Abstract
NadA is a trimeric autotransporter protein of Neisseria meningitidis belonging to the group of oligomeric coiled-coil adhesins. It is implicated in the colonization of the human upper respiratory tract by hypervirulent serogroup B N. meningitidis strains and is part of a multiantigen anti-serogroup B vaccine. Structure prediction indicates that NadA is made by a COOH-terminal membrane anchor (also necessary for autotranslocation to the bacterial surface), an intermediate elongated coiled-coil-rich stalk, and an NH2-terminal region involved in cell interaction. Electron microscopy analysis and structure prediction suggest that the apical region of NadA forms a compact and globular domain. Deletion studies proved that the NH2-terminal sequence (residues 24 to 87) is necessary for cell adhesion. In this study, to better define the NadA cell binding site, we exploited (i) a panel of NadA mutants lacking sequences along the coiled-coil stalk and (ii) several oligoclonal rabbit antibodies, and their relative Fab fragments, directed to linear epitopes distributed along the NadA ectodomain. We identified two critical regions for the NadA-cell receptor interaction with Chang cells: the NH2 globular head domain and the NH2 dimeric intrachain coiled-coil α-helices stemming from the stalk. This raises the importance of different modules within the predicted NadA structure. The identification of linear epitopes involved in receptor binding that are able to induce interfering antibodies reinforces the importance of NadA as a vaccine antigen.
Neisseria meningitidis serotype B (MenB) strains are mostly responsible for septicemia and meningitis in developed countries (6, 18, 19). In silico analysis of the genome of a virulent N. meningitidis B strain (MC58) allowed the identification of the 45-kDa Neisseria adhesin A (NadA) (3, 14). NadA was found to be expressed in ∼50% of N. meningitidis strains isolated from patients but in only ∼5% of strains from healthy individuals, and therefore it may be a risk factor for the development of meningococcal disease (4).
NadA is also a good immunogen, able to induce a bactericidal immune response, and is a component of a multiple anti-MenB vaccine at present under development (3, 10).
In vitro observations indicated that NadA may also be important in mucosal colonization by N. meningitidis B: (i) its expression in Escherichia coli enhances bacterial association with Chang epithelial cells (a human conjunctiva cell line widely used in meningococcal pathogenesis studies) (2); (ii) a NadA knockout mutant of N. meningitidis shows a partial, yet significant, decrease in cell adhesion and invasion compared to a wild-type strain, suggesting that NadA cooperates with other factors in mediating bacterium-cell interactions (2); (iii) a soluble recombinant form of NadA (NadA with a deletion of residues 351 to 405 [NadAΔ351-405]), lacking the membrane anchor region, binds to specific receptor sites with an apparent affinity of 3 μM on Chang cells (2, 9).
Other studies suggest that NadA, besides its role at the level of the mucosal epithelium, also exerts an immune-modulatory action on myeloid cells. Indeed, NadA-specific receptors were observed also on monocytes, macrophages, and monocyte-derived dendritic cells (9, 13). NadA may stimulate antimeningococcal defenses by augmenting the immune response of dendritic cells (self-adjuvant effect) and by increasing antigen presentation by macrophages engaged in antimicrobial activity (9, 13, 17). Immune-stimulatory effects of NadA were strongly synergized by meningococcus-specific outer membrane components (17).
For all of these reasons, NadA appears to be an important determinant in the host-pathogen interaction accompanying meningococcal infection. Consequently, understanding the structural determinants of NadA-cell interaction may help reveal ways to neutralize early meningitidis and fatal meningococcal sepsis.
Structure prediction and homology comparison show that NadA is an oligomeric coiled-coil adhesin (OCA), like YadA of Yersinia enterocolitica (7), UspA2 of Moraxella catarrhalis (5), and BadA of Bartonella henselae (16), belonging to the group of homotrimeric auto-transporter adhesins (TAAs) (5). OCAs are made by two main structural-functional parts: (i) a conserved COOH-terminal membrane anchor, having a β-barrel structure, necessary for the export of the remaining part of the adhesin (passenger domain) on the cell surface; and (ii) an extracellular passenger domain generally formed by an intermediate stalk with a high propensity to form coiled-coil α-helices and by an NH2-terminal region, predicted to have a globular structure and necessary for binding to host cell factors (1, 7, 12). Important exceptions are represented by HadA of Haemophilus influenzae, in which the globular head is missing (15), and by UspA1, where, in addition to a binding site located in the head region, there is a second binding site within the stalk, specific for another target (11).
Concerning NadA, previous studies showed that the deletion of the region of amino acids (aa) 24 to 87, corresponding to the putative receptor binding domain, totally abolishes adhesion of NadA-expressing E. coli cells to Chang cells (2). Attempts to further map the region(s) necessary to cell binding were unsuccessful because deletion mutants lacking the predicted subdomains of aa 24 to 42, 43 to 70, and 71 to 87 were all defective in mediating bacterial cell binding. These results were interpreted assuming either that the whole region of aa 24 to 87 is involved in receptor binding or, alternatively, that each separate deletion alters the structure of the remaining parts of this compact fold. In addition, structure prediction studies suggest that intrachain coiled-coil α-helices apparently located in the stalk might be involved in the formation of the receptor binding site, cooperating with the NH2 globular terminal region (11). Indeed, the possible involvement of dimeric intrachain coiled-coil regions in the binding of OCA adhesins to their cell receptors was suggested by studies on HadA of H. influenzae (15). HadA turned out to be an atypical OCA in which the globular head is missing and the adhesion function is performed by the NH2-terminal dimeric coiled-coil structures. In the absence of a crystallographic three-dimensional (3-D) map of NadA, in this study we built on previous data obtained on an E. coli model using Chang epithelial cells, expressing high levels of NadA-specific binding sites (2), to provide a more exhaustive mapping of the cell receptor binding site of NadA. To do so, we developed deletion mutants devoid of various sequences distributed in the coiled-coil region proximal to the NH2-terminal domain (aa 24 to 87) and progressively closer to the outer membrane anchor. Such mutants were analyzed in terms of their ability to form surface oligomers on the E. coli model and for their efficiency in promoting bacterial association to Chang conjunctiva cells. This information was compared with the ability of sera, affinity-purified antibodies ([Abs] and of their relative Fab fragments) to linear epitopes within the NH2-terminal domain and the coiled-coil stalk, to interfere with cell adhesion of NadA-expressing N. meningitidis and E. coli strains.
MATERIALS AND METHODS
Plasmid construction.
Genes coding for full-length NadA and NadAΔ30-88 were obtained as previously described (2). The mutated genes coding for NadAΔ88-149, NadAΔ180-219, NadAΔ219-289, and NadAΔ269-315 were generated using a Gene Taylor mutagenesis kit according to the manufacturer's instructions (Invitrogen). Briefly, the forward primers containing the mutation site (deletion) and the reverse primers were designed on the NadA sequence in order to delete the region of interest. The digested DNA fragments were cloned into the pET21b vector (Novagen). The ligation products were transformed into E. coli DH5α (Invitrogen), and E. coli BL21(DE3) was used as the expression host (Novagen). DNA cloning and E. coli transformation were performed according to standard protocols. E. coli strains were cultured at 37°C in Luria-Bertani broth supplemented with 100 μg/ml ampicillin.
FACS analysis.
For surface detection of NadA in E. coli using fluorescence-activated cell sorter (FACS) analysis, approximately 2 × 106 bacteria were incubated for 1 h with anti-NadAΔ351-405 (1:1,000) and subsequently for 30 min with R-phycoerythrin (PE)-conjugated goat F(ab)2 antibody to rabbit IgG (diluted 1:100; Jackson ImmunoResearch Laboratories). All antibodies were diluted in phosphate-buffered saline (PBS) with 1% fetal bovine serum (FBS). Samples were analyzed with a FACScan flow cytometer (Becton-Dickinson).
Animals.
Male adult New Zealand White rabbits were obtained from Harlan Italy srl, Italy.
NadA peptide synthesis.
A hydrophobicity map of the NadA protein was obtained based on the full-length amino acid sequence of the molecule, using Lasergene (DNASTAR) software. The peptides were synthesized by a solid-phase method on a Wang resin functionalized with the acid-labile 4-hydroxymethylphenoxyacetic acid linker (Novabiochem, Bad Soden, Germany), using an automated peptide synthesizer (model 433; Applied Biosystems, Foster City, CA). The fluorenylmethoxycarbonyl (Fmoc) strategy (8) was used throughout the peptide chain assembly, utilizing 2-(1H-benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) and 1-hydroxybenzotriazole (HOBt) as coupling reagents. Cleavage of the peptides was performed by reacting the peptidyl resins with a mixture containing trifluoroacetic acid (TFA)-H2O-thioanisole-ethanedithiol-phenol (10 ml/0.5 ml/0.5 ml/0.25 ml/750 mg) for 2.5 h. Crude peptides were purified by preparative reverse-phase high-performance liquid chromatography (HPLC). Molecular masses of the peptides were confirmed by mass spectroscopy with direct infusion on a Micromass ZMD-4000 mass spectrometer (Waters-Micromass). The purity of the peptides was in the range of 95 to 98%, as evaluated by analytical reverse-phase HPLC.
Purified peptides were coupled to keyhole limpet hemocyanin (KLH) and used for animal immunization. An aliquot of each peptide was retained for use in enzyme-linked immunosorbent assays (ELISAs) without coupling to KLH.
Peptide carrier conjugation.
Two milligrams of each peptide was conjugated with 2 mg of mariculture KLH (mcKLH; Pierce), in conjugation buffer, following the manufacturer's instructions, and the solutions were maintained under gentle agitation for 2 h at room temperature. Conjugates were subsequently purified by gel filtration by using Sephadex G-25 resin (Sigma). Fractions containing proteins were mixed, divided in aliquots, and frozen in liquid nitrogen.
Immunization and production of polyclonal Abs.
New Zealand White rabbits were immunized by subcutaneous injection with an aliquot (750 μl) of carrier peptide conjugate mixed 1:1 (vol/vol) with complete Freund's adjuvant (Sigma). Three subsequent injections were administered at 14-day intervals, with incomplete Freund's adjuvant (Sigma). Antisera were taken from immunized rabbits on day 68, and antibodies were purified by affinity chromatography using columns containing Sulfolink coupling gel (Pierce) linked to the different peptides through the sulfhydryl group. Briefly, columns were prepared with 2.5 ml of 50% Sulfolink coupling gel, and 2 mg of each peptide, dissolved in coupling buffer (50 mM Tris-HCl, 5 mM EDTA, pH 8.5), was added to the column and incubated at room temperature for 30 min. Columns were then washed with coupling buffer, and the nonspecific binding sites were blocked with a solution of 50 mM cysteine (Sigma) for 30 min at room temperature. Subsequently, columns were washed again extensively, and antiserum diluted 1:1 (vol/vol) with phosphate-buffered saline (PBS) was added to the columns; after extensive washing, antibodies were eluted with 0.2 M glycine, pH 2.8.
Fab generation and purification.
For each sample, 300 μl of immobilized papain-agarose (Sigma) was activated with activation buffer (50 mM Na-P, pH 7.0, 20 mM cysteine, 10 mM EDTA) for 20 min at 37°C; the resin was then washed and incubated with the antibody solution for 4 h at 37°C. To block the reaction, 75 mM iodoacetamide was added. To purify Fab fragments, papain-treated antibodies were incubated with 100 μl of protein A-agarose (Sigma) for 2 h at room temperature, under gentle shaking; the resin was washed three times, and the supernatants containing Fab fragments were recovered.
ELISA.
The day before the experiment, polystyrene plates (Sarstedt) were coated with 100 μl per well of various peptides (20 ng/μl) or with recombinant NadAΔ351-405 (0.5 μg/ml) or with NadA expressed on the surface of E. coli (E. coli NadA) (described in reference 2) (108/ml bacteria). The day of the experiment, wells were washed, blocked with 1% bovine serum albumin (BSA)-PBS, and incubated for 1 h with different antisera or purified anti-peptide antibodies or Fab antibodies, depending on the experiment. After the binding, wells were exhaustively washed and treated with alkaline phosphatase (AP)-conjugated anti-rabbit IgG(H+L) antibodies (Chemicon). ABTS [2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid); Chemicon] was used as a substrate, and absorbance was measured in an automatic ELISA plate reader (Amersham Biosciences).
SDS-PAGE and Western blotting.
Whole and Fab antibodies were resolved by SDS-PAGE and then subjected to Coomassie staining; alternatively, proteins were blotted onto a nitrocellulose membrane and probed with an alkaline phosphatase-goat anti-IgG (heavy or light chain) antibody (Chemicon). Blots were developed with AP buffer (100 mM NaCl, 5 mM MgCl2, 100 mM Tris-Cl [pH 9.2]) supplemented with 1% (vol/vol) 5-bromo-4-chloro-3-indolylphosphate (BCIP) and 1% (vol/vol) nitroblue tetrazolium (NBT) (Sigma).
To check the trimer formation of the various NadA mutants expressed by E. coli, bacteria were grown at 37°C for 14 h and then recovered by centrifugation, resuspended in 1× SDS sample buffer, and boiled for 10 min. Equal amounts of proteins were separated using a NuPAGE Gel System, according to the manufacturer's instructions (Invitrogen). Proteins were blotted onto nitrocellulose membranes, and Western blotting was performed using an anti-NadAΔ351-405 serum (1:2,000) and a secondary peroxidase-conjugated antibody (Dako).
Adhesion assay.
Chang cells were seeded on 24-well tissue culture plates (1 × 105 cells per well), and after 24 h of incubation in an antibiotic-free medium, approximately 3 × 107 (multiplicity of infection [MOI] of 1:100) bacteria were added per well in Dulbecco's modified Eagle's medium (DMEM) supplemented with 1% FBS and incubated for 1 h at 37°C in 5% CO2. After removal of nonadherent bacteria by washing, cells were lysed with 1% saponin (Sigma); to block the reaction 800 μl of DMEM-1% inactivated FBS (FBSi) was added, and serial dilutions of the suspension were plated onto LB agar to calculate the number of CFU.
Inhibition of adherence of E. coli-NadA with anti-NadA peptide Abs/Fab fragments.
Liquid cultures of E. coli-NadA were washed once in PBS and resuspended in DMEM-1% FBSi in the absence or in the presence of different doses of 45 nM Abs/Fab fragments or of various concentrations of affinity-purified Ab or Fab fragments for 1 h at 4°C. Samples were used to infect Chang cell monolayers (MOI of 100) for 1 h at 37°C. From this point on, the adhesion assay was performed as described above. E. coli-pET was used as a negative control.
Inhibition of adherence of N. meningitidis with NadA linear peptide antiserum.
Cultures of the N. meningitidis M58 strain grown on GC agar were resuspended in DMEM-1% FBS in the absence or presence of different concentrations of rabbit antisera obtained by immunizing animals with both the NadA peptides and NadAΔ351-405, as indicated in the figure legends. Preimmune sera were tested as negative controls. After 1 h of incubation at 4°C, bacteria were added to Chang cell monolayers (MOI of 100) for 1 h at 37°C. As described above, after extensive washing, cells lysis, and agar plating, the number of CFU was determined.
RESULTS
The proximal region of aa 88 to 150 of the NadA stalk is important for NadA-mediated adhesion.
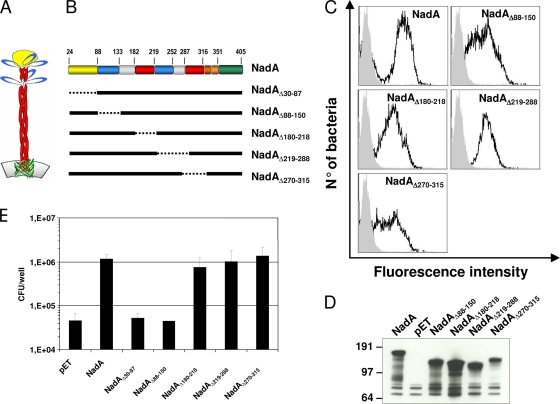
Based on previous studies, it was proposed that residues from aa 24 to 87, corresponding to the predicted NH2-terminal globular head of NadA, are involved in the adhesion to epithelial cells of NadA-expressing bacteria (2). To further investigate the role of NadA head and stalk in the adhesion process, we designed a panel of deletion mutants based on the secondary structure prediction, and we expressed them in E. coli (Fig. 1A and B). According to a prediction algorithm, the stalk region has a high propensity to form α-helices and dimeric coiled-coil tertiary structures (15). FACS and Western blot analyses on whole bacteria showed that all mutants form superficially exposed oligomers. This suggests that the deletion of the intrachain dimeric coiled-coil region does not alter the NadA oligomeric organization, and exposed NadA on the E. coli surface (Fig. 1C and D). Adhesion experiments were performed using each single mutant on Chang epithelial cells. The results shown in Fig. 1E indicate that, in addition to the deletion of the head domain (from aa 30 to 87), the lack of the first dimeric coiled-coil region (from aa 88 to 150) totally abolishes the NadA adhesion property, whereas all the other regions appeared to be irrelevant for bacteria-cell binding. These observations suggest that the dimeric coiled-coil domain (aa 88 to 150) of NadA is necessary for bacterium-cell adhesion together with the previously identified globular NH2 domain (aa 24 to 87).
FIG. 1.
Analysis of NadA binding regions using deletion mutants expressed in E. coli. (A) Proposed three-dimensional organization of NadA protein. Portions of the extracellular passenger domain, which are predicted to form dimeric and trimeric coiled coils, are shown in blue and red, respectively. The profile of different propensities to form dimeric and trimeric coiled-coil supersecondary structures has been calculated using Multicoil software (http://multicoil.lcs.mit.edu/cgibin/multicoil). The NadA globular head is shown in yellow. The α-helix linker region (L2L1) and β-barrel parts within the integral outer membrane translocator domains are shown in orange and green, respectively (15). (B) Schematic representation of the various NadA mutants used in this study. The dimeric and trimeric coiled coils, the α-helix linker, and the beta region are indicated according to the color scheme of panel A. The leader peptide is not indicated. (C) FACS analysis on whole-cell bacteria expressing NadA mutants using anti-NadAΔ351-405 serum. (D) Western blot analysis of the expression of NadA in E. coli. Total cell lysates of the indicated deletion mutants were assayed using an anti-NadAΔ351-405 serum. The mutant NadAΔ30-87 was previously demonstrated to be surface exposed in a trimeric form (2). (E) Adhesion of NadA mutants. Chang monolayers were infected with E. coli expressing full-length NadA or each single deletion mutant (MOI of 100). E. coli-pET, carrying the vector alone, was used as a negative control. Results are reported as the number of CFU per well, and values represent the mean and standard deviation of several experiments performed in triplicate.
Generation of antibodies against NadA peptides recognizing soluble and membrane-associated NadA.
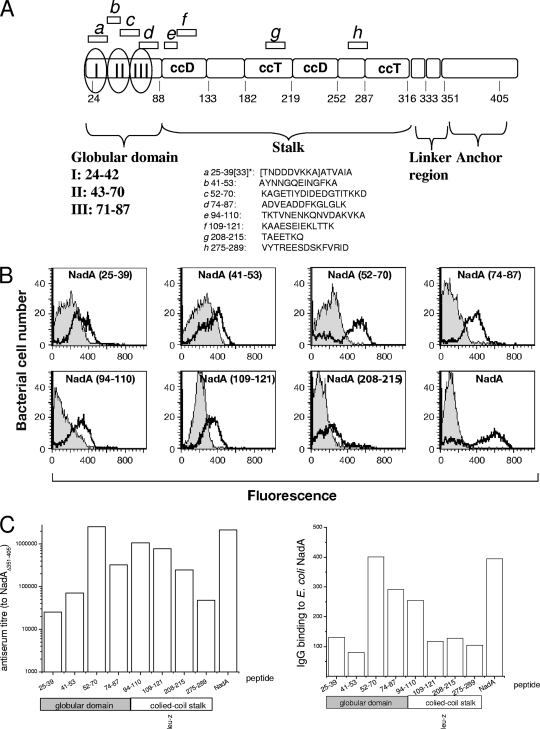
The deletion of the coiled-coil region, aa 88 to 150, could impact NadA adhesion properties either by eliminating a portion directly binding to the NadA receptor or by indirectly affecting the conformation of the real receptor-binding domain. Therefore, we generated antisera to six linear peptides covering these two critical domains (Fig. 2A) and tested their ability to block NadA-mediated cell adhesion. Antisera generated against determinants in the middle of the stalk and closer to the membrane surface (aa 208 to 215 and 275 to 289, respectively), irrelevant for cell binding based on deletion studies with E. coli recombinant strains, were used as negative controls. ELISAs and FACS analysis showed that antipeptide sera specifically recognized the NadAΔ351-405 and the ectodomain of full-length NadA expressed on E. coli (Fig. 2B and C), proving that antilinear epitope antibodies cross-react with the native adhesin. The NadA peptide comprised of residues 52 to 70 (NadA52-70) was apparently the most immunogenic peptide; in fact, the titer of the antiserum was comparable to that induced by native NadA, used as positive control. However, it is also possible that the region of aa 52 to 70 is the more exposed and accessible region of the protein. All preimmune sera gave a negligible signal (data not shown).
FIG. 2.
Cross-reaction of rabbit antisera to linear epitopes of NadA with native NadA in solution and on the E. coli outer membrane. (A) The diagram reports the sequence of NadA linear peptides used to immunize rabbits and their approximate location along the primary structure. In the scheme the tripartite structure of the protein is indicated: the COOH-terminal anchor to the outer membrane with the linker region, the putative coiled-coil intermediate stalk, and the NH2-terminal globular head. Within the last, the three subdomains previously deleted with loss of cell-binding capacity (2) are also indicated as I (aa 24 to 42), II (aa 43 to 70), and III (aa 71 to 87). Antiserum to NadA25-33 (residues in brackets) was also used in this study in addition to antiserum to NadA25-39. The two sera were very similar, and data shown later are therefore primarily relative to antibodies obtained with the sequence at aa 25 to 39. (B) Antisera to linear epitopes of NadA bind to the surface of NadA-expressing E. coli as assessed by FACS analysis. NadA binding antibodies were revealed by PE-labeled secondary anti-rabbit IgG antibodies. (C) ELISA showing the titers of anti-peptide sera, using NadAΔ351-405 or wild-type E. coli NadA as capturing antigens. Columns represent the reciprocal of the serum dilution giving the half-maximal optical density. The location of the sequence targeted within the predicted structure of the adhesin is schematically indicated.
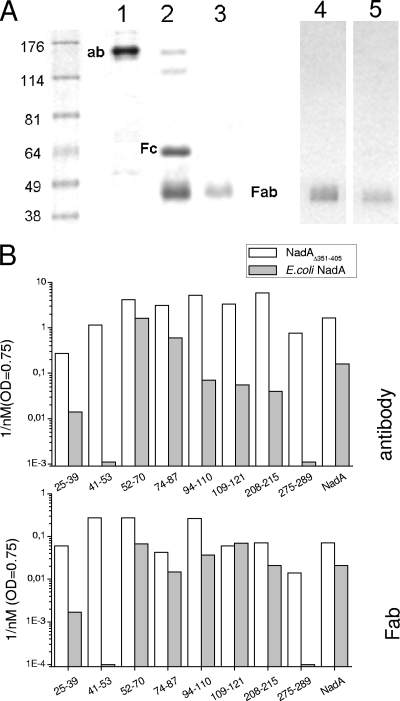
To quantitatively compare the interactions of anti-NadA peptide antibodies, they were affinity purified. In addition, we also produced Fab fragments because these reagents retain the same specificities but have the advantage of being less sterically hindering (∼ 50 kDa) and monovalent (Fig. 3A). Affinity-purified Ab/Fab fragments effectively recognized the peptides used for their generation and did not cross-react with any of the other peptides of our panel (data not shown).
FIG. 3.
Binding of affinity-purified Ab/Fab specific for NadA peptides to soluble or membrane-associated native NadA. (A) Characterization of affinity-purified Abs and Fab fragments. Coomassie staining after SDS-PAGE of a representative example of affinity-purified antipeptide antibody (aa 52 to 70) and after its cleavage by matrix-linked papain to produce Fc and Fab fragments (lanes 1, 2, and 3), and Western blotting of purified Fab fragments with anti-IgG(H+L) antibodies (lanes 4 and 5). (B) ELISAs performed using purified NadAΔ351-405 or E. coli NadA as capturing antigens and different concentrations of affinity-purified Abs and Fab fragments to the indicated NadA peptides. Bound Ab/Fab fragments were revealed by secondary Ab to rabbit IgG conjugated to horseradish peroxidase. After colorimetric development, the reciprocal of the Ab/Fab fragment nanomolar concentration giving an optical density (OD) of 0.75 was calculated from the graphs and plotted.
ELISAs performed with increasing concentrations of purified antipeptide antibodies and Fab fragments allowed characterization of their efficacy in the interaction with purified soluble NadAΔ351-405 and with the protein expressed on the surface of E. coli cells. Data (Fig. 3B) demonstrated that antibodies and Fab fragments bind at similar levels to recombinant soluble NadAΔ351-405. In contrast, when Abs and Fab fragments were challenged with the membrane-anchored NadA, their binding capabilities were clearly differentiated. First of all, anti-NadA25-39 (and anti-NadA24-33 [data not shown]) antibodies and Fab fragments reacted less efficiently to the adhesin expressed on the bacterial surface than the soluble protein. An even stronger decrease in immune reactivity was evident with anti-NadA41-53 Ab/Fab, suggesting that the linear epitope of aa 41 to 53 is poorly accessible when the adhesin is in the membrane environment. The following sequence of aa 52 to 70 remains one of the most accessible sites, confirming the immunogenicity data. On the other hand, antibody reactivity to epitopes from position 52 to 289 decreased progressively moving toward the membrane surface, very likely for steric reasons. In agreement with this interpretation, the corresponding Fab fragments, with a mass equal to one-third that of whole antibodies, showed good immune reactivity also with linear epitopes in positions closer to the bacterial membrane. The only exception was the epitope of aa 275 to 289, scarcely available by both specific Ab and Fab.
Inhibition of NadA-expressing E. coli adhesion to Chang cells by antibodies against NadA linear epitopes.
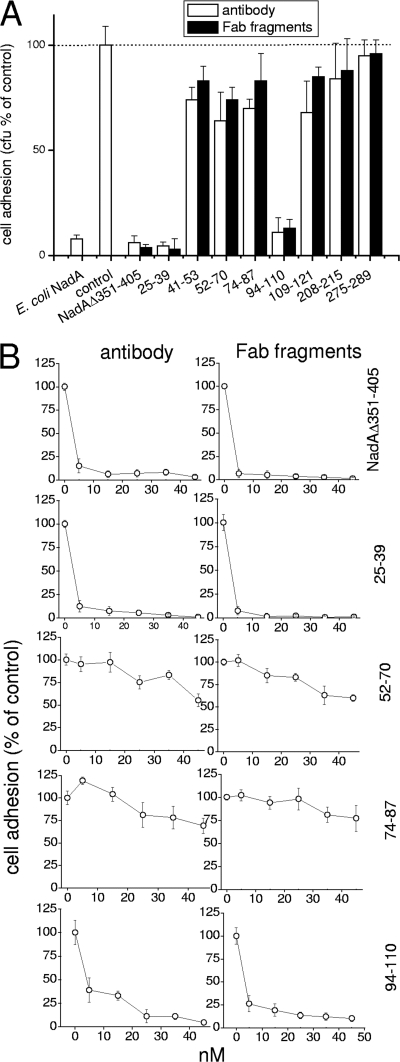
All the Abs and Fab fragments directed against NadA peptides were used to evaluate the contribution of defined linear epitopes to bacterium-cell adhesion mediated by NadA. As shown in Fig. 4A, adhesion of NadA-expressing E. coli to Chang conjunctiva cells was totally abolished by a polyclonal antibody and its relative Fab to the whole extracellular domain of the protein (NadAΔ351-405). Antibodies and Fab fragments to peptides NadA25-39 and NadA94-110 strongly counteracted bacterium-cell adhesion (95% and 86% inhibition, respectively). Antibodies and Fab fragments directed to NadA109-121 and to the remaining part of the globular NH2-terminal domain (NadA aa 41 to 53, 52 to 70, and 74 to 87) were partially inhibitory (∼20 to 30% decrease), while those directed to the stalk region (NadA aa 208 to 215 and 275 to 289) were scarcely neutralizing.
FIG. 4.
Neutralization of NadA-mediated E. coli adhesion to Chang cells by anti-NadA peptide Abs/Fab fragments. Adherence of E. coli NadA to Chang cells was inhibited by the addition of 45 nM (A) or increasing concentrations (B) of affinity-purified Abs/Fab fragments against the specified NadA peptide. Adhesion efficacy is expressed with respect to control sample (E. coli NadA infecting Chang cells in the absence of Ab/Fab fragments), arbitrarily fixed at 100. Data are the means ± standard deviations from several experiments run in triplicate.
When the efficacy of antibodies and Fab fragments able to neutralize NadA-mediated bacterial adhesion was compared with their reactivity with the same antigen by ELISA, it turned out that adhesion neutralization was achieved at subsaturating antigen binding, that is, 5 nM for anti-NadA24-39 and 25 nM for anti-NadA94-110 (Fig. 4B). These results suggest that the regions of aa 24 to 39 and 94 to 110 in the NadA head and stalk, respectively, are essential for the binding activity. Abs/Fab fragments to the interposed region of aa 52 to 87 were confirmed to counteract E. coli-NadA adhesion with reduced efficacy.
Isolated peptides used to immunize animals were also tested for their ability to impair E. coli-NadA adhesion to epithelial cells but were found to be ineffective (data not shown), hinting at the possible involvement of the conformation of NadA modules to exploit their functions.
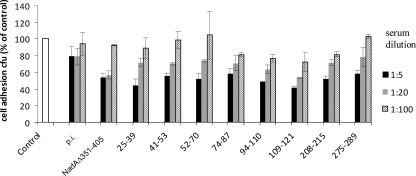
Ability of sera raised against linear NadA epitopes to inhibit N. meningitidis B adhesion to Chang cells.
We tested the inhibitory effect of different dilutions of specific sera against NadA linear peptides on adhesion of NadA-expressing N. meningitidis B strain MC58 to Chang cells. As previously shown, the depletion of the nadA gene in MC58 leads to a partial reduction in the attachment to Chang cells (2). In accordance with this, here we show that a polyclonal anti-NadAΔ351-405 antiserum partially decreased the adhesion of MC58 to Chang cells, giving a maximal inhibition around 40% compared to the control (Fig. 5), suggesting that the NadA contribution to cell adhesion was abolished by specific antibodies. We observed that the whole panel of tested sera exerted a titration-dependent inhibitory effect although antisera to peptides comprised of residues 25 to 39, 94 to 110, and 109 to 121 were the most effective, in agreement with data obtained using the E. coli model. It is noteworthy that the antiserum to NadA109-121, partially inhibitory to E. coli NadA adhesion, is very efficacious in hampering meningococcal cell adhesion. As expected, the serum-specific inhibitory actions were partial and compatible with the neutralization of the NadA contribution to meningococcal cell adhesion. Taken together, these results highlight the key role of the NH2 region of NadA, comprising the head domain (aa 24 to 87) and the adjacent dimeric coiled-coil region (aa 88 to 132) (Fig. 1A and B), in mediating cell interaction.
FIG. 5.
Efficacy of antisera raised against NadA linear peptides to inhibit N. meningitidis adhesion to Chang cells. Adherence to epithelial cells of N. meningitidis serogroup B strain M58 was inhibited using different dilutions of the indicated rabbit sera. Data are expressed compared to control sample (MC58 strain in the absence of serum) arbitrarily fixed at 100. Values represent the means and standard deviations of one representative experiment performed in triplicate. p.i., preimmune serum.
DISCUSSION
The adhesin NadA is an important virulence factor of serogroup B N. meningitidis strains identified by a genomic approach. The discovery of NadA sequences responsible for direct interaction with its cellular receptor may help to elucidate the function of this important N. meningitidis virulence factor. Moreover, since this protein has been proposed to participate in epithelial colonization and possibly cell invasion by N. meningitidis B strains, such information may allow the creation of antibodies able to effectively neutralize any biological function triggered by the formation of the NadA-NadA receptor complex.
Previous data demonstrated that the globular NH2-terminal domain of NadA (aa 24 to 88) is necessary for cell adhesion, in agreement with the general assumption that the apical portion of the adhesin is crucial (2). However, recent evidence obtained with HadA of H. influenzae, an OCA adhesin lacking such a terminal globular domain, and structure prediction suggest that other parts of the adhesin NadA are close to the globular domain and may participate in cell binding (15). On the other hand, a more detailed mapping of the NadA sequences necessary for receptor association within the domain of aa 24 to 88 was without success (2).
In this study, to gain detailed information on the structural determinants of NadA cell binding, we exploited (i) an E. coli strain expressing deletion of NadA and (ii) antibodies directed against specific peptides. These two ways to test the functional involvement of a limited protein region are complementary, being based on different principles. Indeed, the elimination of a sequence may give misleading results if the structure of a protein is consequently affected or, alternatively, if the distance between otherwise stable domains is modified. On the other hand, the binding of an antibody to the same region is supposed to impair functions by impeding or hindering the normal interactions of the target protein with its molecular partners.
Deletion studies showed that a sequence corresponding to the first predicted internal coiled-coil region of NadA is required for cell receptor binding while no other deletion led to defective binding. These observations, combined with previous findings, point to the possibility that the NadA receptor binding site is also formed by α-helices forming coiled coils. Indeed antibodies to a more restricted sequence (aa 94 to 110 and 109 to 121) within this area neutralized (with some difference in efficacy in E. coli and in N. meningitidis) NadA-dependent bacterial adhesion to cells. In contrast, antibodies to a region immediately preceding the second predicted dimeric coiled-coil region were less inhibitory, again in agreement with deletion mutant studies.
Antibodies to the very-NH2-terminal sequence (aa 24 to 39) were also neutralizing, while antibodies directed to stalk regions close to the bacterial outer membrane (aa 208 to 215 and 275 to 289) were poorly effective. This fits with the commonly accepted view that the distal part of the protein is engaged with the cell receptor, while the proximal one serves to protrude this part toward the external region of the bacterial cells. Previous experiments performed with N. meningitidis showed that the expression of NadA partially but significantly contributes to bacterium-epithelial cell association (2). Here, we also confirmed that in this native meningococcal context α-helices in both the NH2-terminal and the dimeric coiled-coil regions are important for NadA-mediated cell adhesion.
Our data suggest that most of the supposed receptor binding site (NadA aa 42 to 88) may not be in close contact with the NadA cell receptor. In fact, not only Fab fragments but also the most neutralizing Abs specific for peptides within this region inhibited bacterial binding to cells less effectively than Abs/Fab fragments to peptides comprised of aa 25 to 39 and 94 to 110. Given the great volume of the whole antibody molecule, such results suggest that the sequence of aa 42 to 88, although close to aa 24 to 42 in the primary structure, is placed in such a way that bound antibodies are oriented in a direction that only partially disturbs the cell-receptor approach. Surprisingly, antibodies and Fab to the neighboring sequences of aa 94 to 110 and 109 to 121 (the latter more efficiently in N. meningitidis B) are again able to neutralize NadA-mediated cell-bacterium adhesion. One possibility accounting for this observation is that the polypeptide chain corresponding to the sequence of aa 94 to 121 returns close to the NH2 terminus (NadA25-39) and that these two regions cooperate to form the complete NadA receptor binding site.
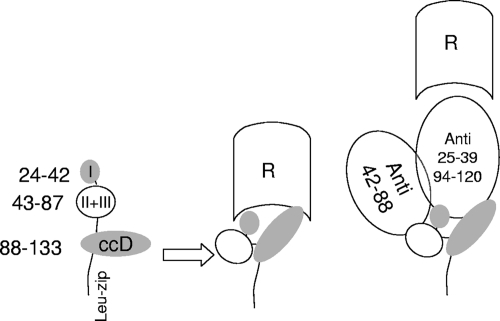
Based on these data and on structure prediction, we propose a comprehensive model of the NH2-terminal domain of NadA (Fig. 6). The subdomain of aa 24 to 39, predicted to form amphipathic α-helices, is assumed to be in direct contact with the NadA receptor, while the two other subdomains are only proximal. In contrast, the following dimeric coiled-coil region is also proposed to provide surface area necessary for receptor binding, in agreement with deletion mutant experiments. Hence, according to this hypothesis, three sets of sequences from aa 24 to 39 and aa 94 to 121 may form the NadA receptor binding site. This model accounts for the less efficient inhibition exerted by antibodies directed to aa 40 to 88: this sequence is, in fact, proposed to be peripheral to the receptor binding region. Such a complex structure could also explain why the separate deletion of all three head subdomains of the head structure altered the proper folding of this region, with a subsequent lack of adhesion properties (2). Poorly neutralizing antibodies may be used to coisolate NadA receptors.
FIG. 6.
Model of NadA-NadA receptor interaction. The picture shows the predicted globular head (aa 24 to 42, subdomain I; aa 43-87, subdomains II and III) and the neighboring intrachain coiled-coil domain (aa 88-133, ccD). It is proposed that the regions around sequences of aa 24 to 39 and 94 to 121 form the surface interacting with the NadA receptor (R), while aa 42 to 88 are not directly associated with NadA receptor. The possible interaction with specific Fab fragments and NadA receptor is also depicted to account for competition studies. A single polypeptide chain of the three forming the adhesin oligomer is shown here for clarity.
Our data provide further evidence on the role not only of the globular NH2 domain but also of the neighboring dimeric coiled-coil structures in NadA-NadA receptor interaction, suggesting that a more complex structure of the protein participates in binding the cellular receptor. Moreover, data on N. meningitidis, showing that anti-NadA antibodies can counteract NadA adhesive functions, further support the efficacy of NadA as an important component of an anti-meningococcus B vaccine.
Acknowledgments
This work was supported by grants from the University of Padua (Progetto di Ateneo 2004 and ex60% 2007 and 2008).
We thank M. Morandi and E. Ciccopiedi (Novartis Vaccine and Diagnostics) for NadAΔ351-405 purification.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Brooks, M. J., J. L. Sedillo, N. Wagner, W. Wang, A. S. Attia, H. Wong, C. A. Laurence, E. J. Hansen, and S. D. Gray-Owen. 2008. Moraxella catarrhalis binding to host cellular receptors is mediated by sequence-specific determinants not conserved among all UspA1 protein variants. Infect. Immun. 76:5322-5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capecchi, B., J. Adu-Bobie, F. Di Marcello, L. Ciucchi, V. Masignani, A. Taddei, R. Rappuoli, M. Pizza, and B. Arico. 2005. Neisseria meningitidis NadA is a new invasin which promotes bacterial adhesion to and penetration into human epithelial cells. Mol. Microbiol. 55:687-698. [DOI] [PubMed] [Google Scholar]
- 3.Comanducci, M., S. Bambini, B. Brunelli, J. Adu-Bobie, B. Aricò, B. Capecchi, M. M. Giuliani, V. Masignani, L. Santini, S. Savino, D. M. Granoff, D. A. Caugant, M. Pizza, R. Rappuoli, and M. Mora. 2002. NadA, a novel vaccine candidate of Neisseria meningitidis. J. Exp. Med. 195:1445-1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Comanducci, M., S. Bambini, D. A. Caugant, M. Mora, B. Brunelli, B. Capecchi, L. Ciucchi, R. Rappuoli, and M. Pizza. 2004. NadA diversity and carriage in Neisseria meningitidis. Infect. Immun. 72:4217-4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cotter, S. E., N. K. Surana, and J. W. St. Geme III. 2005. Trimeric autotransporters: a distinct subfamily of autotransporter proteins. Trends Microbiol. 13:199-205. [DOI] [PubMed] [Google Scholar]
- 6.de Souza, A. L., and A. C. Seguro. 2008. Two centuries of meningococcal infection: from Vieusseux to the cellular and molecular basis of disease. J. Med. Microbiol. 57:1313-1321. [DOI] [PubMed] [Google Scholar]
- 7.El Tahir, Y., and M. Skurnik. 2001. YadA, the multifaceted Yersinia adhesin. Int. J. Med. Microbiol. 291:209-218. [DOI] [PubMed] [Google Scholar]
- 8.Fields, G. B., and R. L. Noble. 1990. Solid phase peptide synthesis utilizing 9-fluorenylmethoxycarbonyl amino acids. Int. J. Pept. Protein Res. 35:161-214. [DOI] [PubMed] [Google Scholar]
- 9.Franzoso, S., C. Mazzon, M. Sztukowska, P. Cecchini, T. Kasic, B. Capecchi, R. Tavano, and E. Papini. 2008. Human monocytes/macrophages are a target of Neisseria meningitidis adhesin A (NadA). J. Leukoc. Biol. 83:1100-1110. [DOI] [PubMed] [Google Scholar]
- 10.Giuliani, M. M., J. Adu-Bobie, M. Comanducci, B. Aricò, S. Savino, L. Santini, B. Brunelli, S. Bambini, A. Biolchi, B. Capecchi, E. Cartocci, L. Ciucchi, F. Di Marcello, F. Ferlicca, B. Galli, E. Luzzi, V. Masignani, D. Serruto, D. Veggi, M. Contorni, M. Morandi, A. Bartalesi, V. Cinotti, D. Mannucci, F. Titta, E. Ovidi, J. A. Welsch, D. Granoff, R. Rappuoli, and M. Pizza. 2006. A universal vaccine for serogroup B meningococcus. Proc. Natl. Acad. Sci. U. S. A. 103:10834-10839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hill, D. J., A. M. Edwards, H. A. Rowe, and M. Virji. 2005. Carcinoembryonic antigen-related cell adhesion molecule (CEACAM)-binding recombinant polypeptide confers protection against infection by respiratory and urogenital pathogens. Mol. Microbiol. 55:1515-1527. [DOI] [PubMed] [Google Scholar]
- 12.Hoiczyk, E., A. Roggenkamp, M. Reichenbecher, A. Lupas, and J. Heesemann. 2000. Structure and sequence analysis of Yersinia YadA and Moraxella UspAs reveal a novel class of adhesins. EMBO J. 19:5989-5999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mazzon, C., B. Baldani-Guerra, P. Cecchini, T. Kasic, A. Viola, M. de Bernard, B. Arico, F. Gerosa, and E. Papini. 2007. IFN-gamma and R-848 dependent activation of human monocyte-derived dendritic cells by Neisseria meningitidis adhesin A. J. Immunol. 179:3904-3916. [DOI] [PubMed] [Google Scholar]
- 14.Pizza, M., V. Scarlato, V. Masignani, M. M. Giuliani, B. Arico, M. Comanducci, G. T. Jennings, L. Baldi, E. Bartolini, B. Capecchi, C. L. Galeotti, E. Luzzi, R. Manetti, E. Marchetti, M. Mora, S. Nuti, G. Ratti, L. Santini, S. Savino, M. Scarselli, E. Storni, P. Zuo, M. Broeker, E. Hundt, B. Knapp, E. Blair, T. Mason, H. Tettelin, D. W. Hood, A. C. Jeffries, N. J. Saunders, D. M. Granoff, J. C. Venter, E. R. Moxon, G. Grandi, and R. Rappuoli. 2000. Identification of vaccine candidates against serogroup B meningococcus by whole-genome sequencing. Science 287:1816-1820. [DOI] [PubMed] [Google Scholar]
- 15.Serruto, D., T. Spadafina, M. Scarselli, S. Bambini, M. Comanducci, S. Hohle, M. Kilian, E. Veiga, P. Cossart, M. R. Oggioni, S. Savino, I. Ferlenghi, A. R. Taddei, R. Rappuoli, M. Pizza, V. Masignani, and B. Arico. 2009. HadA is an atypical new multifunctional trimeric coiled-coil adhesin of Haemophilus influenzae biogroup aegyptius, which promotes entry into host cells. Cell. Microbiol. 11:1044-1063. [DOI] [PubMed] [Google Scholar]
- 16.Szczesny, P., D. Linke, A. Ursinus, K. Bar, H. Schwarz, T. M. Riess, V. A. Kempf, A. N. Lupas, J. Martin, and K. Zeth. 2008. Structure of the head of the Bartonella adhesin BadA. PLoS Pathog. 4:e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tavano, R., S. Franzoso, P. Cecchini, E. Cartocci, F. Oriente, B. Arico, and E. Papini. 2009. The membrane expression of Neisseria meningitidis adhesin A (NadA) increases the proimmune effects of MenB OMVs on human macrophages, compared with NadA− OMVs, without further stimulating their proinflammatory activity on circulating monocytes. J. Leukoc. Biol. 86:143-153. [DOI] [PubMed] [Google Scholar]
- 18.Tzeng, Y. L., and D. S. Stephens. 2000. Epidemiology and pathogenesis of Neisseria meningitidis. Microbes Infect. 2:687-700. [DOI] [PubMed] [Google Scholar]
- 19.van Deuren, M., P. Brandtzaeg, and J. W. van der Meer. 2000. Update on meningococcal disease with emphasis on pathogenesis and clinical management. Clin. Microbiol. Rev. 13:144-166. [DOI] [PMC free article] [PubMed] [Google Scholar]