Abstract
Aeromonas hydrophila secretes a number of protein toxins across the outer membrane via the type II secretion system (T2SS). Assembly of the secretion channel ExeD secretin into the outer membrane is dependent on the peptidoglycan binding domain of ExeA. In this study, the peptidoglycan binding domain PF01471 family members were divided into a prokaryotic group and a eukaryotic group. By comparison of their sequence conservation profiles and their representative crystal structures, we found the prokaryotic members to have a highly conserved pocket(s) that is not present in the eukaryotic members. Substitution mutations of nine amino acids of the pocket were constructed in ExeA. Five of the substitution derivatives showed greatly decreased lipase secretion, accompanied by defects in secretin assembly. In addition, using in vivo cross-linking and in vitro cosedimentation assays, we showed that these mutations decreased ExeA-peptidoglycan interactions. These results suggest that the highly conserved pocket in ExeA is the binding site for its peptidoglycan ligand and identify residues critical for this binding.
Aeromonas hydrophila uses the type II secretion system (T2SS) to transport aerolysin and other protein toxins across the outer membrane (14). The T2SS, also known as the main terminal branch of the general secretion pathway (GSP), is conserved in numerous Gram-negative bacteria (for a recent review, see reference 15). The trans-envelope apparatus is comprised of more than 14 Gsp proteins, with a distal outer membrane secretion channel composed of a 12- to 14-member multimer of GspD, called the secretin (4, 9, 24). In A. hydrophila, the Gsp homologues are termed Exe proteins (14). ExeA and ExeB, although their homologues are not conserved in all T2SSs, are required for both the transport of ExeD into the outer membrane and its multimerization into the functional secretin of A. hydrophila (2). ExeA and ExeB form a complex in the inner membrane, with the C-terminal domain of the ATPase ExeA and the majority of ExeB exposed on the periplasmic side (11, 26, 27). A peptidoglycan binding motif is present in the periplasmic domain of ExeA, and it has been proposed that interactions between ExeA and peptidoglycan are involved in the function of the ExeAB complex in the assembly of the ExeD secretin (12). In support of this hypothesis, mutagenesis studies have indicated that the peptidoglycan binding motif is required for ExeA-peptidoglycan interactions in vivo and in vitro, which result in multimerization of ExeA as well as ExeD secretin assembly and protein secretion (12, 18).
The putative peptidoglycan binding domain (Pfam number PF01471) is present in a variety of proteins related to bacterial cell wall functions (7). Up to the present, 2,310 proteins have been shown to contain this domain, including eight proteins with crystal structures available (Pfam number PF01471). Although two research groups have reported physical interactions between the peptidoglycan binding domain and peptidoglycan, the specific peptidoglycan ligand is still unknown (3, 18).
Seven of the eight crystal structures containing the PF01471 domain (gelatinase A, gelatinase B, Prommp-1, Prommp-2-Timp-2 complex, human type IV collagenase precursor, human collagenase 3, and fibroblast stromelysin-1) are eukaryotic, and one (hydrolase metallo [Zn] DD-peptidase) is prokaryotic (Pfam number PF01471). The domain features a three-α-helix structure that does not resemble the peptidoglycan binding sites in known crystal structures of transpeptidases (21), transglycosylases (20), lytic transglycosylases (25), lysozymes (29), or OmpA-like peptidoglycan binding proteins (16). All of the seven eukaryotic proteins are matrix metalloproteinases (MMP), which degrade the extracellular matrix (28). They contain an N-terminal domain that structurally resembles the peptidoglycan binding domain of hydrolase metallo (Zn) DD-peptidase but do not likely have a specific peptidoglycan binding function, although they may bind to different carbohydrate ligands. The prokaryotic PF01471 domains are usually present as noncatalytic N- or C-terminal modules, as, for example, in the hydrolase metallo (Zn) DD-peptidase of Streptomyces albus G, N-acetylmuramidase (lysozyme) of Clostridium acetobutylicum, and N-acetylmuramoyl-l-alanine amidase CwlA of Bacillus subtilis (7). The PF01471 members are also present in Gram-negative bacteria, including, for example, Escherichia coli, Vibrio cholerae, Pseudomonas aeruginosa, and A. hydrophila (Pfam database). The ∼80-amino-acid residue peptidoglycan binding domain features two repeats, each 16 residues long, connected by a heptapeptide. In the crystal structure of hydrolase metallo (Zn) DD-peptidase, the residues of the two repeats superimpose remarkably well (7). It was proposed that both of the repeats are required for binding a ligand of repeated structure (7).
In this study, the sequences of the prokaryotic and eukaryotic PF01471 members were compared and found to be quite different in conservation profiles, indicating that they may bind to different ligands. We reasoned that the residues conserved in the prokaryotic group but not in the eukaryotic group may be responsible for specific recognition of peptidoglycan. Nine conserved residues, which cluster and form a pocket on the surface, were chosen for substitution mutagenesis in ExeA. Their effects on lipase secretion, secretin assembly, and interactions between ExeA and peptidoglycan were examined. The effects of the mutations have allowed us to identify critically important residues of the peptidoglycan binding site in ExeA and will facilitate our efforts to identify the specific peptidoglycan ligand.
MATERIALS AND METHODS
Bacterial strains, plasmids, and culture conditions.
A. hydrophila wild-type strain Ah65 and exeA::Tn5-751 insertion mutant strain C5.84 were used in this study. The strains containing the appropriate plasmids were grown in buffered Luria-Bertani (LB) medium supplemented with 1.25 μg/ml chloramphenicol and 50 μg/ml kanamycin at 30°C as previously described (12). A total of 0.04 mM IPTG (isopropyl-β-d-thiogalactopyranoside) was added to induce plasmid-encoded ExeAB. See Table 1 for a complete list of strains and plasmids.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain/plasmid | Genotype/phenotype/descriptionb | Reference or source |
|---|---|---|
| A. hydrophila strains | ||
| Ah65 | Wild type | 14 |
| C5.84 | Ah65 exeA::Tn5-751; Kmr | 13 |
| Plasmids | ||
| pMMB207 | tac promoter, wide-host-range vector; Cmr | 22 |
| pKRJ50.2 | 2.5-kb exeAB BstXI in SmaI of pBluescript II KS(+); Ampr | 13 |
| pKRJ50.2RE | pKRJ50.2 with silent mutations for RsrII, AfilII and BspEIa | This study |
| pRJ31.1 | 2.5-kb exeAB BstXI in SmaI of pMMB207; Cmr | 13 |
| pRJ31.1 F476A | PRJ31.1 encoding ExeA F476A | This study |
| pRJ31.1 K481A | pRJ31.1 encoding ExeA F487A | This study |
| pRJ31.1I F487A | pRJ31.1 encoding ExeA F487A | This study |
| pRJ31.1Q488A | pRJ31.1 encoding ExeA Q488A | This study |
| pRJ31.1D496A | pRJ31.1 encoding ExeA D496A | This study |
| pRJ31.1G497A | pRJ31.1 encoding ExeA G497A | This study |
| pRJ31.1I498W | pRJ31.1 encoding ExeA I498W | This study |
| pRJ31.1T503A | pRJ31.1 encoding ExeA T503A | This study |
| pRJ31.1L507A | pRJ31.1 encoding ExeA L507A | This study |
| pET30a | Expression vector; T7lac promoter; Kmr | Novagen |
| pP-ExeA | N-His P-ExeA fragment in NdeI/XhoI of pET30a | 18 |
| pP-ExeAF476A | pP-ExeA encoding P-ExeAF476A | This study |
| pP-ExeAK481A | pP-ExeA encoding P-ExeAK487A | This study |
| pP-ExeAF487A | pP-ExeA encoding P-ExeAF487A | This study |
| pP-ExeAD496A | pP-ExeA encoding P-ExeAD496A | This study |
See Table 2 for locations of the three restriction sites.
Cmr, chloramphenicol resistance; Ampr, ampicillin resistance; Kmr, kanamycin resistance.
DNA preparation, manipulation, and analysis.
Minipreparations of plasmid DNA were obtained using the alkaline lysis method described previously (23). DNA fragments were isolated from 0.8% agarose using the QIAquick gel extraction kit (Qiagen). Plasmids were transformed into electrocompetent E. coli cells using electroporation as described previously (23). Wide-host-range pMMB207 plasmids were transferred into A. hydrophila strains by conjugation via an E. coli S17-1 donor (12).
Site-directed mutagenesis.
The D496A substitution mutant of exeA was constructed by sequential PCR steps to amplify the exeAB fragments as described previously (6), using pKRJ50.2 as a template. To facilitate introducing more substitution mutations, an SphI/XcmI fragment of exeA was synthesized (Integrated DNA Technologies) with silent mutations to introduce RsrII, AfilII, and BspEI restriction sites and transferred into SphI/XcmI of pKRJ50.2 to produce pKRJ50.2RE. F476A, K481A, F487A, Q488A, G497A, I498W, T503A and L507A mutants were constructed by insertion of a hybridized complementary pair or pairs of oligonucleotides containing desired mutations into the appropriate introduced sites of pKRJ50.2RE. The AsiSI/HindIII or BsiWI/HindIII fragments of exeA containing the constructed mutations were exchanged for the same fragment of pRJ31.1 for functional studies. DNA sequencing was used to rule out the possibility of unwanted mutations. See Table 2 for a list of oligonucleotides used and the locations of the three silent-mutation restriction sites.
TABLE 2.
Oligonucleotides used for in vitro mutagenesis
| Description | Oligonucleotidesa |
|---|---|
| D496A | 5′-GGGCTGAACCCGCACGGCATCGCCGGCAG-3′ F and complement |
| 5′-TGTAAAACGACGGCCAGT-3′ upstream | |
| 5′-CAGGAAACAGCTATGAC-3′ downstream | |
| F476A | 5′-GACCGCAAGGTGCGTCGCGCCGACGCCGAGC-3′ F |
| 5′-TTAAGCTCGGCGTCGGCGCGACGCACCTTGCG-3′ R | |
| K481A | 5′-GACCGCAAGGTGCGTCGCTTCGACGCCGAGCTTGCGAACAAGTTG-3′ F |
| 5′-CAGCAGTTCCAGCGCGAGCAGGGGCTGAAT-3′ F | |
| 5′-CCGGATTCAGCCCCTGCTCGCGCTGGAACTGCTGCAACTTGTTC-3′ R | |
| 5′-GCAAGCTCGGCGTCGAAGCGACGCACCTTGCG-3′R | |
| F487A | 5′-TTAAGAACAAGTTGCAGCAGGCCCAGCGCGAGCAGGGGCTGAAT-3′ F |
| 5′-CCGGATTCAGCCCCTGCTCGCGCTGGGCCTGCTGCAACTTGTTC-3′ R | |
| Q488A | 5′-TTAAGAACAAGTTGCAGCAGTTCGCCCGCGAGCAGGGGCTGAAT-3′ F |
| 5′-CCGGATTCAGCCCCTGCTCGCGGGCGAACTGCTGCAACTTGTTC-3′ R | |
| G497A | 5′-CCGGACGCGATCGCCGGCAGCAATACCCTG-3′ F |
| 5′-TTGCGCCTCAACGTGATGGCCGGTGAGCCCATGCCG-3′ F | |
| 5′-GGCATGGGCTCACCGGCCATCACGTT-3′ R | |
| 5′-GAGGCGCAACAGGGTATTGCTGCCGGCGATCGCGT-3′ R | |
| I498W | 5′-CCGGACGGCTGGGCCGGCAGCAATACCCTG-3′ F |
| 5′-TTGCGCCTCAACGTGATGGCCGGTGAGCCCATGCCG-3′ F | |
| 5′-GGCATGGGCTCACCGGCCATCACGTT-3′ R | |
| 5′-GAGGCGCAACAGGGTATTGCTGCCGGCCCAGCCGT-3′ R | |
| T503A | 5′-CCGGACGGCATCGCCGGCAGCAATGCCCTG-3′ F |
| 5′-TTGCGCCTCAACGTGATGGCCGGTGAGCCCATGCCG-3′ F | |
| 5′-GGCATGGGCTCACCGGCCATCACGTT-3′ R | |
| 5′-GAGGCGCAACAGGGCATTGCTGCCGGCGATGCCGT-3′ R | |
| L507A | 5′-CCGGACGGCATCGCCGGCAGCAATACC-3′ F |
| 5′-CTGTTGCGCGCCAACGTGATGGCCGGTGAGCCCATGCCG-3′ F | |
| 5′-GGCATGGGCTCACCGGCCATCACGTT-3′ R | |
| 5′-GGCGCGCAACAGGGTATTGCTGCCGGCGATGCCGT-3′ R |
The substituted nucleotides are in bold; the nucleotides that match the restriction sites RsrII, AfilII, BspEI, and XcmI in pKRJ50.2RE are underlined; F, forward; R, reverse.
Lipase assay.
Lipase activity was measured by assaying the release of p-nitrophenol from p-nitrophenol caprylate (1). Cells were grown to an optical density at 600 nm (OD600) of 1.5. A total of 100 μl of culture supernatant was added to 900 μl of substrate buffer containing 1 mM p-nitrophenol caprylate (pNPC), 100 mM Tris (pH 8.0), and 0.2% Triton X-100, and the reaction mixture was incubated for 30 min at room temperature, during which the OD410 was measured at 5-min intervals. One unit of lipase activity equals 1 nmol pNPC hydrolyzed per minute.
ExeD immunoblot assays.
Whole-cell samples expressing ExeA derivatives were separated on 3 to 8% Criterion precast polyacrylamide Tris-acetate gradient gels (Bio-Rad) and transferred to polyvinylidene difluoride (PVDF) membranes for anti-ExeD immunoblotting (12). A chemiluminescent ECL Advance kit (GE Healthcare) was used for signal development.
Chemical cross-linking.
In vivo cross-linking of ExeA to peptidoglycan was performed as described previously (12). After incubation with 0.5 mM 3′-dithiobis[sulfosuccinimidylpropionate] (DTSSP) at room temperature for 5 min, the peptidoglycan samples were extracted and treated with 5% β-mercaptoethanol before sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). In vitro cross-linking of P-ExeA in the presence of peptidoglycan fragments was performed as described previously (18), except that 0 to 2 mM DTSSP was used.
Construction and purification of P-ExeA variants.
The soluble periplasmic domain of ExeA (P-ExeA) of the wild type and constructed mutant variants were prepared as follows. DNA fragments encoding N-His P-ExeA variants were amplified from pRJ31.1 substitution mutation plasmids by PCR with 5′-CATATGCACCATCATCATCATCATCAGTTCTTCGGCTTCTTCCCC and 5′-CTCGAGTCAGGAAGCCTCCTCCGACAATGTG as primers and inserted into the NdeI/XhoI restriction sites of the pET30a vector to construct pP-ExeA substitution mutation plasmids. All plasmids were sequenced to rule out the possibility of unwanted mutations before being electroporated into E. coli BL21(DE3) for protein expression. P-ExeAs were purified from the cell lysates by His tag affinity chromatography and ion-exchange chromatography as described previously (18). Protein concentrations were determined by measuring UV absorbance at 280 nm (UV280) with theoretical extinction coefficients.
Cosedimentation.
Peptidoglycan was prepared from A. hydrophila C.5.84 cells by a method described previously (8). Purified P-ExeA proteins (1.25 to 5 nM) were used in the cosedimentation assay as described previously (18). The pellet samples were applied to SDS-PAGE gels and immunoblotted with ExeA antiserum.
Native PAGE.
Native-gradient PAGE was performed as described previously (18). Purified P-ExeA proteins (1 mg/ml) were incubated at 37°C for 1 h before being applied to the gel. The proteins were visualized by Coomassie brilliant blue (CBB) R-250 staining as for SDS-PAGE gels.
RESULTS
Separation of the prokaryotic and eukaryotic members of the PF01471 family.
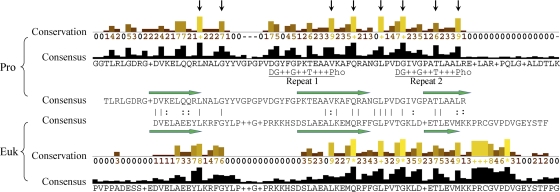
The putative peptidoglycan binding domain family PF01471 contains 2,310 members, with 239 of them designated seed sequences (Pfam database). The 239 seed sequences include 52 eukaryotic and 183 prokaryotic sequences (1 archaea, 6 phages, and 176 bacteria). The two groups of sequences were aligned separately by ClustalW2 (17) and viewed by Jalview (5). The consensus sequences of the two groups were also aligned for comparison of conserved amino acid residues (Fig. 1). We chose the crystal structures of gelatinase A (PDB entry 1CK7) and hydrolase metallo (Zn) DD-peptidase (PDB entry 1LBU) as models for the eukaryotic structure and prokaryotic structure, respectively. Both of the groups show two conserved peptide regions linked by a nonconserved region of variable length. They share apparent similarity in amino acid conservation profiles, as well as the same arrangement of three α-helical regions, which is the characteristic fold of the PF01471 family (Fig. 1). However, the two groups also show significant variation in many conserved amino acids and different lengths of the two conserved peptide regions. It is likely that the residues conserved between both groups are important in the formation of the three-α-helix fold, with the residue variations leading to their different binding functions, including, for example, recognition of peptidoglycan in the prokaryotic group and another ligand (possibly carbohydrate) in the eukaryotic group.
FIG. 1.
Alignments of prokaryotic and eukaryotic members of the putative peptidoglycan binding domain family. The 183 seed sequences of prokaryotic members (top) and 52 seed sequences of eukaryotic members (bottom) of the PF01471 family were aligned separately by ClustalW2 (15) and viewed by Jalview for conservation profiles (5). The respective consensus sequences were aligned (center). The residues with conservation indexes of 5 and above that were conserved in both groups are indicated with vertical arrows. The three α-helical regions are indicated by green arrows, using the crystal structures of hydrolase metallo (Zn) DD-peptidase (PDB entry 1LBU) and gelatinase A (PDB entry 1CK7) as models for the prokaryotic group and eukaryotic group, respectively. The characteristic two-repeat sequences identified previously (7) are underlined. Pho, hydrophobic residues.
Selection and mapping of prokaryote-specific residues.
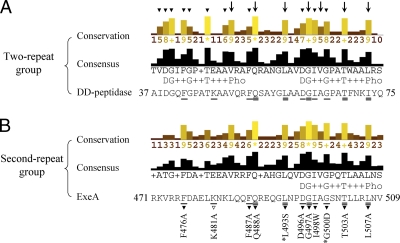
We hypothesize that the residues conserved in the prokaryotic group but not in the eukaryotic group (prokaryote-specific residues) are required for peptidoglycan binding. However, we found that many prokaryote-specific residues are not conserved in ExeA. In particular, only one of the two conserved repeats (DG++G++T+++Pho, where Pho indicates hydrophobic residues), which have been reported as a characteristic feature of the peptidoglycan binding domain family (7), is present in ExeA. Phylogenetic tree analysis indicated that the prokaryotic PF01471 family members could be divided into three major groups (see Fig. S1 in the supplemental material). The two-repeat group, which includes hydrolase metallo (Zn) DD-peptidase, has the characteristic two-repeat sequence. The other two groups contain either only the first repeat (the first-repeat group) or only the second-repeat sequence (the second-repeat group). ExeA belongs to the second-repeat group. The 49 two-repeat members and 34 second-repeat members were aligned separately in Fig. 2 (see Fig. S1 and Table S1 in the supplemental material for a complete list of the sequences). Comparison of the consensus profiles of the two-repeat and second-repeat groups shows that only 7 of 11 prokaryote-specific residues in the two-repeat group are conserved in the second-repeat group, and of these only five are conserved in ExeA.
FIG. 2.
Alignments of the two-repeat group members and the second-repeat group members of the PF01471 family. (A) Alignment of the 49 two-repeat group members. (B) Alignment of the 34 second-repeat group members. The sequences of hydrolase metallo (Zn) DD-peptidase of S. albus G (accession number P00733) and ExeA of A. hydrophila (accession number P45754) are indicated. The common residues that are also conserved in the eukaryotic group with conservation indexes of 5 and above are indicated with arrows. The 11 prokaryote-specific residues with conservation indexes of 5 and above in the two-repeat group are indicated with arrowheads. The five prokaryote-specific residues and five commonly conserved residues chosen for mutagenesis in ExeA are indicated by one line or two lines of underlining, respectively. The non-highly conserved residue K481 subjected to mutagenesis is indicated with an unfilled arrowhead. *L493S and *G500D have been studied previously and are not included in this study. The 11 residues are mapped in the crystal structure of hydrolase metallo (Zn) DD-peptidase in Fig. 3.
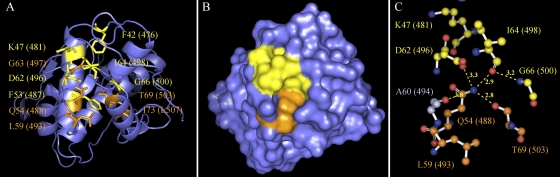
The five prokaryote-specific residues conserved in ExeA are F476, F487, D496, I498, and G500. These correspond to F42, F53, D62, I64, and G66 in hydrolase metallo (Zn) DD-peptidase (Fig. 2). When the five residues were mapped on the crystal structure of hydrolase metallo (Zn) DD-peptidase, they were found to cluster together (Fig. 3 A). More interestingly, these prokaryote-specific residues plus five commonly conserved residues (Q54, L59, G63, T69, and I73) form a pocket on the surface, featuring a centered amine group of Q54 surrounded by the oxygen atoms of D62, I64, T69, and A60. The distances between the nitrogen and oxygen atoms suggest that hydrogen bonds may be formed to maintain the local fold (Fig. 3B and C). As reported previously, hydrolase metallo (Zn) DD-peptidase contains another almost identical pocket that is comprised mainly of the first-repeat residues (7). We propose that the pocket(s) is the peptidoglycan binding site and the prokaryote-specific residues play critical roles in peptidoglycan recognition. To further confine the binding site, we also chose a non-highly conserved residue at the edge of the pocket in the following mutagenesis studies. This residue corresponds to K47 in hydrolase metallo (Zn) DD-peptidase and K481 in ExeA.
FIG. 3.
Mapping of a highly conserved pocket in the crystal structure of hydrolase metallo (Zn) DD-peptidase. The six prokaryote-specific residues F42, K47, F53, D62, I64, and G66 (yellow) and five commonly conserved residues Q54, G63, L59, T69, and I73 (orange) are mapped on the structure of hydrolase metallo (Zn) DD-peptidase (PDB entry 1LBU). The corresponding residues in ExeA are indicated in parentheses. (A) Cartoon view of the structure with the above residues illustrated as sticks. (B) Surface view of the structure. (C) Composition of the putative peptidoglycan binding pocket. Nitrogen atoms and oxygen atoms are marked in blue and red, respectively. The possible hydrogen bonds involved in the composition of the pocket are illustrated with the distances indicated in Å. F42, F53, G63, and I73 are buried beneath the pocket and are not shown in panel C. The images were generated by PyMOL Molecular Graphics System, version 1.3 (Schrödinger, LLC).
Mutagenesis of the pocket-forming residues in ExeA.
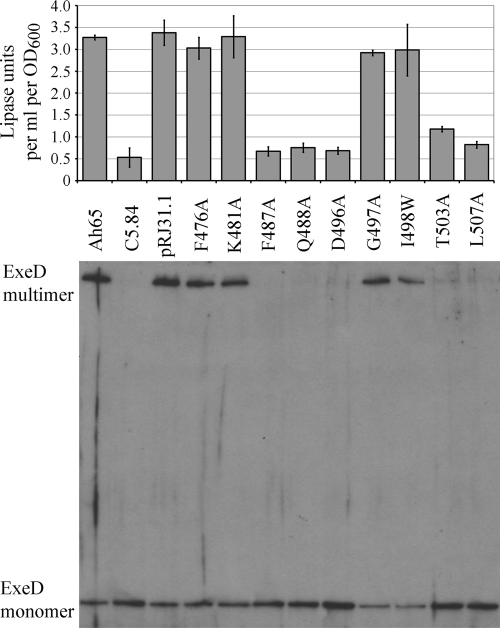
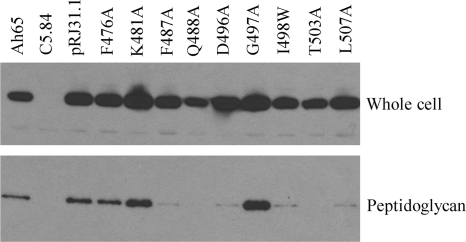
In previous studies, we have constructed F487S, L393S, and G500D in ExeA (12). All of the mutations abrogated the normal function of ExeA in its interactions with peptidoglycan, resulting in an inability to assemble the secretin and a defect in aerolysin secretion. In this study, the rest of the residues identified above were subjected to in vitro mutagenesis to create F476A, K481A, Q488A, D496A, G497A, I498W, T503A, and L507A derivatives (their locations are shown in Fig. 3). Since the previous F487S mutation displayed greatly reduced stability (data not shown), we also constructed F487A. Plasmids encoding the mutant derivatives were introduced into exeA mutant A. hydrophila C5.84 cells. The function of the type II secretion system was analyzed by assessing the amounts of lipase released into the media by the cells. As shown in Fig. 4, lipase secretion was reduced dramatically in strains expressing the F487A, Q488A, D496A, T503A, and L507A mutants, whereas strains expressing the other four substitution mutations showed wild-type secretion levels. These strains were also assayed for assembly of the ExeD secretin. Assembled ExeD is resistant to SDS/heating and runs as a high-molecular-weight (MW) band barely entering gradient SDS-PAGE gels (12). Figure 4 shows that in strains expressing the F487A, Q488A, D496A, T503A, and L507A ExeA mutants, ExeD migrated as a monomer band near the bottom of the gel, indicating that ExeD failed to assemble into the multimeric secretin and therefore explaining the secretion defect of the five strains. The I498W strain, although it achieved wild-type secretion levels, showed an approximately 2-fold decrease in secretin assembly. This is consistent with our finding that full lipase secretion does not require full secretin assembly (data not shown).
FIG. 4.
Lipase secretion and ExeD secretin assembly by A. hydrophila strains expressing substitution mutation derivatives of ExeA. Lipase activity in the culture supernatant of A. hydrophila wild-type strain Ah65 and exeA mutant strain C5.84 complemented with wild-type ExeA (pRJ31.1) or substitution derivatives were assayed as described in Materials and Methods (top). The values of lipase unit per ml per OD600 culture supernatant represent the average obtained from three independent experiments for each strain, with standard deviation indicated. The whole-cell samples were analyzed for secretin assembly on 3 to 8% gradient SDS-PAGE gels and immunoblotted with anti-ExeD serum (bottom). The ExeD multimers and monomers are indicated.
We used in vivo cross-linking to examine interactions between the ExeA mutants and peptidoglycan as described in our previous studies (12). The cells were incubated with cleavable cross-linker DTSSP and subjected to peptidoglycan extraction. Figure 5 shows that wild-type ExeA and three mutants, the F476A, K481A, and G497A mutants, were present in the peptidoglycan samples isolated after the cross-linking. However, the F487A, Q488A, D496A, I498W, T503A, and L507A mutants were cross-linked to peptidoglycan at a much lower level, consistent with their defect in ExeD secretin assembly.
FIG. 5.
In vivo cross-linking of ExeA substitution mutation derivatives to peptidoglycan. A. hydrophila Ah65 cells and exeA mutant C5.84 cells expressing wild-type ExeA (pRJ31.1) or substitution mutants were incubated with cleavable cross-linker DTSSP, and peptidoglycan was extracted with an SDS boiling method as described previously (12). The peptidoglycan samples were treated with reducing reagent to release cross-linked proteins before SDS-PAGE and anti-ExeA immunoblotting. The un-cross-linked whole-cell samples were also applied as controls for the ExeA expression level.
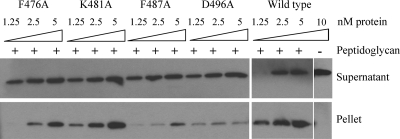
To confirm the in vivo cross-linking results, we constructed and purified the periplasmic domains of ExeAs (P-ExeAs) that contain F476A and K481A (which did not disrupt the normal function of ExeA) and F487A and D496A (which disrupted its normal function). In cosedimentation assays, the P-ExeAs were incubated with peptidoglycan purified from A. hydrophila and centrifuged to pellet the peptidoglycan sacculi and therefore the bound protein, which was detected by anti-ExeA immunoblotting (Fig. 6). Wild-type P-ExeA bound to peptidoglycan, as seen previously (18), whereas the F487A and D496A derivatives showed much-decreased binding, confirming the in vivo cross-linking results.
FIG. 6.
Cosedimentation of P-ExeA substitution mutation derivatives with peptidoglycan. P-ExeA wild-type protein and substitution mutation derivatives (1.25 to 5 nM) were incubated with A. hydrophila peptidoglycan and centrifuged to pellet the peptidoglycan. The supernatant and pellet samples were applied to SDS-PAGE gels and immunoblotted with ExeA antiserum. The wild-type protein (10 nM) was also centrifuged in the absence of peptidoglycan as a control.
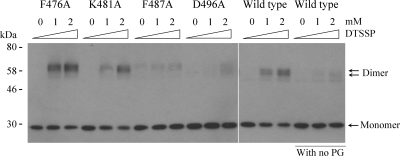
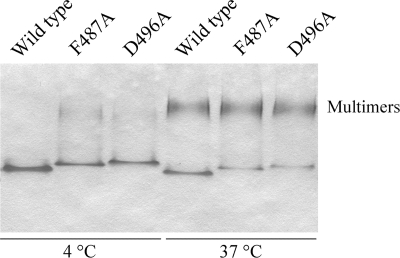
We have previously shown that interaction with peptidoglycan fragments induces P-ExeA to form large multimers, which can be cross-linked by DTSSP into bands of apparent dimers (18). The wild-type and four mutant P-ExeAs were incubated with mutanolysin-hydrolyzed peptidoglycan fragments at 4°C, followed by cross-linking with 0 to 2 mM DTSSP. The mixtures were analyzed by anti-ExeA immunoblotting. Figure 7 shows that the F476A and K481A proteins were cross-linked into the distinctive closely spaced double bands in a manner similar to that of the wild-type protein, whereas the F487A and D496A derivatives were cross-linked only at the background level. This indicates that these two residues are required for the peptidoglycan-induced multimerization. The observed result could be due to either loss of peptidoglycan binding as suggested by the in vivo cross-linking data or alternatively loss of specific residues required for multimerization. We have previously shown that the requirement of peptidoglycan for multimerization was abrogated when P-ExeA was incubated at an elevated temperature (30°C and above) (18). The F487A and D496A P-ExeA derivatives were examined for the temperature-induced multimerization in a native PAGE gel (Fig. 8). The data show that the two mutants are still able to multimerize, confirming that the two residues are required for peptidoglycan binding rather than for multimerization and also that peptidoglycan binding precedes and promotes multimerization at low temperature, as found previously (18).
FIG. 7.
Cross-linking analysis of interactions between P-ExeA substitution mutation derivatives and peptidoglycan fragments. P-ExeA wild-type protein and substitution mutation derivatives were incubated with mutanolysin-digested peptidoglycan fragments and cross-linked with 0 to 2 mM DTSSP. The wild-type protein was also cross-linked in the absence of peptidoglycan (PG) as a control. The cross-linked samples were applied to SDS-PAGE gels and immunoblotted with ExeA antiserum. The apparent dimers are indicated.
FIG. 8.
Native PAGE analysis of temperature-induced multimerization of P-ExeA variants. Purified P-ExeA proteins were incubated at 37°C for 1 h and applied to native PAGE gels as previously described (18). Samples without incubation were also applied as controls. The gel was stained with Coomassie brilliant blue R-250. The multimers are indicated.
DISCUSSION
In this study, we have conducted the first systematic mutagenesis of the peptidoglycan binding pocket of a member of the Pfam PF01471 family. In comparing the sequences of the prokaryotic and eukaryotic members of the family, we found dramatic differences in the residue conservation profiles, suggesting that they may recognize different ligands (as is also suggested by their different functions). It should be noted that the differences between the two groups are not absolute. A few atypical members of the prokaryotic group have the characteristic conserved residues of the eukaryotic members (see Fig. S1 in the supplemental material), and they therefore may bind to the eukaryotic ligand(s). None of the eukaryotic members has the characteristic sequence conserved in the prokaryotic group. When the five residues conserved in the prokaryotic group but not in the eukaryotic group were mapped in the crystal structure of hydrolase metallo (Zn) DD-peptidase, they clustered at one location and formed a pocket along with five commonly conserved residues, centered by the amine of Q54, which appears to form hydrogen bonds with surrounding oxygen atoms (Fig. 3). It is likely that the highly conserved pocket is the peptidoglycan binding site. It is also interesting to find that ExeA and many of the PF01471 members contain only one of the two identical pockets seen in hydrolase metallo (Zn) DD-peptidase, which have been proposed to bind to a repeated peptidoglycan structure (7). The presence of only one binding site in ExeA may have greatly facilitated our mutagenesis studies, since mutation of only one of two binding sites would presumably have much less of an effect on binding.
To better understand the putative peptidoglycan binding site within ExeA, we created substitution mutations for nine residues of the pocket. Mutants expressing F487A, Q488A, D496A, T503A, and L507A alleles abrogated lipase secretion, accompanied by defects in ExeD secretin assembly. These mutants also showed much-decreased interactions with peptidoglycan in vivo and in vitro (Fig. 4 to 7). Although the I498W derivative was able to fully support lipase secretion, much less secretin was assembled compared to the wild-type cells (Fig. 4). Consistently, this derivative showed much-decreased cross-linking to peptidoglycan, suggesting that this residue is involved in peptidoglycan binding but not absolutely required. Indeed, a substitution of this residue to alanine did not have any effects on the function of the protein (data not shown). In addition, this residue is not conserved in the first DG++G++T+++Pho repeat in the two-repeat members of the family (Fig. 2). The two P-ExeA proteins containing the F487A and D496A substitutions still formed multimers at elevated temperature in vitro (Fig. 8), ruling out the possibility that these mutations caused globular folding defects. These data confirm our previous finding that the ExeA-peptidoglycan interactions are involved in the role of ExeAB in the ExeD secretin assembly (12) and also identify the highly conserved pocket as critically important for these interactions. The F476A, K481A, and G497A mutants had a wild-type phenotype, although the F476A mutant showed reduced peptidoglycan binding at a low protein concentration in vitro (Fig. 6). F476 and G497 are embedded in the protein structure, whereas non-highly conserved K481 is located at the edge of the binding site (Fig. 3).
The identification of the six critical residues F487A, Q488A, D496A, I498, T503A, and L507A in this study, together with previously identified L493 and G500, demonstrates the importance of the highly conserved pocket in peptidoglycan binding. Importantly, both the prokaryote-specific residues and the commonly conserved residues are required for the function and/or the folding of the pocket. However, the specific peptidoglycan ligand is still unknown. Does the domain bind to a universal peptidoglycan ligand, or do different members of the PF01471 family bind to different peptidoglycan structures? In an attempt to gain further insight into the relative importance of the identified conserved residues and nonconserved residues of the peptidoglycan binding pocket of the prokaryotic PF01471 motif, we performed molecular docking analysis using AutoDock Vina, version 1.1.1 (30). We were unable to successfully dock the structure of the muropeptide GlcNAc-1,6-anhydroMurNAc-l-Ala-γ-d-Glu-m-A2pm-d-Ala (19) onto either the modeled pocket of the PF0741 region of ExeA or the predicted pocket of hydrolase metallo (Zn) DD-peptidase. The most energetically favorable associations obtained yielded predicted binding energies of −3.5 kcal/mol or less and were all peripheral associations involving few polar contacts.
Biochemical and crystallographic studies aimed at identifying the specific peptidoglycan structure bound by the site in ExeA are in progress.
Supplementary Material
Acknowledgments
This research was supported by grants from the Natural Sciences and Engineering Research Council of Canada and the Canadian Institutes of Health Research. Structural analysis of P-ExeA was supported by a group grant from the Saskatchewan Health Research Foundation to the Molecular Design Research Group, University of Saskatchewan.
We thank Louis Delbaere (deceased) for helpful discussions concerning hydrogen bonding in the peptidoglycan binding site.
Footnotes
Published ahead of print on 22 October 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Aragon, V., S. Kurtz, A. Flieger, B. Neumeister, and N. P. Cianciotto. 2000. Secreted enzymatic activities of wild-type and pilD-deficient Legionella pneumophila. Infect. Immun. 68:1855-1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ast, V. M., I. C. Schoenhofen, G. R. Langen, C. W. Stratilo, M. D. Chamberlain, and S. P. Howard. 2002. Expression of the ExeAB complex of Aeromonas hydrophila is required for the localization and assembly of the ExeD secretion port multimer. Mol. Microbiol. 44:217-231. [DOI] [PubMed] [Google Scholar]
- 3.Briers, Y., G. Volckaert, A. Cornelissen, S. Lagaert, C. W. Michiels, K. Hertveldt, and R. Lavigne. 2007. Muralytic activity and modular structure of the endolysins of Pseudomonas aeruginosa bacteriophages phiKZ and EL. Mol. Microbiol. 65:1334-1344. [DOI] [PubMed] [Google Scholar]
- 4.Chen, L. Y., D. Y. Chen, J. Miaw, and N. T. Hu. 1996. XpsD, an outer membrane protein required for protein secretion by Xanthomonas campestris pv. campestris, forms a multimer. J. Biol. Chem. 271:2703-2708. [DOI] [PubMed] [Google Scholar]
- 5.Clamp, M., J. Cuff, S. M. Searle, and G. J. Barton. 2004. The Jalview Java alignment editor. Bioinformatics 20:426-427. [DOI] [PubMed] [Google Scholar]
- 6.Cormack, B. 2001. Directed mutagenesis using the polymerase chain reaction. Curr. Protoc. Mol. Biol. Chapter 8:Unit 8.5. [DOI] [PubMed]
- 7.Ghuysen, J. M., J. Lamotte-Brasseur, B. Joris, and G. D. Shockman. 1994. Binding site-shaped repeated sequences of bacterial wall peptidoglycan hydrolases. FEBS Lett. 342:23-28. [DOI] [PubMed] [Google Scholar]
- 8.Glauner, B. 1988. Separation and quantification of muropeptides with high-performance liquid chromatography. Anal. Biochem. 172:451-464. [DOI] [PubMed] [Google Scholar]
- 9.Hardie, K. R., A. Seydel, I. Guilvout, and A. P. Pugsley. 1996. The secretin-specific, chaperone-like protein of the general secretory pathway: separation of proteolytic protection and piloting functions. Mol. Microbiol. 22:967-976. [DOI] [PubMed] [Google Scholar]
- 10.Reference deleted.
- 11.Howard, S. P., H. G. Meiklejohn, D. Shivak, and R. Jahagirdar. 1996. A TonB-like protein and a novel membrane protein containing an ATP-binding cassette function together in exotoxin secretion. Mol. Microbiol. 22:595-604. [DOI] [PubMed] [Google Scholar]
- 12.Howard, S. P., C. Gebhart, G. R. Langen, G. Li, and T. G. Strozen. 2006. Interactions between peptidoglycan and the ExeAB complex during assembly of the type II secretin of Aeromonas hydrophila. Mol. Microbiol. 59:1062-1072. [DOI] [PubMed] [Google Scholar]
- 13.Jahagirdar, R., and S. P. Howard. 1994. Isolation and characterization of a second exe operon required for extracellular protein secretion in Aeromonas hydrophila. J. Bacteriol. 176:6819-6826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang, B., and S. P. Howard. 1991. Mutagenesis and isolation of Aeromonas hydrophila genes which are required for extracellular secretion. J. Bacteriol. 173:1241-1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johnson, T. L., J. Abendroth, W. G. Hol, and M. Sandkvist. 2006. Type II secretion: from structure to function. FEMS Microbiol. Lett. 255:175-186. [DOI] [PubMed] [Google Scholar]
- 16.Kojima, S., K. Imada, M. Sakuma, Y. Sudo, C. Kojima, T. Minamino, M. Homma, and K. Namba. 2009. Stator assembly and activation mechanism of the flagellar motor by the periplasmic region of MotB. Mol. Microbiol. 73:710-718. [DOI] [PubMed] [Google Scholar]
- 17.Larkin, M. A., G. Blackshields, N. P. Brown, R. Chenna, P. A. McGettigan, H. McWilliam, F. Valentin, I. M. Wallace, A. Wilm, R. Lopez, J. D. Thompson, T. J. Gibson, and D. G. Higgins. 2007. ClustalW and ClustalX version 2. Bioinformatics 23:2947-2948. [DOI] [PubMed] [Google Scholar]
- 18.Li, G., and S. P. Howard. 2010. ExeA binds to peptidoglycan and forms a multimer for assembly of the type II secretion apparatus in Aeromonas hydrophila. Mol. Microbiol. 76:772-781. [DOI] [PubMed] [Google Scholar]
- 19.Lim, J. H., M. S. Kim, H. E. Kim, T. Yano, Y. Oshima, K. Aggarwal, W. E. Goldman, N. Silverman, S. Kurata, and B. H. Oh. 2006. Structural basis for preferential recognition of diaminopimelic acid-type peptidoglycan by a subset of peptidoglycan recognition proteins. J. Biol. Chem. 281:8286-8295. [DOI] [PubMed] [Google Scholar]
- 20.Lovering, A. L., M. Gretes, and N. C. Strynadka. 2008. Structural details of the glycosyltransferase step of peptidoglycan assembly. Curr. Opin. Struct. Biol. 18:534-543. [DOI] [PubMed] [Google Scholar]
- 21.Macheboeuf, P., C. Contreras-Martel, V. Job, O. Dideberg, and A. Dessen. 2006. Penicillin binding proteins: key players in bacterial cell cycle and drug resistance processes. FEMS Microbiol. Rev. 30:673-691. [DOI] [PubMed] [Google Scholar]
- 22.Morales, V. M., A. Bäckman, and M. Bagdasarian. 1991. A series of wide-host-range low-copy-number vectors that allow direct screening for recombinants. Gene 97:39-47. [DOI] [PubMed] [Google Scholar]
- 23.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 24.Sandkvist, M. 2001. Biology of type II secretion. Mol. Microbiol. 40:271-283. [DOI] [PubMed] [Google Scholar]
- 25.Scheurwater, E., C. W. Reid, and A. J. Clarke. 2008. Lytic transglycosylases: bacterial space-making autolysins. Int. J. Biochem. Cell Biol. 40:586-591. [DOI] [PubMed] [Google Scholar]
- 26.Schoenhofen, I. C., C. Stratilo, and S. P. Howard. 1998. An ExeAB complex in the type II secretion pathway of Aeromonas hydrophila: effect of ATP-binding cassette mutations on complex formation and function. Mol. Microbiol. 29:1237-1247. [DOI] [PubMed] [Google Scholar]
- 27.Schoenhofen, I. C., G. Li, T. G. Strozen, and S. P. Howard. 2005. Purification and characterization of the N-terminal domain of ExeA: a novel ATPase involved in the type II secretion pathway of Aeromonas hydrophila. J. Bacteriol. 187:6370-6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seiki, M. 1999. Membrane-type matrix metalloproteinases. APMIS 107:137-143. [DOI] [PubMed] [Google Scholar]
- 29.Strynadka, N. C., and M. N. James. 1996. Lysozyme: a model enzyme in protein crystallography. EXS 75:185-222. [DOI] [PubMed] [Google Scholar]
- 30.Trott, O., and J. O. Arthur. 2010. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J. Comput. Chem. 31:455-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.