Abstract
Porphyromonas gingivalis, a periodontal pathogen, expresses a group of surface proteins with a common C-terminal domain (CTD) that are exported by a novel secretion system to the surface, where they are covalently attached. Using RgpB as a model CTD protein, we have produced a series of site-directed mutations in the CTD sequence at conserved residues and at residues that may be modified and, hence, surface attached. The mutant RgpB proteins were expressed in a P. gingivalis host lacking functional RgpB and RgpA Arg-specific proteases. The RgpB mutants produced were Y674F, Y674F Y718F, T675Q S679Q T682Q T684Q, T693Q, F695A, D696A, N698A, G699P, G716P, T724Q, T728Q T730Q, and K732Q and a protein with a deletion of residues 692 to 702 (Δ692-702). The mutants were characterized for cell-associated Arg-specific protease activity and for cellular distribution using anti-Rgp antibodies and Western blotting of culture fractions. All the mutants exhibited cell-associated Arg-specific activity similar to that of the positive control except for the D696A and Δ692-702 mutants. For all mutants, except D696A and Δ692-702, the RgpB proteins were found modified and attached to the cell surface, which was the same profile found in the positive-control strain. Only trace amounts of the precursor form of the Δ692-702 mutant were detected in the outer membrane, with none detected in the periplasm or culture fluid although cell transcript levels were normal. The results suggest that residues 692 to 702 of the CTD, in particular, residue D696, have an important role in the attachment of RgpB at the cell surface and that without attachment secretion does not occur.
Porphyromonas gingivalis is an anaerobic bacterium found in subgingival dental plaque and has been implicated as a major pathogen in the initiation and progression of chronic periodontitis (3). There is evidence to suggest that individuals with periodontitis may be more susceptible to cardiovascular diseases (11), and the disease in pregnant women has been linked to preterm birth and low birth weight of their infants (8, 21). The cysteine proteinases, called gingipains, are major virulence factors of P. gingivalis and include the Arg-gingipains (RgpA and RgpB derived from the genes rgpA and rgpB, respectively) which are specific for hydrolysis of arginyl peptide bonds, and Lys-gingipain (Kgp derived from kgp), which is specific for hydrolysis of lysyl peptide bonds (5, 36). Two protein forms are derived from rgpB: a soluble discrete enzyme RgpB and a cell surface-attached form which represents a posttranslationally modified form of the protein. The surface-attached form of RgpB contains heterogeneously modified isoforms that migrate as a diffuse band of 70 to 90 kDa on SDS-PAGE (20, 40) and has been suggested to contain up to 30% carbohydrate (4, 5). In contrast, the soluble RgpB that has a truncated C-terminal domain (CTD) migrates as a discrete band of 50 kDa (10).
The outer membrane proteome of P. gingivalis has been characterized (35, 40), and a common C-terminal domain of approximately 70 to 80 amino acyl residues in length was identified in 34 P. gingivalis proteins (35). These CTD sequences were aligned to reveal a consensus sequence that can be divided into five regions (A to E) (Fig. 1) (35) despite little other sequence similarity. These proteins were described as the C-terminal domain sequence-related family (CTD family). Alignment of the CTD of these P. gingivalis proteins with the RgpB CTD revealed that there are several well-conserved amino acid residues, with just two fully conserved residues (equivalent to G699 and G716 of RgpB) (Fig. 1) (35). Genetic manipulation of RgpB to remove the CTD resulted in accumulation of the precursor form within the periplasmic space and loss of posttranslational modification, surface attachment, and function of the enzyme (35), suggesting that the CTD is important for RgpB export through the outer membrane and that modification was associated with functional protein export.
FIG. 1.
Alignment of the C termini of experimentally identified P. gingivalis proteins with homology to the RgpB C terminus. The sequences are described using TIGR annotation, with PG0506 representing RgpB. The CTD is divided into five regions designated with the letters A to E above the alignment. The black shading indicates identical residues (>40% identity), light gray shading indicates nonpolar substitutions (A, V, F, P, M, I, L, W, and Y), and dark gray shading indicates polar (S, T, Y, H, C, N, G, and Q), acidic (D and E), or basic (R and K) substitutions. Residues that do not have >40% identity but have >70% conservative substitutions are also shaded.
More recently, it has been demonstrated that truncation of the last two RgpB residues (VK) from the C terminus was sufficient to prevent transit of the RgpB precursor form out of the periplasm (19). Notably, however, VK is not a conserved residue pair at the C termini of many other CTD family proteins, occurring only in the CTD of the closely related HagA and Kgp (Fig. 1). The recombinant RgpB precursor polypeptide lacking VK was inactive and lacked the posttranslational modification seen in wild-type surface-attached RgpB (19). Again, these results show that modification is associated with functional export of RgpB.
Monoclonal antibody (MAb) 1B5 has been shown to recognize modified (surface-attached) RgpB but not the C-terminally truncated soluble form of the protein found in the culture supernatant, which lacks the CTD (5, 6). Recently, Paramonov et al. (25) identified the MAb 1B5 epitope as a phosphorylated branched mannan (Manα1-2Manα1-phosphate), which is also a component of a novel anionic lipopolysaccharide (A-LPS) of P. gingivalis. The basic structure of LPS is lipid A covalently linked to a core oligosaccharide, which is further linked to a polysaccharide comprising repeating units. Two different polysaccharides have been characterized in P. gingivalis LPS, designated O-LPS and A-LPS. O-LPS contains O-polysaccharide (O-antigen) tetrasaccharide repeating units (24, 26), whereas A-LPS contains anionic polysaccharide (APS) repeating units of phosphorylated branched mannan (25, 29). The epitope recognized by MAb 1B5 was localized to the Manα1-2Manα1-phosphate side chain in APS and is not present in O-LPS or capsule polysaccharide (25).
The increase in the molecular weight (MW) and diffuse nature of the modified RgpB protein on SDS-PAGE gels is consistent with a glycolipid modification such as LPS. This, together with MAb 1B5 recognizing A-LPS, has led to the suggestion that the CTD of RgpB is essential not only for outer membrane export but also for covalent attachment to the cell surface A-LPS anchor (27). Interestingly, MAb 1B5 also binds to other CTD-containing proteins, namely, RgpAA4 (one of four related RgpA adhesins), P59 (PG2102), and P27 (PG1795), indicating that these proteins also have Manα1-2Manα1-phosphate-associated modifications (40). Hence, based on this reactivity with MAb 1B5, it has been proposed that the CTD proteins are covalently attached to A-LPS on the cell surface through a modified CTD (27). Peptide mass fingerprinting (PMF) analyses of the mature forms of these proteins has not identified any peptide from the CTDs (35, 40), which may be because the modification blocks identification and/or because the CTD, upon secretion, is cleaved either when or before the protein is covalently attached to A-LPS. The CTD is also present in proteins from a range of bacteria in the Bacteroidetes phylum, and the proteins that have been characterized are also attached to the cell surface (9, 39, 41). These results suggest that the CTD is a component of a unique protein secretion and cell surface attachment system in a range of pathogenic bacteria.
To further investigate the function of specific residues of the CTD in protein export and surface attachment through potential sites of O- and N-linked modification, we have created a series of site-directed mutants and a motif B (Fig. 1) deletion mutant within the RgpB CTD and investigated the cellular distribution of these recombinant RgpB (rRgpB) mutant proteins. We show that although substitution of most residues had no effect on rRgpB activity, a D696A mutation and a motif B (residues 692 to 702) deletion mutation abolished secretion and surface attachment of the protein.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
Bacterial strains and plasmids used in this study are listed in Table 1. P. gingivalis strains were grown as described previously (35). P. gingivalis allele exchange mutants were selected on horse blood agar (HBA; Oxoid, Basingstoke, United Kingdom) plates in the presence of 10 μg ml−1 erythromycin and 10 μg ml−1 chloramphenicol. Subsequently, brain heart infusion (BHI; Oxoid) broth cultures were supplemented with 5 μg ml−1 erythromycin and 5 μg ml−1 chloramphenicol. Escherichia coli strains were grown aerobically at 37°C on Luria-Bertani (LB) agar or in LB broth supplemented with 100 μg ml−1 ampicillin.
TABLE 1.
Bacterial strains and plasmids
| Plasmid or strain | Descriptiona | Reference or source |
|---|---|---|
| Plasmids | ||
| pAL30 | ermF ligated into pGem-TEasy | 7 |
| pBH1.1 | pGem-TEasy::rgpB (nt 493-3166) | 20 |
| pUCtetQ | pUC18::tetQ | This study |
| pJY674F | pBH1.1-rgpB(Y674F) | This study |
| pJY674Y718F | pBH1.1-rgpB(Y674F Y718F) | This study |
| pJD696A | pBH1.1-rgpB(D696A) | This study |
| pKTG699P | pBH1.1-rgpB(G699P) | This study |
| pKTG716P | pBH1.1-rgpB(G716P) | This study |
| pKTK732Q | pBH1.1-rgpB(K732A) | This study |
| pKTT728QT730Q | pBH1.1-rgpB(T728Q T730Q) | This study |
| pKTTSTT | pBH1.1-rgpB(T675Q S679Q T682Q T684Q) | This study |
| pKTT724Q | pBH1.1-rgpB(T724Q) | This study |
| pKTERM1 | pUCtetQ::ermF | This study |
| pKTG6 | pKTERM1::rgpB(G699P) | This study |
| pKTG7 | pKTERM1::rgpB(G716P) | This study |
| pKTK1 | pKTERM1::rgpB(K732Q) | This study |
| pKTTT1 | pKTERM1::rgpB(T728Q T730Q) | This study |
| pKTB1 | pKTERM1::rgpB(nt 493-3166) | This study |
| pKTY1 | pKTERM1::rgpB(Y674F) | This study |
| pKTD1 | pKTERM1::rgpB(D696A) | This study |
| pKTYY | pKTERM1::rgpB(Y674F Y718F) | This study |
| pKTTSTT | pKTERM1::rgpB(T675Q S679Q T682Q T684Q) | This study |
| pKTT | pKTERM1::rgpB(T724Q) | This study |
| pCM1 | pGem-TEasy::rgpB(nt 493-2961)-SfoI | This study |
| pCM2 | pCM1::rgpB (nt 3015-3166) | This study |
| pCM3 | pKTERM::rgpBΔ(692-702) | This study |
| pCMT693Q | pKTERM1::rgpB(T693Q) | This study |
| pCMF695A | pKTERM1::rgpB(F695A) | This study |
| pCMN698A | pKTERM1::rgpB(N698A) | This study |
| pAL34 | 934-bp 5-prime fragment of pg0553 ligated between ApaI and AatII sites of pAL30 | This study |
| pAL34.1 | 1,122-bp 3-prime fragment of pg0553 ligated between SpeI and SalI sites of pAL34 | This study |
| P. gingivalis strains | ||
| W50AB | W50 rgpA::catrgpB::tetQ; Cmr Tcr | 40 |
| W50AB-Pos | W50 rgpA::catrgpB::tetQ::rgpB; Cmr Emr | This study |
| W50AB-Neg | W50 rgpA::catrgpB::tetQ::ermF; Cmr Emr | This study |
| W50AB-G699P | W50 rgpA::catrgpB::tetQ::ermF-rgpB(G699P); Cmr Emr | This study |
| W50AB-G716P | W50 rgpA::catrgpB::tetQ::ermF-rgpB(G716P); Cmr Emr | This study |
| W50AB-K732Q | W50 rgpA::catrgpB::tetQ::ermF-rgpB(K732P); Cmr Emr | This study |
| W50AB-T728T730Q | W50 rgpA::catrgpB::tetQ::ermF-rgpB(T728QT730Q); Cmr Emr | This study |
| W50AB-D696A | W50 rgpA::catrgpB::tetQ::ermF-rgpB(D696A); Cmr Emr | This study |
| W50AB-Y674F | W50 rgpA::catrgpB::tetQ::ermF-rgpB(Y674F); Cmr Emr | This study |
| W50AB-Y674Y718F | W50 rgpA::catrgpB::tetQ::ermF-rgpB(Y674F Y718F); Cmr Emr | This study |
| W50AB-T724Q | W50 rgpA::catrgpB::tetQ::ermF-rgpB(T724Q); Cmr Emr | This study |
| W50AB-TSTT | W50 rgpA::catrgpB::tetQ::ermF-rgpB(T675Q S679Q T682Q T684Q); Cmr Emr | This study |
| W50AB-T693Q | W50 rgpA::catrgpB::tetQ::ermF-rgpB(T693Q); Cmr Emr | This study |
| W50AB-F695A | W50 rgpA::catrgpB::tetQ::ermF-rgpB(F695A); Cmr Emr | This study |
| W50AB-N698A | W50 rgpA::catrgpB::tetQ::ermF-rgpB(N698A); Cmr Emr | This study |
| W50AB-Δ692-702 | W50 rgpA::catrgpB::tetQ::ermF-rgpBΔ(692-702); Cmr Emr | This study |
| ECR179 | E. coli JM109, pAL34 | This study |
| ECR179.1 | E. coli JM109, pAL34.1 | This study |
| ECR190 | P. gingivalis W50 pg0553::ermF | This study |
Tcr, tetracycline resistance; Cmr, chloramphenicol resistance; Emr, erythromycin resistance. Nucleotides (nt) are numbered according to GenBank accession number AF007124.
Electroporation of P. gingivalis.
Electroporation-competent P. gingivalis cells were prepared as follows. An overnight-grown starter culture (1.5 ml) of P. gingivalis was inoculated into 200 ml of BHI broth and incubated overnight to an optical density at 650 nm (OD650) of 0.3 to 0.7. Cells were then centrifuged (at 8,000 × g for 20 min at 4°C), and the pellet was washed in 200 ml of ice-cold electroporation buffer (EP buffer; 10% [vol/vol] glycerol, 1 mM MgCl2). The cells were then centrifuged (at 8,000 × g for 20 min at 4°C), and the pellet was suspended in 400 μl of ice-cold EP buffer. Cells (80 μl) were aliquoted to cold microcentrifuge tubes, and 300 to 800 ng of linearized plasmid was added; the mixture was then incubated on ice for 5 min before being transferred into a 0.1-cm gap cuvette (E. coli pulser cuvette; Bio-Rad Laboratories Inc., CA) and electroporated at 1.8 kV, with a capacitance of 25 μF and resistance of 200 Ω. One ml of BHI broth containing 5 μg ml−1 hemin and 0.5 mg ml−1 cysteine was added, and cells were incubated overnight anaerobically before selection of transformants on HBA plates containing 10 μg ml−1 erythromycin or 10 μg ml−1 erythromycin and 5 μg ml−1 chloramphenicol, as appropriate, with anaerobic incubation at 37°C for up to 10 days.
Construction of P. gingivalis RgpB CTD site-directed mutants.
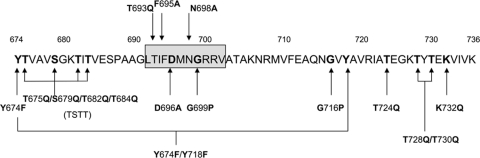
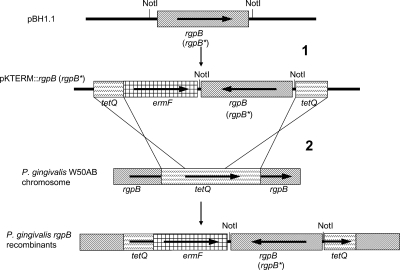
The various plasmids produced in the course of this work are described in Table 1, the amino acids within the RgpB CTD that were subject to mutation or deletion are shown in Fig. 2, and an overview of the recombination strategy is shown in Fig. 3.
FIG. 2.
Site-directed and deletion mutagenesis map of the RgpB CTD. Arrows point to the sites of mutation, with the amino acid changes indicated. Double and quadruple mutations are indicated by joined arrows. Numbering is from the rgpB primary translation. Amino acids 692 to 702 (boxed) were deleted in the W50AB-Δ692-702 mutant.
FIG. 3.
Introduction of recombinant rgpB into P. gingivalis by allele exchange. In step 1, wild-type sequence (rgpB) or mutated rgpB (rgpB*) was excised from pBH1.1-based plasmids by NotI digestion and ligated into the NotI site of pKTERM1. In step 2, the cassette was linearized and electroporated into P. gingivalis W50AB to allow double-crossover recombination events. Horizontal arrows indicate open reading frames and their orientations. The name of each gene is indicated below the arrow. tetQ, tetracycline resistance-encoding gene; ermF, erythromycin resistance gene; rgpB, arginine gingipain B gene.
P. gingivalis W50AB (40) has rgpA and rgpB inactivated by the insertion of cat (chloramphenicol resistance cassette) and tetQ (tetracycline resistance cassette), respectively. The tetQ cassette was selected as the homologous recombination target for the insertion of recombinant rgpB into the P. gingivalis W50AB chromosome. The recombination cassette to be used, pKTERM1, was generated as follows: tetQ was excised from pNJR12 (16) using EcoICRI digestion and ligated into the SmaI site of pUC18, generating plasmid pUCtetQ. The ermF gene (erythromycin resistance) was excised from pAL30 (7) using EcoICRI and SacII, blunt ends were generated using T4 DNA polymerase, and the insert was ligated into EcoRV-digested pUCtetQ. The resultant recombinant plasmid in which ermF was in the same orientation as tetQ was designated pKTERM1.
A QuikChange II site-directed mutagenesis kit (Stratagene) was employed to produce a series of plasmids encoding rRgpBs with CTD mutations, using pBH1.1 as a template and the various primers listed in Table 2. The encoded mutations were the following: Y674F, Y674F Y718F, T675Q S679Q T682Q T684Q (termed TSTT), T693Q, F695A, D696A, N698A, G699P, G716P, T724Q, T728Q T730Q, K732Q, and K732Q (Fig. 2). Each mutated rgpB gene was fully sequenced to confirm that only the desired mutations were generated. A 2.7-kbp fragment containing rgpB was excised from each mutated plasmid and pBH1.1 using NotI digestion and ligated to NotI-digested pKTERM1 (Table 1; Fig. 3). Recombinant plasmids (Table 1) in which the rgpB genes were in the reverse orientation to tetQ and ermF were used to transform P. gingivalis. The cassettes were introduced into P. gingivalis W50AB by electroporation for allele exchange with tetQ (schematically represented in Fig. 3) with transformants selected on HBA plates supplemented with erythromycin and chloramphenicol. The positive control (W50AB-Pos) had rgpB of wild-type sequence and ermF inserted into tetQ, while the negative control (W50AB-Neg) had only ermF inserted into tetQ. All P. gingivalis allele exchanges were confirmed by testing for tetracycline sensitivity and by extensive PCR analysis. Only those constructs in which the orientation of rgpB was opposite to that of ermF were used for further analyses in order to eliminate the possibility of differential polar effects on rgpB expression.
TABLE 2.
Oligonucleotide primers used to construct CTD mutation(s) in rgpBa
| Primer name | Sequence (5′→3′) | Primer location (nt)b | Description |
|---|---|---|---|
| M13 universal forward | TGTAAAACGACGGCCAGT | ||
| M13 universal reverse | CAGGAAACAGCTATGACC | ||
| dmngrForSfoI | GTCCTGCTGCCGGCGCCGAGAGCTCCCAACGCG | 2968-2978 | rgpB(nt 2986-2978)-SfoI-pGemTEasy (nt 103-118) |
| dmngrRevSfoI | CGCGTTGGGAGCTCTCGGCGCCGGCAGCAGGAC | 2978-2968 | Reverse complement of dmngrForSfoI |
| dmngrRevCM | CCCGGCAGCAGGACTTTCTAC | 2981-2961 | rgpB(nt 2981-2961) |
| dmngrForCM | GCTACTGCTAAAAACCGCATGGTATTC | 3015-3041 | rgpB(nt 3015-3041) |
| Gly699Profor | CTGACGATCTTCGATATGAACCCCCGTCGTGTAGCTAC | 2982-3019 | RgpB with Gly699 → Pro |
| Gly699Prorev | GTAGCTACACGACGGGGGTTCATATCGAAGATCGTCAG | 3019-2982 | Reverse complement of Gly699Profor |
| Gly716Profor | GAAGCACAAAACCCTGTGTATGCCGTTCG | 3042-3070 | RgpB with Gly716 → Pro |
| Gly716Prorev | CGAACGGCATACACAGGGTTTTGTGCTTC | 3070-3042 | Reverse complement of Gly716Profor |
| HSMUT2for | ATCGCTCAAGAAGGCAAGCAGTATCAAGAAGTTATAGTG | 3072-3113 | RgpB with Thr728 → Gln and Thr730 → Gln |
| HSMUT2rev | CACTATAACCTTTTCTTGATACTGCTTGCCTTCTTGAGCGAT | 3113-3072 | Reverse complement of HSMUT2for |
| HSMUT3for | GGCAAGACGTATACAGAACAGGTTATAGTGAAGTAATTC | 3084-3122 | RgpB with Lys732 → Gln |
| HSMUT3rev | GAATTACTTCACTATAACCTGTTCTGTATACGTCTTGCC | 3122-3084 | Reverse complement of HSMUT3for |
| S679T684for | CTGTAGCTGTACAAGGTAAGACGATACAAGTAGAAAGTCCTGCTG | 2932-2976 | RgpB with Ser679 → Gln and Thr684 → Gln |
| S679T684rev | CAGCAGGACTTTCTACTTGTATCGTCTTACCTTGTACAGCTACAG | 2976-2932 | Reverse complement of S679T684for |
| T675T682for | GATAAGCCTTATCAAGTAGCTGTACAAGGTAAGCAGATACAAGTAGAAAGTC | 2919-2970 | RgpB with Thr675 → Gln and Thr682 → Gln |
| T675T682rev | GACTTTCTACTTGTATCTGCTTACCTTGTACAGCTACTTGATAAGGCTTATC | 2970-2919 | Reverse complement of T675T682for |
| T724Q-kt-for | GCCGTTCGCATCGCTCAAGAAGGCAAGACGTATAC | 3063-3097 | RgpB with Thr724 → Gln |
| T724Q-kt-rev | GTATACGTCTTGCCTTCTTGAGCGATGCGAACGGC | 3097-3063 | Reverse complement of T724Q-kt-for |
| RgpBD696A-for | GGCTGACGATCTTCGCTATGAACGGCCGTCG | 2980-3010 | RgpB with Asp696 → Ala |
| RgpBD696A-rev | CGACGGCCGTTCATAGCGAAGATCGTCAGCC | 3010-2980 | Reverse complement of RgpBD696A-for |
| RgpBY718F-for | CACAAAACGGCGTGTTTGCCGTTCGCATCGC | 3046-3076 | RgpB with Tyr718 → Phe |
| RgpBY718F-rev | GCGATGCGAACGGCAAACACGCCGTTTTGTG | 3076-3046 | Reverse complement of RgpBY718F-for |
| RgpBY674F-for | CGTAGCCAATGATAAGCCTTTTACTGTAGCTGTATCAGG | 2909-2947 | RgpB with Tyr674 → Phe |
| RgpBY674F-rev | CCTGATACAGCTACAGTAAAAGGCTTATCATTGGCTACG | 2947-2909 | Reverse complement of RgpBY674F-for |
| RgpBT693Qfor | CCTGCTGCCGGGCTGCAAATCTTCGATATG | 2970-2999 | RgpB with Thr693 → Gln |
| RgpBT693Qrev | CATATCGAAGATTTGCAGCCCGGCAGCAGG | 2999-2970 | Reverse complement of RgpBT693Qfor |
| RgpBF695Afor | CCGGGCTGACGATCGCAGATATGAACGGCCG | 2977-3007 | RgpB with Phe695 → Ala |
| RgpBF695Arev | CGGCCGTTCATATCTGCGATCGTCAGCCCGG | 3007-2977 | Reverse complement of RgpBF695Afor |
| RgpBN698Afor | CGATCTTCGATATGGCCGGCCGTCGTGTAGC | 2986-3016 | RgpB with Asn698 → Ala |
| RgpBN698Arev | GCTACACGACGGCCGGCCATATCGAAGATCG | 3016-2986 | Reverse complement of RgpBN698Afor |
All the primers were purchased from GeneWorks, Pty. Ltd., Australia.
Primer alignment location in rgpB of P. gingivalis W50 according to the rgpB sequence of GenBank accession number AF007124. nt, nucleotides.
Construction of P. gingivalis RgpB(Δ692-702) deletion mutant.
Nucleotides encoding amino acids (aa) 692 to 702 (LTIFDMNGRRV) (Fig. 2) were deleted from rgpB using the following stepwise strategy. The primers dmngrRevCM and M13 universal forward (Table 2) were used to amplify a 2.7-kbp region of pBH1.1, containing rgpB nucleotides 493 to 2981 in the GenBank entry AF007124, and the amplicon was ligated to pGEM-TEasy (Promega). An SfoI restriction site was engineered into the construct at the 3′ junction of the rgpB codons and the multiple-cloning site using site-directed mutagenesis (as previously described) with dmngrForSfoI and dmngrRevSfoI to produce pCM1. Nucleotides 3015 to 3166 of rgpB (encoding 34 C-terminal amino acids and 3′ flanking regions) were then amplified as part of a 250-bp amplicon by PCR using pBH1.1 template and oligonucleotides dmngrForCM and M13 universal reverse. The PCR amplicon was treated with T4 DNA polymerase and then digested with NsiI and ligated to SfoI-NsiI-digested pCM1, producing pCM2. The mutated rgpB gene was then excised from pCM2 using AatII and NsiI digestion and ligated to AatII/NsiI-digested pKTERM1, producing pCM3. Following nucleotide sequencing to confirm integrity, pCM3 DNA was electroporated into P. gingivalis, with recombinants isolated as outlined above.
Construction of P. gingivalis W50 PG0553 isogenic mutant.
To make the pg0553 inactivation cassette, a 934-nucleotide 5′ region of pg0553 of P. gingivalis W50 was PCR amplified using Pfu DNA polymerase (Promega, Madison, WI) and primer pair PG0553-AatII-For (TCCGATGACGTCGTTGCCACGGAAGATGGCC) and PG0553-ApaI-Rev (CAAATCGGGCCCGCAGTCAGGACTGGCTGC). The amplicon was digested with AatII and ApaI and then ligated 3′ to ermF in AatII- and ApaI-digested pAL30 (7) to produce pAL34 (Table 1). A 1,122-nucleotide 3′ region of pg0553 was then PCR amplified using PG0553-SalI-For2 (AATTGAGTCGACACCATGTTCCGGCCTGCCA) and PG0553-SpeI-Rev2 (AACGATACTAGTCCTCTGCGCTATCGTGTCT) digested with SalI and SpeI and ligated 5′ to ermF in SalI-SpeI-digested pAL34 to produce pAL34.1 (Table 1). Plasmid pAL34.1 was linearized with ScaI and transformed into P. gingivalis strain W50 with recombinants selected on HBA plates supplemented with 10 μg ml−1 erythromycin. Confirmation of DNA integration was performed by PCR using the primer pairs PG0553KO-For (GCAGTCAGGACTGGCTGCC) and PG0553KO-Rev (CGGAGGGAGTTGTCACCTTC). This created the P. gingivalis W50 PG0553 isogenic mutant (ECR190) (Table 1).
P. gingivalis cell fraction preparation.
P. gingivalis cells grown in BHI broth (40 ml) were harvested at an OD650 of 0.61 (cell density of 2.5 × 109 cells per ml) by centrifugation (8,000 × g for 20 min at 4°C). The supernatant was retained, and the pellet was suspended in 4 ml of TC150 buffer (50 mM Tris-Cl, pH 8.0, 150 mM NaCl, 5 mM CaCl2, 20 mM cysteine-HCl). An aliquot of the cells (2 ml) was centrifuged, and the pellet was collected, suspended in 200 μl of TC150 buffer in the presence of 10 mM Nα-p-tosyl-l-lysine-chloromethyl ketone (TLCK; Sigma), and stored at −70°C for later use in Western immunoassays. Culture supernatants were subjected to ultracentrifugation (300,000 × g for 30 min) to pellet outer membrane vesicles, and the clarified supernatant was retained. For Western blotting the proteins in 2 ml of supernatant were precipitated using 10% (vol/vol) trichloroacetic acid (TCA) (35) and suspended in 15 μl of loading dye. P. gingivalis Sarkosyl-insoluble outer membranes and TCA-precipitated periplasmic protein fractions were prepared as previously described (35).
Arginine amidolytic activity assay.
Whole cells (108 in 20 μl) plus 340 μl of TC150 buffer or 360 μl of vesicle-free supernatant were assayed for Arg-X amidolytic activity using N-benzoyl-dl-arginine-p-nitroanilide (BApNA; Sigma) as previously described (35) using a temperature-controlled Cary 50 Bio UV-visible spectrophotometer (Varian Australia Pty Ltd., Victoria, Australia). BApNA assays were performed on triplicate samples from three separate cultures, and activity is expressed as μmol of BApNA/min/1011 cells.
Western blot analyses.
Proteins of whole cells, outer membrane preparations, concentrated culture supernatant (5 μl), and periplasmic preparations were subjected to SDS-PAGE using NuPAGE Novex Bis-Tris gels (Invitrogen Corporation) and 1× NuPAGE morpholinepropanesulfonic acid (MOPS)-SDS running buffer, following the manufacturer's instructions, with the SeeBlue Plus2 prestained protein standard (Invitrogen Corporation). Gels were stained using Coomassie blue R250 (Bio-Rad, Hercules, CA) or SYPRO Ruby (Molecular Probes Inc., Eugene, OR), or proteins were electrotransferred onto nitrocellulose membranes. Membranes were blocked and probed with anti-rRgpAcat (where RgpAcat indicates the catalytic domain of RgpA) mouse antibodies (anti-Rgp antibodies) diluted 1 in 500 in TNT buffer (25 mM Tris-HCl [pH 7.5], 500 mM NaCl, 0.1% [vol/vol] Triton X-100) as described previously (35). The immunoreactive proteins were detected using SuperSignal West Pico substrate chemiluminescence (Pierce Biotechnology, Inc., Rockford, U. S. A.) and a Las 3000 instrument (Fujifilm) for image capture.
Protein identification.
Proteins were identified in SDS-PAGE gels using in-gel digestion and mass spectrometry (MS) using matrix-assisted laser desorption ionization-two-stage time of flight mass spectrometry (MALDI-TOF/TOF MS) as described previously (18). MS spectra were used to search a P. gingivalis protein sequence database for protein identification, as described previously (40).
Statistical analysis.
Arg-specific proteolytic activities as measured using BApNA as a substrate were statistically analyzed using one-way analysis of variance (ANOVA) with a Scheffe a posteriori multiple comparison using an SPSS statistical software program (version 17; Chicago, IL).
RESULTS
Generation of site-directed mutations of the RgpB CTD.
To investigate the possible role of specific residues of the CTD of RgpB in the secretion and cell surface attachment of the protein, residues within the RgpB CTD were mutated by site-directed mutagenesis or deletion, and the rRgpBs produced in P. gingivalis were characterized for Arg-X amidolytic activity and by Western blotting using anti-Rgp antibodies. Thirteen rRgpBs with CTD mutations were produced: Y674F, Y674F Y718F, T675Q S679Q T682Q T684Q (termed TSTT), T693Q, F695A, D696A, N698A, G699P, G716P, T724Q, T728Q T730Q, and K732Q and a mutant with a deletion of residues 692 to 702 (Δ692-702) (Fig. 2). RgpB residues Y674 and Y718 are reasonably conserved in the CTD sequences and, hence, were mutated to the structurally conservative phenylalanine to investigate whether they are involved in O-linked modification. The eight Ser/Thr residues were mutated in four separate mutants. Ser/Thr residues of the N-terminal cluster were targeted in the same mutant in RgpB(T675Q S679Q T682Q T684Q), and the other Thr residues were mutated in RgpB(T693Q), RgpB(T724Q), and RgpB(T728Q T730Q) in order to investigate whether these residues are potential sites for O-linked modification.
Alignment of the RgpB CTD with CTD sequences of other CTD family proteins revealed that RgpB D696, G699, G716, and K732 are highly conserved residues (Fig. 1) (35), and thus these were also targeted for mutation in this study. D696 was mutated to alanine, and K732 was mutated to glutamine to test the role of residue polarity and charge, respectively, on secretion and attachment. The conserved RgpB G699 and G716 residues may play a role in the conformational stability of the CTD; therefore, these residues were mutated to proline in an attempt to perturb CTD structure.
Analysis of RgpB mutants. (i) Arginine amidolytic activity.
No Arg-X amidolytic activity could be detected in clarified, unconcentrated culture supernatants of any of the strains, including the positive control (W50AB-Pos) expressing the wild-type RgpB enzyme. In contrast, all strains exhibited measurable whole-cell Arg-X amidolytic activity except for the D696A and Δ692-702 mutants, where no activity could be detected. The whole-cell Arg-X amidolytic activity of the W50 positive control (W50AB-Pos) was 4.7 ± 0.3 μmol of BApNA/min/1011 cells, and the whole-cell activities of the mutants Y674F, Y674F Y718F, T675Q S679Q T682Q T684Q, T693Q, F695A, N698A, G699P, G716P, T724Q, T728Q T730Q, and K732Q were not significantly different from the activity of W50AB-Pos.
(ii) Cellular distribution and modification of RgpB mutants.
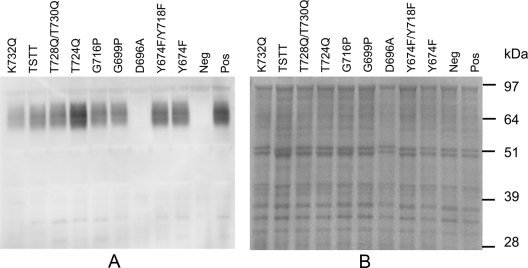
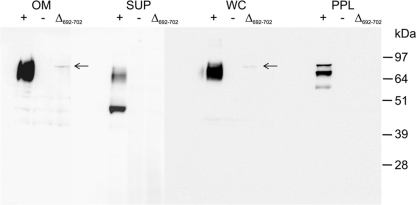
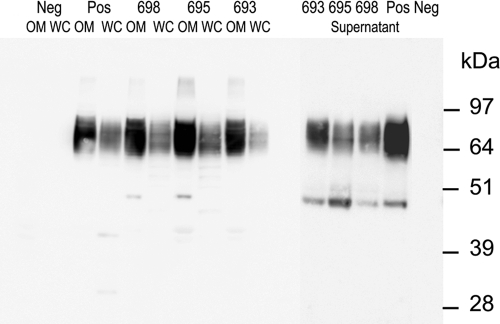
To assess the cellular distribution and modification of the CTD-mutated RgpB proteins, outer membrane, periplasmic protein fractions, clarified supernatant, and whole-cell lysates of the recombinant strains were subjected to SDS-PAGE and Western blot analyses using anti-Rgp antibodies. Western blot analysis of outer membrane fractions of all RgpB CTD mutant strains using the anti-Rgp antibodies showed that the characteristic diffuse band at 70 to 90 kDa, corresponding to modified RgpB (cell surface-attached isoform), was readily observed in all RgpB CTD mutant strains except for the motif B deletion mutant RgpB(Δ692-702), the RgpB(D696A) mutant, and the negative-control strain W50AB-Neg (Fig. 4, 5, and 6). In mutant Δ692-702 the Western blot of the outer membrane fraction revealed a trace amount of a single band of 80 kDa, corresponding to the size of the unprocessed precursor form of the mutated RgpB containing the prodomain, catalytic domain, and mutated CTD (Fig. 5). Repeated Western blotting of outer membrane preparations of the Y674F, Y674F Y718F, T675Q S679Q T682Q T684Q, T693Q, F695A, N698A, G699P, G716P, T724Q, T728Q T730Q, and K732Q mutants and the positive control (W50AB-Pos) indicated no significant differences in the intensities of the 70- to 90-kDa (cell surface attached) RgpB isoform. Western blots of clarified, concentrated supernatant proteins showed similar profiles, with all strains except the Δ692-702, D696A, and W50AB-Neg strains producing trace amounts of a 50-kDa fully processed form of RgpB in the clarified and concentrated culture supernatant (Fig. 5, 6, and 7). The preparations used for Fig. 5 and 6 were contaminated with membrane vesicles so that the membrane-attached isoform (70- to 90-kDa band) was also present in the culture supernatant samples. Repeated Western blotting showed no significant differences between the intensities of the 50-kDa RgpB isoform of the positive control and that of any of the mutants except for Δ692-702 and D696A. Western blots of periplasmic-enriched protein fractions showed no accumulation of mutated RgpB in the periplasm of the Δ692-702 mutant (Fig. 5) or D696A mutant (data not shown).
FIG. 4.
Western blot analyses of outer membrane proteins of P. gingivalis allele exchange mutants probed with anti-Rgp. (A) Western blot. (B) The corresponding SDS-PAGE gel stained with Coomassie blue. Pos, positive control (wild-type sequence RgpB); Neg, negative control. Other recombinant RgpB proteins are indicated by their mutations.
FIG. 5.
Western blot analyses of protein fractions of the P. gingivalis RgpB(Δ692-702) mutant probed with anti-Rgp. +, positive control; −, negative control. Fractions probed with anti-Rgp are indicated as follows: OM, outer membrane; SUP, supernatant; WC, whole cell; PPL, periplasmic. The arrows indicate the faint 80-kDa band corresponding to precursor RgpB.
FIG. 6.
Western blot analyses of protein fractions of P. gingivalis allele exchange mutants probed with anti-Rgp. Neg and Pos are the negative and positive controls, respectively. Fractions probed with anti-Rgp are indicated as follows: 698, RgpB(N698A); 695, RgpB(F695A); 693, RgpB(T693Q); OM, outer membrane; WC, whole cell.
FIG. 7.
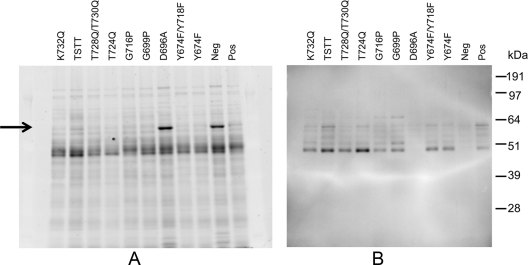
Western blot analyses of clarified and concentrated supernatant proteins of P. gingivalis allele exchange mutants probed with anti-Rgp. (A) The SDS-PAGE gel stained with Coomassie blue. (B) The corresponding Western blot. Pos, positive control (wild-type sequence RgpB); Neg, negative control. Other recombinant Rgp proteins are indicated by their mutations. The arrow indicates the abundant PG0553 which is found in the D696A and negative control.
In the protein profiles of the culture supernatants of the negative control and the Δ692-702 and D696A mutants, a prominent band of approximately 63 kDa appeared that was not present in any of the other strains tested that contained RgpB active enzyme (Fig. 7A; also data not shown). These bands were excised from the gel, digested with trypsin, and analyzed by MALDI-TOF/TOF mass spectrometry. Peptide mass fingerprinting (PMF) indicated that the major protein present in the bands from the three strains was PG0553 (Table 3). This assignment was verified by tandem MS (MS/MS) of four of the observed peptides (Table 3). One of these peptides (m/z 1,494) could not be identified using the default parameters; however, it matched the peptide QISFGGEPLSFSSR with the N-terminal glutamine converted to pyroglutamine. This glutamine is the predicted N terminus of the protein after removal of the signal peptide. Mass spectrometric analysis of this protein revealed that it was a C-terminally truncated form of PG0553 (a predicted endopeptidase) (Table 3).
TABLE 3.
Mass spectrometric analysis data identifying PG0553
| Sample/peptidea | Experimentb | Mascot score | Expect value |
|---|---|---|---|
| W50AB-Neg | PMF | 73 | 1.1E−04 |
| W50AB-D696A | PMF | 81 | 1.9E−05 |
| 28QISFGGEPLSFSSR41 | MS/MS | 60 | 5.5E−05 |
| 56LTPDFNPEDLIAQSR70 | MS/MS | 85 | 1.2E−08 |
| 119AITLYYDAFNIPEGGR134 | MS/MS | 71 | 2.3E−07 |
| 329SDGLLLQLNDEVPLR343 | MS/MS | 52 | 1.8E−05 |
The identities of the four peptides were independently confirmed by MS/MS using the negative-control sample. All peptides matched to within 50 ppm of the calculated mass.
PMF experiments were conducted on the 63-kDa bands excised from the blot shown in Fig. 7A.
To investigate the link between PG0553 and RgpB processing and activity, a P. gingivalis W50 pg0553 isogenic mutant was constructed. The disruption of pg0553 was confirmed using PCR (data not shown). The Arg-X amidolytic cell surface activity of the P. gingivalis W50 pg0553 mutant (ECR190) was not significantly different from that of the W50 wild type. Furthermore, RgpB was present as the cell-attached 70- to 90-kDa isoform in the PG0553 mutant, indicating that the PG0553 protein was not essential for secretion and attachment of RgpB.
Western blotting was repeated for all culture/cell fractions of the Δ692-702 and D696A mutants, including whole cell, inner and outer membrane preparations, and periplasmic and culture fluid fractions; however, RgpB could not be detected in any fraction. To eliminate the possibility of a secondary mutation elsewhere in the cell causing the Δ692-702 and D696A phenotype, a second mutant strain for each was generated by transformation of the background strain W50AB once again with pCM3 and pJD696A, respectively. The phenotype of depressed Rgp(Δ692-702) and RgpB(D696A) production was reproduced in the second mutants (data not shown), and so the rgpB(Δ692-702) and rgpB(D696A) recombinant loci were amplified by PCR and subjected to DNA sequencing, whereupon no errors in nucleotide sequences were detected. To address the possibility that the reduced RgpB(Δ692-702) and RgpB(D696A) production could result from reduced gene transcription, the expression levels of rgpB(Δ692-702) and rgpB(D696A) relative to the control wild-type rgpB gene were determined. Real-time reverse transcription-PCR of total RNA preparations showed no significant difference between the levels of wild-type rgpB positive-control transcript and rgpB(Δ692-702) and rgpB(D696A) transcripts (results not shown). There was no transcript detected in the RgpB negative-control strain, W50AB-Neg.
DISCUSSION
In the present study, S679, T675, T682, T684, T693, N698, T724, T728, T730, Y674, and Y718 residues of the RgpB CTD were selected to screen as potential site(s) of modification and membrane attachment. Whole-cell enzyme activities and Western blot analyses of RgpB with the TSTT mutation, RgpB(T724Q), RgpB(Y674F), RgpB(Y693F), and RgpB(Y674F Y718F) in outer membrane fractions showed the diffuse band of 70 to 90 kDa, as found in the positive control, indicating surface attachment of the mutated proteins. The lack of obvious change in the attachment profiles of these mutated RgpBs suggests that these residues are not essential for secretion or surface attachment of the protein. Whole-cell enzyme activities and Western blotting of whole-cell, outer membrane, periplasm, and supernatant fractions of RgpBs mutated at the conserved residues G699, G716, and K732 also indicated that the protein was attached to the cell surface. G699 and G716 were mutated to prolines in an attempt to perturb the structure of the conserved CTD regions B and D, respectively. In light of the results, the proline substitution was not sufficient to abolish the function of the CTD with respect to secretion and attachment. It was considered possible that perturbation of the structure of RgpB CTD may affect the processing/folding of the protein and, therefore, specific activity of the secreted RgpB. Mikolajczyk et al. (17) have demonstrated that sequential autolytic processing of the prodomain and CTD is required for full specific activity of RgpB, such that changes to the CTD sequence may affect this sequential autolytic processing. However, in this current study, substitution of Pro for conserved Gly residues of the CTD did not affect secretion, attachment, or activity of RgpB.
In a separate study of the RgpB CTD, the last 13 residues of the RgpB CTD were truncated in five separate mutants, and the last five residues were individually mutated to alanines to investigate the function of the CTD (19). The only common residue that was targeted in that study (19) and in the current study was the conserved K732 (K503 in the mature RgpB). In the current study K732 was targeted to investigate the importance of this charged residue located only five amino acids from the C terminus of the CTD. We postulated that the positive charge of the lysine in close proximity to the C terminus may interact with the negatively charged membrane for export of RgpB to the surface. The importance of charged residues involved in sorting of bacterial extracellular proteins can be seen with the Gram-positive sortases (34). Sortases are bacterial enzymes responsible for the covalent attachment of secreted proteins to the cell wall in Gram-positive bacteria. Proteins destined for sortase-mediated attachment contain an N-terminal Sec leader and a C-terminal cell wall sorting signal (CWS) that is characterized by an LPXTG motif, a hydrophobic region, and C-terminal positive charge (34). Sortase cleaves the CWS motif between the threonine-glycine bond and subsequently catalyzes the transpeptidation of the threonine carbonyl to an amine of a pentaglycine cross-bridge, tethering the C terminus of the protein to the bacterial cell wall (2). Mutational analysis of the two positive residues positioned only a few residues from the C terminus of the CWS in staphylococcal protein A showed that this positive charge is required for retention of the polypeptide within the secretory pathway (34).
In the current study, the conserved RgpB Lys732 was mutated to glutamine, which is the most structurally similar uncharged residue to lysine, to remove charge but maintain integrity of the polar structure. The RgpB(K732Q) mutant exhibited Arg-specific amidolytic activity and a Western blot profile similar to that of the positive-control strain, with the presence of the modified 70- to 90-kDa RgpB in outer membrane fractions. These results differ from those observed previously (19) with an RgpB(K732A) mutant, where mutation of Lys732 to an Ala resulted in an accumulation of two distinct bands of approximately 65 and 52 kDa in the periplasm, in addition to the modified 70- to 90-kDa form of RgpB in the outer membrane fraction, albeit at lower intensity than that of the positive control (19). Further, the K732A mutant showed smaller amounts of active enzyme (54% compared to the positive control). These results of smaller amounts of attached RgpB suggest a problem with secretion in this mutant (19). Alanine is a small amino acid with aliphatic hydrophobic properties, which differ significantly from lysine, which has a large side chain and positive charge. Collectively, the results indicate that the positive charge of K732 is not essential for export or attachment of RgpB to the outer membrane; however, it appears that the lysine, possibly because of its polarity and/or the bulk of its side chain, may be important for proper recognition by the secretion system.
The most significant change in RgpB surface attachment in the site-directed mutants was observed with the D696A mutation, where the conserved Asp696 was mutated to Ala. Western blot analyses revealed that Rgp(D696A) could not be detected in whole cells or in any fraction (periplasm, inner membrane, and outer membrane) of the cell or supernatant. As the levels of rgpB transcript in W50-D696A and W50AB-Pos did not differ, and as the RgpB(D696A) precursor could not be detected in the periplasm or culture fluid of the mutant, these findings suggest that the protein may be blocked in the secretion apparatus, which results in the downregulation of translation and/or the upregulation of proteolysis.
Removal of the RgpB CTD motif B that contains Asp696 resulted in a trace amount of unprocessed precursor being detected in the outer membrane fraction of the bacterium but no detection of the protein in any other fraction, including the periplasm and culture fluid. The trace levels and the precursor form of the protein are consistent with the lack of enzyme activity in any fraction (whole cells, supernatant, clarified supernatant, and periplasm) of the mutant. As transcription was also not affected in this mutant, it appears that translation of the RgpB(Δ692-702) protein again may have been downregulated and/or that proteolysis was upregulated due to the protein being trapped in the outer membrane secretory apparatus. In both the Asp696Ala mutant and the motif B deletion mutant, RgpB(Δ692-702), a 63-kDa protein was substantially increased in abundance, and analysis of this protein by mass spectrometry indicated that it was the predicted endopeptidase PG0553. The high level of abundance of this protein, together with the possible downregulated translation of the mutated RgpB proteins (Asp696 and motif B), may account for the small amount of RgpB protein that could be detected in any fractions of these mutants relative to that of the wild type.
To investigate a possible link between the PG0553 proteinase and cell surface processing of RgpB, an isogenic mutant of P. gingivalis W50 lacking PG0553 was created. The cell surface Arg-X amidolytic activity of the pg0553 mutant was not significantly different from that of the control. Further, the cell surface-attached isoform (70 to 90 kDa) of RgpB was present in the pg0553 mutant, suggesting that PG0553 does not have an essential role in the processing of RgpB. As PG0553 was also found substantially increased in abundance in the negative-control strain W50AB-Neg not expressing either RgpA or RgpB, as well as in the D696A and Δ692-702 mutants, it is possible that the high abundance of PG0553 (a cell surface/culture fluid protease) may be related to the lack of the RgpA and RgpB proteases.
The Δ692-702 deletion mutant and the D696A mutant phenotypes were different from those of the RgpB mutants lacking the entire CTD (35) or the last 2 to 13 C-terminal residues of the CTD (19) as the latter mutated RgpB proteins accumulated in the periplasm. This can be explained as the CTD truncated mutants (19, 35) would not be recognized by the outer membrane secretion system since they lacked the signal or complete signal for outer member secretion and, hence, accumulated in the periplasm. The Δ692-702 deletion mutant and the D696A mutant still contained the signal for outer membrane secretion but were unable to be modified (attached) and, hence, became trapped in the secretion system. These results suggest that regions D plus E (Fig. 1) involving the last 22 residues of the RgpB CTD act as the recognition signal for the CTD secretion system, whereas regions B plus C, involving D696, in motif B are the attachment recognition sequence. The results further suggest that secretion and attachment of RgpB are coordinated.
With the exception of Asp696, none of the site-directed mutations performed in this study, including others in motif B, significantly affected enzyme presentation on the surface of the bacterium, and so it seems likely that none of these residues are sites of modification and therefore surface attachment. It can be concluded, therefore, that Asp696 of motif B is important for CTD function. The most likely role of Asp696 and motif B is in recognition by a protein of the outer membrane secretion system that cleaves the CTD and/or covalently attaches the CTD onto a sugar moiety of its membrane anchor A-LPS to form the membrane-modified isoform of the protein. Hence, when Asp696 is missing, the CTD protein cannot be processed and becomes trapped in the outer membrane secretion system. Asp696 does not appear to be essential as a small number of CTD proteins have a Ser, Thr, or Asn residue in its place. This suggests that a small polar residue will fulfill the role of Asp696 in motif B. For example, PG0553, the endopeptidase upregulated in mutants not expressing Rgp enzyme activity, is a CTD protein with a Ser residue in place of RgpB's Asp696 (Fig. 1). Interestingly, PG0553 was found predominantly as a C-terminally truncated isoform in the culture supernatant of the Rgp mutant strains not expressing Arg-X activity. The presence of PG0553 and RgpB in the culture supernatant without the CTD sequence suggests that cleavage of the attached CTD sequence may be the mechanism for release of the surface-attached protein into the culture fluid. As the entire CTD sequence is missing in the culture supernatant isoforms, it is tempting to speculate that the cleavage site of the CTD for ultimate release of the fully processed protein into the culture fluid may involve region A (Fig. 1).
Localization of proteins to the surface of the cell of Gram-negative bacteria requires export of the proteins across the inner membrane, the periplasm, and the outer membrane. Six types of outer membrane protein export systems have been identified that fall into two main categories, those dependent on the Sec translocon in the inner membrane and those independent of Sec (1, 15). Proteins that are Sec dependent have N-terminal sequences that serve as targets for SecB or the signal recognition particle (SRP) that function to target the nascent polypeptide chain to the Sec translocon in the inner membrane (28, 38). After inner membrane transit, passage of Sec-dependent proteins into or across the outer membrane involves transport complexes of the type II system (30a), the chaperone-usher pathway (33), or the type V system, where the polypeptide facilitates its own export by insertion of the carboxyl domain into the outer membrane to form a β-barrel pore in a mechanism referred to as autotransport (13). P. gingivalis has no proteins with sequence similarity to these known Sec-dependent, outer membrane export system proteins (42). Therefore, it appears that the CTD family proteins are exported across the outer membrane by a new type of transport system.
The P. gingivalis proteins PorT (32), PG27 (14), and Sov (30), which have been demonstrated to be required for the secretion of RgpB, RgpA, and Kgp, share the same bacterial species distribution as the CTD and appear to be part of a novel Bacteroidetes-specific secretion system. Recently, Sato et al. (31) reported a list of 55 proteins that share this same species distribution and demonstrated the involvement of some, including PorK, PorL, PorM, PorN, PorP, PorW, and PorU (PG26) in RgpB, RgpA, and Kgp secretion. The system was designated the Por secretion system (PorSS), and some of the proteins of the system are orthologues of gliding motility proteins of Flavobacterium johnsoniae, thus providing a link between the novel protein translocation system and a motility apparatus in the Bacteroidetes phylum (31).
Recently, we have shown in P. gingivalis isogenic mutants lacking PG27 or PorT that both CTD proteins and A-LPS accumulate in the periplasm, indicating that secretion and attachment of the CTD protein to the A-LPS anchor are coordinated (Y.-Y. Chen, B. Peng, M. D. Glew, et al., submitted for publication). The results of the current study with RgpB also suggest that secretion and attachment are coordinated. This coordination could be achieved by the secretion of the CTD protein and translocation of the A-LPS being linked through the covalent attachment of the CTD or cleaved CTD to the A-LPS anchor. Given the range and extent of the mutations performed in this study, in particular all the conserved residues of the CTD, the results suggest that the Ser/Thr/Tyr/Asn residues of the RgpB CTD domain are not the sites of modification and surface attachment. It seems likely that the CTD acts as a site of recognition by a P. gingivalis processing enzyme(s), possibly a novel sortase-like enzyme that cleaves the CTD sequence and attaches the C-terminal carbonyl to a sugar amine of A-LPS. Although sortase activity was initially ascribed to Gram-positive microorganisms only, more recently, sortase homologues have been identified in Gram-negative bacteria (12, 22, 23). Organisms can contain several sortase-like proteins in their genomes (23), and the level of sequence identity is low for orthologous proteins (<25%), making identification difficult (23). Further research is required to identify the protein(s) involved in the coordinated secretion and cell surface attachment of the CTD proteins. In conclusion, the results of the current study show that the CTD motif B containing Asp696 is involved in the cell surface attachment of RgpB in P. gingivalis and that without attachment secretion of the protein does not occur.
Acknowledgments
We acknowledge James Edward Fielding and Brigitte Hoffmann for their technical support.
This study is supported by National Health and Medical Research Council grant number 3500427 and the Cooperative Research Centre for Oral Health Science.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Albers, S. V., Z. Szabo, and A. J. Driessen. 2006. Protein secretion in the Archaea: multiple paths towards a unique cell surface. Nat. Rev. Microbiol. 4:537-547. [DOI] [PubMed] [Google Scholar]
- 2.Bentley, M. L., H. Gaweska, J. M. Kielec, and D. G. McCafferty. 2007. Engineering the substrate specificity of Staphylococcus aureus sortase A. The β6/β7 loop from SrtB confers NPQTN recognition to SrtA. J. Biol. Chem. 282:6571-6581. [DOI] [PubMed] [Google Scholar]
- 3.Christersson, C. E., R. G. Dunford, P. O. Glantz, and R. E. Baier. 1989. Effect of critical surface tension on retention of oral microorganisms. Scand. J. Dent. Res. 97:247-256. [DOI] [PubMed] [Google Scholar]
- 4.Curtis, M. A., J. Aduse-Opoku, and M. Rangarajan. 2001. Cysteine proteases of Porphyromonas gingivalis. Crit. Rev. Oral Biol. Med. 12:192-216. [DOI] [PubMed] [Google Scholar]
- 5.Curtis, M. A., H. K. Kuramitsu, M. Lantz, F. L. Macrina, K. Nakayama, J. Potempa, E. C. Reynolds, and J. Aduse-Opoku. 1999. Molecular genetics and nomenclature of proteases of Porphyromonas gingivalis. J. Periodontal Res. 34:464-472. [DOI] [PubMed] [Google Scholar]
- 6.Curtis, M. A., A. Thickett, J. M. Slaney, M. Rangarajan, J. Aduse-Opoku, P. Shepherd, N. Paramonov, and E. F. Hounsell. 1999. Variable carbohydrate modifications to the catalytic chains of the RgpA and RgpB proteases of Porphyromonas gingivalis W50. Infect. Immun. 67:3816-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dashper, S. G., C. S. Ang, P. D. Veith, H. L. Mitchell, A. W. Lo, C. A. Seers, K. A. Walsh, N. Slakeski, D. Chen, J. P. Lissel, C. A. Butler, N. M. O'Brien-Simpson, I. G. Barr, and E. C. Reynolds. 2009. Response of Porphyromonas gingivalis to heme limitation in continuous culture. J. Bacteriol. 191:1044-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dortbudak, O., R. Eberhardt, M. Ulm, and G. R. Persson. 2005. Periodontitis, a marker of risk in pregnancy for preterm birth. J. Clin. Periodontol. 32:45-52. [DOI] [PubMed] [Google Scholar]
- 9.Duchaud, E., M. Boussaha, V. Loux, J. F. Bernardet, C. Michel, B. Kerouault, S. Mondot, P. Nicolas, R. Bossy, C. Caron, P. Bessieres, J. F. Gibrat, S. Claverol, F. Dumetz, M. Le Henaff, and A. Benmansour. 2007. Complete genome sequence of the fish pathogen Flavobacterium psychrophilum. Nat. Biotechnol. 25:763-769. [DOI] [PubMed] [Google Scholar]
- 10.Eichinger, A., H. G. Beisel, U. Jacob, R. Huber, F. J. Medrano, A. Banbula, J. Potempa, J. Travis, and W. Bode. 1999. Crystal structure of gingipain R: an Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 18:5453-5462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geismar, K., K. Stoltze, B. Sigurd, F. Gyntelberg, and P. Holmstrup. 2006. Periodontal disease and coronary heart disease. J. Periodontol. 77:1547-1554. [DOI] [PubMed] [Google Scholar]
- 12.Haft, D. H., I. T. Paulsen, N. Ward, and J. D. Selengut. 2006. Exopolysaccharide-associated protein sorting in environmental organisms: the PEP-CTERM/EpsH system. Application of a novel phylogenetic profiling heuristic. BMC Biol. 4:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Henderson, I. R., F. Navarro-Garcia, M. Desvaux, R. C. Fernandez, and D. Ala'Aldeen. 2004. Type V protein secretion pathway: the autotransporter story. Microbiol. Mol. Biol. Rev. 68:692-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ishiguro, I., K. Saiki, and K. Konishi. 2009. PG27 is a novel membrane protein essential for a Porphyromonas gingivalis protease secretion system. FEMS Microbiol. Lett. 292:261-267. [DOI] [PubMed] [Google Scholar]
- 15.Kostakioti, M., C. L. Newman, D. G. Thanassi, and C. Stathopoulos. 2005. Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 187:4306-4314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maley, J., N. B. Shoemaker, and I. S. Roberts. 1992. The introduction of colonic-Bacteroides shuttle plasmids into Porphyromonas gingivalis: identification of a putative P. gingivalis insertion-sequence element. FEMS Microbiol. Lett. 72:75-81. [DOI] [PubMed] [Google Scholar]
- 17.Mikolajczyk, J., K. M. Boatright, H. R. Stennicke, T. Nazif, J. Potempa, M. Bogyo, and G. S. Salvesen. 2003. Sequential autolytic processing activates the zymogen of Arg-gingipain. J. Biol. Chem. 278:10458-10464. [DOI] [PubMed] [Google Scholar]
- 18.Ngo, L. H., P. D. Veith, Y. Y. Chen, D. Chen, I. B. Darby, and E. C. Reynolds. 2010. Mass spectrometric analyses of peptides and proteins in human gingival crevicular fluid. J. Proteome Res. 9:1683-1693. [DOI] [PubMed] [Google Scholar]
- 19.Nguyen, K. A., J. Travis, and J. Potempa. 2007. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of gram-negative bacteria? J. Bacteriol. 189:833-843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.O'Brien-Simpson, N. M., R. A. Paolini, B. Hoffmann, N. Slakeski, S. G. Dashper, and E. C. Reynolds. 2001. Role of RgpA, RgpB, and Kgp proteinases in virulence of Porphyromonas gingivalis W50 in a murine lesion model. Infect. Immun. 69:7527-7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Offenbacher, S., V. Katz, G. Fertik, J. Collins, D. Boyd, G. Maynor, R. McKaig, and J. Beck. 1996. Periodontal infection as a possible risk factor for preterm low birth weight. J. Periodontol. 67:1103-1113. [DOI] [PubMed] [Google Scholar]
- 22.Pallen, M. J., R. R. Chaudhuri, and I. R. Henderson. 2003. Genomic analysis of secretion systems. Curr. Opin. Microbiol. 6:519-527. [DOI] [PubMed] [Google Scholar]
- 23.Pallen, M. J., A. C. Lam, M. Antonio, and K. Dunbar. 2001. An embarrassment of sortases—a richness of substrates? Trends Microbiol. 9:97-102. [DOI] [PubMed] [Google Scholar]
- 24.Paramonov, N., D. Bailey, M. Rangarajan, A. Hashim, G. Kelly, M. A. Curtis, and E. F. Hounsell. 2001. Structural analysis of the polysaccharide from the lipopolysaccharide of Porphyromonas gingivalis strain W50. Eur. J. Biochem. 268:4698-4707. [DOI] [PubMed] [Google Scholar]
- 25.Paramonov, N., M. Rangarajan, A. Hashim, A. Gallagher, J. Aduse-Opoku, J. M. Slaney, E. Hounsell, and M. A. Curtis. 2005. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 58:847-863. [DOI] [PubMed] [Google Scholar]
- 26.Paramonov, N. A., J. Aduse-Opoku, A. Hashim, M. Rangarajan, and M. A. Curtis. 2009. Structural analysis of the core region of O-lipopolysaccharide of Porphyromonas gingivalis from mutants defective in O-antigen ligase and O-antigen polymerase. J. Bacteriol. 191:5272-5282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pathirana, R. D., N. M. O'Brien-Simpson, and E. C. Reynolds. 2010. Host immune responses to Porphyromonas gingivalis antigens. Periodontol. 2000 52:218-237. [DOI] [PubMed] [Google Scholar]
- 28.Pohlschroder, M., W. A. Prinz, E. Hartmann, and J. Beckwith. 1997. Protein translocation in the three domains of life: variations on a theme. Cell 91:563-566. [DOI] [PubMed] [Google Scholar]
- 29.Rangarajan, M., J. Aduse-Opoku, N. Paramonov, A. Hashim, N. Bostanci, O. P. Fraser, E. Tarelli, and M. A. Curtis. 2008. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 190:2920-2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saiki, K., and K. Konishi. 2007. Identification of a Porphyromonas gingivalis novel protein Sov required for the secretion of gingipains. Microbiol. Immunol. 51:483-491. [DOI] [PubMed] [Google Scholar]
- 30a.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 31.Sato, K., M. Naito, H. Yukitake, H. Hirakawa, M. Shoji, M. J. McBride, R. G. Rhodes, and K. Nakayama. 2010. A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc. Natl. Acad. Sci. U. S. A. 107:276-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato, K., E. Sakai, P. D. Veith, M. Shoji, Y. Kikuchi, H. Yukitake, N. Ohara, M. Naito, K. Okamoto, E. C. Reynolds, and K. Nakayama. 2005. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 280:8668-8677. [DOI] [PubMed] [Google Scholar]
- 33.Sauer, F. G., K. Futterer, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 34.Schneewind, O., D. Mihaylova-Petkov, and P. Model. 1993. Cell wall sorting signals in surface proteins of gram-positive bacteria. EMBO J. 12:4803-4811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seers, C. A., N. Slakeski, P. D. Veith, T. Nikolof, Y. Y. Chen, S. G. Dashper, and E. C. Reynolds. 2006. The RgpB C-terminal domain has a role in attachment of RgpB to the outer membrane and belongs to a novel C-terminal-domain family found in Porphyromonas gingivalis. J. Bacteriol. 188:6376-6386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Slakeski, N., P. S. Bhogal, N. M. O'Brien-Simpson, and E. C. Reynolds. 1998. Characterization of a second cell-associated Arg-specific cysteine proteinase of Porphyromonas gingivalis and identification of an adhesin-binding motif involved in association of the prtR and prtK proteinases and adhesins into large complexes. Microbiology 144:1583-1592. [DOI] [PubMed] [Google Scholar]
- 37.Reference deleted.
- 38.Veenendaal, A. K., C. van der Does, and A. J. Driessen. 2004. The protein-conducting channel SecYEG. Biochim. Biophys. Acta 1694:81-95. [DOI] [PubMed] [Google Scholar]
- 39.Veith, P. D., N. M. O'Brien-Simpson, Y. Tan, D. C. Djatmiko, S. G. Dashper, and E. C. Reynolds. 2009. Outer membrane proteome and antigens of Tannerella forsythia. J. Proteome Res. 8:4279-4292. [DOI] [PubMed] [Google Scholar]
- 40.Veith, P. D., G. H. Talbo, N. Slakeski, S. G. Dashper, C. Moore, R. A. Paolini, and E. C. Reynolds. 2002. Major outer membrane proteins and proteolytic processing of RgpA and Kgp of Porphyromonas gingivalis W50. Biochem. J. 363:105-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xie, G., D. C. Bruce, J. F. Challacombe, O. Chertkov, J. C. Detter, P. Gilna, C. S. Han, S. Lucas, M. Misra, G. L. Myers, P. Richardson, R. Tapia, N. Thayer, L. S. Thompson, T. S. Brettin, B. Henrissat, D. B. Wilson, and M. J. McBride. 2007. Genome sequence of the cellulolytic gliding bacterium Cytophaga hutchinsonii. Appl. Environ. Microbiol. 73:3536-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yen, M.-R., C. R. Peabody, S. M. Partovi, Y. Zhai, Y.-H. Tseng, and M. H. S. Saier, Jr. 2002. Protein-translocating outer membrane porins of Gram-negative bacteria. Biochim. Biophys. Acta 1562:6-31. [DOI] [PubMed] [Google Scholar]