Abstract
Mycobacterium bovis BCG strains are live, attenuated vaccines generated through decades of in vitro passage. Because in vitro growth does not select for interaction with the host, it has been hypothesized that genetic loci lost from BCG code for virulence determinants that are dispensable for growth in the laboratory, as exemplified by Region of Difference 1 (RD1), which was lost during the original derivation of BCG between 1908 and 1921. Region of Difference 2 (RD2) was lost during the ongoing propagation of BCG between 1927 and 1931, a time that coincides with reports of the ongoing attenuation of the vaccine. In this study, RD2 has been disrupted in M. tuberculosis H37Rv to test whether its loss contributed to the further attenuation of BCG. The deletion of RD2 did not affect in vitro growth; in contrast, the mutant manifested a decrease in pulmonary and splenic bacterial burdens and reduced pathology in C57BL/6 mice at early time points. This attenuated phenotype was complemented by reintroducing the genes Rv1979c to Rv1982 (including mpt64) but not Rv1985c to Rv1986. In RAW 264.7 macrophages, H37Rv:ΔRD2 showed a decreased proliferation and impaired modulation of the host innate immune response; both observations were complemented with Rv1979c to Rv1982. To test the effect of RD2 disruption on innate immunity, Rag−/− mice were infected; H37Rv:ΔRD2 had increased survival times compared those of H37Rv. These findings support the notion that the safety profile of certain BCG vaccines stems from multiple attenuating mutations, with the RD2 deletion resulting in a less-virulent organism through the impaired bacterial manipulation of the host innate immune response.
Tuberculosis (TB) has been, and continues to be, one of the most widespread bacterial infections worldwide (www.who.int/tb/publications/global_report/2009/). On an individual level, the management of TB cases can be achieved in settings where prompt microbiologic diagnosis and appropriate treatment are provided. However, on a global public health level, this approach has had limited success, indicating a need to consider alternative strategies for TB control, including immunization.
For nearly a century, Mycobacterium bovis BCG strains have been used as vaccines against TB. The efficacy observed in a number of different BCG trials has provided proof-of-principle data that TB should, in theory, be a disease that is preventable through vaccination. However, variable estimates of efficacy have diminished the enthusiasm for BCG immunization. While the effectiveness of BCG vaccines has been the subject of controversy, their safety profile generally has not been disputed; about 1 in 100,000 vaccinees suffers disseminated BCG disease, and those with BCG-osis typically have severe immunologic defects, either genetic (e.g., SCID) or acquired (e.g., AIDS) (4, 5, 37). Thus, BCG vaccines provide an example of a profound attenuation, enabling researchers to uncover virulence determinants that are present in M. tuberculosis and virulent M. bovis but absent from BCG strains.
The study of TB pathogenesis through BCG vaccines is supported by theoretical considerations and experimental observations. In theory, an intracellular host-adapted bacterium like M. tuberculosis is equipped with virulence determinants that are critical for interaction with the eukaryotic cell in which it resides. By propagating the bacteria in the laboratory without host selection for more than five decades, as was the case with BCG Pasteur, mutants are expected to emerge based on enhanced fitness in vitro, with no pressure to maintain bacterial factors required during infection. Previous work has confirmed this prediction through the study of Region of Difference 1 (RD1), a 9.8-kb genomic deletion that occurred during the original derivation of BCG between 1908 and 1921 (19). This deletion was shown to contribute to the attenuation of BCG by both gene disruption (18) and complementation experiments (24). Interestingly, the complementation of BCG Pasteur with RD1 did not fully restore pathogenicity in a murine model, thus it was hypothesized that other genes are involved in virulence (25). BCG Pasteur not only lacks RD1 but also has incurred other mutations, including Region of Difference 2 (RD2), which was lost between the years 1927 and 1931, a time when vaccinologists reported the further attenuation of the BCG vaccine (14, 42). To evaluate the role of RD2-associated virulence, we engineered a targeted knockout within the M. tuberculosis reference strain H37Rv and conducted a series of in vitro and in vivo investigations. Here, we demonstrate that the loss of RD2 leads to a reduction in virulence, providing evidence that the safety profile of BCG Pasteur stems from the cumulative effect of multiple attenuating mutations.
MATERIALS AND METHODS
Bacterial strains.
Mycobacterium tuberculosis strains H37Rv, H37Rv:ΔRD2, H37Rv:ΔRD2::pMV306, H37Rv:ΔRD2::p7982, H37Rv:ΔRD2::p8586, H37Rv:ΔRD1, BCG Pasteur, and BCG Russia were cultured in Middlebrook 7H9 liquid medium (Becton Dickinson) containing 10% ADC (albumin dextrose complex; Becton Dickinson), 0.5% glycerol, and 0.08% Tween 80 (Sigma-Aldrich) or Middlebrook 7H10 agar containing 10% OADC (oleic acid plus ADC; Becton Dickinson), 0.5% glycerol, and PANTA (polymyxin B, amphotericin B, nalidixic acid, trimethoprim, and azlocillin; Becton Dickinson). Hygromycin or kanamycin (Sigma-Aldrich) was added to the medium when needed.
Mutant construction and complementation.
The M. tuberculosis H37Rv:ΔRD2 and H37Rv:ΔRD1 strains were prepared using homologous recombination and sucrose counterselection as previously described (18). Specifically, the RD2 deletion in BCG Pasteur is separated by only 335 bp from a region present in M. tuberculosis but deleted from virulent M. bovis, known as RD6 (11). Therefore, to delete the RD2 region from M. tuberculosis, we amplified DNA flanking the region (Rv1977 to Rv1978 and Rv1988 to Rv1989) into the suicide vector pKO for introduction into H37Rv, selection for hygromycin-resistant mutants, and then counterselection on sucrose to screen out clones harboring the intact sacB gene. Details on construct design and the primers utilized are provided in Table 1.
TABLE 1.
Oligonucleotide primers used in this study
| Primer | Sequence |
|---|---|
| SigAF | TGCAGTCGGTGCTGGACA |
| SigAR | CGCGCAGGACCTGTGAGC |
| Rv1985cF | GAAATGCGCTACCTACCAGTG |
| Rv1985cR | GTGAACCCGTCGGATAGATG |
| Rv1986F | ACCGTCGTGTTGCTAGGC |
| Rv1986R | ATACCGCACTGGCTGTGAC |
| 7982F | GTAAGCTTCACTACGTGCCTACAGTCCC |
| 7982R | CATTCTAGAGCTGACCGATTGATATTC |
| KanF | AACAAGCCAGGGATGTAACG |
| KanR | CCATAAAACCGCCCAGTCTA |
| rpl 21 | GGCTATGCCAAGGTGATAGC |
| rpl 22 | CCGATGATCTTCTGTTTGAC |
| rpl 23 | GGTGGTCGCCGTGGAGTTGC |
| rpl 24 | AGTTCGGCGAGACGCAGCAG |
To assess complementation with selected portions of the RD2 region, the integrating plasmid pMV306-kan was used (a gift of P. Domenech) (36). Inserts were amplified with AccuPrime Taq DNA polymerase high fidelity (Invitrogen), and PCR products were confirmed by sequencing at the McGill University and Génome Québec Innovation Centre, Montreal, Canada. Primers were designed to span Rv1979c to Rv1982 (the plasmid was designated p7982) and Rv1985c to Rv1986 (the plasmid was designated p8586) from H37Rv genomic DNA (Fig. 1), with the empty vector used as a complementation control, as has been described previously (6). Primers used in this study are listed in Table 1. Plasmids were electroporated into M. tuberculosis H37Rv:ΔRD2, kanamycin-resistant clones were selected, and the presence of the plasmid was assessed by PCR as has been previously described (39).
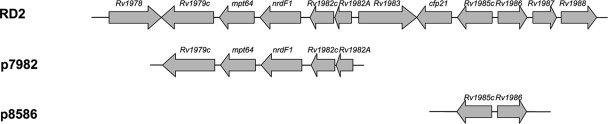
FIG. 1.
RD2 region and constructs for complementation. RD2 region encompasses genes Rv1978 to Rv1988. Amplicons of Rv1979c to Rv1982 and Rv1985c to Rv1986 were cloned into the integrative plasmid pMV306 for complementation and were designated p7982 and p8586, respectively.
Verification of complementation.
For H37Rv:ΔRD2::p7982, we verified the expression of the genes by testing the production and secretion of MPT64, coded for by Rv1980 (www.molepi.mcgill.ca). The immunoblotting of the culture filtrate was performed as described by Charlet et al. (6). The procedure was carried out with the following modification: after bacteria were grown in containment in Middlebrook 7H9 medium, the supernatant was separated by centrifugation and filter sterilized using a 0.22-μm filter (Millipore) prior to separation under reducing conditions on a 12% SDS-PAGE gel. Gel transfer and blotting then were performed by following established protocols.
For H37Rv:ΔRD2::p8586, we determined the expression of both Rv1985c and Rv1986 in the wild-type and complemented strains using quantitative reverse transcription-PCR (qRT-PCR) by following procedures described elsewhere (39). Briefly, RNA was extracted from cultures in log-phase growth (optical density at 600 nm [OD600] of 0.4 to 0.6) using a modified phenol-chloroform extraction method, which has been described previously (6), and levels of Rv1985c and Rv1986 were normalized to the amount of sigA RNA (www.molepi.mcgill.ca). Primers used to in the qRT-PCR experiments are listed in Table 1.
In vitro studies of mutants.
To investigate the presence or absence of phthiocerol dimycocerosate (PDIM) of the H37Rv parental strain and the RD2 deletion mutant, bacteria were grown in Middlebrook 7H9 medium and radiolabeled with [1-14C]propionic acid or [1-14C]acetic acid in the manner described by Reed and colleagues (28). Once bacteria had reached log-phase growth, whole lipids were extracted and separated by thin-layer chromatography (TLC) as previously described (28). To examine if the deletion of RD2 led to the dysregulation of other genes outside this region, bacterial RNA from culture at log-phase growth was analyzed by microarray. RNA was isolated and analyzed using established laboratory protocols (21).
Macrophage preparation.
Macrophages were prepared in a manner similar to that described by Divangahi et al. (9). RAW 264.7 cells were pelleted and resuspended in RPMI 1640 medium plus 10% (vol/vol) fetal bovine serum (Wisent), HEPES (Wisent), 100 U/ml penicillin, and 100 μg/ml streptomycin (Wisent). Macrophages were added into 6- or 12-well culture plates and allowed to adhere overnight at 37°C with 5% CO2.
Macrophage infection.
Bacteria were grown to mid-log phase before being pelleted and resuspended in RPMI 1640 medium plus 10% fetal bovine serum and HEPES. Macrophages (1 × 106 cells per well) were infected with different bacterial strains at a multiplicity of infection (MOI) of 5 for 4 h. Following this, cells were washed three times with warm RPMI, and fresh medium was added. Cells were incubated at 37°C with 5% CO2. At desired time points, triplicate wells for each bacterial strain were examined. Supernatant was removed from the wells and added to a conical tube containing PBS plus 1% Triton X-100 (Sigma) to lyse dead cells containing mycobacteria. Following this, the remaining macrophages were lysed with three washes of PBS plus 1% Triton X-100 and then pelleted and resuspended in Middlebrook 7H9 medium. Bacterial numbers were determined through plating on Middlebrook 7H10 with PANTA, and colony counts were determined after 3 weeks.
Cytokines.
RAW 264.7 macrophages (2 × 105 cells per well) were infected with the strains at multiplicities of infection of 1 and 5 (1:1 and 5:1, respectively) as previously described (9). The macrophages were incubated in the presence of the bacteria for 24 h at 37°C in 5% CO2. After 24 h, the culture supernatant from three wells was pooled and collected and then centrifuged in 0.22-μm durapore tubes (Millipore) to ensure the removal of bacteria and cells, and then they were assayed using enzyme-linked immunosorbent assays (ELISA; R&D Systems) for tumor necrosis factor alpha (TNF-α). For each strain, OD readings were taken from three wells and averaged.
Apoptosis.
In vitro apoptosis was measured using enzyme-linked immunosorbent assays (Cell Death Detection ELISAPlus; Roche Applied Science). Cytoplasmic histone-associated DNA fragments were quantified by following the instructions provided by the manufacturer. Relative apoptosis was calculated as a ratio of the absorbance of infected macrophages to the absorbance of uninfected macrophages.
Animal studies.
Mice were aerosolized with ∼150 CFU of the various bacterial strains mentioned above in accordance with methods previously described (28). Prior to infection, bacterial cultures were adjusted to mid-log-phase growth and stored in 20% glycerol at −80°C. For aerosol infection these stocks were diluted 1:100 in PBS-Tween 20 (0.05%). Eight-week-old C57BL/6 mice (Harlan Laboratories) were aerosolized for a total of 10 min using a Lovelace nebulizer, model 01-100 (In Tox Products). This method resulted in the implantation of approximately 150 to 200 CFU per lung, confirmed at 24 h postinfection by homogenizing lungs from three mice per group in 7H9-ADC and determining bacterial counts. Mice were sacrificed at 1, 2, 3, and 6 weeks after infection, at which point lungs and spleens were extracted, homogenized, serially diluted, and plated to evaluate CFU present in each organ. Mice aerosol infected with complemented strains were sacrificed at 3 weeks. For the time-to-death study, 7- to 8-week-old Rag1tm1Mom Tg(TIE2GFP)287Sato/J mice (Jackson Laboratory) were infected via aerosolization, and bacterial burdens at 24 h postinfection were determined as described above. Mice were weighed weekly and sacrificed if a veterinarian blinded to their infecting strain judged that they manifested the following symptoms: lethargy, trembling, or weight loss of more than 15% of their body weight. Lungs and spleens were harvested prior to fixation in 10% neutral-buffered formalin for histopathological analysis.
Histomorphology.
Immediately following collection, lung and spleen tissue was preserved in 10% neutral-buffered formalin and processed (RIC, Plateforme d'Histologie, Universite de Montreal). To evaluate tissue morphology, sections were cut 4 to 5 μm thick from paraffin-embedded sections and underwent routine hematoxylin and eosin (H&E) staining. Representative slides were evaluated for pathology and granuloma size by a pathologist and a second reader blinded to the bacterial strain and were rated according to the following scale: 0, no pathology; 1, minimal pathology; 2, mild pathology; 3, moderate pathology; 4, severe pathology.
Figures and statistics.
Differences in Gaussian data collected were tested for significance using the student's t test, with P ≤ 0.05 being considered statistically significant. Differences in semiquantitative data (histopathology) were tested for significance using the rank-sum test. Survival data were assessed using the log-rank test. Figures were generated using GraphPad Prism 4.0b.
RESULTS
In vitro assessment.
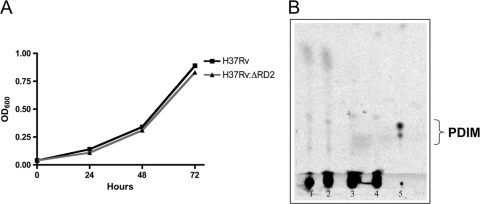
In vitro growth kinetics were similar for H37Rv and H37Rv:ΔRD2 (Fig. 2 A), indicating that the loss of RD2 did not lead to an evident growth defect in the organism, which is consistent with the original RD2 deletion having occurred in vitro. Complemented strains also showed similar growth patterns (data not shown). TLC analysis indicated that the H37Rv parent strain and the RD2 deletion mutant both were lacking PDIM (Fig. 2B). As both H37Rv and the RD2 deletion mutant were negative for PDIM, we concluded that any subsequent differences in virulence were not associated with the loss of this lipid. Microarray analysis to look for the potential effect of RD2 disruption on the expression of genes outside RD2 did not uncover any genes reproducibly dysregulated outside RD2 (data not shown).
FIG. 2.
In vitro characterization of the deletion mutant. (A) In vitro growth kinetics of the H37Rv and H37Rv:ΔRD2 as determined by OD600 readings. The experiment was repeated in triplicate, and data are from one representative experiment. (B) Thin-layer chromatography plate of apolar lipids from H37Rv and H37Rv:ΔRD2. Lanes 1 and 3 represent lipids from the H37Rv strains used in this experiment, labeled with [1-14C]proprionic acid and [1-14C]acetic acid, respectively. Lanes 2 and 4 represent lipids from H37RvΔRD2, labeled with [1-14C]proprionic acid and [1-14C]acetic acid, respectively. Lane 5 is the apolar lipids labeled with [1-14C]proprionic acid from a strain of H37Rv known to produce PDIM.
Analysis of bacteria burden and histopathology in murine infections.
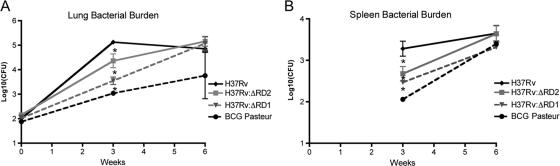
In a first experiment, we tested whether RD2 disruption had a measurable effect on in vivo virulence in the murine model. The murine aerosol infection model allows for the evaluation of lung bacterial burden, an indicator of TB pathogenesis, and splenic burden, an indicator of bacterial dissemination from the site of infection (18). After the aerosol infection of C57BL/6 mice with H37Rv and H37Rv:ΔRD2, we observed steady bacterial growth in the H37Rv-infected mice for the first 3 weeks followed by a plateau, which is consistent with the findings reported by other laboratories (18). In comparison, H37Rv:ΔRD2 showed a 0.75-log reduction in bacterial levels in the lungs and a 0.6-log reduction in the spleen throughout weeks 2 (not shown) and 3 (Fig. 3 A and B). The greatest difference in bacterial burden was observed at 3 weeks postinfection (Fig. 3A and B). For comparison, these data were evaluated against aerosol infections with the attenuated strains H37Rv:ΔRD1 and BCG Pasteur (18). While H37Rv:ΔRD2 showed reduced bacterial burden at 3 weeks postinfection, it should be noted that the decrease was not as severe as those of H37Rv:ΔRD1 and BCG Pasteur. Notably, by week 6, H37Rv:ΔRD2 had achieved a bacterial burden comparable to that of H37Rv, as has been observed previously in the case of H37Rv:ΔRD1 (18).
FIG. 3.
In vivo assessment of virulence. Bacterial growth in the lungs (A) and spleens (B) of C57BL/6 mice. Six- to 8-week-old mice were used, and they were infected via aerosolization with about 150 CFU of H37Rv (solid black line), H37Rv:ΔRD2 (solid gray line), H37Rv:ΔRD1 (dashed gray line), or BCG Pasteur (dashed black line). Data are the means ± standard errors of the means of 10 mice per strain from two separate infections for H37Rv and H37Rv:ΔRD2 and 4 mice per strain from one infection for H37Rv:ΔRD1 and BCG Pasteur. *, P < 0.05.
Complementation experiments.
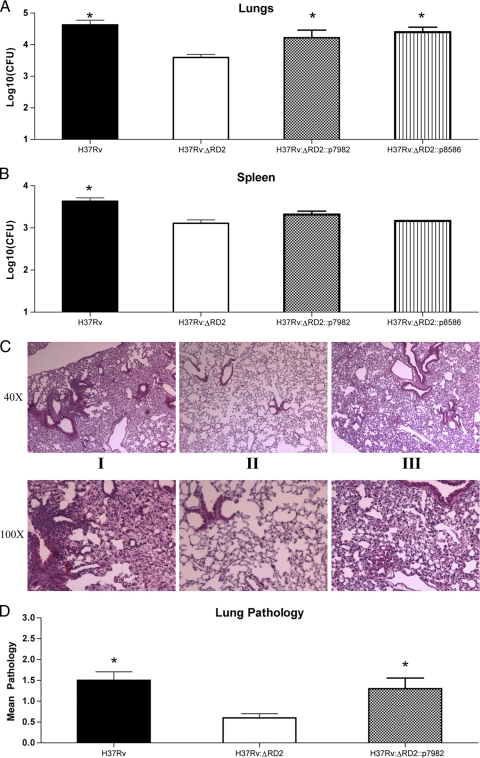
To complement H37Rv:ΔRD2, we considered the 11 genes within this region as candidates and selected those most likely to have a role in virulence based largely on considerations from published literature (1, 13, 33, 40). We excluded Rv1988 from consideration, as this gene has been shown to play a role in resistance to macrolides (23). Tsolaki and colleagues have demonstrated that Rv1983 to Rv1984c were deleted from a clinical strain of M. tuberculosis, demonstrating that these genes are dispensable for full virulence to humans (38). While Rv1978 and Rv1987 were considered candidates, we decided not to pursue the study of these genes, as their predicted functions (hypothetical unknown and hypothetical chitinase) did not suggest an obvious role in virulence, and because their topology in the genome suggested the need for the complementation of individual genes. From this, we were left with two candidate intervals: Rv1979c to Rv1982, which includes the gene coding for the antigen MPT64, and Rv1985c to Rv1986, a putative LysR-like transcriptional regulator and adjacent LysE-like transporter. Since the most-marked difference in bacterial burden was observed 3 weeks postinfection, we selected this time point to test our complemented strains. As shown in Fig. 4 A and B, lung bacterial burden was partially restored in mice infected with either H37Rv:ΔRD2::p7982 or H37Rv:ΔRD2::p8586; however, the partial restoration of splenic bacterial numbers was seen only with H37Rv:ΔRD2::p7982. As we also had noted a difference in histopathology between H37Rv and H37Rv:ΔRD2, we examined the effect these differently complemented strains had on our pathology phenotype. As shown in Fig. 4C, mice infected with H37Rv:ΔRD2::p7982 exhibited a pathological profile that was indistinguishable from that seen in H37Rv-infected mice. This phenotype was observed by both qualitative and quantitative methods, as H37Rv:ΔRD2 had lower pathological scores than H37Rv or H37Rv:ΔRD2::p7982 (Fig. 4D). Conversely, mice infected with H37Rv:ΔRD2::p8586 showed reduced pathology, as seen with H37Rv:ΔRD2 (data not shown). These findings, combined with the observation of splenic CFU, suggested that Rv1985 and Rv1986c do not contribute to full virulence early after infection, perhaps because these genes have a role in the more-chronic phase of infection, as has been suggested by the study of the enduring hypoxic response (32). This strain subsequently was set aside for future investigations. As our results indicated that complementation with Rv1979c to Rv1982 restored most of the attenuation phenotype, subsequent experiments were carried out using this strain alone.
FIG. 4.
Complementation of virulence phenotype. Bacterial burden in the lungs (A) and spleen (B) at 3 weeks postinfection. Six- to 8-week-old C57BL/6 mice were infected by aerosol with H37Rv (black bar), H37Rv:ΔRD2 (white bar), H37Rv:ΔRD2::p7982 (gray bar), or H37Rv:ΔRD2::p8586 (striped bar). Data represent the means ± standard errors of the means of six mice, where an asterisk indicates P < 0.05. (C) Histology of lung lesions from infected mice with H37Rv (i), H37Rv:ΔRD2 (ii), or H37Rv:ΔRD2::p7982 (iii) at 3 weeks postinfection. Five mice per group were used and lungs were stained with H&E. Micrographs (magnification, ×40 and ×100) shown are representative lung sections from each group. (D) Average pathological scores from mice infected with H37Rv (black bar), H37Rv:ΔRD2 (gray bar), or H37Rv:ΔRD2::p7982 (gray bar) at 3 weeks postinfection. Pathology was evaluated from H&E-stained lung sections by two blinded readers and scored on a scale of 0 (absent) to 4 (very severe). Bars represent means ± standard errors of the means, where an asterisk indicates a P value of ≤0.05.
Bacterial growth in macrophages.
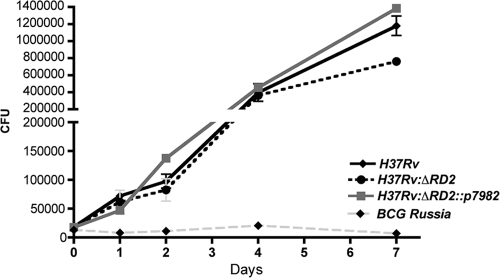
As the phenotype was seen early after infection, when adaptive immune responses have had little opportunity to modulate the course of infection, we tested whether the presence of RD2 affects interaction between M. tuberculosis and its host cell. To investigate this, we infected RAW 264.7 cells at a multiplicity of infection (MOI) of 5:1 and evaluated bacterial growth during a 7-day time period. As a control for severe attenuation, we infected macrophages in parallel with BCG Russia based on published observations of this strain in macrophages (18). Compared to the growth of BCG Russia, a strain of BCG in which the RD2 region is intact, each strain of M. tuberculosis grew better, with no distinguishable differences during the first days of infection. However, by day 7, there was a modest relative decrease in H37Rv:ΔRD2 compared to levels of H37Rv, whereas H37Rv:ΔRD2::p7982 bacterial numbers were similar to those of H37Rv (Fig. 5).
FIG. 5.
Characterization of bacterial mutants during macrophage infection. Bacterial growth in murine macrophages during a 7-day period. RAW 264.7 macrophages were infected with H37Rv (black line), H37Rv:ΔRD2 (dashed black line), H37Rv:ΔRD2::p7982 (solid gray line), or BCG Russia (dashed gray line). Data from each time are taken from the means of wells infected in triplicate, and the experiment was performed twice. Error bars represent ± standard errors of the means, where an asterisk indicates P < 0.05.
Cytokine expression in RAW macrophages.
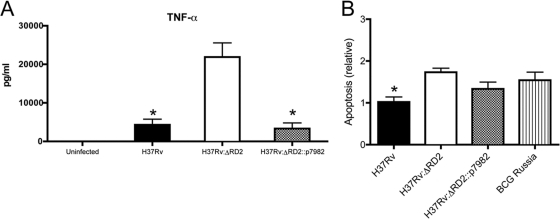
Our data suggested that RD2 modulates the innate immune response to M. tuberculosis. The early control of mycobacterial infection is achieved through the innate immune response, whereby reduced or absent levels of TNF-α or other cytokines have been associated with increased mycobacterial virulence (10, 15, 31, 35). To evaluate innate immunity, cytokine production in a mouse macrophage cell line, RAW 264.7, was performed. These macrophages were infected at an MOI of 1, and cell supernatant was harvested after 24 h. H37Rv produced significantly lower levels of TNF-α compared to that of H37Rv:ΔRD2. However, in macrophages infected with H37Rv:ΔRD2::p7982, TNF-α levels were reduced to a level comparable to that of the wild-type strain H37Rv (Fig. 6 A). These results demonstrate that the loss of RD2 impairs the ability of the bacterium to modulate host cytokines that are important for innate defense against mycobacteria.
FIG. 6.
Effect of RD2 deletion on interaction with macrophages. (A) RAW 264.7 macrophages were infected with H37Rv, H37Rv:ΔRD2, or H37Rv:ΔRD2::p7982 at an MOI of 1, and supernatant was harvested 24 h postinfection for TNF-α determination by ELISA. Data represent the means ± standard errors of the means. (B) Relative apoptosis levels in RAW 264.7 macrophages at 96 h postinfection. Macrophages were infected at an MOI of 5 for 4 h with H37Rv (black bar), H37Rv:ΔRD2::p7982 (gray bar), H37Rv:ΔRD2 (white bar), or BCG Russia (striped bar). Wells were infected in triplicate and apoptosis was evaluated by ELISA. Error bars represent means ± standard errors of the means, where an asterisk indicates P ≤ 0.05.
Apoptosis.
Previously, Mustafa and colleagues described an inverse association between MPT64 expression and apoptosis in biopsy samples from mycobacterial lymphadenitis (22). Our finding that H37Rv:ΔRD2 manifests decreased survival in macrophages combined with the elevated levels of TNF-α led us to suspect that RD2 decreases host cell apoptosis, in which case the presence of this element might be associated with a deviation away from an apoptotic host cell toward necrosis (8). To investigate differences in apoptosis, we measured cytoplasmic histone-associated DNA fragments by ELISA. As a control for apoptosis, BCG Russia was used to infect macrophages, as it is known to induce higher levels of apoptosis than H37Rv (29). At 48 h postinfection, levels of apoptosis were comparable between cells infected with the different strains (data not shown). However, at 4 days postinfection, the levels of relative apoptosis were higher in cells infected with H37Rv:ΔRD2 (Fig. 6B). Cells infected with H37Rv:ΔRD2::p7982 showed levels of apoptosis that were lower than those for H37Rv:ΔRD2; however, this finding failed to achieve statistical significance. The lower bacterial burden seen at the subsequent time point likely was due to higher levels of apoptosis, and the loss of RD2 reduced the ability of the bacteria to prevent programmed cell death.
Survival.
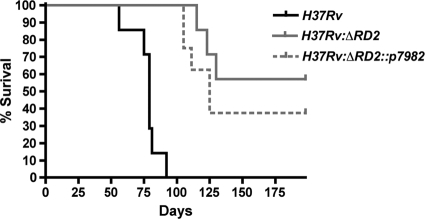
As another experiment to test whether the deletion of RD2 affects the innate immune response to M. tuberculosis, we conducted the aerosol infection of H37Rv, H37Rv:ΔRD2, and H37Rv:ΔRD2::p7982 into Rag1tm1Mom Tg(TIE2GFP)287Sato/J mice that lack an adaptive immune response. As shown in Fig. 7, infection with H37Rv resulted in a median survival of 79 days, and no H37Rv-infected animals survived to the termination of this experiment after 28 weeks. In contrast, only 37.5% of mice infected with H37Rv:ΔRD2 had succumbed to infection by week 28, and therefore no median survival time could be assigned. The H37Rv:ΔRD2::p7982 strain presented an intermediate phenotype, with a median survival time of 125 days.
FIG. 7.
Survival experiment. Survival in C57BL/6 mice infected with different M. tuberculosis strains. Mice were aerosol infected with H37Rv (black line), H37Rv:ΔRD2 (gray line), or H37Rv:ΔRD2::p7982 (dashed line). Approximately 200 CFU were implanted per lung. H37Rv had a median survival of 79 days, H37Rv:ΔRD2::p7982 had a median survival of 125 days, and H37Rv:ΔRD2 failed to achieve a median survival time point. Eight mice per group were used. This is a representative figure of two independent experiments.
DISCUSSION
The genomic differences between BCG strains are well established (3, 17). This has led to the hypothesis that the ongoing evolution of BCG has resulted in the loss of regions involved in virulence and has caused the vaccine to become overattenuated (21). Despite the plethora of genomic data, the phenotypic consequence of mutations in BCG strains has not been convincingly demonstrated beyond the original loss of the RD1 region (18, 24, 34). Intriguingly, Raghavan and colleagues demonstrated that the transcription of genes within the RD2 region is influenced by the RD1-associated secreted virulence factor EspR, presenting a conceptual link between the deletion of these two elements from BCG (26). Our data demonstrate that RD2 plays a role in mycobacterial virulence, and that its deletion from M. tuberculosis leads to a decrease in bacterial growth in both a macrophage and a murine model. Furthermore, the infection of macrophages resulted in increased levels of TNF-α and increased apoptosis with RD2 disruption, suggesting that a portion of RD2 containing the gene coding for MPT64 has an antiapoptotic effect on the host cell.
These findings are consistent with, and expand upon, previous investigations that have explored the role of certain portions of the RD2 region. Mustafa and colleagues demonstrate that the antigen MPT64 (encoded by RD2) may decrease TNF-α production in the cells within a granuloma (22). This antigen is secreted only by actively dividing cells and has been shown to bind plasminogen (40, 41). The treatment of monocytes with TNF-α, followed by plasminogen, was found to decrease the number of cells undergoing apoptosis (20). These observations make it tempting to speculate that MPT64, perhaps in concert with other RD2 proteins, interacts with the host cell to modulate apoptosis through plasminogen binding. Furthermore, if MPT64 is involved in suppressing TNF-α production within the granuloma, and given that TNF-α is crucial for granuloma formation (16, 30), this would fit well with our observation of decreased pathology and reduced bacterial burden in mice infected with H37Rv:ΔRD2.
The attenuation seen in the RD2 knockout strains could extend beyond the subversion of host responses to also affect survival and growth within the macrophage. Rv1982c is predicted to encode a toxin-antitoxin of the VapBC family; this family of proteins has been shown to inhibit translation through RNase activity (27). It has been proposed that toxin-antitoxin systems function in M. tuberculosis to slow growth during unfavorable conditions (2). Thus, it is tempting to speculate that the loss of Rv1982c prevents H37Rv:ΔRD2 from arresting growth within the phagosome that might otherwise promote its survival.
Since tuberculosis is a disease of multiple stages (i.e., initial infection, early logarithmic bacterial growth, the establishment of persistent infection, the eventual induction of active disease, and transmission to a new host), it is likely that different genes of the organism have their greatest importance at different phases of the disease. Therefore, it follows that complete virulence requires a collective effect of many genes, and that the attenuation seen when 11 genes are deleted is unlikely to be reversed fully with the introduction of a few. Consistent with this idea, neither of our complemented strains fully restored all aspects of virulence. It has been shown previously that Rv1985c is upregulated as part of the response to the hypoxic conditions meant to mimic latency (32). It is possible that these genes are involved in virulence at a later time point or in some manner are not revealed by our experiments. Additionally, other genes within the region also may be involved in virulence and warrant further study. For example, the second antigen within this region, CFP21, has been shown to be strongly immunogenic in mice and may well play a role in infection (12).
Finally, Rv1979c is predicted to be an amino acid transporter, although the substrate is unknown (http://genolist.pasteur.fr/TubercuList/). Chen et al. demonstrated that the Rv1979c-containing BCG Japan was able to grow in media where arginine, histidine, lysine, or proline was the sole nitrogen source, while BCG Pasteur was unable to grow under the same conditions (7). Rv1979c therefore may be a transporter for one or more of these amino acids, and the loss of this gene may restrict growth in the macrophage.
Our results suggest that multiple genes within the RD2 region contribute to virulence and are consistent with the view that multiple genomic deletions contributed to the safety profile associated with contemporary BCG vaccines. Ongoing work is assessing whether the addition of these genes into a natural RD2 mutant, BCG Pasteur, increases in the immunogenicity and protective efficacy of the vaccine.
Acknowledgments
We thank the members of the Behr, Reed, and Schurr laboratories for their input. We thank Pilar Domenech and Vlad Kamenski for their invaluable assistance during animal experiments.
R.K. was supported through an MUHC studentship. M.B. is a Chercheur National of the FRSQ and a William Dawson Scholar of McGill University. Work was funded by an operating grant from the Canadian Institutes for Health Research (MOP-79309).
Editor: J. L. Flynn
Footnotes
Published ahead of print on 25 October 2010.
REFERENCES
- 1.Andersen, P., D. Askgaard, L. Ljungqvist, M. W. Bentzon, and I. Heron. 1991. T-cell proliferative response to antigens secreted by Mycobacterium tuberculosis. Infect. Immun. 59:1558-1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arcus, V. L., P. B. Rainey, and S. J. Turner. 2005. The PIN-domain toxin-antitoxin array in mycobacteria. Trends Microbiol. 13:360-365. [DOI] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Casanova, J. L., S. Blanche, J. F. Emile, E. Jouanguy, S. Lamhamedi, F. Altare, J. L. Stephan, F. Bernaudin, P. Bordigoni, D. Turck, A. Lachaux, M. Albertini, A. Bourrillon, J. P. Dommergues, M. A. Pocidalo, F. Le Deist, J. L. Gaillard, C. Griscelli, and A. Fischer. 1996. Idiopathic disseminated bacillus Calmette-Guerin infection: a French national retrospective study. Pediatrics 98:774-778. [PubMed] [Google Scholar]
- 5.Casanova, J. L., E. Jouanguy, S. Lamhamedi, S. Blanche, and A. Fischer. 1995. Immunological conditions of children with BCG disseminated infection. Lancet 346:581. [DOI] [PubMed] [Google Scholar]
- 6.Charlet, D., S. Mostowy, D. Alexander, L. Sit, H. G. Wiker, and M. A. Behr. 2005. Reduced expression of antigenic proteins MPB70 and MPB83 in Mycobacterium bovis BCG strains due to a start codon mutation in sigK. Mol. Microbiol. 56:1302-1313. [DOI] [PubMed] [Google Scholar]
- 7.Chen, J. M., D. C. Alexander, M. A. Behr, and J. Liu. 2003. Mycobacterium bovis BCG vaccines exhibit defects in alanine and serine catabolism. Infect. Immun. 71:708-716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Divangahi, M., M. Chen, H. Gan, D. Desjardins, T. T. Hickman, D. M. Lee, S. Fortune, S. M. Behar, and H. G. Remold. 2009. Mycobacterium tuberculosis evades macrophage defenses by inhibiting plasma membrane repair. Nat. Immunol. 10:899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Divangahi, M., S. Mostowy, F. Coulombe, R. Kozak, L. Guillot, F. Veyrier, K. S. Kobayashi, R. A. Flavell, P. Gros, and M. A. Behr. 2008. NOD2-deficient mice have impaired resistance to Mycobacterium tuberculosis infection through defective innate and adaptive immunity. J. Immunol. 181:7157-7165. [DOI] [PubMed] [Google Scholar]
- 10.Freeman, S., F. A. Post, L. G. Bekker, R. Harbacheuski, L. M. Steyn, B. Ryffel, N. D. Connell, B. N. Kreiswirth, and G. Kaplan. 2006. Mycobacterium tuberculosis H37Ra and H37Rv differential growth and cytokine/chemokine induction in murine macrophages in vitro. J. Interferon Cytokine Res. 26:27-33. [DOI] [PubMed] [Google Scholar]
- 11.Gordon, S. V., B. Heym, J. Parkhill, B. Barrell, and S. T. Cole. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881-892. [DOI] [PubMed] [Google Scholar]
- 12.Grover, A., M. F. Ahmed, I. Verma, P. Sharma, and G. K. Khuller. 2006. Expression and purification of the Mycobacterium tuberculosis complex-restricted antigen CFP21 to study its immunoprophylactic potential in mouse model. Protein Expr. Purif 48:274-280. [DOI] [PubMed] [Google Scholar]
- 13.Harboe, M., S. Nagai, M. E. Patarroyo, M. L. Torres, C. Ramirez, and N. Cruz. 1986. Properties of proteins MPB64, MPB70, and MPB80 of Mycobacterium bovis BCG. Infect. Immun. 52:293-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jensen, K. A. 1946. Practice of the Calmette vaccination. Acta Tuberc. Scand. 20:1-45.
- 15.Keane, J., H. G. Remold, and H. Kornfeld. 2000. Virulent Mycobacterium tuberculosis strains evade apoptosis of infected alveolar macrophages. J. Immunol. 164:2016-2020. [DOI] [PubMed] [Google Scholar]
- 16.Kindler, V., A. P. Sappino, G. E. Grau, P. F. Piguet, and P. Vassalli. 1989. The inducing role of tumor necrosis factor in the development of bactericidal granulomas during BCG infection. Cell 56:731-740. [DOI] [PubMed] [Google Scholar]
- 17.Leung, A. S., V. Tran, Z. Wu, X. Yu, D. C. Alexander, G. F. Gao, B. Zhu, and J. Liu. 2008. Novel genome polymorphisms in BCG vaccine strains and impact on efficacy. BMC Genomics 9:413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis, K. N., R. Liao, K. M. Guinn, M. J. Hickey, S. Smith, M. A. Behr, and D. R. Sherman. 2003. Deletion of RD1 from Mycobacterium tuberculosis mimics bacille Calmette-Guerin attenuation. J. Infect. Dis. 187:117-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell, J. W., N. Baik, F. J. Castellino, and L. A. Miles. 2006. Plasminogen inhibits TNFalpha-induced apoptosis in monocytes. Blood 107:4383-4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mostowy, S., A. G. Tsolaki, P. M. Small, and M. A. Behr. 2003. The in vitro evolution of BCG vaccines. Vaccine 21:4270-4274. [DOI] [PubMed] [Google Scholar]
- 22.Mustafa, T., H. G. Wiker, O. Morkve, and L. Sviland. 2007. Reduced apoptosis and increased inflammatory cytokines in granulomas caused by tuberculous compared to non-tuberculous mycobacteria: role of MPT64 antigen in apoptosis and immune response. Clin. Exp. Immunol. 150:105-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nash, K. A. 2003. Intrinsic macrolide resistance in Mycobacterium smegmatis is conferred by a novel erm gene, erm(38). Antimicrob. Agents Chemother. 47:3053-3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pym, A. S., P. Brodin, R. Brosch, M. Huerre, and S. T. Cole. 2002. Loss of RD1 contributed to the attenuation of the live tuberculosis vaccines Mycobacterium bovis BCG and Mycobacterium microti. Mol. Microbiol. 46:709-717. [DOI] [PubMed] [Google Scholar]
- 25.Pym, A. S., P. Brodin, L. Majlessi, R. Brosch, C. Demangel, A. Williams, K. E. Griffiths, G. Marchal, C. Leclerc, and S. T. Cole. 2003. Recombinant BCG exporting ESAT-6 confers enhanced protection against tuberculosis. Nat. Med. 9:533-539. [DOI] [PubMed] [Google Scholar]
- 26.Raghavan, S., P. Manzanillo, K. Chan, C. Dovey, and J. S. Cox. 2008. Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454:717-721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramage, H. R., L. E. Connolly, and J. S. Cox. 2009. Comprehensive functional analysis of Mycobacterium tuberculosis toxin-antitoxin systems: implications for pathogenesis, stress responses, and evolution. PLoS Genet. 5:e1000767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed, M. B., P. Domenech, C. Manca, H. Su, A. K. Barczak, B. N. Kreiswirth, G. Kaplan, and C. E. Barry III. 2004. A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84-87. [DOI] [PubMed] [Google Scholar]
- 29.Riendeau, C. J., and H. Kornfeld. 2003. THP-1 cell apoptosis in response to Mycobacterial infection. Infect. Immun. 71:254-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roach, D. R., A. G. Bean, C. Demangel, M. P. France, H. Briscoe, and W. J. Britton. 2002. TNF regulates chemokine induction essential for cell recruitment, granuloma formation, and clearance of mycobacterial infection. J. Immunol. 168:4620-4627. [DOI] [PubMed] [Google Scholar]
- 31.Rojas, M., M. Olivier, P. Gros, L. F. Barrera, and L. F. Garcia. 1999. TNF-alpha and IL-10 modulate the induction of apoptosis by virulent Mycobacterium tuberculosis in murine macrophages. J. Immunol. 162:6122-6131. [PubMed] [Google Scholar]
- 32.Rustad, T. R., M. I. Harrell, R. Liao, and D. R. Sherman. 2008. The enduring hypoxic response of Mycobacterium tuberculosis. PLoS One 3:e1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherman, D. R., K. M. Guinn, M. J. Hickey, S. K. Mathur, K. L. Zakel, and S. Smith. 2004. Mycobacterium tuberculosis H37Rv: delta RD1 is more virulent than M. bovis bacille Calmette-Guerin in long-term murine infection. J. Infect. Dis. 190:123-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Spira, A., J. D. Carroll, G. Liu, Z. Aziz, V. Shah, H. Kornfeld, and J. Keane. 2003. Apoptosis genes in human alveolar macrophages infected with virulent or attenuated Mycobacterium tuberculosis: a pivotal role for tumor necrosis factor. Am. J. Respir. Cell Mol. Biol. 29:545-551. [DOI] [PubMed] [Google Scholar]
- 36.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 37.Talbot, E. A., M. D. Perkins, S. F. Silva, and R. Frothingham. 1997. Disseminated bacille Calmette-Guerin disease after vaccination: case report and review. Clin. Infect. Dis. 24:1139-1146. [DOI] [PubMed] [Google Scholar]
- 38.Tsolaki, A. G., A. E. Hirsh, K. DeRiemer, J. A. Enciso, M. Z. Wong, M. Hannan, Y. O. Goguet de la Salmoniere, K. Aman, M. Kato-Maeda, and P. M. Small. 2004. Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc. Natl. Acad. Sci. U. S. A. 101:4865-4870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Veyrier, F., B. Said-Salim, and M. A. Behr. 2008. Evolution of the mycobacterial SigK regulon. J. Bacteriol. 190:1891-1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang, Z., B. M. Potter, A. M. Gray, K. A. Sacksteder, B. V. Geisbrecht, and J. H. Laity. 2007. The solution structure of antigen MPT64 from Mycobacterium tuberculosis defines a new family of beta-grasp proteins. J. Mol. Biol. 366:375-381. [DOI] [PubMed] [Google Scholar]
- 41.Xolalpa, W., A. J. Vallecillo, M. Lara, G. Mendoza-Hernandez, M. Comini, R. Spallek, M. Singh, and C. Espitia. 2007. Identification of novel bacterial plasminogen-binding proteins in the human pathogen Mycobacterium tuberculosis. Proteomics 7:3332-3341. [DOI] [PubMed] [Google Scholar]
- 42.Zeyland, J., and E. Piasecka-Zeyland. 1936. Sur la vitalite du BCG dans l'organisme vaccine. Ann. Institut Pasteur 56:46-51. [Google Scholar]