texas Red filter set. Bright-field and fluorescent images were taken of representative animals by using a Nikon DXM 1200 digital camera.
P. gingivalis growth within infected Drosophila animals.
Thirty-nine Drosophila flies per group were infected (as described above) with the following P. gingivalis strains: W83, ATCC 49417, 381, ATCC 33277, A7A1, and 23A4. Three animals per group were ground in 80 μl of PBS with a Teflon pestle at the following time points (in hours) postinfection: 0, 4, 8, 12, 24, 36, 48, 60, 72, 84, 96, 108, and 120. Viable P. gingivalis cell counts were determined on BBA plates containing gentamicin (8 μg/ml), amphotericin B (3 μg/ml), and vancomycin (1 μg/ml). The experiment was repeated, and the results were averaged.
Statistical methods. (i) Sample size.
Power calculations based on pilot data estimated that a sample size of 136 Drosophila flies per group would be sufficient to detect a relative risk of mortality (RR) of at least 2.0 at an α-level of 0.05 with 90% power when comparing different infections. A sample size of 150 animals per group was used unless otherwise noted. Depending on the number of experimental groups involved, each experiment was divided into three, four, or five parts for feasibility.
(ii) Data analysis.
Survival data were analyzed using the SAS statistical software package (SAS Institute, Cary, NC). A Cox proportional hazards (P-H) model was fitted to the survival data. Likelihood ratio tests were performed, and RR values were obtained from the fitted Cox P-H model and adjusted for the individual “experiments” and “parts.” RR values with P values of <0.05 were considered significant.
RESULTS
P. gingivalis pathogenesis in the Drosophila killing model.
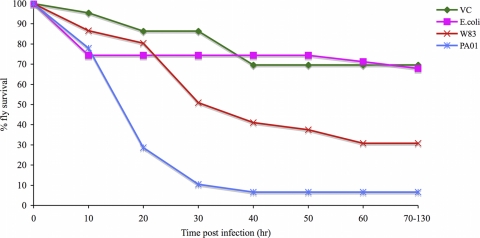
To determine whether P. gingivalis is pathogenic in Drosophila, groups of adult female Drosophila flies were infected with P. gingivalis strain W83, a P. aeruginosa strain (PA01) that was previously shown to be highly pathogenic (19), an E. coli strain (DH5α) that was previously shown to be nonpathogenic (15), or mock infected. The survival curves of infected and mock-infected Drosophila are shown in Fig. 1. Drosophila animals infected with P. gingivalis strain W83 were significantly more likely to die than mock-infected animals (RR for W83 versus mock, 3.58; P = 0.0038) and E. coli DH5α-infected animals (RR for W83 versus DH5α, 2.78; P = 0.01) but significantly less likely to die than P. aeruginosa-infected animals (RR for W83 versus PA01, 0.39; P = 0.0012).
FIG. 1.
Survival curves of adult, female Drosophila flies infected with P. gingivalis and other bacterial species. Eighteen Drosophila flies per group were infected with P. gingivalis (W83), P. aeruginosa (PAO1), or E. coli DH5α or mock infected, and the number of dead animals was recorded every 10 h for a period of 130 h. The experiment was repeated for a total sample size of 36 animals per group.
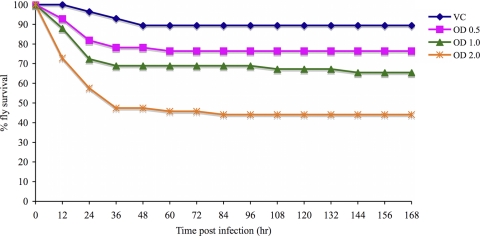
To determine whether P. gingivalis kills Drosophila in a dose-dependent manner, groups of Drosophila flies were infected with increasing concentrations of P. gingivalis strain W83. The survival curves of Drosophila flies infected with different amounts of strain W83 are shown in Fig. 2. An increase in the amount of P. gingivalis that was inoculated into Drosophila resulted in an increase in the number of animals that were killed during the time course of the experiment.
FIG. 2.
Survival curves of adult female Drosophila flies infected with different amounts of P. gingivalis (W83). Thirty Drosophila flies per group were infected with strain W83 at an OD600 of 0.5, 1.0, or 2.0 or were mock infected. The experiment was repeated for a total sample size of 60 animals per group.
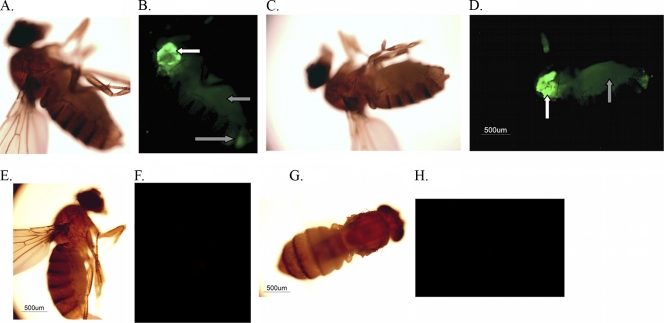
To determine whether P. gingivalis spreads systemically once introduced into Drosophila, animals were infected with FITC-labeled strain W83 or mock infected and examined using fluorescence microscopy 2 h postinfection. As there were no fluorescent P. gingivalis reference strains, FITC labeling was used. Representative bright-field and FITC images of P. gingivalis- and mock-infected animals are shown in Fig. 3. By 2 h postinfection strong FITC fluorescence was observed at the site of inoculation (white arrows), and the fluorescent bacteria were detected throughout the abdomen of the animals (Fig. 3B and D, gray arrows), as has been observed with other bacterial infections in Drosophila (22, 55). This was not surprising given the open circulatory system of Drosophila and the direct inoculation of the bacteria into the hemocoel. FITC fluorescence was not observed in mock-infected animals (Fig. 3F and H).
FIG. 3.
Location of FITC-labeled P. gingivalis within infected Drosophila animals. Drosophila animals were infected with FITC-labeled P. gingivalis strain W83 or mock infected and observed 2 h postinfection by using bright-field and fluorescence microscopy. (A to D) P. gingivalis-infected animals; (E to H) mock-infected flies. White arrows indicate sites of septic injury in the thorax. Gray arrows indicate FITC-labeled P. gingivalis within the abdomens of the infected animals.
Virulence of P. gingivalis strains in the Drosophila killing model.
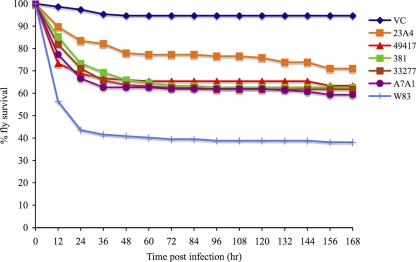
To compare the virulence of several clinically prevalent P. gingivalis strains, Drosophila animals were infected with strains W83, ATCC 49417, ATCC 33277, 381, A7A1, or 23A4 or mock infected. The P. gingivalis strains that were used in this experiment are commonly used lab strains that were originally isolated from individuals with oral infections, and they were selected for this study because they were identified in previous epidemiologic studies as being prevalent in human populations (30, 49). The survival curves for animals infected with each of the P. gingivalis test strains are shown in Fig. 4. The RR values for pairwise comparisons of the P. gingivalis strain infections are shown in Table 2. Based on the RR values, Drosophila animals were significantly more likely to die from infection with any of the P. gingivalis test strains than from a mock infection. Drosophila animals were significantly more likely to die from a W83 infection than from an infection with any of the other P. gingivalis test strains, and they were also more likely to die from an A7A1 infection than from a 23A4 infection.
FIG. 4.
Survival curves of adult female Drosophila flies infected with several P. gingivalis strains. Drosophila animals were infected with P. gingivalis strain W83, ATCC 49417, ATCC 33277, 381, A7A1, or 23A4 or were mock infected.
TABLE 2.
Relative risk of mortality for pairwise comparisons of P. gingivalis infections in Drosophilaa
| Infection | RR of mortality (P value) for pairwise comparison |
|||||
|---|---|---|---|---|---|---|
| W83 | A7A1 | ATCC 33277 | ATCC 49417 | 381 | 23A4 | |
| A7A1 | 1.7 (0.0014) | |||||
| ATCC 33277 | 1.86 (0.0002) | 1.10 (0.621) | ||||
| ATCC 49417 | 1.93 (0.0001) | 1.13 (0.503) | 1.03 (0.859) | |||
| 381 | 1.93 (0.0001) | 1.13 (0.498) | 1.04 (0.855) | 1.00 (0.996) | ||
| 23A4 | 2.62 (<0.0001) | 1.54 (0.031) | 1.41 (0.093) | 1.36 (0.133) | 1.36 (0.133) | |
| VCb | 15.81 (<0.0001) | 9.30 (<0.0001) | 8.49 (<0.0001) | 8.21 (<0.0001) | 8.21 (<0.0001) | 6.04 (<0.0001) |
Values are for pairwise comparisons of infections with groups listed horizontally versus those listed vertically. Results are shown in bold when P was <0.05.
VC, mock-infected animals.
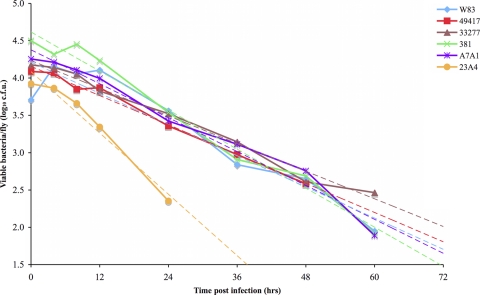
To examine the growth of P. gingivalis strains within infected Drosophila, colony counts were used to determine the level of viable P. gingivalis present in infected animals at specific time points postinfection. P. gingivalis colony-forming units that were recovered per animal at several time points are shown in Fig. 5. Based on the number of bacteria recovered immediately after infection (time point zero), between 5.10 × 103 CFU and 3.17 × 104 CFU of P. gingivalis were inoculated into the animals. Although small increases in the amount of P. gingivalis recovered per Drosophila were sometimes observed, there was an overall downward trend in the amount of bacteria recovered from the animals over time. A similar trend was observed when real-time PCR was used to quantitate P. gingivalis DNA within the animals (data not shown). Significant numbers of viable P. gingivalis could persist in the animals postinfection, which was not surprising, as the bacterium exhibits a high degree of aerotolerance and shows no loss of viability after exposure to air for at least 5 h (54). Viable P. gingivalis could be recovered from the Drosophila up to 24 h postinfection for strain 23A4, up to 48 h postinfection for strain ATCC 49417, and up to 60 h postinfection for strains W83, 381, ATCC 33277, and A7A1.
FIG. 5.
Growth of P. gingivalis in adult female Drosophila flies. Drosophila flies were infected with P. gingivalis strain W83, ATCC 49417, ATCC 33277, 381, A7A1, or 23A4, and viable cell counts were determined on BBA. Each data point represents the log10 value of the average CFU per animal recovered at a specific time point postinfection. Trend lines are shown as dashed lines.
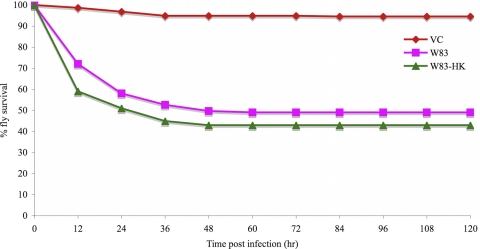
To investigate the mechanism of P. gingivalis killing of Drosophila, the effect of P. gingivalis infection on the viability of Drosophila tissues was examined, and the survival of animals infected with live versus heat-killed bacteria was compared. Animals infected with strain W83 were dissected, and the viability of their tissues was assessed using the vital dye trypan blue and a LIVE/DEAD cytotoxicity assay. No gross differences in tissue integrity were observed between P. gingivalis- and mock-infected animals (data not shown). W83 was heat killed at 60°C for 1 h, which also inactivates the proteases (gingipains) (64). When the survival rates of Drosophila animals infected with live and heat-killed W83 were compared (Fig. 6), no differences were observed in the survival of the two groups of animals.
FIG. 6.
Survival curves of adult female Drosophila flies infected with live or heat-killed (HK; by exposure to 60°C for 1 h) P. gingivalis strain W83.
Effects of P. gingivalis putative virulence factor gene knockouts on Drosophila survival.
To determine whether similar mechanisms are involved in P. gingivalis virulence in mammals and in Drosophila and whether multiple P. gingivalis factors are involved in Drosophila killing, the virulence levels of the following P. gingivalis mutants were compared to their wild-type (wt) parental strains using the Drosophila model: rgpA rgpB deficient (arginine-specific proteases), kgp deficient (lysine-specific protease), fimA deficient (fimbrillin, major fimbriae), mfa1 deficient (minor fimbriae), capsule deficient, and mgl-deficient (l-methione-α-deamino-γ-mercaptomethane-lyase) (Table 1). Groups of wt Drosophila were infected with the P. gingivalis mutant strains, the corresponding wt strains, or mock infected, and the survival rates of the infected animals were compared.
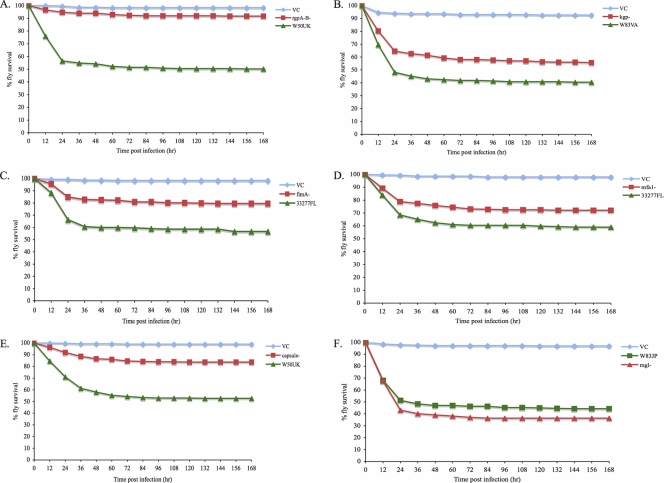
Survival curves for wt Drosophila infected with the wt or mutant P. gingivalis strains are shown in Fig. 7. The RR values for pairwise comparisons of infections with wt versus mutant strains of P. gingivalis are shown in Table 3. An RR value greater than 1 indicates that Drosophila animals were more likely to die from infection with the wt P. gingivalis strain than from an infection with the mutant strain. Drosophila animals infected with P. gingivalis rgpA rgpB, kgp, fimA, mfa1, and capsule mutants were less likely to die than Drosophila animals infected with the corresponding wt strains (Fig. 7A to E and Table 3). Drosophila flies infected with the P. gingivalis mgl mutant were as likely to die as Drosophila flies infected with the wt strain (Fig. 7F and Table 3). The experiments were repeated using a 1:2 dilution of each wt and mutant strain, and similar results were observed (data not shown).
FIG. 7.
Survival curves of wt Drosophila animals infected with wt or mutant strains of P. gingivalis. Green curves, wt-infected animals; red curves, mutant-infected animals; blue curves, mock-infected animals. (A) Survival of Drosophila flies infected with strain W50UK (wt) or an rgpA rgpB (arginine-specific proteases) mutant or mock infected (VC). (B) Survival of Drosophila flies infected with strain W83VA (wt), a kgp (lysine-specific protease) mutant, or mock infected (VC). (C) Survival of Drosophila flies infected with strain 33277FL (wt), a fimA (major fimbriae) mutant, or mock infected (VC). (D) Survival of Drosophila flies infected with strain 33277FL (wt), an mfa1 (minor fimbriae) mutant, or mock infected (VC). (E) Survival of Drosophila flies infected with strain W50UK (wt), a capsule-deficient mutant, or mock infected (VC). (F) Survival of Drosophila flies infected with strain W83JP (wt), an mgl (l-methionine-α-deamino-γ-mercaptomethane-lyase) mutant, or mock infected (VC).
TABLE 3.
Relative risk of mortality for pairwise comparisons of wt versus mutant P. gingivalis infections in Drosophila
|
P. gingivalis strains |
RR (P value) for wt vs mutanta | Findings in other animal model studies (reference[s]) for: |
||
|---|---|---|---|---|
| Mutant | wt | Periodontal bone loss | Abscess and death | |
| rgpA rgpB mutant (arginine-specific proteases) | W50UK | 7.69 (<0.0001) | −b | Correlation (15) |
| kgp mutant (lysine-specific protease) | W83VA | 1.63 (<0.0001) | − | Correlation (15) |
| fimA mutant (fimbrillin, major fimbriae) | 33277FL | 2.13 (<0.0001) | Correlation (14, 46) | − |
| mfa1 mutant (minor fimbriae) | 33277FL | 1.56 (0.0018) | Correlation (46) | − |
| Capsule mutant | W50UK | 2.99 (<0.0001) | − | − |
| mgl mutant | W83JP | 0.87 (0.2) | − | No correlation (47) |
RR values with P values of <0.05 are shown in bold.
−, the strains have not been compared in the indicated model.
DISCUSSION
The goal of this study was to examine the viability of Drosophila melanogaster as a model for examining P. gingivalis-host interactions. This model has been used to examine other bacteria-host interactions; however, P. gingivalis and other oral pathogens are obligate anaerobes and grow optimally at 37°C. The first step was to determine whether P. gingivalis was pathogenic in Drosophila. When the survival of animals infected with P. gingivalis (represented by strain W83) was compared to the survival of animals infected with a known pathogen (P. aeruginosa PA01) (19) and of a nonpathogen (E. coli DH5α) (15) for Drosophila, the P. gingivalis-infected animals were significantly more likely to die than E. coli DH5α-infected animals, although they were significantly less likely to die than P. aeruginosa-infected animals. A dose response was observed for P. gingivalis killing of Drosophila (Fig. 2), indicating that the bacteria could be more pathogenic at higher infective doses; however, at higher concentrations (ODs greater than 2.0) the bacteria clumped on the needle, making the delivery of higher doses unfeasible.
All P. gingivalis test strains were pathogenic to some degree in the Drosophila killing model (Table 2). Strain W83 was significantly more pathogenic than the five other P. gingivalis strains in this model, which correlates with the observations from studies using rodent models, in which W83 was also more pathogenic than the five other P. gingivalis strains (24, 28, 43, 56). W83 is also the strain that is most strongly associated with human periodontitis (30). These results suggest that there is a correlation between P. gingivalis pathogenesis in mammals and in Drosophila.
P. gingivalis is normally exposed to temperatures that range from 37°C to 38.8°C in the oral cavity (2) and grows optimally at 37°C in vitro, and unlike other bacteria studied using the Drosophila model, P. gingivalis is an obligate anaerobe. Thus, we determined whether P. gingivalis could grow in Drosophila. Colony counts were used to examine the growth of the P. gingivalis strains in the animals, and P. gingivalis DNA recovered from infected Drosophila was measured by real-time PCR. The results from the growth experiments indicated that although P. gingivalis does not multiply effectively in the Drosophila body cavity, the bacterium can persist for up to 60 h postinoculation in the animals. The aerobic environment of the Drosophila hemolymph and the temperature at which the infected animals are incubated are the likely factors that prevented the bacterium from growing optimally. The infected animals were incubated at 30°C, as their physiological processes deteriorate at temperatures higher than 30°C (42). When W83 growth at 30°C and 37°C was compared, the bacterium grew poorly at 30°C on solid medium (BBA) and in TSB (data not shown).
The six P. gingivalis factors tested for a role in Drosophila killing have been demonstrated to contribute to the bacterium's virulence in mammals (43, 59, 70, 72). In the current study, five of the components were shown to contribute to P. gingivalis virulence in the Drosophila model. The gingipains are proteases encoded by rgpA (arginine-specific protease), rgpB (arginine-specific protease), and kgp (lysine-specific protease) genes. They are considered to be major P. gingivalis virulence factors due to their ability to degrade a large variety of host proteins (reviewed in references 37, 58, and 60), and they are important to various degrees for pathogenesis in the mouse abscess model of infection (59). P. gingivalis major fimbriae (fimA) are involved in the invasion of gingival epithelial cells (71), and both major and minor (mfa1) fimbriae are important for P. gingivalis autoaggregation (50, 57) and biofilm formation with Streptococcus gordonii (44, 45). mfa1 and fimA mutants are impaired in the ability to induce periodontal bone loss in a rat model of infection (70). The capsule, which is present on some strains of P. gingivalis, is important for virulence in the mouse abscess model of infection, as infection with encapsulated strains results in more soft tissue destruction and death than infection with unencapsulated strains (43). l-Methione-α-deamino-γ-mercaptomethane-lyase (mgl) catalyzes the conversion of l-methionine to the volatile sulfur compound methyl mercaptan (72). Methyl mercaptan is elevated in the mouths (68) and crevicular air (12) of patients with active periodontitis, and exposure to methyl mercaptan has been shown to decrease DNA (38) and protein (38, 46) synthesis in gingival fibroblasts and to inhibit the migration of periodontal ligament cells (46). In the mouse abscess model, the mgl mutant was less able to cause soft tissue destruction and death than the wt strain (72). The results of this study demonstrate that several P. gingivalis factors (capsule, major and minor fimbriae, and gingipains) that are important for pathogenesis in mammals are also involved in the killing of Drosophila, suggesting that there is a strong correlation between P. gingivalis pathogenesis in mammals and in Drosophila. However, the noninvolvement of the mgl gene product in P. gingivalis killing of Drosophila suggests that the requirement for a particular virulence factor by P. gingivalis depends on the host involved. The observed differences in the Drosophila-killing abilities of the P. gingivalis strains suggest that additional bacterial factors are involved in pathogenesis in this model.
In contrast to other bacteria that have been studied in this model, P. gingivalis does not multiply effectively in Drosophila, yet the bacterium still kills the animals. Additionally, no gross differences in tissue integrity were observed between infected and mock-infected animals, indicating that the pathobiology of P. gingivalis in this model is not due to uncontrolled growth of the bacterium or overt damage to Drosophila tissues. Our observation that there was no difference in the survival of Drosophila infected with live- versus heat-killed strain W83 (Fig. 6) suggests that the pathology caused by the bacterium in this model is due to direct killing of the animals by a P. gingivalis component(s) or indirect killing by the host's own response. Schneider et al. observed that pathogens can kill Drosophila via mechanisms that are directly toxic or host mediated (63). The observation that several P. gingivalis factors play a role in Drosophila killing suggests that killing is multifactorial and not due to a single factor, e.g., a toxin. Host-mediated damage in Drosophila can occur either by the hyperactivation of the immune response, or the activation upon infection, of responses that are deleterious to the animals (63). It has been shown that overactivation of the Toll (26) and Imd (8) pathways (major regulators of Drosophila immune response genes), excessive production of nitric oxide (10) and reactive oxygen species (32), and the activation of the JNK pathway via Eiger (cytokine) signaling following Salmonella enterica serovar Typhimurium (9) or Mycobacterium marinum (63) infection results in Drosophila lethality. The molecular mechanisms by which these immune responses result in Drosophila lethality are currently unknown; however, Schneider et al. and others have suggested that they may be either energetically wasteful or directly toxic (26, 63). P. gingivalis major and minor fimbriae, the capsule, and gingipains have been demonstrated to be proinflammatory in mammals (16, 33, 34, 69), and it is known that the bacterium releases outer membrane vesicles (27, 53) that allow pathogen-associated molecular patterns (PAMPs) to be detected at sites distant from the bacterium. As P. gingivalis spreads systemically when inoculated into Drosophila, PAMPs present in cell surface-associated structures like fimbriae, gingipains, and capsular polysaccharide may induce a systemic hyperactivation of Drosophila immune responses, which is deleterious to the bacterium but also harms the animals. Although not systemic, an exaggerated host immune response is primarily responsible for the damage that occurs in periodontitis (61, 67) and in response to P. gingivalis infection in other animal models (4, 11, 35).
Despite the bacterium's inability to multiply effectively in Drosophila, P. gingivalis was still capable of killing the animals, and several lines of evidence suggest that P. gingivalis killing of Drosophila is due to specific interactions between the bacterium and the host. First, P. gingivalis (W83)-infected Drosophila animals were more likely to die than E. coli DH5α-infected Drosophila animals (Fig. 1) that received an equivalent inoculum. Second, P. gingivalis (W83) killing of Drosophila was dose dependent (Fig. 2). Third, strains of P. gingivalis exhibited different killing abilities when inoculated into Drosophila (Fig. 4). Fourth, several P. gingivalis virulence gene mutants are attenuated in the Drosophila killing model (Fig. 7). Finally, several immune response-defective Drosophila mutants show increased susceptibility to killing by P. gingivalis (36).
In this study we have demonstrated that Drosophila melanogaster is a viable new animal model for examining P. gingivalis-host interactions. The introduction of this highly genetically manipulatable, high-throughput animal model with an innate immune system that is remarkably similar to mammals increases the repertoire of tools available to study P. gingivalis-host interactions, especially as the damage that occurs with periodontitis is primarily due to host effects. Although Drosophila is not a natural host for P. gingivalis, the results of this study suggest that there are many aspects of P. gingivalis pathogenesis that are evolutionarily preserved between mammals and Drosophila. Therefore, the Drosophila killing model could be used to identify P. gingivalis factors that are involved in pathogenesis, which is important, as the roles of a large number of P. gingivalis genes are unknown. This model could also be used to identify host factors that fight P. gingivalis infection or contribute to P. gingivalis-induced pathology and to examine the host-pathogen interactions of other oral pathogens. As it is clear that periodontitis is a polymicrobial infection, the Drosophila killing model could be exploited to examine the interactions among different oral species and their interactions with host defenses in a mixed bacterial inoculum.
Acknowledgments
We thank Amanda Simcox (Ohio State University) for kindly supplied Drosophila husbandry materials, advice, and demonstrations. We thank Donna Cain (Ohio State University) for advice on Drosophila husbandry protocols. We thank Prajakta Valavalkar for help with statistical analysis.
This study was supported by grant DE10467 from the National Institutes of Health.
Editor: R. P. Morrison
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Alarco, A. M., A. Marcil, J. Chen, B. Suter, D. Thomas, and M. Whiteway. 2004. Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J. Immunol. 172:5622-5628. [DOI] [PubMed] [Google Scholar]
- 2.Amano, A., A. Sharma, H. T. Sojar, H. K. Kuramitsu, and R. J. Genco. 1994. Effects of temperature stress on expression of fimbriae and superoxide dismutase by Porphyromonas gingivalis. Infect. Immun. 62:4682-4685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apidianakis, Y., L. G. Rahme, J. Heitman, F. M. Ausubel, S. B. Calderwood, and E. Mylonakis. 2004. Challenge of Drosophila melanogaster with Cryptococcus neoformans and role of the innate immune response. Eukaryot. Cell 3:413-419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baker, P. J., M. Dixon, R. T. Evans, L. Dufour, E. Johnson, and D. C. Roopenian. 1999. CD4(+) T cells and the proinflammatory cytokines gamma interferon and interleukin-6 contribute to alveolar bone loss in mice. Infect. Immun. 67:2804-2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker, P. J., R. T. Evans, and D. C. Roopenian. 1994. Oral infection with Porphyromonas gingivalis and induced alveolar bone loss in immunocompetent and severe combined immunodeficient mice. Arch. Oral Biol. 39:1035-1040. [DOI] [PubMed] [Google Scholar]
- 6.Baker, P. J., L. Howe, J. Garneau, and D. C. Roopenian. 2002. T cell knockout mice have diminished alveolar bone loss after oral infection with Porphyromonas gingivalis. FEMS Immunol. Med. Microbiol. 34:45-50. [DOI] [PubMed] [Google Scholar]
- 7.Baumgartner, J. C., W. A. Falkler, Jr., and T. Beckerman. 1992. Experimentally induced infection by oral anaerobic microorganisms in a mouse model. Oral Microbiol. Immunol. 7:253-256. [DOI] [PubMed] [Google Scholar]
- 8.Bischoff, V., C. Vignal, B. Duvic, I. G. Boneca, J. A. Hoffmann, and J. Royet. 2006. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2:e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brandt, S. M., M. S. Dionne, R. S. Khush, L. N. Pham, T. J. Vigdal, and D. S. Schneider. 2004. Secreted bacterial effectors and host-produced Eiger/TNF drive death in a Salmonella-infected fruit fly. PLoS Biol. 2:e418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Broderick, K. E., J. Feala, A. McCulloch, G. Paternostro, V. S. Sharma, R. B. Pilz, and G. R. Boss. 2006. The nitric oxide scavenger cobinamide profoundly improves survival in a Drosophila melanogaster model of bacterial sepsis. FASEB J. 20:1865-1873. [DOI] [PubMed] [Google Scholar]
- 11.Burns, E., G. Bachrach, L. Shapira, and G. Nussbaum. 2006. Cutting edge. TLR2 is required for the innate response to Porphyromonas gingivalis: activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J. Immunol. 177:8296-8300. [DOI] [PubMed] [Google Scholar]
- 12.Coli, J. M., and J. Tonzetich. 1992. Characterization of volatile sulphur compounds production at individual gingival crevicular sites in humans. J. Clin. Dent. 3:97-103. [PubMed] [Google Scholar]
- 13.D'Argenio, D. A., L. A. Gallagher, C. A. Berg, and C. Manoil. 2001. Drosophila as a model host for Pseudomonas aeruginosa infection. J. Bacteriol. 183:1466-1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Gregorio, E., P. T. Spellman, G. M. Rubin, and B. Lemaitre. 2001. Genome-wide analysis of the Drosophila immune response by using oligonucleotide microarrays. Proc. Natl. Acad. Sci. U. S. A. 98:12590-12595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Lima Pimenta, A., P. Di Martino, E. Le Bouder, C. Hulen, and M. A. Blight. 2003. In vitro identification of two adherence factors required for in vivo virulence of Pseudomonas fluorescens. Microbes Infect. 5:1177-1187. [DOI] [PubMed] [Google Scholar]
- 16.d'Empaire, G., M. T. Baer, and F. C. Gibson III. 2006. The K1 serotype capsular polysaccharide of Porphyromonas gingivalis elicits chemokine production from murine macrophages that facilitates cell migration. Infect. Immun. 74:6236-6243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dionne, M. S., N. Ghori, and D. S. Schneider. 2003. Drosophila melanogaster is a genetically tractable model host for Mycobacterium marinum. Infect. Immun. 71:3540-3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eke, P. I., V. O. Rotimi, and B. E. Laughon. 1996. Experimental model for Porphyromonas gingivalis infection in animals. Afr. J. Med. Med. Sci. 25:31-39. [PubMed] [Google Scholar]
- 19.Erickson, D. L., J. L. Lines, E. C. Pesci, V. Venturi, and D. G. Storey. 2004. Pseudomonas aeruginosa relA contributes to virulence in Drosophila melanogaster. Infect. Immun. 72:5638-5645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Evans, R. T., B. Klausen, N. S. Ramamurthy, L. M. Golub, C. Sfintescu, and R. J. Genco. 1992. Periodontopathic potential of two strains of Porphyromonas gingivalis in gnotobiotic rats. Arch. Oral Biol. 37:813-819. [DOI] [PubMed] [Google Scholar]
- 21.Ezzo, P. J., and C. W. Cutler. 2003. Microorganisms as risk indicators for periodontal disease. Periodontol. 2000 32:24-35. [DOI] [PubMed] [Google Scholar]
- 22.Fauvarque, M. O., E. Bergeret, J. Chabert, D. Dacheux, M. Satre, and I. Attree. 2002. Role and activation of type III secretion system genes in Pseudomonas aeruginosa-induced Drosophila killing. Microb. Pathog. 32:287-295. [DOI] [PubMed] [Google Scholar]
- 23.Foley, E., and P. H. O'Farrell. 2004. Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2:e203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Genco, C. A., C. W. Cutler, D. Kapczynski, K. Maloney, and R. R. Arnold. 1991. A novel mouse model to study the virulence of and host response to Porphyromonas (Bacteroides) gingivalis. Infect. Immun. 59:1255-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibson, F. C., III, and C. A. Genco. 2001. Prevention of Porphyromonas gingivalis-induced oral bone loss following immunization with gingipain R1. Infect. Immun. 69:7959-7963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gordon, M. D., M. S. Dionne, D. S. Schneider, and R. Nusse. 2005. WntD is a feedback inhibitor of Dorsal/NF-κB in Drosophila development and immunity. Nature 437:746-749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Grenier, D., and D. Mayrand. 1987. Functional characterization of extracellular vesicles produced by Bacteroides gingivalis. Infect. Immun. 55:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Grenier, D., and D. Mayrand. 1987. Selected characteristics of pathogenic and nonpathogenic strains of Bacteroides gingivalis. J. Clin. Microbiol. 25:738-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Griffen, A. L., M. R. Becker, S. R. Lyons, M. L. Moeschberger, and E. J. Leys. 1998. Prevalence of Porphyromonas gingivalis and periodontal health status. J. Clin. Microbiol. 36:3239-3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Griffen, A. L., S. R. Lyons, M. R. Becker, M. L. Moeschberger, and E. J. Leys. 1999. Porphyromonas gingivalis strain variability and periodontitis. J. Clin. Microbiol. 37:4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gyurko, R., G. Boustany, P. L. Huang, A. Kantarci, T. E. Van Dyke, C. A. Genco, and F. C. Gibson III. 2003. Mice lacking inducible nitric oxide synthase demonstrate impaired killing of Porphyromonas gingivalis. Infect. Immun. 71:4917-4924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ha, E. M., C. T. Oh, J. H. Ryu, Y. S. Bae, S. W. Kang, I. H. Jang, P. T. Brey, and W. J. Lee. 2005. An antioxidant system required for host protection against gut infection in Drosophila. Dev. Cell 8:125-132. [DOI] [PubMed] [Google Scholar]
- 33.Hajishengallis, G., H. Sojar, R. J. Genco, and E. DeNardin. 2004. Intracellular signaling and cytokine induction upon interactions of Porphyromonas gingivalis fimbriae with pattern-recognition receptors. Immunol. Invest. 33:157-172. [DOI] [PubMed] [Google Scholar]
- 34.Hiramine, H., K. Watanabe, N. Hamada, and T. Umemoto. 2003. Porphyromonas gingivalis 67-kDa fimbriae induced cytokine production and osteoclast differentiation utilizing TLR2. FEMS Microbiol. Lett. 229:49-55. [DOI] [PubMed] [Google Scholar]
- 35.Holzhausen, M., L. C. Spolidorio, R. P. Ellen, M. C. Jobin, M. Steinhoff, P. Andrade-Gordon, and N. Vergnolle. 2006. Protease-activated receptor-2 activation: a major role in the pathogenesis of Porphyromonas gingivalis infection. Am. J. Pathol. 168:1189-1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Igboin, C. O., K. P. Tordoff, M. L. Moeschberger, A. L. Griffen, and E. J. Leys. 2011. Porphyromonas gingivalis-host interactions in a Drosophila melanogaster model. Infect. Immun. 79:449-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Imamura, T., J. Travis, and J. Potempa. 2003. The biphasic virulence activities of gingipains: activation and inactivation of host proteins. Curr. Protein Pept. Sci. 4:443-450. [DOI] [PubMed] [Google Scholar]
- 38.Johnson, P. W., W. Ng, and J. Tonzetich. 1992. Modulation of human gingival fibroblast cell metabolism by methyl mercaptan. J. Periodontal Res. 27:476-483. [DOI] [PubMed] [Google Scholar]
- 39.Kastelein, P., T. J. van Steenbergen, J. M. Bras, and J. de Graaff. 1981. An experimentally induced phlegmonous abscess by a strain of Bacteroides gingivalis in guinea pigs and mice. Antonie Van Leeuwenhoek 47:1-9. [DOI] [PubMed] [Google Scholar]
- 40.Katz, J., D. C. Ward, and S. M. Michalek. 1996. Effect of host responses on the pathogenicity of strains of Porphyromonas gingivalis. Oral Microbiol. Immunol. 11:309-318. [DOI] [PubMed] [Google Scholar]
- 41.Klausen, B. 1991. Microbiological and immunological aspects of experimental periodontal disease in rats: a review article. J. Periodontol. 62:59-73. [DOI] [PubMed] [Google Scholar]
- 42.Krebs, R. A., and M. E. Feder. 1998. Hsp70 and larval thermotolerance in Drosophila melanogaster: how much is enough and when is more too much? J. Insect Physiol. 44:1091-1101. [DOI] [PubMed] [Google Scholar]
- 43.Laine, M. L., and A. J. van Winkelhoff. 1998. Virulence of six capsular serotypes of Porphyromonas gingivalis in a mouse model. Oral Microbiol. Immunol. 13:322-325. [DOI] [PubMed] [Google Scholar]
- 44.Lamont, R. J., C. A. Bevan, S. Gil, R. E. Persson, and B. Rosan. 1993. Involvement of Porphyromonas gingivalis fimbriae in adherence to Streptococcus gordonii. Oral Microbiol. Immunol. 8:272-276. [DOI] [PubMed] [Google Scholar]
- 45.Lamont, R. J., A. El-Sabaeny, Y. Park, G. S. Cook, J. W. Costerton, and D. R. Demuth. 2002. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology 148:1627-1636. [DOI] [PubMed] [Google Scholar]
- 46.Lancero, H., J. Niu, and P. W. Johnson. 1996. Exposure of periodontal ligament cells to methyl mercaptan reduces intracellular pH and inhibits cell migration. J. Dent. Res. 75:1994-2002. [DOI] [PubMed] [Google Scholar]
- 47.Leclerc, V., and J. M. Reichhart. 2004. The immune response of Drosophila melanogaster. Immunol. Rev. 198:59-71. [DOI] [PubMed] [Google Scholar]
- 48.Lemaitre, B., and J. Hoffmann. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25:697-743. [DOI] [PubMed] [Google Scholar]
- 49.Leys, E. J., J. H. Smith, S. R. Lyons, and A. L. Griffen. 1999. Identification of Porphyromonas gingivalis strains by heteroduplex analysis and detection of multiple strains. J. Clin. Microbiol. 37:3906-3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lin, X., J. Wu, and H. Xie. 2006. Porphyromonas gingivalis minor fimbriae are required for cell-cell interactions. Infect. Immun. 74:6011-6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Malek, R., J. G. Fisher, A. Caleca, M. Stinson, C. J. van Oss, J. Y. Lee, M. I. Cho, R. J. Genco, R. T. Evans, and D. W. Dyer. 1994. Inactivation of the Porphyromonas gingivalis fimA gene blocks periodontal damage in gnotobiotic rats. J. Bacteriol. 176:1052-1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mansfield, B. E., M. S. Dionne, D. S. Schneider, and N. E. Freitag. 2003. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell. Microbiol. 5:901-911. [DOI] [PubMed] [Google Scholar]
- 53.Mayrand, D., and D. Grenier. 1989. Biological activities of outer membrane vesicles. Can. J. Microbiol. 35:607-613. [DOI] [PubMed] [Google Scholar]
- 54.Nakayama, K. 1994. Rapid viability loss on exposure to air in a superoxide dismutase-deficient mutant of Porphyromonas gingivalis. J. Bacteriol. 176:1939-1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Needham, A. J., M. Kibart, H. Crossley, P. W. Ingham, and S. J. Foster. 2004. Drosophila melanogaster as a model host for Staphylococcus aureus infection. Microbiology 150:2347-2355. [DOI] [PubMed] [Google Scholar]
- 56.Neiders, M. E., P. B. Chen, H. Suido, H. S. Reynolds, J. J. Zambon, M. Shlossman, and R. J. Genco. 1989. Heterogeneity of virulence among strains of Bacteroides gingivalis. J. Periodontal Res. 24:192-198. [DOI] [PubMed] [Google Scholar]
- 57.Nishiyama, S., Y. Murakami, H. Nagata, S. Shizukuishi, I. Kawagishi, and F. Yoshimura. 2007. Involvement of minor components associated with the FimA fimbriae of Porphyromonas gingivalis in adhesive functions. Microbiology 153:1916-1925. [DOI] [PubMed] [Google Scholar]
- 58.O'Brien-Simpson, N. M., P. D. Veith, S. G. Dashper, and E. C. Reynolds. 2003. Porphyromonas gingivalis gingipains: the molecular teeth of a microbial vampire. Curr. Protein Pept. Sci. 4:409-426. [DOI] [PubMed] [Google Scholar]
- 59.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Potempa, J., A. Sroka, T. Imamura, and J. Travis. 2003. Gingipains, the major cysteine proteinases and virulence factors of Porphyromonas gingivalis: structure, function and assembly of multidomain protein complexes. Curr. Protein Pept. Sci. 4:397-407. [DOI] [PubMed] [Google Scholar]
- 61.Preshaw, P. M., R. A. Seymour, and P. A. Heasman. 2004. Current concepts in periodontal pathogenesis. Dent. Update 31:570-572, 574-578. [DOI] [PubMed] [Google Scholar]
- 62.Schneider, D., and M. Shahabuddin. 2000. Malaria parasite development in a Drosophila model. Science 288:2376-2379. [DOI] [PubMed] [Google Scholar]
- 63.Schneider, D. S., J. S. Ayres, S. M. Brandt, A. Costa, M. S. Dionne, M. D. Gordon, E. M. Mabery, M. G. Moule, L. N. Pham, and M. M. Shirasu-Hiza. 2007. Drosophila Eiger mutants are sensitive to extracellular pathogens. PLoS Pathog. 3:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stathopoulou, P. G., J. C. Galicia, M. R. Benakanakere, C. A. Garcia, J. Potempa, and D. F. Kinane. 2009. Porphyromonas gingivalis induce apoptosis in human gingival epithelial cells through a gingipain-dependent mechanism. BMC Microbiol. 9:107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stroschein-Stevenson, S. L., E. Foley, P. H. O'Farrell, and A. D. Johnson. 2006. Identification of Drosophila gene products required for phagocytosis of Candida albicans. PLoS Biol. 4:e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stuart, L. M., J. Boulais, G. M. Charriere, E. J. Hennessy, S. Brunet, I. Jutras, G. Goyette, C. Rondeau, S. Letarte, H. Huang, P. Ye, F. Morales, C. Kocks, J. S. Bader, M. Desjardins, and R. A. Ezekowitz. 2007. A systems biology analysis of the Drosophila phagosome. Nature 445:95-101. [DOI] [PubMed] [Google Scholar]
- 67.Tatakis, D. N., and P. S. Kumar. 2005. Etiology and pathogenesis of periodontal diseases. Dent. Clin. North Am. 49:491-516. [DOI] [PubMed] [Google Scholar]
- 68.Tonzetich, J., P. W. Johnson, and S. K. Ng. 1979. Pathways of cystine metabolism in human saliva. Arch. Oral Biol. 24:35-39. [DOI] [PubMed] [Google Scholar]
- 69.Uehara, A., T. Imamura, J. Potempa, J. Travis, and H. Takada. 2008. Gingipains from Porphyromonas gingivalis synergistically induce the production of proinflammatory cytokines through protease-activated receptors with Toll-like receptor and NOD1/2 ligands in human monocytic cells. Cell. Microbiol. 10:1181-1189. [DOI] [PubMed] [Google Scholar]
- 70.Umemoto, T., and N. Hamada. 2003. Characterization of biologically active cell surface components of a periodontal pathogen. The roles of major and minor fimbriae of Porphyromonas gingivalis. J. Periodontol. 74:119-122. [DOI] [PubMed] [Google Scholar]
- 71.Weinberg, A., C. M. Belton, Y. Park, and R. J. Lamont. 1997. Role of fimbriae in Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 65:313-316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yoshimura, M., Y. Nakano, Y. Yamashita, T. Oho, T. Saito, and T. Koga. 2000. Formation of methyl mercaptan from L-methionine by Porphyromonas gingivalis. Infect. Immun. 68:6912-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]