Abstract
Transmembrane helix 2 (TM2) of the Tar chemoreceptor undergoes an inward piston-like displacement of 1 to 3 Å upon binding aspartate. This signal is transmitted to the kinase-control module via the HAMP domain. Within Tar, the HAMP domain forms a parallel four-helix bundle consisting of a dimer of two amphipathic helices connected by a flexible linker. In the nuclear magnetic resonance structure of an archaeal HAMP domain, residues corresponding to the MLLT sequence between Arg-214 at the end of TM2 and Pro-219 of Tar are an N-terminal helical extension of AS1. We modified this region to test whether it behaves as a continuous helical connection between TM2 and HAMP. First, one to four Gly residues were inserted between Thr-218 and Pro-219. Second, the MLLT sequence was replaced with one to nine Gly residues. Third, the sequence was shortened or extended with residues compatible with helix formation. Cells expressing receptors in which the MLLT sequence was shortened to MLL or in which the MLLT sequence was replaced by four Gly residues performed good aspartate chemotaxis. Other mutant receptors supported diminished aspartate taxis. Most mutant receptors had biased signal outputs and/or abnormal patterns of adaptive methylation. We interpret these results to indicate that a strong, permanent helical connection between TM2 and the HAMP domain is not necessary for normal transmembrane signaling.
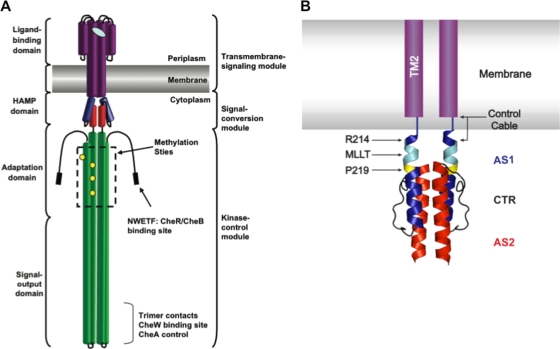
The HAMP domain is a structural motif commonly found in histidine kinases (HKs), adenylate cyclases, methyl-accepting chemotaxis proteins (MCPs), and phosphatases (2). In Escherichia coli and Salmonella enterica MCPs, the HAMP domain is located between a transmembrane-sensing module composed of ligand-binding and transmembrane regions and a kinase-control module composed of adaptation and kinase-activating regions (Fig. 1A) (19). Therefore, HAMP domains are responsible for bidirectional signal transduction between these modules.
FIG. 1.
Domain architecture of the aspartate chemoreceptor. (A) The cartoon, based on a figure from Hazelbauer et al. (2008) (19), illustrates the architecture of the aspartate chemoreceptor. Protein structural domains are labeled on the left, and functional modules are labeled on the right. (B) Schematic of TM2 and the control cable region attached to a ribbon diagram of the solution NMR structure of the Af1503 HAMP domain four-helix bundle (22). TM2 is shown in purple within the membrane. The control cable of TarEc consists of 5 amino acyl residues (Gly-Ile-Arg-Arg-Met) that connect TM2 and AS1 of HAMP. AS1 is shown in blue, AS2 is shown in red, and the 14-residue AS1-AS2 connector (CTR) is shown in black. The residue equivalent to Arg-214 in TarEc is also highlighted in blue, the conserved Pro residue (Pro-219 in TarEc) is highlighted in yellow, and residues equivalent to the MLLT sequence between TM2 and Pro-219 in TarEc are highlighted in cyan.
The determination of a high-resolution three-dimensional structure of a HAMP domain from an MCP remains elusive. However, a solution nuclear magnetic resonance (NMR) structure of a HAMP domain has been determined for the Af1503 protein of unknown function from the archaeal thermophile Archeoglobus fulgidus (22). The domain forms a parallel four-helix bundle, with two amphipathic helices, AS1 and AS2, being contributed by each subunit (Fig. 1B). In this structure, the helices pack in an unusual x-da configuration, commonly referred to as knobs-to-knobs packing, in which the large hydrophobic x residues stabilize both intrasubunit and intersubunit interactions.
Evidence for the existence of a four-helix HAMP bundle within intact receptors comes from disulfide cross-linking experiments with Salmonella enterica Tar (TarSe) (41) and the E. coli Aer redox receptor (AerEc) (44). In vivo genetic studies (1, 48) are also consistent with the existence of a four-helix bundle in the E. coli Tsr receptor (TsrEc).
TarEc functions as the aspartate chemoreceptor in E. coli (39). Each monomer within the homodimeric (15, 29) receptor possess a periplasmic ligand-binding domain composed of four antiparallel α helices that form four-helix bundles (8). The transmembrane regions that flank this periplasmic domain (transmembrane helix 1 [TM1] and TM2) are extensions of the periplasmic helices PD1 and PD4 (27, 33, 37, 40). Aspartate binds at either of two rotationally symmetrical sites at the dimer interface. Each binding site contains residues from PD1 of one subunit and PD1′ and PD4′ of the other. Aspartate binding generates a small (∼1- to 3-Å) vertical displacement into the cytoplasm of one contiguous PD4-TM2 helix relative to the other (14).
E. coli Tar (TarEc) and other chemoreceptors normally activate the histidine protein kinase CheA (5), which is coupled to the receptors via the adapter protein CheW. CheA autophosphorylates, and the phosphoryl group is subsequently transferred to the response regulator CheY (21). CheY-P interacts with FliM within the flagellar motor to promote clockwise (CW) rotation of the flagella (36, 46). Counterclockwise (CCW) motor rotation allows the flagellar filaments to coalesce into a bundle that propels the cell in a run (38). CW rotation of one or more flagella disrupts the bundle and generates a tumble (43). Therefore, the relative activities of CheA and the CheY-P phosphatase, CheZ, establish the ratio of CheY to CheY-P within the cell and hence the frequency of tumbling (11, 21).
The conformational changes induced by aspartate convert TarEc from a stimulator of CheA activity into an inhibitor (6). The resulting drop in CheY-P activity, which is accelerated by CheZ, suppresses tumbling and lengthens the average run. Inhibition of CheA activity is reversed by covalent methylation of the cognate receptor (17). Methylation is facilitated by a transient decrease in the level of the active, phosphorylated form of the CheB methylesterase (26), which is another substrate for phosphotransfer from CheA (21). The well-studied properties of this system, and the possibility of monitoring several different in vivo parameters, make it amenable for examining signal transduction between TM2 and the adjoining HAMP domain.
The apical region of AS1 in TarEc is composed of the tetrapeptide Met-Leu-Leu-Thr, which connects residue Arg-214 at the end of TM2 with the conserved Pro-219 residue within AS1 of the HAMP domain (Fig. 1B). The corresponding sequence is Thr-Ile-Thr-Arg in the Af1503 HAMP domain. In the NMR structure of the isolated Af1503 HAMP, the corresponding four residues (Thr-Ile-Thr-Arg) comprise an unpaired helical extension of the N terminus of AS1 before Pro-283 (9, 22), but it is unclear how these residues may pack in an intact membrane-spanning protein. In the Af1503 HAMP, Pro-283 packs against residues Glu-311 and Ile-312, which form the N terminus of AS2. The residues at the equivalent positions in TarEc are Glu-246 and Met-247 (Fig. 1B).
We modified the length and residue composition of the region between Arg-214 and Pro-219 in TarEc in different ways and monitored the ability of the mutant proteins to support chemotactic migration, to generate the clockwise flagellar rotation that reflects CheA activation, and to regulate adaptive methylation. The results support a model in which the structural tension exerted by TM2 on AS1 controls its signaling state, as proposed by Zhou et al. (48). In the context of that model, the results suggest that when the receptor is in the kinase-inhibiting state, the HAMP domain is in a stable four-helix bundle.
MATERIALS AND METHODS
Bacterial strains and plasmids.
Strains HCB436 [Δtsr7021 Δtrg100 Δ(tar-cheB)2234], RP3098 [Δ(flhD-flhB)4], and VB13 (thr+ eda+ Δtsr7021 trg::Tn10 Δtar-tap5201) are derived from E. coli K-12 strain RP437 (35, 45, 47). All chemoreceptor genes are deleted from strain VB13, as well as strain HCB436, which also carries a ΔcheRB deletion. All in vivo assays of receptor activity were conducted using these two strains. Strain RP3098 contains a deletion of the master regulator flhDC, and it therefore fails to produce any Che proteins. Plasmid pGW100, a derivative of pMK113 (16), was used to express wild-type and mutant tar genes constitutively. Our insertion of a BamHI site downstream of the promoter resulted in a serendipitous 26-nucleotide deletion that reduced tar expression levels to nearly physiological levels (C. Adase, personal communication). In addition, an in-frame coding sequence encoding a seven-residue linker (GGSSAAG) (10) and a C-terminal V5 epitope tag (GKPIPNPLLGLDST) (3) were added to the 3′ end of tar. Plasmid pRD300 (12) is a derivative of pBAD18 (18) that expresses tar with the same C-terminal linker and V5 epitope tag upon induction with l-arabinose. Strain RP3098 was used with pRD300 as previously described (12) to produce standards for the in vivo methylation assay. Mutations in tar were introduced via site-directed mutagenesis (Stratagene).
Chemotactic migration assay.
Aspartate semisolid motility agar was made with 3.25 g/liter Bacto agar (Difco), Che salts [10 mM potassium phosphate (pH 7.0) 1 mM (NH4)2SO4, 1 mM MgSO4, 1 mM glycerol], 1 mM MgCl2, 90 mM NaCl, 0.1 mM l-aspartate, 1 μg/ml thiamine, 50 μg/ml ampicillin, and 20 μg/ml each of l-histidine, l-leucine, l-methionine, and l-threonine. Plates were inoculated with isolated colonies of strain VB13 expressing the various Tar receptors from pGW100 and incubated at 30°C. Chemotactic ring diameters were measured after 8 h and every 4 h thereafter to 24 h. The rate of expansion was calculated in mm/h.
Observation of tethered cells.
HCB436 and VB13 cells producing the various Tar proteins expressed from plasmid pGW100 were grown overnight at 30°C in tryptone broth containing 50 μg/ml ampicillin. Overnight cultures were back-diluted 1:100 into the same medium and grown at 30°C with agitation to an optical density at 600 nm (OD600) of ∼0.6. Cells were then harvested by centrifugation and resuspended in tethering buffer (10 mM potassium phosphate [pH 7.0], 0.1 M NaCl, 0.01 mM EDTA, 0.02 mM l-methionine, 20 mM sodium dl-lactate). Chloramphenicol was added at 20 μg/ml to prevent growth of flagellar filaments after shearing. Cells were sheared in a Waring blender during eight repetitions of 7 s of shearing at high speed with interspersed 13-s pauses to prevent overheating. Cells were collected by centrifugation, washed three times with tethering buffer, and finally resuspended in tethering buffer. A 20-μl aliquot of a 200-fold dilution of antiflagellar filament antiserum was added to 20 μl of these cells. Apiezon-L grease was carefully added around the edge of 12-mm round glass coverslips that had been soaked in fuming nitric acid for 1 h and then thoroughly washed with distilled water. A 40-μl aliquot of the cell-antibody mix was added to the center of the coverslip. The coverslips were incubated in a humidity chamber for 30 min at 30°C. After incubation, the coverslips were affixed to a flow chamber (4), and nontethered cells were washed from the solution. Cells were observed under phase contrast at ×1,000 magnification using an Olympus BH-2 microscope. Rotating cells were videotaped, and 100 cells for each specific receptor were analyzed during 30 s of playback.
RESULTS
Inserting Gly residues between Thr-218 and Pro-219.
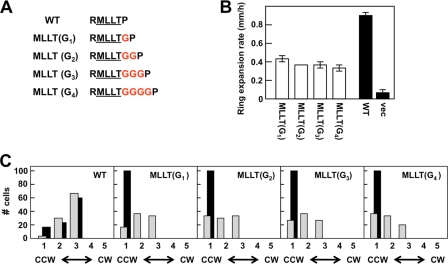
We first examined the effect of inserting one to four Gly residues [the MLLT(Gn) series of receptors] between Thr-218 and Pro-219 (Fig. 2A). In these proteins, the MLLT sequence remains intact and the Leu-216 residue, whose equivalent is important for signaling in the related TsrEc chemoreceptor (48), is retained. Because Gly residues have a low helix-forming propensity (20, 30, 32), these insertions should decrease the strength of any helical connection between the MLLT sequence and Pro-219, as well as increase its length. Also, Gly-rich peptides have been shown to form a structure that is more elongated than the traditional α helix and more compact than a β sheet (31). Therefore, the insertion of multiple Gly residues should make this region much more conformationally flexible.
FIG. 2.
Effects of inserting multiple Gly residues between Thr-218 and Pro-219 of AS1 [MLLT(Gn) mutants] on the signaling state of Tar. (A) Illustration of the MLLT(Gn) series of mutants. Inserted Gly residues are shown in red. (B) Aspartate chemotaxis ring expansion rates, measured in mm/h, for VB13 cells expressing wild-type or mutant [the MLLT(Gn) series] receptors. The error bars represent the standard errors of the mean, with n = 3. (C) Rotational biases of VB13 and HCB436 cells expressing wild-type or mutant receptors. The cells were placed into one of five categories on the basis of their CW/CCW rotational ratios, from left to right: 1, CCW locked; 2, CCW biased; 3, frequently reversing; 4, CW biased; and 5, CW locked. One hundred cells were analyzed for each mutant. WT, wild type.
The plasmid-encoded mutant proteins were first expressed from pGW100 in strain HCB436 (ΔMCP ΔcheRB) to analyze the expression level and to ensure that migration during SDS-PAGE was similar to that of wild-type Tar (see Fig. S1 in the supplemental material). The levels of wild-type Tar were about 3-fold higher than the levels produced from the chromosomal tar gene. Since Tar constitutes about 30% of the receptor population in wild-type cells (25), the total receptor abundance is approximately at the normal physiological level. Steady-state levels of all four MLLT(Gn) receptors were at 60% or higher of the level of wild-type Tar (Table 1). On the basis of these results, the steady-state level of receptor should be sufficient to generate a response to chemoeffectors. Because the other chemoreceptor genes are deleted within strains HCB436 and VB13, any observed responses can be attributed to the receptors expressed from pGW100.
TABLE 1.
Relative levels of mutant Tar receptors
| Variant group and Tar receptora | Protein levelb |
|---|---|
| MLLT(Gn) variants | |
| Wild type | 1.00 |
| MLLT(G1) | 0.68 |
| MLLT(G2) | 0.64 |
| MLLT(G3) | 0.60 |
| MLLT(G4) | 0.60 |
| Variants with different nos. of Gly residues | |
| Wild type | 1.00 |
| G0 | 0.31 |
| G1 | 0.68 |
| G2 | 0.71 |
| G3 | 0.70 |
| G4 | 0.75 |
| G5 | 0.77 |
| G6 | 0.88 |
| G7 | 0.93 |
| G8 | 0.99 |
| G9 | 0.94 |
| Variants by helical connection size | |
| Wild type | 1.00 |
| −3 | 0.30 |
| −2 | 0.32 |
| −1 | 0.33 |
| +1 | 0.30 |
| +2 | 0.30 |
| +3 | 0.29 |
| +4 | 0.30 |
| +5 | 0.25 |
The nomenclature for the mutant Tar receptors is as in the text.
Protein levels were determined by quantitative immunoblotting of V5-tagged receptors from equal numbers of cells grown to the same OD600 and are expressed as a fraction of the level of wild-type V5-tagged Tar from cell extracts processed in the same way.
When these proteins were expressed in the cheR+ cheB+ strain VB13, aspartate chemotaxis rings formed by cells producing MLLT(G1) Tar expanded at 50% of the rate of aspartate rings formed by cells producing wild-type Tar. The MLLT(G2) to MLLT(G4) proteins were only slightly less effective in supporting aspartate taxis (Fig. 2B). There was no indication of a helical periodicity, which would reveal itself as a repeat pattern of approximately 3.6 residues. In addition, the migratory rings formed by strains producing any of the MLLT(Gn) Tar variants were sharper than the rings seen with wild-type Tar (data not shown). Such exceptionally sharp rings may indicate that cells accumulate in regions in which the metabolism of aspartate by the expanding colony creates a very steep concentration gradient (12).
Flagellar rotation in VB13 cells expressing any of the MLLT(Gn) Tar variants was more CCW biased than that in VB13 cells expressing wild-type Tar (Fig. 2C), and the biases were comparable among them. Flagellar rotation in HCB436 cells expressing any of the MLLT(Gn) variants was CCW locked (Fig. 2C). Apparently, the QEQE forms of these receptors are unable to support enough CheA kinase activity to support CW flagellar rotation in the absence of compensatory methylation.
To test whether increased methylation of the MLLT(Gn) proteins compensates for their inherent CCW output bias, the basal state of covalent modification of the V5-tagged MLLT(Gn) receptors was measured in VB13 cells (see Fig. S1 in the supplemental material). Previous work (7) has shown that the V5-tagged proteins behave much like their untagged variants in vivo. This result is consistent with the work of Lai and Hazelbauer (23), who found that C-terminal extensions beyond the CheR binding NWETF motif had a greater effect on methylation in vitro than in vivo. All of the MLLT(Gn) receptors had a higher degree of covalent modification in the absence of chemoeffectors, demonstrating the compensatory effect of increased covalent modification on the inherent signaling bias within the receptors. These proteins showed no substantial change in methylation pattern upon addition of aspartate or NiSO4 (data not shown). However, subtle changes in methylation level that are not clearly detected by our assay are probably sufficient to support some level of chemotaxis in semisolid agar.
The conclusion from these experiments is that weakening the α-helical structure in the connection between TM2 and AS1 diminishes the CheA-stimulating activity of TarEc (Fig. 2C). Increased methylation offsets the resultant CCW output bias, allowing transmission of an aspartate-binding signal, to some degree (Fig. 2B).
Replacing the MLLT sequence with various numbers of Gly residues.
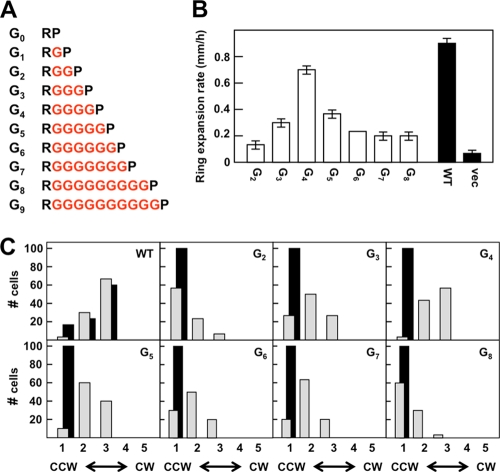
One could argue that the Gly residues introduced into the MLLT(Gn) series of proteins simply loop out of a continuous α helix between TM2 and Pro-219. Therefore, we decided to test the effect of completely replacing the MLLT sequence with four Gly residues. We also added and subtracted Gly residues from this construct to generate the G0 to G9 series of mutant receptors (Fig. 3A). These runs of consecutive Gly residues should have low helix-forming propensities (20, 30, 32).
FIG. 3.
Effects of replacing the helical connector between TM2 and HAMP with multiple Gly residues. (A) Illustration of the Gn constructs. The inserted Gly residues are shown in red. (B) The aspartate chemotaxis-ring expansion rates were measured in VB13 (cheR+ cheB+) cells expressing wild-type or mutant (the Gn series) receptors. The error bars represent the standard errors of the mean, with n = 3. (C) Rotational biases of VB13 and HCB436 cells expressing wild-type or mutant receptors. The cells were categorized as described in the legend to Fig. 2C.
When expressed from pGW100 in HCB436, all receptors within this mutant set were present in amounts 68% or more of wild-type Tar (Table 1), except for the G0 variant (31%). In assays for aspartate taxis, the G4 protein made a chemotaxis ring that expanded at ∼80% of the rate of the ring made by cells expressing wild-type Tar (Fig. 3B). The ring was of the more diffuse type seen with cells expressing wild-type Tar rather than the tight rings seen with the MLLT(Gn) proteins. The diameter of the chemotaxis rings decreased as Gly residues were subtracted or added (Fig. 3B), and the rings became sharper. The colonies formed by cells expressing the more extreme forms, G0, G1, and G9, were indistinguishable from the colonies formed by cells containing only the vector plasmid; therefore, they were not examined further.
The CW/CCW ratio of flagellar rotation in tethered VB13 cells expressing the G4 Tar variant was very similar to that of cells expressing wild-type Tar (Fig. 3C). As Gly residues were added or subtracted from the G4 construct, flagellar rotation became increasingly CCW biased (Fig. 3C). HCB436 cells expressing the QEQE forms of all of the Gn Tar variants were CCW locked (Fig. 3C).
All of the Gn variants were overmethylated relative to wild-type Tar (see Fig. S1 in the supplemental material). None of the proteins showed increased covalent modification in response to the addition of aspartate, but the G4 protein formed a new, slower-migrating band after addition of NiSO4 (data not shown). Again, we may have failed to detect subtle changes in methylation level that support some true aspartate taxis in semisolid agar.
The surprisingly good chemotaxis seen with cells expressing the G4 variant suggests that the connection between Arg-214 and Pro-219 does not need to be highly structured for TarEc to transmit transmembrane signals in response to aspartate binding. The monotonic decrease in chemotaxis seen as Gly residues were deleted from or added to the G4 construct also suggests that the residue length of the TM2-HAMP connection is an important factor in communicating the transmembrane signal in response to aspartate.
Shortening or lengthening helical connection between Arg-214 and Pro-219.
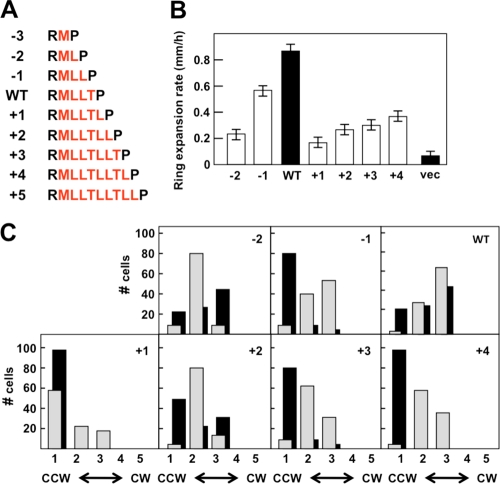
The results described thus far suggest that a highly structured connection between Arg-214 and Pro-219 is not a prerequisite for transmembrane signaling in response to aspartate. However, they do not rule out the possibility that the helical phasing of the MLLT connection is an important factor in signaling. To examine this possibility, we added or subtracted residues from the helical connection to generate the −3 to +5 series of Tar variants (Fig. 4A). In this series, the connection ranged from only the Met-215 residue interposed between Arg-214 and the Pro-219 (the −3 variant) to the sequence MLLTLLTLL between the Arg and Pro residues (the +5 variant). Variants in which the helix was extended even farther were apparently completely unstable, as no protein could be detected (data not shown).
FIG. 4.
Effects of shortening and lengthening the helical connection between TM2 and HAMP. (A) The helix-extending and -shortening constructs are shown. The residues between Arg-214 and Pro-219 are shown in red. (B) Aspartate chemotaxis-ring expansion rates, measured in mm/h, for VB13 (cheR+ cheB+) cells expressing wild-type or mutant (the −2 through +4 series) receptors. The error bars represent the standard errors of the mean, with n = 3. (C) Rotational biases of VB13 and HCB436 cells expressing wild-type or mutant receptors. The cells were categorized as described in the legend to Fig. 2C.
Under our conditions, wild-type and mutant Tar variants were expressed from pGW100 in HCB436, and the mutant receptors were present at steady-state levels ∼30% of the level seen with wild-type Tar (Table 1). VB13 (cheR+ cheB+) cells expressing the −3 and +5 variants were indistinguishable from the vector-only control in aspartate chemotaxis and therefore were not examined further. Cells expressing the −2 and −1 variants had aspartate chemotaxis rings that expanded at 26% and 64%, respectively, of the rate of rings produced by cells expressing wild-type Tar, thereby demonstrating an increasing chemotactic ability conferred by the −2 and −1 receptors (Fig. 4B). Cells expressing the +1, +2, +3, and +4 variants formed aspartate rings that expanded at 18%, 31%, 34%, and 41% of the rate of the rings formed by cells expressing wild-type Tar, respectively. The increasing performance of cells expressing the −2 and −1 variants or the +1, +2, +3, and +4 variants was noticeable and reproducible. Therefore, we observed two sets of Tar receptors that facilitated improved aspartate chemotaxis with an approximately helical repeat. The ring formed by the −1 variant was diffuse, like the rings made by cells expressing wild-type Tar, rather than the ultrasharp rings formed by cells producing the −2 and +1 through +4 variants.
The flagellar rotation of VB13 cells expressing the −2, −1, and wild-type Tar proteins was increasingly CW (Fig. 4C), a result consistent with the improvement in aspartate ring formation (Fig. 4B). A similar pattern was observed for cells expressing the +1 through +4 variants of Tar. Therefore, an indication of helicity in rotational bias is seen in the presence of a complete (cheR+ cheB+) chemotactic circuit. In HCB436 cells that lack covalent modification, this pattern was not observed (Fig. 4C); cells expressing the +1 and +4 receptors were CCW locked and cells expressing the +2 and +3 receptors were CCW biased relative to cells expressing wild-type Tar.
In the absence of chemoeffector, −2, −1, and wild-type Tar receptors were all methylated to a similar extent in strain VB13 (see Fig. S1 in the supplemental material). The −2 receptor did not change its methylation pattern upon addition of aspartate or Ni2+, but the −1 receptor did show increased covalent modification after addition of aspartate (see Fig. S3 in the supplemental material), behaving in that regard much like wild-type Tar. The +1 and +4 proteins were much more highly methylated than wild-type Tar, but the +2 and +3 variants migrated primarily as a single band at the position of the QEQE standard, although there were also fainter bands made by faster- and slower-migrating species. The modification states of the +1 through +4 proteins did not change upon addition of aspartate or NiSO4.
DISCUSSION
Bacterial chemoreceptors can usefully be thought of as transmembrane allosteric enzymes (28). As such, chemoeffectors binding to the extracellular domain (the periplasmic domain in Gram-negative bacteria) act as allosteric ligands to control the activity of the CheA kinase that is coupled to the membrane-distal portion of the cytoplasmic domain (Fig. 1A). Binding of chemoeffectors must generate conformational changes in the extracellular domain that are transmitted to the kinase-control domain. The distance that the conformational signal must traverse is in excess of 300 Å (30 nm).
Because this long-distance propagation of structural rearrangements must be driven by the energy of chemoeffector binding, chemoreceptors must necessarily be in a metastable state. The simplest model proposes that the receptors are two-state systems that exist either in a kinase-stimulating (on) or kinase-inhibiting (off) mode (28). In this view, intermediate levels of kinase stimulation would be generated by a population of receptors that are in the on and off conformations in different proportions, with attractants shifting this equilibrium to the off state and repellents shifting the equilibrium toward the on state. Recent genetic analyses suggest that some mutant proteins can assume signaling states that lie outside the range of conformations assumed by wild-type receptors under physiological conditions (34, 48).
The yin-yang model (42) proposes that control of CheA kinase activity depends on the relative packing states of the adaptation and signal-output modules (Fig. 1A). If the adaptation module is tightly packed and stable, the signal-output domain is more loosely packed and flexible, a conformation that favors stimulation of CheA kinase activity. Conversely, a more loosely packed adaptation module allows tighter packing of the signal-output domain and inhibits kinase-stimulating activity.
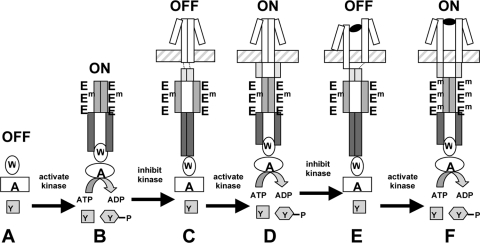
This idea can be extended to describe how the different components of the receptor may work together in signal transduction (summarized in the cartoon in Fig. 5). The CheA kinase by itself, when it is not associated with a receptor, is in the off state (Fig. 5A). When CheA is coupled to an unconstrained kinase-control domain via the CheW adapter protein, it is shifted into the on state, because tight packing of the adaptation domain dominates (Fig. 5B). Addition of a stable HAMP domain, presumably in the form of a parallel four-helix bundle (Fig. 1B), switches the receptor/CheA complex into an off state (Fig. 5C). However, when the stability of the HAMP domain is compromised by its attachment to TM2, the packing of the adaptation domain again dominates and the receptor is in the on state (Fig. 5D). Binding of an attractant ligand would decrease the destabilizing effect of the TM2 connection on HAMP, allowing a stable HAMP bundle to form and putting the receptor in the off state (Fig. 5E). Finally, covalent methylation of the adaptation module would favor its tighter packing, the signal-output domain would again be in its less tightly packed on configuration, and the HAMP domain would again become less stable (Fig. 5F). This scenario is the essence of the dynamic bundle model for HAMP function proposed by Zhou et al. (48).
FIG. 5.
Model of domain interactions within the TarEc chemoreceptor. The periplasmic and transmembrane domains are shown as white rectangles, the HAMP domain is shown as light gray triangles, the adaptation module of the kinase-control domain is shown in medium gray, and the signal-output module of the kinase-control domain is shown in dark gray. The cytoplasmic membrane is represented by the horizontal hatched rectangle. CheA, CheW, and CheY are denoted A, W, and Y, respectively. Aspartate is shown as a black oval, and 3 of the 4 Glu residues per receptor monomer are denoted E when they are unmethylated and Em when they are present as methyl esters. (A) In the absence of receptor, CheA activity is very low. (B) In the presence of the kinase-control domain, the adaptation module is tightly packed, the signal-output module is loosely packed, and CheA activity is stimulated. (C) In the absence of tight coupling to TM2 (short dotted lines), the HAMP domain becomes a tightly packed four-helix bundle that destabilizes the packing of the adaptation module. The loose packing of the adaptation module allows the signal-output module to become tightly packed, and CheA activity is inhibited. (D) In the ligand-free wild-type receptor, TM2 exerts tension on HAMP that destabilizes the four-helix bundle. As a result, the adaptation module can pack tightly, the signal-output module is less tightly packed, and CheA activity is stimulated. (E) Binding of aspartate displaces TM2 downward and relieves the tension on the HAMP domain. The HAMP domain packs tightly, the adaptation module becomes destabilized, and the signal-output domain packs tightly and inhibits CheA activity. (F) After adaptive methylation, the adaptation module packs tightly, even though ligand is still bound. Tight packing of the adaptation module destabilizes both the HAMP domain and the signal-output domain, CheA activity is once again stimulated, and TM2 returns to its original position, as has been observed previously (24).
Considerable evidence (28) suggests that binding of an attractant ligand produces a 1- to 3-Å shift of TM2 toward the cytoplasm. Rotational and tilting motions of TM2 have also been proposed. Whatever the exact nature of the signal—and it could, of course, consist of multiple parameters—a critical question is how movements of TM2 are transmitted to HAMP. Rotational and tilting models are easier to envision if the TM2-HAMP connection is a continuous helix. The dynamic-bundle model, in contrast, which envisions TM2 being connected to HAMP via control cables that can exert increased (destabilizing) or decreased (stabilizing) tension on HAMP, does not require a strong helical connection.
We changed the length and residue character of the TM2-HAMP connection in order to gain a better understanding of how the structure of this region affects transmembrane signaling. We interpret our results to favor the dynamic bundle model, because neither a strong helical structure nor the precise helical register of the TM2-HAMP connection appears to be essential for normal function in the TarEc receptor. The evidence can be summarized as follows.
(i) Insertion of 1 to 4 additional Gly residues between the MLLT sequence preceding Pro-219 [the MLLT(Gn) series] produces mutant receptors that are strongly biased toward the CCW-signaling, off state and that are highly overmethylated. This result is the one predicted if the tension between TM2 and HAMP is decreased and HAMP is allowed to assume a more stable conformation (Fig. 5). The insertion of Gly residues would also be expected to interfere with helical packing of this segment (20, 30, 32), although we have no direct structural information that this is the case. It could be argued that the inserted Gly residues simply loop out of a stable helix or that they are forced into a helical conformation. The latter possibility seems unlikely, because the insertions of 1 to 4 Gly residues have about the same effect (Fig. 2), although the helical register would be different in each case if the Gly residues are in a helical conformation. Although these receptors are strongly CCW biased, adaptive methylation does restore some amount of CW signaling that allows cells expressing them to form expanding rings in aspartate semisolid agar.
(ii) Replacement of the MLLT sequence with 2 to 8 Gly residues also produces receptors that confer a strongly CCW-signaling bias. However, the Tar G4 receptor, which has the same number of residues in the TM2-HAMP connection present in the wild type, can be compensated for by increased adaptive methylation to the point that it performs nearly as well as wild-type TarEc in supporting ring expansion in aspartate semisolid agar and CW flagellar rotation in tethered cells (Fig. 3). This result suggests that it is the length rather than the residue composition of the TM2-HAMP connection that is important. Again, however, we cannot be certain that the 4 Gly residues do not assume a helical structure in the context of the intact receptor, although the strength of the helix must be less than that in the wild-type protein (20, 31). One result that is not consistent with the control cable idea (48) is that decreasing the number of Gly residues to 3 or 2, which should increase the tension exerted on HAMP, does not produce a more CW-biased signal output. At present, we cannot explain this seeming discrepancy from our expectations, although it may reflect some global change in the structure of the TM2-HAMP connection that allows a stable HAMP domain to form.
(iii) Shortening or lengthening the TM2-HAMP connection by removing or adding L and T residues from the normal LLT pattern produced the most unpredictable results. A general phenomenon was that all of these mutant proteins, unlike any of the previous two sets of Gly mutants, were present in substantially lower steady-state levels of the receptor protein (see Fig. S1 in the supplemental material). Adding residues gave clear evidence of the importance of helical register in signaling, since the +3 and +4 receptors, in which the connection became MLLTLLT and MLLTLLTL, respectively, retained better function in supporting expansion of chemotactic rings in aspartate semisolid agar (Fig. 4). Insertion of 3 or 4 residues would come closest to restoring the original helical phasing. However, removing a single residue to generate the MLL (−1) receptor did not have the same effect as adding 1 residue to make the MLLTL receptor, which was highly defective in function. The −1 receptor was quite proficient at supporting expansion of chemotactic rings in aspartate semisolid agar. Furthermore, although its inherent signaling state was strongly CCW biased, adaptive methylation restored an essentially wild-type rotational bias (Fig. 4C), and addition of aspartate led to a further increase in the level of methylation (see Fig. S3 in the supplemental material). The simplest interpretation of these results is that altering the helical phasing of the connection by adding residues has a negative effect on receptor function, whereas decreasing or eliminating helicity of the connection by removing 1 residue does not, per se, drastically disrupt receptor function.
The −2 (ML) receptor was exceptional in that, in the QEQE state, it showed an essentially wild-type rotational bias (Fig. 4C). In the presence of the adaptive-methylation machinery, its level of covalent modification decreased (see Fig. S2 in the supplemental material) and its CCW-signaling bias increased (Fig. 4C). It may be that the HAMP domain in this protein is contorted into one of the destabilizing conformations proposed by Zhou et al. (48) and Parkinson (34), in which the normal coupling between rotational bias and adaptive methylation is compromised.
It is perhaps instructive to compare the effect of changing the length of the helical linker with the movement of the WY aromatic anchor at the cytoplasmic end of TM2 (13). Moving the WY pair in single-residue steps toward the N terminus to generate the −1 through −3 set of anchor-shifted receptors produced increasingly CCW-biased signaling that could be compensated for by increased methylation only in the −1 and −2 receptors. If the anchor remains at the nonpolar/polar interface of the cytoplasmic face of the membrane, these changes would displace Pro-219 farther from the membrane. By this criterion, the −1 through −3 series of WY movements would be the equivalent of the +1 through +3 TM2-HAMP connection mutations. However, the WY mutations should not change the helical register, whereas the +1 through +3 TM2-HAMP mutations should. This comparison suggests that an improper helical register of the TM2-HAMP connection is detrimental to receptor function.
Moving the WY pair in single-residue steps toward the C terminus to generate the +1 through +3 set of aromatic anchor-shifted receptors should pull Pro-219 closer to the membrane. These mutations produced increasingly CW-biased signaling that could be compensated for by decreased covalent modification only in the +1 receptor. The helical register of the TM2-HAMP connection should be preserved in all of these proteins as well. In terms of the displacement of Pro-219, these receptors would be the equivalent of the −1 through −3 TM2-HAMP connection mutations. Again, there is a lack of correspondence in phenotypes, because the −1 TM2-HAMP mutant receptor was CCW biased and somewhat overmethylated, whereas the −2 TM2-HAMP mutant receptor supported a wild-type CW/CCW ratio. This protein, however, evoked a decrease in covalent modification that shifted its signaling bias toward CCW. The conclusion from this comparison is that other factors, such as maintenance of proper helical register, in addition to the separation between the HAMP and the membrane, influence the signaling state of the receptor.
The preponderance of evidence from this work supports the dynamic-bundle model, because the residue length of the connection is clearly important. However, the results do not preclude any of the other models for TM2-HAMP communication that have been proposed, because we do not know the actual structural changes conferred by any of our manipulations of the TM2-HAMP connection. We predict that the various Gly insertions should reduce the propensity of the connection to assume a stable helical connection, but that is only an inference.
One factor that we feel has been underemphasized in the discussion of HAMP function is the strong conservation of the Pro residue (Pro-219 in TarEc). This residue near the beginning of AS1 packs up against the N terminus of AS2 in the published structure of the Af1503 HAMP four-helix bundle. A mutational analysis of the HAMP domain of the Tsr receptor of E. coli (48) has shown that most residue substitutions at the equivalent Pro-221 residue impair or abrogate receptor function. However, the P221K replacement yields a receptor with essentially wild-type function. This result is difficult, but not impossible, to interpret if packing up against AS2 is the primary function of the conserved Pro residue. Why a Lys residue at this position is apparently capable of functioning nearly as well as Pro is not clear. However, a conformationally flexible fulcrum at this position is what might be expected if the control cable model for signal input into a dynamic HAMP four-helix bundle is correct. We eagerly anticipate biophysical/structural studies to test this model experimentally.
Supplementary Material
Acknowledgments
We especially thank J. S. “Sandy” Parkinson for his generosity in sharing his ideas and results in advance of publication. This work has been improved substantially by his involvement. Discussions with J. M. “Marty” Scholtz, Nick Pace, Brian Matthews, Rick Dahlquist, and Joe Falke have all been valuable in developing our ideas and interpreting our data. Alexandra Bryson contributed substantially by performing DNA minipreps, assisting with methylation assays, and monitoring tethered cells. We kindly thank the Glaser lab (Stockholm University) as well as the Chazin lab (Vanderbilt University) for use of their protein electrophoretic equipment. Lily Bartoszek helped assemble and format the references and gave a final proofreading to the manuscript before submission.
Early stages of this work were funded by NIH grant GM 39736. More recent funding has come from the Bartoszek Fund for Basic Biological Science. R.R.D. was supported by a Kirschstein National Research Service Award from the National Institutes of Health (AI075773) and by the German Research Foundation (SFB 807, Transport and Communication across Biological Membranes). R.L.C. was supported by a Chemical-Biology Interface Training Grant from the National Institutes of Health (grant GM065086).
Footnotes
Published ahead of print on 24 September 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ames, P., Q. Zhou, and J. S. Parkinson. 2008. Mutational analysis of the connector segment in the HAMP domain of Tsr, the Escherichia coli serine chemoreceptor. J. Bacteriol. 190:6676-6685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aravind, L., and C. P. Ponting. 1999. The cytoplasmic helical linker domain of receptor histidine kinase and methyl-accepting proteins is common to many prokaryotic signalling proteins. FEMS Microbiol. Lett. 176:111-116. [DOI] [PubMed] [Google Scholar]
- 3.Baty, D. U., J. A. Southern, and R. E. Randall. 1991. Sequence comparison between the hemagglutinin neuraminidase genes of simian, canine and human isolates of simian virus-5. J. Gen. Virol. 72:3103-3107. [DOI] [PubMed] [Google Scholar]
- 4.Berg, H. C., and S. M. Block. 1984. A miniature flow cell designed for rapid exchange of media under high-power microscope objectives. J. Gen. Microbiol. 130:2915-2920. [DOI] [PubMed] [Google Scholar]
- 5.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. U. S. A. 86:1208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borkovich, K. A., and M. I. Simon. 1990. The dynamics of protein phosphorylation in bacterial chemotaxis. Cell 63:1339-1348. [DOI] [PubMed] [Google Scholar]
- 7.Bormans, A. F. 2005. Intradimer and interdimer methylation response by bacterial chemoreceptors to attractant stimulus. Ph.D. dissertation. Texas A&M University, College Station, TX.
- 8.Bowie, J. U., A. A. Pakula, and M. I. Simon. 1995. The three-dimensional structure of the aspartate receptor from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 51:145-154. [DOI] [PubMed] [Google Scholar]
- 9.Butler, S. L., and J. J. Falke. 1998. Cysteine and disulfide scanning reveals two amphiphilic helices in the linker region of the aspartate chemoreceptor. Biochemistry 37:10746-10756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Stewart, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cluzel, P., M. Surette, and S. Leibler. 2000. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science 287:1652-1655. [DOI] [PubMed] [Google Scholar]
- 12.Draheim, R. R., A. F. Bormans, R. Z. Lai, and M. D. Manson. 2005. Tryptophan residues flanking the second transmembrane helix (TM2) set the signaling state of the Tar chemoreceptor. Biochemistry 44:1268-1277. [DOI] [PubMed] [Google Scholar]
- 13.Draheim, R. R., A. F. Bormans, R. Z. Lai, and M. D. Manson. 2006. Tuning a bacterial chemoreceptor with protein-membrane interactions. Biochemistry 45:14655-14664. [DOI] [PubMed] [Google Scholar]
- 14.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falke, J. J., and D. E. Koshland, Jr. 1987. Global flexibility in a sensory receptor: a site-directed cross-linking approach. Science 237:1596-1600. [DOI] [PubMed] [Google Scholar]
- 16.Gardina, P., C. Conway, M. Kossman, and M. Manson. 1992. Aspartate and maltose-binding protein interact with adjacent sites in the Tar chemotactic signal transducer of Escherichia coli. J. Bacteriol. 174:1528-1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goy, M. F., M. S. Springer, and J. Adler. 1977. Sensory transduction in Escherichia coli: role of a protein methylation reaction in sensory adaptation. Proc. Natl. Acad. Sci. U. S. A. 74:4964-4968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman, L. M., D. Belin, M. J. Carson, and J. Beckwith. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 177:4121-4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hazelbauer, G. L., J. J. Falke, and J. S. Parkinson. 2008. Bacterial chemoreceptors: high-performance signaling in networked arrays. Trends Biochem. Sci. 33:9-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heinz, D. W., W. A. Baase, X. J. Zhang, M. Blaber, F. W. Dahlquist, and B. W. Matthews. 1994. Accommodation of amino-acid insertions in an alpha-helix of T4 lysozyme—structural and thermodynamic analysis. J. Mol. Biol. 236:869-886. [DOI] [PubMed] [Google Scholar]
- 21.Hess, J. F., K. Oosawa, P. Matsumura, and M. I. Simon. 1987. Protein phosphorylation is involved in bacterial chemotaxis. Proc. Natl. Acad. Sci. U. S. A. 84:7609-7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hulko, M., F. Berndt, M. Gruber, J. U. Linder, V. Truffault, A. Schultz, J. Martin, J. E. Schultz, A. N. Lupas, and M. Coles. 2006. The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126:929-940. [DOI] [PubMed] [Google Scholar]
- 23.Lai, W. C., and G. L. Hazelbauer. 2005. Carboxyl-terminal extensions beyond the conserved pentapeptide reduce rates of chemoreceptor adaptational modification. J. Bacteriol. 187:5115-5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lai, W. C., M. L. Peach, T. P. Lybrand, and G. L. Hazelbauer. 2006. Diagnostic cross-linking of paired cysteine pairs demonstrates homologous structures for two chemoreceptor domains with low sequence identity. Protein Sci. 15:94-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, M., and G. L. Hazelbauer. 2004. Cellular stoichiometry of the components of the chemotaxis signaling complex. J. Bacteriol. 186:3687-3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lupas, A., and J. Stock. 1989. Phosphorylation of an N-terminal regulatory domain activates the CheB methylesterase in bacterial chemotaxis. J. Biol. Chem. 264:17337-17342. [PubMed] [Google Scholar]
- 27.Lynch, B. A., and D. E. Koshland. 1991. Disulfide cross-linking studies of the transmembrane regions of the aspartate sensory receptor of Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 88:10402-10406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Manson, M. D. 2010. Bacterial chemoreceptors as membrane-spanning allosteric enzymes, p. 107-149. In S. Spiro and R. Dixon (ed.), Sensory mechanisms in bacteria: molecular aspects of signal recognition. Caister Academic Press, Norfolk, United Kingdom.
- 29.Milligan, D. L., and D. E. Koshland, Jr. 1988. Site-directed cross-linking. Establishing the dimeric structure of the aspartate receptor of bacterial chemotaxis. J. Biol. Chem. 263:6268-6275. [PubMed] [Google Scholar]
- 30.Myers, J. K., C. N. Pace, and J. M. Scholtz. 1997. Helix propensities are identical in proteins and peptides. Biochemistry 36:10923-10929. [DOI] [PubMed] [Google Scholar]
- 31.Ohnishi, S., H. Kamikubo, M. Onitsuka, M. Kataoka, and D. Shortle. 2006. Conformational preference of polyglycine in solution to elongated structure. J. Am. Chem. Soc. 128:16338-16344. [DOI] [PubMed] [Google Scholar]
- 32.Pace, C. N., and J. M. Scholtz. 1998. A helix propensity scale based on experimental studies of peptides and proteins. Biophys. J. 75:422-427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pakula, A. A., and M. I. Simon. 1992. Determination of transmembrane protein-structure by disulfide cross-linking—the Escherichia coli Tar receptor. Proc. Natl. Acad. Sci. U. S. A. 89:4144-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Parkinson, J. S. 2010. Signaling mechanisms of HAMP domains in chemoreceptors and sensor kinases. Annu. Rev. Microbiol. 64:101-122. [DOI] [PubMed] [Google Scholar]
- 35.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ravid, S., P. Matsumura, and M. Eisenbach. 1986. Restoration of flagellar clockwise rotation in bacterial envelopes by insertion of the chemotaxis protein CheY. Proc. Natl. Acad. Sci. U. S. A. 83:7157-7161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Scott, W. G., and B. L. Stoddard. 1994. Transmembrane signalling and the aspartate receptor. Structure 2:877-887. [DOI] [PubMed] [Google Scholar]
- 38.Silverman, M., and M. Simon. 1974. Characterization of Escherichia coli flagellar mutants that are insensitive to catabolite repression. J. Bacteriol. 120:1196-1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Springer, M. S., M. F. Goy, and J. Adler. 1977. Sensory transduction in Escherichia coli: two complementary pathways of information processing that involve methylated proteins. Proc. Natl. Acad. Sci. U. S. A. 74:3312-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stoddard, B. L., J. D. Bui, and D. E. Koshland, Jr. 1992. Structure and dynamics of transmembrane signaling by the Escherichia coli aspartate receptor. Biochemistry 31:11978-11983. [DOI] [PubMed] [Google Scholar]
- 41.Swain, K. E., and J. J. Falke. 2007. Structure of the conserved HAMP domain in an intact, membrane-bound chemoreceptor: a disulfide mapping study. Biochemistry 46:13684-13695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swain, K. E., M. A. Gonzalez, and J. J. Falke. 2009. Engineered socket study of signaling through a four-helix bundle: evidence for a yin-yang mechanism in the kinase control module of the aspartate receptor. Biochemistry 48:9266-9277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Turner, L., W. S. Ryu, and H. C. Berg. 2000. Real-time imaging of fluorescent flagellar filaments. J. Bacteriol. 182:2793-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watts, K. J., M. S. Johnson, and B. L. Taylor. 2008. Structure-function relationships in the RAMP and proximal signaling domains of the aerotaxis receptor Aer. J. Bacteriol. 190:2118-2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weerasuriya, S., B. M. Schneider, and M. D. Manson. 1998. Chimeric chemoreceptors in Escherichia coli: signaling properties of Tar-Tap and Tap-Tar hybrids. J. Bacteriol. 180:914-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Welch, M., K. Oosawa, S. Aizawa, and M. Eisenbach. 1993. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc. Natl. Acad. Sci. U. S. A. 90:8787-8791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wolfe, A. J., and H. C. Berg. 1989. Migration of bacteria in semisolid agar. Proc. Natl. Acad. Sci. U. S. A. 86:6973-6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhou, Q., P. Ames, and J. S. Parkinson. 2009. Mutational analyses of HAMP helices suggest a dynamic bundle model of input-output signalling in chemoreceptors. Mol. Microbiol. 73:801-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.