Abstract
Accumulation of certain nonmetabolizable sugar-phosphates (including α-methyl glucoside-6-phosphate) in Escherichia coli is growth inhibitory and elicits the glucose-phosphate stress response. The transcription factor SgrR activates transcription of the small RNA SgrS under stress conditions. SgrS represses translation of mRNAs encoding sugar transporters. The sgrR and sgrS genes are located directly upstream of setA, and this gene organization is conserved in numerous enteric species, prompting the hypothesis that SetA contributes to the glucose-phosphate stress response. SetA is a proton motive force-driven efflux pump capable of transporting various sugars and sugar analogs in vitro. This study demonstrates that setA expression is induced in response to glucose-phosphate stress, and this requires SgrR. Under stress conditions, setA is cotranscribed with sgrS from the sgrS promoter. A setA mutant exhibits a growth defect under stress conditions that can be complemented by setA in trans, suggesting that SetA contributes to the optimal cellular recovery from stress. Despite previous in vitro evidence that SetA can promote efflux of the stress-causing glucose analog α-methyl glucoside, in vivo data in this study indicate that SetA is not the major efflux pump responsible for removal of α-methyl glucoside under stress conditions.
The genome of Escherichia coli encodes three members of the SET (sugar efflux transporter) family, which was identified 10 years ago as a subfamily of the major facilitator superfamily (16). These are SetA, SetB, and SetC, which are also encoded in numerous other enterobacterial genomes (16). A fourth member, SotA, is encoded in the Dickeya dadantii (formerly Erwinia chrysanthemi) genome (24, 30). SetA was originally identified in E. coli as a multicopy suppressor of the IPTG (isopropyl-β-d-thiogalactopyranoside)-induced toxicity associated with overproduction of a membrane protein, and this suppression was due to SetA pumping IPTG out of the cells (16). In vitro studies of SetA revealed that it is a proton motive force-driven efflux pump capable of transporting a wide range of substrates, including various sugars and sugar analogs (17). Despite this information regarding its biochemistry, prior to this study, nothing was known about the conditions that induce the expression of setA or its true physiological function in vivo. Previously, the only in vivo information regarding SET family members was limited to sotA of D. dadantii: the transcription of sotA is activated by the cyclic AMP (cAMP) receptor protein (CRP) and weakly repressed by the 2-keto-3-deoxygluconate repressor KdgR (4, 29). Nevertheless, based on the biochemical characterization (17), it was plausible to speculate that SetA and its homologs might function in vivo as sugar efflux pumps with roles in carbohydrate metabolism or metabolic stress.
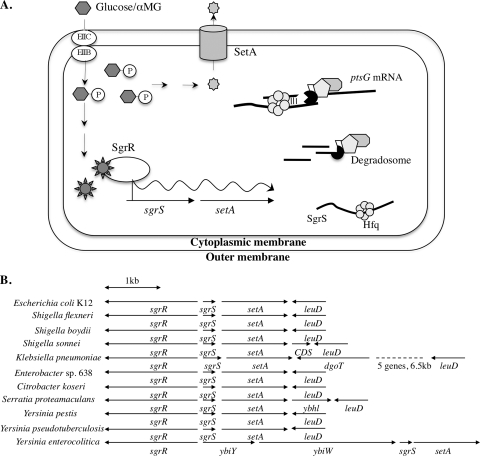
One example of carbohydrate-related metabolic stress in E. coli is referred to as glucose-phosphate stress (7, 9-12, 15, 32, 40, 42), which can be induced by exposing wild-type (WT) E. coli cells to the nonmetabolizable glucose analog α-methyl glucoside (αMG). Under glucose-phosphate stress conditions, E. coli cells initiate a unique cellular response mediated by the transcription factor SgrR, which activates transcription of a small RNA (sRNA), SgrS (Fig. 1 A) (32-34). When expressed under stress, SgrS base pairs with the ptsG mRNA, which encodes EIICBGlc, the phosphoenolpyruvate phosphotranferase system (PTS) protein responsible for transport of both glucose and αMG. The base-pairing interaction results in translation inhibition and degradation of the ptsG mRNA (13, 14, 21, 32, 33). The physiological significance of this regulation is that SgrS stops new synthesis of sugar transporters under conditions where sugar-phosphates are not being metabolized (32). SgrS also encodes the small protein SgrT, which inhibits uptake of glucose via an unknown mechanism (37).
FIG. 1.
Involvement of SgrS and SetA in the glucose-phosphate stress response. (A) Model of glucose-phosphate stress response in E. coli. (B) Organization of genes in the sgrR-sgrS-setA chromosomal region in organisms related to E. coli K-12. The directions of gene transcription are indicated by the arrows.
Stress-responsive activation of sgrS requires the transcription factor SgrR, which is encoded by the sgrR gene divergently transcribed from sgrS (Fig. 1B) (34). The sgrR and sgrS genes are located upstream of setA on the chromosome, and this gene organization is conserved among numerous enteric species (Fig. 1B) (8), suggesting that SetA may play a role in the glucose-phosphate stress response. The very short intergenic region between sgrS and setA (27 nucleotides [nt] in the E. coli K-12 genome) also suggested the possibility that the two genes might form an operon.
In the present study, we sought to characterize the regulation and physiological function of SetA in the glucose-phosphate stress response. We determined that under stress conditions, setA is coexpressed with sgrS in an SgrR-dependent manner. The setA mutant has a growth defect under stress conditions that is medium dependent. The growth defect can be complemented by SetA in trans, confirming that SetA contributes to optimal cellular recovery from stress under some circumstances. The setA mutation also affects the stress-responsive activation of sgrS in a medium-specific manner, suggesting that SetA activity affects signaling through SgrR. Efflux assays using radiolabeled αMG indicate that SetA does not play a significant role in directly pumping out this sugar analog. Together, these data suggest that SetA plays a complex role in the stress response.
MATERIALS AND METHODS
Strain and plasmid construction.
Most strains used for this study are derivatives of DJ480 (D. Jin, National Cancer Institute) and are listed in Table 1. The sequences of all oligonucleotides (Integrated DNA Technologies) used in the construction of mutant strains and plasmids are listed in Table 2.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| MG1655 | Wild-type E.coli K-12 | D. Jin (NCI) |
| DJ480 | MG1655 Δlac X74 | D. Jin (NCI) |
| BAH100 | DJ480 imm21sgrS′-lacZ | This study |
| BAH101 | DJ480 imm21sgrS′-lacZ, ΔsgrR::cm | This study |
| CS168 | DJ480 λattB::lacIqtetR Spr | This study |
| CV107 | DJ480 ΔptsG ΔmanXYZ ΔsgrS | C. K. Vanderpool (NIH) |
| CV700 | DJ480 ΔsgrR::cm | 33 |
| ST101 | DJ480 ΔsetA::cm | This study |
| YS131 | DJ480 setA′-lacZ | This study |
| YS133 | DJ480 setA′-lacZ ΔsgrR::cm | This study |
| YS135 | DJ480 sub1 | This study |
| YS137 | DJ480 sub2 | This study |
| YS140 | DJ480 setA′-lacZ sub1 | This study |
| YS142 | DJ480 setA′-lacZ sub2 | This study |
| YS143 | DJ480 λattB::lacIqtetR Spr ΔsetA::cm | This study |
| YS144 | DJ480 λattB::lacIqtetR SprsgrS1 | This study |
| YS214 | DJ480 imm21sgrS′-lacZ ΔsetA::cm | This study |
| YS215 | DJ480 imm21sgrS′-lacZ ΔsgrS | This study |
| YS219 | DJ480 setA′-lacZ ΔkdgR::FRT | This study |
| YS224 | DJ480 imm21sgrS′-lacZ sgrS1 | This study |
| YS225 | DJ480 imm21sgrS′-lacZ ΔkdgR::FRT | This study |
| YS228 | DJ480 imm21sgrS′-lacZ Δcrp::cm | This study |
| YS231 | DJ480 setA′-lacZ Δcrp::cm | This study |
| Plasmids | ||
| pZE21 | Vector control for plasmid pZEYS1 | 18 |
| pZEYS1 | pZE21 with setA under the control of the PLtetO-1 promoter | This study |
| pHDB3 | Vector control for plasmid pBRBAH3 | 31 |
| pBRBAH3 | pHDB3 with sgrR1 | This study |
TABLE 2.
Oligonucleotides used in this study
| Name | Sequence (5′-3′) |
|---|---|
| O-BAH102 | CCCGGATCCCATCGTTAGGAATTATTGACTTAATATAGG |
| O-CV142 | CCCGAATTCAAAAGAAACGCCAGTGAAGCGGTG |
| O-CV170 | TTTTATTCTCGCCGCGCTAAAAAGGGAACGTATGATCTGGATACCTGTGACGGAAGATCACTTCGC |
| O-CV171 | TTTAACCTTTGCGGTTAAAAATAATGCGACAACAGAAATAACCTTATCACTTATTCAGGCGTAGCACC |
| O-CV193 | CCCGGATCCGTATTGGGCTTACCTTGCAGCACGAC |
| O-YS101 | ACCGCAATTCTGAAAGTTGACTTGCC |
| O-YS102 | GAGCCATCGTCATTATCCAGATCATACG |
| O-YS107 | CGCCCCGGCCACCCCCATCATAAAA |
| O-YS109 | ATAGCATTCACCGTATAAAAGA |
| O-YS110 | CCTGACTGTCAGAACGTTTTGC |
| O-YS125 | CATTTATGCTGGTCGCTTTTATGATGGGGGTGGCCGGGGCGTGTAGGCTGGAGCTGCTTC |
| O-YS126 | ACCGCAATTACCCAGTAGACAGCAAAAGTGCCCCCCAACTTTATTCCGGGGATCCGTCGACC |
| O-YS130 | CCGGTACCGTGTAAAATCACCCGCCAGCAGATT |
| O-YS134 | AACCAGCACAACTTCGCTGTCGCGGTAAAATAGTGCTGATAAAACTGACGCATGGGGCACCCCCTTGCTTCATCGTTAGGAGTTGTTGACTTAATATAGGGAAAAAT |
| O-YS135 | GAAAATAAAATTGCTGTCTTTTGCACAGGAGTTCGGGAATTATGCCATCTGCTCGTCTGCAACAACAGTTCATCCGCCTGTGGCAA |
| O-YS138 | GCCGCGTAAACACCGTTCATACGGC |
| For PCR-RACE | GATATGCGCGAATTCCTGTAGAACGAAC |
| ΔyabNcm1 | GCTCGTCTGCAACAACAGTTATCCGCCTGTGGCAATGCTGCCTGTGACGGAAGATCACTTCGC |
| ΔyabNcm2 | GGGTATTCATGCGCAGGCCGCGCATACTGCGTTGCCCTGTTATCACTTATTCAGGCGTAGCACC |
The sgrS′-lacZ transcriptional reporter fusion was constructed by B. Hussain in our laboratory via previously described methods (33). Briefly, the sgrS promoter fragment was amplified by primers O-BAH102 and O-CV142 (Table 2) containing EcoRI or BamHI sites, respectively, and cloned into plasmid pRS1553. The fusion was crossed into the λattB site on the host chromosome (33), resulting in BAH100. The setA′-lacZ transcriptional fusion in YS131 was constructed using the λ-Red and FLP-mediated site-specific recombination method described previously (6). Briefly, a kanamycin cassette flanked by FLP recombination target (FRT) sites was amplified from template pKD13 (6) using primers O-YS125 and O-YS126 (Table 2) and integrated into the chromosome by λ-Red recombination at the setA locus. The remaining steps were as described in reference 6.
BAH101 was created by B. Hussain (in our laboratory) by moving a ΔsgrR::cm allele into the BAH100 host via P1 transduction. CS168 was created by C. S. Wadler in our laboratory by P1 transduction of λattB::lacIq Spr tetR from donor DH5αZ1 (18) into recipient DJ480. The ΔsetA::cm deletion-insertion mutation in ST101 was constructed by S. Tsang in our laboratory using the λ-Red recombination system (43) and primers O-CV170 and O-CV171 (Table 2). YS133 is derived from YS131 and contains a ΔsgrR::cm allele that was constructed by homologous recombination (43) with a PCR product obtained using primers ΔyabNcm1 and ΔyabNcm2 (Table 2). To construct YS135, a single-stranded oligonucleotide, O-YS135 (Table 2), was transformed into CV1300 to replace the cat-sacB cassette carried by the latter in the SgrR-binding site. The setA′-lacZ fusion was then introduced into YS135 by the same method used for YS131 construction, resulting in strain YS140. YS137 was derived from CS100 (38) by looping out the cat-sacB cassette in the −10 region of the sgrS promoter using the oligonucleotide O-YS134 (Table 2). YS142 was subsequently made by constructing the setA′-lacZ fusion in the YS137 background. YS143 was created by moving the ΔsetA::cm allele into CS168 via P1 transduction. YS144 is derived from CS123, which contains the sgrS1 mutation (38), and was constructed by transducing the λattB::lacIq Spr tetR allele into CS123. YS214 was created by transducing the ΔsetA::cm allele into BAH100. YS215 is derived from CS104 (38), which contains a ΔsgrS allele, and was constructed by inserting the sgrS′-lacZ transcriptional fusion in single copy into the λattB site as described previously (33). YS219 was made first by P1 transduction of a ΔkdgR::FRT-kan-FRT allele from donor JW1816 (1) to recipient DJ480; the kanamycin cassette was then removed by a previously described method (6), and the setA′-lacZ fusion was moved in by P1 transduction. YS224 is derived from CS123 and was created by inserting the sgrS′-lacZ transcriptional fusion in single copy into the λattB sites of the respective strains as described previously (33). Using the same method, the sgrS′-lacZ fusion was inserted into YS219, resulting in strain YS225. YS228 was constructed by moving a Δcrp::cm allele into BAH100 through P1 transduction. YS231 was created by moving a Δcrp::cm allele to YS131 by P1 transduction.
DH5α (Invitrogen) was used for routine cloning procedures. Plasmid pZE21 was described in a previous study (18) and was kindly provided by J. Cronan. To construct plasmid pZEYS1, the setA gene was amplified by PCR using primer pairs O-YS130 and O-CV193 (Table 2), which contain BamHI and KpnI sites, and subsequently cloned into the BamHI- and KpnI-cut vector pZE21. Plasmid pBRBAH3 was created by B. Hussain in our laboratory using the XL1-Red system (Stratagene). Briefly, plasmid pLCV3 carrying the wild-type sgrR gene (33) was transformed into the E. coli mutator strain XL1-Red, and the transformants were grown over many generations using serial passages. Pools of mutagenized plasmids generated by this method were then transformed into host strain CV5230, which contains an sgrS′-lacZ fusion and a ΔsgrR mutation on the chromosome (33). The transformants were screened on lactose MacConkey indicator plates for the Lac+ phenotype, and plasmid pBRBAH3 obtained from the screening was then sequenced to identify the locations of mutations in the sgrR gene.
β-Galactosidase assays.
Strains were grown overnight in LB (100 μg/ml ampicillin was added to the medium when working with strains carrying plasmids pHDB3 and pBRBAH3) or minimal MOPS (morpholinepropanesulfonic acid) medium (Teknova) supplemented with 0.4% glycerol or 0.2% fructose and subcultured 1:200 in fresh medium. The cultures were grown to an optical density at 600 nm (OD600) of ∼0.5 and split into two separate cultures. αMG (Sigma) was added to one of the cultures to the indicated final concentrations. For testing differences in growth under stress conditions, αMG was used at a final concentration of 0.5%. We had shown previously that wild-type and ΔsgrS strains have distinctly different growth characteristics at this αMG concentration (37, 38). For experiments testing signaling leading to transcriptional activation of sgrS′-lacZ, a lower concentration of αMG was used (0.001%) because the sgrS promoter is very sensitive to induction under stress conditions and higher concentrations saturate the activation. For testing signaling through activation of setA′-lacZ, a final concentration of 0.1% αMG was used because this fusion is less sensitive to induction than the sgrS′-lacZ fusion. Samples were taken 45 min after the addition of αMG to the appropriate cultures and assayed for β-galactosidase activity as described previously (3).
RT-PCR.
Strains were grown overnight in LB medium and subcultured 1:200 in fresh medium. Total RNA was extracted by the hot-phenol method (33) both before and 10 min after exposure of the cells to 0.5% αMG. Reverse transcription (RT) was performed with 5 μg of Turbo DNase (Ambion)-treated RNA using the Superscript III reverse transcriptase (Invitrogen) and oligonucleotide O-YS110 (Table 2). The PCR products were amplified from cDNA using the Go Taq (Promega) PCR system with primers O-YS101 and O-YS102 (Table 2).
5′ RACE.
5′ Rapid amplification of cDNA ends (RACE) analysis of the setA transcript was performed as described previously with and without tobacco acid pyrophosphatase to detect transcription initiation sites versus processed ends (2) with RNA isolated from mid-log-phase wild-type (DJ480) cells grown in minimal MOPS medium supplemented with 0.4% glycerol 10 min following exposure of the cells to 0.5% αMG. Oligonucleotide O-YS109 was used for reverse transcription, primers O-YS107 and ForPCR-RACE were used for PCR, and O-YS138 was used for sequencing (Table 2).
Growth experiments.
Strains were grown overnight in minimal MOPS medium supplemented with the indicated carbon source (0.4% glycerol or 0.2% fructose) and 25 μg/ml kanamycin and subcultured (OD600, ∼0.01) into fresh medium containing 1 ng/ml anhydrotetracycline (aTc) (Sigma). The cultures were then grown at 37°C with shaking to an OD600 of ∼0.1, and αMG was added to the medium at a concentration of 0.5%. Growth was monitored hourly by measuring the OD600.
Efflux assays.
Strains were grown overnight in minimal MOPS medium supplemented with 0.4% glycerol and subcultured 1:200 into fresh medium. The cultures were grown at 37°C to an OD600 of ∼0.1. The cells were incubated with [14C]αMG (ARC; 3.3 μM; 1μCi/ml) at room temperature for 20 min and then diluted 200-fold with fresh medium. At the indicated times, 20 ml of the culture was filtered (Fisher; 25 mm; filter pore size, 0.45 μm), and the radioactivity of the filtered cells was determined with a scintillation counter.
RESULTS
setA expression is induced in an SgrR-dependent manner under glucose-phosphate stress conditions.
In order to determine the involvement of setA in the glucose-phosphate stress response, we first examined the expression of setA under stress conditions. A setA′-lacZ fusion was constructed on the chromosome at the setA locus in a Δlac host strain as described in Materials and Methods. In the presence of αMG, the setA′-lacZ fusion was activated ∼10 fold over the nonstress level (Fig. 2 B, WT), demonstrating that setA is indeed induced in response to the stress.
FIG. 2.
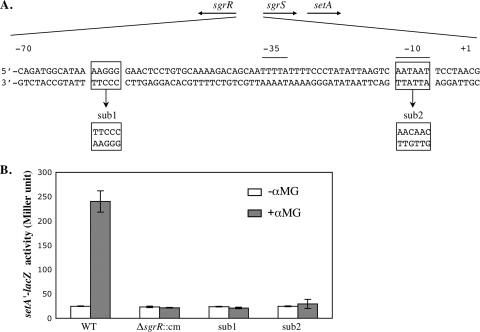
SgrR-dependent activation of setA expression. (A) Genetic organization of the sgrR-sgrS region. The directions of transcription of the sgrR and sgrS genes are depicted by the arrows at the top. The −35 and −10 promoter elements are indicated by horizontal lines above the nucleotide sequence. The boxed nucleotides show the positions where mutations sub1 and sub2 were constructed. The substituted sequences are shown below the wild-type sequence. (B) Cells were grown in LB, and 0.1% αMG was added to half of the cultures. β-Galactosidase activity was assayed 45 min after the addition of αMG. The error bars indicate the standard deviations of three independent experiments.
The gene located 27 nt upstream of setA on the E. coli K-12 chromosome and transcribed in the same orientation is sgrS, which encodes the sRNA required for recovery from glucose-phosphate stress. In many organisms that contain SgrS homologs, setA is present in this position relative to sgrS (Fig. 1B). Our previous work (33) and data in Fig. 2B (WT) demonstrated that expression of sgrS and setA is induced under the same stress conditions. Based on what is known about sgrS regulation, we predicted that SgrR (34), the transcriptional activator of sgrS, would also regulate setA. To elucidate the role of SgrR in regulation of setA, a ΔsgrR::cm allele was introduced into the strain carrying the setA′-lacZ fusion. As shown in Fig. 2B (ΔsgrR::cm), the absence of SgrR did not affect the basal level of setA′-lacZ activity in the absence of stress but abolished activation following the exposure of cells to αMG. This result demonstrated that SgrR is required for stress-dependent induction of setA transcription.
Previous studies defined a conserved region between positions −70 and −55 relative to the start of sgrS transcription as required for the SgrR-dependent activation of sgrS (32, 33). To determine whether the same region is needed for the SgrR-mediated activation of setA, a 5-bp substitution in this region on the chromosome was constructed in the context of the setA′-lacZ chromosomal fusion. This 5-bp substitution (Fig. 2A, sub1) was shown previously (32) to disrupt the interaction between SgrR and the sgrS promoter and, consequently, to eliminate the SgrR-dependent activation of sgrS. Compared with the wild-type fusion, the mutant fusion had a similar basal activity in the absence of the stress. The sub1 mutant fusion was not activated upon exposure of cells to αMG (Fig. 2B, sub1), indicating that this SgrR-binding site is crucial for stress-dependent induction of setA. Similarly, point mutations in the −10 region of the sgrS promoter (Fig. 2A, sub2) that abrogate sgrS activation in response to glucose-phosphate stress (data not shown) also prevented the activation of the setA′-lacZ fusion under stress conditions (Fig. 2B, sub2). Together, these results suggested that the stress-responsive induction of setA was due to SgrR-dependent activation at the sgrS promoter.
setA is coexpressed with sgrS under glucose-phosphate stress conditions.
Based on the setA′-lacZ fusion data (Fig. 2), we predicted that sgrS and setA are cotranscribed under glucose-phosphate stress. To confirm this, RT-PCR was performed on total RNA from wild-type cells in the presence and absence of αMG. Using an sgrS-specific primer and a setA-specific primer (Fig. 3 A), an amplicon of the expected size of ∼150 bp, encompassing the sgrS-setA intergenic region, was produced (Fig. 3B). Levels of this product were reproducibly higher in samples from wild-type stressed cells than in nonstressed cells (Fig. 3B, WT, compare −αMG and +αMG). Though this is semiquantitative RT-PCR, the fact that an sgrS-setA product was detected at high levels in stressed wild-type cells was consistent with the setA′-lacZ fusion results (Fig. 2). This indicated that sgrS and setA are cotranscribed and both are upregulated under glucose-phosphate stress conditions. To further support the results of setA′-lacZ fusions, RT-PCR was performed on total RNA from the sgrR mutant (Fig. 3B, ΔsgrR::cm), as well as the two sgrS promoter mutants (Fig. 3B, sub1 and sub2). The results showed lower abundance of the sgrS-setA product in stressed cells carrying the sgrR sub1 or sub2 mutation compared with stressed wild-type cells, supporting the notion that the stress-responsive activation of setA requires SgrR and sgrS promoter determinants.
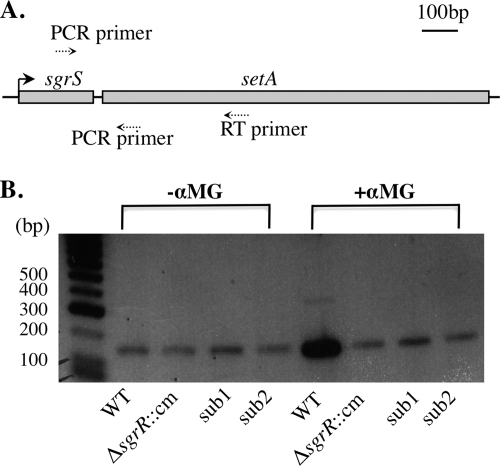
FIG. 3.
RT-PCR analysis of sgrS and setA. (A) Schematic diagram showing the positions of the RT primer and PCR primers within the sgrS-setA region. (B) Wild-type (DJ480), ΔsgrR::cm mutant (CV700), and sgrS promoter mutant sub1 (YS135) and sub2 (YS137) cells were grown in LB medium at 37°C to an OD600 of ∼0.5. Total RNA was prepared from cells before and 10 min after exposure to 0.5% αMG, reverse transcribed, and amplified by PCR as described in Materials and Methods. Control experiments were also performed on each sample without addition of the reverse transcriptase, and no PCR product was detected in any samples (data not shown).
Transcriptional fusions (Fig. 2) and RT-PCR (Fig. 3) demonstrate that when transcription at the sgrS promoter is activated, some transcripts initiating at PsgrS read through the sgrS terminator into setA. However, it was also possible that an additional setA-specific promoter was responsible for the basal transcription of setA′-lacZ. To identify any additional setA transcription initiation sites, 5′ RACE (2) was performed on RNA harvested from stressed (+αMG) and unstressed (−αMG) wild-type cells. The only setA-specific product obtained in this experiment was one that initiated at the previously mapped SgrS 5′ end (data not shown and reference 33). Therefore, if another setA-specific promoter exists, these transcripts must be present at levels too low to detect by 5′ RACE. Together, these analyses indicate that under glucose-phosphate stress, setA is cotranscribed with sgrS from the sgrS promoter, and these two genes form an operon. The basal level of fusion activity (Fig. 2B) and the product observed in RT-PCR (Fig. 3B) in samples from the nonstressed cells may originate from a transcription start site that could not be detected in our experiments or may represent the basal level of expression from the sgrS promoter.
crp and kdgR mutations affect signaling through the glucose-phosphate stress response.
The experiments described above established that transcription of the sgrS-setA bicistronic transcript initiates at the sgrS promoter under glucose-phosphate stress conditions. In studies of sotA, a SET family member in D. dadantii, the global regulators CRP and KdgR were implicated in transcriptional regulation. CRP activated expression of sotA (4), while KdgR, which represses transcription of many genes involved in transport and catabolism of 2-keto-3-deoxygluconate in E. coli (22, 27), was shown to weakly repress the expression of sotA in D. dadantii (29). We therefore tested the effects of CRP and KdgR on regulation of sgrS and setA in E. coli. Given the evidence presented above regarding the coregulation of sgrS and setA (Fig. 2 and 3), we reasoned that CRP and KdgR might regulate setA through one of two mechanisms. First, these regulators might directly bind upstream of the sgrS promoter and directly activate or repress sgrS-setA expression. Second, these regulators could alter sgrS-setA expression indirectly by modulating the signal that results in SgrR-mediated activation at the sgrS promoter. To determine whether CRP and KdgR were involved in one of these modes of regulation, activities of setA′-lacZ and sgrS′-lacZ fusions were monitored in two different host strains: host 1, an sgrR+ host in which stress was induced by addition of αMG to cultures, and host 2, a ΔsgrR host complemented with a stress signal-independent sgrR allele (i.e., an sgrR mutant that does not require stress to activate the sgrS promoter). This sgrR1 allele encodes a glycine-to-glutamate substitution at position 118 of SgrR; the sgrR1 plasmid promotes sgrS transcription independent of glucose-phosphate stress conditions (Fig. 4 D, WT). If crp or kdgR mutations have an effect on regulation of fusions in the context of both host 1 and host 2, it would suggest direct regulation. On the other hand, if crp or kdgR mutations have an effect on regulation of fusions in host 1 but not host 2, it would indicate an indirect mechanism of regulation via effects on signaling through SgrR.
FIG. 4.
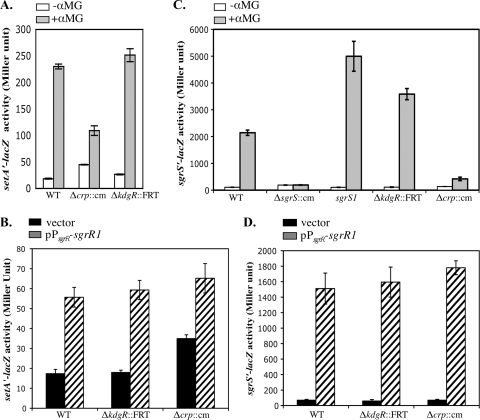
Effects of CRP and KdgR on setA and sgrS expression. (A) Cells were grown in LB to an OD600 of ∼0.5, and 0.1% αMG was added to half of the cultures. β-Galactosidase activity was assayed 45 min after the addition of αMG. (B) Cells were grown in LB supplemented with 100 μg/ml ampicillin to an OD600 of ∼0.5 and assayed for β-galactosidase activity. (C) Cells were grown in LB to an OD600 of ∼0.5, and 0.001% αMG was added to half of the cultures. β-Galactosidase activity was assayed 45 min after the addition of αMG. (D) Cells were grown in LB supplemented with 100 μg/ml ampicillin to an OD600 of ∼0.5 and assayed for β-galactosidase activity. The error bars indicate standard deviations from three independent experiments.
Δcrp::cm and ΔkdgR::FRT alleles were introduced into the hosts described above carrying the setA′-lacZ fusion. In the sgrR+ host, the kdgR mutation did not cause a significant change in the basal level of setA expression (Fig. 4A, −αMG). The level of setA′-lacZ activity in stressed cells (Fig. 4A, +αMG) was reproducibly slightly higher in the ΔkdgR mutant than in the kdgR+ strain, suggesting that KdgR may play a very modest role in regulating setA expression. The crp mutation resulted in a basal level of setA′-lacZ activity that was 2.4-fold higher than in the wild-type strain (Fig. 4A, −αMG), suggesting that CRP might repress setA expression under nonstress conditions. Upon exposure of the Δcrp mutant cells to αMG, setA′-lacZ expression increased ∼2.4-fold compared to nonstressed Δcrp mutant cells, though the induced level of setA′-lacZ activity in the Δcrp mutant was lower than in the wild-type strain (Fig. 4A, +αMG). These results suggested that during glucose-phosphate stress, CRP might exert a slight effect on stress-responsive activation of setA expression.
To address whether the slight effects of the crp and kdgR mutations on setA′-lacZ expression were direct or through effects on signaling through SgrR, we repeated the experiments described above in the ΔsgrR host carrying the stress signal-independent sgrR1 plasmid. In the wild-type background, the sgrR1 plasmid caused a 3.2-fold increase in the level of setA′-lacZ activity over the vector control levels. sgrR1 plasmid-containing cells had ∼3.3-fold-increased setA′-lacZ activity compared to vector control-containing cells in the ΔkdgR mutant background (Fig. 4B, ΔkdgR::FRT and pPsgrR-sgrR1). Consistent with our previous observation (Fig. 4A, −αMG), setA′-lacZ activity was ∼2.2-fold higher in the Δcrp mutant than in the wild type when both strains carried the vector control, i.e., the basal level of activity was higher in the absence of CRP (Fig. 4B, Δcrp::cm/vector). sgrR1 plasmid-containing cells had ∼2-fold-increased setA′-lacZ activity compared to vector control-containing cells in the Δcrp mutant background (Fig. 4B, Δcrp::cm and pPsgrR-sgrR1). Importantly, cells carrying the sgrR1 plasmid had similar levels of setA′-lacZ expression in wild-type and ΔkdgR and Δcrp mutant backgrounds independent of glucose-phosphate stress. Together, these results suggest that while CRP may repress basal levels of setA transcription, neither CRP nor KdgR is likely to exert any direct effect on setA transcription under stress conditions.
Since expression of setA under stress conditions depends on the sgrS promoter, we also tested the effects of KdgR and CRP on an sgrS′-lacZ promoter fusion harbored at the λattB site. In addition to wild-type and kdgR and crp mutant strains, the activity of the sgrS′-lacZ fusion was also monitored in ΔsgrR and sgrS1 (38) mutant hosts. The SgrS1 molecule is defective for regulation of ptsG under glucose-phosphate stress (13) and is more sensitive to αMG than the wild-type strain. Upon exposure to αMG, the sgrS′-lacZ fusion in the wild-type strain was induced ∼20-fold (Fig. 4C, WT), whereas the ΔsgrR::cm mutation completely abolished induction, as observed previously (33). As expected, fusion activity in the sgrS1 mutant was significantly higher than that of the wild-type strain (induced ∼47-fold) (Fig. 4C), because this mutant cannot reduce uptake of αMG by downregulating the glucose transporter ptsG mRNA. Interestingly, the induced level of sgrS′-lacZ activity in the ΔkdgR mutant was also higher than in the wild-type strain (induced ∼32-fold) (Fig. 4C), although the basal expression levels were similar in the two strains, suggesting that KdgR functions as a repressor for the stress-dependent induction of sgrS expression. For the crp mutant, basal levels of expression of sgrS′-lacZ were similar to those in the wild-type strain. When stress was induced with αMG, sgrS transcription was induced by 3-fold in the crp mutant compared to 20-fold in the wild-type strain (Fig. 4C). This result suggested that CRP is required for optimal induction of sgrS under stress conditions.
To investigate whether KdgR and CRP directly or indirectly regulate sgrS promoter activity, we again utilized the plasmid carrying the sgrR1 mutant allele. In the wild-type strain carrying the sgrR1 plasmid, sgrS′-lacZ activity was increased ∼22-fold compared to the vector control strain (Fig. 4D, WT). Both basal (vector control) and induced (sgrR1) levels of sgrS′-lacZ activity in the kdgR and crp mutants were very similar to those in the wild-type cells (Fig. 4D). Collectively, these results suggested that under glucose-phosphate stress conditions, KdgR represses the activation of sgrS (and very slightly of setA) indirectly by altering signaling through the stress response system and therefore activity of SgrR. Similarly, the effect of CRP on the induced level of setA and sgrS transcription was mediated through SgrR-dependent signaling. Interestingly, CRP had an SgrR-independent effect on the basal level of setA transcription, but not on sgrS. This suggested that there might be a small direct effect of CRP on regulation of an unidentified promoter present in the setA′-lacZ but not the sgrS′-lacZ fusion.
A setA mutation causes a defect in recovery from growth inhibition by αMG.
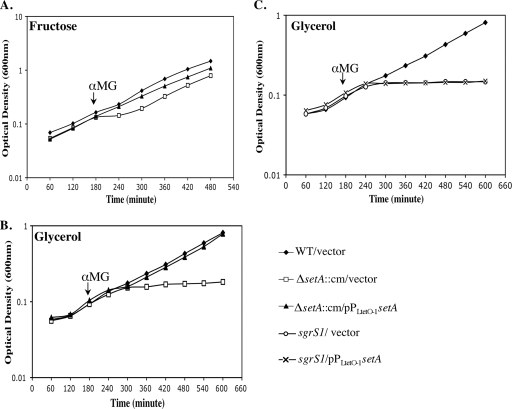
After establishing that setA is activated during glucose-phosphate stress, its role in the stress response was investigated. The growth of wild-type (CS168) and ΔsetA::cm mutant (YS143) strains was monitored before and after they were exposed to αMG. No growth differences between the two strains were apparent in LB medium with or without αMG (data not shown). We have noticed in the course of other studies that wild-type and sgrS mutant strains have more pronounced growth defects when stress is induced in minimal medium compared with LB (C. S. Wadler and C. K. Vanderpool, unpublished data). Therefore, the stress phenotypes of wild-type and ΔsetA::cm cells were also tested in minimal MOPS medium with fructose or glycerol as a carbon source. In minimal MOPS-fructose medium, the ΔsetA::cm mutant strain carrying the vector control (Fig. 5 A, ΔsetA::cm/vector) had a subtle but reproducible phenotype: a longer lag following addition of αMG to the culture than the wild-type strain (Fig. 5A, WT/vector). The setA+ plasmid (pZEYS1, where expression of setA was controlled by a Ptet promoter) eliminated the lag and restored the growth of the ΔsetA::cm strain to a pattern very similar to that of wild-type cells (Fig. 5A, ΔsetA::cm/pPLtetO-1-setA). In minimal MOPS medium with glycerol as the carbon source, the growth defect of the ΔsetA::cm mutant compared with the wild type was more pronounced (Fig. 5B) and was similar to the sgrS1 mutant (Fig. 5C), which is defective in base pairing and subsequent regulation of ptsG during glucose-phosphate stress (13). After stress was induced, the setA mutant strain failed to recover to the wild-type level even after being cultured overnight. The effect of the setA mutation under these conditions was bacteriostatic rather than bactericidal (data not shown). Again, the plasmid carrying PLtetO-1-setA complemented the stress-related growth defect of the setA mutant (Fig. 5B). These data supported our hypothesis that, at least under some growth conditions, SetA plays a role in allowing cells to recover from glucose-phosphate stress. On the other hand, in both media, the setA+ plasmid failed to complement the stress phenotypes of the sgrS1 mutant (Fig. 5C). This indicates that the sgrS1 phenotype is not due to polarity on setA and, further, that the role of SgrS in the stress response cannot be substituted for by the activity of SetA.
FIG. 5.
Growth of wild-type and ΔsetA strains in the presence of αMG. Cells were grown at 37°C in minimal MOPS medium supplemented with 1 ng/ml aTc, 25 μg/ml kanamycin, and 0.2% fructose (A) or 0.4% glycerol (B and C) to an OD600 of ∼0.1, and αMG was added to the cultures to a final concentration of 0.5%. Each graph is representative of three independent trials.
The setA mutation affects signaling through SgrR.
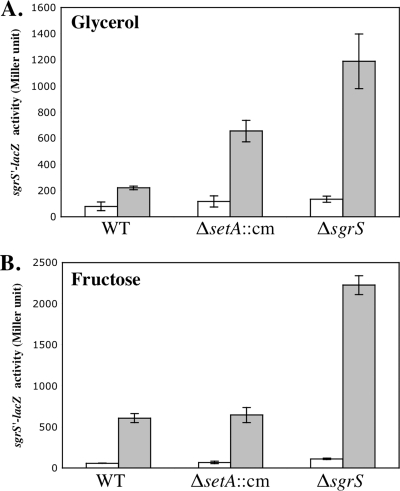
Our original hypothesis was that under stress conditions, SetA facilitates removal of nonmetabolizable sugars, for example, αMG-6-phosphate, by pumping them out. If this were true, we would expect to see differences in signaling through SgrR in a setA mutant compared to the wild-type strain. Since sgrS transcription is controlled by SgrR (33, 34) (Fig. 4C), we measured signaling through SgrR using the sgrS′-lacZ fusion. The activation of the fusion in wild-type and ΔsetA::cm backgrounds was compared to activation in a ΔsgrS strain, where stress and sgrS′-lacZ transcription are increased because the stress response is defective. This experiment was performed in minimal MOPS medium with fructose or glycerol as a carbon source. In minimal MOPS-glycerol medium, basal expression levels (−αMG) of the sgrS′-lacZ fusion were similar in the wild type and the ΔsetA mutant. In wild-type cells, sgrS′-lacZ was induced by ∼2.8-fold upon exposure to αMG, while induction in the ΔsetA mutant was ∼5.6-fold (Fig. 6 A). The activation of sgrS′-lacZ was induced by ∼8.8-fold in the ΔsgrS mutant. This suggested that the ΔsetA mutation impairs the stress response and therefore increases stress signaling through SgrR, though to a lesser degree than the ΔsgrS mutation. In minimal MOPS-fructose medium, however, both the basal and induced levels of sgrS′-lacZ expression in the ΔsetA::cm mutant were very similar to those of the wild type, while the sgrS mutant showed higher induction (Fig. 6B). The differential phenotypes in the two media were reminiscent of the growth experiments (Fig. 5) in that the effect of the setA mutation was more dramatic in minimal glycerol medium than in minimal fructose medium. Together, these results suggested that the setA mutation affects signaling through SgrR in a medium-specific manner.
FIG. 6.
Effects of setA mutation on sgrS′-lacZ activity in response to αMG. Cells were grown in minimal MOPS medium supplemented with 0.4% glycerol (A) or 0.2% fructose (B), and 0.001% αMG was added to half of the cultures. β-Galactosidase activity was assayed after 45 min of αMG addition. White bars represent the absence of αMG, while gray bars represent the presence of αMG. The error bars indicate the standard deviations of three independent experiments.
Efflux of αMG by the cells is independent of SetA.
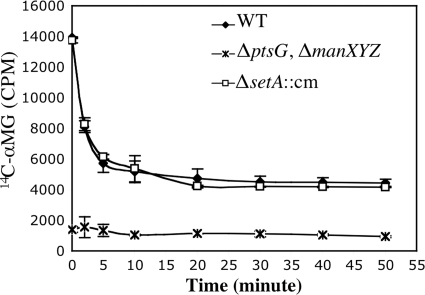
To directly test the hypothesis that SetA promotes efflux of αMG under stress conditions, the efflux of 14C-labeled αMG from wild-type and ΔsetA::cm mutant cells was monitored using an assay described previously (41). αMG was implicated as an in vitro substrate for SetA in a previous study (17). We reasoned that if SetA is the major transporter responsible for removing the nonmetabolizable sugar from the cells during stress, the setA mutant would exhibit an elevated steady-state intracellular αMG level compared to that of the wild-type strain and would also show slowed kinetics of αMG efflux. Since ManXYZ and PtsG are the two main transporters responsible for αMG uptake in E. coli (26), a ΔmanXYZ ΔptsG mutant was also included in the experiment as a negative control. All strains were grown in minimal MOPS-glycerol medium and stressed with [14C]αMG. These are the conditions under which the setA mutant had the strongest growth phenotype (Fig. 5B). After 20 min of incubation, the cells were diluted into fresh medium lacking [14C]αMG to promote its efflux. As shown in Fig. 7, the steady-state level of cell-associated radioactivity in wild-type cells at time zero (immediately upon dilution) was ∼10-fold higher than in the αMG uptake mutant (ΔptsG ΔmanXYZ). In wild-type cells, cell-associated counts decreased rapidly in the first 10 min following dilution and leveled out at a new, lower steady state. This result resembles the pattern reported in other, similar SetA efflux studies (5). The ΔsetA::cm mutant showed a steady-state level of cell-associated radioactivity that was nearly identical to the level in wild-type cells. Moreover, the pattern of [14C]αMG efflux for the ΔsetA::cm mutant was also very similar to that of the wild-type cells, suggesting that SetA does not function as the major αMG efflux pump under these stress conditions.
FIG. 7.
Effects of setA mutation on the efflux of [14C]αMG. Wild-type (DJ480), ΔsetA::cm mutant (ST101), and ΔmanXYZ::kan ΔptsG::cm mutant (CV107) strains were grown at 37°C in minimal MOPS medium supplemented with 0.4% glycerol to an OD600 of ∼0.1. The cells were incubated with [14C]αMG (3.3 μM; 1 μCi/ml) at room temperature for 20 min and then diluted 200-fold with fresh minimal MOPS-glycerol medium. Radioactivity was examined at the indicated times as described in Materials and Methods. The error bars indicate the standard deviations of three independent experiments.
DISCUSSION
In the current study, we have demonstrated that the sugar efflux transporter SetA contributes to the cellular response to nonmetabolizable sugar-phosphates. Under glucose-phosphate stress conditions, setA is coexpressed with sgrS (Fig. 2 and 3), which encodes the small RNA effector of the stress response (32-34). Our data are consistent with an operonic arrangement for sgrS and setA, with transcription initiating upstream of sgrS at a promoter that is activated under stress conditions by the transcription factor SgrR. Most well-characterized sRNA genes to date are transcribed independently (39), but a few cases have been documented where sRNA expression seems to be related to expression of flanking protein-encoding genes. For example, the recently identified spd-sr37 and ccnA in Streptococcus pneumoniae (31) and pel/sagA in group A streptococcus (19, 23) are cotranscribed with downstream protein-coding genes. In E. coli, a previous study identified several sRNAs, e.g., SroC, that may be products of transcriptional attenuation of downstream genes and other sRNAs, e.g., SroD and SroE, that are likely processed from longer mRNAs (36). 6S RNA in E. coli is also processed from a bicistronic RNA, but it is a protein-binding sRNA (39) rather than a base-pairing sRNA like SgrS. The promoter of ryhB, which encodes an Hfq-dependent sRNA in E. coli that regulates various genes involved in iron metabolism (20), was implied to control the expression of the downstream gene yhhX (35). This gene organization is perhaps most analogous to the arrangement between sgrS and setA. However, we have clearly demonstrated here that, unlike YhhX, which encodes a putative d-galactose-1-dehydrogenase (28) whose functional link to RyhB is unclear, SetA contributes to the same stress response system as SgrS (Fig. 5).
The presence of a putative Rho-independent sgrS terminator strongly suggests that bicistronic sgrS-setA transcripts are generated by readthrough of the sgrS terminator. This is reminiscent of the case reported in group A streptococcus where a Rho-independent terminator between the sRNA coding gene pel/sagA and the downstream gene allowed and regulated the readthrough (23). Northern blots using a setA-specific probe did not yield a signal at the size predicted for the sgrS-setA transcript even under stress conditions (data not shown), whereas a strong ∼200-nt signal was seen for SgrS using an SgrS-specific probe under stress conditions (31). Real-time PCR experiments revealed that under glucose-phosphate stress, steady-state levels of the sgrS transcript are ∼600-fold higher than those of the sgrS-setA bicistronic transcript (data not shown), highlighting the significantly greater abundance of the SgrS RNA than of the bicistronic transcript. Collectively, these results suggest that termination usually occurs at the 3′ end of SgrS and rather infrequently transcriptional readthrough produces the bicistronic transcript. Alternatively, the sgrS-setA RNA may be very unstable relative to SgrS.
We cannot rule out an additional setA promoter either within sgrS or further upstream. A basal level of activity for the setA′-lacZ fusion independent of the known sgrS promoter was observed (Fig. 2). Two transcriptional regulators, CRP and KdgR, which influence the expression of sotA, encoding a SetA paralog in D. dadantii (4, 29), were tested for their roles in regulating setA expression. A crp mutation increases the basal level of setA′-lacZ activity (Fig. 4A and B), but not the activity of sgrS′-lacZ, suggesting there may be another promoter controlling setA expression that is repressed by CRP. Further analysis of transcriptional regulation of setA may provide additional clues to its role in cell physiology.
Both KdgR and CRP had modest effects on the activation of setA′-lacZ and more significant effects on sgrS′-lacZ under stress conditions (Fig. 4A and C). The results of experiments using the plasmid-encoded signal-independent SgrR (Fig. 4B and D) implied that the crp and kdgR mutations alter the ability of wild-type SgrR to sense and respond to the stress signal. In the case of kdgR, its deletion is known to cause derepression of eda, which encodes 2-keto-3-deoxy-6-phosphogluconate (KDPG) aldolase that cleaves KDPG to pyruvate and triose-3-phosphate (22). Interestingly, pyruvate was reported previously to abrogate the degradation of ptsG in a Δpgi mutant (14), usually indicative of altered SgrS levels or function. Furthermore, addition of pyruvate to the growth medium increases sgrS′-lacZ fusion activity in stressed wild-type cells (M. Patel, G. Richards, and C. K. Vanderpool, unpublished data). It is therefore possible that increased endogenous pyruvate concentrations caused by the kdgR deletion alter signaling through SgrR and, subsequently, stress-dependent activation of sgrS.
This study showed that SetA is required for optimal recovery from glucose-phosphate stress under certain growth conditions (Fig. 5). The hypothesis that SetA directly pumps out nonmetabolizable sugars (at least αMG) was not supported by our efflux experiments (Fig. 7). Consistent with this result, the stress phenotype exhibited by the sgrS mutant could not be bypassed by ectopic expression of setA (from PLtetO-1-setA) (Fig. 5C). This demonstrates two important points: (i) that the growth defect of an sgrS mutant is not due to polarity on setA and (ii) that the role of SgrS in the stress response cannot be replaced by the activity of SetA, suggesting that SgrS and SetA affect stress recovery by fundamentally different mechanisms. SgrS inhibits production and activity of the αMG transporter PtsG (28, 29) and therefore inhibits intracellular αMG6P accumulation. Since SetA cannot compensate for the lack of SgrS activity, it follows that SetA acts at a level in the stress response that does not directly impact intracellular αMG6P levels. This line of reasoning is consistent with our efflux data (Fig. 7).
SetA is not fully conserved in enterobacterial genomes that encode SgrS (8); for example, it is absent in some pathogenic E. coli and Salmonella strains. While we do not currently understand why setA is absent in these organisms, it may be due to potentially redundant functions of the other two SET proteins, namely, SetB and SetC, at least one of which is encoded in these genomes that lack setA. SetA shares a high degree of amino acid sequence similarity with SetB and SetC (16). Similar to SetA, SetB was also capable of transporting glucose and lactose in vitro, although it had a narrower substrate range (17). The substrate of SetC remains unidentified. Like setA, the genomic locations of these other SET family members suggest involvement in carbohydrate metabolism. In the E. coli K-12 genome, setB is located adjacent to the fruBKA operon encoding the major fructose transporter (26), while setC is in close proximity to genes encoding a putative galactose-pentose-hexuronide transporter (25). While transcription of setB and setC in E. coli K-12 was not induced by glucose-phosphate stress (data not shown), it is possible that in organisms that lack setA, SetB or SetC functionally replaces SetA and contributes in a similar fashion to the glucose-phosphate stress response.
Future studies are required to more fully elucidate the role of SetA in the glucose-phosphate stress response in E. coli. The growth defect and differences in signaling through SgrR (for activation of sgrS transcription) in the setA mutant under certain conditions confirm its connection to the stress response. The conditional nature of the SetA requirement, as well as medium-dependent differences in the severity of the stress, may provide important clues to two major unresolved questions: (i) what is the actual inducing signal and (ii) what is the nature of the metabolic problem experienced by stressed cells? Experiments are under way to determine the bona fide in vivo substrate of SetA, and its identification will no doubt shed new light on the physiology of the glucose-phosphate stress response.
Acknowledgments
We sincerely thank John Cronan and the members of his laboratory for providing the plasmid pZE21 and for use of the scintillation counter. We also thank Basil Hussain, Stephanie Tsang, and Caryn Wadler for construction of several strains used in our study. We are grateful to Susan Gottesman, James Imlay, Gregory Richards, and James Slauch for critical reading of the manuscript and helpful comments; to members of the Slauch laboratory and the members of the Vanderpool laboratory, including Divya Balasubramanian, Maksym Bobrovskyy, Richard Horler, Chelsea Lloyd, and Jennifer Rice, for useful discussions and suggestions; and to Lauren Endriukaitis and Sarah Lai for technical assistance.
This work was supported by the University of Illinois at Urbana-Champaign, the American Cancer Society (research scholar grant ACS2008-01868), and the American Heart Association (scientist development grant 0835355N).
Footnotes
Published ahead of print on 22 October 2010.
REFERENCES
- 1.Baba, T., T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner, and H. Mori. 2006. Construction of Escherichia coli K-12 in-frame, single-gene knock-out mutants—the Keio collection. Mol. System Biol. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed]
- 2.Bensing, B. A., B. J. Meyer, and G. M. Dunny. 1996. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc. Natl. Acad. Sci. U. S. A. 93:7794-7799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clepet, C., I. L. Clainche, and M. Caboche. 2004. Improved full-length cDNA production based on RNA tagging by DNA ligase. Nucleic Acids Res. 32:e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Condemine, G. 2000. Characterization of SotA and SotB, two Erwinia chrysanthemi proteins which modify isopropyl-β-D-thiogalactopyranoside and lactose induction of the Escherichia coli lac promoter. J. Bacteriol. 182:1340-1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dippel, R., and W. Boos. 2005. The maltodextrin system of Escherichia coli: metabolism and transport. J. Bacteriol. 187:8332-8339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ellermeier, C. D., A. Janakiraman, and J. M. Slauch. 2002. Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290:153-161. [DOI] [PubMed] [Google Scholar]
- 7.Englesberg, E., R. L. Anderson, R. Weinberg, N. Lee, P. Hoffee, G. Huttenhauer, and H. Boyer. 1962. L-arabinose-sensitive, L-ribulose 5-phosphate 4-epimerase-deficient mutants of Escherichia coli. J. Bacteriol. 84:137-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Horler, R. S., and C. K. Vanderpool. 2009. Homologs of the small RNA SgrS are broadly distributed in enteric bacteria but have diverged in size and sequence. Nucleic Acids Res. 37:5465-5476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huber, R. E., J. Lytton, and E. B. Fung. 1980. Efflux of β-galactosidase products from Escherichia coli. J. Bacteriol. 141:528-533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huber, R. E., R. Pisko-Dubienski, and K. L. Hurlburt. 1980. Immediate stoichimetric appearance of β-galactosidase products in the medium of Escherichia coli cells incubated with lactose. Biochem. Biophys. Res. Commun. 96:656-661. [DOI] [PubMed] [Google Scholar]
- 11.Irani, M. H., and P. K. Maitra. 1977. Properties of Escherichia coli mutants deficient in enzymes of glycolysis. J. Bacteriol. 132:398-410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kadner, R. J., G. P. Murphy, and C. M. Stephens. 1992. Two mechanisms for growth inhibition by elevated transport of sugar phosphates in Escherichia coli. J. Gen. Microbiol. 138:2007-2014. [DOI] [PubMed] [Google Scholar]
- 13.Kawamoto, H., Y. Koide, T. Morita, and H. Aiba. 2006. Base-pairing requirement for RNA silencing by a bacterial small RNA and acceleration of duplex formation by Hfq. Mol. Microbiol. 61:1013-1022. [DOI] [PubMed] [Google Scholar]
- 14.Kimata, K., Y. Tanaka, T. Inada, and H. Aiba. 2001. Expression of the glucose transporter gene, ptsG, is regulated at the mRNA degradation step in response to glycolytic flux in Escherichia coli. EMBO J. 20:3587-3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee, A. T., and A. Cerami. 1987. Elevated glucose 6-phosphate levels are associated with plasmid mutations in vivo. Proc. Natl. Acad. Sci. U. S. A. 84:8311-8314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, J. Y., P. F. Miller, M. Gosink, and E. R. Olson. 1999. The identification of a new family of sugar efflux pumps in Escherichia coli. Mol. Microbiol. 31:1845-1851. [DOI] [PubMed] [Google Scholar]
- 17.Liu, J. Y., P. F. Miller, J. Willard, and E. R. Olson. 1999. Functional and biochemical characterization of Escherichia coli sugar efflux transporters. J. Biol. Chem. 274:22977-22984. [DOI] [PubMed] [Google Scholar]
- 18.Lutz, R., and H. Bujard. 1997. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 25:1203-1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mangold, M., M. Siller, B. Roppenser, B. J. M. Vlaminckx, T. A. Penfound, R. Klein, R. Novak, R. P. Novick, and E. Charpentier. 2004. Synthesis of group A streptococcal virulence factors is controlled by a regulatory RNA molecule. Mol. Microbiol. 53:1515-1527. [DOI] [PubMed] [Google Scholar]
- 20.Massé, E., and S. Gottesman. 2002. A small RNA regulates the expression of genes involved in iron metabolism in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:4620-4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morita, T., W. El-Kazzaz, Y. Tanaka, T. Inada, and H. Aiba. 2003. Accumulation of glucose 6-phosphate or fructose 6-phosphate is responsible for destabilization of glucose transporter mRNA in Escherichia coli. J. Biol. Chem. 278:15608-15614. [DOI] [PubMed] [Google Scholar]
- 22.Murray, E. L., and T. Conway. 2005. Multiple regulators control expression of the Entner-Doudoroff aldolase (Eda) of Escherichia coli. J. Bacteriol. 187:991-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nizet, V., B. Beall, D. J. Bast, V. Datta, L. Kilburn, D. E. Low, and J. C. S. D. Azavedo. 2000. Genetic locus for streptolysin S production by group A Streptocccus. Infect. Immun. 68:4245-4254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pao, S. S., I. T. Paulsen, and M. H. Saier, Jr. 1998. Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62:1-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Poolman, B., J. Knol, C. van der Does, P. J. Henderson, W. J. Liang, G. Leblanc, T. Pourcher, and I. Mus-Veteau. 1996. Cation and sugar selectivity determinants in a novel family of transport proteins. Mol. Microbiol. 19:911-922. [DOI] [PubMed] [Google Scholar]
- 26.Postma, P. W., J. W. Lengeler, and G. R. Jacobson. 1993. Phosphoenolpyruvate: carbohydrate phosphotransferase systems of bacteria. Microbiol. Rev. 57:543-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pouyssegur, J., and F. Stoeber. 1974. Genetic control of the 2-keto-3-deoxy-D-gluconate metabolism in Escherichia coli K-12: kdg regulon. J. Bacteriol. 17:641-651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reed, J. L., T. D. Vo, C. H. Schilling, and B. O. Palsson. 2003. An expanded genome-scale model of Escherichia coli K-12 (iJR904 GSM/GPR). Genome Biol. 4:R54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rodionov, D. A., M. S. Gelfand, and N. Hugouvieux-Cotte-Pattat. 2004. Comparative genomics of the KdgR regulon in Erwinia chrysanthemi 3937 and other gamma-proteobacteria. Microbiology 150:3571-3590. [DOI] [PubMed] [Google Scholar]
- 30.Saier, M. H., J. T. Beatty, A. Goffeau, K. T. Harley, W. H. Heijne, S. C. Huang, D. L. Jack, P. S. Jahn, K. Lew, J. Liu, S. S. Pao, I. T. Paulsen, T. T. Tseng, and P. S. Virk. 1999. The major facilitator superfamily. J. Mol. Microbiol. Biotechnol. 1:257-279. [PubMed] [Google Scholar]
- 31.Tsui, H. C., D. Mukherjee, V. A. Ray, L. T. Sham, A. L. Feig, and M. E. Winkler. 2010. Identification and characterization of noncoding small RNAs in Streptococcus pneumoniae serotype 2 strain D39. J. Bacteriol. 192:264-279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vanderpool, C. K. 2007. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 10:146-151. [DOI] [PubMed] [Google Scholar]
- 33.Vanderpool, C. K., and S. Gottesman. 2004. Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol. Microbiol. 54:1076-1089. [DOI] [PubMed] [Google Scholar]
- 34.Vanderpool, C. K., and S. Gottesman. 2007. The novel transcription factor SgrR coordinates the response to glucose-phosphate stress. J. Bacteriol. 189:2238-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vassinova, N., and D. Kozyrev. 2000. A method for direct cloning of fur-regulated genes: identification of seven new fur-regulated loci in Escherichia coli. Microbiology 146:3171-3182. [DOI] [PubMed] [Google Scholar]
- 36.Vogel, J., V. Bartels, H. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. H. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wadler, C. S., and C. K. Vanderpool. 2007. A dual function for a bacterial small RNA: SgrS performs base pairing-dependent regulation and encodes a functional polypeptide. Proc. Natl. Acad. Sci. U. S. A. 104:20454-20459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wadler, C. S., and C. K. Vanderpool. 2009. Characterization of homologs of the small RNA SgrS reveals diversity in function. Nucleic Acids Res. 37:5477-5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Waters, L. S., and G. Storzs. 2009. Regulatory RNAs in bacteria. Cell 136:615-628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wilson, T. H., and E. R. Kashket. 1969. Isolation and properties of thiogalactoside transacetylase-negative mutants of Escherichia coli. Biochim. Biophys. Acta 173:501-508. [DOI] [PubMed] [Google Scholar]
- 41.Winkler, H. H. 1971. Efflux and steady stae in α-methylglucoside transport in Escherichia coli. J. Bacteriol. 106:362-368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yarmolinsky, M. B., H. Wiesmeyer, H. M. Kalckar, and E. Jordon. 1959. Hereditary defects in galactose metabolism in Escherichia coli mutants. II. Galactose-induced sensitivity. Proc. Natl. Acad. Sci. U. S. A. 45:1786-1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu, D. G., H. M. Ellis, E. C. Lee, N. A. Jenkins, N. G. Copeland, and D. L. Court. 2000. An efficient recombination system for chromosome engineering in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 97:5978-5983. [DOI] [PMC free article] [PubMed] [Google Scholar]