Abstract
SecA is the ATPase that acts as the motor for protein export in the general secretory, or Sec, system of Escherichia coli. The tetrameric cytoplasmic chaperone SecB binds to precursors of exported proteins before they can become stably folded and delivers them to SecA. During this delivery step, SecB binds to SecA. The complex between SecA and SecB that is maximally active in translocation contains two protomers of SecA bound to a tetramer of SecB. The aminoacyl residues on each protein that are involved in binding the other have previously been identified by site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy; however, that study provided no information concerning the relative orientation of the proteins within the complex. Here we used our extensive collection of single-cysteine variants of the two proteins and subjected pairwise combinations of SecA and SecB to brief oxidation to identify residues in close proximity. These data were used to generate a model for the orientation of the two proteins within the complex.
The general secretory, or Sec, system of Escherichia coli exports proteins from the cytoplasm across the inner membrane to the periplasm or to the outer membrane (4, 13). The system is able to export only proteins that have not achieved their final stably folded state. SecB is a small tetrameric cytoplasmic chaperone organized as a dimer of dimers that captures the precursors of some exported proteins before they acquire stable, folded structures and delivers them to SecA for export (for a review, see the work by Randall and Hardy [15]). In the course of delivery, SecB, with the precursor bound, forms a ternary complex with SecA. SecA is the translocation ATPase that interacts with the translocon SecYEG, a pore through the inner membrane. Using the energy of binding and hydrolysis of ATP, SecA initiates the translocation of precursors to the periplasm (5, 6). The complex that is maximally efficient in coupling ATP hydrolysis to translocation comprises two protomers of SecA and one tetramer of SecB (11).
The interaction of SecA and SecB involves multiple areas of contact. One binding site is between the two carboxy-terminal regions of SecA comprising 21 residues that contain zinc and the flat eight-stranded β-sheets on each of the dimers of SecB (referred to as the side sites) (7, 14, 19). A second binding interaction is between the 13 carboxy-terminal residues of SecB (referred to as the tail sites) and the amino terminus of SecA (14, 16). The active wild-type complex between SecA and SecB has a stoichiometry of two protomers of SecA bound to a tetramer of SecB (referred to as the A2:B4 complex). If interaction at the side sites is eliminated by mutation or specific competition, the two proteins still bind to each other, but the stoichiometry is now one protomer of SecA bound to a tetramer of SecB, i.e., A1:B4 (14), a complex which is inefficient in translocation (11). The loss of only one protomer of SecA from the A2:B4 complex when the side sites are eliminated shows that the binding between these two symmetrical proteins is asymmetric. Previous studies using site-directed spin labeling and electron paramagnetic resonance (EPR) spectroscopy have identified regions of the surface of each protein that interact within the complex (1, 2). However, despite all of these findings we were not able to deduce unambiguously how the two proteins are oriented relative to each other. Here we have used the extensive collection of single-cysteine variants that was created for the spin-labeling studies to generate a model for the relative orientation of the two proteins within the complex.
MATERIALS AND METHODS
Mutagenesis and protein purification.
The single-cysteine variants of SecA and SecB were made by standard recombinant DNA techniques (QuikChange, Stratagene) as described previously (1, 2). The cysteine substitutions were constructed in two different species of SecA. One, SecAC4 (obtained from Donald Oliver), has all four native cysteines replaced with serines and forms a complex with SecB containing a single protomer of SecA and a tetramer of SecB (A1:B4) (14). The other, SecAC98S, retains the three native cysteines that coordinate zinc and has only the native cysteine at position 98 replaced with serine. Thus, SecAC98S forms a complex with SecB containing two protomers of SecA (A2:B4). The cysteines in SecAC98S which coordinate the zinc did not form cross-links with any of the single-cysteine variants of SecB (data not shown). Proteins were purified as previously described (1, 14) and were stored at −80°C in 10 mM HEPES (KOH), 300 mM potassium acetate (KOAc), 2 mM tris(2-carboxyethyl)phosphine (TCEP), pH 7.6. Concentrations of the proteins were determined spectrophotometrically using extinction coefficients of 78,900 M−1cm−1 for the SecA monomer and 47,600 M−1cm−1 for the SecB tetramer.
Assays of activity.
The cysteine variants of SecA and SecB used here were constructed for previous EPR studies and were tested for activity as published (1-3). Here we briefly summarize that characterization. All variants of SecB and all variants of SecA were subjected to analyses by high-performance liquid chromatography (HPLC) on a TSK3000SW (TosoHaas) size exclusion column to ascertain that introduction of cysteine did not disrupt structure or function (data not shown). The absolute molar mass of proteins was determined directly using static light scatter by passing the column eluent through a multiangle laser static light scatter detector followed by a differential refractometer (DAWN-EOS and Optilab rEX, respectively; Wyatt Technology, Inc.). All SecA variants were folded, demonstrated a monomer-dimer equilibrium, and were shown to bind SecB (see the work by Cooper et al. for examples [1]). All species of SecB eluted as a uniform peak of tetrameric SecB (molar mass of ∼70 kDa). SecB species were shown to form complexes with a natural ligand, unfolded precursor galactose-binding protein, as well as with SecA. All complexes were of the expected molar mass.
Oxidative cross-linking of SecA and SecB.
The reducing agent, TCEP, was removed from the single-cysteine variants of SecB and SecA, and the buffer was exchanged for 10 mM HEPES (KOH), 300 mM NaCl, pH 7.6, by using PD SpinTrap G-25 columns (GE Healthcare). Mixtures of the two proteins (15 μl) were prepared on ice to give concentrations of 7 μM SecB tetramer and 7 μM SecA protomer for SecAC4 variants, which form the complex A1:B4, or concentrations of 7 μM SecB tetramer and 14 μM SecA protomer for SecAC98S variants, which form the complex A2:B4. A sample of the mixture (4 μl) was removed into 36 μl of nonreducing sample buffer containing 1% SDS, 8 mM N-ethylmaleimide, and 12 mM EDTA for SDS-polyacrylamide gel electrophoresis. Copper phenanthroline was added to the remainder of the mixture to a final concentration of 0.1 mM. After 30 s on ice, 4.5 μl was pipetted into 36 μl nonreducing sample buffer with EDTA to terminate the reaction. Experiments showed that by 30 s of oxidation with copper phenanthroline the quantity of cross-linked complex had reached a maximum, and if 12 mM EDTA was added before the oxidizing agent, no cross-linking was observed. The samples were incubated at 100°C for 5 min and analyzed by SDS-polyacrylamide (13%) gel electrophoresis. It is important to carry out the oxidative cross-linking as soon as possible after removal of TCEP. Storage overnight in the absence of TCEP often resulted in formation of cysteine sulfoxide or other oxidized products that cannot form disulfide bonds.
SDS-polyacrylamide gel electrophoresis and immunoblotting.
All electrophoresis was carried out on 13% polyacrylamide gels (17). Immunoblotting using antisera to SecB and to SecA and the chromogenic dye 4-chloro-1-naphthol for detection was carried out to confirm the identity of the cross-linked proteins.
Analyses of the results.
After staining and destaining, the gels were photographed using a Kodak EDAS 290 digital camera. The bands containing SecA were quantified using the 1D gel analysis program in TotalLab from Nonlinear Dynamics, Ltd. The quantity of stain in the band containing the cross-linked complex was expressed as a percentage of the total stain in all the bands containing SecA. We can neglect the amount of stain in the cross-linked adduct that is due to SecB: the molar mass of SecA is six times that of SecB, and each unit weight of SecA binds more than twice the amount of Coomassie blue as does SecB. Thus, the fraction of stain due to SecB in the adduct band is less than 8%. Recovery of stain in bands containing SecA in the oxidized mixture averaged 91% of that in the band containing SecA in the lane for which the sample had not been exposed to copper phenanthroline.
Generation of models of orientation.
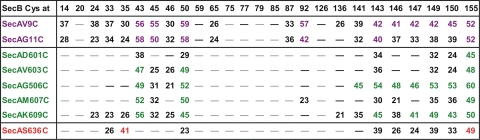
In generating models of the orientation of SecA and SecB in complex, we considered pairs that displayed cross-links of 40% or more. This empirical criterion was used to avoid including random, collisionally induced cross-links. We did not use the results of cross-linking to residue 155 in generating a model of relative orientation of the two proteins, since this residue on each protomer is at the end of a flexible region that if extended could reach as far as 35 Å. This long tether to the sulfhydryl could result in the apparent lack of specificity in cross-linking (Fig. 1).
FIG. 1.
Cross-linking of complexes between cysteine variants of SecA and SecB. Cross-linking values were estimated as described in Materials and Methods. Values of 40% cross-linking or more are shown in purple if the cross-linking partner lies in the SecA amino-terminal region, in green if the partner is located in the SecA linker helix, and in red if the partner lies in the SecA helical scaffold domain. Values between 21% and 39% are black, and those less than 20% are indicated by “—.” Values between 16% and 20% are dark black dashes, and those less than 15% are light gray dashes. All values are the average of between 2 and 5 cross-linking experiments. The average number of cross-linking experiments used to calculate each percentage greater than 40 shown in the figure was 3.1 (41 values obtained from 129 cross-linking experiments).
Generation of movie.
The movie in the supplemental material was generated using the eMovie plugin (8) within the PyMOL molecular graphics system (http://www.pymol.org).
RESULTS AND DISCUSSION
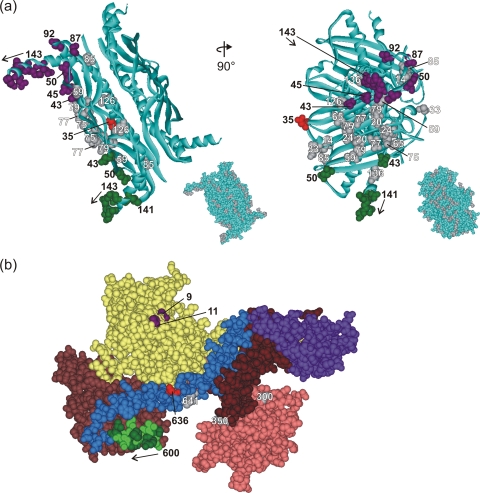
The cytoplasmic chaperone SecB is a tetramer of identical polypeptides, each 155 residues in length (Fig. 2 a). SecA in solution equilibrates between monomer and dimer. The protomers each contain 901 residues (Fig. 2b). The subdomains of SecA include two nucleotide binding folds, NBF1 (Fig. 2b, yellow, residues 1 to 220 and 378 to 420) and NBF2 (Fig. 2b, light brown, residues 421 to 599), a precursor binding domain (PBD) (Fig. 2b, pink, residues 221 to 377), a long α-helix, the helical scaffold domain (HSD) (Fig. 2b, blue, residues 610 to 668), a helical wing domain (Fig. 2b, purple, residues 669 to 755), an intramolecular regulator, IRA1 (Fig. 2b, dark brown, residues 756 to 835), and a short 10-residue α-helix (residues 600 to 609), the linker helix (Fig. 2b, green), that connects NBF2 to HSD. The amino-terminal residues 1 to 8 were deleted from the protein that was crystallized, and the carboxy-terminal domain, residues 836 to 901, was not resolved in the structure. EPR spectroscopy studies have shown that upon binding to SecB, several amino acid residues of SecA become constrained in their mobility, indicating close contact between the two proteins in those regions. The constrained residues lie near the amino terminus as well as in the linker helix and in the helical scaffold domain (1). Here we used pairwise combinations of single-cysteine variants of the two proteins to attempt to produce a disulfide cross-link between them. If such a cross-link was readily and abundantly formed, the two sulfhydryl groups in the proteins were deemed to be in close proximity within the complex. The data obtained, together with the X-ray structures of the proteins, were used to deduce the orientation of SecB relative to SecA in the complex.
FIG. 2.
Structure of SecA and SecB. (a) Structure of tetrameric SecB in ribbon representation from Haemophilus influenzae (Protein Data Bank [PDB] code 1FX3). The Haemophilus structure is used here instead of that of E. coli SecB to illustrate the C-terminal residues 141 to 155, which are not resolved in the E. coli structure (PDB code 1QYN). The numbered CPK models of side chains are those residues used in this study. The residues that showed cross-linking are colored corresponding to the cross-linking partner in SecA. The gray residues showed no cross-linking. The inset shows SecB in CPK representation with all residues used in this study shown in gray. The images on the right correspond to a 90° rotation around the y axis of the images on the left. (b) Structure of SecA in CPK representation with the subdomains in color as described in the text. The residues that showed cross-links are shown in purple (V9 and G11), light green (601, 603, 605, 607, and 609), and red (636). The gray residues did not cross-link. Residue 827 is not shown because it lies in the IRA1 domain (brown) and is masked by the helical wing domain (purple). PDB code 2FSF with the PBD model based on the B. subtilis SecA PDB code 1TF5.
The 26 single-cysteine variants of SecB selected for these studies are distributed over the entire surface of the protein (Fig. 2a; note that SecB is a tetramer; thus, 104 surface positions were tested). The 12 single-cysteine variants of SecA included representatives from several domains as shown in Fig. 2b. To ascertain that introduction of cysteine did not disrupt structure or function, each variant of both SecA and SecB was shown to behave normally when subjected to analyses by column chromatography in combination with static light scatter (data not shown; see Materials and Methods for details).
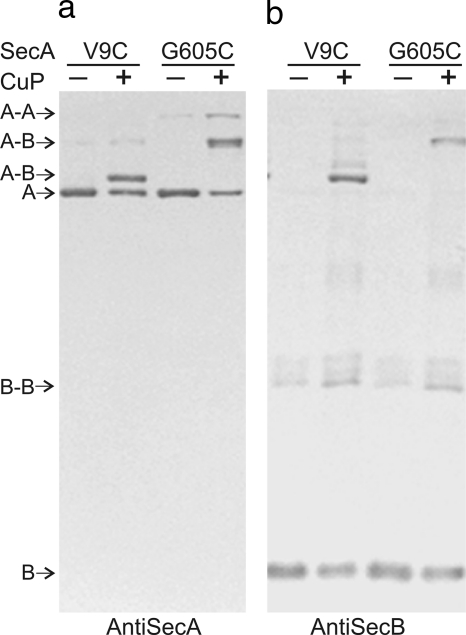
It was important, when attempting to form disulfide bonds between SecA and SecB in complex, to avoid random collisional disulfide bond formation that would lead to misinterpretation of the distance between cysteines. To this end, the oxidative reaction catalyzed by copper phenanthroline was carried out on ice and terminated after 30 s. The products were subjected to SDS-polyacrylamide gel electrophoresis and immunoblotting to identity the cross-linked species. An example of an immunoblot is shown in Fig. 3. Complexes between two variants of SecA, V9C and G605C, and one variant of SecB, Q50C, were oxidized, and the resulting gels were blotted with either antiserum to SecA or antiserum to SecB. The bands that were detected by both antisera are the cross-linked species of interest. Small quantities of dimers of both SecA and SecB were also identifiable as were unreacted protomers of each protein. The mobility of the cross-linked product between SecA and SecB depends on the position of the single cysteine within SecA. SecB cross-linked near the extreme N terminus of SecA, at positions 9 or 11, forms an adduct that displays a higher mobility than does SecB cross-linked near the middle of SecA (compare V9C with G605C, Fig. 3). This is a consequence of the latter having a greater hydrodynamic volume and thereby a lower mobility.
FIG. 3.
Identification of cross-linked species. Immunoblots of unoxidized (−) and oxidized (+) mixtures of single-cysteine variant Q50C of SecB and either variant V9C or variant G605C of SecA. (a) Blotted with antiserum to SecA; (b) blotted with antiserum to SecB. The positions of protomers and dimers of SecA (“A”) and SecB (“B”) as well as those of cross-linked species of SecA (“A-A”), SecB (“B-B”), and SecA with SecB (“A-B”) are indicated.
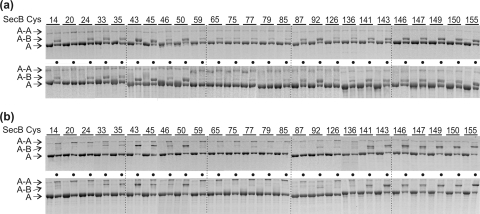
To determine whether SecA and SecB interact with the same relative orientation in the complexes having different stoichiometries, we used single-cysteine variants of two species of SecA, one of which forms the A2:B4 complex and the other of which forms A1:B4. The same pattern of cross-linking with all the variants of SecB was seen when the equivalent variant of each species of SecA was used to form complexes with SecB. Two examples are shown in Fig. 4. Thus, we conclude that a SecB tetramer interacts in the same orientation with a protomer of SecA whether or not the complex formed contains one or two protomers of SecA. EPR studies support this conclusion by showing that spin-labeled residues in SecA responded in the same way to the addition of SecB whether the complex formed was A1:B4 or A2:B4.
FIG. 4.
The same pattern of cross-linking is obtained from both A1:B4 and A2:B4 complexes. (a) SecA variant V9C in an A1:B4 complex (top) or an A2:B4 complex (bottom) with every variant of SecB without (left lane of numbered lanes, no dot) or with (right lane of numbered lanes, dot) oxidation. (b) SecA variant G605C subjected to the same treatment. Only the portion of the gels that has the SecA-containing bands is shown. The positions of the protomer of SecA (“A”), cross-linked protomers of SecA and SecB (“A-B”), and the cross-linked dimer of SecA (“A-A”) are indicated.
All possible pairwise combinations of SecB and SecA single-cysteine variants were subjected to oxidative cross-linking. Four of the SecA variants (S300C, S350C, I641C, and S827C) did not form detectable amounts of cross-links with any of the SecB variants (data not shown). The data for the remaining eight SecA variants are summarized in Fig. 1. The extent of cross-linking is estimated by calculating the quantity of stain in the adduct as the percentage of the total stain in the bands containing SecA. Values of less than 20% cross-linking are indicated by a dash. The data clearly show that some pairs are more readily cross-linked than others, indicating greater proximity of the two cysteines in the complex of SecB with SecA.
We used only pairs of variants that displayed robust cross-linking, greater than 40% (Fig. 1, purple and green), to generate the models of the orientations of SecB on SecA that are shown in Fig. 5. Many of the pairs of cysteines that undergo abundant cross-linking involve at least one cysteine that lies within a flexible region of the protein (Fig. 1). For example, the last 13 residues of SecB (residues 143 to 155) that participate in 23 of the 41 values of 40% lie in regions that are intrinsically disordered as shown by nuclear magnetic resonance (18) and EPR studies (3, 10). The amino terminus of SecA, which participates in 16 of the most abundant cross-links, is likely to be flexible since in the three X-ray structures of SecA from Bacillus subtilis the N-terminal 15 residues have different conformations. In one, the sequence lies at the dimer interface (9), in a different dimer one protomer has the N terminus extended, and in the other protomer it is unresolved (20). In the structure of B. subtilis SecA as a monomer, the N terminus forms two turns of a helix and is then extended (12). The observed cross-linking between the flexible amino terminus of SecA and the flexible carboxyl terminus of SecB agrees with our recent demonstration by isothermal titration calorimetry of their direct interaction (16). This study used a synthetic peptide corresponding to residues 2 through 11 of the SecA amino terminus and a series of SecB species with progressively longer deletions of the C terminus to demonstrate interaction of the peptide with the C-terminal 13 amino acid residues of SecB. The data presented here are the first demonstration that the C-terminal residues of SecB have a second site of interaction: the short linker helix of SecA, comprising residues 600 to 609. This stretch of residues is also likely to be flexible. The equivalent sequence in the crystal structure of the SecA dimer from Bacillus subtilis exists in one protomer as an α-helix and as a β-strand in the other protomer (20), indicating sufficient flexibility to assume different conformations.
FIG. 5.
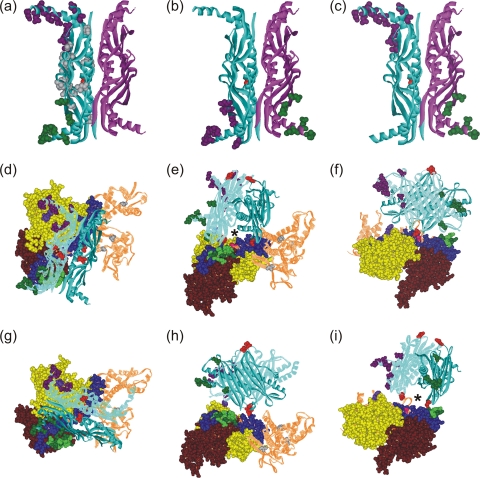
Models of the orientation of SecB and SecA within a complex. SecB residues that cross-link to SecA shown as CPK models colored according to the cross-linking partner on SecA: purple for SecAV9 and G11, green for residues on SecA between 600 and 609. (a) SecB with both protomers involved in contact on the same dimer; (b and c) each of the dimers contains one protomer involved in contacts, directly across the dimer interface (b) or diagonally opposite (c). Two orientations that satisfy all cross-links are shown: one in panels d to f and a second in panels g to i. The SecA protomer has the subdomains that cross-link to SecB in CPK representation: nucleotide binding folds NBF1 (yellow) and NBF2 (light brown), the precursor binding domain (PBD; pink), the helical scaffold domain (HSD; blue), and the linker helix (green). The remaining domains that show no contacts, the helical wing domain and the intramolecular regulator, are shown as brown lined ribbons. The residues that show cross-links are purple (V9 and G11), light green (601, 603, 605, 607, and 609), and red (636). A second protomer could be positioned on the upper surface through symmetrically related residues on the protomers of SecB. The red CPK residues (SecB35) are the sites that contact the long blue helix at residue SecA636 (red CPK). An asterisk marks the channel that would be occupied by a precursor polypeptide as described in the text.
The 4-fold symmetry of SecB, which is a tetramer organized as a dimer of dimers, introduces difficulty in determining the orientation of SecB bound to SecA. In a complex with a stoichiometry of A2:B4, we assume that one protomer of SecA interacts with two protomers of a SecB tetramer (A1:B2). Most of the residues on SecB that cross-link to greater than 40% (residues 43 and 50 and residues 143 through 150) have multiple binding partners. Any one of the residues on SecB shows cross-linking both to residues near the amino terminus (residues 9 and 11, purple numbers in Fig. 1) and to residues in the linker helix (residues 600 to 609, green numbers in Fig. 1). Since the two areas of SecA are well separated in space (Fig. 2b), it is most likely that the cross-linking partners on SecB lie in different protomers. However, the two protomers of SecB that are involved in contacts can be arranged in three possible ways (Fig. 5a, b, and c). They could both be in the same dimer of the SecB dimer of dimers (Fig. 5, one dimer is blue ribbon, and the other is magenta ribbon), or each of the dimers could contain one protomer that makes contacts with SecA, and these SecB protomers could be either directly across the dimer-dimer interface (Fig. 5b) or diagonally opposite each other (Fig. 5c). In each of these possible arrangements, the cysteines that have been shown to cross-link are depicted as Corey-Pauling-Koltun (CPK) models in purple if the cross-linked partner is in the N-terminal region of SecA and in green if the partner lies in the linker helix of SecA (Fig. 5d).
Determination of the orientation of SecA bound to SecB is made difficult not only by the 4-fold symmetry of SecB but also by the flexibility and multiple binding partners of the C-terminal tails of SecB. A crucial observation in developing a model is that of all the residues tested that are in well-structured regions and that cross-linked to greater than 40%, only residue 35 in SecB, which is at the edge of the eight-stranded β-sheet, has a single binding partner, residue 636 of SecA, which is in the long helical scaffold domain (Fig. 5, both residues are shown as red CPK models). This singularity provides a reference point for orienting a SecB dimer on a SecA protomer. Using the contact between residues 636 of SecA and 35 of SecB as a pivot point, we rotated the tetramer of SecB relative to SecA to bring the C termini of the SecB protomers into close proximity to their observed cross-linking partners. There are two possible orientations that satisfy all the proximities deduced from the cross-linking data (Fig. 5d to f and g to i; the movie in the supplemental material depicts the orientation of the model in Fig. 5d). In one, the C-terminal tails of SecB that are involved in contact are on the same dimer (Fig. 5d). In the other, the protomers involved in contact are directly across the dimer interface (Fig. 5g). Comparison of Fig. 5d and g shows that the axis of the dimer-of-dimer interface of SecB is rotated 90° relative to the underlying SecA in the two orientations.
Figure 5e and i show that in a complex of SecA2:SecB4, a second protomer of SecA could bind on the upper surface of SecB in either orientation through the identical symmetrically related contacts on the unoccupied protomers of SecB. The only residue on these protomers shown as a CPK representation is residue 35 (red), which is the contact to residue 636 of SecA. It is possible that in a SecA2:SecB4 complex one protomer of SecA might bind as illustrated in Fig. 5d and the second as illustrated in Fig. 5g, resulting in a 90° rotation of one protomer relative to the other. It is clear that in all of the proposed models no dimer interface could form between the protomers of SecA since they are separated by SecB, which is bound between them.
The documented contact between the zinc-containing motif in the C-terminal 21 aminoacyl residues of SecA and the flat eight-stranded β-sheet on the dimer of SecB (7, 14, 19) is not illustrated in Fig. 5 because the last 65 residues of SecA were not resolved in the crystal structure. Furthermore, because of the length of the missing region, knowledge of the contact provides no information that is useful in establishing a unique orientation.
Our previous work has shown that the contacts between the two symmetric proteins are asymmetric (14). One protomer of SecA must lack some contacts that are present between the second protomer and SecB. Work is in progress to identify the source of the asymmetry.
During protein export, precursors must be transferred from SecB to SecA and subsequently through the translocon. Studies using site-directed spin labeling and EPR spectroscopy show that in the complex between SecB and a precursor, the polypeptide is wrapped around the surface of SecB. It occupies a channel at the interface of the dimer of dimers, crosses over the ends or the flat sides of SecB, and binds in the channel on the opposite side (3, 10). Either of the orientations of SecB illustrated in Fig. 5 would facilitate the transfer of the precursor from SecB to SecA in a ternary complex. In both orientations the crucial cross-link between SecB residue 35, which lies on the edge of the ligand-binding channel at the dimer interface, and SecA residue 636 in the helix scaffold domain positions the ligand-binding channel of SecB directly above the linker helix and the portion of the helix scaffold domain (residues 636 through 645) which were shown to provide the binding surface for polypeptide ligands (1). This channel can be seen in Fig. 5e and i (marked with an asterisk). An interesting question that remains is how the transfer of the polypeptide from contact with SecB to the binding site on SecA and further through the SecYEG translocon is achieved.
Supplementary Material
Acknowledgments
We thank Jennine M. Crane and Virginia F. Smith for critically reading the manuscript. We are grateful to Anastassios Economou for providing the coordinates of E. coli SecA with the PBD modeled. We thank Angela A. Lilly for construction of the single-cysteine mutants of SecA and SecB and Wing Cheung Lai for the movie included in the supplemental material.
This work was supported by an endowment from the Hugo Wurdack Trust at the University of Missouri and grant number GM29798 from the National Institutes of Health (NIH), National Institute of General Medical Sciences (NIGMS). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or NIGMS.
Simon J. S. Hardy is Honorary fellow of the Department of Biology, University of York, York YO10 5DD, United Kingdom.
Footnotes
Published ahead of print on 29 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Cooper, D. B., V. F. Smith, J. M. Crane, H. C. Roth, A. A. Lilly, and L. L. Randall. 2008. SecA, the motor of the secretion machine, binds diverse partners on one interactive surface. J. Mol. Biol. 382:74-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crane, J. M., C. Mao, A. A. Lilly, V. F. Smith, Y. Suo, W. L. Hubbell, and L. L. Randall. 2005. Mapping of the docking of SecA onto the chaperone SecB by site-directed spin labeling: insight into the mechanism of ligand transfer during protein export. J. Mol. Biol. 353:295-307. [DOI] [PubMed] [Google Scholar]
- 3.Crane, J. M., Y. Suo, A. A. Lilly, C. Mao, W. L. Hubbell, and L. L. Randall. 2006. Sites of interaction of a precursor polypeptide on the export chaperone SecB mapped by site-directed spin labeling. J. Mol. Biol. 363:63-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Driessen, A. J., and N. Nouwen. 2008. Protein translocation across the bacterial cytoplasmic membrane. Annu. Rev. Biochem. 77:643-667. [DOI] [PubMed] [Google Scholar]
- 5.Economou, A., J. A. Pogliano, J. Beckwith, D. B. Oliver, and W. Wickner. 1995. SecA membrane cycling at SecYEG is driven by distinct ATP binding and hydrolysis events and is regulated by SecD and SecF. Cell 83:1171-1181. [DOI] [PubMed] [Google Scholar]
- 6.Economou, A., and W. Wickner. 1994. SecA promotes preprotein translocation by undergoing ATP-driven cycles of membrane insertion and deinsertion. Cell 78:835-843. [DOI] [PubMed] [Google Scholar]
- 7.Fekkes, P., J. G. de Wit, A. Boorsma, R. H. Friesen, and A. J. Driessen. 1999. Zinc stabilizes the SecB binding site of SecA. Biochemistry 38:5111-5116. [DOI] [PubMed] [Google Scholar]
- 8.Hodis, E., G. Schreiber, K. Rother, and J. L. Sussman. 2007. eMovie: a storyboard-based tool for making movies. Trends Biochem. Sci. 32:199-204. [DOI] [PubMed] [Google Scholar]
- 9.Hunt, J. F., S. Weinkauf, L. Henry, J. J. Fak, P. McNicholas, D. B. Oliver, and J. Deisenhofer. 2002. Nucleotide control of interdomain interactions in the conformational reaction cycle of SecA. Science 297:2018-2026. [DOI] [PubMed] [Google Scholar]
- 10.Lilly, A. A., J. M. Crane, and L. L. Randall. 2009. Export chaperone SecB uses one surface of interaction for diverse unfolded polypeptide ligands. Protein Sci. 18:1860-1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mao, C., S. J. S. Hardy, and L. L. Randall. 2009. Maximal efficiency of coupling between ATP hydrolysis and translocation of polypeptides mediated by SecB requires two protomers of SecA. J. Bacteriol. 191:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Osborne, A. R., W. M. Clemons, Jr., and T. A. Rapoport. 2004. A large conformational change of the translocation ATPase SecA. Proc. Natl. Acad. Sci. U. S. A. 101:10937-10942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Papanikou, E., S. Karamanou, and A. Economou. 2007. Bacterial protein secretion through the translocase nanomachine. Nat. Rev. Microbiol. 5:839-851. [DOI] [PubMed] [Google Scholar]
- 14.Randall, L. L., J. M. Crane, A. A. Lilly, G. Liu, C. Mao, C. N. Patel, and S. J. S. Hardy. 2005. Asymmetric binding between SecA and SecB two symmetric proteins: implications for function in export. J. Mol. Biol. 348:479-489. [DOI] [PubMed] [Google Scholar]
- 15.Randall, L. L., and S. J. S. Hardy. 2002. SecB, one small chaperone in the complex milieu of the cell. Cell. Mol. Life Sci. 59:1617-1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Randall, L. L., and M. T. Henzl. 2010. Direct identification of the site of binding on the chaperone SecB for the amino terminus of the translocon motor SecA. Protein Sci. 19:1173-1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Randall, L. L., T. B. Topping, D. Suciu, and S. J. S. Hardy. 1998. Calorimetric analyses of the interaction between SecB and its ligands. Protein Sci. 7:1195-1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Volkert, T. L., J. D. Baleja, and C. A. Kumamoto. 1999. A highly mobile C-terminal tail of the Escherichia coli protein export chaperone SecB. Biochem. Biophys. Res. Commun. 264:949-954. [DOI] [PubMed] [Google Scholar]
- 19.Zhou, J., and Z. Xu. 2003. Structural determinants of SecB recognition by SecA in bacterial protein translocation. Nat. Struct. Biol. 10:942-947. [DOI] [PubMed] [Google Scholar]
- 20.Zimmer, J., W. Li, and T. A. Rapoport. 2006. A novel dimer interface and conformational changes revealed by an X-ray structure of B. subtilis SecA. J. Mol. Biol. 364:259-265. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.