Abstract
Xanthomonas campestris pv. campestris is an epiphytic bacterium that can become a vascular pathogen responsible for black rot disease of crucifers. To adapt gene expression in response to ever-changing habitats, phytopathogenic bacteria have evolved signal transduction regulatory pathways, such as extracytoplasmic function (ECF) σ factors. The alternative sigma factor σE, encoded by rpoE, is crucial for envelope stress response and plays a role in the pathogenicity of many bacterial species. Here, we combine different approaches to investigate the role and mechanism of σE-dependent activation in X. campestris pv. campestris. We show that the rpoE gene is organized as a single transcription unit with the anti-σ gene rseA and the protease gene mucD and that rpoE transcription is autoregulated. rseA and mucD transcription is also controlled by a highly conserved σE-dependent promoter within the σE gene sequence. The σE-mediated stress response is required for stationary-phase survival, resistance to cadmium, and adaptation to membrane-perturbing stresses (elevated temperature and ethanol). Using microarray technology, we started to define the σE regulon of X. campestris pv. campestris. These genes encode proteins belonging to different classes, including periplasmic or membrane proteins, biosynthetic enzymes, classical heat shock proteins, and the heat stress σ factor σH. The consensus sequence for the predicted σE-regulated promoter elements is GGAACTN15-17GTCNNA. Determination of the rpoH transcription start site revealed that rpoH was directly regulated by σE under both normal and heat stress conditions. Finally, σE activity is regulated by the putative regulated intramembrane proteolysis (RIP) proteases RseP and DegS, as previously described in many other bacteria. However, our data suggest that RseP and DegS are not only dedicated to RseA cleavage and that the proteolytic cascade of RseA could involve other proteases.
Bacteria often encounter diverse and rapidly changing environments. To overcome harmful situations, they must be capable of sensing external changes and transmitting this information across biological membranes into the cell, which results in the appropriate redirection of gene expression to prevent or repair cellular damages caused by stress. Extracytoplasmic function (ECF) σ factors provide one common means of bacterial signal transduction to regulate gene expression in response to various extracellular changes (65). ECF σ factors represent the largest and most diverse subfamily of σ70 proteins. They generally recognize a −35 box with a clear bias toward a GAAC in their target promoters, while the −10 region tends to be highly variable between ECF subfamily members (65). One of the best-studied ECF σ factors is the key regulator of the extracytoplasmic stress response factor σE from Escherichia coli, encoded by the rpoE gene (56). ECF proteins were recently divided into 43 major phylogenetically distinct groups named ECF01 to ECF43 (65). RpoE-like ECF σ factors are part of one predominant subgroup found in most bacterial phyla and comprise ECF01 to -04 proteins. RpoE-like ECF σ factors are autoregulated and are required for a wide range of functions. For instance, the E. coli σE factor is essential for growth and promotes the expression of factors that help to preserve and/or restore cell envelope integrity (2). Salmonella enterica serovar Typhimurium σE is required for protection against reactive oxygen species and antimicrobial peptides and for stationary-phase survival (20, 67). Bacillus subtilis σW seems to constitute an antibiosis regulon acting against cell envelope stress (34). S. Typhimurium σE, Pseudomonas aeruginosa AlgU, and Vibrio cholerae σE are required for virulence (5). ECFs can thus be considered models to understand how bacteria sense and respond to their environment both during their interaction with their host and in their free-living state.
RpoE-like ECF σ factors are tightly regulated in order to coordinate their activation with the appropriate environmental cues. In most cases, the σE factor is cotranscribed with a cognate transmembrane anti-σ factor possessing an extracytoplasmic domain and an intracellular σ-binding domain. In the absence of stimulus, the membrane-bound anti-σ binds tightly to the σ factor, thereby keeping it inactive (33). Upon receiving a proper signal, the anti-σ factor is inactivated by regulated intramembrane proteolysis (RIP), resulting in the release and subsequent activation of the σE factor. This mechanism has been well studied for the anti-σ factors RseA, MucA, and RsiW, regulating the activity of E. coli σE, P. aeruginosa AlgU, or B. subtilis σW, respectively (1, 32, 75). In E. coli, the accumulation of C-terminal domains of unfolded porins is the activating signal of the RpoE response by triggering the activation of the inner-membrane-anchored protease, DegS (site 1 protease), and the subsequent cleavage of RseA within its periplasmic domain by DegS. The resulting truncated anti-σ factor is then a suitable substrate for a second inner-membrane protease, RseP/YaeL (site 2 protease), which cleaves RseA near the cytoplasmic face of the inner membrane, releasing an RseAcyto-σE complex into the cytoplasm, where the remaining RseA fragment is degraded by cytoplasmic proteases, resulting in the active σE (1). Another important mediator of the extracytoplasmic stress response is the periplasmic protease DegP, also known as HtrA and DO in E. coli or MucD in P. aeruginosa (22, 55). DegP binds to and degrades misfolded proteins and acts as a chaperone to direct the proper folding of some envelope proteins (66). As such, this family of proteases regulates the σE stress response system by removing misfolded proteins in the periplasm that could activate the degradation pathway of the anti-σE factor, even in the absence of stress (27).
The Gram-negative phytopathogenic bacterium Xanthomonas campestris pv. campestris is an epiphytic bacterium that can become a vascular pathogen, causing black rot disease of crucifers (52). The bacterium produces a large amount of extracellular polysaccharide (EPS) that plays an important role during bacterial infection, and X. campestris pv. campestris has been used as a model organism for investigating the mechanism of bacterial pathogenesis. X. campestris pv. campestris flourishes in and adapts to a wide range of habitats: during epiphytic life, X. campestris pv. campestris is exposed to harsh stresses, such as oligotrophic conditions, desiccation, or large changes in temperature. Upon entry into plant tissues, X. campestris pv. campestris cells must face defense reactions of the host, including oxidative conditions. Finally, the natural life cycle of X. campestris pv. campestris includes long periods of survival on seeds or plant scraps or in the soil, where again it must survive a variety of stressful conditions before it can infect a new host plant. Its ability to manage variable and often lethal external conditions can be partly attributed to its large repertoire of alternative σ factors. Of the 4,179 open reading frames (ORFs) comprising the large 5.1-Mb X. campestris pv. campestris strain ATCC 33913 genome, 15 ORFs encode characterized or putative σ factors, 10 of which belong to the ECF subfamily (23). Little is known about which σ factors are required for the survival of X. campestris pv. campestris under stress and the contribution of these factors to virulence. The classification of ECF σ factors strongly suggested that the XCC1267 gene encodes the σE factor of X. campestris pv. campestris (65). Moreover, previous work by Cheng et al. (17) described the biochemical characterization of the σE factor of X. campestris pv. campestris strain 11 and suggested that it could have a role in the heat shock response. Therefore, we aimed at deciphering the roles and regulation mechanisms of the extracytoplasmic stress response regulator σE in X. campestris pv. campestris.
In the present work we characterized the rpoE operon genes, rpoE, rseA, and mucD. Using primer extension and lacZ transcriptional reporter fusions, we show that rpoE transcription is autoregulated and that RseA and MucD are negative regulators of σE activity. We identified 45 putative members of the σE regulon by a transcriptome analysis, including the heat stress σ factor σH and a number of periplasmic or membrane proteins. We provided evidence that σE is an important regulator of stress responses in X. campestris pv. campestris, since it has a role in heat adaptation, resistance to cadmium, and stationary-phase survival. Furthermore, our results strongly suggest that σE is regulated by a RIP mechanism involving RseP (XCC1366) and DegS (XCC3898) putative proteases, as in many other bacteria. However, our data suggest that the RIP proteases RseP and DegS are not only dedicated to RseA cleavage and that the proteolytic cascade of RseA could involve other proteases.
MATERIALS AND METHODS
Strains and growth conditions.
The X. campestris pv. campestris strains, plasmids, and oligonucleotides used or generated in this study are listed in Table 1. X. campestris pv. campestris cells were grown at 30°C in MOKA (yeast extract, 4 g/liter; Casamino Acids, 8 g/liter; K2HPO4, 2 g/liter; MgSO4·7H2O, 0.3 g/liter) (9) or in KADO (MOKA plus 1% sucrose) (39) medium. E. coli cells were grown at 37°C in LB medium (44). Antibiotics were used at the following concentrations for X. campestris pv. campestris: rifampin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 5 μg/ml. Antibiotics were used at the following concentrations for E. coli: ampicillin, 50 μg/ml; kanamycin, 50 μg/ml; tetracycline, 10 μg/ml. The following supplements were added when required: sucrose, 1%, and isopropyl-β-d-thiogalactopyranoside (IPTG), 0.5 mM.
TABLE 1.
X. campestrispv. campestris strains, plasmids, and oligonucleotides used or generated in this study
| Strain, plasmid, or oligonucleotide | Sequence (5′ to 3′) | Features or purpose | Source or reference |
|---|---|---|---|
| Plasmids | |||
| pVO155 | pUC119 derivative containing the promoterless gus (uidA) reporter gene encoding β-glucuronidase; used for insertion mutagenesis; Kmr Ampr | 51 | |
| pFAJ1700 | pTR102-derived expression vector containing a multiple-cloning site and transcriptional terminators in both orientations; Tetr Ampr | 25 | |
| pCZ750 | pFAJ1700 containing the KpnI-AscI lacZ gene from the pCZ367 plasmid; Tetr Ampr | 9 | |
| p917 | pFAJ1700 derivative containing 2,094 bp of pSC150 with lacI, tacp promoter, and T7 terminator; Tetr Ampr | 11 | |
| p917-lacI | p917 derivative containing 1,526 bp of pMF533 with lacI, tacp promoter, ribosome binding site, and T7 terminator; used for protein overexpression; Tetr Ampr | This study | |
| pCZ-rpoEp | pCZ750 derivative; rpoEp-lacZ; Tetr Ampr | This study | |
| pCZ-rseAp | pCZ750 derivative; rseAp-lacZ; Tetr Ampr | This study | |
| pCZ-rpoHp | pCZ750 derivative; rpoHp-lacZ; Tetr Ampr | This study | |
| pCZ-prcp | pCZ750 derivative; prcp-lacZ; Tetr Ampr | This study | |
| pCZ-ompWp | pCZ750 derivative; ompWp-lacZ; Tetr Ampr | This study | |
| pCZ-hrpFp | pCZ750 derivative; hrpFp-lacZ; Tetr Ampr | This study | |
| pCZ-xcc0401p | pCZ750 derivative; xcc0401p-lacZ; Tetr Ampr | This study | |
| p917-rpoE | p917-lacI derivative; tacp-rpoE; Tetr Ampr | This study | |
| pMF533 | pMALc2E (New England Biolabs) containing malE-gadE fusion | 16 | |
| pK18mobsacB | Mobilizable cloning vector containing a modified sacB gene from B. subtilis; used for gene disruption; Kanr | 61 | |
| pK18-rpoEU | pK18 containing ∼1 kb upstream of the rpoE gene of X. campestrispv.campestris (up to 15 nucleotides downstream of the ATG start codon) | This study | |
| pK18-rpoEU+D | pK18 containing ∼1 kb upstream and ∼1 kb downstream of the rpoE gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| pK18-rseAU | pK18 containing ∼1 kb upstream of the rseA gene of X. campestrispv.campestris (up to 15 nucleotides downstream of the ATG start codon) | This study | |
| pK18-rseAU+D | pK18 containing ∼1 kb upstream and ∼1 kb downstream of the rseA gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| pK18-mucDU | pK18 containing ∼1 kb upstream of the mucD gene of X. campestrispv.campestris (up to 15 nucleotides downstream of the ATG start codon) | This study | |
| pK18-mucDU+D | pK18 containing ∼1 kb upstream and ∼1 kb downstream of the mucD gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| pK18-rpoEUrseAD | pK18 containing ∼1 kb upstream of the rpoE gene (up to 15 nucleotides downstream of the ATG start codon) and ∼1 kb downstream of the rseA gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| pK18-rpoEUmucDD | pK18 containing ∼1 kb upstream of the rpoE gene (up to 15 nucleotides downstream of the ATG start codon) and ∼1 kb downstream of the rseA gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| pK18-degSU | pK18 containing ∼1 kb upstream of the degS gene of X. campestrispv.campestris (up to 15 nucleotides downstream of the ATG start codon) | This study | |
| pK18-degSU+D | pK18 containing ∼1 kb upstream and ∼1 kb downstream of of the degS gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| pK18-rsePU | pK18 containing ∼1 kb upstream of the rseP gene of X. campestrispv.campestris (up to 15 nucleotides downstream of the ATG start codon) | This study | |
| pK18-rsePU+D | pK18 containing ∼1 kb upstream and ∼1 kb downstream of the rseP gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| pK18-xcc1664U | pK18 containing ∼1 kb upstream of the XCC1664 gene of X. campestrispv.campestris (up to 15 nucleotides downstream of the ATG start codon) | This study | |
| pK18-xcc1664U+D | pK18 containing ∼1 kb upstream and ∼1 kb downstream of the XCC1664 gene of X. campestrispv.campestris (from 15 nucleotides upstream of the stop codon) | This study | |
| Strains | |||
| E. coli | |||
| DH5α | λ− φ80dlacZΔM15 Δ(lacZYA-argF)U169 recA1 endA1 hsdR17(rK− mK−) supE44 thi-1 gyrA relA1 | Laboratory collection | |
| X. campestrispv. campestris | |||
| 568 | Wild-type strain; rifampin-resistant derivative of X. campestrispv.campestris LMG568/ATCC33913 | 9 | |
| XcPB1 | ΔrpoE; Rifr | This study | |
| XcPB2 | ΔrseA; Rifr | This study | |
| XcPB3 | ΔmucD; Rifr | This study | |
| XcPB4 | ΔrpoE-rseA; Rifr | This study | |
| XcPB5 | ΔrpoE-rseA-mucD; Rifr | This study | |
| XcPB6 | Δxcc1664; Rifr | This study | |
| XcPB7 | ΔdegS; Rifr | This study | |
| XcPB8 | ΔrseP; Rifr | This study | |
| rpoE::pVO | XCC1267::pVO155; Rifr Kmr | This study | |
| Oligonucleotidesa | |||
| rpoE F-Sma | TTTCCCGGGCATACGGCGCGGGATAGTGTTC | rpoE gene deletion | |
| rpoE R-Xba | TTTTCTAGAGACTTCGGCCATGTCGGG | ||
| rpoE F-Xba | TTTTCTAGACACCGTGTATGACCAATAACC | ||
| rpoE R-Hind | TTTAAGCTTGGCCGAACATCTGGGTGCGG | ||
| rseA F-Sma | TTTCCCGGGTGACATCGCCCAGTTCGAG | rseA gene deletion | |
| rseA R-Xba | TTTTCTAGAGTTATTGGTCATACACGGTG | ||
| rseA F-Xba | TTTTCTAGACCGCAGGACTGATGTTCTCGCC | ||
| rseA R-Hind | TTTAAGCTTGAAAGCCAGGTCGATCGGGATC | ||
| mucD F-Eco (B) | TTTGAATTCGAAAGCGCTACCCGTGAGCGAC | mucD gene deletion | |
| mucD R-Xba | TTTTCTAGAGCGGGGATTCATCAGGTTG | ||
| mucD F-Xba | TTTTCTAGAGCGGGCGGCTGAGACGCAGGG | ||
| mucD R-Hind | TTTAAGCTTGGGTGTCGCCGACCGGCGCGCC | ||
| rseP F-Sma | TTTCCCGGGACCCAGGCGCATGCCGGTGATC | rseP gene deletion | |
| rseP R-Xba | TTTTCTAGAGAAATCACCCATGGATGCAAC | ||
| rseP F-Xba | TTTTCTAGAGTTCCACGATGAAGCTGCTCC | ||
| rseP R-Hind | TTTAAGCTTGAAGATGTCGCCGGCCTTGGGG | ||
| degS F-Eco | TTTGAATTCAACGCTGTTCTTGGCCACCAC | degS gene deletion | |
| degS R-Xba | TTTTCTAGACAGCGGTCGCATGCAACGGATTC | ||
| degS F-Xba | TTTTCTAGACTCATGCGTTGATCCGGCGTG | ||
| degS R-Hind | TTTAAGCTTCATGGCGCCGAATTTCATGGG | ||
| xcc1664 F-Eco | TTTGAATTCGCCGCCCAGATCGGGCGTG | XCC1664 gene deletion | |
| xcc1664 R-Bam | TTTGGATCCCCCACCGGGCACTGCATGATTTC | ||
| xcc1664 F-Bam | TTTGGATCCGCGCCATGACCACGCCCTG | ||
| xcc1664 R-Hind | TTTAAGCTTGGAAAGCCATCCAGGCGC | ||
| rpoE RT-F (A) | GCGGCGTTCGATGTGTTGG | ||
| rpoE-EXT (I) | CCAGCTCCTGAGGTGTATC | Mapping of rpoEp | |
| mucD RT-F (D) | CCATCACCCGCAAGGACGCC | ||
| mucD-EXT (G) | GCCAACGGCAGGGTCATGG | ||
| mucD(2)-EXT (K) | GAACATCTGGGTGCGGATG | ||
| mucD(3)-EXT (H) | GGAAATGATGAAGCCCGAAC | ||
| rseA-EXT (E) | CTTTCGGTCTCCAGCAGAGG | ||
| rseA(2)-EXT (J) | GGACATGTCAGGGTTATTG | Mapping of rseAp | |
| rseA(3)-EXT (F) | GTTCGCGGGACACGAACAAG | ||
| rseA RT-F (C) | GACGAAGAGTTGGCCGGCTG | ||
| lacI-FseI | TTTGGCCGGCCATCGAATGGTGC | ||
| PtaqNcoBam | TTGGATCCATGGGCTATGGTCCTTGTTG | ||
| P1268-Hind | TTTAAGCTTCGGTGGCAGACAGG | Upstream region of rseA (fusion to lacZ) | |
| P1268-Xba | TTTCTAGATGGCCGACCGGAGTTC | ||
| P1267-Hind | TTTAAGCTTGGGGCAGGGCAGCTCGG | Upstream region of rpoE (fusion to lacZ) | |
| P1267-Xba | TTTTCTAGAAGTCGGGCAATGAGACC | ||
| ORF1267-Nco | TTTCCATGGCCGAAGTCGATACACC | rpoE gene | |
| ORF1267-Hind | TTTAAGCTTCATACACGGTGTCGCTGAC | overexpression | |
| xcc1535-EXT | GTTCACCACGTCCGGTGAGCTC | Mapping of XCC1535p | |
| xcc0964-EXT | CTCGGCCAGCATAATCTTGCC | Mapping of XCC0964p | |
| hrpF-EXT | CCTCGCAGTGACAGAGCAG | Mapping of hrpFp | |
| hpa1-EXT | CTGCAGGTTGATGAAGTTGG | Mapping of hpaIp | |
| ompW-EXT | GGAAATGGAACGCATGAGGG | Mapping of ompWp | |
| pqqB-EXT | CCGATCCCAAAACGATGATG | Mapping of pqqAp | |
| xcc0401-EXT | GTCGGTCTGGATCTGGATCTG | Mapping of XCC0401p | |
| prc-EXT | CCAGCAATGCCATCGGAGTG | Mapping of prcp | |
| xcc4186-EXT | GAAGCTGCGTACTGCGTTGCAG | Mapping of XCC4186p | |
| rpoH-EXT | GATTGTTTGCCACAAGGGCAGTC | Mapping of rpoHp | |
| xcc3227-EXT | GGAAGGAGCGGCGGTGCGTTC | Mapping of XCC3227p | |
| xcc1246-EXT | GGCAAGTCTGATCCTCTTGG | Mapping of XCC1246p | |
| P0539-Hind | TTTAAGCTTGGCCGGGCTGGTCGAGGG | Upstream region | |
| P0539-Xba | TTTTCTAGATTGTTAGGCCCTTGGAGTGG | of ompW (fusion to lacZ) | |
| PhrpF-Hind | TTTAAGCTTCGGGGTCCAATCCAAGCC | Upstream region | |
| PhrpF-Xba | TTTCTAGAGCAGTGGCGGAGCTG | of hrpF (fusion to lacZ) | |
| Pprc-Hind | TTTAAGCTTGTCACCCTGCGCGACCTG | Upstream region | |
| Pprc-Xba | TTTTCTAGATGAAAGAAGGGCGGTGATC | of prc (fusion to lacZ) | |
| P0401-Hind | TTTAAGCTTCGTCCTGCGCCACCGCAG | Upstream region | |
| P0401-Xba | TTTTCTAGATTCGATTCTGCGACAGCC | of XCC0401 (fusion to lacZ) | |
| P3771-Hind | TTTAAGCTTCAATGCGGTGCTGACGGTGG | Upstream region | |
| P3771-Xba | TTTTCTAGAACTATTGGACTGCTGGGTAC | of rpoH (fusion to lacZ) | |
| P1230-Hind | TTTAAGCTTCGGCATCGGCGTCCTCTTC | Upstream region of XCC1230 (fusion to lacZ) | |
| P1230-Xba | TTTTCTAGATTGCTGCACCCCCATTCTG |
Recombinant DNA procedures.
Genomic DNA from X. campestris pv. campestris was extracted using the DNeasy Blood and Tissue kit according to the instructions of the manufacturer (Qiagen). Plasmid DNA and PCR products were purified with the Qiagen mini-plasmid purification kits and PCR purification kits, respectively. E. coli strain DH5α was used for cloning. Restriction enzymes, T4 DNA ligase, T4 polynucleotide kinase, and Phusion High-Fidelity DNA polymerase were used as specified by the manufacturer (New England Biolabs).
Construction of in-frame deletion mutant strains in X. campestris pv. campestris.
The two-step recombination system (59), based on the inability of X. campestris pv. campestris carrying the sacB gene to grow in media with high sucrose concentrations, was used for the chromosomal inactivation of the rpoE, rseA, mucD, rseP, degS, and XCC1664 genes of X. campestris pv. campestris. For each planned inactivation experiment, a mobilizable X. campestris pv. campestris integration vector was constructed, which contained two 1,000-bp fragments on each side of the gene to be deleted, comprising the first and the last 18 nucleotides of the selected gene, thus providing two homology regions for recombination.
Genomic DNA of X. campestris pv. campestris strain 568 and primer pairs rpoE F-Sma/rpoE R-Xba and rseA F-Xba/rpoE R-Hind were used to obtain two PCR products of ∼1 kb from the region upstream of rpoE and downstream of rpoE, respectively. The products were sequentially cloned into the appropriate sites of the pK18mobsacB vector, starting with the upstream region, to finally obtain the mobilizable plasmid pK18-rpoEU+D. The same cloning procedure was used for the other genes, using the primer pairs indicated in Table 1. The resulting plasmids were verified by sequencing from both ends with standard primers. The plasmids were transformed into E. coli DH5α and mobilized into X. campestris pv. campestris by conjugation, as described previously (68). Successful first recombinants (chromosomal integration mutants) were selected by plating cells on MOKA containing kanamycin. For selection of the second recombination event, the integration mutants were plated on MOKA containing 5% (wt/vol) sucrose. Clones that had lost kanamycin resistance were screened by colony PCR using primers outside the cloned regions. Two of the confirmed transconjugants were randomly chosen for further study.
Construction of rpoE-overexpressing X. campestris pv. campestris strains for complementation.
To construct the overexpression plasmid for the rpoE gene, an FseI-BamHI fragment encompassing the lacI gene and the tacp promoter was amplified by PCR using plasmid pMF533 (16) as a template, together with primers lacI-FseI and tacp NcoBam. The fragment generated was subsequently cloned into the pCZ917 plasmid (11) to yield p917-lacI. The resulting plasmid contains the lacI gene and the tacp promoter, followed by a ribosome binding site and the restriction sites for NcoI and BamHI. For complementation of deletion mutants of X. campestris pv. campestris, a 621-bp DNA fragment containing the entire rpoE gene was amplified by PCR using total DNA of X. campestris pv. campestris strain 568 as a template and primers ORF1267-Nco and ORF1267-Hind and cloned into the p917-lacI plasmid using the NcoI and HindIII enzymes. The resulting plasmid, p917-rpoE, was verified by sequencing and subsequently transferred to X. campestris pv. campestris by triparental conjugation. For the complementation experiments, different concentrations of IPTG were tested, and in order to avoid deleterious effects of σE overexpression, we chose not to add IPTG because the expression of σE from the leaky tacp promoter was sufficient.
Construction of promoter-reporting plasmids and β-galactosidase assays.
The promoter regions (∼500 bp upstream of the ATG) of genes rpoE, rseA, prc, ompW, hrpF, XCC0401, rpoH, and XCC1230 were PCR amplified with primer sets shown in Table 1, using genomic DNA from X. campestris pv. campestris strain 568 as a template. These promoter regions were cloned as HindIII-XbaI fragments into the pCZ750 plasmid (9) upstream of a promoterless lacZ gene. The resulting plasmids were confirmed by sequencing and introduced into X. campestris pv. campestris strains by triparental conjugation. Overnight cultures were diluted to an optical density at 600 nm (OD600) of 0.05 and grown at 30°C for 9 h (∼3 generations). β-Galactosidase activities were assayed as described previously (44). The data shown are the averages of at least two independent cultures, each measured in triplicate. Negligible β-galactosidase activity was derived from the control promoterless plasmid pCZ750 (less than 9 Miller units) (data not shown).
RNA purification.
Overnight cultures were diluted to an OD600 of 0.05 and grown at 30°C for 9 h (∼3 generations) to reach an OD600 of 0.4. If the cells required a temperature upshift, it was done for 60 min at 35°C. For all experiments, 10-ml culture samples were harvested by immediately adding 1.25 ml of ice-cold 5% water-saturated phenol in ethanol and centrifugation at 5,000 rpm. The cell pellets were flash frozen in liquid N2 and stored at −80°C. Total RNA was extracted from the cell pellets by use of TRIzol reagent following the manufacturer's specifications (Invitrogen). A further treatment with 0.03 U RQ1 DNase I (Promega) per μg of RNA for 30 min at 37°C, followed by phenol extraction and ethanol precipitation, was carried out. RNA was evaluated for quantity and quality using the NanoDrop1000 spectrophotometer (NanoDrop Technologies) and agarose gel electrophoresis. The absence of DNA contamination was confirmed by PCR.
RT-PCR.
One microgram of total RNA was reverse transcribed into cDNA by using PrimeScript reverse transcriptase (RT) (Takara Bio) with random hexamers according to the manufacturer's instructions, and 0.5 μl of each retrotranscription reaction mixture was subjected to PCR using GoTaq DNA polymerase (Promega). Positive controls were performed with genomic DNA, and negative controls were performed with RNA that had not been subjected to retrotranscription (data not shown). The cycling conditions used were 95°C for 2 min and 35 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 1 min 30 s, followed by incubation at 72°C for 5 min.
Microarray data collection and statistical analyses.
Briefly, total RNA was extracted from four sets of identically treated batch cultures of ΔrpoE (no σE) and ΔrseA (overactivation of σE) mutant X. campestris pv. campestris strains grown up to mid-exponential phase in MOKA medium at 30°C. Total RNA (10 μg) was reverse transcribed in the presence of aminoallyl dUTP using Superscript II (Invitrogen) and random hexamers for priming, according to the manufacturer's instructions. The resulting amine-modified cDNA was then chemically labeled at the aminoallyl group using Alexa Fluor 555 and 647 reactive dyes (Invitrogen). Hybridization took place underneath a coverslip in 60 μl of warmed DIG Easy Hybridization buffer (Roche) at 42°C for 16 h in a sealed humidified chamber.
Xcc5kOLI microarrays (62) based on the genome sequence of X. campestris pv. campestris strain B100 (70) were used for hybridizations. The array contains 4,441 50-mer to 70-mer oligonucleotides representing the predicted protein-encoding genes. In addition, it contains 15 stringency controls of the genes gapA, rpsA, rpsB, rpsL, and rpmI (70%, 80%, and 90% identity to the native sequence), 12 alien DNA oligonucleotides, and 5 spiking control oligonucleotides. Each probe was spotted in three replicates.
After being washed, the hybridized microarray slides were scanned using an Axon GenePix 4100A scanner (Molecular Devices). The acquired microarray images were analyzed with GenePix Pro (version 3.0.6.90). Preprocessing of raw data and statistical analyses were performed using the Bioconductor and LIMMA packages in the R programming environment (64). Spots marked as “bad” (flags ≤ −49) by GenePix were excluded from further analysis. Intensity normalization was performed using the robustspline method (within arrays) and the aquantile method (between arrays). Within array replicates, correlation was estimated and incorporated in the linear model was inferred by LIMMA, which was further used to build an empirical Bayes moderated t test statistic to assess differential expression. P values were adjusted for multiple testing using the method of Benjamini and Hochberg (8) to control the false-discovery rate at a level of 0.05, and the statistical significance threshold to decide differential expression was set to 0.05.
Primer extension analyses.
For these experiments, we used one of the four RNA sample sets that were used for the microarray experiments. Primer extension experiments were performed at 50°C using PrimeScript Reverse Transcriptase (Takara Bio) and primers hybridizing in the 5′ region of the coding sequence of the respective genes (Table 1). Ten micrograms of total RNA and 1 pmol of 32P end-labeled primers were used per reaction. The extension products were loaded on a 6% denaturing polyacrylamide gel adjacent to a sequencing ladder obtained with the 32P-labeled universal cycle primer and pUC18 plasmid provided in the Thermo Sequenase cycle-sequencing kit (USB).
Microarray data accession numbers.
Fully annotated microarray data from this study have been deposited in ArrayExpress under accession no. E-MEXP-2935. The array design is available in ArrayExpress under accession no. A-MEXP-1909.
RESULTS
rpoE genomic organization.
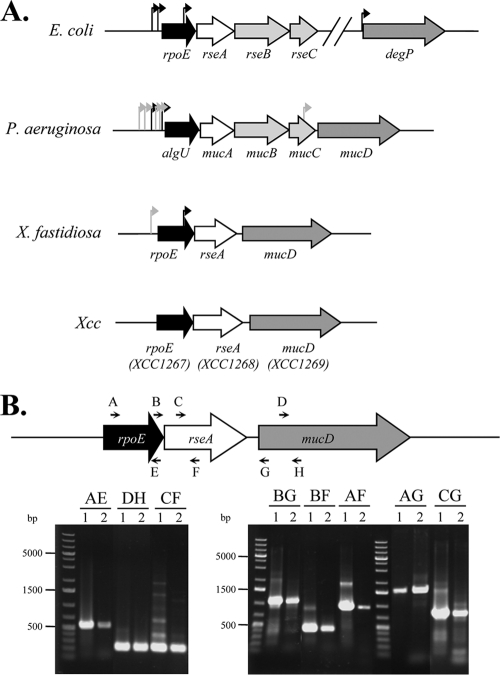
The rpoE (XCC1267) genomic organization found in the X. campestris pv. campestris genome is similar to that of other gammaproteobacteria, such as E. coli, P. aeruginosa, and Xanthomonas fastidiosa (Fig. 1 A). The gene immediately downstream of rpoE is predicted to encode an alanine-rich protein of 286 amino acids that contains an N-terminal anti-σE protein RseA domain (pfam03872; RseA_N). Taken together with the fact that the genes encoding the σE factors are contiguous to a coding region specifying an anti-σ factor, this strongly suggested that XCC1268 was the putative anti-σE factor, and XCC1268 has been renamed rseA. The third gene of the rpoE cluster is the mucD orthologue XCC1269. MucD is a periplasmic serine protease of P. aeruginosa that belongs to the HtrA protein family and alleviates periplasmic stress by degrading misfolded outer membrane proteins (OMPs) selectively (75). Hence, MucD is an indirect negative regulator of σE activity. Interestingly, P. aeruginosa and X. fastidiosa carry a mucD gene in the rpoE operon, but E. coli does not (Fig. 1A). In other bacteria, there may be one or two genes downstream of rseA (namely, rseB-mucB and rseC-mucC), which encode accessory σE regulatory proteins (Fig. 1A). RseB is a negative regulator of σE that binds to the periplasmic domain of RseA, probably in order to block the access of DegS to the cleavage site of RseA (40). The function of RseC is generally unknown, but it has been reported to be both a positive and a negative regulator (10, 46). Strikingly, there are no orthologues of rseB and rseC in the X. campestris pv. campestris genome (Fig. 1A), suggesting that σE activity might be regulated only by RseA binding in this bacterium.
FIG. 1.
Transcriptional organization of the X. campestris pv. campestris rpoE region. (A) Schematic (to scale) showing the organization of the rpoE region in E. coli, P. aeruginosa, X. fastidiosa, and X. campestris pv. campestris. The arrows indicate experimentally demonstrated promoter start sites (black, σE-dependent promoter; gray, σE-independent promoter). (B) Organization of the rpoE region showing the primer pairs used for the amplifications (Table 1 shows primer sequences) and agarose gel of the RT-PCR amplification products. For each primer pair (named according to the letter code of each primer), two lanes are shown (lane 1, positive control using genomic DNA templates; lane 2, RT-PCR using RNA extracted from cells in exponential phase). The molecular size marker is the O'GeneRuler 1-kb Plus DNA Ladder (Fermentas). In each case, the main extension product migrated at the expected size: AE, 524 bp; DF, 228 bp; CF, 235 bp; BG, 1,058 bp; BF, 393 bp; AF, 912 bp; CG, 900 bp; and AG, 1,577 bp.
To investigate whether rpoE-rseA-mucD constituted a single transcription unit, RT-PCR experiments were carried out using appropriate primers located within the rpoE-rseA-mucD region. As shown in Fig. 1B, we could identify specific transcripts encompassing rpoE and rseA (Fig. 1B, primer pairs AF and BF), rseA and mucD (Fig. 1B, primer pair CG), and rpoE and mucD (Fig. 1B, primer pairs AG and BG). These data strongly suggest that rpoE-rseA-mucD form a single transcription unit.
Transcriptional regulation of the rpoE operon.
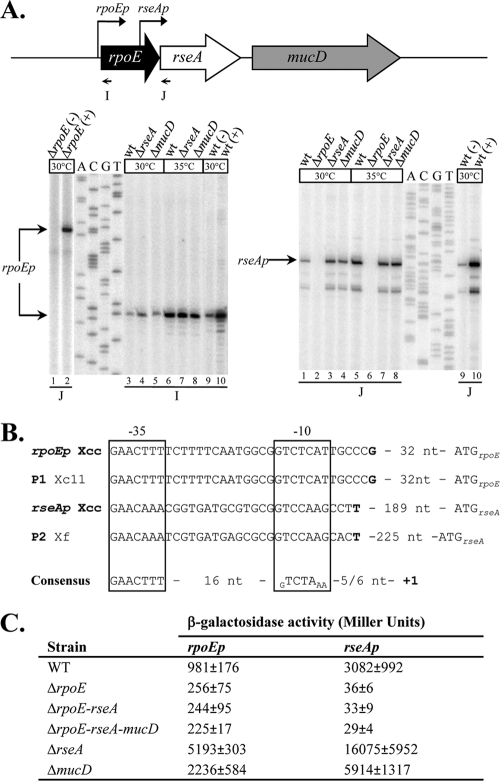
A typical σE-dependent promoter has been identified upstream of the rpoE gene in X. campestris pv. campestris strain 11 (Fig. 2 B) (17). Since this promoter sequence is conserved in X. campestris pv. campestris, we postulated that rpoE was autoregulated in X. campestris pv. campestris. A single transcription start site upstream of the rpoE gene (rpoEp) was identified by primer extension (Fig. 2A, left, lane 3), and it matched P1 of X. campestris pv. campestris strain 11 (Fig. 2B). To test the transcriptional regulation of rpoEp, we conducted primer extension experiments using RNAs purified from strains with rpoE, rseA, or mucD deleted (chromosomal unmarked in-frame deletions; see Materials and Methods for details). The growth of the mutants was comparable to that of the parental strain in rich medium (MOKA) at 30°C, evidencing a doubling time of ∼3 h (data not shown), suggesting that rpoE plays a nonessential role under ordinary growth conditions. As expected, rpoEp expression was strongly reduced when rpoE was inactivated (Fig. 2A, left, lane 1), and conversely rpoEp expression was increased when rseA or mucD was deleted (Fig. 2A, left, compare lanes 4 and 5 with lane 3). Note that the effect of mucD deletion is minor but reproducible. Further, the wild-type (WT) or ΔrpoE mutant strain was complemented with plasmid p917-rpoE, carrying the rpoE gene under the control of the IPTG-inducible promoter tacp. Overexpression of σE from plasmid p917-rpoE significantly increased the amount of rpoEp expression in the wild-type strain (Fig. 2A, left, lanes 9 and 10) and restored rpoEp activity, while no activity was seen when the control plasmid p917 was introduced into the ΔrpoE mutant strain (Fig. 2A, left, lanes 1 and 2).
FIG. 2.
Expression of the rpoE operon genes. (A) Determination of the transcription start site of the rpoE operon genes by primer extension. The schematic (to scale) shows the primers used for transcriptional start site mapping experiments, and the black arrows indicate the positions of the two identified σE-dependent promoters. Total RNAs from the WT, ΔrpoE, ΔrseA, ΔmucD, and WT or ΔrpoE strains containing the control plasmid p917 (−) or the σE-overexpressing plasmid p917-rpoE (+) were used as templates in primer extension experiments when they were suited. Total RNAs were obtained from cells incubated at 30°C or after a 60-min shift at 35°C. The arrows indicate the bands corresponding to the observed start site. (B) Sequence alignment depicting the relevant features of rpoEp and rseAp compared to σE-binding sites of P1 of X. campestris pv. campestris strain 11 (Xc11) and P2 of X. fastidiosa (Xf) (17, 24). The transcription start sites identified by primer extension are indicated in boldface, and the putative −10 and −35 regions are boxed. The consensus ECF02 group of ECF σ factor sites is indicated below (65). (C) Determination of rpoEp and rseAp activities in different strains. Plasmids containing a transcriptional fusion of the upstream region of rpoEp or rseAp to the lacZ gene were transferred into X. campestris pv. campestris strains. Overnight cultures of these strains grown in MOKA medium were diluted in the same medium and grown for 9 h before determination of β-galactosidase activity. The results represent the mean values of at least two independent experiments, each performed in triplicate, with the standard errors.
These results were confirmed by the analysis of the β-galactosidase activity expressed from the reporter plasmid pCZ-rpoEp carrying a fusion between a DNA fragment containing rpoEp and the lacZ gene (see Materials and Methods). This plasmid was introduced into the X. campestris pv. campestris wild-type strain and the ΔrpoE, ΔrseA, ΔmucD, ΔrpoE-rseA, and ΔrpoE-mucD mutants. We observed that deletion of rpoE caused an ∼4-fold reduction in rpoEp-driven β-galactosidase activity (Fig. 2C), showing that the expression of σE is autoregulated in X. campestris pv. campestris, as in many other bacteria and as previously reported in X. campestris pv. campestris strain 11 (17). However, significant β-galactosidase activity remained when rpoE was deleted (∼256 Miller units) (Fig. 2C), suggesting that the rpoE operon could also be controlled by a σE-independent promoter. This is consistent with the regulation of rpoE transcription by a combination of σE-dependent and σE-independent promoters in other bacteria (Fig. 1). Nevertheless, we were unable to map such a promoter using primer extension, possibly because its expression was too low under our experimental conditions. The deletion of rseA or mucD caused ∼5-fold and ∼2-fold activation, respectively, of rpoEp expression (Fig. 2C), indicating that RseA and MucD are negative regulators of σE-dependent activity. The modest effect of mucD deletion on the transcriptional activation of rpoEp compared with the impact of rseA deletion (Fig. 2A, left, and C) was in agreement with the predicted role of MucD as an indirect negative regulator of σE that acts by removing misfolded proteins that activate proteases for degradation of anti-σE.
To check the effect of unfolding stress on rpoEp transcription, we tested the effect of temperature stress, since σE has been shown to be involved in cell survival after a heat shock stress in X. campestris pv. campestris strain 11 and the closely related bacterium X. fastidiosa (17, 24). More generally, σE is involved in the transcription of a set of heat shock response genes and the heat shock sigma factor σH in several bacterial species (3, 28, 69). We conducted primer extension experiments using RNAs extracted from X. campestris pv. campestris strains that had been shifted from 30°C to 35°C. As shown in Fig. 2A (left), there was a strong increase in rpoEp transcription upon heat treatment (compare lane 6 with lane 3). Similar transcription levels were obtained in WT, ΔrseA, and ΔmucD strains (compare lanes 7 and 8 with lane 6). These results showed that rpoE expression was induced by temperature stress. Moreover, these strongly suggest that RseA and MucD are negative regulators of σE in the absence of activating signal, which could be the presence of nonfolded proteins in the periplasm.
To go further into the regulation of the rpoE operon, we tested for the presence of other promoters using primer extension experiments with primers located at the 3′ end of rpoE and the 5′ end of rseA or mucD. As for X. fastidiosa and E. coli, a typical σE-dependent promoter was found within the rpoE coding sequence and was named rseAp (Fig. 2A, right, and B). Note that we used a primer further upstream to show that the upper band corresponded to the transcription start site of rseAp (data not shown). The rseAp regulation pattern followed that of rpoEp, being activated by σE (Fig. 2A, right, compare lane 10 to lane 9), repressed by RseA and MucD (Fig. 2A, right, compare lanes 3 and 4 to lane 1), and induced by heat stress (Fig. 2A, right, compare lane 5 to lane 1). As a negative control, we checked that there was no transcript corresponding to rseAp when RNAs purified from the ΔrpoE strain were used (Fig. 2A, right, lanes 2 and 6). Moreover, we could not detect any independent transcription start site for mucD if we used a primer located in the mucD gene, while we could still see a band corresponding to the rseAp start site (see Fig. S1 in the supplemental material). Taken together with the RT-PCR results (Fig. 1B), this strongly suggested that the mucD gene was transcribed from both rpoEp and rseAp. Moreover, as shown in Fig. 2C, the β-galactosidase activity driven from a plasmidic rseAp-lacZ fusion was higher than that of rpoEp (∼3 fold) and was strictly dependent on σE, since almost no activity was detected in the absence of rpoE. Our results suggest that σE positively autoregulates itself and upregulates RseA and MucD. Since RseA and MucD are negative regulators of σE activity, this could set up a negative feedback loop to ensure rapid downregulation of the σE-dependent response for a return to the baseline level after a protein-folding stress condition or during normal growth.
σE is involved in stationary-phase survival and in response to diamide and cadmium.
Several reports have indicated that rpoE mutants of Gram-negative bacteria are more susceptible to environmental stresses (60). To evaluate the role of σE in the physiology and environmental stress response of X. campestris pv. campestris, bacterial growth and the resistance levels of isogenic WT, ΔrpoE, ΔrpoE-rseA, ΔrpoE-rseA-mucD, ΔrseA, and ΔmucD strains against different stresses were determined. All of the strains exhibited mucoid phenotypes when grown on KADO plates, suggesting that the production of exopolysaccharides was not affected by the absence or the overactivation of σE (data not shown), as previously described in X. campestris pv. campestris strain 11 (17). Moreover, the morphology of the mutant cells (determined by phase-contrast microscopy) grown on MOKA medium was unaffected in stationary phase, but ΔrseA mutant cells were significantly smaller than WT cells in exponential phase (data not shown). This change in cell shape could indicate a link between σE and peptidoglycan assembly, as suggested for E. coli (31).
Bacteria were cultivated aerobically in MOKA medium and exposed to different treatments during mid-exponential growth for several periods of time, and viable-cell counts were determined. Resistance to acidic pH, 0.1% SDS, alkaline stress caused by 40 mM NaOH, oxidative stress caused by 5 mM H2O2, and hyperosmotic stress caused by 1.5 M NaCl was assayed. No statistically significant differences between the WT and ΔrpoE mutant strains were observed (data not shown). In addition, the different strains were tested for survival upon oxidative stress (1 M diamide and 200 mM paraquat) and against cell wall-active antibiotics (50 mg/ml vancomycin, 10 mg/ml polymyxin B sulfate, and 100 mM chlorpromazine) using a disk diffusion assay. Briefly, X. campestris pv. campestris cultures in mid-exponential phase were plated on MOKA agar, and sterile paper disks saturated with 10 μl of chemicals were layered on top prior to incubation at 30°C. Strains with rpoE deleted showed no significant differences (data not shown) apart from increased sensitivity to diamide: the diameter of growth inhibition for ΔrpoE, ΔrpoE-rseA, and ΔrpoE-rseA-mucD mutants was 2.85 cm (±0.20 cm) compared with 1.9 cm (±0.02 cm) for the WT strain. This suggests that σE is involved in the oxidative-stress response via a thiol oxidation pathway in X. campestris pv. campestris.
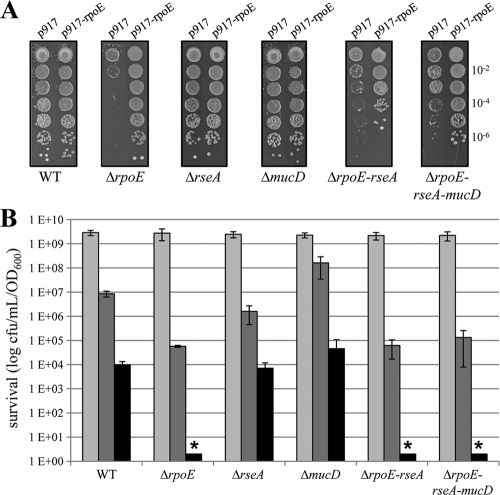
Further, σE factors have been reported to be required for metal resistance in E. coli (26). We submitted X. campestris pv. campestris cells to cadmium stress by spotting serial dilutions of cultures of WT and mutant strains in the late exponential phase of growth on MOKA agar plates containing cadmium (40 μM) and incubating them at 30°C for 72 h. The results of a representative spot dilution experiment are shown in Fig. 3 A. We checked that control plasmid p917 had no impact on growth in the different X. campestris pv. campestris strains (data not shown). The deletion of rpoE was detrimental for resistance to cadmium, since there was a clear growth defect compared to the WT strain. Normal growth was restored when σE was overexpressed from plasmid p917-rpoE. These results indicate that σE is required for full adaptation to cadmium stress in X. campestris pv. campestris, as in E. coli.
FIG. 3.
Stress sensitivity of ΔrpoE mutants. (A) Influence of cadmium on growth of X. campestris pv. campestris WT and mutant strains containing control plasmid p917 or σE-overexpressing plasmid p917-rpoE. Serial 10-fold dilutions of late-exponential-phase bacteria were spotted on MOKA plates containing 40 μM cadmium and incubated at 30°C for 72 h. Each experiment was repeated three times. (B) Effect of rpoE deletion on stationary-phase survival of X. campestris pv. campestris. Overnight cultures in MOKA medium were diluted to an OD600 of 0.05 (time zero), and survival was monitored by viable-cell counting 24 h (light-gray bars), 48 h (dark-gray bars), and 96 h (black bars) postinoculation. The data are the mean values from four experiments, with the error bars representing standard deviations. The asterisks indicate no detectable cells.
It has been shown that σE activity increases upon entry into stationary phase in E. coli (19) and that S. Typhimurium σE is required for stationary-phase survival (67). To monitor the stationary-phase survival of X. campestris pv. campestris WT and mutant strains, the cells were grown at 30°C on a continuously shaking platform, and when the cells reached stationary phase, viable-cell counts were determined periodically by plating cells onto MOKA plates. As shown in Fig. 3B, the survival of ΔrpoE mutants was severely impaired, since the mutants displayed a 100-fold reduction in viability with respect to the WT after 48 h in stationary phase and there was no survival after 72 h, whereas the ΔrseA mutant did not show any appreciable phenotype under the conditions tested. These results strongly suggest that σE is required for stationary-phase survival of X. campestris pv. campestris.
Heat sensitivity of the X. campestris pv. campestris ΔrpoE mutant.
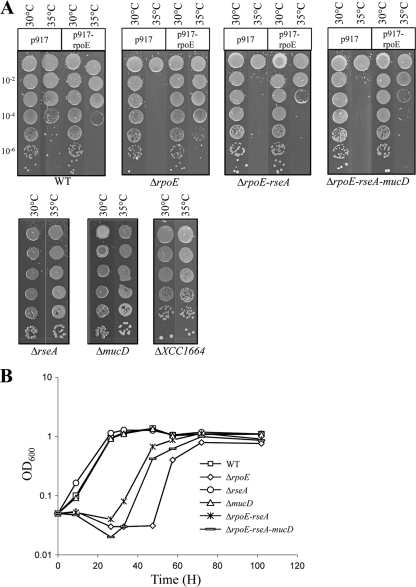
Since we had shown that exposure to heat (35°C) activated σE, we tested the effect of temperature stress in isogenic WT, ΔrpoE, ΔrpoE-rseA, ΔrpoE-rseA-mucD, ΔrseA, and ΔmucD strains. Heat shocks from 42°C to 50°C were applied to exponentially growing cultures for several time points up to 30 min. Viable-cell counts were determined for all strains, and all the mutants gave similar responses to heat-killing treatment compared to the WT (data not shown), indicating that σE is not essential for survival after a heat shock in X. campestris pv. campestris. To check if σE could be involved in adaptation to heat stress, the growth of rpoE mutants was compared to that of the WT strain by spotting serial dilutions of bacterial cultures in the late exponential phase of growth on MOKA agar plates and incubating them at 30°C and 35°C. The results of a representative experiment are shown in Fig. 4 A. Growth at 35°C was strongly impaired for the ΔrpoE mutants (ΔrpoE, ΔrpoE-rseA, and ΔrpoE-rseA-mucD), while there was no or little effect on growth for mutants that overactivate σE (ΔrseA) or for mutants of other ECF σ factors (ΔXCC1664). Conversely, there was no difference between the WT and mutant strains in plating efficiency when the strains were incubated for 72 h at 30°C. However, it must be noted that the colonies formed at 35°C with the ΔmucD mutant are reproducibly smaller than those formed at 30°C. This could be due to its putative protective role during protein folding stress by degrading misfolded proteins in the periplasm.
FIG. 4.
The ΔrpoE mutant of X. campestris pv. campestris is sensitive to heat and ethanol. (A) Heat sensitivities of X. campestris pv. campestris WT and mutant strains containing no plasmid, control plasmid p917, or σE-overexpressing plasmid p917-rpoE. Serial 10-fold dilutions of late-exponential-phase bacteria were spotted on plates and incubated at 35°C for 72 h. Each experiment was repeated three times. (B) Growth curve of X. campestris pv. campestris WT and mutant strains in the presence of 1.5% ethanol. Cells were precultured in MOKA medium overnight and then diluted to an OD600 of 0.05 in MOKA medium containing 1.5% ethanol. One representative experiment out of three independent replicates is shown.
Complementation of the ΔrpoE mutants with plasmid p917-rpoE partially restored growth at 35°C (Fig. 4A). When we introduced the control plasmid p917 into the WT strain, we observed a small decrease in growth at 35°C compared to 30°C that could be the result of the combination of heat and antibiotic stresses (since we added tetracycline in the plates to maintain the plasmid). This inhibitory effect was even stronger when plasmid p917-rpoE was introduced into the WT strain, probably due to the toxicity of σE overexpression, as has been described for E. coli (58). This could explain why complementation of ΔrpoE-rseA and ΔrpoE-rseA-mucD mutants is less efficient than that of the ΔrpoE mutant, since the lack of appropriate posttranslational regulation of σE could be detrimental to bacterial fitness. We also tested the effect of σE on adaptation to cold stress at 14°C, and there was no difference between the WT and mutant strains (data not shown). Our results show that deletion of rpoE makes X. campestris pv. campestris cells extremely vulnerable to temperature adaptation at 35°C.
Given that ethanol is an amphiphilic compound that mimics the effects of high-temperature stress and that σE is required for growth in the presence of ethanol in several bacteria, such as X. fastidiosa or V. cholerae (24, 42), we tested the growth of WT and mutant X. campestris pv. campestris strains in the presence of 1.5% ethanol. As shown in Fig. 4B, the deletion of the rpoE gene strongly impaired growth, while deletion of rseA or mucD had no significant effect. Overall, these results indicate that σE contributes to cell envelope stress adaptation of X. campestris pv. campestris.
Identification of genes regulated by the σE factor.
To gain insight into the functions regulated by σE in X. campestris pv. campestris, a global approach was chosen by comparing transcriptomes of the ΔrpoE strain (no σE activity) to that of the ΔrseA strain (overactivation of σE) using Xcc5kOLI microarrays (62). Given that there are nine ECFs in X. campestris pv. campestris and that they recognize close promoter sequences (65), we chose these setups in order to mimic a physiological activation of σE and to minimize nonspecific promoter recognition by σE. In the microarray analyses, any gene with a P value of ≤0.05 showing an increase of expression of 1.3-fold or more and a reduction in expression of 1.5-fold or more in the ΔrseA strain in comparison to the ΔrpoE strain was defined as being regulated by σE, either directly or indirectly (Tables 2 and 3). We have chosen relatively low thresholds to identify the significantly σE-regulated genes, since we have done four independent biological replicates, including a dye swap experiment. A total of 45 genes comprising 37 putative transcription units (TUs) were induced (Table 2), and 20 genes were repressed (Table 3). As expected, the rpoE and mucD transcripts were included in the group of upregulated genes. Nevertheless, the signal for the rseA transcript was in the range of the background under the conditions employed (data not shown), suggesting that there could be a problem with the detection of the oligonucleotide probe.
TABLE 2.
Genes with increased expression in the ΔrseA strain compared with the ΔrpoE strain
| ID | Gene | Ratioa | Description of gene product | Putative TUb | Characteristics |
|---|---|---|---|---|---|
| Regulatory function | |||||
| XCC1267 | rpoE2 | 4.28 | RNA polymerase σ factor σE | XCC1267, -68, -69 | Transcription initiation |
| XCC3771 | rpoH | 1.52 | Heat shock RNA polymerase factor σH | Response to heat; transcription initiation | |
| XCC3348 | 1.48 | Putative sensor-response regulator hybrid | XCC3347, -48 | Two-component signal transduction system (phosphorelay) | |
| Metabolism | |||||
| XCC2432 | fadE2 | 3.42 | Putative acyl-coenzyme A dehydrogenase | XCC2430, -31, -32 | Oxidation reduction |
| XCC3937 | baf | 3.14 | Putative type III pantothenate kinase | XCC3938, -37, -36 | Positive regulation of transcription |
| XCC3047 | bioI | 1.45 | Putative cytochrome P450 hydroxylase | Heme b metabolic process | |
| (NC_003902: 3492579-3492659, plus strandc) | pqqA | 1.41 | Putative coenzyme PQQ biosynthesis protein A | XCC2937, -38, -39, -40, -41 | Pyrroloquinoline quinone biosynthetic process |
| XCC1588 | 1.32 | Putative sulfite oxidase subunit YedY | Electron carrier activity | ||
| XCC3906 | 1.31 | Putative cytochrome B561 | XCC3905, -06 | Respiratory electron transport chain | |
| Protein synthesis and fate | |||||
| XCC1269 | mucD | 2.91 | Putative periplasmic protease | XCC1267, -68, -69 | Proteolysis |
| XCC1047 | hspA | 2.02 | Low-molecular-wt heat shock protein | Response to stress | |
| XCC3493-like degradation complex | hslU | 1.83 | Chaperone subunit of a proteasome | XCC3493, -94 | Response to stress |
| XCC3227 | moxR | 1.67 | MoxR-like AAA+ ATPase chaperone | XCC3227, -26, -25, -, 24, -23, -22 | ATPase activity |
| XCC2393 | htpG | 1.53 | Molecular chaperone Hsp90 family | Protein folding | |
| XCC1535 | -d | 1.41 | FKPB-type peptidyl-prolyl cis-trans isomerase | Protein folding | |
| XCC1475 | dnaJ | 1.36 | Molecular chaperone Hsp40 family | XCC1474, -75 | Response to heat; protein folding |
| XCC3450 | prc | 1.34 | Putative carboxyl-terminal processing protease | Proteolysis | |
| XCC1474 | dnaK | 1.31 | Molecular chaperone Hsp70 family | XCC1474, -75 | Response to heat; protein folding |
| T3S, translocation, and regulation machinery | |||||
| XCC1240 | hpa1 | 2.15 | Harpin-like elicitor protein (T3SS-dependent secreted protein) | ||
| XCC1217 | hrpF | 2.04 | Type III translocon protein | Interaction with host via protein secreted by T3SS | |
| XCC1246 | 1.99 | Type III effector protein (XopAL class) | Pathogenesis | ||
| XCC1241 | hpa2d | 1.78 | Lytic transglycosylase-like | ||
| XCC1222 | hrpD6 | 1.64 | Type III secretion system component | XCC1222, -21, -20, -19 | Protein secretion by the T3SS |
| XCC1220 | hpaB | 1.54 | Global T3S chaperone | XCC1222, -21, -20, -19 | T3SS |
| XCC2565 | 1.50 | Leucin-rich-repeat-containing protein/type III effector protein (XopAC class) | Pathogenesis | ||
| XCC4186 | 1.49 | Leucin-rich-repeat-containing protein/type III effector (XopL class) | Pathogenesis | ||
| XCC1221 | hrpE | 1.47 | Type III secretion system pilus protein | XCC1222, -21, -20, -19 | T3SS complex |
| XCC2896 | psvA | 1.42 | Peptidase C48 family/type III effector protein (XopD class) | Proteolysis | |
| XCC1224 | hpaA | 1.34 | Type III secreted virulence factor | XCC1222, -21, -20, -19 | Interaction with host via protein secreted by T3SS |
| XCC1219 | hrpW | 1.33 | Harpin pectate lyase | XCC1222, -21, -20, -19 | Interaction with host via protein secreted by T3SS |
| Cell envelope | |||||
| XCC0539 | ompW3 | 1.39 | Putative outer membrane protein | Cell outer membrane | |
| XCC2266 | pglAd | 1.31 | Putative polygalacturonase | Carbohydrate metabolic process | |
| XCC3925 | ecnAd,e | 1.31 | Putative entericine A | Response to toxin | |
| Unknown function | |||||
| XCC1308 | -d | 2.22 | Hypothetical protein | XCC1306, -07, -08 | |
| XCC0401 | 2.06 | Hypothetical protein/ribosomal protein S30Ae/σ54 modulation protein | Primary metabolic process | ||
| (NC_003902: 3290964-3290833, minus strandc) | 2.05 | Small putative membrane protein | |||
| XCC3224 | 2.03 | Hypothetical membrane protein | XCC3227, -26, -25, -24, -23, -22 | ||
| XCC1244 | 1.95 | Hypothetical protein | |||
| XCC0944 | -d | 1.83 | Conserved hypothetical protein | ||
| XCC3226 | 1.54 | Hypothetical protein | XCC3227, -26, -25, -24, -23, -22 | ||
| XCC3798 | -d | 1.47 | Calcium-binding EF hand motif | Calcium ion binding | |
| XCC3887 | 1.35 | Hypothetical protein | |||
| XCC2566 | 1.35 | Putative carboxymethylenebutenolidase | Hydrolase activity | ||
| XCC0863 | -d,e | 1.33 | Putative membrane protein | ||
| XCC1736 | -d | 1.33 | Putative secreted protein | XCC1737, -36, -35, -34 | Catalytic activity |
Ratio, averaged expression ratio of σE induced (ΔrseA)/no σE (ΔrpoE).
TUs are listed in chromosomal order.
Nonannotated gene in the genome of X. campestrispv.campestris ATCC 33913. Genomic location contains X. campestrispv.campestris ATCC 33913 chromosome accession number followed by the start coordinate, end coordinate, and strand.
A predicted cleavable signal sequence.
Predicted transmembrane helices.
TABLE 3.
Genes with decreased expression in ΔrseA strain, as compared with ΔrpoE
| ID | Gene | Ratioa | Description of gene product | Characteristic |
|---|---|---|---|---|
| Transporter activity | ||||
| XCC2867 | btuBb | −1.97 | TonB-dependent transporter | Transport; membrane |
| XCC3316 | -b | −1.73 | TonB-dependent transporter | Transport; membrane |
| XCC2497 | -b | −1.61 | Pseudo-TonB-dependent transporter/Oar like | Transport; membrane |
| XCC3271 | -b | −1.54 | Pseudo-TonB-Dependent Transporter/Oar like | Transport; membrane |
| XCC1892 | cirAb | −1.39 | TonB-dependent transporter | Transport; membrane |
| Regulatory function | ||||
| XCC3677 | −9.64 | Putative two-component system sensor kinase | Two-component signal transduction system (phosphorelay) | |
| XCC1935 | rpoN2 | −1.54 | RNA polymerase σ54 factor | Transcription initiation |
| XCC1276 | −1.50 | Putative sensor/response regulator hybrid | Two-component signal transduction system (phosphorelay) | |
| Metabolism | ||||
| XCC2410 | −23.07 | Ketoglutarate semialdehyde dehydrogenase | Metabolic process | |
| XCC2295 | −1.68 | Putative polyhydroxyalkanoate synthesis repressor | ||
| XCC3324 | ilvB | −1.57 | Acetolactate synthase | Branched-chain family amino acid biosynthetic process |
| XCC0550 | atpF | −1.55 | F0F1 ATP synthase subunit B | ATP synthesis-coupled proton transport |
| Nucleic acid metabolic process | ||||
| XCC2904 | hsdS | −7.86 | Type I restriction enzyme (specificity chain) homologue | DNA modification |
| XCC0377 | mnmG | −1.38 | Pyridine nucleotide-disulfide oxidoreductase, class II | tRNA processing |
| Catalytic activity | ||||
| XCC0700 | -b | −1.39 | Putative peptidase S15 | Proteolysis |
| Motility and attachment | ||||
| XCC3232 | pilM | −1.35 | Type IV pilus assembly protein PilM | Pilus assembly |
| Cell envelope | ||||
| XCC3017 | ompP6b | −.53 | Outer membrane protein P6 precursor (OmpA family) | Cell outer membrane |
| Oxidative stress function | ||||
| XCC1109 | katE | −5.64 | Catalase | Response to oxidative stress |
| Unknown function | ||||
| XCC1080 | −5.74 | Hypothetical protein | ||
| XCC2823 | −1.37 | Hypothetical protein |
Ratio, averaged expression ratio of σE induced (ΔrseA)/no σE (ΔrpoE).
A predicted cleavable signal sequence.
Of the upregulated genes, σE transcribes an array of biosynthetic enzymes that are involved in fatty acid metabolism (fadE2 and XCC3937), redox metabolic functions (XCC1588), and electron transport systems (bioI and XCC3906). This raised electron-transport system activity could possibly compensate for proton leakage across the membrane when its integrity is compromised or could be a response to the formation of reactive oxygen species generated by a perturbation of the electron transport chain. This is further supported by the induction of pqqA, since the redox cofactor PQQ can act as an antioxidant metabolite to detoxify reactive oxygen species (45).
A large number of σE-upregulated genes bear signal sequences or transmembrane domains, which is consistent with the σE response having a role in monitoring and preserving the membrane during stress. In addition, three genes encode cell envelope proteins. Among them, XCC0539 encodes a predicted member of the OmpW/AlkL family that is found in all Gram-negative bacteria and is involved in the protection of bacteria against various forms of environmental stress (36). pglA encodes a predicted polygalacturonase (PG) to selectively degrade the pectic polymers of the plant cell walls, and PGs are virulence factors in closely related bacterial species (73). Interestingly there is also an overrepresentation of type III secretion (T3S)-related genes within the σE regulon of X. campestris pv. campestris. In plant-pathogenic bacteria, the T3S system (T3SS) is one of the key pathogenicity factors and is encoded by the chromosomal hrp gene cluster. Here, we identified 12 genes whose products are involved in all aspects of T3S machinery and effector proteins that are under the control (at least partial) of σE. In xanthomonads, the hrp cluster is organized into at least six transcriptional units and is under the positive control of HrpG and HrpX (30). In our experiments, only two hrp operons (hrpE and hrpF) were upregulated, suggesting that σE-dependent regulation of T3S genes did not occur via HrpG or HrpX. To check this, we tested the effect of rpoE inactivation on plasmid-driven hrpXp- or hrpGp-lacZ transcriptional fusion reporters, and there was no significant change in their activity (data not shown).
As expected from the known σE-dependent stress responses, a heat shock response was induced in X. campestris pv. campestris, since several upregulated gene products are highly conserved heat shock proteins (HSPs), such as dnaK-dnaJ, hspA, htpG, and hslU, and the heat stress σH factor encoded by rpoH. Not only important during heat stress, many HSPs assist protein folding and homeostasis and are general stress proteins (49). Two other heat shock protease genes, hslV (forming a putative TU, hslU-hslV) and lon, showed a small but statistically significant 1.23-fold induction (data not shown). In many bacteria, most of these HSP genes are transcribed by the σH factor and rpoH expression is regulated by σE (50), and our data strongly suggest that heat stress regulation is the same in X. campestris pv. campestris. This is further supported by the presence of σH promoter elements (CTTGAAN13-14CCCCATNT) (41) within the 300 nucleotides upstream of the start codons of dnaK, hspA, htpG, lon, and hslV (data not shown). Nine σE-regulated genes encode proteins involved in protein quality control in the cytoplasm (e.g., DnaK and HslU) and in the periplasm (e.g., Prc and XCC1535), possibly to cope with the defects in membrane protein insertion, folding, and assembly resulting from σE-activating stress signals. Interestingly, the predicted periplasmic protease Prc has been reported to be implicated in the proteolysis of the anti-σE factors MucA in P. aeruginosa and RsiW in B. subtilis (32, 57). It is also worth noting that we expected to find XCC1535 in our analysis, since it shares the same promoter elements with its orthologue in X. fastidiosa (xf0644), which has been shown to be a direct target of σE (24).
Among the functions encoded by 20 σE-downregulated genes, there was an overrepresentation of proton motive force (PMF) consumers that could compete with putative proton leakage in response to localized disturbances in membrane integrity: (i) TonB-dependent transporters (TBDTs), which are in the outer membrane and are mainly involved in iron, vitamin B12, or plant-derived carbohydrate uptake (9); (ii) ATP synthase; and (iii) flagella, since the transcript for the alternative σ factor RpoN2, which is responsible for the transcription of the flagellar T3S system (76), was downregulated upon σE overactivation. Thus, σE could function to maintain the membrane potential component of the PMF in X. campestris pv. campestris as in S. Typhimurium (7). In addition, the pilM gene was repressed by σE, and type IV pili could also represent an energy burden for the cell under stressful conditions.
One of the most downregulated genes was hsdS, possibly to provide restriction alleviation in response to stress stimuli, in agreement with work showing that the signal of chromosomal DNA damage might be transmitted to the cell surface via activation of the σE regulon (4). Among genes negatively regulated by σE, there was only one OMP. This minor inhibitory effect on OMP biosynthesis contrasts with other bacteria in which the σE response limits the expression of a number of OMPs via small RNAs (54) to prevent the accumulation of misfolded intermediates. This discrepancy may be due to the setup of our experiments, since we did not overexpress σE (instead, σE was constantly activated), but it could also suggest that the σE signaling system is regulated differently in X. campestris pv. campestris.
Validation of microarray experiments and determination of a σE-binding consensus motif.
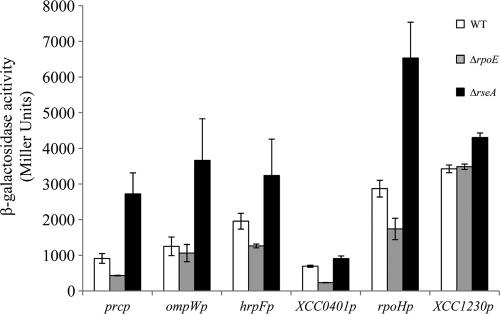
In order to validate the results of the microarray data, a subset of five genes from the σE regulon was randomly chosen for verification of σE dependence in vivo, using lacZ transcriptional reporter fusion assays. The upstream regions (∼500 bp) of the chosen genes were fused to a promoterless lacZ reporter gene in a broad-host-range vector as described in Materials and Methods. We also included in this study a non-σE-dependent promoter region as a negative control. To test the expression of these promoters with different levels of σE activity, the plasmids were transferred to the isogenic WT, ΔrpoE, and ΔrseA strains. As expected, the gene fusions showed dramatically increased expression in the ΔrseA mutant strain containing constitutively high σE activity (Fig. 5). However, there was only a small decrease of β-galactosidase activity upon loss of σE, probably because the cells were grown under nonactivating conditions. As expected, the expression levels of the control gene XCC1230 were similar in all strains.
FIG. 5.
Expression of σE-regulated genes. Reporter plasmids carrying the 5′ ends of selected genes and their upstream promoter regions fused to lacZ were transferred to three X. campestris pv. campestris strains with different σE activities: WT, ΔrpoE, and ΔrseA. Overnight cultures of these strains grown in MOKA medium were diluted in the same medium and grown for 9 h before determination of β-galactosidase activity. The results represent the mean values of at least two independent experiments, each performed in triplicate, and the error bars indicate the standard errors.
To confirm our transcriptome analysis, we also checked some phenotypes associated with predicted σE-regulated functions. In X. campestris pv. campestris, the alternative σ54 factor RpoN2 is responsible for the transcription of the flagellar genes (76). Since RpoN2 was negatively regulated by σE, it was possible that rseA mutants possessed reduced motility. To test this, we measured the motility of the WT, ΔrpoN2, and ΔrseA strains (data not shown). The ΔrpoN2 mutant cells had impaired motility, and ΔrseA mutant motility was delayed, since its swimming zone was reproducibly smaller than that of the WT after 2 days at 30°C. These results are consistent with the microarray analyses. In addition, we tested the ΔrpoE mutants for pathogenicity on a host plant, since σE controlled virulence-associated genes. The X. campestris pv. campestris WT and mutant strains were inoculated into Arabidopsis thaliana Sf-2 ecotype leaves by using the leaf-clipping method. The deletion of rpoE did not reproducibly alter symptom development (data not shown), suggesting that σE is not required for the virulence of X. campestris pv. campestris under these conditions.
To further verify the transcriptome data and to identify σE promoter elements, we selected 11 σE-upregulated genes from nearly all the functional categories identified, and their promoters were mapped by primer extension (Fig. 6; see Fig. S1 in the supplemental material). We added to our study the XCC0964 gene (encoding a putative membrane metalloprotease), since it shares promoter elements with its X. fastidiosa orthologue (xf2594), which has been shown to be a direct target of σE (24). We could not detect any reverse transcription product for XCC1246 unless σE was overexpressed from plasmid p917-rpoE, suggesting that the amount of XCC1246 transcripts was below the detection threshold. For rpoH, two primer extension products (rpoHp1 and rpoHp2) were detected. All primer extension experiments confirmed the trend of changes in transcript levels following σE activation observed in microarray experiments (Fig. 6, compare lanes ΔrseA and ΔrpoE), with the exception of the distal promoter of rpoH (rpoHp2) (see below). Moreover, all promoters except rpoHp2 were induced when σE was overexpressed from the p917-rpoE plasmid [Fig. 6, compare lanes wt(+) and wt(−)]. The proximal promoter rpoHp1 was controlled by σE, since its activity increased when rseA or mucD was deleted and decreased when rpoE was deleted. In addition, the loss of rpoHp1 activity was complemented by the overexpression of σE from plasmid p917-rpoE [Fig. 6, compare lanes ΔrpoE(+) and ΔrpoE(−)]. The distal rpoHp2 was σE independent, since its activity did not vary in mutants of the rpoE operon. Overall, these results indicated good verification of the microarray data.
FIG. 6.
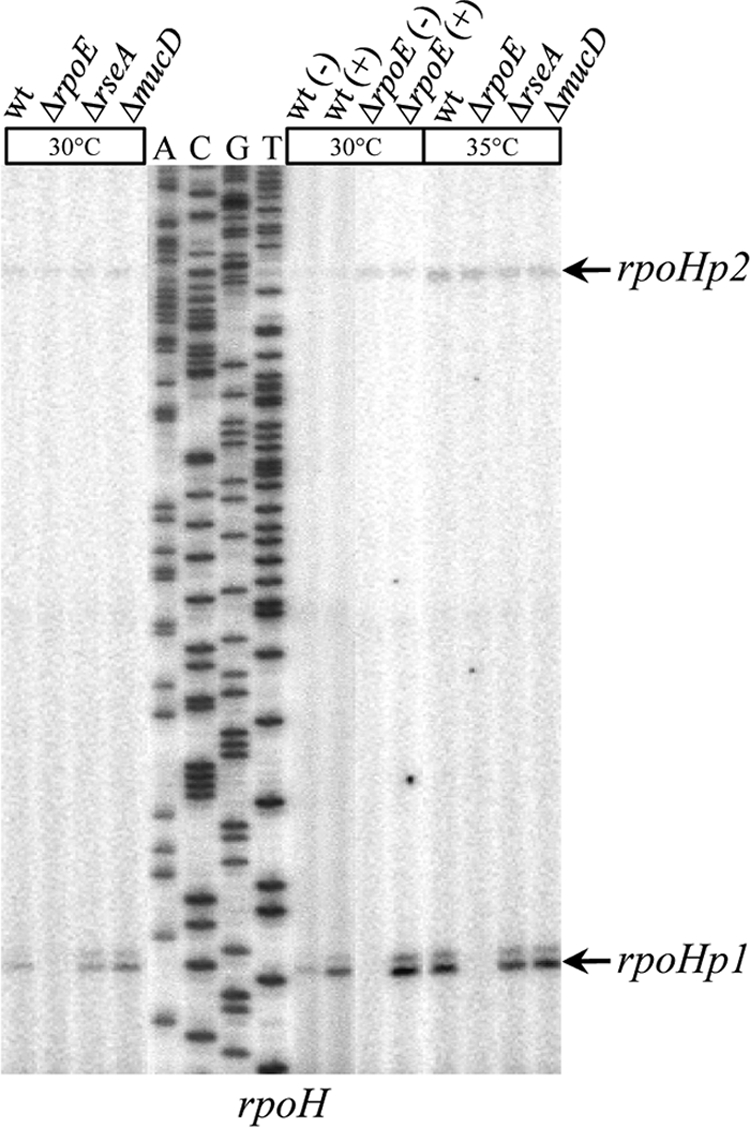
Determination of the transcription start sites of rpoH. Total RNAs from WT, ΔrpoE, ΔrseA, ΔmucD, and WT or ΔrpoE strains containing control plasmid p917 (−) or σE-overexpressing plasmid p917-rpoE (+) were used as templates in primer extension experiments. Total RNAs were obtained from cells incubated at 30°C or after a 60-min shift at 35°C. Primer rpoH-EXT was 5′ end labeled with 32P and extended with reverse transcriptase to map its corresponding gene promoter sequences. The arrows indicate the bands corresponding to the main start sites observed.
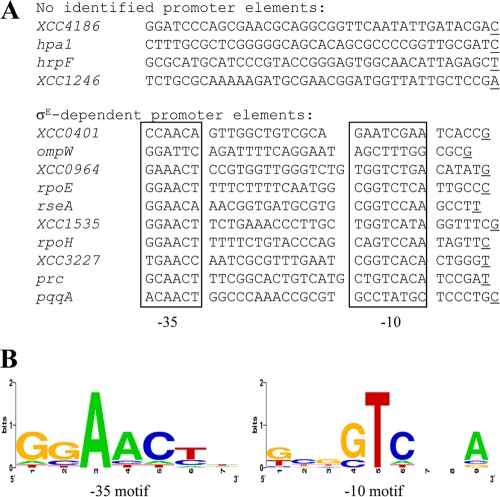
A consensus for the σE-binding motif was obtained by alignment of the 40 nucleotides upstream of the experimentally determined start sites and a search for conserved promoter elements identified from other bacterial σE regulons and for rpoEp and rseAp (65) (Fig. 2B). As shown in Fig. 7 A, 8 out of the 12 selected genes had these conserved promoter elements, indicating that they could be directly regulated by σE. The proposed consensus for X. campestris pv. campestris σE target promoter motifs (GGAACTN15-17GTCNNA) is very similar to the σE-binding sequence of homologous ECF σ factors from the ECF02 group (65) (Fig. 2B and 7B). It harbors the hallmark feature of an ECF-type promoter, the AAC motif in the −35 region, and the −10 region contains highly conserved TC and A residues. Interestingly, only half of the 14 mapped promoters depended solely on σE for their transcription: rpoEp, rseAp, XCC1535p, rpoHp2, XCC3227p, XCC0964p, and prcp (Fig. 2A and 6; see Fig. S1 in the supplemental material). This suggests that partially σE-dependent genes could be controlled by other σ factors that overlap functionally in X. campestris pv. campestris. We were unable to identify σE conserved promoter elements for the four T3S-related genes XCC1246, XCC4186, hpaI, and hrpF, suggesting that the effect of σE on their expression is indirect.
FIG. 7.
Identification of the X. campestris pv. campestris σE promoter recognition sequence. (A) Alignment of σE-dependent promoters identified by primer extension assays. The transcription start site is underlined, and the −35 and −10 motifs are boxed. (B) The aligned promoter sequences were analyzed using the WebLogo program (http://weblogo.berkeley.edu/). The height of a stack indicates sequence conservation (2 = 100% conservation), and the height of each individual nucleotide within the stack indicates its relative frequency at that position.
To gain further insight into σE activity under heat stress conditions and to further validate our microarray data, we extracted total RNAs from bacterial cultures after a shift to 35°C. We included only the genes whose expression was heat inducible (Fig. 6 and data not shown; see Fig. S1 in the supplemental material). The relative levels of rpoH, ompW, XCC3227, hspA, XCC1535, and prc transcripts dramatically increased upon heat treatment, indicating that a subset of the σE regulon was heat responsive. Both promoters of rpoH were induced by a temperature upshift, but only rpoHp1 was upregulated in a σE-dependent manner. We failed to identify conserved promoter elements upstream of the rpoHp2 start site, so we could not predict which σ factor could be in charge of its transcription. The heat inducibility of XCC3227p, XCC1535p, and prcp expression was dependent on σE (see Fig. S1 in the supplemental material), as for rpoEp and rseAp (Fig. 2A). Hence, this suggests that their induction is likely to be accomplished by activation of σE upon heat exposure, probably mediated by anti-σ RseA cleavage. In contrast, ompWp heat induction was σE independent, and this will need further investigation. Interestingly, we could not directly test the impact of σH on gene expression because, despite numerous attempts, we could not inactivate the rpoH gene. It is thus likely (although not formally proven) that rpoH is essential in X. campestris pv. campestris, as it is thought to be in other bacteria (29, 63).
Regulation of σE-mediated response by RseP (XCC1366) and DegS (XCC3898).
Upon cell envelope stress in E. coli, RseA is sequentially cleaved by the RIP proteases DegS and YaeL (1). Specifically, the recognition of the C termini of unfolded OMPs allows DegS to cleave the periplasmic C terminus (site 1) of RseA. This converts RseA into a substrate for RseP, which cleaves the transmembrane segment (site 2) of RseA. To gain insights into the σE activation pathway, we asked whether the σE-activating signal generated in X. campestris pv. campestris also requires DegS and RseP. To find the X. campestris pv. campestris homologues of DegS and RseP, BLASTP (http://blast.ncbi.nlm.nih.gov/Blast.cgi) searches were performed using DegS and RseP homologues from E. coli and P. aeruginosa. The X. campestris pv. campestris genome contains a homologue of RseP (XCC1366, with its predicted amino acid sequence sharing 56% and 62% similarity with E. coli RseP and P. aeruginosa MucP, respectively; XCC1366 will be referred to as RseP protease), and XCC3898, annotated as protease DO, had the highest level of homology to DegS proteins (XCC3898, with its predicted amino acid sequence sharing 61% similarity with E. coli DegS and P. aeruginosa AlgW; XCC3898 will be referred to as DegS protease). In X. campestris pv. campestris, there was also significant similarity (62%) between DegS and MucD, since they belong to the widely conserved family of HtrA proteins (18).
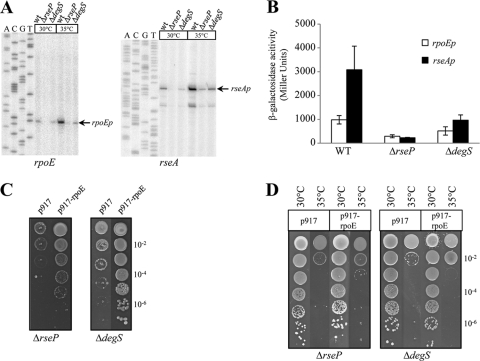
To probe the role of the RseP and DegS proteases in σE-dependent transcription in X. campestris pv. campestris, we constructed chromosomal in-frame deletions of rseP or degS. As shown in Fig. 8 A, upon deletion of degS or rseP, there was a strong decrease in the expression of the σE-dependent promoters rpoEp and rseAp at both 30°C and 35°C compared to the WT levels. Moreover, σE-dependent transcriptional activity from rpoEp-lacZ or rseAp-lacZ in the ΔrseP and ΔdegS mutants was significantly reduced during normal growth (Fig. 8B). These results indicated that degS and rseP are required for the activation of σE, thus suggesting a role for DegS and RseP proteases in cleaving RseA in X. campestris pv. campestris. Interestingly, the two proteases do not have the same impact on σE-dependent activity, since the ΔrseP mutant was the most affected. If the RIP protease cascade is conserved in X. campestris pv. campestris, this raises the possibility that, at least under certain conditions, site 1 proteases other than DegS could initiate the cleavage of RseA or that the second-site cleavage of RseA by RseP could occur independently of a site 1 protease. However, σE-dependent activity was still induced in ΔrseP and ΔdegS mutants following a temperature upshift to 35°C (Fig. 8A), indicating that RseP and DegS had little or no impact on the heat stress response of σE activation.
FIG. 8.
The deletion of rseP or degS impairs σE activation. (A) Expression of σE-dependent promoters using primer extension assays. Total RNA was prepared from the WT, ΔrseP, and ΔdegS mutant strains grown at 30°C or exposed to a temperature upshift at 35°C for 60 min. Primers rpoE-EXT and rseA(2)-EXT were used. (B) Determination of rpoEp and rseAp activities in WT, ΔdegS, and ΔrseP strains. Plasmids containing a transcriptional fusion of the upstream region of rpoEp or rseAp to the lacZ gene were transferred into X. campestris pv. campestris strains. Overnight cultures of these strains grown in MOKA medium were diluted in the same medium and grown for 9 h before determination of β-galactosidase activity. The results represent the mean values of at least two independent experiments, each performed in triplicate, and the error bars indicate the standard errors. (C) Influence of 40 μM cadmium on growth of X. campestris pv. campestris WT, ΔdegS, and ΔrseP strains containing control plasmid p917 or σE-overexpressing plasmid p917-rpoE. Serial 10-fold dilutions of late-exponential-phase bacteria were spotted on MOKA plates containing cadmium and incubated at 30°C for 72 h. (D) Heat sensitivities of X. campestris pv. campestris WT, ΔdegS, and ΔrseP strains containing control plasmid p917 or σE-overexpressing plasmid p917-rpoE. Serial 10-fold dilutions of late-exponential-phase bacteria were spotted on plates and incubated at 35°C for 72 h. For panels C and D, each experiment was repeated three times.
The protease mutants were then tested for σE-dependent stress responses, such as resistance to cadmium (Fig. 8C) and adaptation to increased temperature (Fig. 8D). As expected, deletion of rseP or degS resulted in a strong growth defect in the presence of cadmium, similar to the results obtained with a ΔrpoE mutant (Fig. 3A). The overexpression of σE from plasmid p917-rpoE restored resistance to cadmium, indicating that overexpression of σE bypasses the need for RseP and DegS for metal resistance. These data point to a role of proteolytically active DegS and RseP in the activation of σE in X. campestris pv. campestris during cadmium stress. At 35°C, growth of the ΔdegS and the ΔrseP mutants was strongly impaired, similar to the results obtained with a ΔrpoE mutant (Fig. 4A). However, the overexpression of σE from plasmid p917-rpoE did not fully restore the growth defect phenotype of the protease mutant strains, probably because DegS and RseP are essential for growth at 35°C. These results strongly suggest that DegS and RseP degrade cytoplasmic-membrane-localized substrates (other than RseA) that could be involved in the heat stress response. Furthermore, it must be noted that, as for X. fastidiosa, there are no σE-dependent promoter elements upstream of the degS gene, but instead, its promoter presents a putative σH consensus sequence (data not shown). Unfortunately, we have been unable to experimentally identify degS or rseP promoters, so that the regulation of these genes under stress conditions is still under investigation. All these data show that DegS and RseP are required to activate σE, and this is probably achieved by two-step proteolysis to liberate σE from RseA. However, a residual induction of σE activity could still occur without these proteases, depending on growth conditions.
DISCUSSION
Here, we have combined different approaches to investigate the role and mechanism of σE-dependent activation in X. campestris pv. campestris. We demonstrated that σE is an important regulator of the stress response of X. campestris pv. campestris, since it is involved in stationary-phase survival and resistance to membrane-perturbing stresses. However, we were unable to find any increased sensitivity of the rpoE mutant to a variety of chemical stresses (data not shown). This limited response was not surprising, since there are 9 other ECF σ factors in X. campestris pv. campestris. Each one could respond to a specific stress stimulus and could substitute functionally for the others, since ECFs tend to share overlapping promoter specificity, as has been well described in B. subtilis for σM, σW, and σX (43). Knowledge of the remaining ECF σ factors and their inducing signals is sparse, and it is crucial to address these issues in future studies.
As in X. campestris pv. campestris strain 11 and X. fastidiosa, the rpoE gene of X. campestris pv. campestris is organized as a single transcription unit with the anti-σ gene rseA and the protease gene mucD. However, the transcriptional control of rpoE expression is different in these bacterial species, since it is controlled by the housekeeping σ factor in X. fastidiosa (24), while transcription of rpoE is autoregulated in X. campestris pv. campestris, like most of its known orthologues (our results and reference 17). This indicates that regulatory pathways may have evolved differently in these closely related bacterial species in order to be able to process different environmental parameters, since X. fastidiosa appears to have an in planta-restricted lifestyle while X. campestris pv. campestris is able to survive in the environment between infections. Our results strongly suggest that RseA is the anti-σE factor and that MucD is a negative regulator of σE-dependent activity. The transcription of rseA and mucD is controlled by a highly conserved σE-dependent promoter within the σE gene (24, 59). This negative feedback loop would ensure the tight control of σE-dependent activity in X. campestris pv. campestris in order to avoid the energy cost of an inappropriate σE response, as we observed that the overexpression of σE was deleterious to X. campestris pv. campestris fitness.
We used microarrays to define the X. campestris pv. campestris σE regulon by comparing the levels of the transcripts of all annotated ORFs in the rpoE deletion mutant with those in the rseA deletion mutant in order to be closer to physiological levels of induction of the σE response. σE upregulates at least 45 genes, including rpoE regulon members themselves, and 20 genes are downregulated upon σE overactivation. We identified the X. campestris pv. campestris σE promoter consensus motif (GGAACTN15-17GTCNNA), which has the conserved characteristics of promoter elements of ECF σ factors from the ECF02 group (65). Most σE-dependent genes fall broadly into the same categories previously described for the core and extended σE regulon members (59) with structural components of the cell envelope, OMP assembly, the periplasmic chaperone, fatty acid biosynthesis, and two-component systems. The higher electron transport system activity could possibly compensate for proton leakage across the membrane when its integrity is compromised in order to maintain the PMF. In agreement with this notion, a functional σE regulon was essential for the maintenance of PMF in S. Typhimurium (7), and we observed that genes encoding functions that are PMF-energized processes (ATP synthesis, motility, and active transport) were downregulated when σE was overactive.
In contrast to core σE regulons of enterobacteria, we did not find genes involved in cell envelope biogenesis (59). Variation in growth conditions, treatment conditions, and microarray platforms could be the reasons for this discrepancy. A subset of σE-dependent genes in X. campestris pv. campestris belongs to the highly conserved heat shock regulon involved in aiding protein folding, protein disaggregation, and proteolysis and comprising the heat shock sigma factor (rpoH). We also showed that rpoH transcription was directly regulated by σE under both normal and heat stress conditions, so that σH could in turn direct transcription of the heat shock regulon. The ability of σE to mediate the expression of rpoH has been reported in several other bacteria, particularly in gammaproteobacteria (3, 69). It is noteworthy that the rpoH gene of X. campestris pv. campestris strain 11 has σE promoter elements identical to those in X. campestris pv. campestris, but previous work did not correctly predict the σE-dependent promoter of rpoH in X. campestris pv. campestris strain 11 (37). We have also identified a distal σE-independent promoter for rpoH, and its expression was slightly induced by temperature. These data underline the complex regulation of rpoH, as in many other bacteria, e.g., in E. coli there are five promoters upstream of the rpoH gene, recognized by σ70, σS, σE, and σ54 (38). This ensures that the protective heat shock response can be triggered by several environmental cues or during the cell cycle. In X. campestris pv. campestris it seems particularly relevant, since we could not inactivate the rpoH gene under normal growth conditions (data not shown). Moreover, we observed that rpoEp, rseAp, XCC1535p, XCC3227p, and prcp were directly activated by σE following heat treatment at 35°C. XCC1535 and Prc are predicted to be periplasmic proteins and to have roles, respectively, in protein folding and proteolysis. This suggests that they could prevent aggregation of misfolded periplasmic protein derivatives under increased export stress, which could occur when cellular envelope integrity is compromised. Taken together, our results support the essential role of rpoE in heat stress protection of X. campestris pv. campestris and suggest that the heat shock response in X. campestris pv. campestris is similar to that of E. coli and other gammaproteobacteria.
We also found that σE is involved in the transcriptional activity of a subset of genes encoding proteins involved in the T3S translocation and regulation machinery. Only two of the six hrp operons of X. campestris pv. campestris, hrpE and hrpF, were induced. This is reminiscent of the regulation of the hrpC and hrpE operons by the two-component regulatory system ColR/ColS in X. campestris pv. campestris 8004 (77), suggesting that individual hrp operons might be targeted by alternative regulatory networks integrating diverse environmental signals. Taken together with the contribution of σE to the functionality of T3S systems of the enteropathogens Yersinia pseudotuberculosis and S. Typhimurium (15, 53), this regulation suggests a link between cell envelope perturbation and T3S induction in X. campestris pv. campestris. This could occur during the pathogenesis process in response to host factors (oxidative antimicrobial systems and membrane-targeted host defense peptides) that compromise bacterial membrane integrity. We tested the hypothesis that σE could be involved in X. campestris pv. campestris pathogenesis, but we were unable to see significant differences between the WT and the ΔrpoE mutant when we inoculated them into A. thaliana (data not shown). These results are in agreement with previous studies based on X. campestris pv. campestris strain 11 (17). This lack of a phenotype does not exclude the possibility that σE has a role under natural infection conditions of X. campestris pv. campestris that may be discovered using a different plant model or route of infection. Moreover, T3S is involved not only in pathogenesis, but also in pathogen dissemination. Mutation of genes encoding structural components and regulatory genes of the T3SS of Xanthomonas fuscans subsp. fuscans altered the ability of the bacterium to transmit to bean seeds through both vascular and floral pathways (21). The importance of rpoE in resisting external stresses suggests that σE may be involved in survival in the environment, which constitutes a prerequisite to plant infection and disease development (35). Bacterial epiphytic fitness and seed transmission experiments will be crucial to decipher the role of σE in the dispersion of plant-pathogenic bacteria, colonization, and survival on their hosts.
In E. coli, the σE response is triggered when periplasmic protein folding and assembly are compromised. The accumulation of unassembled OMPs is the activation signal for the initiation of a sequential proteolytic cascade of the membrane-spanning anti-σ RseA by DegS and RseP (72). DegS and RseP homologues have been identified in X. campestris pv. campestris based on sequence homologies. The deletion of their corresponding genes led to a decrease in σE-dependent activity at both 30°C and 35°C. Our results strongly suggest that the predicted proteases DegS and RseP regulate σE by the mechanism of RIP as described in E. coli. However, there were differences in the proteolytic process: when proteases were inactivated, residual σE-dependent transcription under normal growth conditions remained, and there was still induction of the σE response by a temperature upshift. This was rather unexpected, because it would be the first example of σE activation by a temperature upshift that did not require DegS and RseP proteases. Consequently, it must be assumed that RseA/σE cleavage can be performed by other proteases, depending on stress stimuli, in X. campestris pv. campestris. This is supported by the recent findings that acid stress activation of σE-dependent gene expression in S. Typhimurium is independent of the unfolded OMP signal or the DegS protease (48). These observations unravel a new activation pathway for the σE response. In addition, in E. coli, it has been shown that the HtrA family members DegQ and DegS can complement the loss of the periplasmic protease Prc (6). Interestingly, prc was one of the σE target genes identified in our microarray analysis, and we also showed that its expression was heat inducible. Tail-specific proteases like Prc have been reported to be implicated in RIP: CtpB of B. subtilis triggers activation of σK indirectly by cleaving the SpoIVFA protein (14), and Prc of P. aeruginosa is implicated in the AlgU/MucA σ/anti-σ system (57). Hence, it is conceivable that Prc is involved in RseA/σE cleavage in X. campestris pv. campestris under some conditions, and it will be the aim of future work to address these hypotheses. These unusual cleavage features of the RseA/σE system in X. campestris pv. campestris could also be due to the absence of the rseB gene in the rpoE operon. RseB is a negative regulator of the σE-dependent envelope stress response that binds to the periplasmic domain of RseA (1). RseB prevents DegS and RseP from cleaving RseA, suggesting that interaction of RseB with RseA must be altered before the signal transduction cascade is activated. There is no homologue of RseB in the X. campestris pv. campestris genome, and it is unknown if another protein could fulfill its protective function for inappropriate cleavage of RseA in the absence of activating stress.
Other observations were that degS and rseP mutants were strongly affected for growth at 35°C and in the presence of cadmium but that σE overexpression in these mutant strains restored growth only in the presence of heavy metals. These data implied that both DegS and RseP are required for growth at 35°C, possibly because they could have other substrates. It is the case for E. coli RseP, which could use the signal-transducing protein FecR as a substrate (13), and the B. subtilis RseP orthologue, RasP, which can attack the cell division protein FtsL (12). In addition, DegS and MucD belong to the HtrA protein family. In E. coli, the HtrA family member DegQ can rescue the conditional lethality of degP deletion (6, 71). It is tempting to speculate that since there is no homologue of DegQ in X. campestris pv. campestris, DegS could also substitute for the X. campestris pv. campestris DegP orthologue, MucD. This could explain why the mucD mutant is not affected by temperature stress in X. campestris pv. campestris, whereas in most bacterial species degP mutants show a thermosensitive phenotype in vitro (47, 74).
Taken together, the data show that while the general function of σE is regulation of the extracellular stress response in X. campestris pv. campestris, its specific functions and regulation pathways are complex and multifaceted. A lot of questions remain to be answered, and continued work on the role and RIP mechanism of the σE response will be of major importance in ultimately understanding the nature and complexity of the envelope stress response, as these regulatory proteins can be found in a great variety of prokaryotes.
Supplementary Material
Acknowledgments
We are grateful to Claude Bruand for critical reading of the manuscript. We thank Denis Bidot, Manuela Meyer, and Eva Schulte-Berndt for technical assistance.
This work was supported by the Centre National de la Recherche Scientifique (CNRS) and the Université de Toulouse (UPS) and by grants 0313805A (Bundesministerium für Bildung und Forschung, Germany) and SPP 1316 (German Research Foundation) to A.B.
Footnotes
Published ahead of print on 22 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Ades, S. E. 2008. Regulation by destruction: design of the σE envelope stress response. Curr. Opin. Microbiol. 11:535-540. [DOI] [PubMed] [Google Scholar]
- 2.Alba, B. M., and C. A. Gross. 2004. Regulation of the Escherichia coli σ-dependent envelope stress response. Mol. Microbiol. 52:613-619. [DOI] [PubMed] [Google Scholar]
- 3.Aramaki, H., T. Hirata, C. Hara, M. Fujita, and Y. Sagara. 2001. Transcription analysis of rpoH in Pseudomonas putida. FEMS Microbiol. Lett. 205:165-169. [DOI] [PubMed] [Google Scholar]
- 4.Asakura, Y., and I. Kobayashi. 2009. From damaged genome to cell surface: transcriptome changes during bacterial cell death triggered by loss of a restriction-modification gene complex. Nucleic Acids Res. 37:3021-3031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bashyam, M. D., and S. E. Hasnain. 2004. The extracytoplasmic function sigma factors: role in bacterial pathogenesis. Infect. Genet. Evol. 4:301-308. [DOI] [PubMed] [Google Scholar]
- 6.Bass, S., Q. Gu, and A. Christen. 1996. Multicopy suppressors of prc mutant Escherichia coli include two HtrA (DegP) protease homologs (HhoAB), DksA, and a truncated R1pA. J. Bacteriol. 178:1154-1161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Becker, L. A., I. S. Bang, M. L. Crouch, and F. C. Fang. 2005. Compensatory role of PspA, a member of the phage shock protein operon, in rpoE mutant Salmonella enterica serovar Typhimurium. Mol. Microbiol. 56:1004-1016. [DOI] [PubMed] [Google Scholar]
- 8.Benjamini, Y., and Y. Hochberg. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57:289-300. [Google Scholar]
- 9.Blanvillain, S., D. Meyer, A. Boulanger, M. Lautier, C. Guynet, N. Denancé, J. Vasse, E. Lauber, and M. Arlat. 2007. Plant carbohydrate scavenging through tonB-dependent receptors: a feature shared by phytopathogenic and aquatic bacteria. PLoS One 21:e224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Boucher, J. C., M. J. Schurr, H. Yu, D. W. Rowen, and V. Deretic. 1997. Pseudomonas aeruginosa in cystic fibrosis: role of mucC in the regulation of alginate production and stress sensitivity. Microbiology 143:3473-3480. [DOI] [PubMed] [Google Scholar]
- 11.Boulanger, A., G. Déjean, M. Lautier, M. Glories, C. Zischek, M. Arlat, and E. Lauber. 2010. Identification and regulation of the N-acetylglucosamine utilization pathway of the plant pathogenic bacterium Xanthomonas campestris pv. campestris. J. Bacteriol. 192:1487-1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bramkamp, M., L. Weston, R. A. Daniel, and J. Errington. 2006. Regulated intramembrane proteolysis of FtsL protein and the control of cell division in Bacillus subtilis. Mol. Microbiol. 62:580-591. [DOI] [PubMed] [Google Scholar]
- 13.Braun, V., S. Mahren, and A. Sauter. 2006. Gene regulation by transmembrane signaling. Biometals 19:103-113. [DOI] [PubMed] [Google Scholar]
- 14.Campo, N., and D. Z. Rudner. 2006. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol. Cell 23:25-35. [DOI] [PubMed] [Google Scholar]
- 15.Carlsson, K. E., J. Liu, P. J. Edqvist, and M. S. Francis. 2007. Extracytoplasmic-stress-responsive pathways modulate type III secretion in Yersinia pseudotuberculosis. Infect. Immun. 75:3913-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Castanié-Cornet, M.-P., K. Cam, B. Bastiat, A. Cros, P. Bordes, and C. Gutierrez. 2010. Acid stress response in Escherichia coli: mechanism of regulation of gadA transcription by RcsB and GadE. Nucleic Acids Res. 38:3546-3554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng, C. Y., S. Y. Shieh, C. C. Hsu, and M. T. Yang. 2008. Characterization and transcriptional analysis of an ECF sigma factor from Xanthomonas campestris pv. campestris. FEMS Microbiol. Lett. 289:250-257. [DOI] [PubMed] [Google Scholar]
- 18.Clausen, T., C. Southan, and M. Ehrmann. 2002. The HtrA family of proteases: implications for protein composition and cell fate. Mol. Cell 10:443-455. [DOI] [PubMed] [Google Scholar]
- 19.Costanzo, A., and S. E. Ades. 2006. Growth phase-dependent regulation of the extracytoplasmic stress factor, σE, by guanosine 3′,5′-bispyrophosphate (ppGpp). J. Bacteriol. 188:4627-4634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Crouch, M. L., L. A. Becker, I. S. Bang, H. Tanabe, A. J. Ouellette, and F. C. Fang. 2005. The alternative sigma factor σE is required for resistance of Salmonella enterica serovar Typhimurium to anti-microbial peptides. Mol. Microbiol. 56:789-799. [DOI] [PubMed] [Google Scholar]
- 21.Darsonval, A., A. Darrasse, D. Meyer, M. Demarty, K. Durand, C. Bureau, C. Manceau, and M. A. Jacques. 2008. The type III secretion system of Xanthomonas fuscans subsp. fuscans is involved in the phyllosphere colonization process and in transmission to seeds of susceptible beans. Appl. Environ. Microbiol. 74:2669-2678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dartigalongue, C., H. Loferer, and S. Raina. 2001. EcfE, a new essential inner membrane protease: its role in the regulation of heat shock response in Escherichia coli. EMBO J. 20:5908-5918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.da Silva, A. C. R., J. A. Ferro, F. C. Reinach, C. S. Farah, L. R. Furlan, et al. 2002. Comparison of the genomes of two Xanthomonas pathogens with differing host specificities. Nature 417:459-463. [DOI] [PubMed] [Google Scholar]
- 24.da Silva Neto, J. F., T. Koide, S. L. Gomes, and M. V. Marques. 2007. The single extracytoplasmic-function σ factor of Xylella fastidiosa is involved in the heat shock response and presents an unusual regulatory mechanism. J. Bacteriol. 189:551-560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dombrecht, B., J. Vanderleyden, and J. Michiels. 2001. Stable RK2-derived cloning vectors for the analysis of gene expression and gene function in gram-negative bacteria. Mol. Plant Microbe Interact. 14:426-430. [DOI] [PubMed] [Google Scholar]
- 26.Egler, M., C. Grosse, G. Grass, and D. H. Nies. 2005. Role of the extracytoplasmic function protein family sigma factor RpoE in metal resistance of Escherichia coli. J. Bacteriol. 187:2297-2307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ehrmann, M., and T. Clausen. 2004. Proteolysis as a regulatory mechanism. Annu. Rev. Genet. 38:709-724. [DOI] [PubMed] [Google Scholar]
- 28.Erickson, J. W., and C. A. Gross. 1989. Identification of the σE subunit of Escherichia coli RNA polymerase: a second alternative sigma factor involved in high-temperature gene expression. Genes Dev. 3:1462-1471. [DOI] [PubMed] [Google Scholar]
- 29.Grall, N., J. Livny, M. Waldor, M. Barel, A. Charbit, and K. L. Meibom. 2009. Pivotal role of the Francisella tularensis heat-shock sigma factor RpoH. Microbiology 155:2560-2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gürlebeck, D., F. Thieme, and U. Bonas. 2006. Type III effector proteins from the plant pathogen Xanthomonas and their role in the interaction with the host plant. J. Plant Physiol. 163:233-255. [DOI] [PubMed] [Google Scholar]
- 31.Hayden, J. D., and S. E. Ades. 2008. The extracytoplasmic stress factor, σE, is required to maintain cell envelope integrity in Escherichia coli. PLoS One 3:e1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Heinrich, J., K. Hein, and T. Wiegert. 2009. Two proteolytic modules are involved in regulated intramembrane proteolysis of Bacillus subtilis RsiW. Mol. Microbiol. 74:1412-1426. [DOI] [PubMed] [Google Scholar]
- 33.Helmann, J. D. 1999. Anti-sigma factors. Curr. Opin. Microbiol. 2:135-141. [DOI] [PubMed] [Google Scholar]
- 34.Helmann, J. D. 2006. Deciphering a complex genetic regulatory network: the Bacillus subtilis sigmaW protein and intrinsic resistance to antimicrobial compounds. Sci. Prog. 89:243-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hirano, S. S., and C. D. Upper. 1983. Ecology and epidemiology of foliar bacterial plant pathogens. Annu. Rev. Phytopathol. 21:243. [Google Scholar]
- 36.Ho, E. M., H. W. Chang, S. I. Kim, H. Y. Kahng, and K. H. Oh. 2004. Analysis of TNT (2,4,6-trinitrotoluene)-inducible cellular responses and stress shock proteome in Stenotrophomonas sp. OK-5. Curr. Microbiol. 49:346-352. [DOI] [PubMed] [Google Scholar]
- 37.Huang, L. H., Y. H. Tseng, and M. T. Yang. 1998. Isolation and characterization of the Xanthomonas campestris rpoH gene coding for a 32-kDa heat shock sigma factor. Biochem. Biophys. Res. Commun. 244:854-860. [DOI] [PubMed] [Google Scholar]
- 38.Janaszak, A., B. Nadratowska-Wesołowska, G. Konopa, and A. Taylor. 2009. The P1 promoter of the Escherichia coli rpoH gene is utilized by σ70-RNAP or σS-RNAP depending on growth phase. FEMS Microbiol. Lett. 291:65-72. [DOI] [PubMed] [Google Scholar]
- 39.Kado, C. I., and M. G. Heskett. 1970. Selective media for isolation of Agrobacterium, Corynebacterium, Erwinia, Pseudomonas, and Xanthomonas. Phytopathology 60:969-976. [DOI] [PubMed] [Google Scholar]
- 40.Kim, D. Y., E. Kwon, J. Choi, H. Y. Hwang, and K. K. Kim. 2010. Structural basis for the negative regulation of bacterial stress response by RseB. Protein Sci. 19:1258-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Koo, B. M., V. A. Rhodius, E. A. Campbell, and C. A. Gross. 2009. Dissection of recognition determinants of Escherichia coli σ32 suggests a composite −10 region with an ′extended −10′ motif and a core −10 element. Mol. Microbiol. 72:815-829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kovacikova, G., and K. Skorupski. 2002. The alternative sigma factor σE plays an important role in intestinal survival and virulence in Vibrio cholerae. Infect. Immun. 70:5355-5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo, Y., and J. D. Helmann. 2009. Extracytoplasmic function sigma factors with overlapping promoter specificity regulate sublancin production in Bacillus subtilis. J. Bacteriol. 191:4951-4958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miller, J. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 45.Misra, H. S., N. P. Khairnar, A. Barik, K. Indira Priyadarsini, H. Mohan, and S. K. Apte. 2004. Pyrroloquinoline-quinone: a reactive oxygen species scavenger in bacteria. FEBS Lett. 578:26-30. [DOI] [PubMed] [Google Scholar]
- 46.Missiakas, D., M. P. Mayer, M. Lemaire, C. Georgopoulos, and S. Raina. 1997. Modulation of the Escherichia coli σE (RpoE) heat-shock transcription-factor activity by the RseA, RseB and RseC proteins. Mol. Microbiol. 24:355-371. [DOI] [PubMed] [Google Scholar]
- 47.Mo, E., S. E. Peters, C. Willers, D. J. Maskell, and I. G. Charles. 2006. Single, double and triple mutants of Salmonella enterica serovar Typhimurium degP (htrA), degQ (hhoA) and degS (hhoB) have diverse phenotypes on exposure to elevated temperature and their growth in vivo is attenuated to different extents. Microb. Pathog. 41:174-182. [DOI] [PubMed] [Google Scholar]
- 48.Muller, C., I. S. Bang, J. Velayudhan, J. Karlinsey, K. Papenfort, J. Vogel, and F. C. Fang. 2009. Acid stress activation of the σE stress response in Salmonella enterica serovar Typhimurium. Mol. Microbiol. 71:1228-1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nollen, E. A., and R. I. Morimoto. 2002. Chaperoning signaling pathways: molecular chaperones as stress-sensing ‘heat shock’ proteins. J. Cell Sci. 115:2809-2816. [DOI] [PubMed] [Google Scholar]
- 50.Nonaka, G., M. Blankschien, C. Herman, C. A. Gross, and V. A. Rhodius. 2006. Regulon and promoter analysis of the E. coli heat-shock factor, σ32, reveals a multifaceted cellular response to heat stress. Genes Dev. 20:1776-1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Oke, V., and S. R. Long. 1999. Bacterial genes induced within the nodule during the Rhizobium-legume symbiosis. Mol. Microbiol. 32:837-849. [DOI] [PubMed] [Google Scholar]
- 52.Onsando, J. M. 1992. Black rot of crucifers, p. 243-252. In H. S. Chaube, J. Kumar, A. N. Mukhopadhyay, and U. S. Singh (ed.), Plant diseases of international importance. II. Diseases of vegetable and oil seed crops. Prentice Hall, Englewood Cliffs, NJ.
- 53.Osborne, S. E., and B. K. Coombes. 2009. RpoE fine tunes expression of a subset of SsrB-regulated virulence factors in Salmonella enterica serovar Typhimurium. BMC Microbiol. 9:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Papenfort, K., V. Pfeiffer, F. Mika, S. Lucchini, J. C. D. Hinton, and J. Vogel. 2006. σE dependent small RNAs of Salmonella respond to membrane stress by accelerating global omp mRNA decay. Mol. Microbiol. 62:1674-1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qiu, D., V. M. Eisinger, D. W. Rowen, and H. D. Yu. 2007. Regulated proteolysis controls mucoid conversion in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. U. S. A. 104:8107-8112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raivio, T. L., and T. J. Silhavy. 2001. Periplasmic stress and ECF sigma factors. Annu. Rev. Microbiol. 55:591-624. [DOI] [PubMed] [Google Scholar]
- 57.Reiling, S. A., J. A. Jansen, B. J. Henley, S. Singh, C. Chattin, M. Chandler, and D. W. Rowen. 2005. Prc protease promotes mucoidy in mucA mutants of Pseudomonas aeruginosa. Microbiology 151:2251-2261. [DOI] [PubMed] [Google Scholar]
- 58.Rezuchova, B., and J. Kormanec. 2001. A two-plasmid system for identification of promoters recognized by RNA polymerase containing extracytoplasmic stress response σE in Escherichia coli. J. Microbiol. Methods 45:103-111. [DOI] [PubMed] [Google Scholar]
- 59.Rhodius, V. A., W. C. Suh, G. Nonaka, J. West, and C. A. Gross. 2006. Conserved and variable functions of the σE stress response in related genomes. PLoS Biol. 4:e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rowley, G., M. Spector, J. Kormanec, and M. Roberts. 2006. Pushing the envelope: extracytoplasmic stress responses in bacterial pathogens. Nat. Rev. Microbiol. 4:383-394. [DOI] [PubMed] [Google Scholar]
- 61.Schäfer, A., A. Tauch, W. Jäger, J. Kalinowski, G. Thierbach, and A. Pühler. 1994. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145:69-73. [DOI] [PubMed] [Google Scholar]
- 62.Serrania, J., F.-J. Vorhölter, K. Niehaus, A. Pühler, and A. Becker. 2008. Identification of Xanthomonas campestris pv. campestris galactose utilization genes from transcriptome data. J. Biotechnol. 135:309-317. [DOI] [PubMed] [Google Scholar]
- 63.Slamti, L., J. Livny, and M. K. Waldor. 2007. Global gene expression and phenotypic analysis of a Vibrio cholerae rpoH deletion mutant. J. Bacteriol. 189:351-362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Smyth, G. K. 2004. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat. Appl. Genet. Mol. Biol. 3:3. [DOI] [PubMed] [Google Scholar]
- 65.Staroń, A., H. J. Sofia, S. Dietrich, L. E. Ulrich, H. Liesegang, and T. Mascher. 2009. The third pillar of bacterial signal transduction: classification of the extracytoplasmic function (ECF) sigma factor protein family. Mol. Microbiol. 74:557-581. [DOI] [PubMed] [Google Scholar]
- 66.Strauch, K. L., and J. Beckwith. 1988. An Escherichia coli mutation preventing degradation of abnormal periplasmic proteins. Proc. Natl. Acad. Sci. U. S. A. 85:1576-1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Testerman, T. L., A. Vazquez-Torres, Y. Xu, J. Jones-Carson, S. J. Libby, and F. C. Fang. 2002. The alternative sigma factor σE controls antioxidant defences required for Salmonella virulence and stationary-phase survival. Mol. Microbiol. 43:771-782. [DOI] [PubMed] [Google Scholar]
- 68.Turner, P., C. E. Barber, and M. J. Daniels. 1985. Evidence for clustered pathogenicity genes in Xanthomonas campestris pv. campestris. Mol. Gen. Genet. 199:338-343. [Google Scholar]
- 69.Vanaporn, M., P. Vattanaviboon, V. Thongboonkerd, and S. Korbsrisate. 2008. The rpoE operon regulates heat stress response in Burkholderia pseudomallei. FEMS Microbiol. Lett. 284:191-196. [DOI] [PubMed] [Google Scholar]
- 70.Vorhölter, F. J., S. Schneiker, A. Goesmann, L. Krause, T. Bekel, O. Kaiser, B. Linke, T. Patschkowski, C. Rückert, J. Schmid, V. K. Sidhu, V. Sieber, A. Tauch, S. A. Watt, B. Weisshaar, A. Becker, K. Niehaus, and A. Pühler. 2008. The genome of Xanthomonas campestris pv. campestris B100 and its use for the reconstruction of metabolic pathways involved in xanthan biosynthesis. J. Biotechnol. 134:33-45. [DOI] [PubMed] [Google Scholar]
- 71.Waller, P. R., and R. T. Sauer. 1996. Characterization of degQ and degS, Escherichia coli genes encoding homologs of the DegP protease. J. Bacteriol. 178:1146-1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walsh, N. P., B. M. Alba, B. Bose, C. A. Gross, and R. T. Sauer. 2003. OMP peptide signals initiate the envelope-stress response by activating DegS protease via relief of inhibition mediated by its PDZ domain. Cell 113:61-71. [DOI] [PubMed] [Google Scholar]
- 73.Wang, L., W. Rong, and C. He. 2008. Two Xanthomonas extracellular polygalacturonases, PghAxc and PghBxc, are regulated by type III secretion regulators HrpX and HrpG and are required for virulence. Mol. Plant Microbe Interact. 21:555-563. [DOI] [PubMed] [Google Scholar]
- 74.Wood, L. F., and D. E. Ohman. 2006. Independent regulation of MucD, an HtrA-like protease in Pseudomonas aeruginosa, and the role of its proteolytic motif in alginate gene regulation. J. Bacteriol. 188:3134-3137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wood, L. F., and D. E. Ohman. 2009. Use of cell wall stress to characterize σ22 (AlgT/U) activation by regulated proteolysis and its regulon in Pseudomonas aeruginosa. Mol. Microbiol. 72:183-201. [DOI] [PubMed] [Google Scholar]
- 76.Yang, T. C., Y. W. Leu, H. C. Chang-Chien, and R. M. Hu. 2009. Flagellar biogenesis of Xanthomonas campestris requires the alternative sigma factors RpoN2 and FliA and is temporally regulated by FlhA, FlhB, and FlgM. J. Bacteriol. 191:2266-2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Zhang, S. S., Y. Q. He, L. M. Xu, B. W. Chen, B. L. Jiang, J. Liao, J. R. Cao, D. Liu, Y. Q. Huang, X. X. Liang, D. J. Tang, G. T. Lu, and J. L. Tang. 2008. A putative colR(XC1049)-colS(XC1050) two-component signal transduction system in Xanthomonas campestris positively regulates hrpC and hrpE operons and is involved in virulence, the hypersensitive response and tolerance to various stresses. Res. Microbiol. 159:569-578. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.