Abstract
The Escherichia coli rluD gene encodes a pseudouridine synthase responsible for the pseudouridine (Ψ) modifications at positions 1911, 1915, and 1917 in helix 69 of 23S rRNA. It has been reported that deletion of rluD in K-12 strains of E. coli is associated with extremely slow growth, increased readthrough of stop codons, and defects in 50S ribosomal subunit assembly and 30S-50S subunit association. Suppressor mutations in the prfB and prfC genes encoding release factor 2 (RF2) and RF3 that restore the wild type-growth rate and also correct the ribosomal defects have now been isolated. These suppressors link helix 69 Ψ residues with the termination phase of protein synthesis. However, further genetic analysis reported here also reveals that the slow growth and other defects associated with inactivation of rluD in E. coli K-12 strains are due to a defective RF2 protein, with a threonine at position 246, which is present in all K-12 strains. This is in contrast to the more typical alanine found at this position in most bacterial RF2s, including those of other E. coli strains. Inactivation of rluD in E. coli strains containing the prfB allele from E. coli B or in Salmonella enterica, both carrying an RF2 with Ala246, has negligible effects on growth, termination, or ribosome function. The results indicate that, in contrast to those in wild bacteria, termination functions in E. coli K-12 strains carrying a partially defective RF2 protein are especially susceptible to perturbation of ribosome-RF interactions, such as that caused by loss of h69 Ψ modifications.
Pseudouridine (Ψ) is among the most common posttranscriptional modifications occurring in rRNA and has been found at conserved regions of the rRNAs in organisms from all three kingdoms (28, 29). In Escherichia coli, there are 11 Ψ residues in 16S and 23S rRNAs, and the 7 synthases responsible for these modifications have now been identified (30). The functions of specific Ψ residues have been investigated by deletion of the corresponding Ψ synthase genes (rsuA, rluA, rluB, rluC, rluD, rluE, and rluF) and characterization of the resulting deletion mutants. With the notable exception of rluD, deletion of individual Ψ synthase genes has little effect on cell growth (29). In contrast, inactivation or deletion of rluD has been reported to have profound effects on cell growth, 50S subunit assembly, and subunit association (14, 32). RluD is responsible for Ψ1911, Ψ1915, and Ψ1917 modifications in helix 69 (h69) of 23S rRNA. This functionally important stem-loop structure forms a bridge between the 30S and 50S ribosomal subunits and interacts with translation factors and tRNAs at multiple stages of protein synthesis (39).
The location of Ψ residues in h69 and the phenotypes of the ΔrluD mutants lacking h69 Ψ residues suggest that these posttranscriptional modifications are important for ribosome function. Strains lacking RluD are extremely unstable and rapidly give rise to faster-growing, suppressor-containing derivatives (12, 14). Some insights into h69 Ψ functions were provided by the identification of one such ΔrluD suppressor carrying a missense mutation in the prfB gene, encoding release factor 2 (RF2) (9). The same study also showed that ΔrluD mutants had elevated levels of stop codon readthrough and that readthrough was reduced to wild-type levels in the ΔrluD prfB double mutant. These data link h69 Ψ modifications with the activities of release factors at termination, consistent with the now-known sites of RF-ribosome interactions, revealed by X-ray crystallography of termination complexes (16, 17, 38).
In this study, we have isolated and characterized 21 further ΔrluD suppressors. In addition to 8 different amino acid substitutions in RF2, we also recovered suppressor mutations in the prfC gene encoding release factor RF3. In all cases, stop codon readthrough was restored to near-wild type levels in the ΔrluD prfB and ΔrluD prfC double mutants. We have also examined the contribution of different E. coli strain backgrounds to the ΔrluD phenotype: all reported genetic and biochemical analyses of RluD function have been carried out with K-12 strains of E. coli. While the K-12 strain has been used as a wild-type model organism, it carries a variant and defective RF2 protein, with a Thr residue at position 246, in contrast to all other bacteria (including other E. coli strains) which have Ala or Ser at this position. We have examined the effect of Thr246 RF2 on the phenotypes conferred by the ΔrluD mutation, by replacing the K-12 (Thr246) prfB allele with the E. coli B (Ala246) allele. Surprisingly, deletion of the rluD gene in a strain carrying the E. coli B RF2 (Ala246) allele had no effect on growth, ribosomal subunit association, or stop codon readthrough. Moreover, deletion of the rluD gene in Salmonella enterica, which also carries the typical bacterial Ala246 RF2 had negligible effects on these same parameters. These data indicate that while h69 Ψ residues contribute to RF-ribosome interactions, the severe phenotypes of ΔrluD mutants reported in the literature are largely the result of a defective RF2 protein in the E. coli K-12 strains used for these analyses and are not typical of bacteria carrying fully active RF2 proteins.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The bacterial strains used in this study are listed in Table 1. The pSG and pLG series of lacZ plasmids used here to monitor stop codon readthrough have been described previously (25, 27). pSG3/4 UGA and pSG34-11 carry UGA mutations; pLG12-6, pSG12-6, and pSG163 UAG carry UAG stop codons; and pSG25 and pSG853 carry a wild-type lacZ and a UAA stop codon.
TABLE 1.
Bacterial strains
| Straina | Relevant genotype/RF2 or RF3 alteration | Source or reference |
|---|---|---|
| E. coli strains | ||
| BL21(DE3) | E. coli B F−ompT gal dcm lon hsdSB λ(DE3 [lacI lacUV5-T7 gene 1 ind1 sam7 nin5]) | Laboratory collection |
| CSH142 | ara Δ(gpt-lac)5 thi | 23 |
| MG1655 | F− λ−ilvG rfb-50 rph-1 | Laboratory collection |
| LE392 | glnV44 supF58 lac galK2 galT22 metB1 trpR55 hsdR514 | Laboratory collection |
| ME 97 | MG1655 rluD::cat (“dust”) | |
| JW2880-1 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− ΔserA764::kan rph-1 Δ(rhaD-rhaB)568 hsdR514 | CGSC |
| MC279 | Δ(lac-pro) thi leu-48 (UGA) | Laboratory collection |
| MC359 | MC279 prfB (E. coli B) | This work |
| MC360 | MG1655 prfA1 Tn10 (linked) ΔserA764::kan | Laboratory collection |
| MC361 | CSH142 prfB (E. coli B) | This work |
| MC362 | CSH142 prfB (E. coli B) ΔrluD::cat | This work |
| MC363 | LE392 ΔrluD::cat | This work |
| MC364 | MG1655 ΔrluD::cat | This work |
| MC365 | MC279 thr43::Tn10 | Laboratory collection |
| MC366 | MC279 serA764::Kan | Laboratory collection |
| MC367 | MG1655 ΔrluD::cat prfB12 (RF2 D131Y) | This work |
| MC368 | MG1655 ΔrluD::cat prfB13 (RF2 E157K) | This work |
| MC369 | MG1655 ΔrluD::cat prfB14 (RF2 E167K) | This work |
| MC370 | MG1655 ΔrluD::cat prfB15 (RF2 E172A) | This work |
| MC371 | MG1655 ΔrluD::cat prfB16 (RF2 Q290K) | This work |
| MC372 | MG1655 ΔrluD::cat prfB17 (RF2 A293T) | This work |
| MC373 | MG1655 ΔrluD::cat prfB18 (RF2 A293E) | This work |
| MC374 | MG1655 ΔrluD::cat prfB19 (RF2 A293V) | This work |
| MC375 | MG1655 ΔrluD::cat rds-2; not RF1/2/3 | This work |
| MC376 | MG1655 ΔrluD::cat prfC20 (RF3 T99I) | This work |
| MC377 | MG1655 ΔrluD::cat prfC21 (RF3 V119F) | This work |
| MC378 | MG1655 ΔrluD::cat prfC22 (RF3 V119G) | This work |
| MC379 | MG1655 ΔrluD::cat rds-2 prfC23 (RF3 L126P) | This work |
| MC380 | MG1655 ΔrluD::cat prfC24 (RF3 T30I) | This work |
| MC381 | MG1655 ΔrluD::cat prfC25 (RF3 D313Y) | This work |
| MC382 | MC279 prfB12 (RF2 D131Y) | This work |
| MC383 | MC279 prfB13 RF2 E157K | This work |
| MC384 | MC279 prfB14 (RF2 E167K) | This work |
| MC385 | MC279 prfB15 (RF2 E172A) | This work |
| MC386 | MC279 prfB16 (RF2 Q290K) | This work |
| MC387 | MC279 prfB17 (RF2 A293T) | This work |
| MC389 | MC279 prfB18 (RF2 A293E) | This work |
| MC390 | MC279 prfB19 (RF2 A293V) | This work |
| MC391 | MC279 prfC20 (RF3 T99I) | This work |
| MC392 | MC279 prfC21 (RF3 V119F) | This work |
| MC393 | MC279 prfC22 (RF3 V119G) | This work |
| MC394 | MC279 prfC23 (RF3 L126P) | This work |
| MC395 | MC279 prfC24 (RF3 T30I) | This work |
| MC396 | MC279 prfC25 (RF3 D313Y) | This work |
| MC397 | MC279 ΔrluD::cat | This work |
| MC398 | MC279 ΔrluD::cat prfB (E. coli B) | This work |
| MC399 | MC279 ΔrluD::cat prfB12 (RF2 D131Y) | This work |
| MC400 | MC279 ΔrluD::cat prfB 13(RF2 E157K) | This work |
| MC401 | MC279 ΔrluD::cat prfB14 (RF2 E167K) | This work |
| MC402 | MC279 ΔrluD::cat prfB15 (RF2 E172A) | This work |
| MC403 | MC279 ΔrluD::cat prfB16 (RF2 Q290K) | This work |
| MC404 | MC279 ΔrluD::cat prfB17 (RF2 A293T) | This work |
| MC405 | MC279 ΔrluD::cat prfB18 (RF2 A293E) | This work |
| MC406 | MC279 ΔrluD::cat prfB19 (RF2 A293V) | This work |
| MC407 | MC279 ΔrluD::cat prfC20 (RF3 T99I) | This work |
| MC408 | MC279 ΔrluD::cat prfC21 (RF3 V119F) | This work |
| MC409 | MC279 ΔrluD::cat prfC22 (RF3 V119G) | This work |
| MC410 | MC279 ΔrluD::cat prfC23 (RF3 L126P) | This work |
| MC411 | MC279 ΔrluD::cat prfC24 (RF3 T30I) | This work |
| MC412 | MC279 ΔrluD::cat prfC25 (RF3 D313Y) | This work |
| MC413 | CSH142 prfB (E. coli B) rluD::cat (from ME97) | This work |
| S. enterica strains | ||
| SL4213 | hsdL6 hsdSA29 (rLT− mLT+ rS− mS+) metA22 metE551 ilv-452 trpB2 xyl-404 rpsL120 galE496 Fels2−nml | Laboratory collection |
| TrpE91 | trpE91 | Laboratory collection |
| MS60 | trpE91 ΔrluD::cat | This work |
Unless stated, all E. coli strains are K-12, and S. enterica serovar Typhimurium strains are derived from LT2.
Media and standard genetic and molecular techniques.
Rich and minimal medium preparation, transductions with P1 and KB1 phages, preparation of competent cells, transformations, and Hfr crosses were carried out as described previously (4, 6, 23). Antibiotics were used at the following concentrations (in μg/ml): ampicillin at 200, kanamycin at 25, chloramphenicol at 25, and tetracycline at 12.5. Growth rates were carried out in LB medium at 37°C. β-Galactosidase activities were determined on cultures grown in minimal medium with necessary supplements and tetracycline as described previously (26). Electroporations were carried out in 0.1-cm cuvettes and a MicroPulser electroporator (Bio-Rad). Electrocompetent cells were prepared according to the literature supplied by the manufacturer. PCR amplifications followed standard protocols and were carried out with Taq polymerase (New England Biolabs, Ipswich, MA). Reactions were purified on Wizard columns (Promega, Madison WI), and DNA sequencing was carried out at the DNA core facility at the University of Missouri, Columbia, MO.
Detergent lysates of cells were prepared as described previously (11). 30S and 50S subunits were separated from 70S ribosomes and polysomes by centrifugation through linear 10 to 40% sucrose gradients in a Beckman SW28 rotor for 18 h at 17,000 rpm. The gradients were analyzed on an ISCO gradient fractionator connected to an ISCO UV5 detector.
Deletion of the rluD gene.
The entire rluD coding region was deleted in both E. coli and S. enterica using the method of Datsenko and Wanner (5). Plasmids pKD3 or pKD4 were employed as templates in PCRs using 70-mer primers that carried 50 bases of homology to the sequences upstream and downstream of the rluD coding region and 20 bases of complementarity to the FLP recombination target (FRT)-flanked antibiotic resistance elements in pKD3 and pKD4. PCR products generated with E. coli-derived primers were electroporated into either LE392 or MG1655 cells that had previously been transformed with the λ Red plasmid, pKD46, and grown in the presence of 10 mM arabinose to induce λ Red expression. Transformants were selected on chloramphenicol or kanamycin, as appropriate. The S. enterica strain SL4213 transformed with pKD46 was grown in the presence of 10 mM arabinose and electroporated with PCR products carrying Salmonella sequences and transformants selected as described above. Following purification, colonies were screened for replacement of rluD sequences with an antibiotic resistance element by whole-cell PCR, using primers flanking the rluD sequences used for recombination of antibiotic resistance elements. The deletion mutations were moved into new strains by transduction, using phages P1 and KB1, for E. coli and S. enterica, respectively.
Selection and identification of ΔrluD suppressors.
Faster-growing derivatives of the E. coli K-12 MG1655 ΔrluD::cat strain were isolated from platings of cultures that had been grown to saturation in rich broth. In all, 21 cultures were grown, and a single, faster-growing colony was recovered from each culture. The prfB gene was amplified from each faster-growing isolate and sequenced. Hfr mapping of faster-growing isolates that had wild-type prfB sequences was carried out as described, using the Hfr::Tn10 kit (37). Exconjugants were selected on medium containing both chloramphenicol and tetracycline. Exconjugants that had lost the suppressor allele were readily identified, based on the slow growth of these colonies on LB agar plates. Transductions with P1 lysates made on a thr-43::Tn10 strain were used to test if any suppressors mapped to the 99-min region of the chromosome, where prfC is located (3). Again, loss of the suppressor phenotype was scored by slow growth of the tetracycline-resistant transductants on LB agar plates. The prfC gene was then amplified and sequenced from all isolates that had wild-type prfB sequences.
All prfB and prfC mutations were transferred into the MC279 strain background by P1-mediated transduction. For prfB mutations, MC366, a derivative of MC279, which carries the serA764::Kan mutation linked to prfB, was transduced to serine independence with phage grown on each prfB mutant. For prfC mutations, the thr43::Tn10 strain MC365 was transduced to threonine independence with phage grown on each prfC mutant. Individual transductants were screened for the presence of the prfC or prfB mutations by determining if they yielded tiny or large colonies (the wild-type or ΔrluD suppressor phenotypes, respectively) in a subsequent transduction with phage prepared on a ΔrluD::cat strain. The prfC or prfB gene was then amplified and sequenced from putative (rluD+) suppressor-containing transductants to verify the presence of the mutation. Each single prfB or prfC mutant verified by DNA sequencing was subsequently transduced to chloramphenicol resistance with phage prepared on a ΔrluD::cat strain to generate isogenic ΔrluD prfB and ΔrluD prfC double mutants, respectively.
RESULTS
Isolation and identification of ΔrluD suppressors.
The entire coding region of rluD was deleted in strains MG1655 and LE392 and replaced with a chloramphenicol acetyltransferase (cat) cassette (5). Both ΔrluD::cat strains grew slowly on solid medium, and faster-growing derivatives invariably arose during growth in liquid medium. The faster growth of such derivatives appears to be due to suppressor mutations, since the rluD gene is still deleted in these strains and previous work has shown that faster-growing ΔrluD derivatives still lack h69 Ψ residues (13, 32). One such ΔrluD suppressor has been identified by Ejby et al. (9) as a mutation in prfB. However, since h69 interacts with many translation factors, it seemed possible that alterations in one or more of these other factors might also suppress the slow growth of ΔrluD strains. In an effort to explore the interaction of h69's Ψ residues with its ligands, 21 further ΔrluD suppressors were isolated. Of the strains carrying these mutations, 20 had wild-type growth rates, while the remaining suppressor mutant (MC375) (Table 2) grew faster than the parental MG1655 ΔrluD::cat strain but considerably slower than wild-type MG1655.
TABLE 2.
Effects of rluD and altered RF2 and RF3 termination factors on growth and stop codon readthrougha
| RF or RluD strain description | DT (min) | Readthrough (Miller units of β-galactosidase)b |
||||
|---|---|---|---|---|---|---|
| pSG34-11 UGA | pSG163 UAG | pSG853 UAA | pSG12-6 UAG | pSG25 WT | ||
| E. coli strains | ||||||
| WT (K-12) | 33 ± 3 | 131 ± 16 | 119 ± 20 | 9 ± 1 | 58 ± 2 | 31,517 ± 4,056 |
| RF2 D131Y | 26 ± 1 | 93 ± 8 | 109 ± 8 | 6 ± 1 | 85 ± 8 | 31,885 ± 3,052 |
| RF2 E157K | 31 ± 2 | 79 ± 3 | 105 ± 3 | 6 ± 1 | 75 ± 7 | 28,441 ± 2,273 |
| RF2 E167K | 30 ± 3 | 122 ± 21 | 113 ± 13 | 6 ± 1 | 70 ± 7 | 28,133 ± 1,982 |
| RF2 E172A | 32 ± 1 | 63 ± 4 | 104 ± 5 | 5 ± 1 | 76 ± 7 | 33,373 ± 2,838 |
| RF2 Q290K | 33 ± 3 | 84 ± 3 | 108 ± 11 | 6 ± 1 | 78 ± 11 | 29,232 ± 1,912 |
| RF2 A293T | 33 ± 2 | 115 ± 5 | 104 ± 10 | 7 ± 1 | 82 ± 11 | 30,610 ± 1,454 |
| RF2 A293E | 32 ± 1 | 468 ± 69 | 115 ± 20 | 11 ± 2 | 90 ± 10 | 31,375 ± 2,271 |
| RF2 A293V | 28 ± 2 | 104 ± 14 | 117 ± 7 | 6 ± 1 | 95 ± 15 | 28,329 ± 3,486 |
| RF2 A246 [B] | 32 ± 1 | 75 ± 14 | 107 ± 8 | 5 ± 1 | 57 ± 8 | 26,839 ± 3,471 |
| RF3 T30I | 33 ± 2 | 158 ± 20 | 316 ± 15 | 6 ± 1 | 189 ± 26 | 37,778 ± 4,182 |
| RF3 T99I | 30 ± 2 | 143 ± 6 | 192 ± 5 | 8 ± 1 | 90 ± 11 | 31,145 ± 4,858 |
| RF3 V119F | 27 ± 2 | 200 ± 14 | 238 ± 20 | 9 ± 1 | 182 ± 42 | 41,583 ± 816 |
| RF3 V119G | 31 ± 1 | 139 ± 29 | 201 ± 12 | 8 ± 1 | 101 ± 2 | 35,238 ± 1,997 |
| RF3 L126P | 28 ± 1 | 164 ± 7 | 178 ± 8 | 10 ± 2 | 98 ± 11 | 35,904 ± 2,951 |
| RF3 D313Y | 29 ± 2 | 163 ± 29 | 288 ± 16 | 9 ± 1 | 226 ± 20 | 33,311 ± 4,802 |
| ΔrluD::cat | 138 ± 13 | 1,997 ± 126 | 904 ± 80 | 95 ± 19 | 148 ± 14 | ND |
| RF2 D131Y ΔrluD::cat | 31 ± 2 | 39 ± 5 | 128 ± 10 | 6 ± 1 | 29 ± 2 | 39,999 ± 2,179 |
| RF2 E157K ΔrluD::cat | 39 ± 4 | 51 ± 5 | 176 ± 13 | 8 ± 1 | 35 ± 10 | 39,369 ± 3,567 |
| RF2 E167K ΔrluD::cat | 38 ± 1 | 62 ± 14 | 143 ± 5 | 6 ± 1 | 30 ± 3 | 42,370 ± 3,344 |
| RF2 E172A ΔrluD::cat | 38 ± 1 | 48 ± 5 | 160 ± 5 | 9 ± 1 | 32 ± 7 | 38,288 ± 3,731 |
| RF2 E290K ΔrluD::cat | 39 ± 3 | 71 ± 7 | 150 ± 4 | 9 ± 1 | 27 ± 1 | 41,604 ± 9,100 |
| RF2 A293T ΔrluD::cat | 37 ± 3 | 100 ± 13 | 232 ± 35 | 8 ± 2 | 33 ± 3 | 33,534 ± 6,247 |
| RF2 A293E ΔrluD::cat | 37 ± 2 | 426 ± 53 | 171 ± 6 | 6 ± 1 | 29 ± 3 | 27,631 ± 2,515 |
| RF2 A293V ΔrluD::cat | 37 ± 3 | 159 ± 15 | 263 ± 29 | 15 ± 1 | 41 ± 5 | 41,295 ± 8,044 |
| RF2 A246 [B] ΔrluD::cat | 32 ± 2 | 49 ± 5 | 133 ± 11 | 5 ± 1 | 26 ± 3 | 38,385 ± 2,687 |
| RF3 T30I ΔrluD::cat | 51 ± 4 | 196 ± 13 | 659 ± 60 | 15 ± 1 | 92 ± 11 | 34,934 ± 4,213 |
| RF3 T99I ΔrluD::cat | 40 ± 2 | 72 ± 6 | 274 ± 40 | 12 ± 2 | 27 ± 9 | 32,225 ± 4,354 |
| RF3 V119F ΔrluD::cat | 41 ± 4 | 141 ± 9 | 432 ± 20 | 12 ± 1 | 52 ± 14 | 32,526 ± 1,306 |
| RF3 V119G ΔrluD::cat | 34 ± 1 | 68 ± 5 | 263 ± 40 | 7 ± 1 | 30 ± 7 | 27,569 ± 2,019 |
| RF3 L126P ΔrluD::cat | 28 ± 2 | 88 ± 8 | 307 ± 13 | 9 ± 1 | 26 ± 5 | 29,208 ± 1,951 |
| RF3 D313Y ΔrluD::cat | 46 ± 4 | 179 ± 12 | 507 ± 30 | 18 ± 2 | 86 ± 5 | 28,229 ± 1,076 |
| S. enterica strains | ||||||
| WT | 32 ± 3 | 116 ± 7 | 361 ± 13 | 9 ± 2 | 83 ± 6 | 34,967 ± 2,116 |
| ΔrluD::cat | 36 ± 2 | 91 ± 4 | 291 ± 20 | 11 ± 3 | 36 ± 3 | 33,270 ± 2,260 |
[B] indicates the E. coli B prfB allele (RF2 A246). WT, wild type; DT, doubling time; ND, not determined (the slow-growing ΔrluD::cat strain was unable to maintain the wild-type lacZ plasmid pSG25).
Values are means ± standard deviations.
The prfB gene was sequenced in all 21 suppressor strains. Fifteen of the strains had single base changes in the coding region, resulting in amino acid changes at 6 different positions in RF2 (Table 1 and Fig. 1). Single isolates of E157K, A293T, and A293V mutant RF2, two independent isolates of each of the D131Y, E167K, E172A, and Q290K variants, and four independent isolates of the A293V mutant RF2 were recovered. To map the remaining suppressors, two suppressors with wild-type prfB genes were used as recipients in Hfr crosses. These experiments indicated that both strains carried suppressor mutations in the 98- to 6-min arc of the chromosome, between the origins of transfer of HfrC and HrfP4X (19). The prfC gene, encoding release factor RF3, is located in this region (99.3 min), close to the thr gene cluster at 0 min (3). To test if any of the remaining suppressors might be prfC alleles, 4 suppressor strains with wild-type prfB genes were used as recipients in P1 transductional crosses with a thr-43::Tn10 donor. Approximately 44% of the tetracycline-resistant transductants showed loss of the suppressor phenotype. Sequencing of the prfC gene from the 6 suppressor strains carrying wild-type prfB genes showed that 5 of these carried single base changes in the prfC coding region, resulting in amino acid changes at 4 different RF3 positions (T30I, T99I, V119F, V119G, and D313Y) (Table 1 and Fig. 1). The remaining suppressor strain, MC375, which grew at an intermediate rate, had wild-type prfA, prfB and prfC sequences, indicating that none of the termination factors was altered in this strain. The suppressor mutation in this strain was designated rds, for rluD suppressor. The weak suppressor phenotype of the MC375 strain prevented its use as a reliable marker in Hfr crosses, and the identity of this rds suppressor mutation has not yet been determined. However, a derivative of MC375 arose spontaneously on agar plates that had a completely wild-type growth rate, and sequencing of the prfB and prfC genes from this strain (MC379) showed that it carried an RF3 protein with an L126P substitution.
FIG. 1.
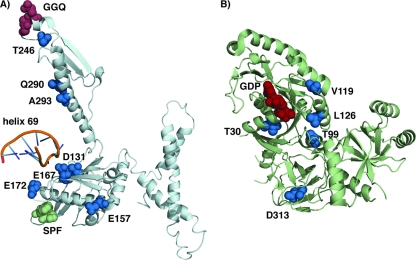
Crystal structures of RF2 bound to the T. thermophilus 70S ribosome (A) and RF3 (B). Residues altered in mutant factors are shown as blue spheres. The 23S rRNA helix 69 (orange) is indicated in relationship to RF2. The GGQ and SPF motifs in RF2 are shown as red and green spheres, respectively. GDP bound to RF3 is shown as red spheres. Figures were rendered with PyMol using pdb entries 2wh1 (A) and 2h5e (B).
All prfB and prfC mutations were transferred to a clean genetic background by P1-mediated transduction, and the prfB or prfC genes were sequenced to verify the presence of the mutant alleles in these strains. Subsequently, the ΔrluD::cat mutation was introduced into each prfB and prfC mutant to reconstruct the ΔrluD prfB and ΔrluD prfC double mutants. Growth rate determinations showed that while the ΔrluD::cat strain had a doubling time of at least 138 min, most of the ΔrluD prfB double mutants had doubling times close to the wild-type doubling time of 33 ± 3 min (Table 2). Four of the six RF3 mutants had doubling times slightly longer (40 to 51 min) than that of the wild type. Curiously, while the altered RF3 with the L126P substitution arose as a secondary suppressor, in the reconstructed strain, it suppressed the ΔrluD growth defect as well as any of the strongest prfB or prfC suppressor alleles (Table 2). All of the prfB and prfC single mutants grew at rates close to that of the wild type.
Inactivation of rluD has been show to affect 50S-subunit assembly and 30S-50S subunit association (14). Analysis of the subunit association properties of the ribosomes from wild-type and ΔrluD::cat strains and from the ΔrluD prfB and ΔrluD prfC double mutants on sucrose gradients showed that while ribosomes from the ΔrluD::cat strain had substantially increased levels of free subunits (Fig. 2), ribosomes from all of the ΔrluD prfB and ΔrluD prfC double mutants had wild-type levels of free subunits and 70S ribosomes (Fig. 2 and data not shown).
FIG. 2.
Ribosome profiles of wild-type strain, ΔrluD::cat strain, and ΔrluD::cat strains carrying representative prfB (RF2) and prfC (RF3) suppressor mutations or the prfB allele [B] from E. coli B. The direction of sedimentation is from left to right. The positions of 30S and 50S subunits and 70S ribosomes are indicated on the ΔrluD::cat strain profile.
Effects of mutant RFs on termination.
The effects of rluD, prfB, and prfC mutations on termination were examined by measuring the levels of stop codon readthrough, using a series of lacZ nonsense mutants. Readthrough of all three stop codons was increased substantially in the ΔrluD::cat mutant, relative to the wild-type strain (Table 2 and Fig. 3). The extent of the increases depended on the individual stop codon and the surrounding codon context. In the ΔrluD prfB and ΔrluD prfC double mutants, readthrough of all stop codons was less than that observed with the ΔrluD::cat strain and in many instances was close to or even less than that seen with wild-type cells.
FIG. 3.
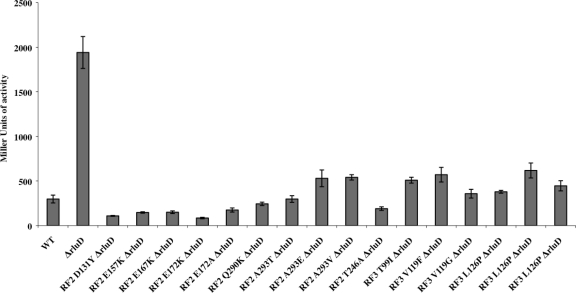
Effects of h69 Ψ modification and mutant release factors on UGA readthrough. Wild-type and mutant strains were transformed with plasmid pSG3/4 UGA, carrying a UGA mutation in the 5′ end of the coding region of lacZ. Synthesis of β-galactosidase requires readthrough of this UGA stop codon. Bars represent (Miller) units of β-galactosidase activity.
X-ray crystallographic and cryo-electron microscopy (EM) analyses of termination complexes have shown that release factors RF1, RF2, and RF3 interact with h69. Alterations to h69 structure thus have the potential to affect RF-ribosome interactions, and it is not hugely surprising that readthrough of all three stop codons is affected by the ΔrluD::cat mutation. RF3 recycles both RF1 and RF2 from the posttermination ribosome and effectively increases their availability to bind to termination complexes. Thus, the effects of prfC mutations on termination at all three stop codons in the ΔrluD prfC double mutants reported in Table 2 are consistent with the known interactions of RF3 with both RF1 and RF2. However, the effects of mutant RF2 on readthrough of UAG codons in ΔrluD prfB double mutants is unexpected. RF2 recognizes both UAA and UGA, but not UAG codons. Nonetheless, the readthrough of UAG, as well as UGA and UAA, codons seen with the ΔrluD::cat mutant is decreased substantially by altered RF2 in the ΔrluD prfB double mutants.
The effects of altered RF2 on UAG readthrough in ΔrluD prfB double mutants raises the possibility that these substitutions may alter recognition of the cognate stop codon by the mutant RF2. Consistent with this proposal, several of the RF2 mutations isolated by us and by Ejby et al. (9) as ΔrluD suppressors have previously been shown to affect recognition of cognate termination codons, when the mutant factors are overexpressed from high-copy-number plasmids (35). We have addressed the possible effects of altered RF2 factors on termination at UAG codons by asking if the mutant RF2s can assume UAG-decoding functions and render RF1 dispensable. The prfA1 (R137P) mutation renders RF1 temperature sensitive, and the mutant fails to grow at 42°C (15, 33). A series of prfA1 prfB double mutants was constructed by introducing prfB mutations into the prfA1 strain MC360. However, both the parental prfA1 strain and the prfA1 prfB derivatives were equally unable to grow at 42°C. We conclude that none of the mutant RF2s studied here has sufficient UAG-decoding activity to render RF1 dispensable.
We next asked if UAG decoding by a suppressor tRNA was affected by the altered RF2 proteins. Since decoding of a stop codon by a suppressor tRNA involves competition between the tRNA and a release factor, we reasoned that any UAG decoding by the mutant RF2 should decrease UAG reading by the tRNASer-derived suppressor, SupD. However, measurements of β-galactosidase activities in a series of isogenic strains carrying supD67, the UAG lacZ reporter plasmid pLG12-6, and selected prfB alleles showed that the UAG suppressor activity of supD67 was unaffected by any of the prfB mutations examined (data not shown). While these results cannot completely exclude near-cognate decoding by altered RF2, they suggest that at least when expressed from single-copy, chromosomal genes, any near-cognate decoding by the mutant RF2s occurs at a low level. Other explanations for the effects of altered RF2 on UAG readthrough are considered in Discussion.
Effects of strain background on the ΔrluD phenotype.
The experiments described above implicate termination as the main ribosomal activity affected by loss of h69 Ψ residues. However, the E. coli K-12 strains in which all the analyses of rluD function have been carried out already carry a variant RF2 with a threonine residue at position 246. This is in contrast to class I RFs in other bacteria, including other E. coli strains, that carry serine or alanine at this position (8). Moreover, the K-12 Thr246 variant of RF2 is distinctly less effective in termination, both in vivo and in vitro, than the Ala246 RF2 (8, 24). To address any potential contribution of the K-12 Thr246 RF2 to the ΔrluD phenotype, we have constructed K-12 derivatives carrying the E. coli B prfB allele (Ala246 RF2) and then inactivated the rluD gene in this genetic background. Strains MC279 and MC359 are isogenic, differing only in the identities of their prfB alleles. Growth of MC359, which carries the E. coli B prfB allele, is indistinguishable from that of MC279, which carries the K-12 prfB allele. Termination at UGA codons is detectably more efficient in MC359 than in MC279, as has previously been reported (24). However, in distinct contrast to MC279, introduction of the ΔrluD::cat mutation into the MC359 strain background had no effect on growth and did not promote increased readthrough of stop codons (Table 2). In addition, cell lysates from the Ala246 RF2 ΔrluD::cat strain showed normal levels of free subunits and 70S ribosomes (Fig. 2). In effect, the wild-type E. coli B prfB allele is as good a suppressor of the ΔrluD phenotype as any of the selected prfB or prfC suppressor mutations. To confirm this result, the K-12 prfB allele was replaced with the E. coli B prfB allele in two other K-12 strains, CSH142 and BW25113 (the parental strain of the Keio knockout collection [2]) to give strains MC361 and MC415, respectively. Inactivation of the rluD gene in MC361 or MC415 by introduction of the ΔrluD::cat allele constructed here (to give strains MC362 and MC416, respectively) similarly did not affect growth or ribosomal subunit association (not shown). A subsequent genetic cross showed that the slow-growth phenotype, characteristic of ΔrluD mutants could be regenerated in the ΔrluD::cat strain MC362 by reintroduction of the K-12 prfB allele. When MC362 was transduced to kanamycin resistance with phage prepared on a K-12 ΔserA764::kan strain (serA and prfB are tightly linked), approximately equal numbers of large and small transductants were recovered. The prfB gene from four small and four large transductants was amplified and sequenced. All four of the small transductants had inherited the K-12 prfB allele along with the linked serA764::kan marker, while the four large transductants had retained the E. coli B prfB allele. This experiment confirms that a fully functional (Ala246) RF2 protein alone can rescue the slow growth of rluD-inactivated strains and that no additional suppressor mutations are present in strain MC262.
While the pseudouridlylation of h69 in the ΔrluD::cat strains constructed here has not been examined directly, it has been examined previously with strains carrying an rluD gene interrupted by a cat cassette (32). The rluD::cat disruption allele studied by Raychaudhuri et al. (32) is also present in strain ME97 (9) and was introduced by P1 transduction into strains MC41 and MC361, carrying the E. coli K-12 and B prfB alleles, respectively. All MC41 chloramphenicol-resistant transductants were small and slow-growing and gave rise to faster-growing derivatives, while the MC361 transductants grew at a wild-type rate. Moreover, the effects of the rluD::cat insertion allele on ribosomal subunit association and readthrough of stop codons were identical to that promoted by the ΔrluD::cat deletion allele constructed in this study (data not shown). We conclude from these experiments that the ΔrluD::cat deletion allele constructed here, as well as the previously characterized rluD::cat disruption allele, interact identically with the K-12 and B prfB alleles. Together, these results suggest that the slow growth and ribosomal defects associated with the ΔrluD mutants reported here and by other laboratories are largely a consequence of a defective RF2 protein, present in K-12 strains of E. coli.
Deletion of rluD in Salmonella enterica.
To examine the effects of rluD loss in other bacteria, the rluD gene was inactivated in Salmonella enterica serovar Typhimurium. Like most bacteria, S. enterica has an RF2 with Ala at position 246. Isogenic rluD+ and ΔrluD::cat S. enterica strains both grew at similar rates in liquid medium (Table 2) and displayed similar levels of ribosomal subunits and 70S ribosomes on sucrose gradients (not shown). Readthrough of stop codons in both strains occurred at similar levels, with the exception of UAG readthrough, which appeared to be slightly more efficient in the ΔrluD::cat strain, at least in one UAG sequence context (pSG12-6) (Table 2). The lack of any strong phenotype associated with rluD inactivation in Salmonella strains resembles what is observed with E. coli strains carrying the Ala246 RF2 protein. Together, these results suggest that loss of h69 Ψ residues has little consequence in bacteria with fully active termination factors and that the previously reported phenotypes of ΔrluD mutants are a peculiarity of E. coli K-12 strains carrying a partially defective RF2 protein.
DISCUSSION
Pseudouridines have been detected, or inferred, at conserved positions in h69 in bacterial, archaeal, and eukaryal ribosomes. The number and locations of these modifications vary between organisms; E. coli, Bacillus subtilis, and Deinococcus radiodurans have Ψ at positions 1911, 1915, and 1917, and Ψ1915 is methylated in at least E. coli and D. radiodurans. Thermus thermophilus has Ψ at positions 1911 and 1917, Haloarcula marismortui has Ψ at 1915 and 1917, while Sulfolobus acidocaldarius has a single Ψ at position 1917 (7, 20, 22, 28). In Saccharomyces cerevisiae cytoplasmic ribosomes, Ψs are found at positions 1915 and 1917, as well as in the stem region of h69 at positions 1921 and 1923 (E. coli numbering) (18). Spectroscopic analyses of h69 oligonucleotides suggest that Ψ residues facilitate conformational changes in the RNA and influence its stability (1). Investigations by Gutgsell and colleagues suggested that h69 Ψ modifications are important for ribosome biogenesis and function (14). The identification of a suppressor mutation in the prfB gene by Ejby et al. (9) that suppressed all of the defects associated with rluD inactivation provided a link between termination of translation and h69 Ψ modifications. In the work reported here, the isolation of additional suppressor mutations in prfB and the identification of a distinct class of suppressors in the prfC gene encoding RF3, supports and extends the link between termination and h69 Ψ residues. However, the demonstration that the slow growth, impaired subunit association, and increased readthrough phenotypes associated with rluD inactivation are limited to E. coli K-12 strains carrying a partially defective RF2 protein suggests that wild-type ribosomes can tolerate loss of h69 Ψ modifications without compromising function. Thus, as is observed for the other Ψ modifications in 23S and 16S rRNAs, h69 Ψ modifications also appear to be largely dispensable in wild-type bacteria.
Effects of altered RF2 and RF3 on termination functions.
The altered RF2 (E172K) isolated by Ejby et al. (9) that suppressed the growth defects caused by RluD loss displayed more efficient termination of UGA codons than the corresponding wild-type factor, in rluD+ as well as ΔrluD::cat backgrounds. This result was interpreted in terms of a gain-of-activity mutation in RF2 that made the factor inherently more effective in termination on wild-type as well as on hypomodified ribosomes. All of the altered RF2 and RF3 release factors isolated here restore the stop codon to near-wild-type levels in the ΔrluD::cat strain, consistent with the proposal that defective termination underlies most, if not all, of the defects associated with loss of h69 Ψ residues. While most of the altered RF2 factors isolated here also terminate at UGA more effectively than wild-type RF2 in rluD+ strains, the three mutant RF2s with changes at Ala293 are distinct and display higher levels of UGA readthrough. In addition, in rluD+ strains, all of the mutant RF3 factors show modest increases in UAG and UGA readthrough levels. Thus, while these mutant RF2 and RF3 factors can improve the defective termination associated with undermodified ribosomes, not all the factors are inherently more efficient in termination functions on wild-type ribosomes. This suggests that these mutant factors may have a differential interaction with modified versus hypomodified ribosomes or may ameliorate a step in the termination reaction that is uniquely rate limiting in undermodified ribosomes.
Defective termination has also been proposed to underlie the defects in rRNA processing and ribosome assembly that result in the small amounts of unstable 70S ribosomes and the accumulation of precursor particles observed with ΔrluD strains (9). According to this model, increased readthrough affects synthesis of RNase III and the translational coupling mechanism regulating ribosomal protein synthesis. Our observation that ribosomal subunit assembly and association are restored to wild-type levels in the suppressor-containing strains is consistent with such a model.
The class II release factor RF3 removes the class I release factors RF1 and RF2 from the ribosome, following peptide release. In so doing, RF3 increases the amounts of class I factors available for binding to pretermination ribosomes and enhances the overall efficiency of termination. The crystal structure of RF3·GDP reveals that the protein is folded into three distinct domains (10). Domain I (residues 3 to 278) consists of a GTPase domain and “an EF-G-like G′ subdomain.” Five of the six mutants isolated here (the T30I, T99I, V119F, V119G, and L126P mutants) affect residues located in domain I, while the remaining mutant carries an alteration (D313Y) in a residue that is located in the interface between domains II and III. RF3 mutants with alterations in the G domain and domain III that suppressed the temperature sensitivity of both RF1 and RF2 mutants have been isolated by Matsumura et al. (21). Suppression of RF1 and RF2 temperature sensitivity likely occurs through enhanced recycling of class I release factors off the ribosome by the altered RF3, and this mechanism may also underlie suppression of the termination defects in the ΔrluD::cat strain. The most recent model for RF3-dependent recycling of class I release factors posits that GDP is exchanged for GTP upon binding of RF3·GDP to the ribosome. GTP binding affects the conformation of both RF3 and the ribosome and ultimately results in the release of the class I release factors (10). The location of many of the ΔrluD::cat suppressor mutations in domain I of RF3 suggests that these alterations may affect transitions between nucleotide-free, GDP- and GTP-bound forms of RF3 on the ribosome. The D313Y substitution analyzed here lies in the domain II-III interface, and Gao et al. (10) also showed that the adjacent H311A substitution affected GDP release from ribosome-bound RF3. Thus, the D313Y mutation is also positioned to affect nucleotide-RF3 interactions. Alteration of the GDP-to-GTP exchange rates on RF3 has the potential to affect its recycling of class I factors and thus the efficiency of termination.
Release factor RF2 is a four-domain protein. Domain 1 contributes to interactions with RF3, while domain 2 carries the SPF “tripeptide anticodon” that contributes to stop codon recognition. Domains 2 and 4 pack against one another to form a central core of the protein. Extending from this central core is domain 3 which contains the invariant GGQ motif that interacts with the peptidyltransferase center of the 50S subunit and is required for triggering hydrolysis (31). Residues D131, E157, E167, and E172 described here are in domain 2, while Q290 and A293 are in domain 3. None of the RF2 residues mutated here is seen to make direct contact with h69 in the crystal structure of T. thermophilus RF2 bound to the ribosome (38). The closest approaches are by E167 and D131, which are 6.63 and 6.36 Å away from Ψ1911 and C1914, respectively. However, P160 (equivalent to E172 of E. coli RF2) contacts R125 of protein S13. The same S13 residue, in turn, makes a backbone contact with A1913 in h69, and A1913 is believed to play a pivotal role in connecting stop codon recognition in the decoding center with activation of hydrolysis by the peptidyltransferase center during termination (16, 17). Thus, while none of the domain 2 residues studied here appears to make direct contacts with h69, they have the potential to perturb S13-RF2 contacts and, consequently, the interaction between S13 and h69.
In solution, the class I RFs can assume both closed and extended conformations (36). However, only in the extended conformation is it possible for the “tripeptide anticodon” and GGQ motifs of RF1 or RF2 to interact simultaneously with the ribosomal decoding and peptidyltransferase centers, respectively. X-ray crystal structures of RF1 or RF2 bound to the ribosome show that h69 interacts with a small “switch” loop (residues 308 to 322 in E. coli RF2), connecting domains 3 and 4. Two of the RF2 residues described here, Q290 and A293, lie in an adjacent helix (α7), and mutations in these residues conceivably may affect the orientation of the switch loop or the GGQ motif at either end of the α7 helix, upon ribosome binding.
UAG decoding in ΔrluD and ΔrluD prfB strains.
The increased readthrough of stop codons observed in ΔrluD::cat strains (Table 2) (9) indicates that posttranscriptional modifications of h69 affect the efficiency of termination, most likely by altering RF-ribosome interactions. However, among the 20 suppressors of the growth defect caused by rluD inactivation, no mutations affecting RF1 were recovered. Surprisingly, suppressor mutations in the release factor RF2 restored wild-type readthrough at the near-cognate UAG codon as well as the cognate UAA and UGA codons in ΔrluD::cat strains. While our genetic data cannot completely exclude the possibility that the mutant RF2s decode UAG codons, the data suggest that any such near-cognate reading occurs at low levels.
Mora et al. (24) have argued that one of the consequences of the Thr246 variant RF2 in K-12 strains is that UAA codons in these strains are read predominantly by RF1. If the interaction of the Thr246 RF2 with the ribosome is compromised by loss of h69 Ψ modifications, the burden of UAA decoding may shift even further onto RF1. Since UAA codons are the most commonly used termination codons, particularly in highly expressed genes, titration of the available RF1 at UAA codons will decrease the amount of RF1 available at the relatively rare UAG codons, promoting their readthrough by near-cognate tRNAs. Restoration of efficient RF2-dependent termination in our suppressor-containing strains would also have the effect of redistributing the RF1 available for UAG decoding. In summary, the Thr246 RF2 present in K-12 strains, because of its inherently weaker termination activity, may be uniquely sensitive to perturbation of ribosome-RF interactions, such as might be caused by loss of h69 Ψ residues. In addition, rluD inactivation in K-12 strains may also indirectly affect UAG readthrough, through titration of limiting RF1 by abundant UAA codons. The RF2 with Ala at position 246, as well as the wild-type RF1 and RF3 factors, are fully active and are thus more resilient to disruption of RF-ribosome interactions.
The prfB alleles in different strains of E. coli affect the phenotypes of rluD inactivation.
The K-12 strain of E. coli has been used extensively for genetic and biochemical studies since the 1940s. The prfB allele present in K-12 strains, encoding Thr at position 246, appears limited to K-12 strains and absent from other strains of E. coli, including the B strains and the C6 strain MRE600, which has been used extensively for biochemical analyses (8). Mora et al. (24) have argued that the truly wild-type E. coli prfB allele, such as that found in B strains, has an Ala at position 246 and that the K-12 prfB allele should be considered mutant. In in vitro termination assays, the Thr246 RF2 is less effective than the Ala246 RF2 (8). Here, we show that the previously reported growth and functional defects associated with rluD inactivation are eliminated when the Thr246 RF2 in K-12 strains is replaced with the Ala246 RF2 from E. coli B. In addition, inactivation of rluD in Salmonella, which also carries the more typical bacterial Ala246 RF2, does not affect growth, stop codon readthrough, or ribosomal subunit association. These results lead us to conclude that inactivation of rluD in bacteria carrying fully active release factors has little effect on cell physiology. It is of interest to note that Sato and Iino (34) have reported recently that inactivation of rluD in Bifidobacterium bifidum does not affect growth of this Gram-positive bacterium, consistent with our conclusion that rluD loss is tolerated in wild-type bacteria.
The systematic analyses of rRNA Ψ synthases in E. coli by Ofengand and Campo indicated that, with the notable exception of rluD, each of the rRNA Ψ synthase genes could be individually inactivated with little obvious effect on phenotype (29). In addition, a strain lacking multiple synthases (the ΔrluB ΔrluC ΔrluE ΔrluF strain) is only modestly affected in growth rate (29). The work presented here shows that when truly wild-type bacterial strains are used, rluD can also be inactivated with little apparent consequence, and this suggests that it may be now possible to construct strains lacking all rRNA Ψ residues. Experiments to test this possibility are in progress.
Acknowledgments
We are indebted to Michael Sørensen for supplying strains and to John Wertz of the Coli Genetic Stock Center for strains and assistance with allele numbers. Thanks are due to Jennifer Carr, Steen Pedersen, Jaanus Remme, and Michael Sørensen for their comments on the manuscript.
This work was supported by grant no. MCB0745025 from the National Science Foundation (to M.O.).
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Abeysirigunawardena, S. C., and C. S. Chow. 2008. pH-dependent structural changes of helix 69 from Escherichia coli 23S ribosomal RNA. RNA 14:782-792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berlyn, M. K. 1998. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boro, H., and J. E. Brenchley. 1971. A new generalized transducing phage for Salmonella typhimurium LT2. Virology 45:835-836. [DOI] [PubMed] [Google Scholar]
- 5.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis, R. W., D. Botstein, and J. R. Roth. 1980. Advanced bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 7.Del Campo, M., C. Recinos, G. Yanez, S. C. Pomerantz, R. Guymon, P. F. Crain, J. A. McCloskey, and J. Ofengand. 2005. Number, position, and significance of the pseudouridines in the large subunit ribosomal RNA of Haloarcula marismortui and Deinococcus radiodurans. RNA 11:210-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dinçbas-Renqvist, V., A. Engstrom, L. Mora, V. Heurgue-Hamard, R. Buckingham, and M. Ehrenberg. 2000. A post-translational modification in the GGQ motif of RF2 from Escherichia coli stimulates termination of translation. EMBO J. 19:6900-6907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ejby, M., M. A. Sorensen, and S. Pedersen. 2007. Pseudouridylation of helix 69 of 23S rRNA is necessary for an effective translation termination. Proc. Natl. Acad. Sci. U. S. A. 104:19410-19415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gao, H., Z. Zhou, U. Rawat, C. Huang, L. Bouakaz, C. Wang, Z. Cheng, Y. Liu, A. Zavialov, R. Gursky, S. Sanyal, M. Ehrenberg, J. Frank, and H. Song. 2007. RF3 induces ribosomal conformational changes responsible for dissociation of class I release factors. Cell 129:929-941. [DOI] [PubMed] [Google Scholar]
- 11.Gregory, S. T., C. A. Brunelli, J. S. Lodmell, M. O'Connor, and A. E. Dahlberg. 1998. Genetic selection of rRNA mutations. Methods Mol. Biol. 77:271-281. [DOI] [PubMed] [Google Scholar]
- 12.Gutgsell, N., N. Englund, L. Niu, Y. Kaya, B. G. Lane, and J. Ofengand. 2000. Deletion of the Escherichia coli pseudouridine synthase gene truB blocks formation of pseudouridine 55 in tRNA in vivo, does not affect exponential growth, but confers a strong selective disadvantage in competition with wild-type cells. RNA 6:1870-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gutgsell, N. S., M. Del Campo, S. Raychaudhuri, and J. Ofengand. 2001. A second function for pseudouridine synthases: a point mutant of RluD unable to form pseudouridines 1911, 1915, and 1917 in Escherichia coli 23S ribosomal RNA restores normal growth to an RluD-minus strain. RNA 7:990-998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gutgsell, N. S., M. P. Deutscher, and J. Ofengand. 2005. The pseudouridine synthase RluD is required for normal ribosome assembly and function in Escherichia coli. RNA 11:1141-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ito, K., M. Uno, and Y. Nakamura. 2000. A tripeptide ‘anticodon’ deciphers stop codons in messenger RNA. Nature 403:680-684. [DOI] [PubMed] [Google Scholar]
- 16.Korostelev, A., H. Asahara, L. Lancaster, M. Laurberg, A. Hirschi, J. Zhu, S. Trakhanov, W. G. Scott, and H. F. Noller. 2008. Crystal structure of a translation termination complex formed with release factor RF2. Proc. Natl. Acad. Sci. U. S. A. 105:19684-19689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Laurberg, M., H. Asahara, A. Korostelev, J. Zhu, S. Trakhanov, and H. F. Noller. 2008. Structural basis for translation termination on the 70S ribosome. Nature 454:852-857. [DOI] [PubMed] [Google Scholar]
- 18.Liang, X. H., Q. Liu, and M. J. Fournier. 2007. rRNA modifications in an intersubunit bridge of the ribosome strongly affect both ribosome biogenesis and activity. Mol. Cell 28:965-977. [DOI] [PubMed] [Google Scholar]
- 19.Low, K. B. 1987. Hfr strains of Escherichia coli K-12, p. 1134-1137. In F. C. Neidhardt, J. L. Ingraham, K. B. Low, B. Magasanik, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella typhimurium: cellular and molecular biology, vol. 2. American Society for Microbiology, Washington, DC. [Google Scholar]
- 20.Massenet, S., I. Ansmant, Y. Motorin, and C. Branlant. 1999. The first determination of pseudouridine residues in 23S ribosomal RNA from hyperthermophilic Archaea Sulfolobus acidocaldarius. FEBS Lett. 462:94-100. [DOI] [PubMed] [Google Scholar]
- 21.Matsumura, K., K. Ito, Y. Kawazu, O. Mikuni, and Y. Nakamura. 1996. Suppression of temperature-sensitive defects of polypeptide release factors RF-1 and RF-2 by mutations or by an excess of RF-3 in Escherichia coli. J. Mol. Biol. 258:588-599. [DOI] [PubMed] [Google Scholar]
- 22.Mengel-Jørgensen, J., S. S. Jensen, A. Rasmussen, J. Poehlsgaard, J. J. Iversen, and F. Kirpekar. 2006. Modifications in Thermus thermophilus 23 S ribosomal RNA are centered in regions of RNA-RNA contact. J. Biol. Chem. 281:22108-22117. [DOI] [PubMed] [Google Scholar]
- 23.Miller, J. H. 1991. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Mora, L., V. Heurgue-Hamard, M. de Zamaroczy, S. Kervestin, and R. H. Buckingham. 2007. Methylation of bacterial release factors RF1 and RF2 is required for normal translation termination in vivo. J. Biol. Chem. 282:35638-35645. [DOI] [PubMed] [Google Scholar]
- 25.O'Connor, M. 2007. Interaction between the ribosomal subunits: 16S rRNA suppressors of the lethal DeltaA1916 mutation in the 23S rRNA of Escherichia coli. Mol. Genet. Genomics 278:307-315. [DOI] [PubMed] [Google Scholar]
- 26.O'Connor, M., and A. E. Dahlberg. 1995. The involvement of two distinct regions of 23 S ribosomal RNA in tRNA selection. J. Mol. Biol. 254:838-847. [DOI] [PubMed] [Google Scholar]
- 27.O'Connor, M., C. L. Thomas, R. A. Zimmermann, and A. E. Dahlberg. 1997. Decoding fidelity at the ribosomal A and P sites: influence of mutations in three different regions of the decoding domain in 16S rRNA. Nucleic Acids Res. 25:1185-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ofengand, J., and A. Bakin. 1997. Mapping to nucleotide resolution of pseudouridine residues in large subunit ribosomal RNAs from representative eukaryotes, prokaryotes, archaebacteria, mitochondria and chloroplasts. J. Mol. Biol. 266:246-268. [DOI] [PubMed] [Google Scholar]
- 29.Ofengand, J., and M. D. Campo. 2004. Modified nucleosides in Escherichia coli ribosomal RNA. In R. Curtiss, EcoSal—Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [DOI] [PubMed]
- 30.Ofengand, J., A. Malhotra, J. Remme, N. S. Gutgsell, M. Del Campo, S. Jean-Charles, L. Peil, and Y. Kaya. 2001. Pseudouridines and pseudouridine synthases of the ribosome. Cold Spring Harb. Symp. Quant. Biol. 66:147-159. [DOI] [PubMed] [Google Scholar]
- 31.Petry, S., A. Weixlbaumer, and V. Ramakrishnan. 2008. The termination of translation. Curr. Opin. Struct. Biol. 18:70-77. [DOI] [PubMed] [Google Scholar]
- 32.Raychaudhuri, S., J. Conrad, B. G. Hall, and J. Ofengand. 1998. A pseudouridine synthase required for the formation of two universally conserved pseudouridines in ribosomal RNA is essential for normal growth of Escherichia coli. RNA 4:1407-1417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rydén, S. M., and L. A. Isaksson. 1984. A temperature-sensitive mutant of Escherichia coli that shows enhanced misreading of UAG/A and increased efficiency for some tRNA nonsense suppressors. Mol. Gen. Genet. 193:38-45. [DOI] [PubMed] [Google Scholar]
- 34.Sato, T., and T. Iino. 2010. Genetic analyses of the antibiotic resistance of Bifidobacterium bifidum strain Yakult YIT 4007. Int. J. Food Microbiol. 137:254-258. [DOI] [PubMed] [Google Scholar]
- 35.Uno, M., K. Ito, and Y. Nakamura. 2002. Polypeptide release at sense and noncognate stop codons by localized charge-exchange alterations in translational release factors. Proc. Natl. Acad. Sci. U. S. A. 99:1819-1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vestergaard, B., S. Sanyal, M. Roessle, L. Mora, R. H. Buckingham, J. S. Kastrup, M. Gajhede, D. I. Svergun, and M. Ehrenberg. 2005. The SAXS solution structure of RF1 differs from its crystal structure and is similar to its ribosome bound cryo-EM structure. Mol. Cell 20:929-938. [DOI] [PubMed] [Google Scholar]
- 37.Wanner, B. L. 1986. Novel regulatory mutants of the phosphate regulon in Escherichia coli K-12. J. Mol. Biol. 191:39-58. [DOI] [PubMed] [Google Scholar]
- 38.Weixlbaumer, A., H. Jin, C. Neubauer, R. M. Voorhees, S. Petry, A. C. Kelley, and V. Ramakrishnan. 2008. Insights into translational termination from the structure of RF2 bound to the ribosome. Science 322:953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yusupov, M. M., G. Z. Yusupova, A. Baucom, K. Lieberman, T. N. Earnest, J. H. Cate, and H. F. Noller. 2001. Crystal structure of the ribosome at 5.5 A resolution. Science 292:883-896. [DOI] [PubMed] [Google Scholar]