Abstract
Serum response factor (SRF) recruits members of two families of signal-regulated coactivators, the extracellular signal-regulated kinase (ERK)-regulated ternary complex factors (TCFs) and the actin-regulated myocardin-related transcription factors (MRTFs), to its target genes through its DNA-binding domain. Whether coactivator association is required for SRF function in vivo and whether particular SRF functions reflect specific coupling to one or the other signal pathway have remained largely unexplored. We show that SRF is essential for thymocyte positive selection and thymic Treg and NK T-cell development but dispensable for early thymocyte development and negative selection. Expression of wild-type SRF, or mutants lacking the N-terminal phosphorylation sites or C-terminal transcriptional activation domain, restores positive selection in SRF null thymocytes. In contrast, SRF.V194E, which cannot recruit TCF or MRTF family members, is inactive, although it is recruited to target genes. Fusion of a TCF C-terminal activation domain to SRF.V194E effectively restores ERK-dependent single-positive (SP) thymocyte development. The resulting SP thymocytes exhibit normal surface marker expression and proliferation following T-cell receptor cross-linking. Thus, ERK signaling through the TCF pathway to SRF is necessary and sufficient for SRF function in thymocyte positive selection.
The transcription factor serum response factor (SRF) interacts with two families of regulatory cofactors, the ternary complex factors (TCFs; SAP-1, Elk-1, and Net) subfamily of Ets proteins, and the myocardin family (myocardin, MRTF-A/MAL/Mkl1, and MRTF-B/Mkl2), integrating signals required for differentiation and cytoskeletal gene expression in multiple cell types (43, 47). The TCFs are controlled by mitogen-activated protein kinase phosphorylation of their C-terminal regulatory domains (7, 52), and the myocardin-related transcription factors (MRTFs) are controlled by Rho GTPase signaling through their actin-binding RPEL domains (38, 46). In one system, in vitro cardiomyogenesis, cofactor recruitment has been shown to be essential for SRF function (42). Although the different cofactor families compete for a common site on the SRF DNA-binding domain (38, 54, 59), cofactor recruitment is gene specific (38, 54). As a result, SRF target genes exhibit preferential coupling to TCF- or MRTF-linked signals (18, 54), but the molecular basis for this remains imperfectly understood.
The extent to which TCF- or MRTF-linked signaling is necessary or sufficient for a particular SRF function has not been extensively studied. SRF-null embryos fail at gastrulation (2), and tissue-specific inactivation of SRF has revealed diverse developmental roles in multiple different tissues (1, 16, 44). SRF-null cells exhibit aberrant cytoskeletal gene expression and dynamics (49, 51), and the similar phenotypes of SRF- and MRTF-deficient cells show that Rho-actin-MRTF signaling is necessary for the adhesive, motile, and morphogenetic behaviors of hippocampal neurites (26, 40, 50) and cancer cells (37). In contrast, Ras-extracellular signal-regulated kinase (ERK) signaling through the TCFs to SRF plays an important role in thymocyte positive selection (12, 13, 16). In this Ras-ERK-dependent process, the self-peptide-major histocompatibility complex (MHC) interaction with the newly rearranged T-cell receptor (TCR) induces immature CD4+ CD8+ double-positive (DP) thymocytes to differentiate into CD4 and CD8 single-positive (SP) cells (9, 15). Positive selection also requires the Egr family of zinc finger proteins (5, 12, 28, 30), which appear to be the principal TCF-SRF targets in this system (12).
It has remained unclear whether SRF is essential for thymocyte development. Residual positive selection persists in cells lacking all three TCFs, and processes such as β-selection and Treg development remain unaffected by TCF inactivation (12, 13, 55), even though they are pre-TCR/TCR and/or ERK dependent (4, 8, 39). Here we show that SRF is essential for thymocyte positive selection and Treg development, and we report our results of a structure-function analysis of SRF, utilizing retroviral transduction to reexpress SRF mutants in SRF-null cells. We demonstrate that cofactor recruitment is essential for SRF function and, using an SRF-TCF fusion protein incapable of recruiting endogenous SRF cofactors, show that ERK signaling to SRF target genes can functionally substitute for SRF in thymocyte development.
MATERIALS AND METHODS
Mice.
Srff/f C57/BLJ mice (44) were crossed to C57/BLJ CD2-Cre transgenic mice (14), generating Srff/+ hCD2cre mice, which were subsequently backcrossed with Srff/f to generate Srff/f hCD2cre animals (referred to here as SrfhCD2cre). SrfhCD2cre animals were crossed as appropriate to TCR-αβ transgenic strains carrying the OT-II TCR (3) (from Bill Heath, WEHI, Melbourne, Australia), the F5 TCR (33), and the HY TCR (25). For reconstitution, 8- to 10-week-old Rag2−/− mice who received acidified water for 1 week were exposed to two doses of 500 rads separated by 3 h. Bone marrow from the femurs of 6- to 8-week-old retrovirally transduced animals (see below) was injected into the tail veins (0.5 ×106 to 1.0 ×106 cells/mouse), and analysis was performed 6 weeks later. Animals were maintained under specific-pathogen-free conditions in the Cancer Research UK Biological Resources Unit. Animal experimentation, approved by the Cancer Research UK Animal Ethics committee, was carried out under Home Office license PPL 80/2152.
Flow cytometry.
Cell preparations and analysis were as described previously (12, 13, 55). For analysis we used a FACSCalibur or LSR-II apparatus (BD Biosciences, San Jose, CA) with CellQuest or FloJo software. A MoFlo cell sorter (DakoCytomation, Fort Collins, CO) was used to isolate DP thymocytes, defined by cell surface marker expression, to >97% purity (fluorescence-activated cell sorter [FACS] analysis). Antibodies (12, 55) were conjugated to either fluorescein isothiocyanate (FITC), phycoerythrin (PE), allophycocyanin (APC), or biotin (revealed using streptavidin-peridinin chlorophyll protein [PerCP], streptatvidin-APC, or streptatvidin-Alexa Fluor 750). mCD1d/PBS57 tetramers were generously supplied by the National Institute of Allergy and Infectious Disease MHC Tetramer Core Facility. In the scatter plots, bars show the means and standard errors of the means for each data set. Statistical significance was assessed using the unpaired Student t test. For retroviral reconstitution experiments, the nonpaired Mann-Whitney U test was used because it was unclear whether the data point distribution was Gaussian.
Cell stimulation and signaling analysis.
Cell stimulation was either by spinning cells onto anti-CD3-coated plates or by addition of anti-CD3 followed by goat anti-hamster IgG, as specified in the figures. Flow cytometric analysis of ERK signaling and intracellular protein expression was as described previously (12). For assessment of DP-APC cell conjugation, purified B cells were incubated overnight with various concentrations of ovalbumin (Ova) peptide and 30 μg/ml of lipopolysaccharide (LPS; Sigma), washed four times in warm medium, and mixed with total thymocytes at a ratio of 2:1. Conjugate formation was monitored by flow cytometry for B220+ Thy1+ cells, gating on the DP population. For calcium signaling, 107 thymocytes were incubated with Indo-1 (1 μM) for 45 min at 37°C in 1 ml complete RPMI, then washed in serum-free medium and stained on ice in phosphate-buffered saline with 1% fetal calf serum (FCS) and 2 mM EDTA for markers as appropriate together with unlabeled anti-CD3. Cells were washed and divided into 0.5-ml aliquots in complete RPMI. Washed cells were warmed to 37°C and analyzed for 30 to 45 s before cross-linking of cell-bound anti-CD3 with goat anti-hamster IgG (Jackson Immunoresearch), following which data acquisition was continued. For analysis, cells were gated on the DP population, and the ratio of emission at 410 nm (bound Indo-1) versus 485 nm (unbound Indo-1) was determined as a measure of the intracellular [Ca2+]. For long-term activation of SP thymocytes, cells were stimulated by addition of anti-CD3 and anti-CD28, with analysis 48 h later for CD25 and CD69 expression as described above and for cell cycle entry by a 20-h pulse of bromodeoxyuridine (BrdU).
Gene expression analysis.
Target gene expression was analyzed in sorted DP thymocytes. For analysis of reconstituted virally transduced cells, bulk thymocytes were rested overnight pending initial analysis prior to sorting for the transduced green fluorescent protein-positive (GFP+) DP population. Analysis using SYBR green-based real-time PCR (Invitrogen, Carlsbad, CA) with normalization was to Rps16 expression (12), using the Applied Biosystems 7500 machine; standard curves for each primer set were determined using genomic DNA. Primers were as described previously (12), in addition to the following: Actb intron (fwd), 5′-CTGAGACATGCAAGGAGTGCAA-3′; Actb intron (rev), 5′-AGTAGGTCTAAGTGGAGCCCCTGT-3′; Coro1c intron (fwd), 5′-CGCAGAGCGTGCTTATTCG-3′; Coro1c intron (rev), 5′-TGCCAACCATTTCCAAAACTAA-3′; Fos (fwd), 5′-TGACTGGAGGTCTGCCTGAGGCTT-3′; Fos (rev), 5′-GCTCCAAGGATGGCTTGGGCTC-3′; Gata3 (fwd), 5′-CGTTTTTCGCAGGAGCAGTATC-3′; Gata3 (rev), 5′-ACCACCTCGAGCTCCTTTGAA-3′; Myh9 (fwd), 5′-CCGCAAGTCACCATGGCTCAGCA-3′; Myh9 (rev), 5′-ACTTCTTGGCAGCCCAGTCAGCT-3′; Nr4a1 (fwd), 5′-GCTTGGCACCCACCTCTCCGA-3′; Nr4a1 (rev), 5′-CACGGGTGCGTCCAGAATGCC-3′; Srf (fwd), 5′-CAGGGACACAGGTGCTGTT-3′; Srf (rev), 5′-TGATCATGGGCTGCAGTTT-3′; Vcl intron (fwd), 5′-GATCCTGGTGTCTGTCGCTTCT-3′; Vcl intron (rev), 5′-TGAGCAAAATGCCCCGAA-3′.
Gene deletion analysis.
The efficiency of Srf gene inactivation was determined by comparing the relative abundance of Srf exon 2 with exon 7. Quantitative real-time PCR with primers specific for exon 2 [a and b; Srf (fwd) and Srf (rev), above] and exon 7 (c, 5′-GGTTGGAGGGAACCACTGT-3′, and d, 5′-CTGGGAGAAGGGGGAAGAC-3′) (see Fig. S1A in the supplemental material), in conjunction with mouse universal probes 49 and 69 from Roche (Roche Fast Chemistry; 04913957001), was conducted following the Roche protocol on an Applied Biosystems 7500 QPCR machine. For quantification, wild-type genomic DNA was used to determine the standard curve for each exon.
Chromatin immunoprecipitation and gel mobility shift assays.
Chromatin immunoprecipitation was performed as described previously (12, 38), with quantitation of the percent input DNA recovered. Antibodies were the following: anti-SRF (Santa Cruz Biotechnology sc-335) and anti-FLAG (Sigma F7425). Control experiments to determine backgrounds assessed glyceraldehyde 3-phosphate dehydrogenase (GAPDH) promoter recovery with specific antibodies and target gene recovery with irrelevant antibodies. Additional primers used were Actb (fwd), 5′-AGCGAGATTGAGGAAGAGGATGA-3′, and Actb (rev), 5′-GGCGGAGGCTATTCCTGTACAT-3′. Gel mobility shift assays, with supershifts using anti-FLAG M2, anti-SRF (Sigma), and anti-SAP-1 antibodies were conducted as described previously (13, 59).
Retroviruses and reconstitution.
SRF derivatives were expressed using the pMIG/MSCV-IRES-GFP vector as N-terminally tagged FLAG fusions. SRF mutants were as follows: SRF52Δ114 and SRF265Δ (21) and SRF.V194E (32). SRF.V194E-ElkC and V194E-ElkNA contain Elk-1 residues 307 to 428 joined C-terminal to SRF residue 265, with or without the ERK phospho-acceptor residues mutated to alanine (S324A, S336A, T353A, T363A, T368A, S383A, S389A, T417A, and T422A) (41). GP2-293 cells were contransfected with retroviral constructs and vesicular stomatitis virus G protein by using Fugene under containment conditions. Viral supernatants (from 48 h and 72 h) were then used to infect Phoenix cells by spin infection. Expression of SRF derivatives was monitored by immunoprecipitation with FLAG M2 antibody (Sigma) and Western blotting for anti-FLAG M2 antibody conjugated to horseradish peroxidase (Sigma).
Donor SRFhCD2cre mice were injected intraperitoneally with 0.5 mg fluorouracil (David Bull Laboratories, Lidcomb, New South Wales, Australia) 3 days before bone marrow harvesting. After red blood cell lysis, bone marrow cells were incubated for 4 days in prestimulation mixture (Iscove's modified Dulbecco's medium [IMDM], 2% FCS, 0.03% Primaton RL [Sigma Aldrich], 5 μg/ml pork insulin, 10% WEHI supernatant, 5 ng/ml interluekin-3 [IL-3], 10 ng/ml IL-6, 100 ng/ml stem cell factor [Biosource]). For infection, medium was replaced with freshly prepared viral supernatant supplemented with 4 μg/ml Polybrene. Infection was performed by centrifugation for 45 min at 2,000 × g at room temperature. After 1 h, prestimulation mixture was added to the viral supernatant. A second spin-infection was performed 4 h later, and after 1 h, viral supernatant was replaced with prestimulation mixture overnight. Twenty-four hours later, a third spin-infection was performed. Cells were cultured in prestimulation mixture for a further 3 days, followed by intravenous injection into irradiated Rag2−/− hosts. Tissue analysis was performed 6 to 8 weeks later.
RESULTS
SRF is essential for positive selection.
Inactivation of the Srf gene at the DP stage of thymocyte development leads to a substantial reduction in SP thymocyte numbers, and peripheral T cells lacking SRF are not detectable (16). In contrast, even deletion of all three TCF cofactors does not abolish SP thymocyte development or compromise peripheral T-cell viability (12, 13). To examine the role of SRF in thymocyte development in more detail, we used an Srf allele in which exon 2 is flanked by loxP sites (Srff/f) (see Fig. S1A in the supplemental material) (44) in conjunction with an hCD2-cre transgene (14) to inactivate Srf at the early CD4− CD8− double-negative (DN) stage of thymocyte development (referred to here as SrfhCD2cre). Comparison of the ratio of Srf exon 2 to exon 7 in genomic DNA and total cellular RNA by PCR revealed virtually quantitative deletion of the gene within the DN and DP thymocyte subsets (see Fig. S1B and C), while analysis of whole-thymocyte protein extracts by electrophoretic mobility shift assay showed that SRF DNA-binding activity was reduced to below the detection limit in both subsets (see Fig. S1D).
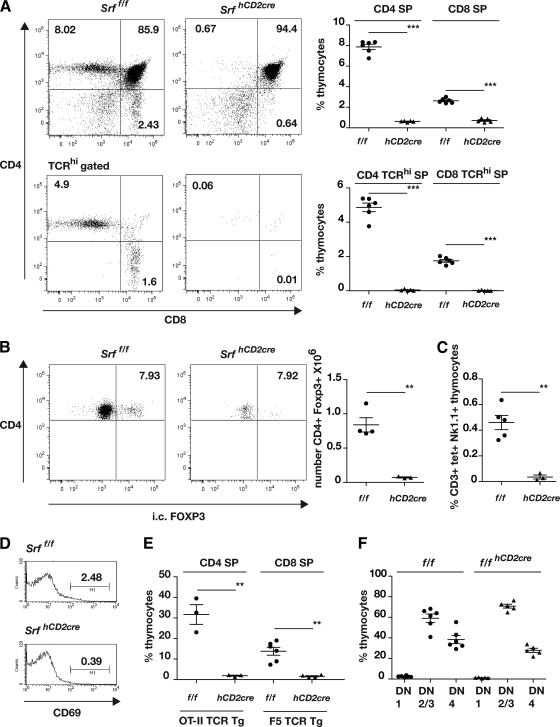
Srf deletion led to no significant change in thymus cellularity (Srff/f, [184 ± 14.5] × 106, n = 6; SrfhCD2cre, [179.2 ± 17.04] × 106, n = 5), but total CD4 and CD8 SP thymocyte numbers were reduced by 92% and 73%, respectively. Mature TCRhi SP thymocytes were virtually absent (Fig. 1A), and PCR analysis of the residual TCRhi SP populations showed that these cells contained at least one undeleted Srff allele (see Fig. S1B in the supplemental material), indicating that an intact Srf gene is essential for SP thymocyte maturation (see Discussion). Thymic Treg (CD4+ FOXP3+) cells, and NKT (NK1.1+ CD3+ CD1d/PBS57 tetramer+) cells were also virtually absent from SrfhCD2cre animals (Fig. 1B and C). The hCD2-cre transgene did not affect thymocyte development in mice carrying an intact Srf allele (see Fig. S2A in the supplemental material). SrfhCD2cre thymuses exhibited substantially reduced numbers of DP cells expressing the selection marker CD69, consistent with a block in positive selection (Fig. 1D). This cannot result from failure of TCR rearrangement, since selection of OT-II or F5 TCR-αβ transgenic thymocytes (3, 33) is also blocked by Srf inactivation (Fig. 1E; see also Fig. S2B). The early DN1, DN2/3, and DN4 subpopulation stages of thymocyte development, as defined by expression of the CD44 and CD25 markers, were largely unaffected by Srf inactivation, and TCR expression was upregulated normally upon transition to the CD44lo CD25lo DN4 population (Fig. 1F; see also Fig. S2C). Within the DN4 population the proportion of γδ TCR+ cells was somewhat elevated, and this was reflected in an ∼5-fold increase in the number of mature HSA- γδ TCR+ thymocytes seen in SrfhCD2cre animals (see Fig. S2D). As observed with TCF mutants, SrfhCD2cre male animals also maintained negative selection of the HY transgene (see Fig. S2D) (12, 25).
FIG. 1.
Thymocyte development in Srf mutant mice. (A) SRF is essential for thymocyte positive selection. Left, representative CD4/CD8 profiles of Srff/f and SrfhCD2cre thymocytes, without or with gating for TCRβhi cells. Right, summary of the data as scatter plots. (B) SRF is essential for the generation of thymic regulatory T cells. Left, representative CD4/FOXP3 profiles of Srff/f and SrfhCD2cre thymocytes. Right, numbers of CD4+ FOXP3+ cells, shown as scatter plots. (C) SRF is essential for NK T-cell development. Proportions of CD3+ Nk1.1+ CD1d/PBS57 tetramer (tet)+ cells are summarized as scatter plots. (D) Reduced CD69 expression in SrfhCD2cre thymocytes. Data are plotted as histograms. (E) Defective selection of OT-II and F5 TCR-αβ transgenic thymocytes in SrfhCD2cre mice. Data are summarized as scatter plots; for primary data, see Fig. S2B in the supplemental material. (F) DN thymocyte distribution is unperturbed in the absence of SRF. Data are summarized as scatter plots; for primary data, see Fig. S2C.
Taken together, these results show that inactivation of Srf does not impair early thymocyte development but leads to a virtually complete block in thymocyte development at a point subsequent to the DN4 stage, without affecting negative selection, at least in the HY model. Consistent with this finding, peripheral T-cell populations in the spleen and lymph nodes contained substantially reduced numbers of T cells (see Fig. S3 in the supplemental material), of which the majority of these also harbored an intact Srf allele, suggesting that only cells that have escaped Srf inactivation can exit and be maintained in the periphery (see Fig. S1B in the supplemental material).
Srf-null DP cells signal normally but exhibit defective IE gene induction.
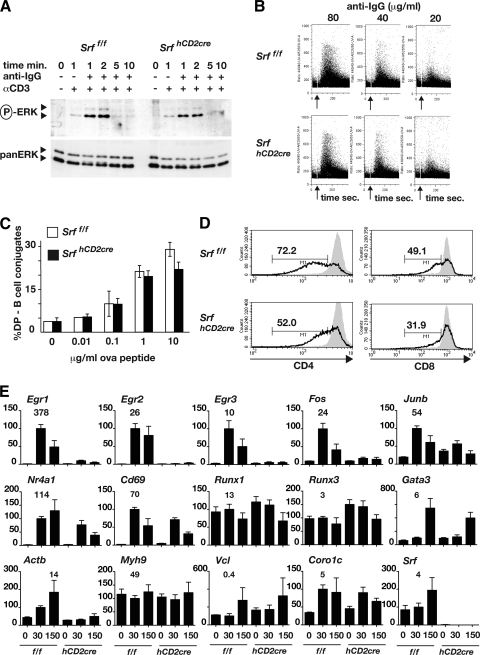
Thymocyte positive selection is absolutely dependent on ERK signaling (15), and calcineurin/NFAT activity is essential for thymocytes to become competent to activate ERK (17). TCR cross-linking on DP thymocytes from Srff/f and SrfhCD2cre animals resulted in similar extents and kinetics of ERK activation (Fig. 2A; see also Fig. S4A in the supplemental material) and calcium signaling (Fig. 2B). Moreover, while SRF is intimately involved in cell motility and adhesion (43), SrfhCD2cre DP thymocytes from OT-II TCR-αβ transgenic animals were not significantly impaired in their ability to form conjugates with LPS-activated B cells presenting Ova peptide (Fig. 2C). SrfhCD2cre thymocytes also exhibited a reduced ability to downregulate the CD4 and CD8 coreceptors following TCR cross-linking in vitro (Fig. 2D).
FIG. 2.
SRF-null thymocytes are competent for TCR-induced signaling but defective for immediate-early gene transcription. (A) ERK activation. Immunoblot analysis of active (phospho-) and total ERK in Srff/f and SrfhCD2cre DP thymocytes following cross-linking of anti-CD3 with goat anti-hamster IgG. (B) Calcium flux in anti-CD3-stimulated DP thymocytes, measured by Indo-1 fluorescence. Events were recorded for 350 s, and the arrow indicates time of cross-linker addition. (C) Formation of peptide-specific conjugates between OT-II TCR-αβ transgenic Srff/f or SrfhCD2cre DP thymocytes and LPS-activated B cells, detected as B220+ Thy1+ cells. (D) Downregulation of CD4 and CD8 coreceptors in Srff/f or SrfhCD2cre DP thymocytes following TCR cross-linking. Solid gray, unstimulated; black line, 22 h after TCR cross-linking via plate-bound anti-CD3. (E) Reverse transcription-PCR analysis of SRF target gene transcripts in Srff/f (f/f) and SrfhCD2cre (hCD2cre) DP cells at the indicated times (in minutes) following TCR cross-linking. Transcripts were quantitated relative to the Rps16 RNA. On the histograms, transcript levels at 30 min after anti-CD3 stimulation were set to 100; the absolute level of each transcript relative to Rps16 is shown above the 30-min time point. Note that the probes for Actb, Coro1c, and Vcl detected intron transcripts, not mRNA.
The preceding results suggest that SrfhCD2cre thymocytes are defective in execution of TCR-induced signaling, and so we examined activation of SRF target genes. TCR-induced activation of the Egr1, Egr2, Egr3, Fos, and Junb genes, which is strongly ERK-TCF dependent in thymocytes (12, 13), was reduced to background levels in SrfhCD2cre cells (Fig. 2E; see also Fig. S4B in the supplemental material). Activation of the Nr4a1 gene, which in thymocytes is also a target for Ca2+-MEF2 signaling (56-58), was less severely affected (Fig. 2E). Despite the low levels of CD69 expression seen in SrfhCD2cre thymocytes in vivo, Cd69 transcription was induced in SrfhCD2cre DP thymocytes following TCR cross-linking, albeit somewhat less strongly than in wild-type cells (Fig. 2E). Expression of Gata3, Runx1, and Runx3, which are strongly implicated in SP thymocyte maturation (for recent reviews, see references 9 and 10), appeared normal in SrfhCD2cre DP thymocytes (Fig. 2E). Transcription of the cytoskeletal SRF targets Vcl, Myh9, and Coro1c was unaffected, but the inducibility of Actb was impaired (Fig. 2E).
SRF function requires cofactor recruitment.
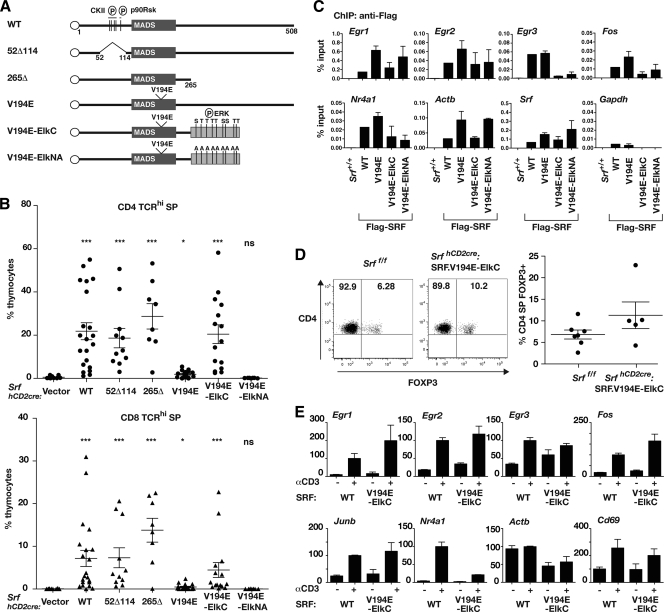
We next investigated the functional domains of SRF required for its activity in thymocyte development. Previous studies of SRF have defined N-terminal sites for phosphorylation by CKII (S77, S79, S83, and S85) (23, 34, 35) and p90rsk (S103) (48), which affect its DNA-binding kinetics; specific residues within the DNA-binding domain essential for its interaction with the TCF and MRTF coactivators (32, 59); and a C-terminal activation domain (20, 21, 24, 31) (Fig. 3A). Recombinant pMIG-SRF-IRES-GFP retroviruses were generated (see Fig. S5 in the supplemental material) and used for infection of SrfhCD2cre bone marrow, which was then used to reconstitute defective hematopoiesis in Rag2−/− animals; the coexpressed GFP marker was used to isolate transduced cell populations for analysis.
FIG. 3.
Structure-function analysis of SRF in SrfhCD2cre thymocytes. (A) Structures of the SRF derivatives analyzed. The SRF DNA-binding domain is shown as a box with the V194E mutation indicated, the FLAG epitope as a circle, the Elk-1 C-terminal sequences as a gray bar, and intact or alanine-substituted phosphorylation sites as vertical lines. WT, wild type. (B) Positive selection in Rag2−/− animals reconstituted with SrfhCD2cre bone marrow transduced with SRF mutant retroviruses. Scatter plots show the proportions of CD4 SP TCRhi (upper) and CD8 SP TCRhi (lower) cells in each virally transduced (GFP+) thymocyte population. (C) SRF derivatives bind SRF targets in the transduced DP thymocyte population. SRF chromatin immunoprecipitation (ChIP) was performed using anti-FLAG on total thymocytes from wild-type animals (+/+) or from Rag2−/− animals reconstituted with SrfhCD2cre bone marrow transduced with the indicated SRF derivatives. Recovery of target promoters was expressed as the percentage of input DNA. Controls were ChIP recovery of the Gapdh promoter, which is not an SRF target, and anti-FLAG ChIP results with wild-type cells (left-most lanes). Each virally transduced population analyzed contained at least 50% GFP+ cells. (D) SRF.V194E-ElkC promotes thymic regulatory T-cell development. (Left) CD4/FOXP3 thymocyte profiles from Srff/f animals and from Rag2−/− animals reconstituted with SrfhCD2cre:SRF.V194E-ElkC bone marrow, the latter gated on virally transduced (GFP+) cells. (Right) Summary of the data as scatter plots. (E) SRF target gene induction in sorted WT DP thymocytes or virally transduced GFP+ DP thymocytes from Rag2−/− animals reconstituted with SrfhCD2cre:SRF.V194E bone marrow (V194E-ElkC). Cells were analyzed either without stimulation (−) or 30 min after TCR cross-linking (+). Transcripts were analyzed by reverse transcription-PCR, quantitated relative to Rps16 RNA, and displayed with levels 30 min after anti-CD3 stimulation set to 100.
Expression of wild-type SRF efficiently restored positive selection of mature CD4 and CD8 TCRhi thymocytes, which were produced in even greater numbers than in wild-type animals (Fig. 3B). Positive selection was restored equally effectively by expression of SRF derivatives lacking the N-terminal phosphorylation sites or C-terminal activation domain; in contrast, thymocytes expressing SRF.V194E, which are unable to recruit either TCF or MRTF coactivators (32, 59), remained virtually completely defective for positive selection (Fig. 3B; see also Fig. S6 in the supplemental material). Ecotopically expressed wild-type SRF, but not SRF.V194E, could form complexes with endogenous thymocyte SAP-1 in gel mobility shift assays with transduced thymocyte extracts (see Fig. S7 in the supplemental material). Retrovirally transduced SRF was expressed at a substantially higher level than endogenous SRF in wild-type thymocytes (see Fig. S7), and this perhaps accounts for the increased efficiency of positive selection seen in the SRF-transduced population. Moreover, effective recruitment of both wild-type SRF and SRF.V194E to SRF target genes, including the Egr family, Fos, Actb, and Srf itself, was directly detectable in transduced thymocyte populations by chromatin immunoprecipitation (Fig. 3C). These data indicate that the major role of SRF in thymocytes is to provide a platform for the recruitment of cofactors responsible for the activation of genes involved in T-cell development and that SRF's phosphorylation and transcriptional activation functions are dispensable in this context.
ERK-TCF signaling to SRF is sufficient for thymocyte positive selection.
We next tested whether ERK signaling through the TCFs is sufficient for SRF function. ERK signaling through the TCFs plays a major role in thymocyte selection, and the SAP-1 and Elk-1 TCFs are functionally equivalent in this system (12, 13). We substituted the SRF C-terminal activation domain of SRF.V194E, which cannot itself recruit coactivators, with the Elk-1 C terminus, with its ERK phosphorylation sites either intact or mutated to alanine (SRF.V194E-ElkC and SRF.V194E-ElkNA) (Fig. 3A; see also Fig. S5 in the supplemental material). Retroviral transduction of SRF.V194E-ElkC, but not SRF.V194E-ElkNA, could effectively restore the ability of SrfhCD2cre bone marrow to generate CD4 and CD8 TCRhi thymocytes in reconstituted Rag2−/− animals (Fig. 3B). Within the SrfhCD2cre:SRF.V194E-ElkC CD4 SP population, FOXP3+ thymocytes were present at levels comparable to the wild type (Fig. 3D). Binding of both SRF.V194E-ElkC and SRF.V194E-ElkNA derivatives to SRF target promoters was readily detectable in SrfhCD2cre DP thymocytes by chromatin immunoprecipitation, although recovery at Egr3 was poor (Fig. 3D) (see Discussion). We isolated viable GFP+ SrfhCD2cre:SRF.V194E-ElkC DP thymocytes and tested their ability to activate SRF targets genes in response to TCR cross-linking. Although the purification protocol substantially reduced the strength of the transcriptional response, SRF.V194E-ElkC restored inducible expression of the Egr1, Egr2, Fos, and Junb genes, although it had little effect on the inducibility of the Egr3, Nr4a1, and Actb genes (Fig. 3E) (see Discussion).
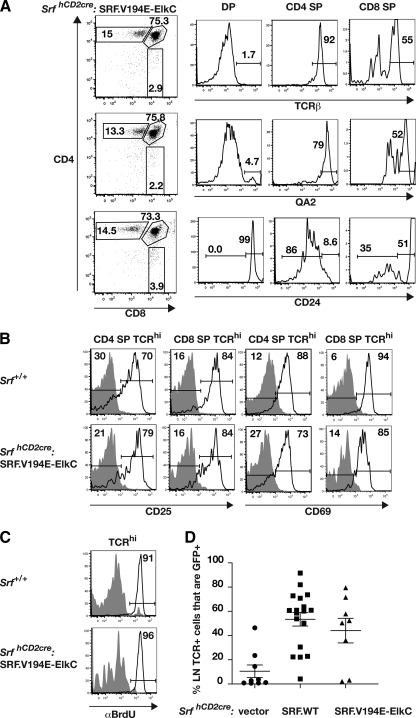
Upon differentiation to CD4 SP and CD8 SP cells in vivo, SrfhCD2cre:SRF.V194E-ElkC cells upregulated TCR-β and QA2 and downregulated CD24 expression, as do wild-type thymocytes (Fig. 4A). Upon long-term stimulation by anti-CD3/anti-CD28 in vitro, SrfhCD2cre:SRF.V194E-ElkC SP thymocytes upregulated cell surface expression of CD25 and CD69 as efficiently as wild-type cells within 48 h, although the level of CD69 expression in activated SrfhCD2cre:SRF.V194E-ElkC CD8 SP cells was somewhat lower (Fig. 4B). Finally, SrfhCD2cre:SRF.V194E-ElkC thymocytes were competent to enter the cell cycle following anti-CD3/anti-CD28 stimulation, as assessed by BrdU incorporation 72 h later (Fig. 4C). Thus, activation of SRF targets in response to TCR-induced ERK signaling can substitute for SRF function in generation of functional SP thymocytes. Examination of lymph nodes in Rag2−/− animals reconstituted with wild-type SRF- or SRF.V194E-ElkC-transduced SrfhCD2cre bone marrow, indicated that SRF.V194E-ElkC T cells are capable of migration to and establishment in the periphery (Fig. 4D).
FIG. 4.
The SRF-Elk fusion protein generates mature thymocytes that can be activated through the T-cell receptor. SrfhCD2cre:SRF.V194E-ElkC thymocytes were analyzed from reconstituted Rag2−/− animals by gating on GFP+ cells. (A) TCR-β, QA2, and CD25 expression during thymocyte development. SrfhCD2cre:SRF.V194E-ElkC thymocytes were gated on DP, CD4 SP, and CD8 SP cells and analyzed for expression of the indicated markers. (B) Upregulation of CD25 and CD69 following TCR cross-linking. Wild-type (top panels) or SrfhCD2cre:SRF.V194E-ElkC thymocytes were analyzed before (gray shading) or 48 h after (black lines) stimulation by anti-CD3/anti-CD28. (C) TCR cross-linking induces proliferation of SrfhCD2cre:SRF.V194E-ElkC thymocytes. Proliferation was assessed by incorporation of BrdU during a 20-h pulse either without stimulation (gray shading) or 48-h after stimulation by anti-CD3 and anti-CD28. (Top) Wild-type thymocytes; (bottom) SrfhCD2cre:SRF.V194E-ElkC thymocytes. (D) Proportions of GFP+ (i.e., retrovirally transduced) lymph node TCR+ cells in animals reconstituted with SrfhCD2cre cells transduced by vector, wild-type SRF, or SRF.V194E-ElkC.
DISCUSSION
In this study we analyzed the role of serum response factor in thymocyte development. We found that inactivation of the Srf gene and depletion of SRF protein at the early stages of thymocyte development completely block thymocyte positive selection and Treg and NK T-cell development but have no discernible effect on early thymocyte development or on negative selection, at least in the HY model. Srf inactivation did not appreciably impair generation of TCR-dependent signals, nor did it affect thymocyte-antigen-presenting cell interactions, at least in vitro. SRF function in this system absolutely requires recruitment of its regulatory cofactors but is independent of presumed SRF N-terminal regulatory phosphorylations and of its C-terminal activation domain. Fusion of a TCF C-terminal regulatory domain to an SRF mutant defective in coactivator recruitment rescues its ability to promote thymocyte differentiation, showing that ERK signaling through TCF to SRF target genes is sufficient to promote thymocyte differentiation. Taken together, the data show that in this system SRF functions primarily as a scaffold to recruit TCF cofactors to SRF targets in thymocyte differentiation.
Our Srf deletion strategy, which inactivated the gene at the early DN stages of thymocyte development, revealed a more pronounced thymocyte selection phenotype than found in a previous study, in which Srf was inactivated only at the DP stage (16). That study reported reduced SP thymocyte abundance; a lack of Srf-deleted T cells in the periphery was attributed to a requirement for SRF activity in peripheral T-cell maintenance. In contrast, we found that Srf is absolutely required for generation of mature thymocytes, and it is likely that the absence of peripheral T cells seen in SrfhCD2cre animals reflects this rather than any role for Srf in peripheral cell survival. Indeed, acute inactivation of Srf in the peripheral T-cell population does not result in significant cell death, suggesting that Srf is not required for viability per se (D. Maurice and R. Treisman, unpublished data). Our data also show that SRF activity is not required for passage through the β-selection checkpoint, in agreement with our previous studies of TCF mutants (12, 55), even though it appears that TCR-inducible gene activation is reduced to background levels. The apparently paradoxical finding that the Egr family genes, which are TCF-SRF targets, also apparently play a role in these processes (8, 39), may perhaps be explained by a requirement for their basal rather than inducible expression. Defective Egr-Id3 signaling may underlie the increased production of γδ thymocytes that we observed in SrfhCD2cre animals (29).
Thymocyte positive selection, while strongly dependent on the SAP-1 TCF, remains at least partly intact upon deletion of all three TCF proteins, and Treg and NK T-cell development is barely affected by TCF inactivation (12, 55). Our results nevertheless show that both processes are completely SRF dependent and can be restored by ERK-dependent activation of SRF target genes. The absence of all three TCFs may allow the MRTF proteins to compete more effectively for normally TCF-dependent targets, allowing partial rescue of their expression by coupling to MRTF-linked signaling; alternatively, it might allow access to TCF-dependent targets by other Ets family proteins, many of which are expressed in any given cell type (6, 11, 22). While our observations suggest that the failure of Treg and NK T-cell development reflects the absence of positive selection in SRF-null cells, the differential effects of TCF signaling on these processes and positive selection remain puzzling. Given that TCF-SRF signaling is required for proliferation in at least some cell contexts (D. Maurice, P. Costello, and R. Treisman, unpublished data), it is conceivable that the marginal effects of TCF deletion on Treg development reflect the fact that these cells are likely to be generated from a less proliferative precursor population than SP thymocytes (45, 53).
The most striking SRF functional requirement revealed by our analysis was for the interaction with TCF. The SRF.V194E mutant, which can recruit neither TCF nor MRTF proteins, was completely inactive in thymocyte development, even though it could be recruited to SRF target genes. An obligate requirement for recruitment of cofactors was also demonstrated for SRF-dependent differentiation of embryonic stem cells to cardiomyocytes in vitro, but the cofactors involved were not conclusively identified (42). We think it likely that the majority of SRF functions, if not all, are dependent on cofactor interactions. The activity of SRF.V194E was restored upon its fusion to the C-terminal regulatory domain of the Elk-1 TCF, and the function of the chimera was dependent on the integrity of the Elk-1 C-terminal phosphorylation sites, consistent with its responding to TCR-generated ERK signals. Thus, direction of TCR-induced ERK signals to SRF target genes is sufficient to promote thymocyte development. The other major nonplant MADS protein family, the MEF2 transcription factors, also interact with multiple regulatory cofactors via their DNA-binding domains (for a review, see reference 36), and it might be interesting to use gene fusion approaches to dissect the roles of their cofactors in vivo.
Previous studies have shown that recruitment of the TCF and MRTF cofactors to SRF targets is to at least some extent gene specific (38, 54), but the ability of the SRF.V194E fusion to rescue thymocyte development indicates that this system can tolerate the misdirection of ERK signals to normally MRTF-specific SRF target genes. We think it is unlikely that the fusion protein will effectively interact with all genomic targets bound by wild-type SRF, however, although we detected its recruitment to all targets that we tested. While SRF binds constitutively to targets such as Fos in vivo (19), its interaction with a large proportion of MRTF-dependent targets is induced by signaling, presumably reflecting cooperative binding with MRTFs (C. Esnault and R. Treisman, unpublished data). Moreover, SRF binds cooperatively with TCF at nonconsensus sites in vitro (27). It is thus likely that the SRF.V194E-ElkC fusion protein, which cannot itself recruit cofactors, will not interact with those SRF targets dependent on such cooperative interactions in wild-type cells. These considerations imply that at least in the thymocyte system, targeting of ERK-TCF signaling to only a subset of SRF targets is sufficient for authentic SRF function.
The SRF N-terminal phosphorylation sites for CKII and Rsk are dispensable for thymocyte development, even though these sites strongly affect in vitro DNA-binding kinetics and are phosphorylated in vivo to high stoichiometry and the Rsk site is signal responsive (23, 34, 35, 48). The SRF C-terminal transcriptional activation domain also did not contribute to SRF activity, even though it registers in transfection assays in vitro (20, 21, 24, 31). These observations may reflect differential regulatory requirements of SRF target gene expression in different situations. It will be interesting to test the roles of these domains, and of the SRF regulatory cofactors, in other biological contexts.
Supplementary Material
Acknowledgments
We thank Facundo Batista, Cyril Esnault, and Caetano Reis e Sousa for helpful discussions, advice, and communication of data prior to publication; Derek Davies and the staff of the LRI Flow Cytometry facility for FACS support; and the LRI Biological Resources Unit for animal husbandry.
Research at LRI is supported by Cancer Research UK through institutional core funding. A.M. was supported by an EMBO long-term fellowship.
A.M. and P.C. devised and executed thymus analysis and retroviral transduction experiments and wrote the paper. R.N. and M.S. devised and performed DNA-binding studies and gene expression and chromatin immunoprecipitation experiments. D.M. performed the analysis of DP-APC conjugation and calcium signaling. D.T. and D.D. provided the Srff/f animals. R.T. conceived of the project, devised experiments, and wrote the paper.
Footnotes
Published ahead of print on 22 November 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Alberti, S., et al. 2005. Neuronal migration in the murine rostral migratory stream requires serum response factor. Proc. Natl. Acad. Sci. U. S. A. 102:6148-6153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arsenian, S., B. Weinhold, M. Oelgeschlager, U. Ruther, and A. Nordheim. 1998. Serum response factor is essential for mesoderm formation during mouse embryogenesis. EMBO J. 17:6289-6299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnden, M. J., J. Allison, W. R. Heath, and F. R. Carbone. 1998. Defective TCR expression in transgenic mice constructed using cDNA-based alpha- and beta-chain genes under the control of heterologous regulatory elements. Immunol. Cell Biol. 76:34-40. [DOI] [PubMed] [Google Scholar]
- 4.Bensinger, S. J., A. Bandeira, M. S. Jordan, A. J. Caton, and T. M. Laufer. 2001. Major histocompatibility complex class II-positive cortical epithelium mediates the selection of CD4(+)25(+) immunoregulatory T cells. J. Exp. Med. 194:427-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettini, M., H. Xi, J. Milbrandt, and G. J. Kersh. 2002. Thymocyte development in early growth response gene 1-deficient mice. J. Immunol. 169:1713-1720. [DOI] [PubMed] [Google Scholar]
- 6.Boros, J., et al. 2009. Overlapping promoter targeting by Elk-1 and other divergent ETS-domain transcription factor family members. Nucleic Acids Res. 37:7368-7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buchwalter, G., C. Gross, and B. Wasylyk. 2004. Ets ternary complex transcription factors. Gene 324:1-14. [DOI] [PubMed] [Google Scholar]
- 8.Carleton, M., et al. 2002. Early growth response transcription factors are required for development of CD4(−)CD8(−) thymocytes to the CD4(+)CD8(+) stage. J. Immunol. 168:1649-1658. [DOI] [PubMed] [Google Scholar]
- 9.Carpenter, A. C., and R. Bosselut. 2010. Decision checkpoints in the thymus. Nat. Immunol. 11:666-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins, A., D. R. Littman, and I. Taniuchi. 2009. RUNX proteins in transcription factor networks that regulate T-cell lineage choice. Nat. Rev. Immunol. 9:106-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Collins, P. J., Y. Kobayashi, L. Nguyen, N. D. Trinklein, and R. M. Myers. 2007. The ets-related transcription factor GABP directs bidirectional transcription. PLoS Genet. 3:e208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costello, P., et al. 2010. Ternary complex factors SAP-1 and Elk-1, but not Net, are functionally equivalent in thymocyte development. J. Immunol. 185:1082-1092. [DOI] [PubMed] [Google Scholar]
- 13.Costello, P. S., R. H. Nicolas, Y. Watanabe, I. Rosewell, and R. Treisman. 2004. Ternary complex factor SAP-1 is required for Erk-mediated thymocyte positive selection. Nat. Immunol. 5:289-298. [DOI] [PubMed] [Google Scholar]
- 14.de Boer, J., et al. 2003. Transgenic mice with hematopoietic and lymphoid specific expression of Cre. Eur. J. Immunol. 33:314-325. [DOI] [PubMed] [Google Scholar]
- 15.Fischer, A. M., C. D. Katayama, G. Pages, J. Pouyssegur, and S. M. Hedrick. 2005. The role of erk1 and erk2 in multiple stages of T cell development. Immunity 23:431-443. [DOI] [PubMed] [Google Scholar]
- 16.Fleige, A., et al. 2007. Serum response factor contributes selectively to lymphocyte development. J. Biol. Chem. 282:24320-24328. [DOI] [PubMed] [Google Scholar]
- 17.Gallo, E. M., et al. 2007. Calcineurin sets the bandwidth for discrimination of signals during thymocyte development. Nature 450:731-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gineitis, D., and R. Treisman. 2001. Differential usage of signal transduction pathways defines two types of serum response factor target gene. J. Biol. Chem. 276:24531-24539. [DOI] [PubMed] [Google Scholar]
- 19.Herrera, R. E., P. E. Shaw, and A. Nordheim. 1989. Occupation of the c-fos serum response element in vivo by a multi-protein complex is unaltered by growth factor induction. Nature 340:68-70. [DOI] [PubMed] [Google Scholar]
- 20.Hill, C. S., et al. 1993. Functional analysis of a growth factor-responsive transcription factor complex. Cell 73:395-406. [DOI] [PubMed] [Google Scholar]
- 21.Hill, C. S., J. Wynne, and R. Treisman. 1994. Serum-regulated transcription by serum response factor (SRF): a novel role for the DNA binding domain. EMBO J. 13:5421-5432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hollenhorst, P. C., A. A. Shah, C. Hopkins, and B. J. Graves. 2007. Genome-wide analyses reveal properties of redundant and specific promoter occupancy within the ETS gene family. Genes Dev. 21:1882-1894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Janknecht, R., R. A. Hipskind, T. Houthaeve, A. Nordheim, and H. G. Stunnenberg. 1992. Identification of multiple SRF N-terminal phosphorylation sites affecting DNA binding properties. EMBO J. 11:1045-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansen, F. E., and R. Prywes. 1993. Identification of transcriptional activation and inhibitory domains in serum response factor (SRF) by using GAL4-SRF constructs. Mol. Cell. Biol. 13:4640-4647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kisielow, P., H. Bluthmann, U. D. Staerz, M. Steinmetz, and H. von Boehmer. 1988. Tolerance in T-cell-receptor transgenic mice involves deletion of nonmature CD4+8+ thymocytes. Nature 333:742-746. [DOI] [PubMed] [Google Scholar]
- 26.Knoll, B., et al. 2006. Serum response factor controls neuronal circuit assembly in the hippocampus. Nat. Neurosci. 9:195-204. [DOI] [PubMed] [Google Scholar]
- 27.Latinkic, B. V., M. Zeremski, and L. F. Lau. 1996. Elk-1 can recruit SRF to form a ternary complex upon the serum response element. Nucleic Acids Res. 24:1345-1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lauritsen, J. P., et al. 2008. Egr2 is required for Bcl-2 induction during positive selection. J. Immunol. 181:7778-7785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lauritsen, J. P., et al. 2009. Marked induction of the helix-loop-helix protein Id3 promotes the γδ T cell fate and renders their functional maturation Notch independent. Immunity 31:565-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lawson, V. J., K. Weston, and D. Maurice. 2010. Early growth response 2 regulates the survival of thymocytes during positive selection. Eur. J. Immunol. 40:232-241. [DOI] [PubMed] [Google Scholar]
- 31.Lee, T. C., Y. Shi, and R. J. Schwartz. 1992. Displacement of BrdUrd-induced YY1 by serum response factor activates skeletal alpha-actin transcription in embryonic myoblasts. Proc. Natl. Acad. Sci. U. S. A. 89:9814-9818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ling, Y., A. G. West, E. C. Roberts, J. H. Lakey, and A. D. Sharrocks. 1998. Interaction of transcription factors with serum response factor. Identification of the Elk-1 binding surface. J. Biol. Chem. 273:10506-10514. [DOI] [PubMed] [Google Scholar]
- 33.Mamalaki, C., et al. 1993. Positive and negative selection in transgenic mice expressing a T-cell receptor specific for influenza nucleoprotein and endogenous superantigen. Dev. Immunol. 3:159-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manak, J. R., N. de Bisschop, R. M. Kris, and R. Prywes. 1990. Casein kinase II enhances the DNA binding activity of serum response factor. Genes Dev. 4:955-967. [DOI] [PubMed] [Google Scholar]
- 35.Marais, R. M., J. J. Hsuan, C. McGuigan, J. Wynne, and R. Treisman. 1992. Casein kinase II phosphorylation increases the rate of serum response factor-binding site exchange. EMBO J. 11:97-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McKinsey, T. A., C. L. Zhang, and E. N. Olson. 2002. MEF2: a calcium-dependent regulator of cell division, differentiation and death. Trends Biochem. Sci. 27:40-47. [DOI] [PubMed] [Google Scholar]
- 37.Medjkane, S., C. Perez-Sanchez, C. Gaggioli, E. Sahai, and R. Treisman. 2009. Myocardin-related transcription factors and SRF are required for cytoskeletal dynamics and experimental metastasis. Nat. Cell Biol. 11:257-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miralles, F., G. Posern, A. I. Zaromytidou, and R. Treisman. 2003. Actin dynamics control SRF activity by regulation of its coactivator MAL. Cell 113:329-342. [DOI] [PubMed] [Google Scholar]
- 39.Miyazaki, T. 1997. Two distinct steps during thymocyte maturation from CD4−CD8− to CD4+CD8+ distinguished in the early growth response (Egr)-1 transgenic mice with a recombinase-activating gene-deficient background. J. Exp. Med. 186:877-885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mokalled, M. H., A. Johnson, Y. Kim, J. Oh, and E. N. Olson. 2010. Myocardin-related transcription factors regulate the Cdk5/Pctaire1 kinase cascade to control neurite outgrowth, neuronal migration and brain development. Development 137:2365-2374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murai, K., and R. Treisman. 2002. Interaction of serum response factor (SRF) with the Elk-1 B box inhibits RhoA-actin signaling to SRF and potentiates transcriptional activation by Elk-1. Mol. Cell. Biol. 22:7083-7092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Niu, Z., et al. 2008. Serum response factor orchestrates nascent sarcomerogenesis and silences the biomineralization gene program in the heart. Proc. Natl. Acad. Sci. U. S. A. 105:17824-17829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Olson, E. N., and A. Nordheim. 2010. Linking actin dynamics and gene transcription to drive cellular motile functions. Nat. Rev. Mol. Cell Biol. 11:353-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Parlakian, A., et al. 2004. Targeted inactivation of serum response factor in the developing heart results in myocardial defects and embryonic lethality. Mol. Cell. Biol. 24:5281-5289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pennington, D. J., et al. 2006. Early events in the thymus affect the balance of effector and regulatory T cells. Nature 444:1073-1077. [DOI] [PubMed] [Google Scholar]
- 46.Pipes, G. C., E. E. Creemers, and E. N. Olson. 2006. The myocardin family of transcriptional coactivators: versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 20:1545-1556. [DOI] [PubMed] [Google Scholar]
- 47.Posern, G., and R. Treisman. 2006. Actin' together: serum response factor, its cofactors and the link to signal transduction. Trends Cell Biol. 16:588-596. [DOI] [PubMed] [Google Scholar]
- 48.Rivera, V. M., et al. 1993. A growth factor-induced kinase phosphorylates the serum response factor at a site that regulates its DNA-binding activity. Mol. Cell. Biol. 13:6260-6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schratt, G., et al. 2002. Serum response factor is crucial for actin cytoskeletal organization and focal adhesion assembly in embryonic stem cells. J. Cell Biol. 156:737-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stern, S., et al. 2009. A nuclear actin function regulates neuronal motility by serum response factor-dependent gene transcription. J. Neurosci. 29:4512-4518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun, Q., et al. 2006. Defining the mammalian CArGome. Genome Res. 16:197-207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Treisman, R. 1994. Ternary complex factors: growth factor regulated transcriptional activators. Curr. Opin. Genet. Dev. 4:96-101. [DOI] [PubMed] [Google Scholar]
- 53.van Santen, H. M., C. Benoist, and D. Mathis. 2004. Number of T reg cells that differentiate does not increase upon encounter of agonist ligand on thymic epithelial cells. J. Exp. Med. 200:1221-1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang, Z., et al. 2004. Myocardin and ternary complex factors compete for SRF to control smooth muscle gene expression. Nature 428:185-189. [DOI] [PubMed] [Google Scholar]
- 55.Willoughby, J. E., et al. 2007. Raf signaling but not the ERK effector SAP-1 is required for regulatory T cell development. J. Immunol. 179:6836-6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Youn, H. D., C. M. Grozinger, and J. O. Liu. 2000. Calcium regulates transcriptional repression of myocyte enhancer factor 2 by histone deacetylase 4. J. Biol. Chem. 275:22563-22567. [DOI] [PubMed] [Google Scholar]
- 57.Youn, H. D., and J. O. Liu. 2000. Cabin1 represses MEF2-dependent Nur77 expression and T cell apoptosis by controlling association of histone deacetylases and acetylases with MEF2. Immunity 13:85-94. [DOI] [PubMed] [Google Scholar]
- 58.Youn, H. D., L. Sun, R. Prywes, and J. O. Liu. 1999. Apoptosis of T cells mediated by Ca2+-induced release of the transcription factor MEF2. Science 286:790-793. [DOI] [PubMed] [Google Scholar]
- 59.Zaromytidou, A. I., F. Miralles, and R. Treisman. 2006. MAL and ternary complex factor use different mechanisms to contact a common surface on the serum response factor DNA-binding domain. Mol. Cell. Biol. 26:4134-4148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.