Abstract
Tannerella forsythia is a Gram-negative oral anaerobe which contributes to the development of periodontitis, an inflammatory disease of the tooth-supporting tissues leading to tooth loss. The mechanisms by which this bacterium colonizes the oral cavity are poorly understood. The bacterium has been shown to express two distinct sialidases, namely, SiaHI and NanH, with the latter being the major sialidase. Bacterial sialidases can play roles in pathogenesis by cleaving sialic acids on host glycoproteins, destroying their integrity, and/or unmasking hidden epitopes on host surfaces for colonization. In the present study, we investigated the roles of the SiaHI and NanH sialidases by generating and characterizing specific deletion mutants. Our results showed that the NanH deficiency resulted in a total loss of sialidase activity associated with the outer-membrane and secreted fractions. On the other hand, the SiaHI deficiency resulted in only a slight reduction in the total sialidase activity, with no significant differences in the levels of sialidase activity in the outer membrane or secreted fractions compared to that in the wild-type strain. The results demonstrated that NanH is both surface localized and secreted. The NanH-deficient mutant but not the SiaHI-deficient mutant was significantly attenuated in epithelial cell binding and invasion abilities compared to the wild-type strain. Moreover, the NanH-deficient mutant alone was impaired in cleaving surface sialic acids on epithelial cells. Thus, our study suggests that NanH sialidase might play roles in bacterial colonization by exposing sialic acid-hidden epitopes on epithelial cells.
Tannerella forsythia is a fastidious Gram-negative anaerobe which grows in the subgingival crevice, a space between the tooth and gum tissue. It is implicated in the development of periodontitis (37), a chronic inflammatory disease resulting in the destruction of connective tissue supporting teeth as well as bone, leading to tooth loss. A number of putative virulence factors have been identified in T. forsythia (33), such as a cell surface-associated leucine-rich-repeat BspA protein (34), a surface S layer (28, 30), an α-d-glucosidase and an N-acetyl-β-glucosaminidase (14), proteases (9, 29), apoptosis-inducing activity (23), and sialidases (3, 22). Sialidases are glycohydrolases which release the terminal sialic acid residues from sialoglycoconjugates. Sialic acids are commonly present as terminal sugars on glycoproteins and glycolipids on the surface of a wide variety of eukaryotic cells and secreted glycoproteins (1, 41). The sialic acid residues modulate a variety of biological functions, such as mediating cell-cell interactions via carbohydrate-lectin interactions, stabilizing conformation of the glycoproteins, or masking receptors for ligand binding (1, 41). With respect to bacterially expressed sialidases, they have been shown to provide pathogenic advantages to bacteria by counteracting the host's innate and adaptive immune responses (6, 25), inducing chemokine release from epithelial cells (19), exposing cryptitopes by unmasking sialic acid-masked epitopes for adhesion (6, 39), promoting biofilm formation (36), and degrading host glycoproteins to obtain nutrients (2, 5, 18, 31, 39).
Previous studies have shown expression of sialidase in T. forsythia isolates, which has also been used for diagnostic identification of the species. T. forsythia has been reported to express two different sialidases, encoded by the siaHI (17) and nanH genes (38). The NanH sialidase appears to be the major sialidase expressed by the bacterium. A recent study has demonstrated that sialic acid released from the sialylated glycoconjugates by NanH sialidase action may serve as a growth factor for biofilm growth (26). These previous studies utilized recombinant expression and gene complementation in Escherichia coli for the characterization of sialidases and the sialic acid uptake system of T. forsythia. In the present study, we generated and characterized siaHI and nanH deletion mutants of T. forsythia to define the roles of the respective sialidases in host cell interactions. T. forsythia has been identified within the buccal epithelium of patients (27) and has been demonstrated to attach to and invade epithelial cells in vitro (16, 28, 30). Since oral bacteria can exploit glycoconjugates and glycolipids on buccal and gingival epithelial cells for mediating attachment, we predicted that T. forsythia sialidases would play roles in such interactions. For this purpose, we tested siaHI and nanH deletion mutants for their ability to attach and invade epithelial cells. Our studies demonstrated that NanH activity plays an important role in bacterial attachment to and invasion into epithelial cells, likely through unmasking sialic acid-hidden bacterial binding epitopes on epithelial cells.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
T. forsythia strains were grown anaerobically (5% CO2, 10% H2, 85% N2) in BF broth or on BF agar plates (12) with or without appropriate antibiotics. E. coli strains were grown in LB medium aerobically at 37°C. E. coli DH5α (Invitrogen, Carlsbad, CA) was used as a host for cloning and plasmid purification, and BL21(DE3) (EMD Bioscience, Gibbstown, NJ) was used as a host for expression and purification of the recombinant proteins. The KB epithelial cell line (CCL17; American Type Culture Collection, Manassas, VA) was maintained in Dulbecco's modified Eagle medium (DMEM) (Gibco, Buffalo, NY) supplemented with 10% fetal bovine serum and 50 μg/ml of gentamicin. The cultures were incubated at 37°C under 5% CO2. KB cells were grown to near-confluence (90 to 95%) for the assays. KB cells were originally thought to be derived from an oral epidermal carcinoma. However, DNA fingerprinting later confirmed that this cell line is derived from the HeLa cell line (human cervical epithelial carcinoma) due to cross-contamination (American Type Culture Collection, Manassas, VA). The human gingival epithelial cell line, OBA-9, was obtained from Denis Kinane (University of Pennsylvania) and maintained at 37°C, 5% CO2, in keratinocyte serum-free medium supplemented with 10 μg/ml of insulin, 5 μg/ml of transferrin, 10 μM 2-mercaptoethanol, 10 μM 2-aminoethanol, 10 nM sodium selenite, 50 μg/ml of bovine pituitary extract, 100 U/ml of penicillin-streptomycin, and 50 ng/ml amphotericin B (Lonza Inc., Allendale, NJ).
Construction of SiaHI and NanH mutants.
siaHI (TF2207) and nanH (TF0035) sequences were retrieved from the Oral Pathogen Sequence Database at Los Alamos, NM, under the previous name of the organism, T. forsythensis (http://www.oralgen.lanl.gov/). Insertional mutants for each of the genes were constructed by an allelic replacement strategy described previously (12). Briefly, DNA fragments containing the ermF gene flanked by upstream and downstream regions of either TF2207 (siaHI) or TF0035 (nanH) were electroporated into T. forsythia ATCC 43037 cells, and transformants were selected on agar-erythromycin plates. The primer sequences used in the study are shown in Table 1. Briefly, for constructing the siaHI deletion mutant, a DNA fragment containing TF2207 with flanking sequences was initially amplified by PCR using primers 1 and 2 from T. forsythia 43037 genomic DNA. This PCR product was then used as a template to amplify the upstream and downstream fragments of TF2207 with primer sets 1 and 3 and 2 and 5, respectively. The ermF fragment (797 bp) was amplified from pVA2198 (8) with primers 4 and 6. Primers 3, 4, 5, and 6 contained overlapping sequences for ermF and TF2207 to allow generation of a fusion fragment by the PCR overlap strategy. Purified PCR products of TF2207 with flanking regions and ermF (797 bp) were used as templates for the overlap PCR (13) using primers 1 and 2. Likewise, a fusion construct was generated by a PCR overlap strategy for constructing a nanH deletion mutant. The primer sets 1N and 3N and 2N and 4N were used to amplify upstream and downstream fragments flanking nanH (TF0035), respectively, and the primers 3N, 4N, 5N, and 6N, containing overlap sequences of ermF and TF0035, were used for constructing the fusion fragment. The fusion product for each gene was then transformed into T. forsythia 43037 by electroporation as previously described (12). Transformants were plated on BF agar plates containing 5 μg/ml of erythromycin and incubated at 37°C anaerobically for 14 days. Following incubation, erythromycin-resistant colonies were screened by PCR and Southern blotting. One representative transformant corresponding to each gene deletion, confirmed by PCR and Southern blotting, was selected and further used for analyses.
TABLE 1.
Primers used in this study
| Function or primer | Sequence (5′ to 3′)a | Description |
|---|---|---|
| Construction of siaHI mutant | ||
| 1 | ATACCAGAAAACCCCACAGC | PCR primer for TF2207 |
| 2 | CCGCAAAGAAGCCATATTTTC | PCR primer for TF2207 |
| 3 | aaaaatttcatccttcgTGAGTATCGACAGACGAAACTTTC | PCR overlap primer |
| 4 | gggcaatttcttttttgtcatGACAATTTTGAGTTAGATATAC | PCR overlap primer |
| 5 | GAAAGTTTCGTCTGTCGATACTCAcgaaggatgaaattttt | PCR overlap primer |
| 6 | GTATATCTAACTCAAAATTGTCatgacaaaaaagaaattgcccgttcgttt | PCR overlap primer |
| Construction of nanH mutant | ||
| 1N | CTATCCGAAGATGATGTGGTCG | PCR primer for TF0035 |
| 2N | CCCTGTTCTTCGTTATAAATACGGC | PCR primer for TF0035 |
| 3N | gggcaatttcttttttgtcatATTTTTTCGAATCGCACTTCC | PCR overlap primer |
| 4N | aaaaatttcatccttcgTGAAGCATCGTTTGTCTTTTC | PCR overlap primer |
| 5N | GAAAAGACAAACGATGCTTCAcgaaggatgaaattttt | PCR overlap primer |
| 6N | GGAAGTGCGATTCGAAAAAATatgacaaaaaagaaattgccc | PCR overlap primer |
| Construction of recombinant NanH | ||
| nanH-F | GCGCGGATCCATGAAAAAGTTTTTTTGGATb | |
| nanH-R | CGCGCGAATTCTCATCGTATCAGGTCTTTGATTTTAACc | |
| RT-PCR | ||
| TF0035-F | TGAAAGCCGTTCGTCTGT | nanH |
| TF0035-R | CAACGGTCGTCACTTCAT | nanH |
| TF0036-F | CTCCGCAACAAAGACGAA | Downstream ORF of nanH |
| TF0036-F | ATCTCCCGTTTCAGCTCT | Downstream ORF of nanH |
| TF2207-F | CAGCGATGAACTTCCTCG | siaHI |
| TF2207-R | ATAACTGCGCTGATGGTG | siaHI |
| TF2206-F | CTTCCTTTGCCGGTGGTT | Downstream ORF of siaHI |
| TF2206-R | TCCCATGTCATGTGCCAG | Downstream ORF of siaHI |
| TF 16S-F | GGGTGAGTAACGCGTATGTAACCT | 16S rRNA |
| TF-16S-R | ACCCATCCGCAACCAATAAA | 16S rRNA |
Small letters indicate ermF.
BamHI site is underlined.
EcoRI site is underlined.
RNA isolation and reverse transcription (RT)-PCR.
Total bacterial RNA was isolated from 2-ml cultures (optical density at 600 nm [OD600] = 0.4) by using the RiboPure total RNA isolation kit (Ambion Inc. Austin, TX), followed by DNase I treatment (1 U/μg RNA for 1 h at 37°C). DNase was inactivated by treatment with a DNase-deactivating resin (Ambion). The total RNA from each of the wild-type and mutant strains was isolated from the mid-log-phase culture. The cDNA was synthesized from 1 μg total RNA by utilizing SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA) with random hexamers (Promega, Madison, WI) according to the supplier's recommendations. Twenty percent of the cDNA preparation or of a preparation without addition of reverse transcriptase was subjected to PCR using gene-specific primers. T. forsythia 16S rRNA-specific primers were utilized for normalization. Primer sequences are listed in Table 1.
Sialidase activity.
The total cell lysate, surface-associated (membrane fraction), and secreted (spent-medium) sialidase activities of the wild type and SiaHI and NanH deletion mutants were compared. The fluorogenic substrate, 2′-(4-methylumbelliferyl)-α-d-N-acetylneuraminic acid (4-MU-NeuNA) (Sigma), was used to assay sialidase activity as described previously (15). Qualitative screening of sialidase activity in intact bacteria, total cell lysate, outer membrane fraction, or spent medium filtered through a 0.2-μm filter was performed in microtiter plate wells. The T. forsythia membrane fraction was prepared as follows. Cells were suspended in Tris buffer (50 mM Tris [pH 7.3], 0.15 M NaCl, and 5 mM MgCl2) and ultrasonicated on ice for 5 min (50% power and a duty cycle of 5). Undisrupted cells and debris were removed by centrifugation (10,000 × g, 4°C, 30 min), and the supernatant was collected. The supernatant was then ultracentrifuged (80,000 × g, 4°C, 1.5 h), and the pellet was washed twice with Tris buffer and finally resuspended in Tris buffer and stored at −70°C. Briefly, for an activity assay, 50 μl of test sample was added to 50 μl of 0.1 M sodium acetate buffer containing 2% Triton X-100 (pH 4.5) in microtiter wells. Following incubation of the mixture at room temperature for 5 min, 100 μl of 50 μM 4-MU-NeuNAc in 0.1 M sodium acetate buffer (pH 4.5) was added, and the mixtures were incubated at 37°C for 15 to 30 min. The fluorescence intensity was then examined under long-wavelength UV light (365 nm) after 15 min at 37°C. Blue-white fluorescence indicated the presence of sialidase activity. Quantitative sialidase assays were performed after stopping the reaction with 100 μl 100 mM Tris-HCl, pH 10.0. The fluorescence intensities were measured in a fluorimeter (Labsystems Thermo, Waltham, MA) with an excitation wavelength of 380 nm and an emission wavelength of 460 nm every 10 min starting from 0 to 1 h. The specific activity of the T. forsythia cell-associated or -secreted sialidase was determined by extrapolating fluorescence intensities to a standard curve generated with a commercial purified Clostridium perfringens sialidase (Sigma, St. Louis, MO) with known enzyme activity.
Expression and purification of recombinant NanH.
The NanH-encoding open reading frame (ORF) (TF0035) was PCR amplified from genomic DNA using the primers nanH-F (engineered BamHI site) and nanH-R (engineered EcoRI site). These primers amplify the NanH ORF from the start codon as previously proposed (38). The PCR fragment digested with BamHI and EcoRI was cloned into the pGEX-4T expression vector (GE Life Sciences) at the same sites. The recombinant plasmid clone with the correct in-frame insert sequence, designated pGEX-nanH, allowing expression of NanH as a fusion protein with N-terminal glutathione-S-transferase, was transformed into E. coli BL21. An E. coli transformant clone grown to an optical density of 0.2 at 600 nm was induced with isopropyl-β-d-thiogalactopyranoside (IPTG) (0.1 mM, final concentration), and the culture was incubated for an additional 3 h with vigorous shaking to express the fusion protein. The cells were harvested by centrifugation at 5,000 × g for 30 min, suspended in phosphate-buffered saline (PBS), and subjected to cell disruption by ultrasonication. The cell lysates were centrifuged at 10,000 × g for 10 min at 4°C to separate the soluble fraction from the debris. The soluble fraction was used for analyzing sialidase activity and was also subjected to SDS-PAGE to detect the fusion glutathione S-transferase (GST)-NanH protein. The recombinant fusion protein was purified from E. coli lysates by affinity purification on glutathione-agarose (Sigma) medium according to a previously described protocol (35). The specific activities of the purified fractions were determined by comparison with Clostridium perfringens sialidase (Sigma). Fusion protein samples with sialidase activities in the range of 10 to 50 U/mg protein were routinely purified.
Attachment and invasion assays.
For both attachment and invasion assays, T. forsythia cells were used at a multiplicity of infection of 100. Epithelial cell monolayers (KB or OBA-9) were incubated for either 1 h or 4 h for attachment or invasion assays, respectively, as described previously (16). Briefly, for attachment assays, monolayers incubated with bacteria were washed three times with DMEM, epithelial cell-associated bacteria were retrieved by lysing monolayers in distilled water, and bacteria were plated for counting. For invasion assays, epithelial cell monolayers incubated with bacteria were washed with DMEM and then treated with gentamicin (300 μg/ml) and metronidazole (200 μg/ml) to kill external bacteria. The monolayers were then washed with DMEM and were lysed by water to allow intracellular bacteria counting. For inhibition assays, bacteria were pretreated with the sialidase inhibitor 2-deoxy-2,3-dehydro-N-acetylneuraminic acid (Neu5Ac2en) at a concentration of 10 mM for 30 min.
Lectin staining.
KB or OBA-9 cells (105) were seeded on poly-d-lysine-coated slides. Infection was carried out with 4 × 108 bacteria for 2 h. Uninfected cells were alternatively treated with recombinant GST-NanH (rGST-NanH) (0.2 U/ml sialidase). Cells were fixed with 3% paraformaldehyde for 15 min. Biotinylated Maackia amurensis lectin (MAL) (Vector Laboratories) was incubated with cells at 2.5 μg/ml, respectively, for 1 h. After washing with PBS, cells were treated with 1 μg/ml DyLight 488-conjugated streptavidin (Vector Laboratories, Burlingame, CA), and nuclei were then stained with 4′,6-diamidino-2-phenylindole (DAPI) fluorescent stain (Vector Laboratories). Fluorescence was determined on mounted slides with Vectashield (Vector Laboratories).
Statistical analysis.
Strain comparisons for sialidase expression and attachment assays were analyzed using the Student t test using Prism 5.0 software (GraphPad Software, San Diego, CA).
RESULTS
NanH- and SiaHI-deficient mutants show significant loss of T. forsythia-associated sialidase activity.
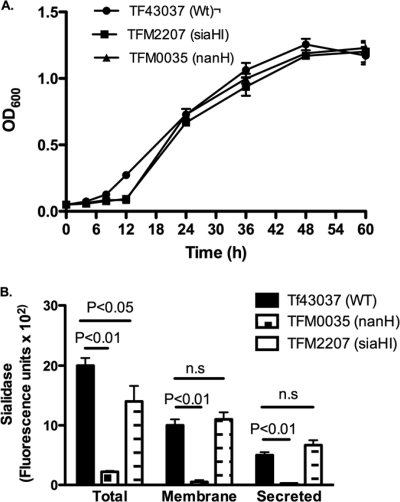
T. forsythia expresses two distinct sialidases, namely, SiaHI (TF2207) (17) and NanH (TF0035) (38). While SiaHI shows no significant sequence similarity to any of the known bacterial sialidases, NanH (TF0035) shows characteristic motifs prototypical of sialidases. NanH is 65% identical to Bacteroides fragilis NanH and is the major sialidase in T. forsythia. In addition, analysis of the nanH-associated operon indicated that the operon contains genes with putative functions in glycoprotein degradation, sialic acid release, and assimilation; the genes include those encoding N-acetylneuraminate lyase (TF0030; NanA), an N-acetylglucosamine epimerase (TF0031; NanE), a transport protein of the major facilitator superfamily (TF0032; MFS), a β-hexosaminidase (TF0036), a putative 9-O-acetylesterase (TF0037), and two proteins with similarity to RagAB-like TonB-dependent receptors (TF0033 and TF0034). To determine the roles of the SiaHI and NanH sialidases, we constructed specific gene deletion mutants for each and characterized the mutants for their epithelial cell binding and invasion abilities. We wanted to determine if sialidases were involved in exposing sialic acid-masked cryptic epitopes on epithelial cells for bacterial interactions. For this purpose, siaHI and nanH deletion mutants were constructed by an allelic replacement strategy with the erythromycin resistance marker, ermF. The double-crossover strategy utilized in the study results in the replacement of sialidase genes (siaHI or nanH) from their translational start to stop codons with the ermF gene, thus expressing the erythromycin resistance marker from the corresponding sialidase gene promoters and maintaining the integrity of the operons. The expected integration events were confirmed by sequencing the PCR generated fragments encompassing the integration sites in the respective transformants (data not shown). A representative mutant for each of the siaHI and nanH genes, named TFM2207 (siaHI) and TFM0035 (nanH), was further characterized. Specifically, with regard to the nanH deletion, since the nanH gene is located downstream of the putative sialic acid uptake locus, the disruption is likely not to effect uptake of free sialic acid as well. Conclusive evidence to rule out any polar effects was obtained by RT-PCR and electrophoretic analysis. The results showed intense PCR products for TF2206 (ORF downstream of siaHI in the siaHI mutant) and TF0036 (ORF downstream of nanH in the nanH mutant) similar to that of the wild-type strain (data not shown). In comparison to the wild-type strain, although the siaHI and nanH mutants showed a slight growth lag in the early log phase, they reached full growth similar to that of the wild-type strain by late log phase (absorbance of 1.2 ± 0.1 at 600 nm) (Fig. 1A) in nutrient-rich BF broth (34). An initial slight lag in mutants during early log phase is likely due to the metabolic burden from expression of the antibiotic resistance (Emr) marker. Thus, the deletion of siaHI or nanH did not affect the overall fitness of the respective mutants in broth cultures.
FIG. 1.
Characterization of sialidase mutants. (A) Growth of strains in BHI broth supplemented with fetal bovine serum and N-acetyl muramic acid as determined by OD600. Wt, wild type. (B) Sialidase activities of bacterial strains measured with the fluorescent substrate 4-methylumbelliferyl-N-acetyl-α-d-neuraminic acid. Sialidase activities associated with total cell extract and membrane and secreted fractions were determined for each strain. Equal amounts of protein fractions (25 to 100 μg) for each strain were assayed. Bars represent fluorescence values (means and standard deviations) from triplicate readings. Data are representative of more than three independent experiments with similar results. WT, wild type.
The loss of sialidase activity in mutants was demonstrated by enzymatic assays. Sialidase activity was determined in total cell extracts, outer membrane fractions, and culture supernatants. The cells and spent culture supernatants from mid-log phase (absorbance of 0.6 at 600 nm) were harvested. Cell pellets were either lysed in 2% Triton X-100 to estimate total enzyme activity or used to prepare outer-membrane fractions as described previously (12). The culture supernatants were concentrated with a Centricon-30 device as per the manufacturer's recommendations. Equal amounts of protein fractions from respective strains were assayed for sialidase activity. The results showed that the total sialidase activities in the siaHI mutant TFM2207 and the nanH mutant TFM0035 were diminished by approximately 10% and 90% of the activity observed in the wild-type strain, respectively (Fig. 1B). Moreover, sialidase activity was detected in the outer-membrane and secreted fractions from the wild type but not in strain TFM0035; TFM2207 did not show a significant loss of activity in either fraction (Fig. 1B). These results indicated that NanH, in accordance with in silico structure predictions, is membrane associated as well as secreted, and SiaHI is likely periplasmic.
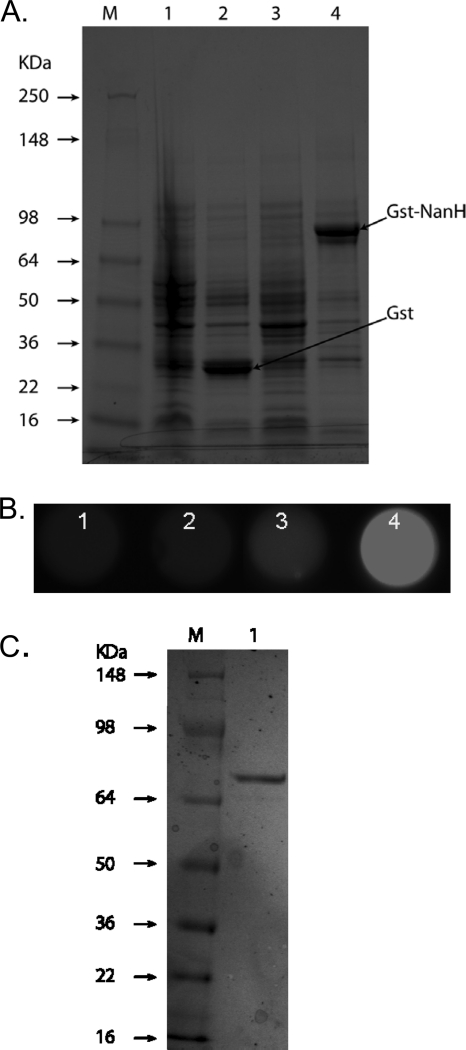
Expression of enzymatically active NanH sialidase in E. coli.
NanH was expressed as a fusion protein with glutathione S-transferase (GST) by the pGEX-4T expression plasmid in E. coli BL21. Consistent with the predicted molecular size of the GST-NanH fusion protein, a band migrating at approximately 85 kDa (27.5-kDa GST plus a predicted molecular mass of 59.7 kDa for NanH) on 10% SDS-PAGE gels was observed in E. coli cell lysates with the recombinant plasmid following IPTG induction (Fig. 2A). E. coli with the control plasmid showed an expected 27.5-kDa GST band following IPTG induction. Moreover, the soluble fraction obtained from a GST-NanH-expressing (BL21/pGEX-nanH) but not a GST alone-expressing (BL21/pGEX-4T) clone showed strong sialidase activity (Fig. 2B) following IPTG induction. These results thus demonstrated that NanH, when expressed in E. coli as a fusion partner with GST, is enzymatically active. The recombinant GST-NanH fusion protein was purified from lysates using glutathione agarose affinity medium to near-homogeneity, indicated by a major band of approximately 85 kDa on a 4-to-12%-gradient SDS-PAGE gel (Fig. 2C). We routinely obtained enzymatically active GST-NanH fusion protein samples with specific activities in the range of 10 to 50 U/mg protein. These samples were utilized as the source for NanH activity for subsequent investigations.
FIG. 2.
Expression of T. forsythia NanH in E. coli as a GST fusion protein. (A). SDS-PAGE of E. coli clones harboring the control (BL21/pGEX-4T; lanes 1 and 2) or recombinant (BL21/pGEX-nanH) plasmid in the presence (lanes 2 and 4) or absence (lanes 1 and 3) of IPTG. E. coli cultures grown to an absorbance of 0.2 at 600 nm were incubated for an additional 3 h in the presence or absence of IPTG. Cell lysates (50 μg protein for each) were separated on 10% gels and stained with Coomassie. (B) Sialidase activity assayed in lysates of corresponding clones in microtiter wells (BL21/pGEX-4T, lanes 1 [uninduced] and 2 [induced]; BL21/pGEX-nanH, lanes 3 [uninduced] and 4 [induced]; 10 μg protein lysate for each) using 4-MU-NeuAc as a substrate. Fluorescence was visualized on a UV transilluminator. (C) SDS-PAGE of purified rGST-NanH protein on a 4-to-12%-gradient polyacrylamide gel. Lane M, protein markers; lane 1, rGst-NanH.
NanH deletion results in reduced bacterial attachment to epithelial cells.
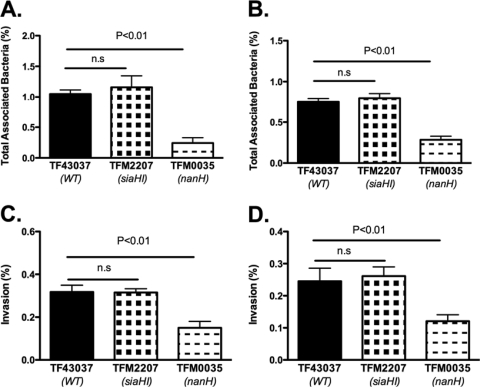
Epithelial cells are decorated with a variety of sialic acid-containing glycoconjugates, and conceivably, the sialic acid residues may provide bacteria a carbon and nitrogen source and/or may modulate bacterial attachment. With respect to the role of sialic acids in bacterial interactions with epithelial cells, terminal sialic acids might serve as ligands for bacterial lectins or alternatively mask hidden epitopes for bacterial binding. We utilized a KB cell model system, described previously, to study the role of sialidases in bacterial interaction with epithelial cells (16). KB cells are a derivative of HeLa cells (human cervical epithelium), and both of these cell types have been extensively used for studying oral pathogen-epithelial cell interactions (10, 28). In addition, we used the OBA-9 cell line as a representative human gingival epithelial cell model (20). Our results showed that the wild-type T. forsythia ATCC 43037 strain and its sialidase-defective mutants TFM2207 (siaHI) and TFM0035 (nanH) adhered to and invaded both KB and OBA-9 cells to different degrees. In comparison to the wild type and TFM2207 (siaHI), there was significant reduction in the total attachment of the TFM0035 (nanH) mutant to KB (Fig. 3A) as well as OBA-9 (Fig. 3B) cells. However, no significant differences were observed between the attachment levels of the wild type and TFM2207 to either KB or OBA-9 cells. In addition, significant reductions in the invasion abilities of TFM0035 in comparison to that of the wild-type or TFM2207 strain with respect to KB (Fig. 3C) and OBA-9 (Fig. 3D) cells was evident.
FIG. 3.
(A and B) Attachment of bacteria to epithelial cells. Epithelial cell monolayers (KB and OBA-9) were incubated for 1 h with bacteria in a CO2 incubator at 37°C. Monolayers were then washed and lysed in distilled water, and total associated bacteria were plated on agar plates for counting. Attachment levels (means and standard deviations of triplicate readings) were expressed as percentages of attached bacteria relative to the total number of input bacteria. (A) KB cells. Data representative of three independent experiments show a significant reduction in the attachment of the nanH mutant strain, TFM0035, compared to that of the wild-type strain, ATCC 43037. The mean attachment levels for TFM0035 were 23.5, 29.4, and 22.5% of that of the wild-type strain, while the TFM2207 attachment levels were not significantly different from that of the wild-type strain in three independent experiments. (B) OBA-9 cells. Reduced attachment of TFM0035 compared to that of the wild-type strain was also observed for OBA-9 cells; mean TFM0035 attachment levels were 37.2 and 30.7% of that of the wild-type strain in two independent experiments, whereas TFM2207 attachment levels were not significantly different from that of the wild-type strain. (C and D) Invasion of epithelial cells. Strains were incubated with epithelial cell monolayers for 4 h, followed by metronidazole treatment for 1 h to kill outside bacteria. The invading bacteria were then counted after epithelial cell lysis and plating on agar plates. (C) KB cells. Data representative of three independent experiments shows significantly reduced invasion (mean and standard deviations) by TFM0035 compared to that by the wild-type strain; mean invasion levels of TFM0035 were 46.5, 42.1, and 38.6% of that of the wild-type strain. In none of the independent experiments did the siaH mutant, TFM2207, show reduced invasion compared to the wild-type strain. (D) OBA-9 cells. Reduced invasion of TFM0035 compared to that of the wild-type strain was also observed for OBA-9 cells; TFM0035 invasion levels were 49.5 and 42.8% of that of the wild-type strain, and TFM2207 invasion levels were not significantly different from that of the wild-type strain in two independent experiments.
Sialidase activity associated with NanH mediates bacterial attachment.
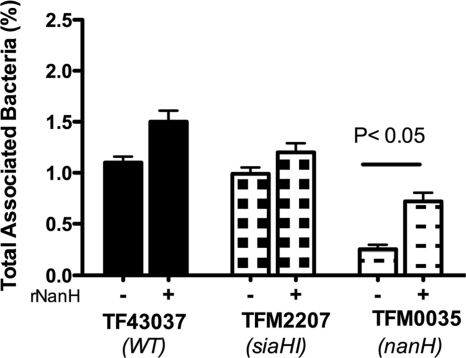
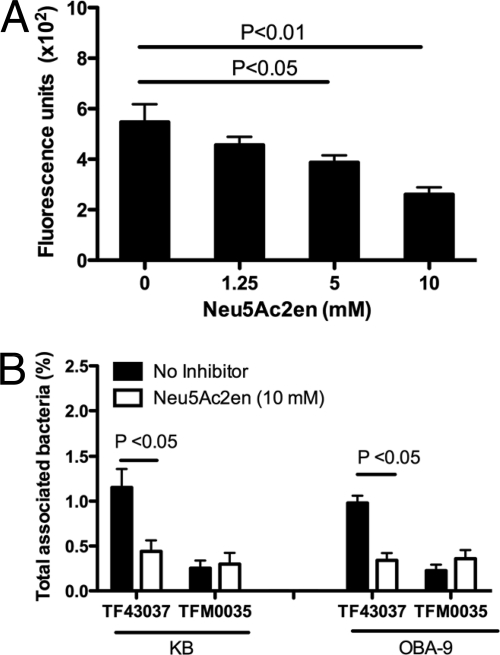
As described above, the deletion of nanH and not siaHI resulted in the impairment of bacterial attachment to epithelial cells. To confirm if a deficiency in sialidase activity in TFM0035 (nanH) was responsible for the observed phenotype, we complemented TFM0035 in trans with purified recombinant sialidase since gene complementation in T. forsythia has so far been unsuccessful. For trans complementation, rGST-NanH (0.02 U/ml) along with bacteria was added to the KB cell monolayers, and attachment abilities were determined. The results demonstrated that complementation with sialidase significantly enhanced the attachment ability of TFM0035 compared to the attachment ability of the noncomplemented controls (Fig. 4). To further confirm the involvement of sialidase activity of NanH in bacterial attachment, we utilized a broad-spectrum sialidase inhibitor, Neu5Ac2en. Neu5Ac2en caused dose-dependent inhibition of T. forsythia-associated sialidase activity (Fig. 5A). For inhibition assays, bacteria were pretreated with Neu5Ac2en (10 mM) for 30 min at 37°C, and the inhibitor (10 mM) was added to the culture medium. The results showed that this treatment significantly reduced the association of the wild-type strain with both KB and OBA-9 cells compared to that of the corresponding nontreated controls (Fig. 5B). Moreover, the inhibitor treatment did not further reduce the total association of the NanH mutant strain TFM0035 with either KB or OBA-9 cells. Taken together, these results confirm that sialidase activity associated with NanH plays a significant role in epithelial cell attachment by T. forsythia.
FIG. 4.
trans complementation of mutants with recombinant sialidase. Epithelial cell monolayers were incubated for 1 h in a CO2 incubator at 37°C with different strains in the presence or absence of recombinant sialidase, and bacterial attachment for each strain was determined. Data represented are from one of three independent experiments with similar results.
FIG. 5.
Effect of sialidase inhibitor Neu5Ac2en on epithelial cell attachment. (A) Dose-dependent inhibition of T. forsythia-associated sialidase activity by Neu5Ac2en. Triton X-100-extracted fractions from T. forsythia wild-type cells were treated with increasing concentrations of sialidase inhibitor for 30 min at 37°C. Sialidase activity was then measured with the fluorescent substrate 4-methylumbelliferyl-N-acetyl-α-d-neuraminic acid. Bars represent fluorescence values (means and standard deviations) of triplicate readings. Data are representative of more than three independent experiments with similar results. (B) Inhibition of T. forsythia wild-type attachment to epithelial cells (KB or OBA-9) by a sialidase inhibitor. T. forsythia wild-type cells were pretreated with 10 mM Neu5Ac2en for 30 min prior to incubation with epithelial cell monolayers. Monolayers with bacteria were incubated for 1 h, and bacterial attachment levels were determined as previously described. Data are representative of three (KB cells) or two (OBA-9 cells) independent experiments with similar results.
T. forsythia desialylates epithelial cells surfaces.
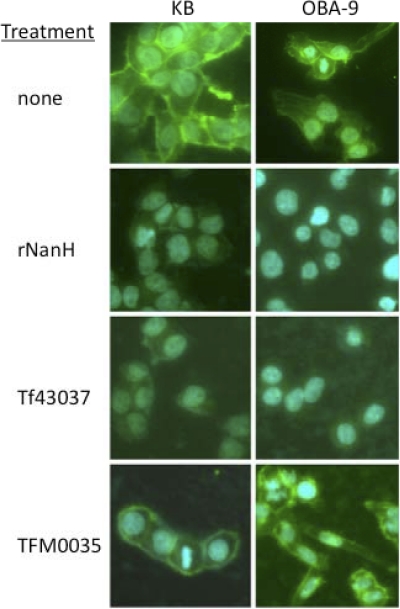
Previous studies have shown that NanH cleaves sialic acid from a variety of glycoconjugates, with a preference for α2-3 glycosidic linkages. In order to validate that T. forsythia indeed cleaves cell surface sialic acids by its NanH activity, we visualized epithelial cell surfaces following infection with different strains by fluorescence microscopy using Maackia amurensis lectin (MAL), which recognizes α2-3-linked sialic acids. The treatment of epithelial cells with rGST-NanH sialidase (0.02 U/ml) significantly reduced the lectin binding compared to that with the nontreated controls (Fig. 6), demonstrating a loss of cell surface α2-3-linked terminal sialic acids by NanH sialidase. Furthermore, α2-3-linked terminal sialic acids were also conspicuously absent from the epithelial cell surfaces treated with the wild-type T. forsythia strain compared to those of cells infected with TFM0035 (Fig. 6). These results thus demonstrated that T. forsythia NanH cleaves α2-3-linked terminal sialic acids from epithelial cell surfaces.
FIG. 6.
T. forsythia cleaves epithelial cell surface sialic acids. Surface carbohydrates of epithelial cells were analyzed by lectin binding after 2 h of treatment with rNanH, T. forsythia 43037 (WT), or TFM0035 (nanH). Cells were fixed with 3% paraformaldehyde for 30 min and then reacted for 1 h with biotinylated Maackia amurensis lectin, specific for α2-3-linked terminal sialic acids. Lectin binding was visualized by staining with fluorescence (DyLight 488)-labeled streptavidin. Nuclei were stained with DAPI.
DISCUSSION
T. forsythia grows subgingivally under anaerobic conditions where it likely utilizes host proteinaceous components for growth. While this suggests predominantly an asaccharolytic physiology, the bacterium produces a variety of glycosidases which can provide a rich source of sugars. The roles of these glycosidases, including recently identified SiaHI and NanH sialidases in bacterial physiology or pathogenesis, have not been fully investigated. Bacterial sialidases have been shown to be important in providing free sialic acid as a nutritional carbon and nitrogen source from glycoconjugates (42) and in contributing to pathogenesis by exposing epitopes for bacterial adherence through cleaving terminal sugars on host glycoconjugates (6). During the colonization process, Capnocytophaga canimorsus surface-localized sialidase has also been shown to initiate host deglycosylation, promoting bacterial growth and persistence in vivo (21); Pseudomonas aeruginosa sialidase has been shown to promote biofilm formation in the lungs of cystic fibrosis patients (36). In addition, Streptococcus pneumoniae surface-anchored sialidase has been shown to promote endothelial cell attachment and invasion (40), and S. pneumoniae sialidases have further been shown to initiate deglycosylation of host proteins, including IgA1 and human secretory component, for growth and persistence in the airway (4). In addition, bacterial sialidases have also been shown to be involved in defense against host immune surveillance. In this regard, deglycosylation of human serum glycoproteins by the combined action of S. pneumoniae sialidases and exoglycosidases has been shown to increase resistance to complement-dependent killing and phagocytosis of bacteria (7).
The current study was undertaken specifically to determine the roles of sialidases produced by T. forsythia. We were interested in determining the role of T. forsythia sialidases in adhesion to and invasion into epithelial cells. Epithelial cells are decorated by a wide variety of glycoconjugates (31), which in many cases have terminal sialic acid residues. These terminal sialic acid residues on host surfaces can serve as ligands for bacterial colonization, maintain a net negative charge repulsive for bacteria, or mask epitopes (cryptitopes) that would otherwise facilitate bacterial adherence. Additionally, removal of these terminal sialic acids may expose other sugars for binding or utilization by bacteria. Thus, we hypothesized that T. forsythia sialidases might be involved in exposing new cryptitopes for bacterial binding. Our study demonstrated a role for T. forsythia surface-associated and secreted NanH sialidase in attachment and invasion of epithelial cells. We observed that in comparison to the wild-type and SiaHI-deficient strains, a NanH-deficient mutant, TFM0035, was significantly attenuated in binding and invasion ability. Furthermore, inactivation of sialidase activity with an inhibitor significantly blocked bacterial attachment to epithelial cells. Since complementation in trans with recombinant enzyme did not restore the epithelial cell binding ability of the NanH mutant to wild-type levels, it remains to be seen if functions other than sialic acid removal might also contribute to the attachment, such as the NanH protein directly interacting as an adhesin with epithelial cells. A role for SiaHI could not be demonstrated in the current study. Our results utilizing a SiaHI-deficient mutant suggested that this sialidase is periplasmic in nature and thus may not be accessible for interactions. It is likely that this enzyme may be involved in scavenging sialic acid from internalized glycoconjugates in the periplasm. We concluded that NanH is the major sialidase utilized by the bacterium for mediating epithelial cell interactions. We and others have shown that other components, including a leucine-rich repeat surface protein, BspA (16), as well as other surface layer proteins (28, 30), are involved in bacterial binding to and invasion into epithelial cells. Since bacterial attachment and invasion constitute a multistep process, it is reasonable to predict that NanH sialidase, along with other attachment and invasion factors, is coordinately regulated. In this process, unmasking of binding epitopes for T. forsythia attachment factors by sialidase activity may be an initial step. Since sialic acid serves as a growth factor only in biofilms and not in planktonic growth (26), the NanH-deficient mutant TFM0035 showed no significant growth defects and reached full growth, like the wild type, in broth cultures. Based on these studies, we predict that the NanH-deficient mutant TFM0035 might be defective in biofilm growth on human glycoconjugated coated surfaces, such as human salivary mucins. We are currently exploring the effects of sialidase deletion on biofilm growth. The role of SiaHI sialidase could not be assessed, since the siaHI deletion did not result in any change in bacterial growth or bacterial ability to attach/invade epithelial cells in vitro. Moreover, since in silico structural predictions suggest NanH to be surface localized and SiaHI to be periplasmic, this suggests that these enzymes might be performing different functions in the pathogenesis and physiology of T. forsythia. In addition to the roles of NanH sialidase in epithelial cell interactions (this study) and in providing sialic acid as a biofilm growth-promoting factor (26), it is possible that a NanH-dependent release of sialic acid could be important for sialylation of surface macromolecules, such as lipopolysaccharide (LPS) or outer membrane proteins important for host immune evasion. For example, Neisseria gonorrhoeae (24) and Haemophilus influenzae (32) sialylated bacterial components have been suggested to allow pathogens to disguise themselves from host immune systems through molecular mimicry and avoid killing by the host immune complement pathway and phagocytes (11).
In the oral cavity, T. forsythia comes in direct contact with sialylated glycoproteins present in saliva, mucin-coated barriers formed on teeth and mucosa, and gingival epithelial cell surfaces. T. forsythia encodes several other glycosidases, such as α-d-glucosidase (TF0094) (encoded by SusB), N-acetyl-β-d-glucosaminidase (TF2925) (encoded by hexA), β-glucosidase (TF0014) (encoded by bglX), α-fucosidase (TF0421) (encoded by fucO), α-arabinofuranosidase (TF0780) (encoded by abfA), β-galactosidase (TF0229, TF1468, and TF2386), and α-1,2-mannosidase (TF0617 and TF1335). In conjunction with NanH and SiaHI, these glycosidases might hydrolyze terminal glycosidic linkages of complex oligosaccharides, proteoglycans, and oligosaccharides associated with the host glycoproteins, affecting their functional integrity, and could promote disease progression and/or help provide nutrient sources for coexisting bacteria residing in the oral cavity. It is also tempting to speculate that NanH and SiaHI sialidases, together with these glycosidases, could be involved in the maintenance of bacterial communities within the oral cavity by providing sugar metabolites as nutrients or providing new cryptitopes to other bacteria. In this regard, the cohabiting bacterium Fusobacterium nucleatum, which binds galactose residues, can benefit from the NanH activity of T. forsythia. Plausibly, NanH might expose penultimate galactose residues on oral mucosal surfaces by removing terminal sialic acid residues for F. nucleatum binding.
In summary, we describe a role for NanH sialidase in epithelial cell attachment by T. forsythia and speculate that sialidase inhibitors might be utilized to block T. forsythia pathogenesis and modulate periodontitis.
Acknowledgments
We thank Howard K. Kuramitsu for critical reading of the manuscript.
This study was supported by a grant (DE014749) from the NIDCR.
Editor: J. N. Weiser
Footnotes
Published ahead of print on 15 November 2010.
REFERENCES
- 1.Angata, T., and A. Varki. 2002. Chemical diversity in the sialic acids and related alpha-keto acids: an evolutionary perspective. Chem. Rev. 102:439-469. [DOI] [PubMed] [Google Scholar]
- 2.Bradshaw, D. J., K. A. Homer, P. D. Marsh, and D. Beighton. 1994. Metabolic cooperation in oral microbial communities during growth on mucin. Microbiology 140:3407-3412. [DOI] [PubMed] [Google Scholar]
- 3.Braham, P. H., and B. J. Moncla. 1992. Rapid presumptive identification and further characterization of Bacteroides forsythus. J. Clin. Microbiol. 30:649-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burnaugh, A. M., L. J. Frantz, and S. J. King. 2008. Growth of Streptococcus pneumoniae on human glycoconjugates is dependent upon the sequential activity of bacterial exoglycosidases. J. Bacteriol. 190:221-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byers, H. L., E. Tarelli, K. A. Homer, and D. Beighton. 1999. Sequential deglycosylation and utilization of the N-linked, complex-type glycans of human alpha(1)-acid glycoprotein mediates growth of Streptococcus oralis. Glycobiology 9:469-479. [DOI] [PubMed] [Google Scholar]
- 6.Corfield, T. 1992. Bacterial sialidases—roles in pathogenicity and nutrition. Glycobiology 2:509-521. [DOI] [PubMed] [Google Scholar]
- 7.Dalia, A. B., A. J. Standish, and J. N. Weiser. 2010. Three surface exoglycosidases from Streptococcus pneumoniae, NanA, BgaA, and StrH, promote resistance to opsonophagocytic killing by human neutrophils. Infect. Immun. 78:2108-2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fletcher, H. M., H. A. Schenkein, and F. L. Macrina. 1994. Cloning and characterization of a new protease gene (prtH) from Porphyromonas gingivalis. Infect. Immun. 62:4279-4286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grenier, D. 1995. Characterization of the trypsin-like activity of Bacteroides forsythus. Microbiology 141:921-926. [Google Scholar]
- 10.Han, Y. W., W. Shi, G. T. Huang, S. Kinder Haake, N. H. Park, H. Kuramitsu, and R. J. Genco. 2000. Interactions between periodontal bacteria and human oral epithelial cells: Fusobacterium nucleatum adheres to and invades epithelial cells. Infect. Immun. 68:3140-3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harvey, H. A., W. E. Swords, and M. A. Apicella. 2001. The mimicry of human glycolipids and glycosphingolipids by the lipooligosaccharides of pathogenic Neisseria and Haemophilus. J. Autoimmun. 16:257-262. [DOI] [PubMed] [Google Scholar]
- 12.Honma, K., S. Inagaki, K. Okuda, H. K. Kuramitsu, and A. Sharma. 2007. Role of a Tannerella forsythia exopolysaccharide synthesis operon in biofilm development. Microb. Pathog. 42:156-166. [DOI] [PubMed] [Google Scholar]
- 13.Horton, R. M., S. N. Ho, J. K. Pullen, H. D. Hunt, Z. Cai, and L. R. Pease. 1993. Gene splicing by overlap extension. Methods Enzymol. 217:270-279. [DOI] [PubMed] [Google Scholar]
- 14.Hughes, C. V., G. Malki, C. Y. Loo, A. C. Tanner, and N. Ganeshkumar. 2003. Cloning and expression of alpha-D-glucosidase and N-acetyl-beta-glucosaminidase from the periodontal pathogen, Tannerella forsythensis (Bacteroides forsythus). Oral Microbiol. Immunol. 18:309-312. [DOI] [PubMed] [Google Scholar]
- 15.Inagaki, S., H. K. Kuramitsu, and A. Sharma. 2005. Contact-dependent regulation of a Tannerella forsythia virulence factor, BspA, in biofilms. FEMS Microbiol. Lett. 249:291-296. [DOI] [PubMed] [Google Scholar]
- 16.Inagaki, S., S. Onishi, H. K. Kuramitsu, and A. Sharma. 2006. Porphyromonas gingivalis vesicles enhance attachment, and the leucine-rich repeat BspA protein is required for invasion of epithelial cells by “Tannerella forsythia.” Infect. Immun. 74:5023-5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishikura, H., S. Arakawa, T. Nakajima, N. Tsuchida, and I. Ishikawa. 2003. Cloning of the Tannerella forsythensis (Bacteroides forsythus) siaHI gene and purification of the sialidase enzyme. J. Med. Microbiol. 52:1101-1107. [DOI] [PubMed] [Google Scholar]
- 18.King, S. J., K. R. Hippe, and J. N. Weiser. 2006. Deglycosylation of human glycoconjugates by the sequential activities of exoglycosidases expressed by Streptococcus pneumoniae. Mol. Microbiol. 59:961-974. [DOI] [PubMed] [Google Scholar]
- 19.Kuroiwa, A., A. Hisatsune, Y. Isohama, and H. Katsuki. 2009. Bacterial neuraminidase increases IL-8 production in lung epithelial cells via NF-kappaB-dependent pathway. Biochem. Biophys. Res. Commun. 379:754-759. [DOI] [PubMed] [Google Scholar]
- 20.Kusumoto, Y., H. Hirano, K. Saitoh, S. Yamada, M. Takedachi, T. Nozaki, Y. Ozawa, Y. Nakahira, T. Saho, H. Ogo, Y. Shimabukuro, H. Okada, and S. Murakami. 2004. Human gingival epithelial cells produce chemotactic factors interleukin-8 and monocyte chemoattractant protein-1 after stimulation with Porphyromonas gingivalis via toll-like receptor 2. J. Periodontol. 75:370-379. [DOI] [PubMed] [Google Scholar]
- 21.Mally, M., H. Shin, C. Paroz, R. Landmann, and G. R. Cornelis. 2008. Capnocytophaga canimorsus: a human pathogen feeding at the surface of epithelial cells and phagocytes. PLoS Pathog. 4:e1000164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moncla, B. J., P. Braham, and S. L. Hillier. 1990. Sialidase (neuraminidase) activity among gram-negative anaerobic and capnophilic bacteria. J. Clin. Microbiol. 28:422-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakajima, T., N. Tomi, Y. Fukuyo, H. Ishikura, Y. Ohno, R. Arvind, T. Arai, I. Ishikawa, and S. Arakawa. 2006. Isolation and identification of a cytopathic activity in Tannerella forsythia. Biochem. Biophys. Res. Commun. 351:133-139. [DOI] [PubMed] [Google Scholar]
- 24.Ram, S., A. K. Sharma, S. D. Simpson, S. Gulati, D. P. McQuillen, M. K. Pangburn, and P. A. Rice. 1998. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J. Exp. Med. 187:743-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reinholdt, J., M. Tomana, S. B. Mortensen, and M. Kilian. 1990. Molecular aspects of immunoglobulin A1 degradation by oral streptococci. Infect. Immun. 58:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Roy, S., C. I. Douglas, and G. P. Stafford. 2010. A novel sialic acid utilization and uptake system in the periodontal pathogen Tannerella forsythia. J. Bacteriol. 192:2285-2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rudney, J. D., R. Chen, and G. J. Sedgewick. 2005. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J. Dent. Res. 84:59-63. [DOI] [PubMed] [Google Scholar]
- 28.Sabet, M., S. W. Lee, R. K. Nauman, T. Sims, and H. S. Um. 2003. The surface (S-) layer is a virulence factor of Bacteroides forsythus. Microbiology 149:3617-3627. [DOI] [PubMed] [Google Scholar]
- 29.Saito, T., K. Ishihara, T. Kato, and K. Okuda. 1997. Cloning, expression, and sequencing of a protease gene from Bacteroides forsythus ATCC 43037 in Escherichia coli. Infect. Immun. 65:4888-4891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sakakibara, J., K. Nagano, Y. Murakami, N. Higuchi, H. Nakamura, K. Shimozato, and F. Yoshimura. 2007. Loss of adherence ability to human gingival epithelial cells in S-layer protein-deficient mutants of Tannerella forsythensis. Microbiology 153:866-876. [DOI] [PubMed] [Google Scholar]
- 31.Severi, E., D. W. Hood, and G. H. Thomas. 2007. Sialic acid utilization by bacterial pathogens. Microbiology 153:2817-2822. [DOI] [PubMed] [Google Scholar]
- 32.Severi, E., G. Randle, P. Kivlin, K. Whitfield, R. Young, R. Moxon, D. Kelly, D. Hood, and G. H. Thomas. 2005. Sialic acid transport in Haemophilus influenzae is essential for lipopolysaccharide sialylation and serum resistance and is dependent on a novel tripartite ATP-independent periplasmic transporter. Mol. Microbiol. 58:1173-1185. [DOI] [PubMed] [Google Scholar]
- 33.Sharma, A. 2010. Virulence mechanisms of Tannerella forsythia. Periodontol. 2000 54:106-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharma, A., H. T. Sojar, I. Glurich, K. Honma, H. K. Kuramitsu, and R. J. Genco. 1998. Cloning, expression, and sequencing of a cell surface antigen containing a leucine-rich repeat motif from Bacteroides forsythus ATCC 43037. Infect. Immun. 66:5703-5710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smith, D. B., and L. M. Corcoran. 1994. Expression and purification of glutathione-S-transferase fusion proteins. Curr. Protoc. Mol. Biol. 28:16.7.1-16.7.7. [DOI] [PubMed] [Google Scholar]
- 36.Soong, G., A. Muir, M. I. Gomez, J. Waks, B. Reddy, P. Planet, P. K. Singh, Y. Kaneko, M. C. Wolfgang, Y. S. Hsiao, L. Tong, and A. Prince. 2006. Bacterial neuraminidase facilitates mucosal infection by participating in biofilm production. J. Clin. Invest. 116:2297-2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tanner, A. C., and J. Izard. 2006. Tannerella forsythia, a periodontal pathogen entering the genomic era. Periodontol. 2000 42:88-113. [DOI] [PubMed] [Google Scholar]
- 38.Thompson, H., K. A. Homer, S. Rao, V. Booth, and A. H. Hosie. 2009. An orthologue of Bacteroides fragilis NanH is the principal sialidase in Tannerella forsythia. J. Bacteriol. 191:3623-3628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tong, H. H., L. E. Blue, M. A. James, and T. F. DeMaria. 2000. Evaluation of the virulence of a Streptococcus pneumoniae neuraminidase-deficient mutant in nasopharyngeal colonization and development of otitis media in the chinchilla model. Infect. Immun. 68:921-924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Uchiyama, S., A. F. Carlin, A. Khosravi, S. Weiman, A. Banerjee, D. Quach, G. Hightower, T. J. Mitchell, K. S. Doran, and V. Nizet. 2009. The surface-anchored NanA protein promotes pneumococcal brain endothelial cell invasion. J. Exp. Med. 206:1845-1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Varki, A. 1997. Sialic acids as ligands in recognition phenomena. FASEB J. 11:248-255. [DOI] [PubMed] [Google Scholar]
- 42.Vimr, E. R., K. A. Kalivoda, E. L. Deszo, and S. M. Steenbergen. 2004. Diversity of microbial sialic acid metabolism. Microbiol. Mol. Biol. Rev. 68:132-153. [DOI] [PMC free article] [PubMed] [Google Scholar]