Abstract
Colicin M (ColM), which is produced by some Escherichia coli strains to kill competitor strains from the same or related species, was recently shown to inhibit cell wall peptidoglycan biosynthesis through enzymatic degradation of its lipid II precursor. ColM-producing strains are protected from the toxin that they produce by coexpression of a specific immunity protein, named Cmi, whose mode of action still remains to be identified. We report here the resolution of the crystal structure of Cmi, which is composed of four β strands and four α helices. This rather compact structure revealed a disulfide bond between residues Cys31 and Cys107. Interestingly, these two cysteines and several other residues appeared to be conserved in the sequences of several proteins of unknown function belonging to the YebF family which exhibit 25 to 35% overall sequence similarity with Cmi. Site-directed mutagenesis was performed to assess the role of these residues in the ColM immunity-conferring activity of Cmi, which showed that the disulfide bond and residues from the C-terminal extremity of the protein were functionally essential. The involvement of DsbA oxidase in the formation of the Cmi disulfide bond is also demonstrated.
Colicins are toxins (bacteriocins) secreted by certain Escherichia coli strains that kill competitor strains from the same or closely related bacterial species (12, 37, 38). All colicins exhibit a similar structural organization comprising three domains, with each domain being responsible for a specific step in the toxicity process: binding to an outer membrane receptor (central domain), translocation through the cell envelope (N-terminal domain), and expression of the toxic activity (C-terminal domain). Colicins are generally classified into two groups, groups A and B, depending on the translocation machinery that they parasitize to enter the cell, namely, the TolABQR system and the TonB/ExbBD system, respectively. These toxins exert their deleterious effects through a broad range of activities: most often, by formation of pores in the cytoplasmic membrane, but also by enzymatic degradation of nucleic acids or inhibition of protein synthesis (12).
Colicin M (ColM) is the smallest colicin (271 amino acid residues) identified to date and the only one known to interfere with the biogenesis of the cell envelope (18, 21, 22, 41). It is imported into cells via the outer membrane receptor FhuA and the TonB translocation system (10, 25, 35). Its precise target and mode of action were elucidated only recently (18). ColM was demonstrated to be an enzyme (phosphodiesterase) catalyzing the hydrolysis of the peptidoglycan lipid II precursor undecaprenyl-pyrophospho-MurNAc(-pentapeptide)-GlcNAc (where MurNAc and GlcNAc represent N-acetylmuramic acid and N-acetylglucosamine, respectively), thereby inhibiting peptidoglycan polymerization steps and provoking cell lysis. The site of cleavage was unambiguously located between the undecaprenyl (C55) and pyrophospho groups of the peptidoglycan precursor (18). More recently, dissection experiments which allowed us to more precisely localize the C-terminal toxicity domain (residues 124 to 271) of ColM were performed (6, 8).
Most colicins are plasmid encoded, and strains are generally protected from the toxin that they produce by concomitant expression of a gene encoding a highly specific immunity protein. In most cases, a third plasmid gene that codes for a lysis protein required for release of the colicin in the external medium is also present (12). Nuclease-type colicins are released in tight complex with their cognate immunity proteins, thereby inhibiting their nuclease activity and preventing cellular autodestruction. The situation is different for ColM: strains producing this colicin do not express a lysis gene; the corresponding immunity protein (Cmi, also named ImM) is located in the periplasm and remains anchored to the outer side of the cytoplasmic membrane (20, 32), i.e., the site where ColM exerts its cytotoxic activity. Cmi is composed of 117 amino acid residues (13.8 kDa) and is anchored in the membrane via its N-terminal transmembrane domain (residues 1 to 23) (34). It was earlier shown that a truncation mutant of Cmi lacking the first 23 amino acids (Cmi Δ1-23) targeted to the periplasm still conferred immunity against exogenously added ColM, although much less efficiently, indicating that immobilization of Cmi in the membrane was not absolutely required (20).
The mechanism by which the Cmi immunity protein protects cells from the cytotoxic activity of ColM still remains to be elucidated. It had been proposed earlier that Cmi inactivated ColM during its import, while it is in the periplasm and before it can reach its target in the cytoplasmic membrane (20). We report here the large-scale purification and X-ray structure of the periplasmic domain of the immunity protein. The structure reveals that it has an α-β fold, which has previously been found in cystatins and amine oxidases. The 2 cysteine residues of Cmi form a disulfide bond that is important for stabilization of the protein structure in the periplasmic space and protection of cells against the peptidoglycan precursor-degrading activity of ColM. Furthermore, we show that residues located at the C-terminal extremity of the Cmi protein are crucial for the immunity-conferring activity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Strains and plasmids are described in Table 1. E. coli strain DH5α (Bethesda Research Laboratories) was used for propagation of plasmids, and strain Rosetta(DE3)(pLysSRARE) (Novagen) was used for overproduction of the Cmi protein. The BW25113 strain was obtained from B. Wanner (16), and its derivatives carrying deletions of the dsbA and dsbC chromosomal genes were from the Keio collection (5). Plasmid vector pTrc99A was obtained from Amersham Biosciences, and the construction of plasmids pTrcHis60 and pET2160 was previously described (11, 36). The pTO4 plasmid carrying the cma gene encoding ColM and the cmi immunity gene has also been previously described (33). 2YT medium (29) was used to grow the cells, and growth was followed by monitoring the optical density of the cultures at 600 nm (OD600) with a Shimadzu UV-1601 spectrophotometer. Ampicillin, kanamycin, and chloramphenicol were used at 100, 50, and 25 μg·ml−1, respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain/plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| DH5α | supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 φ80dlacZΔM15 | BRL |
| Rosetta(DE3) | F−ompT hsdSB (rB− mB−) gal dcm lacY1 (DE3) | Novagen |
| BW25113 | lacIqrrnBT14 ΔlacZWJ16hsdR514 ΔaraBADAH33 ΔrhaBADLD78 | 16 |
| JW1836 | BW25113 ΔyebF::Kanr | 5 |
| JW3832 | BW25113 ΔdsbA::Kanr | 5 |
| JW2861 | BW25113 ΔdsbC::Kanr | 5 |
| Plasmids | ||
| pLysSRARE | Camr | Novagen |
| pTrc99A | Ampr | 3 |
| pTrcHis60 | pTrc99A derivative, Ampr | 36 |
| pET2160 | pET21d derivative, Ampr | 10 |
| pMLD232 | pTrc99A::cmi | 8 |
| pMLD234 | pTrcHis60::cmi | This study |
| pFFG11 | pET2160::cmi | This study |
| pFFG12 | pET2160::cmiΔ1-23 | This study |
| pMLD307 | pTrc99A::cmiΔ117 | This study |
| pMLD308 | pTrc99A::cmiΔ116-117 | This study |
| pMLD309 | pTrc99A::cmiΔ115-117 | This study |
| pMLD310 | pTrc99A::cmiΔ114-117 | This study |
| pMLD311 | pTrc99A::cmiΔ113-117 | This study |
| pMLD312 | pTrc99A::cmiΔ112-117 | This study |
| pMLD314 | pTrcHis60::yebF | This study |
| pMLD316 | pTrcHis60::yebF1-32-cmi29-117 | This study |
| pFFG11-Cys31Alaa | pET2160::cmiCys31Ala | This study |
| pMLD232-Cys31Alaa | pTrc99A::cmiCys31Ala | This study |
For clarity, only plasmids expressing the Cys31A mutated form of Cmi are indicated here, but all the other point mutations mentioned in the text, from Lys27Ala to Tyr114Ala, were similarly introduced in both the pFFG11 and pMLD232 plasmids. Genes encoding C-terminally truncated forms of Cmi (from Δ117 to Δ112-117) were expressed only in the pTrc99A vector. The oligonucleotides used for construction of these different expression vectors and site-directed mutagenesis experiments are shown in Table S1 in the supplemental material.
General DNA techniques.
PCR amplification of genes was performed in a Thermocycler 60 apparatus (Bio-med) using Expand high-fidelity polymerase (Roche). DNA fragments were purified using a Wizard PCR Preps DNA purification kit (Promega), and standard procedures for DNA digestion, ligation, and agarose gel electrophoresis (40) were used. Plasmid isolations were carried out by the alkaline lysis method (40), and E. coli cells were transformed with plasmid DNA by the method of Dagert and Ehrlich (15) or by electroporation.
Construction of expression plasmids.
Different plasmids allowing overexpression of the ColM immunity gene, cmi, were constructed as follows. The cmi gene was amplified by PCR from the pTO4 plasmid using primers Cmi1 and Cmi2 (see Table S1 in the supplemental material), and the amplified material was digested by BspHI and PstI and inserted between the compatible NcoI and PstI sites of the pTrc99A vector, generating plasmid pMLD232 (8) that allows cmi expression under the control of the strong isopropyl-β-d-thiogalactopyranoside (IPTG)-inducible trc promoter. For expression of a C-terminally His6-tagged form of Cmi, the gene was amplified with primers Cmi1 and Cmi3, and the resulting fragment was cut with BspHI and BglII and inserted between the NcoI and BglII sites of pTrcHis60, generating plasmid pMLD234. The latter insert was also cloned between the NcoI and BglII sites of the pET2160 vector, generating plasmid pFFG11. For expression of the soluble form of the Cmi protein lacking the first 23 amino acids (Cmi Δ1-23) in C-terminally His6-tagged form, the truncated cmi gene was amplified using primers Cmi4 and Cmi3, and the resulting DNA was cut with BspHI and BglII and inserted between the NcoI and BglII sites of pET2160, yielding the pFFG12 plasmid. Site-directed mutagenesis of the wild-type and C-terminally His6-tagged forms of the Cmi protein was performed directly on the corresponding expression plasmids, pDML232, pDML234, or pFFG11, by using a QuikChange II XL site-directed mutagenesis kit (Stratagene). Pairs of complementary oligonucleotides that were used for introducing mutations in the gene sequence are shown in Table S1 in the supplemental material. Cmi variants lacking from 1 to 6 of the C-terminal amino acid residues were also generated: these truncated forms of the cmi gene were amplified using Cmi1 and Cmi-stop primers, and the resulting PCR fragments were cut by BspHI and BglII and then cloned into pTrcHis60. A plasmid expressing the E. coli yebF gene was also constructed, as follows: the yebF sequence was amplified by PCR from the E. coli chromosome using the YebF1 and YebF2 primers, and the DNA fragment was cut by BspHI and BamHI and then cloned between the compatible NcoI and BglII sites of pTrcHis60, yielding the pMLD314 plasmid, allowing expression of YebF in C-terminally His6-tagged form. A plasmid expressing a YebF-Cmi chimera protein in which the first 28 amino acid residues (MKVI… EDKG) of Cmi were replaced by the 32 first residues of the E. coli YebF protein (MKKR… SVTF) was constructed as follows: first, the N-terminal sequence of YebF and the N-terminally truncated fragment of Cmi were amplified by PCR using as primers the pairs of oligonucleotides YebF1/Chimrev and Chimfor/Cmi5, respectively; the two resulting products were then hybridized and used as the template for PCR amplification of the complete gene fusion using primers YebF1 and Cmi5. The latter fragment was cut by BspHI and BamHI and inserted within the compatible restriction sites, NcoI and BglII, of pTrcHis60, yielding pMLD316. The sequences of all the cloned inserts and mutagenesis products were verified by DNA sequencing (Eurofins MWG Operon, Germany).
Expression and purification of Cmi Δ1-23.
Rosetta(DE3)(pLysSRARE) cells carrying plasmid pFFG12 were grown at 37°C in 2YT medium supplemented with ampicillin and chloramphenicol (1-liter cultures). When the optical density of the culture reached 0.5, the temperature was decreased to 15°C. Two hours later, IPTG was added at a final concentration of 1 mM and growth was continued overnight at 15°C. Cells were harvested and washed twice with 100 ml 20 mM Tris-HCl buffer, pH 7.5, containing 200 mM NaCl, 10 mM MgCl2, and 10 mM 2-mercaptoethanol (buffer A). Cells were disrupted by means of a French press, and the resulting suspension was centrifuged at 100,000 × g for 30 min in a TL100 Beckman centrifuge. The C-terminally His6-tagged Cmi protein was then purified from the supernatant fraction (under native conditions) by chromatography on nickel-nitrilotriacetate (Ni2+-NTA) agarose, following the manufacturer's recommendations (Qiagen). After binding of the protein to the polymer and extensive washing with buffer A containing 15 mM imidazole, elution of Cmi Δ1-23 was observed with 100 to 400 mM imidazole. The corresponding fractions were pooled, concentrated by ultrafiltration (10-kDa nominal-molecular-mass-cutoff Amicon Ultra-15 centrifugal filter; Millipore), and dialyzed overnight against 100 volumes of buffer A. In order to further purify and determine the oligomerization state of the protein, samples of the purified preparation were loaded onto a Hi-Load 16/60 Superdex 75 column (GE Healthcare) and elution was performed with buffer A at a flow rate of 1 ml·min−1. The Cmi protein was recovered under one main peak, and its elution volume was consistent with the protein being a monomer. Standards used for column calibration were β-amylase, alcohol dehydrogenase, bovine serum albumin, carbonic anhydrase, cytochrome c, and vitamin B12.
Protein monitoring.
SDS-PAGE analysis of proteins was performed as described by Laemmli and Favre (27). The protein concentration was determined by using the Bradford procedure (9) with bovine serum albumin as the standard or by quantitative amino acid analysis with a Hitachi model L8800 analyzer (ScienceTec), after hydrolysis of the samples for 24 h at 105°C in 6 M HCl containing 0.05% 2-mercaptoethanol.
Immunoassay for detection of His6-tagged proteins.
Protein samples (crude cell extracts or the resulting membrane fractions) were separated by SDS-PAGE, and proteins were transferred to Immobilon-P membranes (Millipore) by electrophoresis. Immunological blotting was carried out with anti-His horseradish peroxidase conjugate antibody (Qiagen), used at a dilution of 1:2,000. The protein bands were visualized by chemoluminescent reaction following the manufacturer's recommendations (Perkin-Elmer).
MALDI-TOF mass spectrometry.
Matrix-assisted laser desorption ionization-time-of-flight (MALDI-TOF) mass spectra were recorded in the linear mode with delayed extraction on a PerSeptive Voyager-DE STR instrument (Applied Biosystems) equipped with a 337-nm laser. Buffer was removed from the samples by using a ZipTip C4 pipette tip (Millipore), according to the manufacturer's recommendations with slight modifications. Briefly, the purified Cmi Δ1-23 protein was adsorbed on the ZipTip and, after it was washed with 0.1% trifluoroacetic acid (TFA), was eluted with 7.5 μl of 0.1% TFA in 70% acetonitrile. Subsequently, 1 μl of matrix solution (10 mg·ml−1 sinapinic acid in 0.1% TFA-acetonitrile [70/30, vol/vol]) was deposited on the plate, followed by addition of 0.5 or 1 μl of concentrated protein. After evaporation of the solvents, spectra were recorded in the positive mode at an acceleration voltage of +25 kV and with an extraction delay time of 300 ns. Cytochrome c was used as an external calibrant.
Crystallization trials.
Initial crystallization trials using a commercial vapor diffusion screen identified an initial crystallization condition which was composed of 100 mM morpholineethanesulfonic acid (MES) buffer, pH 6.5, and 12% polyethylene glycol (PEG) 20000 (Classics screen, condition 96; Qiagen) using a protein concentration of approximately 30 mg·ml−1. Further optimization of this condition using an in-house-developed additive screen identified 2 of 96 conditions which yielded an obvious improvement in the crystals: 1 mM FeCl3 and 90 mM CdSO4. Before they were flash frozen in liquid nitrogen, the crystals were briefly washed in cryoprotectant containing 30% glycerol. Data were collected at the Proxima 1 beam line of Synchrotron Soleil, Saint-Aubin, France. In order to obtain phases of Cmi, crystals were treated overnight with a freezing solution in which CdSO4 was replaced by 100 mM ZnCl2, before being frozen in liquid nitrogen.
Structure solution.
Data sets collected at the zinc K absorption edge (1.282 Å) in a single-wavelength anomalous dispersion (SAD) experiment indicated the presence of strong anomalous dispersion. The SHELXD program (42) was able to locate the corresponding zinc substructure, the coordinates, and B factors, and the occupancies of the substructure were then refined with the SHARP program (17). Initial phases, again calculated using SHARP, were improved using the solvent flipping and density modification algorithms implemented by the SOLOMON (1) and DM (14) programs, respectively. The atomic model of Cmi was built and refined initially by the program BUCCANEER (13) and then by the program PHENIX.AUTOBUILD (44), which were able to construct the two molecules in their entirety. Further rounds of refinement and adjustments to the model were performed using the PHENIX.REFINE (2) and COOT (19) programs, respectively. Final refinement was performed with the REFMAC program (30) while applying tight noncrystallographic restraints from residues 28 to 99 between chains A and B and while modeling the TLS (translation libration screw mode) groups as one group for chain A and one group for chain B.
ColM susceptibility assay.
BW25113 and derivative strains (dsb mutants and Cmi-expressing strains) were grown exponentially in 2YT medium at 37°C (25-ml cultures). At the appropriate OD600, pure C-terminally His6-tagged ColM was added at various concentrations and cell growth was followed by monitoring the absorbance at constant time intervals.
Materials and chemicals.
Pure C-terminally His6-tagged colicin M was prepared as described previously (8). IPTG was obtained from Eurogentec, and Ni2+-NTA agarose was obtained from Qiagen. Antibiotics and reagents were from Sigma. DNA ligase and restriction enzymes were obtained from New England Biolabs, and DNA purification kits were from Promega and Macherey-Nagel. Eurofins MWG performed syntheses of oligonucleotides as well as DNA sequencing.
Protein structure accession number.
The atomic coordinates and structure factors corresponding to the structure of Cmi presented here have been deposited in the Protein Data Bank (PDB), Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/), with the accession code 2xgl.
RESULTS AND DISCUSSION
Purification of Cmi.
The cmi open reading frame contains four putative ATG initiation codons (32). Ölschläger and coworkers earlier determined the N-terminal amino acid sequence of the protein expressed in vivo to be MKVIS, designating the third ATG as the actual translation start signal (34). The protein sequence starts with a 23-residue hydrophobic region which likely functions as a signal sequence but which does not end with a typical cleavage site. Cmi is known to be located in the periplasm and to be anchored in the membrane via this N-terminal transmembrane fragment (20). In the present work, repeated attempts to overproduce the full-length Cmi protein from pTrc99A derivative vectors failed because induction of gene expression was toxic and resulted in cell lysis (data not shown). We then focused our attention on the Cmi Δ1-23 truncated variant that remains in the cytoplasm and was therefore a suitable candidate for the purpose of protein structure analysis. Moreover, this truncated form of Cmi was known to be functionally active, when it was targeted to the periplasm (this work and reference 20). No significant expression of the truncated variant was obtained from E. coli host strains BL21 and C43, but attempts were successful when the Rosetta(DE3)(pLysSRARE) strain that is adapted for expression of genes containing rare codons was used instead. SDS-PAGE analysis of induced Rosetta cell extracts showed the accumulation of a protein of about 12 kDa, a value consistent with the calculated molecular mass (12,288 Da, with the C-terminal Arg-Ser-His6 extension included). A significant proportion of the overproduced His6-tagged protein was recovered in the soluble fraction following cell disruption and centrifugation. The protein was then purified by two steps of chromatography on Ni2+-NTA and Superdex 75 (see Fig. S1 in the supplemental material). The final preparation of protein was at least 95% pure, as judged by SDS-PAGE, and the yield was approximately 15 mg of purified protein per liter of culture. MALDI-TOF mass spectrometry confirmed the purity and integrity of the Cmi preparation. Peaks of m/z 6,079.6, 12,153.5, and 24,381.9, corresponding to the [M + 2H]2+, [M + H]+, and [2 M + H]+ ions, respectively, were observed (see Fig. S1 in the supplemental material). These results were in agreement with the molecular mass of the protein without its N-terminal methionine (12,156.5 Da), indicating that the latter residue had been cleaved off during in vivo overexpression of the recombinant protein.
Crystallization and structure determination of Cmi.
Crystallization screening identified an initial crystallization condition composed of 100 mM MES buffer, pH 6.5, and 12% PEG 20000 (see Materials and Methods), which yielded improved crystals with the addition of 90 mM CdSO4. Needle-like crystals (100 μm by 10 μm by 10 μm) grew from 200-nl crystallization drops and yielded diffraction to 2.7-Å resolution (Table 2). The presence of cadmium in the crystallization solution prompted us to attempt single-wavelength anomalous dispersion experiments at long wavelengths. Despite the presence of a significant, albeit weak, anomalous signal, we were unable to solve the structure using the phases that we obtained. Therefore, the crystals were soaked overnight in a solution containing 100 mM ZnCl2. After this treatment, the crystals remained isomorphous with the native samples. After data collection at the wavelength corresponding to the zinc absorption edge, we were then able to locate the heavy atom substructure and to calculate sufficiently good phases to begin constructing the atomic model of Cmi (Table 2).
TABLE 2.
Data collection and refinement statistics
| Parameter | Native (Cd2+ containing) | SAD (Zn2+-soaked crystal) |
|---|---|---|
| Data set | ||
| Wavelength (Å) | 1.907 | 1.282 |
| Unit cell parameters (Å) | a = 52.00, b = 101.10, c = 43.52 | a = 51.52, b = 100.78, c = 43.45 |
| Space group | P21212 | P21212 |
| Resolution limits (Å)a | 50-2.7 (2.8-2.7) | 50.3-2.7 (2.8-2.7) |
| No. of observations measureda | 32,573 (1,957) | 30,164 (3,049) |
| No. of unique reflections measureda | 6,651 (622) | 6,463 (649) |
| Completeness (%)a | 99.1 (94.2) | 97.7 (98.0) |
| 〈I/σ(I)〉a | 14.3 (5.0) | 9.42 (2.62) |
| Rsym (%)a,b | 9.6 (31.1) | 13.9 (60.2) |
| Refinement statistics | ||
| No. of nonhydrogen atoms (protein/water/other) | 1478/15/32 | |
| Resolution range (Å) | 46.2-2.7 | |
| R/Rfreec (%) | 22.2/26.8 | |
| RMSD bonds (Å)/angles (°) | 0.010/1.146 | |
| 〈B〉d (Å2) for protein/water/other | 17.0/16.5/40.7 | |
| Ramachandran plot, preferred/allowed/outliers (%) | 97.73/2.27/0 |
Values in parentheses refer to the highest-resolution shell (2.8 to 2.7Å).
Rsym = ΣhΣi |〈I〉h − Ih,i|/ΣhΣi Ih,i, where 〈I〉h is the mean intensity for reflection Ih, and Ih,i is the intensity of an individual measurement of reflection, Ih.
Rfree was calculated using 308 (4.6%) of the reflections, which were set aside from the refinement. R = Σ|Fo − Fc|/Σ|Fo × 100.
〈B〉, average temperature factor.
Architecture of Cmi.
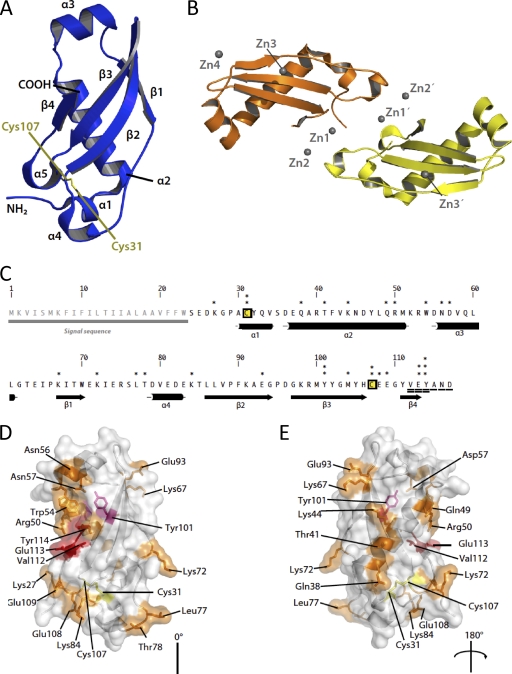
The structure of Cmi consists of four helices (α1 to α4) as well as a four-stranded antiparallel β sheet, which curves around the long α1 helix. The structure contains a disulfide bridge (Cys31-Cys107), which links the N terminus of helix α1 to loop-connecting strands β3 and β4 (Fig. 1 A). The asymmetric unit of the crystals consists of two copies of Cmi, which are related by noncrystallographic symmetry (Fig. 1B). We were able to model residues 25 to 115 of chain A and residues 27 to 115 of chain B into the electron density. The two copies of Cmi are very similar; from residues 28 to 99, the root mean square displacement (RMSD) over all atoms, calculated using the LSQKAB program (4), is 0.08. The termini display additional divergence in their structures relative to each other, since the RMSD increases to 0.27 Å over amino acids 27 to 115 (89 aligned residues). In order to estimate the oligomeric state of Cmi, analysis of the structure with the PISA program (26) was performed. The results indicate that the largest interface area between protomers (defined as the difference between the accessible surface areas of the isolated and interfacing structures, divided by two) in the crystal lattice is of 485 Å2, rather low to be physiologically relevant protein-protein interfaces. However, the crystal contacts between protomers of Cmi are notable because of the participation of metal ions in the protein-protein interfaces: either cadmium in the case of the native crystals or zinc after the ZnCl2 soak used for phasing.
FIG. 1.
(A) Architecture of Cmi. The colicin M immunity protein is shown as blue cartoons. Elements of secondary structure detected by the DSSP program (24) are labeled accordingly, as are the amino and carboxy termini of the protein. The disulfide bridge between Cys31 and Cys107 is also highlighted in yellow stick form. (B) Contents of the asymmetric unit. Four Zn2+ ions (Zn1A, Zn1B, Zn2A, and Zn2B) line the interface between the two monomers. Zn3A and Zn3B are situated approximately centrally on the β sheet, whereas Zn4 is distant from the main zinc-mediated interface. Chain A is depicted in orange, chain B is yellow, and gray spheres represent zinc ions. This figure was prepared using the PyMol program (http://pymol.sf.net). (C) Primary structure of Cmi, along with a schematic representation of the secondary structure below. Cysteine residues are labeled yellow and boxed; asterisks denote residues selected for mutagenesis. Two and three asterisks represent the residues that exhibit a medium phenotype (MIC of ColM, 5 ng·ml−1) and a strong phenotype (MIC of ColM, 1 ng·ml−1), respectively, when they were tested with the pTrc99A plasmid (Table 3). Residues that were C-terminally truncated are underscored, and residues showing a strong phenotype when they are deleted are doubly underlined. (D and E) Side chains of the residues highlighted in panel C are shown as sticks and are color coded as follows: orange, residues which did not yield a strong or medium phenotype when they were mutated or truncated; pink, residues which showed a medium phenotype when they were mutated; red, residues which most severely affected the function of Cmi when they were either truncated or mutated.
Metal-binding sites in the structure.
We identified seven metal-binding sites in the crystal structure of Cmi. Of these, three pairs of sites are structurally analogous and are present in each of the two noncrystallographically related monomers (chains A and B; Fig. 1B), and the seventh site is unrelated to the others. Metal ions Zn1A and Zn1B are both coordinated by Asp82, His106, Glu109, and a chloride ion. Metal ions Zn2A and Zn2B are also related by NCS and are coordinated by side chains from Asp55, Asp79, and Glu113. The third pair of sites, Zn3A and Zn3B, is contacted by Glu71, Asp96, Met100, and a chloride ion. Zn4 is at the only unique site and is coordinated by residues Glu93 from both chains A and B (see Table S3 and Fig. S2 in the supplemental material).
The fold of Cmi is reminiscent of cystatins.
Although there is no significant sequence similarity between Cmi and proteins of known structure in the PDB, a search of the PDB using the DALI algorithm (23) revealed that Cmi shows notable structural similarity to various domains of amine oxidases, phenylethylamine oxidase, latexin, and carboxypeptidase A4, although these proteins do not share significant sequence similarity (see Table S2 and Fig. S3 in the supplemental material). These domains are representatives of the cystatin fold superfamily, as defined by the structural classification of proteins (SCOP) (31). Despite the resemblance between Cmi and cystatins, we find it unlikely that Cmi would inhibit either cysteine proteases (the target of cystatins) or ColM by a similar mechanism, because of (i) the absence of the N-terminal sequence in Cmi, which in stefin B inserts into the active site and forms the major point of contact, and (ii) significant clashes that would probably occur between one helix of Cmi and the active site of the cysteine protease. A superposition of Cmi and stefin B, which has been solved in complex with papain (43), is illustrated in Fig. S4 in the supplemental material. It depicts the nature of such clashes, as well as the nature of the interaction between the N-terminal sequence of stefin B and papain.
Sequence similarity between Cmi and members of the YebF family.
A protein sequence alignment generated by a PSI-BLAST search of the nonredundant GenBank protein sequence database showed that Cmi exhibits significant sequence similarity with a family of proteins of unknown function (YebF; Interpro accession number IPR020236) and similar size (117 to 136 residues). Cmi and the YebF orthologues, which showed from 26% (E. coli YebF) to 35% (YebF from Yersinia species) identity over about 90 aligned residues, shared a series of highly conserved residues (Fig. 2). Interestingly, the 2 cysteine residues that formed a disulfide bond in the crystal structure of Cmi appeared to be conserved in all of the YebF sequences (Fig. 2 and data not shown).
FIG. 2.
Sequence alignment of Cmi and YebF proteins. Amino acid sequences of YebF orthologues from Yersinia pestis CO92 (YERPE), Yersinia enterocolitica 8081 (YEREN), Escherichia coli (ECOLI), and Salmonella enterica serovar Typhimurium LT2 (SALTY) were aligned with the Cmi sequence using the ClustalW2 program (28). The 21-residue sequence signal of E. coli YebF that is removed following translocation of the protein to the periplasm and the 23-residue sequence of Cmi involved in attachment of the immunity protein to the membrane are boxed.
Recent investigations of the YebF protein of E. coli showed that it was released into the extracellular medium following its secretion and maturation (cleavage of an N-terminal 21-residue sec leader sequence) into the periplasm (46). This release was so efficient that YebF, whose function still remains to be identified, was then exploited as a carrier protein for the translocation through the cell envelope and the release in the external medium of a variety of heterologous recombinant proteins. Indeed, YebF-fused passenger proteins of various sizes and hydrophilicities were carried in their active states to the medium, providing a powerful tool to circumvent toxicity and other contamination issues associated with production of proteins in E. coli (46). More recently, a global analysis of the E. coli periplasmic protein content revealed that the abundance of YebF was greatly diminished in dsbA and dsbC mutant strains, demonstrating that YebF was a substrate for the oxidase DsbA and disulfide isomerase DsbC proteins that play an essential role in oxidative protein folding (45). As yebF gene expression was not reduced in these strains, the depletion of YebF was interpreted as an impaired folding of this protein in the periplasmic space. Further analysis of the in vivo redox state of periplasmic YebF was performed using a differential thiol-trapping technique, which confirmed the formation of a disulfide bond involving the 2 cysteine residues (Cys35 and Cys108; Fig. 2) of this newly identified substrate of DsbA (45).
Considering the significant similarity between the Cmi and YebF sequences, we wondered whether the expression of YebF could have any effect on the cell susceptibility to ColM. Neither E. coli mutant strain BW25113 ΔyebF nor yebF-overexpressing strain BW25113(pMLD314) behaved differently from the wild-type strain with respect to susceptibility to ColM (data not shown), demonstrating that YebF did not exhibit any Cmi-like activity. Then, we constructed a Cmi variant in which the first 28 residues (including the sequence from residues 1 to 23 involved in the anchoring of the protein to the membrane) were replaced by the 32 first residues of E. coli YebF (encompassing the 21-residue sec leader sequence). The lengths of the exchanged fragments were chosen so that they extend up to the first invariant residue (proline) revealed by alignments of the Cmi and YebF sequences (Pro29 and Pro33 in Cmi and YebF, respectively; Fig. 2). BW25113(pMLD316) cells expressing this YebF-Cmi chimera protein appeared to be resistant to high doses of ColM (Table 3), indicating that fusion of the N-terminal sequence of YebF allowed expression and translocation to the periplasm of the Δ1-28 truncated form of Cmi, which then proved to be functionally active. As cleavage of the YebF-Cmi chimera sec leader sequence is expected to occur in the periplasm, these results confirmed that immobilization of the Cmi protein in the membrane was not absolutely required for activity.
TABLE 3.
ColM immunity-conferring activity of Cmi mutant proteinsa
| Protein | ColM MIC (ng·ml−1) for plasmid vector: |
|
|---|---|---|
| pET2160 | pTrc99A | |
| None (empty vector) | 1.5 | 1.5 |
| Wild-type Cmi | 15 | >1,500 |
| Gln49Ala, Asn56Ala, Lys72Ala | 15 | >100 |
| Lys44Ala, Lys67Ala, Thr78Ala, Glu93Ala, Glu109Ala | 10 | >100 |
| Lys27Ala, Gln38Ala, Thr41Ala, Arg50Ala, Trp54Ala, Asp57Ala, Leu77Ala, Lys84Ala, Met104Ala, Glu108Ala | 5 | >100 |
| Cys31Ala, Cys107Ala, Tyr101Ala, Cys31Ala/C107Ala, Glu113Ala | 1 | 5 |
| Tyr114Ala | 1 | 1 |
| Δ117 | NDb | >100 |
| Δ116-117 | ND | >100 |
| Δ115-117 | ND | >100 |
| Δ114-117 | ND | 1 |
| Δ113-117 | ND | 1.5 |
| Δ112-117 | ND | 1 |
| YebF | ND | 1.5 |
| YebF1-32-Cmi29-117 | ND | >100 |
Site-directed mutagenesis of selected Cmi residues was performed directly on the cmi-expressing plasmids pFFG11 and pMLD232 (pET2160 and pTrc99A derivative plasmids, respectively). Other plasmids expressing truncated Cmi variants as well as YebF and YebF-Cmi chimera proteins were constructed as described in the text. BW25113 cells carrying these plasmids or the corresponding control vectors were grown as described in Materials and Methods. When the OD600 of cultures reached 0.2, pure ColM was added at various concentrations and ColM MICs were determined. Mutations leading to similar effects on the immunity activity of Cmi, as judged by the susceptibility to ColM of transformants, are grouped together.
ND, not determined (these mutations were introduced only in the pTrc99A derivative plasmid).
Role of cysteine residues in the structure and function of Cmi.
As mentioned above, Cmi and all members of the YebF protein family contained 2 invariant cysteine residues located very close to the N- and C-terminal extremities of the mature forms of these proteins. In fact, these protein sequences did not contain any additional cysteine residues (Fig. 2). The observation that the Cys31 and Cys107 residues formed an intramolecular disulfide bond in the crystal structure of Cmi suggested that they should play an essential role in the stabilization of the protein structure and/or maintenance of a functional conformation. The demonstration that the corresponding cysteine residues of YebF (Cys35 and Cys108; Fig. 2) were mainly detected in oxidized form in the periplasmic space of a wild-type E. coli strain and that YebF abundance was dramatically reduced in a dsbA mutant strain (45) was consistent with this hypothesis. To assess the role of the 2 cysteine residues in the structure and/or immunity-conferring activity of the Cmi protein, mutagenesis experiments targeting the Cys31 and Cys107 residues were performed, as were Cmi expression assays in dsbA and dsbC mutant cell backgrounds.
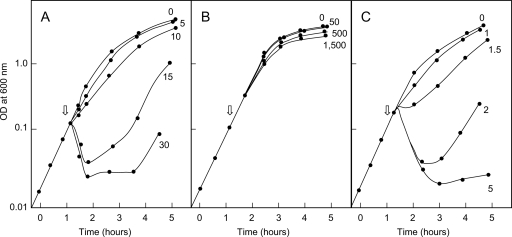
Growth inhibition of reference strain BW25113 was observed with concentrations of ColM higher than 1 ng·ml−1. Basal expression (in the absence of IPTG inducer) of the wild-type cmi gene under the control of the strong trc promoter in plasmid pMLD232 allowed cells to resist the highest concentration of ColM tested (1,500 ng·ml−1; Fig. 3 B). The pET derivative plasmid pFFG11 yielded a lower level of expression of Cmi that could be titrated within a lower range of ColM concentrations, 1 to 15 ng·ml−1 (Fig. 3A). Site-directed mutagenesis of Cmi was thus performed in parallel on the two types of plasmids, allowing us to modulate the level of protein expression and estimate the activity of Cmi variants having either moderately altered or greatly altered ColM immunity-conferring activity.
FIG. 3.
ColM resistance levels conferred by Cmi expression plasmids. BW25113 cells carrying either one of the cmi-expressing plasmids, pFFG11 (A) or pMLD232 (B), or the control vector, pET2160 (C), were grown as described in Materials and Methods. When the OD600 of the cultures reached 0.2, pure colicin M was added at various concentrations (the indicated values are in ng·ml−1).
pFFG11 derivative plasmids expressing Cys31Ala, Cys107Ala, and Cys31Ala/Cys107Ala mutated forms of Cmi protein failed to confer ColM immunity to BW25113 cells (Table 3). When these mutations were introduced into plasmid pMLD232, which allows expression of the Cmi protein to higher levels, very-low-level resistance of transformants to ColM (5 ng·ml−1) was observed. These results showed that the 2 cysteine residues played an essential role in the structure and/or activity of Cmi, which was consistent with the observation that they formed a disulfide bond in the crystal structure. Western blot experiments then showed that the levels of the three cysteine mutants (two with single mutations and one with the double mutation) in cell membranes were much lower than those detected in the wild-type strain (see Fig. S5 in the supplemental material). The failure of these Cmi variants to protect cells against ColM could therefore be explained, at least partly, by their decreased abundance in the cell membranes. The low-level protection conferred by these constructs suggested that the cysteine residues and disulfide bond likely play a crucial role during folding and stabilization of the protein conformation in cell membranes but were not absolutely required for expression of the immunity-conferring activity of this protein.
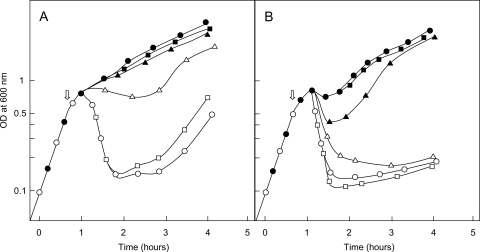
Next, the effects of dsbA and dsbC gene deletions on the susceptibility of BW25113 cells to ColM and on the expression and/or activity of the Cmi immunity protein were investigated. The dsbA mutant strain appeared to be significantly more resistant to ColM than the wild-type and dsbC mutant strains. Indeed, a 4- to 10-fold higher concentration of ColM was required to provoke its lysis at a comparable rate (Fig. 4). Interestingly, Vertommen et al. earlier reported that expression of FhuA, the ColM receptor protein, was significantly reduced in a dsbA genetic background (45). As FhuA contains 4 cysteine residues that normally form two consecutive disulfide bonds, it was speculated that in the absence of DsbA, intermolecular disulfide bonds that prevented FhuA from correctly folding in the outer membrane were formed. The decreased susceptibility to ColM of the dsbA mutant strain observed here was therefore consistent with a reduced uptake of the toxin via FhuA. We then tested whether the dsbA and dsbC mutations had some impact on the expression and/or activity of the Cmi immunity protein. As shown in Fig. 4, the immunity conferred by the pFFG11 plasmid was dramatically diminished in the dsbA mutant strain compared to that in the wild-type and dsbC mutant strains. As the dsbA mutant is intrinsically more resistant to ColM that the two other strains (see above), these data suggested either a decreased abundance, an incorrect folding, or an altered activity of the Cmi protein in dsbA cell membranes. Immunoassays showed that the amounts of wild-type Cmi protein detected in membranes were indeed much lower in the dsbA mutant than in the wild-type strain (see Fig. S5 in the supplemental material), as observed previously for the YebF protein (45). These data clearly showed the involvement of DsbA oxidase in Cmi disulfide bond formation and correct protein folding. The depletion of wild-type Cmi that was observed in a dsbA genetic background was reminiscent of that observed for Cys31Ala/Cys107Ala Cmi mutant proteins in a wild-type background, with both contexts leading to an inhibition of the formation of the disulfide bond of Cmi in the periplasm, either partially (dsbA strain) or totally (cysteine mutants).
FIG. 4.
Effects of dsbA/dsbC mutations on Cmi expression and cell susceptibility to ColM. Cells of BW25113, BW25113 ΔdsbA, and BW25113 ΔdsbC strains carrying either the cmi-expressing plasmid pFFG11 or the empty vector pET2160 were grown as described in Materials and Methods. When the OD600 of the cultures reached 0.6, pure ColM was added at a final concentration of either 10 ng·ml−1 (A) or 40 ng·ml−1 (B). Circles, triangles, and squares, wild-type strain and dsbA and dsbC mutant strains, respectively; closed and open symbols, strains carrying the pFFG11 and pET2160 plasmids, respectively.
Site-directed mutagenesis analysis of Cmi.
The elucidation of the structure of Cmi prompted us to develop a detailed structure-activity relationship analysis of the immunity function. Since no information on residues that a priori could be functionally important was available, we developed an alanine-scanning mutagenesis approach to probe predominantly surface-exposed residues for their involvement in the protection of E. coli against lethal effects of ColM. Some priority in the choice of mutated residues detected as invariant or highly conserved in sequences of YebF proteins was given (Fig. 2). Twenty-three point mutations that targeted the following Cmi residues were generated: Cys31 and Cys107 (see above), as well as Lys44, Gln49, Arg50, Trp54, Asp57, Tyr101, and Tyr114, which appeared as invariant residues, and Lys27, Gln38, Thr41, Asn56, Lys67, Lys72, Leu77, Thr78, Lys84, Glu93, Met104, Glu108, Glu109, and Glu113, which were chosen among the surface-exposed residues.
BW25113 cells carrying pFFG11 derivative plasmids expressing the Lys27Ala, Gln38Ala, Thr41Ala, Lys44Ala, Gln49Ala, Arg50Ala, Trp54Ala, Asn56Ala, Asp57Ala, Lys67Ala, Lys72Ala, Leu77Ala, Thr78Ala, Lys84Ala, and Glu93Ala Cmi mutations were shown to be resistant to ColM concentrations over the range of 5 to 15 ng·ml−1 (Table 3), indicating that these residues did not play a crucial role in the immunity-conferring activity of Cmi. Increasing the level of expression of these Cmi variants (the same mutations listed above introduced in the pMLD232 plasmid) allowed cells to resist ColM at 100 ng·ml−1. In contrast, pFFG11 derivative plasmids expressing the Glu113Ala and Tyr114Ala mutated forms of Cmi did not protect cells against ColM. Transfer of the latter mutations into pMLD232 yielded only a very low level of resistance (5 ng·ml−1 ColM) for cells expressing the Glu113Ala mutant but not for those expressing the Tyr114Ala mutant (Table 3), suggesting that these 2 residues were important for the Cmi activity. Immunoassays were performed and showed that the amounts of mutant protein Tyr114Ala detected in cell membranes were much lower than those of the wild-type protein (see Fig. S5 in the supplemental material), as was also the case for the cysteine mutants. All other mutant proteins, including the Glu113Ala variant, were detected at a wild-type level (see Fig. S5 in the supplemental material; data not shown). Moreover, that the Tyr101Ala mutation gave rise to a reduction in the potency of Cmi was not surprising, given that this tyrosine residue is buried and contributes to the hydrophobic core.
The finding that the few residues the substitution of which yielded a dramatic reduction of Cmi activity were clustered in the C-terminal region of the protein, near and distal to the position of the disulfide bond, suggested a particular involvement of this specific region in ColM immunity (Fig. 1). This prompted us to generate a series of truncated Cmi variants lacking from 1 (Δ117) to 6 (Δ112-117) of the C-terminal protein residues. In vivo expression of these constructs (in the pTrc99A vector) showed that deletion of 1, 2, or 3 residues from the C-terminal extremity did not affect the Cmi activity, with the corresponding strains being resistant to high doses of ColM. Further deletion of the fourth residue or more residues of Cmi then completely abolished the immunity-conferring properties of Cmi (Table 3).
Conclusions.
It was recently demonstrated that ColM was an enzyme (phosphodiesterase) which interferes with the biosynthesis of bacterial cell wall peptidoglycan through the specific degradation of the essential lipid intermediates I and II of this pathway, thereby provoking cell lysis. Although it is well established that Cmi confers to cells immunity against these deleterious effects of ColM, by which mechanism this protection occurs still remains to be elucidated. In their 1991 paper, Ölschläger et al. evoked three possible mechanisms: (i) impedance of ColM uptake, (ii) interaction of Cmi with the ColM targets (later identified as lipids I and II), and (iii) interaction of Cmi with ColM (34). Their results were consistent with only the third of these proposed modes of action. Particularly interesting was the observation that degradation of periplasmic Cmi by trypsin could be partially prevented by the addition of ColM, suggesting a shielding effect likely imparted by the interaction of the two components. Additionally, overexpression of the genes encoding the ColM uptake system (FhuA and TonB machinery) rendered cells expressing Cmi more susceptible to ColM. Overexpression of Cmi compensated for this increased uptake of ColM, indicating that the immunity to ColM action that Cmi conferred was directly titrated, as if a direct interaction between these two proteins indeed took place. However, all attempts to demonstrate such an interaction in vitro have been unsuccessful to date. In particular, no formation of a ColM-Cmi protein complex could be detected by analytical gel filtration and multiangle light scattering (MALS) (see Fig. S6 in the supplemental material). Moreover, no inhibition of the lipid II-degrading activity of ColM could be observed in vitro when a standard ColM assay (8) was performed in the presence of an excess of immunity protein (Cmi/ColM ratio of 20), suggesting that the Cmi protein does not interact efficiently with the ColM protein and/or the lipid II substrate under these assay conditions.
We have now determined the structure of the ColM immunity protein, revealing a fold that is reminiscent of cystatins, the cysteine protease inhibitors, but which is also found in other unrelated proteins. The knowledge of this structure also generates valuable insights into the structure and, potentially, the function of YebF proteins that exhibit significant sequence similarity with Cmi. In particular, the Cmi structure contains a disulfide bridge that is strictly conserved within the YebF family and that plays an essential role for the structure and function of Cmi in vivo. Site-directed mutagenesis of selected conserved and surface-accessible residues of Cmi showed that alteration of only a few side chains located at the C-terminal region of the protein had deleterious effects on the immunity-conferring activity of Cmi. This information will be very useful in the development of further investigations aimed at elucidating the precise mode of action of this immunity protein.
This structure also exemplifies the utility of metal-binding sites which mediate crystal contacts and therefore permit crystal growth. It has been proposed previously that aspartate and glutamate residues be introduced by point mutation at the surface of proteins that are recalcitrant to crystallization, so that they may then be crystallized in the presence of zinc cations (39). Fortunately, Cmi provided sufficient metal-binding sites in its native form for a similar outcome to occur serendipitously, which also allowed us to obtain experimental phases using single-wavelength anomalous dispersion and without the need to resort to more involved procedures, such as metabolic selenium labeling of the protein samples.
By producing ColM, E. coli cells make use of a highly efficient method of eliminating competitor cells from their immediate environment. The pathway on which this bacteriocin acts, i.e., the biosynthesis of peptidoglycan, is the same as that targeted by β-lactam antibiotics, which remain the main class of antibacterial agents in therapeutic use today. ColM has therefore exploited this pathway as an Achilles heel of bacteria, as have humans, in the form of penicillin and variants of it. Because of the rise in the occurrence of multidrug-resistant bacteria, such weaknesses must be targeted to aid future antibacterial drug discovery. That the targets of ColM, the peptidoglycan lipid I and II precursors, are essential, specific to the bacterial world, and found in all types of eubacteria have opened the way to an exploitation of ColM and of its few homologues detected in some Pseudomonas species (7) for the development of antibacterial agents. It remains unclear how Cmi neutralizes the action of ColM, and further work is needed to ascertain how this takes place. However, the elucidation of the X-ray structure of Cmi will greatly facilitate this research.
Supplementary Material
Acknowledgments
Jerôme Cicolari is thanked for performing optimization screens of the Cmi crystals, Bruno Collinet is thanked for performing multiangle light-scattering experiments, and Andrew Thompson is thanked for help with characterizing the anomalous signal exhibited by the cadmium atoms in the native crystals. We thank Philippe Bouloc for providing E. coli strains from the Keio collection and Roland Lloubès and Michel Arthur for helpful discussions.
The beam time at Proxima 1, Synchrotron Soleil, provided in response to proposal 20080498 is gratefully acknowledged. This work was supported by grants from the Agence Nationale de la Recherche (PEPGLYCOL project, ANR-07-MIME-020) and the Centre National de la Recherche Scientifique (UMR 8619).
Footnotes
Published ahead of print on 29 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Abrahams, J. P., and A. G. Leslie. 1996. Methods used in the structure determination of bovine mitochondrial F1 ATPase. Acta Crystallogr. D Biol. Crystallogr. 52:30-42. [DOI] [PubMed] [Google Scholar]
- 2.Afonine, P. V., R. W. Grosse-Kunstleve, P. D. Adams, V. Y. Lunin, and A. Urzhumtsev. 2007. On macromolecular refinement at subatomic resolution with interatomic scatterers. Acta Crystallogr. D Biol. Crystallogr. 63:1194-1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann, E., B. Ochs, and K. J. Abel. 1988. Tightly regulated tac promoter vectors useful for the expression of unfused and fused proteins in Escherichia coli. Gene 69:301-315. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50:760-763. [DOI] [PubMed] [Google Scholar]
- 5.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol. Syst. Biol. 2:2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barnéoud-Arnoulet, A., H. Barreteau, T. Touzé, D. Mengin-Lecreulx, R. Lloubès, and D. Duché. 2010. Toxicity of the colicin M catalytic domain exported to the periplasm is FkpA independent. J. Bacteriol. 192:5212-5219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barreteau, H., A. Bouhss, M. Fourgeaud, J. L. Mainardi, T. Touzé, F. Gérard, D. Blanot, M. Arthur, and D. Mengin-Lecreulx. 2009. Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J. Bacteriol. 191:3657-3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Barreteau, H., A. Bouhss, F. Gérard, D. Duché, B. Boussaid, D. Blanot, R. Lloubès, D. Mengin-Lecreulx, and T. Touzé. 2010. Deciphering the catalytic domain of colicin M, a peptidoglycan lipid II degrading enzyme. J. Biol. Chem. 285:12378-12389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 10.Braun, V., K. Schaller, and H. Wolff. 1973. A common receptor protein for phage T5 and colicin M in the outer membrane of Escherichia coli B. Biochim. Biophys. Acta 323:87-97. [DOI] [PubMed] [Google Scholar]
- 11.Caravano, A., D. Mengin-Lecreulx, J. M. Brondello, S. P. Vincent, and P. Sinaÿ. 2003. Synthesis and inhibition properties of conformational probes for the mutase-catalyzed UDP-galactopyranose/furanose interconversion. Chemistry 9:5888-5898. [DOI] [PubMed] [Google Scholar]
- 12.Cascales, E., S. K. Buchanan, D. Duché, C. Kleanthous, R. Lloubès, K. Postle, M. Riley, S. Slatin, and D. Cavard. 2007. Colicin biology. Microbiol. Mol. Biol. Rev. 71:158-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cowtan, K. 2006. The Buccaneer software for automated model building. 1. Tracing protein chains. Acta Crystallogr. D Biol. Crystallogr. 62:1002-1011. [DOI] [PubMed] [Google Scholar]
- 14.Cowtan, K. 1994. ‘dm’: an automated procedure for phase improvement by density modification. Joint CCP4 ESF-EACBM Newsl. Protein Crystallogr. 31:34-38. [Google Scholar]
- 15.Dagert, M., and S. D. Ehrlich. 1979. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene 6:23-28. [DOI] [PubMed] [Google Scholar]
- 16.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de la Fortelle, E., and G. Bricogne. 1997. Maximum-likelihood heavy-atom parameter refinement in the MIR and MAD methods. Methods Enzymol. 276:472-494. [DOI] [PubMed] [Google Scholar]
- 18.El Ghachi, M., A. Bouhss, H. Barreteau, T. Touzé, G. Auger, D. Blanot, and D. Mengin-Lecreulx. 2006. Colicin M exerts its bacteriolytic effect via enzymatic degradation of undecaprenyl phosphate-linked peptidoglycan precursors. J. Biol. Chem. 281:22761-22772. [DOI] [PubMed] [Google Scholar]
- 19.Emsley, P., and K. Cowtan. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60:2126-2132. [DOI] [PubMed] [Google Scholar]
- 20.Gross, P., and V. Braun. 1996. Colicin M is inactivated during import by its immunity protein. Mol. Gen. Genet. 251:388-396. [DOI] [PubMed] [Google Scholar]
- 21.Harkness, R. E., and V. Braun. 1989. Colicin M inhibits peptidoglycan biosynthesis by interfering with lipid carrier recycling. J. Biol. Chem. 264:6177-6182. [PubMed] [Google Scholar]
- 22.Harkness, R. E., and V. Braun. 1989. Inhibition of lipopolysaccharide O-antigen synthesis by colicin M. J. Biol. Chem. 264:14716-14722. [PubMed] [Google Scholar]
- 23.Holm, L., and C. Sander. 1993. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233:123-138. [DOI] [PubMed] [Google Scholar]
- 24.Kabsch, W., and C. Sander. 1983. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers 22:2577-2637. [DOI] [PubMed] [Google Scholar]
- 25.Kock, J., T. Olschlager, R. M. Kamp, and V. Braun. 1987. Primary structure of colicin M, an inhibitor of murein biosynthesis. J. Bacteriol. 169:3358-3361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krissinel, E., and K. Henrick. 2007. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372:774-797. [DOI] [PubMed] [Google Scholar]
- 27.Laemmli, U. K., and M. Favre. 1973. Maturation of the head of bacteriophage T4. I. DNA packaging events. J. Mol. Biol. 80:575-599. [DOI] [PubMed] [Google Scholar]
- 28.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. Clustal W and Clustal X version 2.0. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 29.Miller, J. H. 1972. Experiments in molecular genetics, p. 431-435. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 30.Murshudov, G. N., A. A. Vagin, and E. J. Dodson. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 53:240-255. [DOI] [PubMed] [Google Scholar]
- 31.Murzin, A. G., S. E. Brenner, T. Hubbard, and C. Chothia. 1995. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 247:536-540. [DOI] [PubMed] [Google Scholar]
- 32.Ölschläger, T., and V. Braun. 1987. Sequence, expression, and localization of the immunity protein for colicin M. J. Bacteriol. 169:4765-4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ölschläger, T., E. Schramm, and V. Braun. 1984. Cloning and expression of the activity and immunity genes of colicins B and M on ColBM plasmids. Mol. Gen. Genet. 196:482-487. [DOI] [PubMed] [Google Scholar]
- 34.Ölschläger, T., A. Turba, and V. Braun. 1991. Binding of the immunity protein inactivates colicin M. Mol. Microbiol. 5:1105-1111. [DOI] [PubMed] [Google Scholar]
- 35.Pilsl, H., C. Glaser, P. Gross, H. Killmann, T. Ölschläger, and V. Braun. 1993. Domains of colicin M involved in uptake and activity. Mol. Gen. Genet. 240:103-112. [DOI] [PubMed] [Google Scholar]
- 36.Pompeo, F., J. van Heijenoort, and D. Mengin-Lecreulx. 1998. Probing the role of cysteine residues in glucosamine-1-phosphate acetyltransferase activity of the bifunctional GlmU protein from Escherichia coli: site-directed mutagenesis and characterization of the mutant enzymes. J. Bacteriol. 180:4799-4803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pugsley, A. P. 1984. The ins and outs of colicins. Part I. Production, and translocation across membranes. Microbiol. Sci. 1:168-175. [PubMed] [Google Scholar]
- 38.Pugsley, A. P. 1984. The ins and outs of colicins. Part II. Lethal action, immunity and ecological implications. Microbiol. Sci. 1:203-205. [PubMed] [Google Scholar]
- 39.Qiu, X., and C. A. Janson. 2004. Structure of apo acyl carrier protein and a proposal to engineer protein crystallization through metal ions. Acta Crystallogr. D Biol. Crystallogr. 60:1545-1554. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, NY.
- 41.Schaller, K., J. V. Höltje, and V. Braun. 1982. Colicin M is an inhibitor of murein biosynthesis. J. Bacteriol. 152:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider, T. R., and G. M. Sheldrick. 2002. Substructure solution with SHELXD. Acta Crystallogr. D Biol. Crystallogr. 58:1772-1779. [DOI] [PubMed] [Google Scholar]
- 43.Stubbs, M. T., B. Laber, W. Bode, R. Huber, R. Jerala, B. Lenarcic, and V. Turk. 1990. The refined 2.4 Å X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 9:1939-1947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Terwilliger, T. C., R. W. Grosse-Kunstleve, P. V. Afonine, N. W. Moriarty, P. H. Zwart, L. W. Hung, R. J. Read, and P. D. Adams. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr. D Biol. Crystallogr. 64:61-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vertommen, D., M. Depuydt, J. Pan, P. Leverrier, L. Knoops, J. P. Szikora, J. Messens, J. C. Bardwell, and J. F. Collet. 2008. The disulphide isomerase DsbC cooperates with the oxidase DsbA in a DsbD-independent manner. Mol. Microbiol. 67:336-349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, G., S. Brokx, and J. H. Weiner. 2006. Extracellular accumulation of recombinant proteins fused to the carrier protein YebF in Escherichia coli. Nat. Biotechnol. 24:100-104. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.