Abstract
A cascade of alternative sigma factors governs the program of developmental gene expression during sporulation in Bacillus subtilis. Little is known, however, about how the early-acting sigma factors are inactivated and replaced by the later-acting factors. Here we identify a small protein, Fin (formerly known as YabK), that is required for efficient switching from σF- to σG-directed gene expression in the forespore compartment of the developing sporangium. The fin gene, which is conserved among Bacillus species and species of related genera, is transcribed in the forespore under the control of both σF and σG. Cells mutant for fin are unable to fully deactivate σF and, conversely, are unable to fully activate σG. Consistent with their deficiency in σG-directed gene expression, fin cells are arrested in large numbers following the engulfment stage of sporulation, ultimately forming 50-fold fewer heat-resistant spores than the wild type. Based in part on the similarity of Fin to the anti-σG factor CsfB (also called Gin), we speculate that Fin is an anti-σF factor which, by disabling σF, promotes the switch to late developmental gene expression in the forespore.
Complex, multistep cell differentiation pathways are typically orchestrated by the activation of sets of regulatory genes in an ordered sequence. In bacteria, such developmental programs are sometimes driven by cascades of RNA polymerase (RNAP) sigma (σ) factors, as in the paradigmatic example of spore formation by Bacillus subtilis (17, 23, 28, 34). Sporulation takes place in a two-compartment sporangium that arises by a process of asymmetric division (Fig 1A). The smaller, forespore compartment develops into the spore, whereas the larger mother cell nurtures the developing forespore. Initially, the forespore and mother cell lie side by side; subsequently, the mother cell engulfs the forespore in a phagocytosis-like process that results in a cell-within-a-cell configuration (Fig. 1A). The engulfed forespore is then encased in protective peptidoglycan cortex and protein coat layers and ultimately released into the environment by lysis of the mother cell.
FIG. 1.
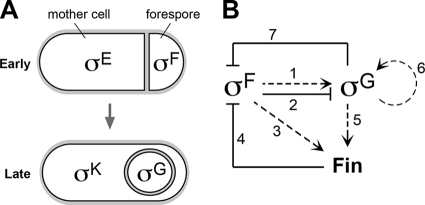
A role for Fin (YabK) in sigma factor switching during sporulation in B. subtilis. (A) Cartoon depicting the sigma factors directing compartment-specific gene expression in sporangia at early (top) and late (bottom) stages of development. At early times, the sigma factors σF and σE direct gene expression in the forespore and mother cell, respectively. At later times, after the forespore is engulfed by the mother cell, σF is replaced by σG and σE is replaced by σK. (B) Model for the switch from σF to σG. To begin, σF activates transcription of the gene (sigG) for σG (arrow 1); however, σG activation is delayed through poorly understood mechanisms that likely involve σF (barred line 2) (see Discussion). To trigger the switch to σG, σF also directs synthesis of its own inhibitor, Fin (arrow 3). Once σF is inactivated by Fin (barred line 4), σG becomes active. This transition is reinforced by two mechanisms. First, σG continues to direct fin synthesis, resulting in sustained σF inhibition (arrow 5). Second, σG autoregulates its own gene, leading to large amounts of the late sigma factor (arrow 6). σG also inhibits σF by an unknown, Fin-independent pathway (barred line 7) (see Discussion). In the absence of Fin, unchecked σF activity prevents σG activation, likely due to the same mechanisms represented by barred line 2. Dashed arrows indicate transcriptional regulation. Lines with barred ends indicate inhibition by currently unknown mechanisms.
Gene expression after asymmetric division is driven chiefly by four compartment-specific sigma factors—σF, σE, σG, and σK—that direct RNAP to distinct sets of developmental genes (15, 32, 40). The σF and σE factors are early-acting regulatory proteins that control gene expression in the forespore and mother cell, respectively. At later times, σG replaces σF in the forespore, whereas σK replaces σE in the mother cell (Fig. 1A). Importantly, this switch to late developmental gene expression requires not only mechanisms to synthesize and activate σG and σK but also mechanisms to inactivate and/or remove σF and σE. The regulation of σG and σK synthesis and activation at the appropriate time and place has been studied extensively and is known in some detail (albeit more for σK than for σG) (reviewed in references 17 and 28). However, it remains poorly understood how σF and σE are inactivated at the transition to late gene expression. Indeed, little overlap between σF and σG activities in the forespore or between σE and σK activities in the mother cell is detected, indicating that one or more controls must exist to temporally segregate them (21). Furthermore, evidence shows that the late-acting sigma factors directly or indirectly trigger negative-feedback loops that inactivate their predecessors: deletion of the gene for σG or σK results in inappropriately sustained σF or σE activity, respectively (4, 6, 13, 20, 43).
Further clues have emerged regarding replacement of σE by σK in the mother cell: the σK-dependent negative-feedback loop appears to operate at the level of transcription of the σE structural gene and specifically requires that σK is transcriptionally active (43, 44). The latter finding, which was obtained using a variant of σK that binds RNAP but is transcriptionally inactive, eliminates a simple model in which the σE-to-σK transition is driven by competition for RNAP (18) and instead indicates that one or more target genes of σK are involved (44). In contrast, almost nothing is known of the nature of the mechanisms that mediate the switch from σF to σG in the forespore.
Here we present evidence that a small, conserved protein that we named Fin (previously annotated YabK) is expressed in the forespore and is required for the efficient transition from σF- to σG-directed gene expression. Remarkably, fin mutant cells are deficient for spore formation and progress slowly, if at all, past the engulfment stage (III) of sporulation, a phenotype consistent with a defect in σG activation. Thus, fin represents a previously unrecognized and uncharacterized sporulation gene. Given the similarity of Fin to the anti-σG factor CsfB (also called Gin) (7, 11, 19, 29), as presented herein, we speculate that Fin functions as an anti-σF factor which, by antagonizing σF, facilitates the switch to σG and promotes the transition to late developmental gene expression in the forespore.
MATERIALS AND METHODS
General methods.
Bacterial strains were propagated in Luria-Bertani medium. When appropriate, antibiotics were included at the following concentrations: chloramphenicol (5 μg/ml), erythromycin plus lincomycin (1 μg/ml and 25 μg/ml, respectively), spectinomycin (100 μg/ml), kanamycin (5 μg/ml), phleomycin (0.4 μg/ml), and ampicillin (100 μg/ml). To measure sporulation efficiency, cells were induced to sporulate by nutrient exhaustion for 24 h at 37°C in Difco (Schaeffer's) sporulation medium (DSM) (27, 30). The number of CFU that survived heat treatment (80°C for 20 min) was determined and normalized to the number of heat-resistant CFU obtained in parallel from the wild-type strain. For all other experiments, sporulation was induced at 37°C by the Sterlini-Mandelstam resuspension method (27, 33). β-Galactosidase activity was measured as previously described (6).
Strain and plasmid construction.
B. subtilis strains used in this study were derived by transformation of the prototrophic laboratory strain PY79 (42) or derivatives thereof with chromosomal DNA, plasmids, or PCR products. The genes utilized to confer resistance of B. subtilis to antibiotics were as follows: cat (chloramphenicol), erm (erythromycin plus lincomycin), spc (spectinomycin), kan (kanamycin), and phleo (phleomycin). Competent B. subtilis cells were prepared as previously described (41). Unless otherwise noted, PY79 chromosomal DNA served as a template for PCR amplification. Plasmids were cloned and propagated in the Escherichia coli strain DH5α. Plasmid mutagenesis was performed with a QuikChange II XL site-directed mutagenesis kit (Stratagene). The genotypes, features, and sources of strains and plasmids used in this study are listed in Table 1. The sequences of primers utilized in strain and plasmid construction are provided in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| B. subtilis strainsa | ||
| PY79 | Prototrophic wild type | 42 |
| RL1275 | ΔsigF::erm | Laboratory stock derived from MO173 (gift of P. Stragier) |
| AHB98 | ΔsigG::kan | 7 |
| AHB199 | ΔcsfB::tet | 7 |
| AHB1931 | Δfin::phleo | This study |
| AHB1983 | Δfin::phleo sacA::Pfin-fin kan | This study |
| AHB1933 | amyE::Pfin-lacZ cat | This study |
| AHB1956 | amyE::Pfin-lacZ cat ΔsigF::erm | This study |
| AHB1934 | amyE::Pfin-lacZ cat ΔsigG::kan | This study |
| AHB2085 | Δfin::phleo amyE::Pfin-gfp-fin spc | This study |
| AHB881 | amyE::PspoIIQ-lacZ cat | 6 |
| AHB1953 | amyE::PspoIIQ-lacZ cat Δfin::phleo | This study |
| AHB1985 | amyE::PspoIIQ-lacZ cat Δfin::phleo sacA::Pfin-fin kan | This study |
| AHB882 | amyE::PspoIIQ-lacZ cat ΔsigG::kan | 6 |
| AHB1954 | amyE::PspoIIQ-lacZ cat ΔsigG::kan Δfin::phleo | This study |
| AHB324 | ywrK::Tn917::amyE::PsspB-lacZ cat | 6 |
| AHB1952 | ywrK::Tn917::amyE::PsspB-lacZ cat Δfin::phleo | This study |
| AHB1984 | ywrK::Tn917::amyE::PsspB-lacZ cat Δfin::phleosacA::Pfin-fin kan | This study |
| AHB1879 | ywrK::Tn917::amyE::PsspB-lacZ cat ΔcsfB::tet | This study |
| AHB2112 | ywrK::Tn917::amyE::PsspB-lacZ cat Δfin::phleo ΔcsfB::tet | This study |
| Plasmids | ||
| pAH247 | phleo in pBluescript KS(+) | This study |
| pAH515 | amyE::Pfin-lacZ cat | This study |
| pAH537 | sacA::Pfin-fin kan | This study |
| pAH585 | amyE::Pfin-gfp-fin spc | This study |
All B. subtilis strains are isogenic with PY79.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′)a | Description |
|---|---|---|
| AH493 | cgtactgcagcagccgttatatg | Δfin LFH-PCR primer (P1) |
| AH494 | caattcgccctatagtgagtcgtgccatacaagggtcctcctgatg | Δfin LFH-PCR primer (P2) |
| AH495 | ccagcttttgttccctttagtgagtaaaaagctttggtgtagacactagacc | Δfin LFH-PCR primer (P3) |
| AH496 | gagagatacttcacgagctcctgatc | Δfin LFH-PCR primer (P4) |
| AH497 | gatcgaattcgacagtcatttacgacgaccttg | Forward primer, Pfin upstream sequence (EcoRI site) |
| AH498 | gatcaagcttcatacaagggtcctcctgatg | Reverse primer, Pfin downstream sequence including RBS and fin start codon (HindIII site) |
| AH499 | gatcggatccctctgtttcctgcgagaacag | Reverse primer, downstream of fin (BamHI site) |
| AH521 | catcaggaggacccttgtatgGCTAGCgctttgcattattattgtcg | Mutagenesis primer to insert NheI site downstream of fin ATG start codon, top strand |
| AH522 | cgacaataataatgcaaagcGCTAGCcatacaagggtcctcctgatg | Mutagenesis primer to insert NheI site downstream of fin ATG start codon, bottom strand |
| AH387 | gatcgctagcagtaaaggagaagaacttttcactggag | Forward primer, gfp upstream sequence (starting with second codon) (NheI site) |
| AH388 | gatcgctagctttgtatagttcatccatgccatgtg | Reverse primer, gfp downstream sequence (omitting stop codon) (NheI site) |
Restriction endonuclease recognition sites are underlined. Insertions in mutagenesis primers are indicated with uppercase letters.
The Δfin::phleo deletion strain (AHB1931) was generated by the long-flanking-homology PCR (LFH-PCR) method (39), using primer pairs AH493/AH494 and AH495/AH496, with the plasmid pAH247 [phleo in pBluescript KS(+); see below] as the source for the phleomycin resistance gene.
Plasmids were constructed as follows. pAH247 [phleo in pBluescript KS(+)] was constructed by subcloning a BamHI/SalI fragment harboring the phleo resistance gene from pKM080 (a gift of D. Rudner) into BamHI/SalI-digested pBluescript KS(+) (Stratagene). pAH515 (amyE::Pfin-lacZ cat) was constructed by cloning a HindIII/EcoRI PCR fragment containing the fin promoter (Pfin), ribosome-binding site (RBS), and start codon (amplified with primers AH497 and AH498) into HindIII/EcoRI-digested pAH124 (amyE::lacZ cat) (6). pAH537 (sacA::Pfin-fin kan) was generated by cloning a HindIII/BamHI PCR product harboring the fin open reading frame (ORF) and its upstream promoter sequences (amplified with primers AH497 and AH499) into HindIII/BamHI-digested pSac-Kan (sacA::kan) (26). pAH585 (amyE::Pfin-gfp-fin spc) was built in three steps. First, a HindIII/BamHI PCR product harboring the fin ORF and its upstream promoter sequences (the same as that used for construction of pAH537; amplified with primers AH497 and AH499) was ligated with HindIII/BamHI-digested pDG1731 (amyE::spc) (16), yielding pAH540. Second, pAH540 was mutagenized with primer pair AH521/AH522 to insert an in-frame NheI restriction site downstream of the fin start codon, yielding pAH581. Finally, an NheI/NheI PCR fragment containing the gfp ORF (amplified from pAC172 [12] with primers AH387 and AH388) was ligated into NheI-digested pAH581, yielding pAH585.
Microscopy.
Cells expressing the gfp-fin fusion gene were collected by brief, gentle centrifugation at hour 3.5 of sporulation and were resuspended in phosphate-buffered saline (PBS) containing 1 μg/ml of the membrane stain FM4-64 (Invitrogen). Fluorescence microscopy was performed with an Olympus BX61 microscope fitted with an Olympus UPlanF1 100× phase-contrast objective. Green fluorescent protein (GFP) fluorescence was visualized using filter set U-M41001 (excitation filter, 455 to 495 nm; dichroic mirror, 505 nm; and emission filter, 510 to 555 nm), while FM4-64 fluorescence was visualized using filter set U-MWG2 (excitation filter, 510 to 550 nm; dichroic mirror, 570 nm; and barrier filter, >590 nm). Images were captured with an Orca-R2 digital charge-coupled device (CCD) camera using Simple PCI imaging software, version 6.0 (Hamamatsu Corporation). Exposure times were typically 500 to 1,000 ms for both GFP and FM4-64. Images were false-colored, overlaid, and adjusted for brightness and contrast by use of ImageJ software (1).
To evaluate the developmental status of fin mutant and control cells, cells were collected by brief, gentle centrifugation at hour 6 of sporulation and resuspended in PBS containing 1 μg/ml FM4-64 and 10 μg/ml Mitotracker green FM (MTG; Invitrogen). The microscope setup and image acquisition/processing procedure were the same as those described above, except that the U-M41001 filter was used to image MTG fluorescence, with a typical exposure time of 200 ms. Dual labeling of cell membranes with FM4-64, which cannot permeate membranes, and MTG, which can permeate membranes, allows for identification of sporangia that have completed forespore engulfment, as previously described (31). In brief, the membranes of fully engulfed forespores stain only with MTG, while those of unengulfed forespores stain with both FM4-64 and MTG. Cells were additionally observed by phase-contrast microscopy to identify sporangia that had progressed to postengulfment stages of sporulation (cortex and coat assembly), as indicated by increased forespore refractility (i.e., “phase-bright” forespores).
Identification and alignment of Fin and CsfB orthologs.
Fin (YabK) and CsfB orthologs were identified by BLAST searches (2; http://www.ncbi.nlm.nih.gov/) with B. subtilis protein sequences. The following proteins were chosen as representative orthologs (GenBank accession numbers are given in parentheses): B. subtilis 168 CsfB (NP_387905), Bacillus licheniformis ATCC 14580 CsfB (YP_077310), Geobacillus kaustophilus HTA426 CsfB (YP_145875), Oceanobacillus iheyensis HTE831 CsfB (NP_690955), Bacillus anthracis Ames CsfB (NP_842595), Bacillus halodurans C-125 CsfB (NP_240906), B. subtilis 168 Fin (NP_387935), B. licheniformis ATCC 14580 YabK (YP_089740), G. kaustophilus HTA426 YabK (YP_145900), O. iheyensis HTE831 YabK (NP_690983), B. anthracis Ames YabK (NP_842620), and Bacillus clausii KSM-K16 YabK (YP_173586). CsfB from B. clausii and YabK from B. halodurans were not found annotated in databases; however, manual inspection of the relevant regions of their respective genomes (B. clausii KSM-K16 and B. halodurans C-125) revealed the presence of their encoding genes.
Multiple sequence alignments of CsfB and YabK/Fin orthologs were generated with ClustalW (37).
RESULTS
fin (yabK) is expressed in the forespore under the control of σF and σG.
The yabK gene, which we renamed fin, is located at an origin-proximal position on the chromosome (5.1° relative to the origin, at 0°/360°), between the genes pth (formerly named spoVC), which encodes peptidyl-tRNA hydrolase (25), and mfd, which encodes a transcription repair coupling factor (3) (Fig. 2A). The spoVT gene, which encodes a regulator of σG-directed gene expression in the forespore (5), is also in close proximity to fin on the chromosome (downstream of mfd). Interestingly, this gene organization (like fin itself [see below]) is conserved among Bacillus species and species of related genera.
FIG. 2.
fin (yabK) is a small gene that is expressed in the forespore under the control of σF and σG. (A) The fin gene (previously annotated yabK), depicted within its chromosomal context. To our knowledge, this gene synteny is conserved in all Bacillus genomes completed to date. The black bar indicates the fin-containing region that was inserted at the sacA locus to complement the mutation of fin. The drawing is to scale (as indicated), except that 2 kb of the mfd gene is not shown. (B) Sequence of the fin coding and upstream promoter regions. The putative −10 and −35 promoter sequences are boxed, with the consensus σF- and σG-recognized sequences shown above (40). Also boxed are the fin ATG start codon and TAA stop codon. The RBS for fin is underlined, while the dashed line indicates the 3′ end of the upstream pth gene, which overlaps the predicted fin promoter elements. (C) The fin promoter (Pfin) is activated by σF and σG during sporulation. The accumulation of β-galactosidase from a Pfin-lacZ reporter gene was measured during sporulation of wild-type cells (WT; ⋄), cells deleted for sigF (ΔsigF; ○), and cells deleted for sigG (ΔsigG; ▵) (strains AHB1933, AHB1956, and AHB1934, respectively). AU, arbitrary units. (D) GFP-Fin localizes diffusely throughout the forespore during early and late sporulation. Cells deleted for the native fin gene and harboring a functional gfp-fin gene fusion at the amyE locus (strain AHB2085) were observed by fluorescence microscopy at hour 3.5 of sporulation. GFP fluorescence is shown in grayscale (GFP-Fin) or false-colored green (merge). Membrane fluorescence from the dye FM4-64 is shown in grayscale (membrane) or false-colored red (merge). White arrowheads indicate a forespore at an early stage of sporulation (immediately following asymmetric division), while the white arrows indicate a forespore at a later stage of sporulation (after engulfment; note that membranes surrounding engulfed forespores are not stained due to membrane impermeability for the FM4-64 dye). Bar = 1 μm.
Close inspection of the fin upstream sequence revealed a nearly perfect match to the consensus −35 and −10 elements of promoters recognized by σF (40) (Fig. 2B). This is consistent with a study that assigned fin to the σF regulon by microarray analysis (32). We note, however, that the same elements are also excellent matches for the σG recognition consensus (40) (Fig. 2B). Another sequence immediately upstream of the fin ORF displays strong similarity to the optimal B. subtilis RBS (38) (Fig. 2B).
To investigate the expression of fin, we fused its upstream sequences, including the putative promoter (Pfin), RBS, and ATG start codon, in frame to the lacZ reporter gene. β-Galactosidase production in cells harboring this Pfin-lacZ fusion (integrated at the nonessential amyE locus) began at hour 2 of sporulation, consistent with the timing of σF activation in the forespore (Fig. 2C). Pfin-lacZ also displayed a second wave of activity at later times (starting after hour 3), most consistent with the timing of σG activity (Fig. 2C). To test the dependence of fin expression on σF and σG, we introduced deletions of their encoding genes (sigF and sigG, respectively) into the Pfin-lacZ strain. As shown in Fig. 2C, deletion of sigG mostly eliminated the late phase of Pfin expression, while deletion of sigF blocked both the early and late phases (note that σG activation requires σF; as such, sigF cells lack σF- and σG-directed gene expression). In all, these findings suggest that fin is activated in the developing forespore first by σF and later by σG. Importantly, the deletion of genes encoding other sporulation sigma factors (including σE and σK), either individually or pairwise with sigF or sigG, yielded results that were consistent with this conclusion (data not shown).
To visualize the compartmentalization of fin expression, we inserted the coding sequence of gfp in frame downstream of the ATG start codon of the full-length fin gene, leaving the upstream promoter and RBS intact. The resulting fusion gene (Pfin-gfp-fin; henceforth called gfp-fin for simplicity) was integrated at the amyE locus in a strain deleted for native fin. Importantly, gfp-fin was able to complement the fin mutant phenotypes (data not shown; see below), indicating that the encoded GFP-Fin fusion protein is functional. As shown in Fig. 2D, GFP-Fin localized diffusely within the forespore compartment of sporangia at both early and late stages of sporulation. Indeed, GFP-Fin was detectable in the forespore compartment beginning very soon after asymmetric septation (Fig. 2D, white arrowheads) and continuing through the completion of forespore engulfment (Fig. 2D, white arrows). We therefore concluded that Pfin is expressed in the forespore compartment throughout development and, moreover, that Fin appears to be a soluble, cytosolic protein.
Fin is conserved among Bacillus species and is similar to the anti-σG factor CsfB.
The fin ORF encodes a small protein of 76 amino acids with a predicted molecular mass of 8.8 kDa. A BLAST search against a nonredundant protein database revealed that Fin is encoded in most, if not all, genomes sequenced to date from Bacillus species and species of other closely related genera (2). One exception appeared to be B. halodurans; however, manual inspection of the pth-mfd intergenic region from this species revealed an unannotated ORF corresponding to fin. In contrast, our searches failed to identify fin in Listeria species, which are closely related to Bacillus but are unable to sporulate, or in the more distantly related endospore formers of the genus Clostridium.
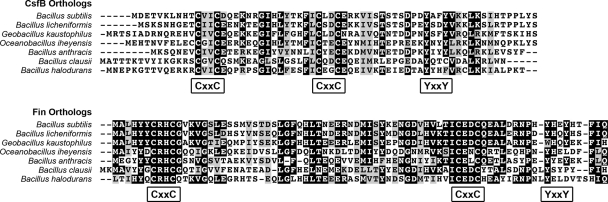
As shown in Fig. 3, the amino acid sequence of Fin is conserved throughout the length of representative orthologs (>30% identity, >50% similarity). Notably, Fin harbors two absolutely conserved Cys-X-X-Cys motifs, often found in zinc-binding proteins, that are spaced ∼40 amino acids apart (Fig. 3). As drawn to our attention by A. Henriques and P. Stragier (personal communications), this makes Fin similar to the anti-σG factor CsfB (also known as Gin) (7, 11, 19, 29). CsfB is also a small, highly conserved protein harboring two invariant Cys-X-X-Cys motifs (Fig. 3). Furthermore, the CsfB and Fin protein families both contain an imperfectly conserved Tyr-X-X-Tyr motif at their C termini (Fig. 3). Like fin, csfB is expressed in the forespore under the control of σF (13). Unlike Pfin, however, PcsfB does not appear to be activated by σG (A. H. Camp and R. Losick, unpublished results). Finally, the csfB gene is located in the same origin-proximal chromosomal region as fin (approximately 25 kb apart). In all, these similarities suggest that Fin may be evolutionarily, structurally, and/or functionally related to CsfB.
FIG. 3.
The Fin protein is conserved among Bacillus species and displays similarity to the anti-σG factor CsfB. Multiple sequence alignments are shown for CsfB orthologs (top) and Fin (YabK) orthologs (bottom) from various Bacillus and related species. The full amino acid sequence for each protein is shown. Identical or similar residues present in more than 50% of the sequences are shaded black or gray, respectively. Conserved Cys-X-X-Cys (CxxC) and Tyr-X-X-Tyr (YxxY) motifs present in Fin and CsfB ortholog families are indicated. Accession numbers for protein sequences are listed in Materials and Methods. Note that CsfB is also conserved more widely in endospore formers, including Clostridium species, and that multiple sequence alignments of the entire CsfB family have been published previously (19, 29).
fin is required for the switch from σF- to σG-directed gene expression.
Given the presence of Fin in the forespore and its similarity to the anti-σG factor CsfB (see above), we wondered whether Fin is a regulator of forespore gene expression. To test this, we first monitored σF activity in the absence of fin by using a lacZ reporter gene under the control of the σF-dependent spoIIQ promoter (PspoIIQ) (22). As shown in Fig. 4A, PspoIIQ-lacZ activity commenced at hour 2 of sporulation in both wild-type and fin cells, consistent with the timing of σF activation in the forespore. Strikingly, however, σF-directed β-galactosidase production in fin cells reached higher overall levels (approximately twice that seen in the wild type) and persisted to later times of sporulation (peaking at hour 4 versus hour 3 in the wild type) (Fig. 4A). Importantly, reintroduction of a wild-type copy of the fin gene at the sacA locus complemented the effect of the mutation, confirming that this phenotype was due to the absence of fin and not a polar effect on the adjacent, downstream gene (mfd) (Fig. 4A). Furthermore, we confirmed that the effect of fin deletion was not specific to PspoIIQ, as fin mutant cells displayed hyperactivity of other σF-dependent promoters, including PcsfB, PcsfC, and PyyaC (data not shown) (13, 32, 40).
FIG. 4.

fin is required for the efficient switch from σF to σG in the forespore. (A) σF remains active at late times during sporulation in the absence of fin. The σF-dependent activation of a PspoIIQ-lacZ reporter gene was monitored during sporulation of wild-type cells (WT; ⋄), cells deleted for fin (Δfin; ▵), and cells deleted for fin and harboring a fin complementation construct at the sacA locus (Δfin + sacA::fin; ○) (strains AHB881, AHB1953, and AHB1985, respectively). (B) Fin and σG additively repress σF at late times during sporulation. σF-Directed β-galactosidase production from a PspoIIQ-lacZ reporter gene was measured during sporulation of wild-type cells (WT; ⋄), cells deleted for fin (Δfin; ▵), cells deleted for sigG (ΔsigG; ⧫), and cells simultaneously deleted for fin and sigG (ΔsigG Δfin; ▴) (strains AHB881, AHB1953, AHB882, and AHB1954, respectively). (C) Fin is required for full σG activation. The σG-dependent activation of a PsspB-lacZ reporter gene was monitored during sporulation of wild-type cells (WT; ⋄), cells deleted for fin (Δfin; ▵), and cells deleted for fin and harboring a fin complementation construct at the sacA locus (Δfin + sacA::fin; ○) (strains AHB324, AHB1952, and AHB1984, respectively). (D) Deletion of csfB partially restores σG activity to fin mutant cells. PsspB-lacZ reporter gene activation by σG was monitored during sporulation of wild-type cells (WT; ⋄), cells deleted for fin (Δfin; ▵), cells deleted for csfB (ΔcsfB; ⧫), and cells simultaneously deleted for fin and csfB (Δfin ΔcsfB; ▴) (strains AHB881, AHB1953, AHB1879, and AHB2112, respectively).
Next, we tested the effect of a fin mutation on σG activity by using a lacZ reporter gene under the control of the sspB promoter (PsspB), which is recognized exclusively by σG (35). Remarkably, σG-directed β-galactosidase production from the PsspB-lacZ reporter was significantly decreased in fin mutant cells (≤25% of wild-type levels by hour 6) (Fig. 4C). As described above, we determined that σG activity was restored to wild-type levels when fin was reintroduced (Fig. 4C). In all, we concluded that Fin facilitates the switch from σF to σG: σF does not shut off and σG does not turn on fully in the absence of fin.
We and others previously reported that deletion of sigG causes overexpression of σF target genes (including spoIIQ) at late times during sporulation, suggesting that σG directly or indirectly inhibits σF (4, 6, 13). This raised the possibility that Fin functions primarily to promote σG activity and that the σF hyperactivity phenotype of cells mutant for fin is caused secondarily, by reduced σG activity. If this were true, then fin should have no effect on σF activity in the absence of sigG (i.e., the effect of a sigG mutation should be epistatic to a fin mutation). However, as shown in Fig. 4B, we found that σF activity was higher in sigG fin mutant cells than in either single mutant alone, indicating that Fin does not influence σF activity indirectly through σG. As such, the simplest interpretation of the results is that Fin is an anti-σF factor which, by antagonizing σF, facilitates the switch to σG. It is on the basis of this phenotype that we renamed the yabK gene fin, for σF inhibitor.
Finally, these results indicate that increased σF activity in the absence of fin interferes with subsequent σG activation. One explanation could be that hyperactive σF directs increased transcription of the gene for the anti-σG factor CsfB (13). Indeed, the csfB promoter (PcsfB), like PspoIIQ, was more active in fin cells (data not shown). To test whether CsfB contributes to the block in σG activation in fin cells, we measured σG activation in cells lacking fin and csfB. Consistent with previous reports (7, 11), deletion of csfB alone did not appreciably affect the timing or level of activation of the σG-dependent reporter PsspB-lacZ (Fig. 4D). However, we found that elimination of csfB significantly increased σG activation in fin mutant cells, albeit not to wild-type levels (>3-fold increase over the level in the fin single mutant) (Fig. 4D). In all, these findings are consistent with a model in which sustained σF activity interferes with σG activation, in part due to increased production of the anti-σG factor CsfB.
fin mutant cells display a sporulation phenotype and are arrested after engulfment.
Our finding that Fin is required for the efficient switch from σF to σG in the forespore immediately raised the question of whether the fin mutant is defective for spore formation. To test this, we determined the number of heat-resistant spores present after 24 h of sporulation in wild-type and fin cells. Strikingly, we found that the fin mutant displayed a 50-fold reduction in spore formation relative to the wild type (Fig. 5). (We note, however, that fin cells still produced a substantial number of heat-resistant spores [∼1 × 107 spores per ml, compared to ∼5 × 108 spores per ml for the wild type].) Sporulation was restored to almost wild-type levels by the reintroduction of fin at the sacA locus (sporulation at ∼60% of wild-type efficiency) (Fig. 5). We presume that the incomplete complementation in this sensitive assay was due to subtle differences in fin expression from an ectopic locus. Finally, we found that deletion of csfB partially rescued spore formation by fin cells (approximately 6-fold) (data not shown), consistent with the partial rescue of σG activation observed in fin csfB cells (see above and Fig. 4D).
FIG. 5.
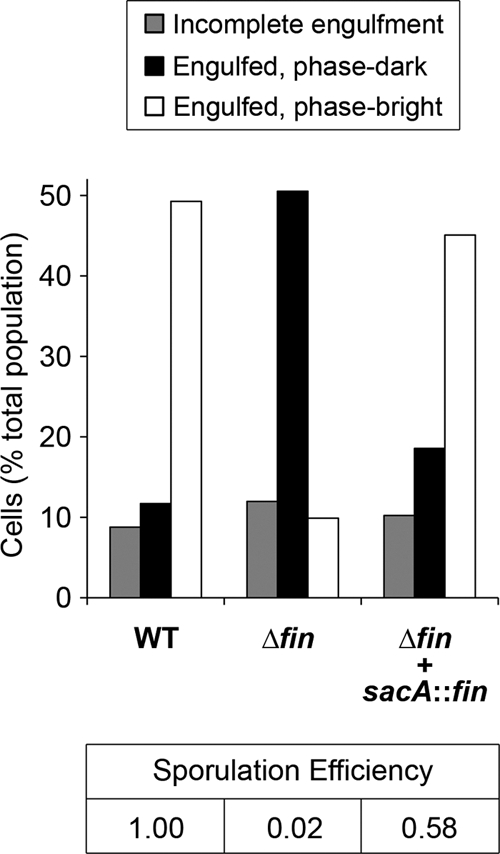
Cells lacking fin are defective for spore formation and are arrested after engulfment. The status of sporulation at hour 6 of sporulation was determined for wild-type cells (WT), cells deleted for fin (Δfin), and cells deleted for fin and harboring a fin complementation construct at the sacA locus (Δfin + sacA::fin) (strains PY79, AHB1931, and AHB1983, respectively), using a combination of fluorescence and phase-contrast microscopy (see Materials and Methods) (n > 190 for each sample). The remaining, nonsporulating ∼25 to 30% of cells from each population are not shown. Also indicated (bottom) are the sporulation efficiencies for the indicated strains. Sporulation efficiency was calculated as the number of heat-resistant spores present after 24 h of growth/sporulation in DSM normalized to the number present in the wild-type sample.
Next, we subjected fin cells to a fluorescence and phase-contrast microscopy-based assay to determine the stage of sporulation at which they are arrested (see Materials and Methods) (31). We found that cells lacking fin entered the sporulation pathway (as indicated by asymmetric septation) and progressed through engulfment in a manner that was nearly indistinguishable from that of the wild type (data not shown). However, we observed a significant defect in the ability of fin cells to subsequently form phase-bright forespores, which is typically indicative of the late, postengulfment events of forespore cortex and coat assembly. As shown in Fig. 5, by hour 6 nearly 50% of wild-type sporangia harbored engulfed, phase-bright forespores. At the same time, however, only 10% of fin sporangia had reached the same morphological stage. Instead, the majority of fin sporangia (51%) harbored engulfed, phase-dark forespores (Fig. 5). Importantly, we found that reintroduction of fin at the sacA locus restored the ability of the fin mutant to progress to later stages of sporulation, similar to the wild type (Fig. 5). In all, these results suggest that fin cells proceed slowly, if at all, past the engulfment stage (III) of sporulation, an interpretation that is consistent with a defect in σG activation.
DISCUSSION
To date, only limited progress has been made in unraveling the mechanisms that mediate the switch from one sigma factor to another during spore formation. Here we discovered a critical role for a small, conserved protein that we named Fin in the transition from early to late gene expression in the forespore. The fin gene is expressed in the forespore throughout development, under σF and σG control. In the absence of fin, the early-acting σF protein fails to shut off appropriately and, conversely, the late-acting σG protein fails to become activated fully. Concomitantly, fin cells are defective for sporulation, producing 50-fold fewer spores than the wild type. During sporulation, fin cells are arrested in large numbers following the engulfment stage (III), consistent with a defect in activation of σG, which is required for postengulfment events, including cortex and coat assembly.
We note that yabK (fin) was previously assigned a role in DNA repair, given that null mutations of the gene were observed to act as suppressors of the recombination defect of certain recombination genes (8, 9). On this basis, yabK was named subA, for suppressor of recU and recB (9). We think it improbable that yabK (fin) plays a role in DNA repair, in light of our evidence that it is expressed during sporulation, its orthologs are present only in endospore-forming species closely related to Bacillus, and it likely functions to inhibit σF (see below). Instead, and given that the previous authors failed to show that the phenotypes attributed to the yabK mutation could be reversed by complementation, it seems likely that the observed suppression was due to an effect on the expression of the downstream gene mfd, which encodes a transcription repair coupling factor (3).
How does Fin participate in the switch from σF to σG? One possibility is that Fin helps to promote σG activation such that it can surmount σF activity in the forespore. In this scenario, deletion of fin causes primarily reduced σG activity, which in turn permits sustained σF activity, as has been observed previously (6). However, this is not supported by our genetic analysis. If Fin were driving the σF-to-σG switch through activation of σG, then fin should not influence σF activity in the absence of σG. In contrast, we found that a fin sigG mutant displayed even more σF activity than the sigG single mutant. As such, we instead favor an alternative explanation in which Fin facilitates the σF-to-σG switch by inhibiting σF. In this model, deletion of fin causes primarily derepression of σF, which in turn interferes with σG activation.
As shown in Fig. 1B, we propose the following comprehensive model for the σF-to-σG transition. To begin, σF activates transcription of the gene (sigG) for σG (arrow 1) (36). However, activation of σG is delayed through poorly understood mechanisms that may include weak transcription of sigG by σF (Camp and Losick, unpublished data), inhibition by the σF-activated anti-σG factor CsfB (13, 19), and/or competition between σF and σG for RNAP (although the latter idea is controversial [10]) (barred line 2). To trigger the switch to σG, σF also turns on the gene encoding its own inhibitor, Fin (arrow 3). Once σF is sufficiently inactivated by Fin (barred line 4), its successor, σG, can become active. (The importance of σF inhibition for σG activation is considered in more detail below.) The transition to σG is then reinforced by two mechanisms. First, σG continues to direct fin synthesis, resulting in sustained σF inhibition (arrow 5). Second, σG autoregulates its own gene, leading to large amounts of the late sigma factor (arrow 6). Finally, we note that Fin cannot account for all observed σF inhibition, given that a fin sigG double mutant displayed more σF derepression than a fin mutant alone. As such, σG must additionally inhibit σF by an unknown, Fin-independent pathway (barred line 7).
A noteworthy feature of this model is that σG is crippled in the presence of sustained σF activity, as is the case for fin cells. This suggests that σF either directly or indirectly inhibits σG (indicated by barred line 2 in Fig. 1B). We speculate that this inhibition is mediated by some of the same mechanisms that ordinarily delay σG activation, including σF-dependent production of the σG inhibitor CsfB and competition between σF and σG for RNAP (see above). Consistent with the former, we found that deleting csfB partially rescued the σG activation and sporulation defects of fin mutant cells. In addition to competing for RNAP, the σF and σG regulons may also compete for the raw materials required for transcription and translation, such as nucleotides and amino acids. The latter idea is especially intriguing given our recent finding that the forespore loses its self-sufficiency to support macromolecular synthesis at around the time of the switch from σF to σG (6). Our “feeding tube” model posits that the mother cell restores the metabolic potential of the forespore at this time by providing critical small molecules through a novel channel apparatus connecting the two cells (7, 14, 24). It is possible, however, that resources may still be limiting in the forespore even in the presence of the feeding tube.
How might Fin inhibit σF? One possible clue comes from the similarity of Fin to the anti-σG factor CsfB. CsfB is a potent inhibitor of σG and is likely to accomplish this by binding to the sigma factor (19, 29), although a direct biochemical interaction between CsfB and σG has not yet been demonstrated. CsfB was proposed to be the key target of an intercellular signaling pathway controlling σG activity in the forespore (19), but other work has convincingly indicated that the anti-σG factor instead plays an auxiliary role in preventing premature σG activation (see above) (7, 11). Like Fin, CsfB is a small protein that harbors two absolutely conserved Cys-X-X-Cys motifs and a less-conserved Tyr-X-X-Tyr motif (19, 29). Importantly, in the case of CsfB, these motifs are critical for σG inhibition and binding to σG in a yeast two-hybrid assay (29). It is tempting to speculate, therefore, that Fin binds to and inhibits σF analogously to the case for CsfB and σG. However, efforts to demonstrate Fin-dependent inhibition of σF activity in vegetative cells engineered to produce both proteins have so far been unsuccessful (Camp and Losick, unpublished results). A similar experiment successfully demonstrated CsfB-mediated inhibition of σG (19, 29). Conceivably, Fin may require one or more factors or conditions present only during sporulation to interact with and inhibit σF. We also cannot exclude a model in which Fin does not bind to σF but instead inhibits it indirectly.
In conclusion, our identification and characterization of fin represent significant steps forward in our understanding of the switch from σF to σG during B. subtilis spore formation. Moreover, the sporulation defect associated with fin mutation underscores the importance of properly regulated transitions in developmental gene expression during sigma factor cascades, as predicted decades ago (23, 34), and raises the possibility that the full list of sporulation genes is not yet complete.
Acknowledgments
We thank A. Henriques and P. Stragier for drawing our attention to YabK and its similarity to CsfB. We also thank members of the Losick laboratory for helpful discussions.
This work was supported in part by a Helen Hay Whitney Foundation postdoctoral research fellowship to A.H.C., a National Science Foundation graduate research fellowship to A.F.W., and a National Institutes of Health grant (GM18568) to R.L.
Footnotes
Published ahead of print on 29 October 2010.
REFERENCES
- 1.Abramoff, M. D., P. J. Magelhaes, and S. J. Ram. 2004. Image processing with ImageJ. Biophotonics Int. 11:36-42. [Google Scholar]
- 2.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 3.Ayora, S., F. Rojo, N. Ogasawara, S. Nakai, and J. C. Alonso. 1996. The Mfd protein of Bacillus subtilis 168 is involved in both transcription-coupled DNA repair and DNA recombination. J. Mol. Biol. 256:301-318. [DOI] [PubMed] [Google Scholar]
- 4.Bagyan, I., L. Casillas-Martinez, and P. Setlow. 1998. The katX gene, which codes for the catalase in spores of Bacillus subtilis, is a forespore-specific gene controlled by σF, and KatX is essential for hydrogen peroxide resistance of the germinating spore. J. Bacteriol. 180:2057-2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bagyan, I., J. Hobot, and S. Cutting. 1996. A compartmentalized regulator of developmental gene expression in Bacillus subtilis. J. Bacteriol. 178:4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Camp, A. H., and R. Losick. 2009. A feeding tube model for activation of a cell-specific transcription factor during sporulation in Bacillus subtilis. Genes Dev. 23:1014-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camp, A. H., and R. Losick. 2008. A novel pathway of intercellular signalling in Bacillus subtilis involves a protein with similarity to a component of type III secretion channels. Mol. Microbiol. 69:402-417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carrasco, B., M. C. Cozar, R. Lurz, J. C. Alonso, and S. Ayora. 2004. Genetic recombination in Bacillus subtilis 168: contribution of Holliday junction processing functions in chromosome segregation. J. Bacteriol. 186:5557-5566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carrasco, B., S. Fernández, K. Asai, N. Ogasawara, and J. C. Alonso. 2002. Effect of the recU suppressors sms and subA on DNA repair and homologous recombination in Bacillus subtilis. Mol. Genet. Genomics 266:899-906. [DOI] [PubMed] [Google Scholar]
- 10.Chary, V. K., P. Xenopoulos, and P. J. Piggot. 2006. Blocking chromosome translocation during sporulation of Bacillus subtilis can result in prespore-specific activation of σG that is independent of σE and of engulfment. J. Bacteriol. 188:7267-7273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chary, V. K., P. Xenopoulos, and P. J. Piggot. 2007. Expression of the σF-directed csfB locus prevents premature appearance of σG activity during sporulation of Bacillus subtilis. J. Bacteriol. 189:8754-8757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chastanet, A., and R. Losick. 2007. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol. Microbiol. 64:139-152. [DOI] [PubMed] [Google Scholar]
- 13.Decatur, A., and R. Losick. 1996. Identification of additional genes under the control of the transcription factor σF of Bacillus subtilis. J. Bacteriol. 178:5039-5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doan, T., C. Morlot, J. Meisner, M. Serrano, A. O. Henriques, C. P. Moran, and D. Z. Rudner. 2009. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 5:e1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eichenberger, P., S. T. Jensen, E. M. Conlon, C. van Ooij, J. Silvaggi, J. E. Gonzalez-Pastor, M. Fujita, S. Ben-Yehuda, P. Stragier, J. S. Liu, and R. Losick. 2003. The σE regulon and the identification of additional sporulation genes in Bacillus subtilis. J. Mol. Biol. 327:945-972. [DOI] [PubMed] [Google Scholar]
- 16.Guerout-Fleury, A. M., N. Frandsen, and P. Stragier. 1996. Plasmids for ectopic integration in Bacillus subtilis. Gene 180:57-61. [DOI] [PubMed] [Google Scholar]
- 17.Hilbert, D. W., and P. J. Piggot. 2004. Compartmentalization of gene expression during Bacillus subtilis spore formation. Microbiol. Mol. Biol. Rev. 68:234-262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ju, J., T. Mitchell, H. Peters, and W. G. Haldenwang. 1999. Sigma factor displacement from RNA polymerase during Bacillus subtilis sporulation. J. Bacteriol. 181:4969-4977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Karmazyn-Campelli, C., L. Rhayat, R. Carballido-Lopez, S. Duperrier, N. Frandsen, and P. Stragier. 2008. How the early sporulation sigma factor σF delays the switch to late development in Bacillus subtilis. Mol. Microbiol. 67:1169-1180. [DOI] [PubMed] [Google Scholar]
- 20.Kunkel, B., L. Kroos, H. Poth, P. Youngman, and R. Losick. 1989. Temporal and spatial control of the mother-cell regulatory gene spoIIID of Bacillus subtilis. Genes Dev. 3:1735-1744. [DOI] [PubMed] [Google Scholar]
- 21.Li, Z., and P. J. Piggot. 2001. Development of a two-part transcription probe to determine the completeness of temporal and spatial compartmentalization of gene expression during bacterial development. Proc. Natl. Acad. Sci. U. S. A. 98:12538-12543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Londoño-Vallejo, J. A., C. Frehel, and P. Stragier. 1997. SpoIIQ, a forespore-expressed gene required for engulfment in Bacillus subtilis. Mol. Microbiol. 24:29-39. [DOI] [PubMed] [Google Scholar]
- 23.Losick, R., and J. Pero. 1981. Cascades of sigma factors. Cell 25:582-584. [DOI] [PubMed] [Google Scholar]
- 24.Meisner, J., X. Wang, M. Serrano, A. O. Henriques, and C. P. Moran, Jr. 2008. A channel connecting the mother cell and forespore during bacterial endospore formation. Proc. Natl. Acad. Sci. U. S. A. 105:15100-15105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menez, J., R. H. Buckingham, M. de Zamaroczy, and C. K. Campelli. 2002. Peptidyl-tRNA hydrolase in Bacillus subtilis, encoded by spoVC, is essential to vegetative growth, whereas the homologous enzyme in Saccharomyces cerevisiae is dispensable. Mol. Microbiol. 45:123-129. [DOI] [PubMed] [Google Scholar]
- 26.Middleton, R., and A. Hofmeister. 2004. New shuttle vectors for ectopic insertion of genes into Bacillus subtilis. Plasmid 51:238-245. [DOI] [PubMed] [Google Scholar]
- 27.Nicholson, W. L., and P. Setlow. 1990. Sporulation, germination, and outgrowth, p. 391-450. In C. R. Harwood and S. M. Cutting (ed.), Molecular biological methods for Bacillus. John Wiley & Sons, New York, NY.
- 28.Piggot, P. J., and D. W. Hilbert. 2004. Sporulation of Bacillus subtilis. Curr. Opin. Microbiol. 7:579-586. [DOI] [PubMed] [Google Scholar]
- 29.Rhayat, L., S. Duperrier, R. Carballido-Lopez, O. Pellegrini, and P. Stragier. 2009. Genetic dissection of an inhibitor of the sporulation sigma factor σG. J. Mol. Biol. 390:835-844. [DOI] [PubMed] [Google Scholar]
- 30.Schaeffer, P., J. Millet, and J. P. Aubert. 1965. Catabolic repression of bacterial sporulation. Proc. Natl. Acad. Sci. U. S. A. 54:704-711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sharp, M. D., and K. Pogliano. 1999. An in vivo membrane fusion assay implicates SpoIIIE in the final stages of engulfment during Bacillus subtilis sporulation. Proc. Natl. Acad. Sci. U. S. A. 96:14553-14558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steil, L., M. Serrano, A. O. Henriques, and U. Völker. 2005. Genome-wide analysis of temporally regulated and compartment-specific gene expression in sporulating cells of Bacillus subtilis. Microbiology 151:399-420. [DOI] [PubMed] [Google Scholar]
- 33.Sterlini, J. M., and J. Mandelstam. 1969. Commitment to sporulation in Bacillus subtilis and its relationship to development of actinomycin resistance. Biochem. J. 113:29-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stragier, P., and R. Losick. 1990. Cascades of sigma factors revisited. Mol. Microbiol. 4:1801-1806. [DOI] [PubMed] [Google Scholar]
- 35.Sun, D., P. Fajardo-Cavazos, M. D. Sussman, F. Tovar-Rojo, R. M. Cabrera-Martinez, and P. Setlow. 1991. Effect of chromosome location of Bacillus subtilis forespore genes on their spo gene dependence and transcription by σF: identification of features of good σF-dependent promoters. J. Bacteriol. 173:7867-7874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun, D. X., R. M. Cabrera-Martinez, and P. Setlow. 1991. Control of transcription of the Bacillus subtilis spoIIIG gene, which codes for the forespore-specific transcription factor σG. J. Bacteriol. 173:2977-2984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vellanoweth, R. L., and J. C. Rabinowitz. 1992. The influence of ribosome-binding-site elements on translational efficiency in Bacillus subtilis and Escherichia coli in vivo. Mol. Microbiol. 6:1105-1114. [DOI] [PubMed] [Google Scholar]
- 39.Wach, A. 1996. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast 12:259-265. [DOI] [PubMed] [Google Scholar]
- 40.Wang, S. T., B. Setlow, E. M. Conlon, J. L. Lyon, D. Imamura, T. Sato, P. Setlow, R. Losick, and P. Eichenberger. 2006. The forespore line of gene expression in Bacillus subtilis. J. Mol. Biol. 358:16-37. [DOI] [PubMed] [Google Scholar]
- 41.Wilson, G. A., and K. F. Bott. 1968. Nutritional factors influencing the development of competence in the Bacillus subtilis transformation system. J. Bacteriol. 95:1439-1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Youngman, P., J. B. Perkins, and R. Losick. 1984. Construction of a cloning site near one end of Tn917 into which foreign DNA may be inserted without affecting transposition in Bacillus subtilis or expression of the transposon-borne erm gene. Plasmid 12:1-9. [DOI] [PubMed] [Google Scholar]
- 43.Zhang, B., and L. Kroos. 1997. A feedback loop regulates the switch from one sigma factor to the next in the cascade controlling Bacillus subtilis mother cell gene expression. J. Bacteriol. 179:6138-6144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang, B., P. Struffi, and L. Kroos. 1999. σK can negatively regulate sigE expression by two different mechanisms during sporulation of Bacillus subtilis. J. Bacteriol. 181:4081-4088. [DOI] [PMC free article] [PubMed] [Google Scholar]