Abstract
Expression of urease is essential for gastric colonization by Helicobacter pylori. The increased level of urease in gastric acidity is due, in part, to acid activation of the two-component system (TCS) consisting of the membrane sensor HP0165 and its response regulator, HP0166, which regulates transcription of the seven genes of the urease gene cluster. We now find that there are two major ureAB transcripts: a 2.7-kb full-length ureAB transcript and a 1.4-kb truncated transcript lacking 3′ ureB. Acidic pH (pH 4.5) results in a significant increase in transcription of ureAB, while neutral pH (pH 7.4) increases the truncated 1.4-kb transcript. Northern blot analysis with sense RNA and strand-specific oligonucleotide probes followed by 5′ rapid amplification of cDNA ends detects an antisense small RNA (sRNA) encoded by the 5′ ureB noncoding strand consisting of ∼290 nucleotides (5′ureB-sRNA). Deletion of HP0165 elevates the level of the truncated 1.4-kb transcript along with that of the 5′ureB-sRNA at both pH 7.4 and pH 4.5. Overexpression of 5′ureB-sRNA increases the 1.4-kb transcript, decreases the 2.7-kb transcript, and decreases urease activity. Electrophoretic mobility shift assay shows that unphosphorylated HP0166 binds specifically to the 5′ureB-sRNA promoter. The ability of the HP0165-HP0166 TCS to both increase and decrease ureB expression at low and high pHs, respectively, facilitates gastric habitation and colonization over the wide range of intragastric pHs experienced by the organism.
Colonization of healthy human and animal stomachs is a property of gastric Helicobacter species, including the human pathogen, Helicobacter pylori. H. pylori maintains a relatively neutral periplasmic pH in the face of a highly intragastric acidic environment, preserving cytoplasmic pH homeostasis and its proton motive force. This acid acclimation is distinct from the acid tolerance/resistance of other neutralophiles that transit but do not colonize the stomach (22, 47, 48). The most important component of acid acclimation is the nickel metaloenzyme urease, which generates the buffers NH3 and HCO3− from the metabolism of ambient urea, maintaining both cytoplasmic and periplasmic pHs to enable the organism to survive and grow in the stomach. The organism expresses very high levels of the urease A and B subunits, more than any other known ureolytic microbe, accounting for as much as 8% of the total bacterial protein (34). The expression of active urease requires six of the seven genes of the urease gene cluster, ureA and ureB for the catalytic subunits and ureE, ureF, ureG, and ureH, necessary for nickel insertion into the apourease, UreA/UreB. The third gene of the urease cluster, ureI, encodes a pH-gated urea channel that increases urea access to intrabacterial urease in acid (51, 71).
Because of the importance of this enzyme for survival in an acidic environment, most attention has been focused on its upregulation under acidic conditions. There is increased urease gene transcription by the activation of the two-component system (TCS) HP0165-HP0166 (ArsRS) (39, 41) and the cytoplasmic histidine kinase HP0244 (74). Although increased urease activity in the presence of acid is beneficial, this activity in the absence of acid and in the presence of urea is lethal to the organism in vitro (14, 33). Gastric pH can climb to as high as pH 6.0 after a meal due to the strong buffering effect of food, and urea is always present in gastric juice. Hence, urease may act as a double-edged sword, being essential for survival and colonization on the highly acidic gastric surface but lethal at relatively neutral intragastric pH. Therefore, H. pylori may downregulate urease activity at a relatively neutral pH to ensure its survival. The transcriptional induction of urease genes (ureA, ureB, and ureI) in response to low pH (pH 5.0) is mediated mainly by the HP0165-HP0166 TCS (39-41). The phosphorylated response regulator HP0166 binds to extended regions overlapping the PureA and PureI promoters (41). In addition to the TCS, transcription of the ureA and ureB genes is positively regulated by NikR in response to increasing concentrations of Ni2+ in the surrounding medium (63, 64). While transcriptional control of gene expression is clearly of primary importance in prokaryotes, these organisms also employ regulatory mechanisms to control translation, initiation, and mRNA stability. To achieve this, many bacteria rely on the expression of small, noncoding RNAs (70). In bacteria, noncoding regulatory RNAs are usually between 50 and 300 nucleotides (nt) in length and are thus known as small RNAs (sRNAs) (67). Most sRNAs that have been characterized act as posttranscriptional regulators by interacting with specific mRNA targets, modulating message stability and/or altering mRNA accessibility to the translational machinery (30). sRNAs are involved in a number of cellular processes, including translational quality control, by blocking or freeing ribosome binding sites (4, 31), acid resistance in Escherichia coli (38, 59), and iron homeostasis (11, 19), and have recently been implicated in regulating the virulence of several pathogens (23, 46). It is now widely accepted that bacterial sRNAs play central roles in gene expression regulation in response to environmental changes.
Many sRNAs act by posttranscriptional regulation of mRNAs via base-pairing interactions (57). Two classes of bacterial base-pairing sRNAs can be distinguished (70). There are cis-encoded sRNAs that are located in the same noncoding DNA region and that are therefore fully complementary to their targets over a long sequence stretch, whereas trans-encoded sRNAs are located in another chromosomal location and are only partially complementary to their target mRNAs. The known regulatory mechanisms employed by cis-encoded antisense sRNAs include transcription attenuation, translation inhibition, inhibition of primer maturation, and promotion or inhibition of mRNA degradation (7). The majority of regulation by known trans-encoded sRNAs is negative (1, 25). Base pairing between the sRNA and its target mRNA usually leads to repression of protein levels through translational inhibition, mRNA degradation, or both. In many cases, the RNA chaperone Hfq is required for trans-encoded sRNA-mediated regulation, presumably to facilitate RNA-RNA interactions due to limited complementarity between the sRNA and target mRNA (1).
Although the majority of these sRNAs were discovered in E. coli, small RNAs appear to be ubiquitous in bacteria (5, 28, 66, 69, 75). A recent primary transcriptome study in H. pylori using a novel dRNA-seq approach (54) identified an unexpected wealth of sRNAs from H. pylori, with ∼60 of them being validated, despite the lack of a conserved Hfq protein. These results show that many sRNA-mediated regulations are yet to be discovered in H. pylori. Many of the small RNAs are transcribed under the control of TCSs (15, 24, 62), providing additional control of expression of genes that are primarily regulated at the transcriptional level via these TCSs (12).
In the current study, we have identified a novel urease regulatory mechanism by an sRNA-mediated downregulation of ureB expression, which is controlled primarily by the HP0165-HP0166 TCS. Shown here is the presence of an antisense sRNA (5′ureB-sRNA) that is cis encoded by the noncoding strand of the 5′ ureB gene, which regulates ureAB expression at a posttranscriptional level by targeted degradation of the sRNA-mRNA duplexes, resulting in a truncated ureAB mRNA leading to reduced translation of functional UreB protein. This 5′ureB-sRNA is downregulated by HP0165 in response to acidic pH; hence, UreB expression increases in acid media. Electrophoretic mobility shift assay (EMSA) shows that a nonphosphorylatable HP0166 response regulator binds to the promoter region of 5′ureB-sRNA, whereas phosphorylated HP0166 does not. Overexpression of 5′ureB-sRNA greatly increases the 1.4-kb transcript level and decreases urease activity. This novel regulatory mechanism maintains a high urease level at acidic pH but permits a relatively rapid decrease of urease activity when the organism is transiting to neutral pH.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
H. pylori strain 43504 was obtained from American Type Culture Collection (ATCC). HP0165-deficient mutant 43504/ΔHP0165::Km was constructed by allelic exchange using a kanamycin resistance gene as described below. Primary plate cultures of H. pylori were grown from glycerol stocks on Trypticase soy agar (TSA) plates with 5% sheep blood (Fisher Scientific) for 2 to 3 days in a microaerobic environment (5% O2, 10% CO2, 85% N2) at 37°C. In preparation for an experiment, bacteria were scraped from the plates, suspended in 1 mM phosphate HP buffer (138 mM NaCl, 5 mM KCl, 1 mM CaCl2, 0.5 mM MgCl2, 10 mM glucose, 1 mM glutamine), pH 7.0 (53), and transferred to fresh plates for 24 h. For exposure to pH 4.5, the overnight culture of H. pylori strain 43504 on TSA plates supplemented with 5% sheep blood was suspended in brain heart infusion (BHI) medium (Difco) to an optical density at 600 nm (OD600) of 0.20 to 0.25. The pH of the BHI medium was adjusted to 7.4 or 4.5 using concentrated HCl, followed by filtration to remove any precipitate before the bacteria were added. H. pylori was incubated in the presence of 5 mM urea with shaking (120 rpm) under microaerobic conditions at 37°C for 30 min. E. coli strains were grown in Luria-Bertani (LB) broth. When necessary, the following antibiotics were added at the indicated final concentrations: ampicillin, 100 μg/ml; kanamycin, 50 μg/ml (for E. coli) or 20 μg/ml (for H. pylori).
Construction of deficient mutant H. pylori strain (43504/ΔHP0165::Km) by allelic exchange mutagenesis.
To construct the H. pylori strain with the mutation for HP0165 deficiency (43504/ΔHP0165::Km), a pBluescript II vector (Stratagene) carrying a kanamycin resistance cassette flanked by a 613-bp fragment, comprising 518 bp of the 3′ region of the HP0166 open reading frame (ORF) and 29 bp of the intergenic region (IGR) upstream of the HP0165 ORF, and a 429-bp fragment, comprising 67 bp of the intergenic region downstream of the HP0165 ORF and 362 bp of the 5′ region of the HP0163 ORF, was introduced into H. pylori strain 43504 by natural transformation. The DNA fragments were amplified by PCR from chromosomal DNA of H. pylori strain 43504 with primer pairs HP0166-5′P(314-346)-XbaI/HP0166-3′P(925-963)-SalI and HP0163-5′P(2210-2245)-BglII/HP0163-3′P(2636-2672)-Acc65I (Table 1), respectively. The resulting strain (43504/ΔHP0165::Km) had the HP0165 gene replaced with a kanamycin resistance cassette. The kanamycin-selected mutant strains were confirmed by PCR.
TABLE 1.
Oligonucleotide primers/probes used in this study
| Name | Sequence (5′ to 3′)a | Siteb | Strand | Positionc |
|---|---|---|---|---|
| HP0166-5′P(314-346) | CATGTAACCAATCTAGATGAGCCATATACCGGC | XbaI | − | 174345-174377 |
| HP0166-3′P(925-963) | CTTTAAAAAAGATAGAGGTCGACATAACCCCTTAACTCC | SalI | + | 173728-273766 |
| HP0163-5′P(2210-2245) | CCCGAAAATTTGAGATCTGTGAGCGGAATGAAGGGG | BglII | − | 172447-172482 |
| HP0163-3′P(2636-2672) | GCCCATGGTCGGTACCTTCACAAAAACACAAATCCGC | Acc65I | + | 172020-172056 |
| pHP0072-0073R-5′P | CATTATCACTCCAATTTTAA | − | 78188-78207 | |
| pHP0071-0067-3′P | GCCTTTTCCTTCCAAACAAA | + | 75336-75355 | |
| HP0073-3′P(580-603) | ACATCGCTTCAATACCCACTTCAT | + | 77698-77721 | |
| HP0072-5′P(1111-1133) | CTACAGGCGATAAAGTGAGATTG | − | 77168-77190 | |
| HP0072-3′P(1563-1586) | TGGAGTGATAGTAGTCGCATTAGT | + | 76715-76738 | |
| HP0072-5′P(2159-2182) | TATGGGTCGTGTGGGTGAAGTTAT | − | 76119-76142 | |
| HP0072-3′P(2653-2673) | CAATGTGAGCGGTAGTGTCGT | + | 75628-75648 | |
| HP0072-5′P(2470-2493) | CCCCACAACCAGTTTATTACAGAG | − | 75808-75831 | |
| HP0072-3′P(2943-2966) | TGCCTTTTCCTTCCAAACAAAAAT | + | 75335-75358 | |
| 5′-ureB 1-1S | CTACAGGCGATAAAGTGAGATTGGGCGATACAGACTTGATCGCTGAAGTAGA | − | 77139-77190 | |
| 5′-ureB 1-2S | ACATGACTACACCATTTATGGCGAAGAGCTTAAATTCGGTGGCGGTAAAACC | − | 77087-77138 | |
| 5′-ureB 1-3S | CTGAGAGAAGGCATGAGCCAATCCAACAACCCTAGCAAAGAAGAATTGGAT | − | 77036-77086 | |
| 5′-ureB 2-1S | CTAATCATCACTAACGCTTTAATCGTGGATTACACCGGTATTTATAAAGCGGATA | − | 76981-77035 | |
| 5′-ureB 2-3S | GCAAGATGGCGTTAAAAACAATCTTAGCGTAGGTCCTGCTACTGAAGCCTTAGCC | − | 76871-76925 | |
| 5′-ureB 3-1S | GGTGAAGGTTTGATCGTAACTGCTGGTGGTATTGACACACACATCCACTTCA | − | 76819-76810 | |
| 5′-ureB 1-2AS | GGTTTTACCGCCACCGAATTTAAGCTCTTCGCCATAAATGGTGTAGTCATGT | + | 77087-77138 | |
| 5′-ureB 2-1AS | CTAATCATCACTAACGCTTTAATCGTGGATTACACCGGTATTTATAAAGCGGATA | + | 76981-77035 | |
| cagBA-IGR-5′P | tttgggtaccTTTTTAATCGTCTCAGGTTCA | KpnI | + | 579301-579321 |
| cagBA-IGR-3′P | ctagccatggGACTATCGGTATCTTATTGGTATCA | NcoI | − | 579869-579892 |
| 5′-ureB sRNA(+)-5′P | CTTGCcAtgGCTGTAGGGATTTGTTGGGGTG | NcoI | + | 76785-76815 |
| 5′-ureB sRNA(+)-3′P | CTGAGAGAAGGCATGAGCCA | − | 77067-77086 | |
| 5′-ureB sRNA(+)-3′P-rev | TGGCTCATGCCTTCTCTCAG | + | 77067-77086 | |
| 5′-ureB sRNA(−)-5′P | AAAACCaTGgGAGAAGGCATGAGCCAATCC | NcoI | − | 77063-77092 |
| 5′-ureB sRNA(−)-3′P | CTGTAGGGATTTGTTGGGGT | + | 76795-76814 | |
| 5′-ureB sRNA(−)-3′P-rev | ACCCCAACAAATCCCTACAG | − | 76795-76814 | |
| HP0092-T1-5′P-rev | AGTTTAATTACCAGTGGATA | + | 98324-98343 | |
| HP0092-T1-5′P | TATCCACTGGTAATTAAACT | − | 98324-98343 | |
| HP0092-T1-3′P | ATTTcTGCaGATCCGCCTTATTTCCTCTCT | PstI | + | 98206-98235 |
| HP0072-5′P(1507-1526) | CTTTTGCAAGCGGTGTAACA | − | 76775-76794 | |
| HP0072-3′P(1605-1623) | CAGCCGCTCTGAGCATCCA | + | 76678-76696 |
Sequences in uppercase letters are derived from the genome sequences of H. pylori 26695 (58). Sequences introduced for cloning purposes are given in lowercase letters, and restriction recognition sites are underlined.
Restriction recognition sites.
Nucleotide positions refer to the genome sequence of H. pylori 26695 (58).
Construction of overexpression strain for 5′ureB-sRNA.
pTM117, which was reported to be a transcriptional reporter and complementation vector in H. pylori (9), is used for the overexpression of 5′ureB-sRNA by insertion of a 593-bp DNA fragment containing the promoter region of cagA at KpnI/NcoI sites to generate pTM-PcagA. The cagA promoter fragment was prepared by PCR from the intergenic region between cagB and cagA (55) of the H. pylori strain 26695 genome with primer pair cagBA-IGR-5′P-KpnI/cagBA-IGR-3′P-NcoI (Table 1). For the plasmid used for overexpression of 5′ureB-sRNA [pTM-PcagA-5′ureB-sRNA(+)], the fusion fragment of 5′ureB-sRNA and a transcriptional terminator identified from H. pylori (HP0092-T1) (10) were prepared in a two-step PCR process by amplification of the 5′ureB-sRNA fragment with primer 5′-ureB sRNA(+)-5′P-NcoI and fusion primer HP0092-T1-5′P-rev-5′-ureB sRNA(+)-3′P (Table 1), and HP0092-T1 fragment with fusion primer 5′-ureB sRNA(+)-3′P-HP0092-T1-5′P and HP0092-T1-3′P-PstI (Table 1). The two fragments were subsequently combined as templates in a PCR sewing with primer pair 5′-ureB sRNA(+)-5′P-NcoI/HP0092-T1-3′P-PstI to enable the fusion, which was followed by cloning of the fusion fragment into pTM-PcagA at NcoI/PstI sites. The plasmid expressing anti-5′ureB-sRNA [pTM-PcagA-5′ureB-sRNA(−)] was constructed by fusion of the complementary 5′ureB-sRNA sequence and the HP0092-T1 terminator, followed by cloning into pTM-PcagA at NcoI/PstI sites downstream of the cagA promoter, in a strategy similar to that described above, with primer pairs (listed in Table 1) 5′-ureB sRNA(−)-5′P-NcoI/HP0092-T1-5′P-rev-5′-ureB sRNA(−)-3′P (for the complementary 5′ureB-sRNA fragment), 5′-ureB sRNA(−)-3′P-HP0092-T1-5′P/HP0092-T1-3′P-PstI (for the HP0092-T1 terminator), and 5′-ureB sRNA(−)-5′P-NcoI/HP0092-T1-3′P-PstI (for PCR sewing). Plasmids pTM-PcagA, pTM-PcagA-5′ureB-sRNA(+), and pTM-PcagA-5′ureB-sRNA(−) were introduced into wild-type H. pylori strain 43504 via natural transformation (49), and the transformants were selected on BHI medium plates containing kanamycin.
RNA preparation.
Total RNA was isolated from the H. pylori strains using TRIzol reagent (Invitrogen, Carlsbad, CA) combined with RNeasy columns (Qiagen, CA). The bacterial pellet was resuspended in 500 μl of TRIzol reagent (Invitrogen) and lysed at room temperature for 5 min before 100 μl of chloroform was added. Following centrifugation at 12,000 × g for 10 min at 4°C, the supernatant was mixed with 250 μl of 100% ethanol and applied to an RNeasy spin column (Qiagen), and RNA purification combined with on-column DNase treatment using an RNase-free DNase set (Qiagen) was processed following the manufacturer's instructions (beginning with the application to the column). The RNA concentration was quantified by determination of the absorbance at 260 nm using an ND-1000 spectrophotometer (NanoDrop Technologies), and the quality was evaluated by capillary electrophoresis using a model 2100 bioanalyzer with an RNA 6000 Nano assay kit (Agilent Technologies).
Preparation of probes for Northern blot analysis.
For preparation of RNA probes, a series of DNA fragments corresponding to the different regions of the ureA and ureB genes, including probes 5′-ureA (510 bp, containing 250 bp of the upstream sequence and 260 bp of the 5′-ureA ORF), 5′-ureB (476 bp), 3′-ureB (515 bp), IGR-ureAB (1,493 bp, containing the complete 967 bp of ureA, 3 bp of the intergenic region between ureA and ureB, and 523 bp of the 5′-ureB ORF), the complete ureB gene (1,563 bp), and IGR-ureBI (497 bp, containing 304 bp of the 3′-ureB and the 193-bp intergenic region between ureB and ureI ORF), were prepared by PCR with primer pairs pHP0072-0073R-5′P/HP0073-3′P(580-603), HP0072-5′P(1111-1133)/HP0072-3′P(1563-1586), HP0072-5′P(2159-2182)/HP0072-3′P(2653-2673), pHP0072-0073R-5′P/ HP0072-3′P(1563-1586), HP0072-5′P(1111-1133)/HP0072-3′P(2653-2673), and HP0072-5′P(2470-2493)/HP0072-3′P(2943-2966), listed in Table 1, respectively, using genomic DNA from H. pylori strain 26695 as the template. These PCR products were cloned into the pCR4-TOPO vector, which has T3 and T7 promoters located at each end of the insert. The orientation of the insert for each construct was determined by sequencing. To synthesize sense RNA probes, the constructs were digested with NotI (for 5′-ureA) or SalI (for IGR-ureBI) and transcribed with T3 RNA polymerase (MAXIscript in vitro transcription kit; Ambion) using α-32P-labeled UTP (MP Biochemicals) or were digested with PstI and transcribed with T7 RNA polymerase (for 5′-ureB, 3′-ureB, ureB, and IGR-ureAB). To synthesize the antisense RNA probes, the constructs were digested with PstI (for 5′-ureA) or KpnI (for IGR-ureBI) and transcribed with T7 RNA polymerase or were digested with NotI and transcribed with T3 RNA polymerase (for 5′-ureB, 3′-ureB, ureB, and IGR-ureAB). For preparation of the DNA probe, the DNA fragment for 5′-ureA (510 bp) generated by PCR as described above was radiolabeled with [α-32P]dCTP using a Prim-It II random primer labeling kit (Stratagene, La Jolla, CA). For preparation of the oligonucleotide probes listed in Table 1, the strand-specific sense oligonucleotides 1-1S (52 nt), 1-2S (52 nt), 1-3S (51 nt), 2-1S (55 nt), 2-3S (55 nt), and 3-1S (52 nt) and the antisense oligonucleotides 1-2AS (52 nt) and 2-1AS (55 nt), corresponding to different regions of 5′ ureB, were synthesized (Eurofins MWG Operon) and 5′ end labeled radioactively by T4 polynucleotide kinase (Promega) and [γ-32P]ATP.
Northern blot analysis.
For detection of the small RNA, total RNAs (5 μg) were fractionated in 10% or 6% polyacrylamide-urea gels (Invitrogen) and electrically transferred to Zeta-Probe GT membranes (Bio-Rad Laboratories). For detection of mRNA species, 15 μg of total RNAs was fractionated on 1.2% agarose-formaldehyde gels and transferred to Nytran membranes (Whatman Schleicher & Schuell) using TurboBlotter rapid downward transfer systems (Whatman Schleicher & Schuell). For verification of equal loading, the RNA on the gel was visualized by ethidium bromide staining and photographed to compare the rRNA band intensities before the RNA was transferred. For the DNA probe, the membrane blots were hybridized with 32P-labeled 5′-ureA DNA probe overnight at 65°C in a buffer containing 0.45 M sodium phosphate (pH 7.2), 7% sodium dodecyl sulfate (SDS), 1% bovine serum albumin (BSA), and 20 mM EDTA (13). The hybridized blots were washed with 0.1 × SSC (1 × SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.1% SDS at 65°C. For RNA probes, the blots were hybridized with 32P-labeled sense or antisense RNA probes for 5′ ureA, 5′ ureB, 3′ ureB, IGR ureAB, complete ureB, or IGR ureBI in ULTRAhyb ultrasensitive hybridization buffer (Ambion) at 68°C and washed with 0.1× SSC-0.1% SDS. For oligonucleotide probes, the blots were hybridized with 32P-labeled strand-specific oligonucleotide probes 5′-ureB 1-1S, 1-2S, 1-3S, 2-1S, 2-3S, 3-1S, or 2-1AS in ULTRAhyb oligonucleotide hybridization buffer (Ambion) at 42°C and then washed with 2× SSC-0.5% SDS. The hybridized blots were autoradiographed using a PhosphorImager 445 SI apparatus (Molecular Dynamics, Sunnyvale, CA). The bands representing each transcript in the hybridized blots and 23S and 16S rRNAs in ethidium bromide-stained gels were quantified using an ImageJ analysis system (available from rsb.info.nih.gov/ij/). The relative expression level for each transcript was normalized to its corresponding total intensity of 23S and 16S rRNA. Each hybridization was repeated at least 3 times with independently prepared RNA samples.
5′ RACE.
A BD Smart RACE cDNA amplification kit (BD Biosciences Clontech, Palo Alto, CA) was used to determine the transcription start site of the antisense sRNA complementary to the 5′ region of ureB in H. pylori by rapid amplification of cDNA ends (RACE)-PCR. According to the manufacturer's manual, 1 μg of total RNA isolated from H. pylori wild-type strain 43504 was reverse transcribed with a strand-specific oligonucleotide sense primer, 5′-ureB 1-3S, which is the most-5′-end sense probe that was able to detect the ∼290-nt antisense sRNA complementary to the 5′ region of the ureB ORF, and BD PowerScript reverse transcriptase (BD Biosciences Clontech), while the BD Smart II A oligonucleotide was also included in the reverse transcription (RT) reaction mixture. When RT reaches the 5′ end of the sRNA template, the Smart II A oligonucleotide attaches to the first-strand cDNA tail and serves as an extended template for RT, resulting in a complete cDNA copy of the original sRNA with the additional Smart sequence at the 5′ end (32, 77). The same primer used for RT, 5′-ureB 1-3S, which corresponds to the 3′ end of the sRNA, was used for 5′ RACE-PCR with Smart universal primer. The RACE-PCR products were cloned into the T/A cloning vector pCR4-TOPO (Invitrogen), and the transcription start site and location in the genome for the sRNA were determined by DNA sequencing in both directions with T7 and M13 reverse primers.
Urease assay.
Urease activity was measured radiometrically (53). Wild-type H. pylori strain 43504, the HP0165-deficient mutant strain (43504/ΔHP0165::Km), and the 5′ureB-sRNA-overexpressing strain [43054/pTM-PcagA-5′ureB-sRNA(+)] were grown on TSA plates, harvested, and suspended in 1 ml of 1 mM phosphate H. pylori buffer, pH 7.0. The bacterial suspensions were diluted 1:100 with 25 mM sodium phosphate buffer containing 5 mM [14C]urea with a specific activity of 10 μCi/μmol and 0.1% detergent octaethylene glycol monododecyl ether (C12E8; Sigma) to fully permeabilize the bacteria to urea. Plastic wells containing 0.5 M KOH-soaked filter paper hung from rubber stoppers were used to collect the liberated 14CO2 that resulted from the hydrolysis of urea by urease. Urease activity was measured for 30 min at 37°C with constant agitation. The reaction was terminated by the addition of 5 N H2SO4 to liberate all labeled CO2 from the incubation medium. The wells containing the filter paper were placed in a scintillation cocktail (HiIonicFluor; Packard Instruments, Meriden, CT), and the radioactivity was measured by scintillation counting (1216 RackBeta; LKB Instruments, Gaithersburg, MD). All experiments were performed in triplicate. The urease activity was reported as the number of micromoles of CO2 released per minute per milligram of protein. The protein concentration was determined by the Lowry method.
EMSAs.
A 116-bp DNA fragment corresponding to the potential promoter region of 5′ureB-sRNA was prepared by PCR with primer pair HP0072-5′P(1507-1526)/HP0072-3′P(1605-1623) (Table 1) and 5′ end labeled radioactively by T4 polynucleotide kinase and [γ-32P]ATP (50 μCi). Recombinant HP0166-His6 (73) was phosphorylated in vitro in a buffer containing 20 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, 1 mM dithiothreitol (DTT), and 10 mM carbamylphosphate over a period of 60 min at 25°C. Binding of HP0166 to DNA was carried out in a 10-μl reaction mixture containing 104 cpm of 33P-labeled DNA, 1 μg of poly(dI-dC) (Sigma), 25 mM NaPO4 (pH 7), 150 mM NaCl, 0.1 mM MgSO4, and 1 mM DTT. The DNA binding reaction was initiated by the addition of phosphorylated HP0166-His6 or unphosphorylated HP0166-D52N-His6 (73), and the reaction mixture was incubated at room temperature for 20 min. Samples were then loaded directly onto a 6% DNA-retardation polyacrylamide gel (Invitrogen). Electrophoresis was pursued for 1 h (14 V/cm) at room temperature, and the gels were then dried and analyzed by autoradiography.
RESULTS
The HP0165-HP0166 two-component system controls ureB gene expression by regulating the level of a 1.4-kb truncated ureAB transcript.
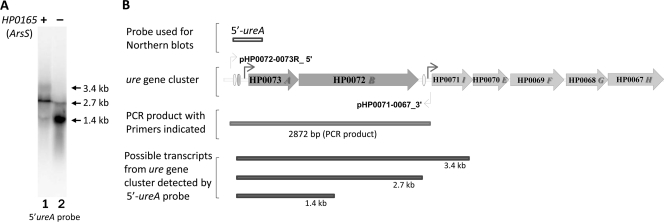
Northern blot analysis with a ureA probe identified a major transcript of 2.7 kb for ureAB (3). Our previous study has shown that this transcript was upregulated about 3- to 4-fold in response to pH 4.5 compared to the response to pH 7.4 due to activation at PureA (74). Northern blot analysis with a 5′-ureA probe in the HP0165-deficient mutants constructed from H. pylori strain 43504 detected an ∼1.4-kb truncated ureAB transcript (Fig. 1 A, lane 2) of greater intensity than in the wild-type strain (Fig. 1A, lane 1). Since the ∼1.4-kb truncated ureAB transcript was detected by the 5′-ureA probe, it is concluded that this truncated transcript contains complete ureA and the 5′ untranslated region of the ureAB gene but lacks the 3′ ureB region (Fig. 1B). The 2.7-kb intact ureAB transcript was still present in the mutant, although at a level much reduced compared to that in the wild type. There were variable amounts of a 3.4-kb transcript corresponding to the ureABI transcript, as described previously (3). These results suggest that at least three different transcripts corresponding to ureAB from the urease gene cluster are present.
FIG. 1.
Truncated ureAB transcripts are detected in HP0165-deficient mutant strain. (A) Total RNA was extracted from wild-type H. pylori strain 43504 (lane 1) and the 43504/ΔHP0165::Km mutant strain (lane 2). The RNA was size fractionated on 1.2% agarose formaldehyde gels and hybridized with the 5′ ureA-specific DNA probe. (B) Structure of the urease gene cluster and its flanking region, probes used for Northern blot analysis, primers (light bent arrows; the primers are listed in Table 1) used for the PCR analysis, the PCR product, and a summary of possible transcripts from the ureAB gene cluster obtained with the 5′-ureA probe. The dark bent arrows denote promoters. The ovals represent HP0166 binding sites.
To determine whether the truncated transcript was due to an artificial change in the ureAB genome, the genomic sequences of ureAB were examined. PCR using a primer (pHP0072-0073R-5′P) corresponding to the intergenic sequence upstream of PureA and a primer (pHP0071-0067-3′P) corresponding to the upstream region of the ureI gene generated a 2,872-bp product (Fig. 1B) with genomic DNA from the HP0165-deficient mutant strain. Sequence analysis of the 2,872-bp PCR product confirmed that no artificial changes in the ureAB genomic structure of the HP0165-deficient mutant strain had occurred. These results suggest that HP0165 also regulates urease gene expression by controlling the expression level of the truncated ureAB transcript. It may be that these truncated ureAB transcripts are due to the posttranscriptional degradation at the 3′ ureB region of the ureAB transcript.
A cis-encoded antisense small RNA transcribed from the 5′ ureB noncoding strand is identified by Northern blot analysis.
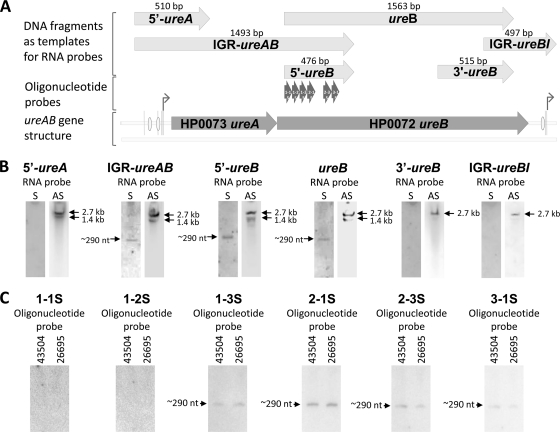
A possible regulatory mechanism for truncation of ureB could be an antisense small RNA that acts posttranscriptionally by targeted degradation/stabilization of the ureAB mRNA. We therefore prepared, by PCR, a series of DNA fragments corresponding to the different regions of the ureA and ureB genes (Fig. 2 A), including 5′-ureA, 5′-ureB, 3′-ureB, IGR-ureAB, complete ureB, and IGR-ureBI. These PCR products were cloned into a pCR4-TOPO vector with T3 and T7 promoters located at each end of the insert. The sense RNA probes were synthesized by in vitro transcription with T3 or T7 RNA polymerase and α-[32P]UTP and were then used for Northern analysis, in which the RNAs were fractionated in 10% polyacrylamide-urea gels.
FIG. 2.
Detection of a cis-encoded antisense small RNA transcribed from the 5′ ureB noncoding strand by Northern analysis. (A) ureAB gene structure and the DNA fragments that were used as templates for RNA probe synthesis by in vitro transcription. The strand-specific oligonucleotide probes are also shown. The bent arrows denote promoters. The ovals represent HP0166 binding sites. (B) The total RNAs were extracted from wild-type H. pylori strain 43504. RNAs were separated in a 10% polyacrylamide-urea gel and then transferred to a Zeta-GT membrane. Sense RNA probes (lanes S) for 5′ ureA, IGR ureAB, 5′ ureB, full ureB, 3′ ureB, and IGR ureBI were used for Northern blot analysis. As a control, antisense RNA probes (lanes AS) were also used for Northern blot analysis. (C) For further characterization of the ∼290-nt antisense sRNA complementary to the 5′ region of ureB, total RNAs isolated from wild-type H. pylori strains 43504 and 26695 were separated in a 6% polyacrylamide-urea gel and then transferred to a Zeta-GB membrane. Six strand-specific oligonucleotide sense probes (1-1S, 1-2S, 1-3S, 2-1S, 2-3S, and 3-1S, each with a size of 51 to 55 nt) corresponding to different regions of the 5′-ureB probe were used in Northern blot analysis.
The sense RNA probes for 5′ ureA, 3′ ureB, and IGR ureBI did not detect any transcript encoded by the ureAB noncoding strand (Fig. 2B). However, the sense RNA probes for IGR ureAB, 5′ ureB, and complete ureB did identify an ∼290-nt transcript (Fig. 2B). Since all of these three probes share the same sequence at the 5′ ureB, it is very likely that they detected an identical transcript encoded by the 5′ ureB noncoding strand. As controls, antisense probes for each of the regions of the ureAB gene detected a similar transcription pattern, as shown in Fig. 1, in which RNAs were separated in a 1.2% agarose gel, suggesting the specificity of detection by sense RNA probes. However, with the antisense RNA probes for 3′ ureB and IGR ureBI, only the 2.7-kb transcript was found, which supports our conclusion that the 1.4-kb truncated ureAB transcript lacks the sequence corresponding to the 3′ end of ureB.
Identification of the ∼290-nt antisense sRNA complementary to the 5′ region of ureB.
To further characterize the ∼290-nt antisense sRNA complementary to the 5′ region of ureB in terms of its specific genomic location and size, we prepared six strand-specific oligonucleotide sense probes (1-1S, 1-2S, 1-3S, 2-1S, 2-3S, and 3-1S, each with a size of 51 to 55 nt) corresponding to different regions of the previously used 5′-ureB probe (476 nt) (Fig. 2A) and used in Northern analysis and 5′ RACE to determine the specific location and the 5′ end of this sRNA in the H. pylori genome.
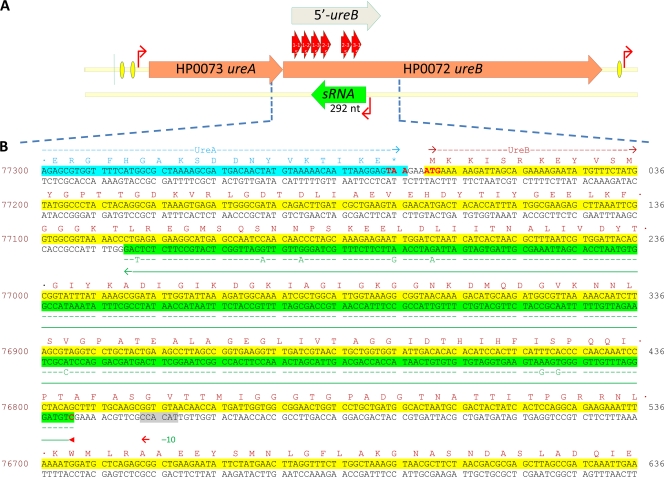
Northern analysis with strand-specific oligonucleotide probes 1-1S and 1-2S did not detect any transcript. However, the ∼290-nt transcript was indeed found with the 1-3S, 2-1S, 2-3S, and 3-1S probes in both H. pylori strain 43504 and H. pylori strain 26695 (Fig. 2C), indicating that this sRNA is expressed at least in these two strains. 5′ RACE experiments with a strand-specific oligonucleotide (1-3S) identified this sRNA as an antisense RNA produced from the complementary strand of ureB in wild-type H. pylori strain 43504 (Fig. 3). After cloning and sequencing of the 5′ RACE product, the length of the sRNA was found to be 292 nt, assuming that the 3′ end of the sRNA is the nucleotide corresponding to the 5′ end of the 1-3S primer. By alignment of the sRNA sequence identified from H. pylori strain 43504 with the corresponding region of the strain 26695 genome sequence, the 5′ end of the sRNA is mapped to nucleotide 76795 in the totally sequenced genome of H. pylori strain 26695. Thus, this sRNA extends from positions 442 to 151 of the coding sequence of ureB (Fig. 3B). The coding sequences for the sRNA from strains 43504 and 26695 share ∼97% identity, with only 8 nucleotides differing between the two strains (Fig. 3B).
FIG. 3.
5′ RACE mapped the antisense sRNA complementary to the 5′ third of ureB. (A) Map of the ureAB gene cluster with an antisense sRNA complementary to the 5′ third of ureB. The 5′-ureB sense RNA probe and the specific oligonucleotide probes (1-1S, 1-2S, 1-3S, 2-1S, 2-3S, and 3-1S) used for identification of sRNA are shown. Red bent arrows denote promoters; orange and green bars with arrows denote ureAB genes and antisense 5′ureB-sRNA genes, respectively. Yellow ovals represent HP0166 binding sites. (B) Gene sequence corresponding to 3′ end of ureA and 5′ region of ureB from H. pylori strain 26695. The coding sequences for ureA and ureB are highlighted in the upper strand in blue and yellow, respectively. The coding sequence for the 5′ureB-sRNA complementary to the 5′ ureB is highlighted in the lower strand in green with an arrowed green line underneath. The −10 promoter sequence is shaded in gray. The amino acid sequences coded by ureA and ureB are also shown on the top of their coding sequences. The alignment of the coding sequence for 5′ureB-sRNA identified from H. pylori strain 43504 is indicated in green letters, and only the nucleotides that are different from the corresponding sequence in strain 26695 are shown. The numbers with regard to the complete genome sequence from strain 26695 are shown on the left side. The numbers with regard to the coding sequence of ureB are shown on the right side.
The expression profiles at pH 7.4 and 4.5 show that antisense sRNA expression is inversely related to the mRNA level of ureAB.
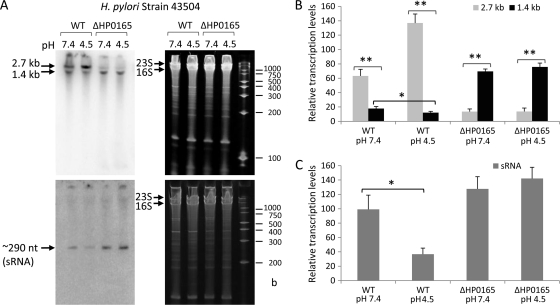
The expression profile of the ∼290-nt 5′ureB-sRNA complementary to 5′-ureB was examined under different pH conditions, given the acid acclimation function of its host genes, ureA and ureB. An overnight culture of wild-type H. pylori (strain 43504) and the HP0165-deficient mutant strain (43504/ΔHP0165::Km) was exposed to pH 7.4 and pH 4.5 for 30 min, and Northern analysis was performed on equal amounts of RNA fractionated in 6% polyacrylamide-urea gels with a strand-specific oligonucleotide sense probe (5′-ureB 2-1S) detecting sRNA and an antisense probe (5′-ureB 2-1AS) detecting ureAB transcripts.
The pH-responsive expression profile (Fig. 4A and B) showed that in the H. pylori wild-type strain, the level of the 1.4-kb truncated ureAB transcript was increased and the level of the intact 2.7-kb transcript was decreased at pH 7.4 compared to the levels at pH 4.5. Meanwhile, at pH 4.5 the level of expression of the 5′ureB-sRNA was significantly decreased compared to that at pH 7.4 (Fig. 4A and C). The increase in the level of the 2.7-kb transcript at pH 4.5 (Fig. 4A and B) is due to activation of PureA by the HP0165-HP0166 TCS.
FIG. 4.
The pH-responsive expression profile of an antisense sRNA in H. pylori by Northern blot analysis. (A) The total RNAs from H. pylori 43504 wild-type (WT) and 43504/ΔHP0165::Km mutant strains were harvested after treatment at pH 7.4 and pH 4.5 for 30 min. RNA samples (5 μg) were separated in 6% polyacrylamide-urea gels and then transferred to a Zeta-Probe GT membrane. The ureAB transcripts were detected with a strand-specific oligonucleotide antisense probe (5′-ureB 2-1AS) (top panels), and the sRNA was detected with an oligonucleotide sense probe (5′-ureB 2-1S) complementary to the 5′ ureB (bottom panels). RNA samples were run alongside RNA Century Marker-Plus size markers (Ambion) with the indicated sizes. The gels (stained with ethidium bromide) are shown as loading controls (right panels). The relative transcript levels normalized to the corresponding intensity of 23S and 16S rRNA for intact 2.7-kb and truncated 1.4-kb ureAB transcripts (B) and ∼290-nt 5′ureB-sRNA (C) from H. pylori 43504 wild-type and 43504/ΔHP0165::Km mutant strains under different pH conditions are shown in the bar graphs. Error bars represent standard deviations from three hybridization experiments with independently prepared RNA samples. *, P < 0.05; **, P < 0.005.
In the HP0165 mutant strain, no pH 4.5-induced increase of the 2.7-kb ureAB transcript level was seen, and the 2.7-kb transcript level was much lower than that in the wild type since there is no activation of PureA by the HP0165-HP0166 TCS. However, the level of the 1.4-kb truncated transcript that results in truncated UreB protein was significantly increased under both pH conditions compared to that in the wild-type strain. As shown in Fig. 4C, the transcript level of 5′ureB-sRNA in the wild-type strain at pH 7.4 was ∼2.5-fold greater than that at pH 4.5, while the transcript level of 5′ureB-sRNA in the HP0165-deficient mutant was significantly higher than that in the wild-type strain at either pH. These results suggest that acid-responsive expression of ureAB is also controlled by a cis-encoded antisense sRNA transcribed from the 5′ ureB noncoding strand, which provides the organism with the ability to downregulate full-length ureAB expression at pH levels that are inimical to survival in the presence of urea. This is primarily regulated at the transcriptional level via the HP0165-HP0166 two-component system.
Effects of 5′ureB-sRNA overexpression on ureAB transcripts.
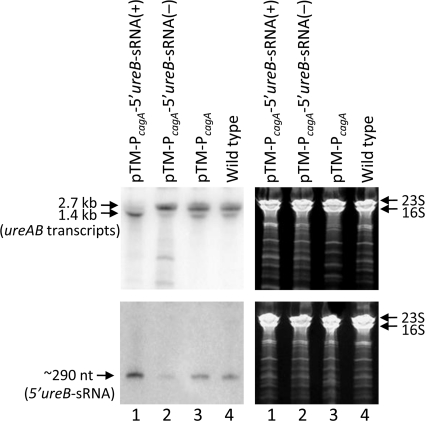
To see if 5′ureB-sRNA has direct effects on the mRNA level of its host gene, ureAB, one of three self-replicating plasmids was introduced into an H. pylori wild-type strain so that normal, high, and very low intracellular levels of 5′ureB-sRNA were generated in H. pylori. To obtain high levels of this antisense sRNA, we transferred a plasmid containing the full sequence of this 5′ureB-sRNA under the control of a relatively strong cagA promoter (55) into the wild-type H. pylori 43504 strain [pTM-PcagA-5′ureB-sRNA(+)]. To deplete this sRNA without introducing changes in the ureAB coding sequence, we also cloned this sRNA in reverse complementary orientation under the control of the cagA promoter and the HP0092-T1 terminator (10) from H. pylori as a signal for the termination of transcription, in essence, expressing an anti-5′ureB-sRNA [pTM-PcagA-5′ureB-sRNA(−)]. The third plasmid served as a control with no additional insertion downstream of cagA promoter (pTM-PcagA). Northern blot analysis verified successful overexpression or downregulation of the 5′ureB-sRNA in transformed cells of H. pylori strain 43504 with pTM-PcagA-5′ureB-sRNA(+) or pTM-PcagA-5′ureB-sRNA(−) (Fig. 5, bottom panels). At pH 7.4, the PcagA-driven 5′ureB-sRNA overexpression resulted in a greatly reduced level of the 2.7-kb ureAB transcript and an increased level of the truncated 1.4-kb transcript (Fig. 5, lanes 1 in top panels). On the other hand, expression of the anti-5′ureB-sRNA led to a decrease in the level of the truncated 1.4-kb ureAB transcript (Fig. 5, lanes 2 in top panels), while the transformed cells with pTM-PcagA showed no significant difference in the levels of 5′ureB-sRNA and ureAB transcripts compared to those for the wild type (Fig. 5, lanes 3 and 4). The 5′ureB-sRNA overexpression and the mutant phenotype in the levels of ureAB transcripts indicated that 5′ureB-sRNA has a direct regulatory effect on ureAB.
FIG. 5.
Effects of increased and decreased 5′ureB-sRNA expression on ureAB mRNA levels. The total RNAs from the H. pylori 43504 wild type, the transformant with pTM-PcagA-5′ureB-sRNA(+) that expressed a high level of 5′ureB-sRNA, the transformant with pTM-PcagA-5′ureB-sRNA(−) that expressed a low level of 5′ureB-sRNA, and control strains with pTM-PcagA were harvested after treatment at pH 7.4 for 30 min. RNA samples (5 μg) were separated in 6% polyacrylamide-urea gels and then transferred to a Zeta-Probe GT membrane. The ureAB transcripts were detected with a strand-specific oligonucleotide antisense probe (5′-ureB 2-1AS) that can also detect anti-5′ureB-sRNA, shown as a lower-molecular-mass band (top panels), and the 5′ureB-sRNA was detected with an oligonucleotide sense probe (5′-ureB 2-1S) complementary to the 5′ ureB (bottom panels). The gels (stained with ethidium bromide) are shown as loading controls (right panels).
Urease regulation by 5′ureB-sRNA.
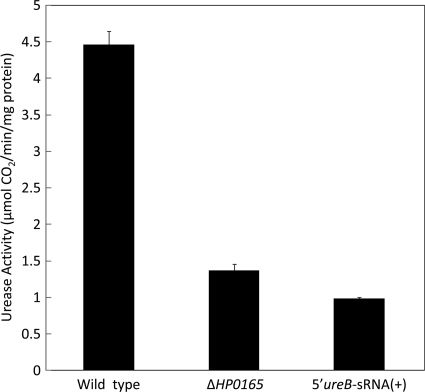
To determine the effect of 5′ureB-sRNA on H. pylori urease activity that is dependent on the transcription products from the ureAB gene, assays for urease from intact bacteria of wild-type H. pylori strain 43504, HP0165-deficient mutant strain (43504/ΔHP0165::Km), and 5′ureB-sRNA-overexpressing strain [43054/pTM-PcagA-5′ureB-sRNA(+)] at neutral pH were done by measuring the amount of 14CO2 released from [14C]urea. As shown in Fig. 6, the overexpression of 5′ureB-sRNA in strain 43054/pTM-PcagA-5′ureB-sRNA(+) resulted in an ∼80% decrease in urease activity compared to that in the wild-type strain. The HP0165 deficiency in mutant strain 43504/ΔHP0165::Km resulted in an ∼70% decrease in urease activity. These results are consistent with those of the Northern blot analysis (Fig. 4 and 5), in which both the mutation for HP0165 deficiency and 5′ureB-sRNA overexpression resulted in a reduced level of the 2.7-kb ureAB intact transcript and an increased level of the truncated 1.4-kb transcript.
FIG. 6.
Urease activity of wild-type H. pylori strain 43503, an HP0165-deficient mutant strain (43504/ΔHP0165::Km), and a 5′ureB-sRNA-overexpressing strain [43054/pTM-PcagA-5′ureB-sRNA(+)]. Intact bacterial cells from each strain were radiometrically assayed at neutral pH for urease activity. Urease activity (micromoles of CO2 liberated per minute per milligram of protein) was calculated by measuring the amount of 14CO2 released from [14C]urea.
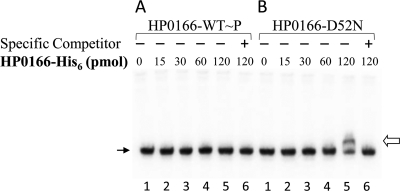
Unphosphorylated HP0166 (HP0166-D52N) binds specifically to the promoter region of 5′ureB-sRNA.
To examine whether a binding site for response regulator HP0166 is present in the promoter region of 5′ureB-sRNA, the purified recombinant HP0166-His6 (73) that was phosphorylated in vitro by carbamylphosphate (HP0166-WT∼P) and a mutated response regulator, HP0166-D52N (73), that is not able to be phosphorylated at the aspartate phosphorylation site were used in the gel mobility shift DNA binding assay with a DNA fragment corresponding to the promoter-regulatory region of 5′ureB-sRNA. As shown in Fig. 7, no gel shift was detected for the phosphorylated HP0166 (HP0166-WT∼P), suggesting that there is no direct interaction between phosphorylated response regulator HP0166 and the promoter region of 5′ureB-sRNA. However, a band with a prominent gel shift was observed when 120 pmol of HP0166-D52N was added to the reaction mixture (Fig. 7B, lane 5). The addition of a 50-fold excess of unlabeled DNA fragment corresponding to the promoter region of 5′ureB-sRNA prevented DNA binding of the radiolabeled probe (Fig. 7B, lane 6). These results show that the unphosphorylated response regulator HP0166 binds specifically to the promoter region of 5′ureB-sRNA.
FIG. 7.
Identification of the direction interaction between response regulator HP0166 and the promoter region of 5′ureB-sRNA. The DNA fragment for the promoter region of 5′ureB-sRNA was labeled with [γ-32P]ATP. Labeled probe was incubated in the absence (lane 1) or presence of increasing amounts of phosphorylated wild-type HP0166-His6 (HP0166-WT∼P) or nonphosphorylatable mutated protein HP0166-D52N (15, 30, 60, and 120 pmol, lanes 2 to 5, respectively) or in the presence of both HP0166-His6 (120 pmol, either HP0166-WT∼P or HP0166-D52N) and a 50-fold excess of unlabeled probe as a specific competitor (lane 6). The open and line arrows indicate the shifted band and free probes, respectively.
DISCUSSION
Transcripts from the urease gene cluster of H. pylori are cleaved to produce several species of mRNA (3). Hence, a pH-dependent posttranscriptional regulatory mechanism for urease gene expression by mRNA decay has been suggested (3). However, no explanation as to how urease gene transcripts are regulated at a posttranscriptional level and how ureAB mRNA decay is facilitated has been offered.
In the current study, we provide evidence, for the first time, for a cis-encoded antisense sRNA (5′ureB-sRNA) acting on the expression of ureAB at the posttranscriptional level by targeted degradation of ureAB mRNA, resulting in truncation of the 3′ region of ureB of the ureAB transcript. Expression of 5′ureB-sRNA is negatively regulated by the HP0165-HP0166 TCS in response to an acidic environment and is positively regulated at neutral pH with converse changes in the length of the ureAB transcript.
Using Northern blot analysis with sense RNA probes and strand-specific oligonucleotide probes followed by a 5′ RACE, we identified a 5′ureB-sRNA that has a specific target in its host gene mRNA because it is fully complementary to the 5′ third of the ureB gene over its full length. The predicted −10 motif sequence (tgtTACACC, lowercase letters indicate more variable nucleotides) of σ80 in H. pylori shows a strong similarity to the extended Pribnow box (tgnTAtaAT) identified in a primary transcriptome study (54). This is located immediately upstream of the transcription start site of the 5′ureB-sRNA (Fig. 3B). Therefore, this sRNA appears to be a primary transcript rather than a processed transcript by imperfect termination. The ureB mRNA is transcribed as the second transcript together with ureA from a single bicistronic operon, ureAB, driven by the promoter PureA that produces a 2.7-kb intact ureAB transcript in exponentially growing H. pylori (3). Overexpression of 5′ureB-sRNA results in a significant increase in the level of the 1.4-kb truncated ureAB transcript and a decrease in the level of the 2.7-kb intact ureAB transcript and leads to reduced urease activity. Apparently, the 5′ureB-sRNA interacts by base pairing with a coding region of the ureB transcript which does not include the Shine-Dalgarno sequence (Fig. 3). Therefore, it is very unlikely that this sRNA interferes with the initiation of translation through competition with 16S rRNA for the Shine-Dalgarno sequence, which is the usual case for trans-encoded regulatory sRNAs in bacteria (6, 26). Here, the transient formation of sRNA-ureB mRNA duplexes makes them a target for selective downstream degradation, which results in a 1.4-kb truncated ureAB transcript that lacks most of the ureB transcript from the 3′ end. Base pairing of the sRNA can target RNase to the region and result in mRNA cleavage or transcriptional termination, leading to reduced levels of downstream gene transcripts (70). Although the final product of the antisense sRNA-ureB mRNA binding is a full duplex that may be degraded by RNase III (29, 65) or RNase E (36), the detailed mechanism that affects mRNA stability by a cis-encoded antisense sRNA has not yet been defined.
The reciprocally regulated transcription level of the 5′ureB-sRNA compared to that of its target mRNA (ureAB) under neutral and acidic pH conditions indicates a targeted degradation of ureB mRNA mediated by the 5′ureB-sRNA. As shown in Fig. 4, the 5′ureB-sRNA is more efficiently transcribed at pH 7.4 in the wild-type H. pylori strain, which initiates the coupled degradation of the ureB transcript, resulting in the observed increase in the level of truncated 1.4-kb ureAB transcript. On the other hand, the transcription level of the 5′ureB-sRNA is decreased at pH 4.5 and the level of the intact 2.7-kb ureAB transcript is increased, partially due to the decrease of targeted degradation of ureB transcripts mediated by the 5′ureB-sRNA. These effects are much exaggerated in the strain with the HP0165 deletion, where the 5′ureB-sRNA level remains high independent of pH. This is associated with an increase in the level of the 1.4-kb truncated ureB and a decrease in the level of the 2.7-kb transcript, as found in the wild type at pH 7.4.
A very limited number of cis-encoded antisense sRNAs in bacteria have been reported, because most methods to identify regulatory RNAs in bacteria used so far (5, 45, 69) have been focused on intergenic regions. Most of the cis-encoded antisense sRNAs were found to be expressed on bacteriophage plasmids and transposons and function to maintain the appropriate copy number of the mobile element (7, 68). However, some of the cis-encoded antisense sRNAs were also found to be expressed on bacterial chromosomes (7). For example, in E. coli, base pairing between the stationary-phase-induced GadY antisense sRNA and the mRNA from acid response gene gadXW leads to cleavage of the duplex between the gadX and gadW genes and increased levels of a gadX transcript by a targeted stabilization (38, 59). In Synechocystis species, the iron stress-repressed IsrR antisense sRNA base pairs with sequences within the isiA coding region of the isiAB transcript, leading to decreased levels of an isiA transcript by a targeted degradation (19).
TCSs allow bacteria to monitor diverse environmental cues and to adjust gene expression accordingly at the transcriptional level (27, 56). Much evidence showing that the expression of a subset of small RNAs is regulated by TCSs has accumulated (61), thus expanding the scope of genetic control exerted by TCSs. For example, posttranscriptional modulation of porin (including OmpF, OmpC, and OmpA) levels in E. coli is finely tuned by a set of sRNAs (including MicF, MicC, and MicA) (2, 12, 16, 42) as a result of different stimuli channeled through the osmosensory EnvZ/OmpR two-component system (76), the SoxR/SoxS system for monitoring oxidative stress (17), and the extracytoplasmic stress-responsive pathway that determines the intracellular levels of the sigma factor σE (18).
The dRNA-seq studies with wild-type H. pylori strain 26695 found that the H. pylori transcriptome is complicated and predicted hundreds of sRNA candidates from intergenic regions, regions antisense to ORFs, and also sense regions within ORFs (54). However, the cis-encoded antisense sRNA from the 5′ ureB region that we find in this study was not found by the dRNA-seq approach either at mid-logarithmic phase or under acid stress (pH 5.2) (54). It is likely that the expression level of this 5′ureB-sRNA is too low in the wild-type H. pylori strain at neutral pH and even lower at acidic pH (Fig. 4) for it to be distinguished from the background level by the global analysis of the transcription start site set that was the basis of the primary transcriptome studies. A significantly higher transcript level of this antisense sRNA is found in HP0165-deficient mutant under both pH conditions (Fig. 4C), because the expression of this sRNA is negatively regulated by the HP0165-HP0166 TCS. Using EMSA, we also identified a direct interaction between the potential promoter region of 5′ureB-sRNA and the unphosphorylated response regulator HP0166. This confirmed that 5′ureB-sRNA is regulated by the HP0165-HP0166 TCS. Furthermore, this is the first time that a specific regulatory role by unphosphorylated response regulator HP0166 was identified, even though previous studies have suggested that unphosphorylated response regulator HP0166 may play an essential role in H. pylori growth (50). Perhaps at neutral pH or in HP0165-deficient mutants most of HP0166 is unphosphorylated and therefore binds the promoter region of 5′ureB-sRNA and consequently induces the expression of 5′ureB-sRNA, leading to a downregulation of urease activity.
The transcriptional induction of urease genes in H. pylori is regulated mainly by the HP0165-HP0166 TCS (39, 41). The histidine kinase sensor senses low pH due to a histidine residue 94 (H94) in the periplasmic input domain (37) and triggers autophosphorylation of HP0165 and the subsequent phosphorylation of its cognate response regulator HP0166. The phosphorylated response regulator then binds to extended regions overlapping the PureA and PureI promoters (41) to positively regulate the transcription of urease genes. Now we have shown that deletion of HP0165 prevents the increased expression of the 2.7-kb intact ureAB transcript at pH 4.5 and elevates the level of the 1.4-kb truncated transcript (Fig. 4A and B). This confirms that this TCS can act both as an activator of urease gene expression and as a repressor of transcription of the 5′ureB-sRNA which base pairs with the 5′ureB region of the ureAB transcript, leading to the posttranscriptional truncation of ureB mRNA.
Much is now known about the response of H. pylori to acidic conditions in the presence of urea. Cytoplasmic urea hydrolysis by urease produces ammonia and carbonic acid. CO2 is formed from this carbonic acid in the cytoplasm and is catalyzed by cytoplasmic β-carbonic anhydrase. The cytoplasmic CO2 produced diffuses rapidly through UreI and less so through the inner membrane into the periplasm where bicarbonate is produced due to periplasmic α-carbonic anhydrase (72). The HCO3− maintains the periplasm at pH 6.1 over a wide range of medium pHs between 6.2 and 2.5 (44, 52, 53) and is aided by buffering of entering protons by NH3. The activity of intrabacterial urease is controlled by the inner membrane pH-gated channel UreI, which increases the access of the substrate, urea, to the cytoplasm of the bacterial cell in response to acidic pH (8, 43, 51, 71). Urease, the channel protein, UreI, and both carbonic anhydrases are essential for gastric colonization in animal models of H. pylori infection (20, 21, 35, 60).
The data presented above show that H. pylori also has a mechanism for downregulation of urease that enables it to adapt to a neutral pH in the face of the constant presence of urea in gastric juice using the same TCS that activates urease gene cluster expression in response to an acidic pH. This adaptation to low acidity is the converse of acid acclimation, defined by the changes induced by transition from neutral to acidic pH.
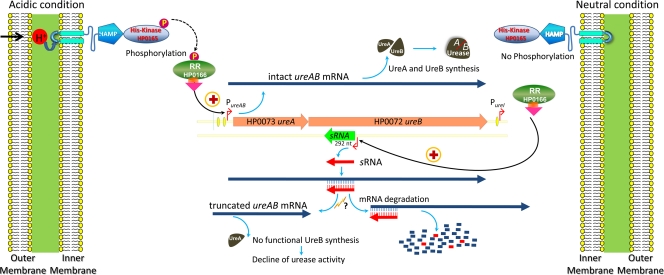
In conclusion, while several different mechanisms that upregulate expression of ureAB have been identified, we have now found a mechanism, also controlled by the HP0165-HP0166 TCS, which downregulates ureB expression mediated by a cis-encoded antisense sRNA transcribed from the 5′ ureB noncoding strand in response to elevated environmental pH. These concepts are presented in the model (Fig. 8). This posttranscriptional regulatory mechanism allows downregulation of urease expression in response to neutral external pH while maintaining sufficient UreA levels to provide a faster change of urease activity when acidic pH is reencountered. This results in a response of urease levels not only to acidic pH but also to relatively neutral pH in H. pylori. This ability of H. pylori to both up- and downregulate urease activity presumably facilitates continued colonization of the human stomach under widely different intragastric pH conditions. Furthermore, this mechanism may also enhance transmission of the infection where neutral pH is encountered prior to gastric access.
FIG. 8.
Simplified model representing the HP0165-HP0166 TCS regulation of ureAB gene expression by unphosphorylated HP0166 on 5′ureB-sRNA under neutral pH conditions and by phosphorylated HP0166 on PureA under acidic conditions. Under neutral conditions, HP0165 is not activated and the response regulator HP0166 is not phosphorylated. The unphosphorylated HP0166 binds to the 5′ureB-sRNA promoter, leading to transcription of 5′ureB-sRNA and consequent truncation of ureB, which results in a decline of urease activity. This reflects adaptation to nonacidic pH. In acidic medium, HP0165 is activated with phosphorylation of HP0166 and the phosphorylated HP0166 and then binds to the PureA promoter to positively regulate the transcription of ureAB genes, which results in upregulation of ureA and ureB with a consequent increase of urease activity, reflecting acid acclimation. Antisense sRNAs are shown in red, and mRNAs are shown in dark blue thick-line arrows. Red bent arrows denote promoters, and orange and green bars with arrows denote the ureAB gene and antisense 5′ureB-sRNA gene, respectively. A lightning sign indicates the action of RNase III or RNase E. Plus signs denote positive regulation; a question mark denotes not yet experimentally confirmed. Yellow ovals represent HP0166 binding sites.
Acknowledgments
This work was supported by U.S. Department of Veterans Affairs and NIH grants DK46917, 53462, and 58333.
Thanks are due to Olga Vagin and Jeffrey Kraut for reading the manuscript.
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Aiba, H. 2007. Mechanism of RNA silencing by Hfq-binding small RNAs. Curr. Opin. Microbiol. 10:134-139. [DOI] [PubMed] [Google Scholar]
- 2.Aiba, H., S. Matsuyama, T. Mizuno, and S. Mizushima. 1987. Function of micF as an antisense RNA in osmoregulatory expression of the ompF gene in Escherichia coli. J. Bacteriol. 169:3007-3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akada, J. K., M. Shirai, H. Takeuchi, M. Tsuda, and T. Nakazawa. 2000. Identification of the urease operon in Helicobacter pylori and its control by mRNA decay in response to pH. Mol. Microbiol. 36:1071-1084. [DOI] [PubMed] [Google Scholar]
- 4.Altuvia, S., A. Zhang, L. Argaman, A. Tiwari, and G. Storz. 1998. The Escherichia coli OxyS regulatory RNA represses fhlA translation by blocking ribosome binding. EMBO J. 17:6069-6075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Argaman, L., R. Hershberg, J. Vogel, G. Bejerano, E. G. Wagner, H. Margalit, and S. Altuvia. 2001. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr. Biol. 11:941-950. [DOI] [PubMed] [Google Scholar]
- 6.Brantl, S. 2002. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta 1575:15-25. [DOI] [PubMed] [Google Scholar]
- 7.Brantl, S. 2007. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 10:102-109. [DOI] [PubMed] [Google Scholar]
- 8.Bury-Mone, S., J. M. Thiberge, M. Contreras, A. Maitournam, A. Labigne, and H. De Reuse. 2004. Responsiveness to acidity via metal ion regulators mediates virulence in the gastric pathogen Helicobacter pylori. Mol. Microbiol. 53:623-638. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, B. M., T. K. McDaniel, J. M. Whitmire, H. Gancz, S. Guidotti, S. Censini, and D. S. Merrell. 2007. Expanding the Helicobacter pylori genetic toolbox: modification of an endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl. Environ. Microbiol. 73:7506-7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castillo, A. R., S. S. Arevalo, A. J. Woodruff, and K. M. Ottemann. 2008. Experimental analysis of Helicobacter pylori transcriptional terminators suggests this microbe uses both intrinsic and factor-dependent termination. Mol. Microbiol. 67:155-170. [DOI] [PubMed] [Google Scholar]
- 11.Chen, Q., and J. H. Crosa. 1996. Antisense RNA, fur, iron, and the regulation of iron transport genes in Vibrio anguillarum. J. Biol. Chem. 271:18885-18891. [DOI] [PubMed] [Google Scholar]
- 12.Chen, S., A. Zhang, L. B. Blyn, and G. Storz. 2004. MicC, a second small-RNA regulator of Omp protein expression in Escherichia coli. J. Bacteriol. 186:6689-6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Church, G. M., and W. Gilbert. 1984. Genomic sequencing. Proc. Natl. Acad. Sci. U. S. A. 81:1991-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clyne, M., A. Labigne, and B. Drumm. 1995. Helicobacter pylori requires an acidic environment to survive in the presence of urea. Infect. Immun. 63:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cui, Y., A. Chatterjee, and A. K. Chatterjee. 2001. Effects of the two-component system comprising GacA and GacS of Erwinia carotovora subsp. carotovora on the production of global regulatory rsmB RNA, extracellular enzymes, and harpinEcc. Mol. Plant Microbe Interact. 14:516-526. [DOI] [PubMed] [Google Scholar]
- 16.Delihas, N., and S. Forst. 2001. MicF: an antisense RNA gene involved in response of Escherichia coli to global stress factors. J. Mol. Biol. 313:1-12. [DOI] [PubMed] [Google Scholar]
- 17.Demple, B. 1996. Redox signaling and gene control in the Escherichia coli soxRS oxidative stress regulon—a review. Gene 179:53-57. [DOI] [PubMed] [Google Scholar]
- 18.Douchin, V., C. Bohn, and P. Bouloc. 2006. Down-regulation of porins by a small RNA bypasses the essentiality of the regulated intramembrane proteolysis protease RseP in Escherichia coli. J. Biol. Chem. 281:12253-12259. [DOI] [PubMed] [Google Scholar]
- 19.Duhring, U., I. M. Axmann, W. R. Hess, and A. Wilde. 2006. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl. Acad. Sci. U. S. A. 103:7054-7058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eaton, K. A., and S. Krakowka. 1994. Effect of gastric pH on urease-dependent colonization of gnotobiotic piglets by Helicobacter pylori. Infect. Immun. 62:3604-3607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ferrero, R. L., V. Cussac, P. Courcoux, and A. Labigne. 1992. Construction of isogenic urease-negative mutants of Helicobacter pylori by allelic exchange. J. Bacteriol. 174:4212-4217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Foster, J. W. 2004. Escherichia coli acid resistance: tales of an amateur acidophile. Nat. Rev. Microbiol. 2:898-907. [DOI] [PubMed] [Google Scholar]
- 23.Geissmann, T., M. Possedko, E. Huntzinger, P. Fechter, C. Ehresmann, and P. Romby. 2006. Regulatory RNAs as mediators of virulence gene expression in bacteria. Handb. Exp. Pharmacol. 2010:9-43. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez, N., S. Heeb, C. Valverde, E. Kay, C. Reimmann, T. Junier, and D. Haas. 2008. Genome-wide search reveals a novel GacA-regulated small RNA in Pseudomonas species. BMC Genomics 9:167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gottesman, S. 2005. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 21:399-404. [DOI] [PubMed] [Google Scholar]
- 26.Gottesman, S. 2004. The small RNA regulators of Escherichia coli: roles and mechanisms. Annu. Rev. Microbiol. 58:303-328. [DOI] [PubMed] [Google Scholar]
- 27.Groisman, E. A., and C. Mouslim. 2006. Sensing by bacterial regulatory systems in host and non-host environments. Nat. Rev. Microbiol. 4:705-709. [DOI] [PubMed] [Google Scholar]
- 28.Hershberg, R., S. Altuvia, and H. Margalit. 2003. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 31:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Huntzinger, E., S. Boisset, C. Saveanu, Y. Benito, T. Geissmann, A. Namane, G. Lina, J. Etienne, B. Ehresmann, C. Ehresmann, A. Jacquier, F. Vandenesch, and P. Romby. 2005. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 24:824-835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Livny, J., and M. K. Waldor. 2007. Identification of small RNAs in diverse bacterial species. Curr. Opin. Microbiol. 10:96-101. [DOI] [PubMed] [Google Scholar]
- 31.Majdalani, N., C. Cunning, D. Sledjeski, T. Elliott, and S. Gottesman. 1998. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl. Acad. Sci. U. S. A. 95:12462-12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matz, M., D. Shagin, D. Shagin, O. Britanova, S. Lukyanov, L. Diatchenko, and A. Chenchik. 1999. Amplification of cDNA ends based on template-switching effect and step-out PCR. Nucleic Acids Res. 27:1558-1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Meyer-Rosberg, K., D. R. Scott, D. Rex, K. Melchers, and G. Sachs. 1996. The effect of environmental pH on the proton motive force of Helicobacter pylori. Gastroenterology 111:886-900. [DOI] [PubMed] [Google Scholar]
- 34.Mobley, H. L., M. D. Island, and R. P. Hausinger. 1995. Molecular biology of microbial ureases. Microbiol. Rev. 59:451-480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mollenhauer-Rektorschek, M., G. Hanauer, G. Sachs, and K. Melchers. 2002. Expression of UreI is required for intragastric transit and colonization of gerbil gastric mucosa by Helicobacter pylori. Res. Microbiol. 153:659-666. [DOI] [PubMed] [Google Scholar]
- 36.Morita, T., K. Maki, and H. Aiba. 2005. RNase E-based ribonucleoprotein complexes: mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 19:2176-2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muller, S., M. Gotz, and D. Beier. 2009. Histidine residue 94 is involved in pH sensing by histidine kinase ArsS of Helicobacter pylori. PLoS One 4:e6930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Opdyke, J. A., J. G. Kang, and G. Storz. 2004. GadY, a small-RNA regulator of acid response genes in Escherichia coli. J. Bacteriol. 186:6698-6705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pflock, M., P. Dietz, J. Schar, and D. Beier. 2004. Genetic evidence for histidine kinase HP165 being an acid sensor of Helicobacter pylori. FEMS Microbiol. Lett. 234:51-61. [DOI] [PubMed] [Google Scholar]
- 40.Pflock, M., N. Finsterer, B. Joseph, H. Mollenkopf, T. F. Meyer, and D. Beier. 2006. Characterization of the ArsRS regulon of Helicobacter pylori, involved in acid adaptation. J. Bacteriol. 188:3449-3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pflock, M., S. Kennard, I. Delany, V. Scarlato, and D. Beier. 2005. Acid-induced activation of the urease promoters is mediated directly by the ArsRS two-component system of Helicobacter pylori. Infect. Immun. 73:6437-6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rasmussen, A. A., M. Eriksen, K. Gilany, C. Udesen, T. Franch, C. Petersen, and P. Valentin-Hansen. 2005. Regulation of ompA mRNA stability: the role of a small regulatory RNA in growth phase-dependent control. Mol. Microbiol. 58:1421-1429. [DOI] [PubMed] [Google Scholar]
- 43.Rektorschek, M., A. Buhmann, D. Weeks, D. Schwan, K. W. Bensch, S. Eskandari, D. Scott, G. Sachs, and K. Melchers. 2000. Acid resistance of Helicobacter pylori depends on the UreI membrane protein and an inner membrane proton barrier. Mol. Microbiol. 36:141-152. [DOI] [PubMed] [Google Scholar]
- 44.Rektorschek, M., D. Weeks, G. Sachs, and K. Melchers. 1998. Influence of pH on metabolism and urease activity of Helicobacter pylori. Gastroenterology 115:628-641. [DOI] [PubMed] [Google Scholar]
- 45.Rivas, E., and S. R. Eddy. 2001. Noncoding RNA gene detection using comparative sequence analysis. BMC Bioinformatics 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Romby, P., F. Vandenesch, and E. G. Wagner. 2006. The role of RNAs in the regulation of virulence-gene expression. Curr. Opin. Microbiol. 9:229-236. [DOI] [PubMed] [Google Scholar]
- 47.Sachs, G., D. L. Weeks, K. Melchers, and D. R. Scott. 2003. The gastric biology of Helicobacter pylori. Annu. Rev. Physiol. 65:349-369. [DOI] [PubMed] [Google Scholar]
- 48.Sachs, G., D. L. Weeks, Y. Wen, E. A. Marcus, D. R. Scott, and K. Melchers. 2005. Acid acclimation by Helicobacter pylori. Physiology (Bethesda) 20:429-438. [DOI] [PubMed] [Google Scholar]
- 49.Salama, N. R., G. Otto, L. Tompkins, and S. Falkow. 2001. Vacuolating cytotoxin of Helicobacter pylori plays a role during colonization in a mouse model of infection. Infect. Immun. 69:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schar, J., A. Sickmann, and D. Beier. 2005. Phosphorylation-independent activity of atypical response regulators of Helicobacter pylori. J. Bacteriol. 187:3100-3109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Scott, D. R., E. A. Marcus, D. L. Weeks, A. Lee, K. Melchers, and G. Sachs. 2000. Expression of the Helicobacter pylori ureI gene is required for acidic pH activation of cytoplasmic urease. Infect. Immun. 68:470-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scott, D. R., E. A. Marcus, D. L. Weeks, and G. Sachs. 2002. Mechanisms of acid resistance due to the urease system of Helicobacter pylori. Gastroenterology 123:187-195. [DOI] [PubMed] [Google Scholar]
- 53.Scott, D. R., D. Weeks, C. Hong, S. Postius, K. Melchers, and G. Sachs. 1998. The role of internal urease in acid resistance of Helicobacter pylori. Gastroenterology 114:58-70. [DOI] [PubMed] [Google Scholar]
- 54.Sharma, C. M., S. Hoffmann, F. Darfeuille, J. Reignier, S. Findeiss, A. Sittka, S. Chabas, K. Reiche, J. Hackermuller, R. Reinhardt, P. F. Stadler, and J. Vogel. 2010. The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 464:250-255. [DOI] [PubMed] [Google Scholar]
- 55.Spohn, G., D. Beier, R. Rappuoli, and V. Scarlato. 1997. Transcriptional analysis of the divergent cagAB genes encoded by the pathogenicity island of Helicobacter pylori. Mol. Microbiol. 26:361-372. [DOI] [PubMed] [Google Scholar]
- 56.Stock, A. M., V. L. Robinson, and P. N. Goudreau. 2000. Two-component signal transduction. Annu. Rev. Biochem. 69:183-215. [DOI] [PubMed] [Google Scholar]
- 57.Storz, G., and S. Gottesman. 2006. The RNA world, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 58.Tomb, J. F., O. White, A. R. Kerlavage, R. A. Clayton, G. G. Sutton, R. D. Fleischmann, K. A. Ketchum, H. P. Klenk, S. Gill, B. A. Dougherty, K. Nelson, J. Quackenbush, L. Zhou, E. F. Kirkness, S. Peterson, B. Loftus, D. Richardson, R. Dodson, H. G. Khalak, A. Glodek, K. McKenney, L. M. Fitzegerald, N. Lee, M. D. Adams, J. C. Venter, et al. 1997. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature 388:539-547. [DOI] [PubMed] [Google Scholar]
- 59.Tramonti, A., M. De Canio, and D. De Biase. 2008. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol. Microbiol. 70:965-982. [DOI] [PubMed] [Google Scholar]
- 60.Tsuda, M., M. Karita, M. G. Morshed, K. Okita, and T. Nakazawa. 1994. A urease-negative mutant of Helicobacter pylori constructed by allelic exchange mutagenesis lacks the ability to colonize the nude mouse stomach. Infect. Immun. 62:3586-3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Valverde, C., and D. Haas. 2008. Small RNAs controlled by two-component systems. Adv. Exp. Med. Biol. 631:54-79. [DOI] [PubMed] [Google Scholar]
- 62.Valverde, C., S. Heeb, C. Keel, and D. Haas. 2003. RsmY, a small regulatory RNA, is required in concert with RsmZ for GacA-dependent expression of biocontrol traits in Pseudomonas fluorescens CHA0. Mol. Microbiol. 50:1361-1379. [DOI] [PubMed] [Google Scholar]
- 63.van Vliet, A. H., E. J. Kuipers, B. Waidner, B. J. Davies, N. de Vries, C. W. Penn, C. M. Vandenbroucke-Grauls, M. Kist, S. Bereswill, and J. G. Kusters. 2001. Nickel-responsive induction of urease expression in Helicobacter pylori is mediated at the transcriptional level. Infect. Immun. 69:4891-4897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Vliet, A. H., S. W. Poppelaars, B. J. Davies, J. Stoof, S. Bereswill, M. Kist, C. W. Penn, E. J. Kuipers, and J. G. Kusters. 2002. NikR mediates nickel-responsive transcriptional induction of urease expression in Helicobacter pylori. Infect. Immun. 70:2846-2852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vogel, J., L. Argaman, E. G. Wagner, and S. Altuvia. 2004. The small RNA IstR inhibits synthesis of an SOS-induced toxic peptide. Curr. Biol. 14:2271-2276. [DOI] [PubMed] [Google Scholar]
- 66.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vogel, J., and E. G. Wagner. 2007. Target identification of small noncoding RNAs in bacteria. Curr. Opin. Microbiol. 10:262-270. [DOI] [PubMed] [Google Scholar]
- 68.Wagner, E. G., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46:361-398. [DOI] [PubMed] [Google Scholar]
- 69.Wassarman, K. M., F. Repoila, C. Rosenow, G. Storz, and S. Gottesman. 2001. Identification of novel small RNAs using comparative genomics and microarrays. Genes Dev. 15:1637-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Waters, L. S., and G. Storz. 2009. Regulatory RNAs in bacteria. Cell 136:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Weeks, D. L., S. Eskandari, D. R. Scott, and G. Sachs. 2000. A H+-gated urea channel: the link between Helicobacter pylori urease and gastric colonization. Science 287:482-485. [DOI] [PubMed] [Google Scholar]
- 72.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2007. HP0165-HP0166 two-component system (ArsRS) regulates the acid-induced expression of HP1186 {alpha}-carbonic anhydrase in Helicobacter pylori by activating the pH-dependent promoter. J. Bacteriol. 189:2426-2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2006. Involvement of the HP0165-HP0166 two-component system in expression of some acidic-pH-upregulated genes of Helicobacter pylori. J. Bacteriol. 188:1750-1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wen, Y., J. Feng, D. R. Scott, E. A. Marcus, and G. Sachs. 2009. The pH-responsive regulon of HP0244 (FlgS), the cytoplasmic histidine kinase of Helicobacter pylori. J. Bacteriol. 191:449-460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wilderman, P. J., N. A. Sowa, D. J. FitzGerald, P. C. FitzGerald, S. Gottesman, U. A. Ochsner, and M. L. Vasil. 2004. Identification of tandem duplicate regulatory small RNAs in Pseudomonas aeruginosa involved in iron homeostasis. Proc. Natl. Acad. Sci. U. S. A. 101:9792-9797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yoshida, T., L. Qin, L. A. Egger, and M. Inouye. 2006. Transcription regulation of ompF and ompC by a single transcription factor, OmpR. J. Biol. Chem. 281:17114-17123. [DOI] [PubMed] [Google Scholar]
- 77.Zhu, Y. Y., E. M. Machleder, A. Chenchik, R. Li, and P. D. Siebert. 2001. Reverse transcriptase template switching: a SMART approach for full-length cDNA library construction. Biotechniques 30:892-897. [DOI] [PubMed] [Google Scholar]