Abstract
Long interspersed element 1s (LINE-1s or L1s) are a family of non-long-terminal-repeat retrotransposons that predominate in the human genome. Active LINE-1 elements encode proteins required for their mobilization. L1-encoded proteins also act in trans to mobilize short interspersed elements (SINEs), such as Alu elements. L1 and Alu insertions have been implicated in many human diseases, and their retrotransposition provides an ongoing source of human genetic diversity. L1/Alu elements are expected to ensure their transmission to subsequent generations by retrotransposing in germ cells or during early embryonic development. Here, we determined that several subfamilies of Alu elements are expressed in undifferentiated human embryonic stem cells (hESCs) and that most expressed Alu elements are active elements. We also exploited expression from the L1 antisense promoter to map expressed elements in hESCs. Remarkably, we found that expressed Alu elements are enriched in the youngest subfamily, Y, and that expressed L1s are mostly located within genes, suggesting an epigenetic control of retrotransposon expression in hESCs. Together, these data suggest that distinct subsets of active L1/Alu elements are expressed in hESCs and that the degree of somatic mosaicism attributable to L1 insertions during early development may be higher than previously anticipated.
The human genome is highly complex in structure, but only ∼1.5% of human DNA has protein coding potential (53). More than 40% of the genome is composed of sequences derived from mobile genetic elements (transposons and retrotransposons) (53). At present, only long interspersed element 1s (LINE-1s or L1s) and some short interspersed elements (SINEs) are actively transposing in the human genome (62). LINE-1 elements (here LINE-1s) are autonomous retrotransposons that constitute ∼17% of human DNA (53), and recent estimates indicate that an average human genome contains around 80 to 100 sequences that are able to transpose, i.e., are retrotransposition-competent LINE1s (here RC-L1s) (19, 71). However, these elements vary dramatically in their retrotransposition activity in cell culture-based retrotransposition assays (19). In addition, allelic heterogeneity in retrotransposition activity (56, 73) and the presence of RC-L1 elements that show the presence or absence of polymorphism between individuals (8, 15, 84) imply that there can be significant variation in RC-L1 activity between individual genomes.
An RC-L1 is ∼6 kb in length (29, 72) and contains an ∼900-bp-long 5′ untranslated region (UTR) with internal promoter activity (78), two open reading frames (ORFs), an ∼150-bp-long 3′ UTR, and a poly(A) tail (72). ORF1 encodes a 40-kDa protein with RNA binding and nucleic acid chaperone activities (38, 40, 52, 59, 60). ORF2 encodes a 150-kDa protein with reverse transcriptase (RT) and endonuclease activities (33, 61). Both proteins are required for the mobilization of L1 within the human genome (65). L1 retrotransposition involves the reverse transcription of an mRNA intermediate by a mechanism termed target-primed reverse transcription (25, 26, 55, 64). The mobilization of SINEs occurs by a similar mechanism (46), through the use of LINE-1-encoded ORF2p (28).
Alu elements are the most successful human SINEs, and they are present at greater than one million copies in the human genome (53). Alu elements are nonautonomous non-long-terminal-repeat retrotransposons derived from human gene 7SL (reviewed in references 9 and 23), and the average human genome contains ∼6,000 active core Alu elements (12). An Alu core is defined as the ∼280-bp region that includes both Alu monomers that are capable of retrotransposing in cultured cells but excludes any flanking genomic 5′ or 3′ regions.
Despite the high prevalence of transposable elements in the human genome and the presence of several LINE and SINE subfamilies in this genome, apparently at present only certain members of each class are active (designated “young,” “human-specific,” or “hot” elements [reviewed in reference 62]). As a consequence, L1 and Alu elements can act as insertional mutagens, and indeed, many cases of human disease have been caused by such insertions (11, 37). In addition to their potential as insertional mutagens, there are many ways in which de novo L1/Alu insertions and L1/L1 or Alu/Alu recombination can impact the human genome (reviewed in references 11, 23, 37, 44, and 50). Overall, it is estimated that 1 in 35 to 45 newborns harbors a de novo L1 or Alu retrotransposition event (24, 31, 42, 49). These new events must occur either in parental germ cells or early in embryonic development, prior to the partitioning of the germ cell lineage. Indeed, through the characterization of human mutagenic insertions and the use of mouse models of L1 retrotransposition, it has been revealed that L1 retrotransposition can occur in germ cells, during early embryonic development, and in particular somatic tissues (3, 7, 18, 27, 35, 47, 66-68, 80). On the other hand, recent studies have revealed that L1 mobilization processes are a source of genomic variation among humans, with particular impact on our somatic genome, as revealed by the identification of several de novo L1 insertions in a cohort of lung tumors (10, 27, 31, 42, 45).
Human embryonic stem cells (hESCs) offer an excellent model to study biological processes during early human development, as they mimic pluripotent cells isolated from the inner cell mass (ICM) of human embryos (79). Several hESC lines and human embryonic carcinoma (hEC) cell lines express L1 retrotransposition intermediates (ribonucleoprotein particles [RNPs; 39, 52, 58]), and a diverse range of L1 mRNAs (representing active and inactive subfamilies) are expressed in these cells (35, 41, 58, 74). Furthermore, several cultured hESC lines can support the retrotransposition of engineered LINE-1 elements using a cultured-cell-based assay (35, 65). There is a growing but disparate set of observations relating to host factors that influence the retrotransposition of Alu and L1, including the differential effect of APOBEC proteins on the mobility of L1 and Alu (recently reviewed in reference 21), the control of L1 expression by DNA methylation in germ cells by DNMT3L, Piwi proteins, and Piwi-interacting RNAs (17, 57), as well as single-stranded retrotransposon DNA degradation by the exonuclease Trex1 (77). These observations suggest that there is a diverse array of host defense systems that can interfere with L1 retrotransposition. Perhaps the most enigmatic feature of these systems is the fact that full-length L1 human-specific (L1Hs) elements contain an active antisense promoter in their 5′ UTR (76). Recently, it was reported that, in conjunction with the sense L1 promoter, transcripts initiated from the antisense promoter could trigger an RNA interference (RNAi) response that attenuates the mobility of L1 in cultured cells (75, 85). Intriguingly, deletion of the L1 antisense promoter enhances retrotransposition in cultured cells (85), but it has been retained in the vast majority of endogenous active elements, suggesting that it has some essential, perhaps regulatory, function.
To characterize a sample of the active retrotransposon “transcriptome” of hESCs, we cloned and sequenced expressed Alu elements and tested their retrotransposition potential in cultured human cells. We also utilized antisense L1 (AS-L1) transcripts, expressed in hESCs, to identify and map expressed L1 elements and their host genes. In addition, we found that the antisense promoter of L1 is robust over evolutionary time and that most expressed L1s are located within genes, suggesting epigenetic control of their expression.
MATERIALS AND METHODS
Cell culture.
All reagents were purchased from GIBCO-Invitrogen, unless otherwise indicated. The cell lines PA-1 (86) and HeLa-HA were grown as previously described (6). Briefly, cells were passaged by standard trypsinization (using a 0.05% stock) and the culture medium was minimum essential medium (MEM) supplemented with 10% heat-inactivated fetal bovine serum, 1× nonessential amino acids, and 1 mM l-glutamine. 2102Ep (4) and N-Tera2D1 cl1 (N-Tera2D1) (5) cells were grown in high-glucose Dulbecco MEM supplemented with 10% fetal bovine serum and 1 mM l-glutamine. hESCs were grown as previously described (35). hESC lines H7, H9, and H13B (WA07, WA09, and WA13) were obtained from Wicell and maintained on gelatin-coated plates using irradiated mouse embryonic fibroblasts (MEFs) from CF-1 mice (Chemicon). Gamma irradiation with a 2100 Cesium source indicator was used to mitotically inactivate MEFs. MEFs were used at a density of 25,000/cm2. The culture medium for hESCs was Dulbecco MEM-knockout supplemented with 4 ng/ml b-FGF, 20% knockout serum replacement, 1 mM l-glutamine, 50 μM β-mercaptoethanol, and 0.1 mM nonessential amino acids. hESCs were manually passage twice a week. Transfected hESCs were grown in Matrigel-coated plates (B&D) using MEF-conditioned medium for 24 h (35). All of the cell lines were grown in a humidified incubator at 37°C with 7% CO2.
Approval from the Spanish National Embryo Ethical Committee was obtained to work with hESCs.
Plasmid DNAs.
Plasmid DNAs were purified using a Midiprep kit from Qiagen, checked for superhelicity by electrophoresis on 0.7% agarose-ethidium bromide gels (only highly supercoiled preparations of DNA [>90%] were used for transfection), and filtered through a 0.22-μm filter. The following plasmids were used. pRL-SV40 is a 4.8-kb plasmid that contains the coding region of Renilla luciferase under the transcriptional control of the early simian virus 40 (SV40) promoter. It is cloned in a modified pBSKS II (Stratagene) plasmid that contains an SV40 late polyadenylation signal. 5S-FF is a 5.7-kb plasmid that contains the 5′ UTR of a human L1Hs element (L1.3) (71) cloned in the sense orientation in plasmid pGL3-basic (Promega). 5AS-FF is a 5.7-kb plasmid that contains the 5′ UTR of a human L1Hs element (L1.3) (71) cloned in the antisense orientation in plasmid pGL3-basic (Promega). Derivatives of plasmids 5S-FF and 5AS-FF but containing the 5′ UTR from older LINE-1s (L1PA2, L1PA3, L1PA4, L1PA6, L1PA7, L1PA8, and L1PA10) were constructed using the same procedure. pCEP-5′UTRORF2NoNeo has been described previously (2). It contains a 5.0-kb NotI-BamHI fragment containing the L1.3 5′ UTR and L1.3 ORF2 cloned in pCEP4 (Invitrogen). pAluNF1-neoIII contains a 2.1-kb fragment containing the 7SL promoter, a copy of the NF1 Alu element (a Ya5 member) (82), a neo3 self-splicing indicator cassette (30), a 33-bp poly(A) tail, and a BC1 transcription termination sequence cloned in pBSKS-II (Invitrogen). An AgeI and a BstZ17I site were introduced into the 5′and 3′ends of Alu NF1, respectively, to help the cloning of Alu elements expressed in hESCs. pCEP-EGFP contains the 0.9-kb coding sequence of the humanized enhanced green fluorescent protein (EGFP), which was derived from plasmid phrGFP-C (Stratagene) cloned in pCEP4 (Invitrogen).
Transfection of cultured cells and assays.
HeLa-HA, 2102Ep, N-Tera2D1, and PA-1 cells were transfected using Fugene6 (Roche) as previously described (6, 83).
hESCs were transfected by nucleofection (Amaxa) exactly as described previously (35), using 4 × 106 cells and 4 μg of purified DNA (2 μg of pRL-SV40 and 2 μg of either 5S-FF or 5AS-FF). The luciferase signal was read using the dual-system kit from Promega.
The Alu trans-retrotransposition assay in HeLa-HA cells was conducted in six-well tissue culture plates as previously described (34). Briefly, HeLa-HA cells were plated at 4 × 104/well in six-well tissue culture plates. We used a full plate per Alu construct to be analyzed. Approximately 14 to 18 h after plating, three wells of the plate were cotransfected with 0.66 μg of a reporter plasmid (pAluNF1-neoIII) and 0.33 μg of a driver L1 that lacks an indicator cassette (pCEP-5′UTRORF2NoNeo). We used 3 μl of Fugene 6 transfection reagent (Roche Biochemical). The remaining three wells were cotransfected with equal amounts of an EGFP reporter plasmid (human Renilla green fluorescent protein [pCEP-EGFP]), a reporter plasmid, and a driver L1. At 72 h posttransfection, this set of wells was trypsinized and subjected to flow cytometry. The percentage of EGFP cells was used to determine the transfection efficiency of each sample. At 72 h posttransfection, cells in the remaining wells were subjected to G-418 selection (400 μg/ml) for 12 days. The retrotransposition efficiency is expressed as the number of G-418-resistant foci divided by the number of transfected (EGFP-positive) cells.
RNA extraction and cDNA synthesis.
Cells were washed twice with 1× phosphate-buffered saline (Invitrogen), and total RNA was extracted using the TRIzol reagent (Invitrogen). To generate cDNAs, 4 μg of total RNA was treated with 100 U of RNase-free DNase I (Promega) for 1 h at 37°C. To prevent contamination with genomic DNA, the DNase treatment was repeated twice. Then, 1 μg of RNA was reverse transcribed with Moloney murine leukemia virus RT (25 U; Promega) primed with a 3′ random amplification of cDNA ends (RACE) primer for 1 h at 42°C by following the manufacturer's instructions. The sequence of the RACE primer is 5′GCGAGCACAGAATTAATACGACTCACTATAGGTTTTTTTTTTTTVN.
Alu element library.
To generate a library of expressed Alu elements, RACE-primed cDNAs were used in a PCR with primers Outer (5′GCGAGCACAGAATTAATACGACT) and Alu_library (5′ GGTGGCTCACGCCTGTAATCCCAG) in triplicate using High Fidelity Expand Taq (Roche). We used a 3′ RACE primer to prevent amplification of exonized Alu elements. The PCR conditions included an initial cycle of 95°C for 2 min, followed by 25 cycles of 30 s at 94°C, 30 s at 54°C, and 30 s at 72°C, with a final step of 72°C for 10 min. Thirty microliters of each PCR product was resolved on 2% agarose gels, the band was excised, and the DNA was extracted using the QIAquick extraction kit (Qiagen). PCR amplification products were cloned in pGEMT-Easy (Promega), and approximately 15 clones per reaction were randomly sequenced using M13 universal primers. Sequences were analyzed by BLAT (51) at http://www.genome.ucsc.edu using the March 2006 human genome assembly. The Alu subfamily was determined using RepeatMasker at http://www.repeatmasker.org.
Antisense-based identification of expressed LINE-1s.
To generate a library of expressed LINEs, RACE-primed cDNAs were used in a PCR with primers Outer and ABIEL_library (5′GTGAGATGAACCCGGTACCTCAG) in triplicate using High Fidelity Expand Taq (Roche). The PCR conditions included an initial cycle of 95°C for 2 min, followed by 30 cycles of 30 s at 94°C, 30 s at 54°C, and 90 s at 72°C, with a final step of 72°C for 10 min. Thirty microliters of each PCR product was resolved on 2% agarose gels, and products were excised and purified in two groups, 100- to 300-bp and 300- to 600-bp sizes, unless otherwise indicated. DNA was extracted using the QIAquick extraction kit (Qiagen), and products were cloned in pGEMT-Easy (Promega). Approximately 30 clones per reaction were randomly sequenced using M13 universal primers. Sequences were first analyzed by RepeatMasker (http://www.repeatmasker.org) to determine the subfamily of LINE-1 that generated the antisense transcript. The unique nonrepeated portion of the sequence was extracted and mapped using BLAT (51) (http:www.genome.ucsc.edu) to the March 2006 human reference sequence (NCBI36.3/hg18) to identify a source LINE-1 locus.
Analysis of L1 and Alu expression.
Total RNA was extracted using TRIzol (Invitrogen) and following the manufacturer's directions. Next, 1 μg was treated with 2 U of RNase-free DNase I (Invitrogen) for 30 min at room temperature. To prevent genomic DNA contamination, this step was repeated twice. Then, a High-Capacity cDNA reverse transcription kit (Applied Biosystems) was used to generate cDNAs.
To determine the L1 expression level, triplicate samples of diluted (1/5 and 1/10) cDNAs were used in a real-time PCR using Platinum SYBR green quantitative PCR SuperMix-UDG (Invitrogen) in an MX3005P real-time PCR machine (Stratagene). We used two sets of oligonucleotide primers (27) to amplify 61-bp and 84-bp amplicons from the 5′ UTR and ORF2 regions, respectively, of a consensus L1Hs element. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was amplified as an internal normalization control as previously described (63). We determined the threshold cycles (CTs) for LINE-1 and GAPDH and performed a melting curve analysis from 50°C to 95°C with readings every 0.2°C to confirm the identities of the amplified products. The CT obtained from the GAPDH PCR was used to normalize the mRNA contents of the samples.
To determine which L1s are expressed in pluripotent cells, a fraction of the cDNAs was subjected to conventional RT-PCR using primers that amplify a 235-bp portion of L1 ORF1 (35, 36). Next, amplified products were cloned into pGEMT-Easy (Promega) and 30 randomly selected clones were sequenced. Upon sequencing and RepeatMasker analysis, we determined the type (i.e., subfamily) of L1 expressed as described previously (35, 36).
Finally, to determine the expression level of Alu subfamilies Y, S, and J, triplicate samples of diluted cDNAs were used in a real-time PCR using the conditions described above. For each Alu subfamily, we designed specific primers (available upon request) for each subfamily by using a database of all known human Alu elements (12). As described above, we also determined the CT for GAPDH, which was used to normalize the mRNA content in the samples. Note that L1/Alu quantification using this procedure may likely amplify other L1/Alu fragments exonized or present in longer transcription units.
DNA extraction and genotyping PCRs.
Genomic DNA was extracted from H13B (grown on Matrigel as described previously [35]) and HeLa cells using the DNeasy Blood Mini kit (Qiagen) by following the manufacturer's instructions. We then used 200 ng of genomic DNA per genotyping PCR using High Fidelity Expand Taq (Roche). The PCR conditions included an initial cycle of 95°C for 4 min, followed by 35 cycles of 30 s at 94°C, 30 s at 54°C, and 60 s at 72°C, with a final step of 72°C for 10 min. Twenty microliters of each PCR product was resolved on 1.5% agarose gels, and the amplification products were excised, purified (using the QIAquick extraction kit [Qiagen]), and cloned into pGEMT-Easy (Promega). We sequenced at least four clones of each PCR product to confirm the identity of the amplified product.
TFBS analyses.
The 5′ UTR of L1PA1, L1PA2, L1PA3, L1PA4, L1PA6, L1PA7, L1PA8, and L1PA10 elements was analyzed using the TF Search at http://www.cbrc.jp/research/db/TFSEARCH.html. Only those transcription factor binding sites (TFBS) that showed a score of >0.93 were considered in the analysis. As a control for false-positive TFBSs, we generated a scrambled sequence of each 5′ UTR sequence (at http://www.molbiol.ru/eng/scripts/01_16.html) that was analyzed using the TF Search. None of the TFBSs identified in the LINE-1 5′ UTRs was identified in the scrambled sequences (see Document 1 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf).
Identification of L1Hs elements in human genomic sequence resources.
To determine the percentage of full-length L1Hs elements within human genes, we performed BLAST searches of four major genomic DNA sequence data sources. These were the GenBank nucleotide database (April 2008), the human genome reference assembly (NCBI36.3/hg18) (53), the Celera Genomics human genome shotgun assembly (AADB/November 2001) (81), and the HuRef diploid human genome sequence (J. Craig Venter Institute whole-genome shotgun assembly [May 2007; http://www.jcvi.org/research/huref/]). This assembly represents a composite haploid version of the diploid genome sequence from a single individual (J. Craig Venter) (54). To be considered for further analysis, the identified L1Hs elements had to have a genomic size of >5,900 bp and show ≥98.5% sequence identity to a known hot L1 (L1.3, accession no. L19088) (71). The 37-bp poly(A) tail located at the 3′ end of the L1.3 element sequence was removed so that L1 hits would not be excluded due to variation in the length of this simple-sequence tract. All L1 sequences meeting these criteria and their flanking sequences were exhaustively compared to remove redundant sequences. This analysis identified a nonredundant set of 533 elements whose insertion points were mapped to the human reference sequence (NCBI36.3/hg18), irrespective of whether the element was present in the reference assembly. The insertion coordinates of the 533 mapped elements were compared to the transcription start and stop coordinates of a nonredundant set of 20,304 human genes derived from the UCSC Genome Browser RefSeq Genes track. Where multiple transcripts were present for a gene, the transcript with the largest genomic size was used. One hundred sixty-four (∼30%) of the 533 L1 elements mapped within RefSeq Gene transcription units by these criteria.
Statistical analyses.
To determine if L1s within genes are preferentially expressed in pluripotent cells, we used a hypergeometric analysis as follows. The total population of N segments was assigned 533, in which n (164) have a particular annotation, X = “located within genes.” In samples, we analyzed k genes with that annotation in a sample of K genes (those expressed L1s). Next, we calculated the probability of the observation using the hypergeometric distribution as follows:
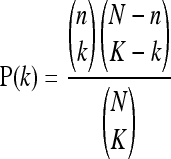 |
where N is the number of segments on the reference list, n is the number of segments on the reference list annotated with X, K is the number of segments on the input list, and k is the number of segments on the input list annotated with X, and
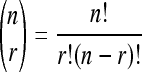 |
To generate a P value, we used the following equation:
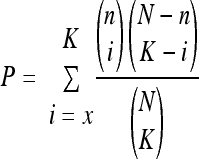 |
RESULTS
Alu elements expressed in hESCs.
To obtain a profile of Alu elements expressed in hESCs, we isolated total RNAs from three undifferentiated hESC lines (H7, H9, and H13B) and employed a 3′ RACE primer to generate a library of cDNAs. These cDNAs were used in PCRs with an outer primer and a primer designed to be able to amplify a broad range of Alu elements (12). We conducted PCRs in triplicate, cloned and sequenced the products, and identified the subfamily of each Alu element sequence using Repeatmasker (http://www.repeatmasker.org/), analyzing more than 100 distinct sequences. We observed that hESC lines express a wide range of Alu elements, including both old and young subfamilies (Fig. 1a; see Table 1 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf). We confirmed the expression of subfamilies Y, S, and J by quantitative RT-PCR using primers specific for each subfamily (Fig. 1b; see Materials and Methods). Subfamilies Y, Sx, and Sp were the most abundant, and there were no large differences between male and female hESCs in the type of Alu elements expressed (see Table 1 posted at the above URL).
FIG. 1.
Alu elements expressed in hESCs. (a) Three pie charts showing the representation and percentage of each Alu subfamily expressed in hESCs (each hESC line is indicated above each pie along with the sex of the cell line). For more details, see Tables 1 and S2 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf. (b) Alu expression in pluripotent cells. Shown are representative quantitative RT-PCR results for Alu RNA expression (subfamilies Y, S, and J) in H9 hESCs and differentiated HeLa cells as a control. The graphs show the n-fold changes in Alu expression with respect to HeLa cells (subfamily J). To normalize results, we determined the amplification CT for GAPDH (see Materials and Methods). (c) Rationale of the Alu mobilization assay in cultured HeLa cells (28). In the assay, cells are cotransfected with two plasmids and selected with G-418 to select Alu trans-retrotransposition events. The cartoon shows the structure of the L1 driver used (left side; see Materials and Methods) that contains only L1.3 ORF2 and a tagged Alu element with a self-splicing-based retrotransposition cassette (30) (right side). The L1 driver produces a functional ORF2p that can mediate the transmobilization of an expressed, tagged Alu RNA that has removed the self-splicing intron, causing activation of a functional neo gene. CMV, cytomegalovirus promoter. (d) Results of transmobilization assay of Alu elements expressed in hESCs. Each image shows representative data from assays conducted in triplicate (see Materials and Methods). The clone name and Alu subfamily are indicated within a white box in each image (see Table 2 and Fig. 1 posted at the above URL). Ya5 and Ya5 with no driver served as positive and negative controls, respectively, as described previously (13, 28, 43). Alu Ya5 is a known active Alu element (82) and was used to normalize the rate of trans retrotransposition of each assayed Alu element cloned from hESC. Its transfection without an L1 driver served as an internal negative control. (e) Quantification of the trans-retrotransposition activities of cloned Alu elements expressed in hESCs. For normalization, the percentage of trans retrotransposition generated by an Alu Ya5 element was designated 100%. The standard deviation of each assay is also indicated.
TABLE 1.
Summary of LINE-1 elements expressed in hEC cellsa
| Cloneb | Chromosome | LINE-1 | Full length | Polymorphic | Gene | Size (bp) | Other | % active |
|---|---|---|---|---|---|---|---|---|
| A1 | 10 (q21.1) | L1Hs | ? | Yes | None | 140 | Repeats | UNd |
| A10 | 8 (q23.3) | L1PA3 | Yes | NA | EST CB852078 | 52 | UN | |
| A11 | 4 (q13.1) | L1PA3 | Yes | NAc | LEC3/LPHN3 | 58 | UN | |
| B1 | 1 (p36.33) | L1Hs | ? | Yes | EST AK125248 | 202 | UN | |
| B4 | 2 (q21.3) | L1PA2 | Yes | NA | EST DA063346 | 67 | UN | |
| C5 | 12 (p13.32) | L1PA2 | Yes | NA | None | 254 | Repeats | UN |
| C8 | 12 (q21.31) | L1PA2 | Yes | NA | EST EG328524 | 64 | UN | |
| C10 | 1 (q25.1) | L1PA2 | Yes | NA | RABGAP1L | 140 | UN | |
| C11 | 4 (p16.1) | L1PA2 | Yes, rearranged | NA | None | 22 | UN | |
| D2 | 1 (q43) | L1Hs | Yes | No | CHRM3 | 87 | UN | |
| D3 | 3 (p24.1) | L1Hs | Yes | No | None | 207 | UN | |
| D4 | 11 (p15.4) | L1PA2 | Yes | NA | EST CD642260 | 400 | UN | |
| D6 | 9 (p21.1) | L1PA3 | Yes | NA | EST CR736801 | 474 | UN | |
| D10 | 18 (q21.2) | L1Hs | ? | Yes | EST AI420530 | 120 | UN | |
| D11 | 3 (q13.12) | L1PA3 | Yes | NA | BBX | 40 | UN | |
| F8 | 1 (p31.1) | L1Hs | ? | Yes | None | 136 | Repeats | UN |
| G4 | X (q22.3) | L1Hs | ? | Yes | IL1RAPL2 | 357 | UN | |
| H1 | 1 (p32.2) | L1Hs | ? | Yes | EST AI345549 | 338 | UN | |
| H3 | 3 (p14.1) | L1PA8 | Yes | NA | SLC25A26 | 85 | UN | |
| H10 | 5 (q22.1) | L1Hs | Yes | No | CAMK4 | 248 | 5-20 |
Column 1 (from the left) indicates the name of the clone. Column 2 indicates the chromosomal position and cytogenetic band. Column 3 indicates the subfamily of L1 according to Repeatmasker (http://www.repeatmasker.org/). Column 4 indicates if a full-length L1 is annotated in the HGRS (March 2006 assembly, http://genome.ucsc.edu/), where ? indicates that a full-length L1 is not annotated in that position. Also indicated is if the full-length L1 is a chimeric element (rearranged). Column 5 indicates if the identified full-length L1 is polymorphic. We only considered L1Hs elements to be polymorphic. Column 6 indicates the name of the gene that contains the full-length L1. Column 7 indicates the size of the unique sequence characterized (not including the size of the antisense portion of the full-length L1). Column 8 indicates other characteristics of the locus that contains the full-length L1 producing the antisense transcript. Column 9 indicates the activity of the L1Hs element as determined by Brouha et al. (19).
Duplicated clones were excluded from the analysis.
NA, not applicable (for older subfamilies).
UN, unassayed.
The core sequence from Alu elements expressed in hESCs is active in cultured cells.
An average human genome contains ∼6,000 active core Alu elements, including the modern Y and older S Alu subfamilies (12). We therefore analyzed cloned each Alu element for the presence of 124 conserved positions that are retained by active core Alu elements (12). We found that ∼70% of the Alu elements expressed in hESCs contain the majority (>80%) of these conserved nucleotides (see Table 2 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf). Thus, hESCs express many Alu elements that contain a potentially active core element, with a modest enrichment of the youngest Alu subfamily, Y (see below).
TABLE 2.
Summary of LINE-1 elements expressed in hESCsa
| Cloneb | Chromosome | LINE-1 | Full length | Polymorphic | Gene | Size (bp) | Other | % active |
|---|---|---|---|---|---|---|---|---|
| 13_RS_F1 | 4 (p16.3) | L1PA4 | Yes | NAc | ZNF595 | 174 | UNd | |
| 13_RS_F2 | 14 (q21.1) | L1PA3 | Yes | NA | BX248273 | 250 | UN | |
| 13_RS_F5 | 10 (q21.3) | L1Hs | ? | Yes | EST BG772213 | 105 | UN | |
| 13_RS_F6 | 10 (q21.1) | L1Hs | ? | Yes | None | 140 | UN | |
| 13_RS_F8 | 5 (p14.1) | L1PA3 | Yes | NA | AK309747 | 96 | UN | |
| 13_RS_F10 | 3 (p26.1) | L1Hs | ? | Yes | GRM7 | 195 | UN | |
| 13_RS_F12 | 2 (q14.1) | L1Hs | Yes | No | EST DA742384 | 121 | 0.1-5 | |
| 13_RS_G1 | 4 (q24) | L1PA5 | Yes | NA | GSTCD | 49 | UN | |
| 13_RS_G2 | 15 (q15.3) | L1PA7 | Yes | NA | FRMD5 | 39 | UN | |
| 13_RS_G5 | 1 (q43) | L1PA3 | Yes | NA | CHRM3 | 79 | UN | |
| 13_RS_G7 | 6 (q21) | L1PA2 | Yes | NA | CR598940 | 100 | UN | |
| 13_RS_G9 | 4 (q13.1) | L1Hs | Yes | No | LPHN3 | 101 | 0.1-5 | |
| 13_RS_H6 | 2 (q36.1) | L1Hs | Yes | No | None | 43 | Repeats | 0.1-5 |
| 13_RS_H9 | 18 (q12.1) | L1PA2 | Yes | No | None | 144 | UN | |
| 13_RS_H12 | 9 (p12) | L1PA5 | Yes, rearranged | NA | DKFZp572C163 | 137 | UN | |
| 13_RS_39 | 4 (q12) | L1Hs | ? | Yes | PDGFRA | 155 | UN | |
| 13_RS_46 | 1 (p31.3) | L1Hs | ? | Yes | EST BI831900 | 137 | UN | |
| 13_RS_48 | 3 (p24.3) | L1Hs | ? | Yes | EST AV682563 | 208 (126.3)e | UN | |
| 13_RL_A2 | 15 (q21.1) | L1Hs | ? | Yes | C15orf33 | 492 | UN | |
| 13_RL_A4 | 12 (p12.1) | L1Hs | Yes | No | SLCO1A2/IAPP | 426 | UN | |
| 13_RL_A9 | 9 (q32) | L1Hs | Yes | No | SNX30 | 441 | UN | |
| 13_RL_A10 | 6 (p21.1) | L1Hs | Yes | No | SUPT3H | 535 | >20 | |
| 13_RL_A11 | 7 (q21.11) | L1Hs | ? | Yes | EST AI631520 | 451 | UN | |
| 13_RL_B7 | X (q26.2) | L1Hs | Yes | No | AA205629 | 299 | 5-20 | |
| 13_RL_C5 | 3 (q13.2) | L1PA3 | Yes | NA | EST AI218267 | 305 (421.3)e | UN | |
| 7_RS_D5 | 19 (q13.31) | L1P1 | Yes, rearranged | No | ZNF227 | 30 | UN | |
| 7_RS_D6 | 2 (q21.3) | L1Hs | Yes | No | EST DA063346 | 62 | 0.1-5 | |
| 7_RS_D7 | 14 (q21.1) | L1PA3 | Yes | NA | BX248273 | 250 | UN | |
| 7_RS_D10 | 3 (p24.3) | L1Hs | Yes | No | EST AW630075 | 47 | UN | |
| 7_RS_D11 | 15 (q15.3) | L1PA7 | Yes | NA | FRMD5 | 39 | UN | |
| 7_RS_E5 | 8 (q21.2) | L1PA5 | Yes | NA | EST BF667587 | 42 | UN | |
| 7_RS_E7 | 13 (q21.1) | L1PA15 | Yes | NA | EST CX788080 | 15 | UN | |
| 7_RS_E11 | 5 (q23.1) | L1PA5 | Yes | NA | None | 49 | UN | |
| 7_RS_E12 | X (q26.2) | L1Hs | Yes | No | EST AA205629 | 299 (99.8)e | 5-20 | |
| 7_RL_F2 | 18 (q12.1) | L1Hs | Yes | No | None | 467 | Repeats | UN |
| 7_RL_F3 | 4 (p16.3) | L1PA4 | Yes | NA | ZNF595 | 174 | UN | |
| 7_RL_F5 | 3 (q13.2) | L1Hs | ? | Yes | EST BE568549 | 365 | UN | |
| 7_RL_F6 | 12 (q15) | L1Hs | ? | Yes | None | 388 | LTR repeat | UN |
| 7_RL_F7 | 4 (q24) | L1Hs | ? | Yes | EST AA307003 | 270 | UN | |
| 7_RL_F9 | 2 (p12) | L1Hs | ? | Yes | EST DA734553 | 456 | UN | |
| 7_RL_F10 | 8 (q21.11) | L1Hs | ? | Yes | AK024242 | 557 | UN | |
| 7_RL_G1 | 7 (q21.11) | L1Hs | ? | Yes | EST AI631520 | 451 | UN | |
| 7_RL_G2 | 11 (q14.3) | L1P1 | Yes, rearranged | NA | None | 101 | Repeats | UN |
| 7_RL_G5 | 7 (q21.12) | L1PA3 | Yes | NA | ADAM22 | 330 | UN | |
| 7_RL_G7 | 4 (q34.3) | L1Hs | Yes, rearranged | No | VEGFC | 412 | UN | |
| 7_RL_H1 | 2 (q31.1) | L1Hs | ? | Yes | EST BX114253 | 170 (345.1)e | UN | |
| 9_RS_H7 | 4 (q24) | L1Hs | ? | Yes | EST AA307003 | 186 | UN | |
| 9_RS_H8 | 1 (q32.3) | L1PA5 | Yes | NA | C1orf97 | 49 | UN | |
| 9_RS_H9 | 10 (q21.1) | L1Hs | ? | Yes | None | 140 | UN | |
| 9_RS_H10 | 2 (q21.3) | L1Hs | Yes | No | EST DA063346 | 90 | UN | |
| 9_RS_91 | X (q13.1) | L1PA4 | Yes | NA | KIF4A | 185 | UN | |
| 9_RS_92 | 14 (q21.1) | L1PA3 | Yes | NA | BX248273 | 250 | UN | |
| 9_RS_94 | 4 (q24) | L1PA5 | Yes | NA | GSTCD | 64 | UN | |
| 9_RS_96 | 10 (q21.1) | L1PA3 | Yes | NA | None | 176 | UN | |
| 9_RS_99 | 5 (q13.1) | L1PA3 | Yes | NA | EST DB235360 | 121 | UN | |
| 9_RS_910 | 8 (q21.11) | L1Hs | ? | Yes | EST BM667133 | 300 | UN | |
| 9_RS_911 | 2 (q14.1) | L1Hs | Yes | No | EST DA742384 | 122 | 0.1-5 | |
| 9_RS_912 | 22 (q12.1) | L1PA4 | Yes | NA | None | 36 | UN | |
| 9_RS_915 | 12 (q21.21) | L1PA3 | Yes | NA | None | 27 | Ultraconserved region | UN |
| 9_RS_917 | 3 (p24.2) | L1PA5 | Yes | NA | THRB | 203 | UN | |
| 9_RS_921 | 5 (q13.1) | L1Hs | ? | Yes | EST CA432586 | 111 | UN | |
| 9_RS_923 | 7 (q21.12) | L1PA3 | Yes | NA | ADAM22 | 322 (87.7)e | UN | |
| 9_RL_929 | 18 (q12.1) | L1Hs | Yes | No | None | 467 | Repeats | UN |
| 9_RL_931 | 4 (p16.3) | L1PA4 | Yes | NA | ZNF595 | 595 | UN | |
| 9_RL_935 | 8 (q23.3) | L1Hs | ? | Yes | EST CB852078 | 293 | UN | |
| 9_RL_938 | 9 (q32) | L1Hs | Yes | No | SNX30 | 467 | UN | |
| 9_RL_940 | 7 (q21.11) | L1Hs | ? | Yes | EST AI631520 | 451 | UN | |
| 9_RL_943 | 1 (p22.2) | L1Hs | Yes | No | HFM1 | 369 | 0.1-5 | |
| 9_RL_944 | 15 (q21.1) | L1Hs | ? | Yes | C15orf33 | 452 | UN | |
| 9_RL_951 | 16 (q21) | L1Hs | Yes, rearranged | No | CDH8 | 344 | UN | |
| 9_RL_952 | 8 (p23.1) | L1PA6 | Yes | NA | FDFT1 | 204 | UN | |
| 9_RL_953 | 14 (q31.3) | L1PA4 | Yes | NA | EST BU687365 | 165 (380.7)e | UN |
For details, see Table 1, footnote a.
Duplicated clones were excluded from the analysis. _RL_ and _RL_ indicate whether the clone was sequenced from a short- or a long-size pool.
NA, not applicable (for older subfamilies).
UN, unassayed.
Average size of sequenced inserts from a short- or a long-size pool.
We next sought to determine whether the core sequences derived from hESC-expressed Alu elements are active in cultured cells (Fig. 1c) (28). To avoid bias, we chose 13 hESC-expressed Alu elements at random and cloned their core sequence into the backbone of plasmid pAluNF1-neoIII (see Materials and Methods). All of these Alu cores contain the conserved G25 nucleotide, which is critical for SRP 9/14 binding, and all but two elements contained the G159 nucleotide, which also is present in the conserved SRP 9/14 binding site (see Fig. 1 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf). Of the 13 randomly selected Alu elements analyzed, 7 belong to the Sx subfamily, 2 each belong to the Sp and Sg subfamilies, 1 belongs to the Sc subfamily, and 1 belongs to the Y subfamily. We then tested these Alu constructs for retrotransposition in HeLa cells (28). In this assay, an untagged driver L1 (that produces ORF2p [2, 13]) is cotransfected with an Alu element tagged with a reporter gene (conferring resistance to G-418 [30]) that can only be activated upon a single round of trans retrotransposition (Fig. 1c). As controls we used a known active Ya5 Alu element (28, 82) which was transfected with or without an L1 driver.
Of the 13 Alu elements analyzed (Fig. 1d and e), 8 (∼61%) had at least 10% of the activity of a Ya5 Alu element (elements A-1_Sx, A-12_Y, A-13_Sx, A-14_Sx, A-15_Sg, A-16_Sc, A-19_Sp, and A-20_Sx, Fig. 1d and e). The remaining five Alu elements had less than 2% of the activity of the Ya5 element (A-5_Sx, A-18_Sp, A-21_Sx, A-32_Sg, and A-34_Sx), likely due to the presence of mutations, deletions, and insertions within the core sequence (see Fig. 1 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf). In agreement with a previous study (12), we found an old Sx Alu element that displayed a high level of trans retrotransposition (clone A-20_Sx, shows ∼70% of the activity of a Ya5 Alu element, Fig. 1d and e). Control experiments indicated that Alu Ya5 mobilization could only be achieved upon cotransfection with a driver L1 (Fig. 1d and e, compare Ya5 and Ya5 with no driver) (28, 43). Thus, undifferentiated hESCs express a wide variety of Alu elements and some of these contain an active core element.
The L1 sense and antisense promoters are active in hESCs and hEC cells.
Endogenous L1 RNPs can be detected in hESC and hEC cell lines, suggesting that the L1 sense promoter is active in pluripotent cells (35, 41, 58, 74). We confirmed these findings by determining the amount of sense L1 transcripts present in pluripotent cells (and compared this to the amount in differentiated cells as a control). Briefly, we designed real-time PCR primer sets annealing to either the 5′ UTR or ORF2 region of a consensus L1 and determined the L1 mRNA contents of hESC, hEC, and HeLa cells (see Materials and Methods). We observed that, on average, pluripotent cells express 10 to 15 times more sense L1 mRNA than differentiated cells (Fig. 2a). In addition, we used conventional RT-PCR and sequencing to determine which L1 is expressed in pluripotent cells as described previously (35, 36). We observed that a wide range of L1s is expressed in hEC cells and hESCs (Fig. 2b).
FIG. 2.
Both LINE-1 5′ UTR promoters are active in hESCs and hEC cells. (a) L1 expression in pluripotent cells. The cartoon is a schematic of a human LINE-1 element. The relative position and amplification lengths (dashed lines) of the two set of primers (L1-52 and L1-22) (27) used in the quantitative RT-PCR are indicated below the schematic. The position of the amplified region is based on the L1.3 sequence (L19088.1 [71]). Below are shown representative quantitative RT-PCR results for L1 RNA expression using the 5′ UTR (L1-52) or ORF2 (L1-22) primer set analyzed in pluripotent cells and differentiated HeLa cells as a control (35, 36). The graphs show the n-fold changes in L1 expression with respect to H9 hESCs. To normalize the results, we determined the amplification CT for GAPDH (see Materials and Methods). (b) Pie charts showing the percentages of L1 subfamilies expressed in hEC cells (2102Ep cells) and hESCs (H9 cells). (c) Cartoon of an RC-L1 (top) with two clones generated that contain the 5′ UTR of L1.3 in the antisense (left, 5AS-FF) or the sense (right, 5S-FF) orientation. The white boxes indicate the ORF coding for firefly luciferase, and black lollipops represent polyadenylation signals. (d) Quantification of the activities of the sense and antisense L1Hs promoters in hESCs and 2102Ep hEC cells. The graph shows the firefly luciferase activity in LUs, normalized to Renilla luciferase activity, which served as an internal control for transfection efficiency (see Materials and Methods), of the sense (5S) and antisense (5AS) promoters in hEC cells (2102Ep, dark bar graph), hESCs (H13B (gray bar graph), and H7 cells (white bar graph) measured in triplicate (the standard deviation of the assay is indicated in each graph). LUs from these assays are shown in Fig. 2 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf. UTF, untransfected.
Next, we examined if the L1 antisense (AS) promoter was active in pluripotent cells and if it could allow the identification of expressed L1s by expression originating from their AS promoter. A previous report characterized an antisense promoter located in the 5′ UTR of L1 (region between bp 400 and 600) (76) and reported human cell-specific transcripts originating from these promoters.
To test the strength of both sense and antisense L1 promoters in pluripotent cells, we cloned the 5′ UTR of an L1Hs element (L1.3) (71) into the firefly luciferase reporter pGL3-basic plasmid (Fig. 2c) in the sense (5S-FF) or antisense orientation (5AS-FF). We then cotransfected these plasmids into cultured cells with a Renilla luciferase internal control (driven by the SV40 promoter). When plasmids were transfected into the male hEC line 2102Ep, we detected active transcription produced from both sense and antisense L1 promoters (Fig. 2d, top graph). We also detected efficient transcription from both L1 promoters in H13B (male) and H7 (female) hESCs (Fig. 2d, middle and bottom graphs). Although hESCs are notoriously difficult to transfect, the observed difference in promoter strength is not likely caused by a technical difficulty, as the level of luciferase units (LUs) is far higher than in the untransfected controls (10- to 100-fold, depending of the construct; see Fig. 2 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf). Notably, the sense promoter was 3 to 13 times more active than the antisense L1Hs promoter in all of the cell lines tested, which is consistent with previous reports (76, 85).
Antisense-based identification of expressed LINE-1s.
Given that the L1 antisense promoter is active in hECs and hESCs, we reasoned that L1s located in different genomic loci could give rise to unique transcripts that could be mapped precisely to the genome (Fig. 3) (76). Thus, we adapted a 3′ RACE protocol to precisely map mRNAs produced by transcription from the L1 antisense promoter into flanking genomic sequences (see Fig. 3 for a simplified example with five L1s). The assay involved the isolation of total RNA from cells (Fig. 3a), the generation of a cDNA library using a 3′ RACE primer (in triplicate, Fig. 3b), and a final PCR (also in triplicate) that used a primer complementary to the 3′ RACE adapter and an L1 AS primer to allow the specific amplification of AS-L1 transcripts (Fig. 3c). PCR products were then cloned and sequenced, and the unique part of L1 AS transcripts was mapped back to the human genome reference sequence (HGRS) using BLAT (51) (Fig. 3d and e). This procedure will identify L1s with active AS transcription and very likely sense transcription.
FIG. 3.
Rationale of the antisense-based identification of expressed LINE-1s. (a) Cartoons of five full-length L1s at different chromosomal locations (indicated by the color of the thin shadowed line that flanks each L1 [blue, red, orange, green, or purple]). In each full-length L1 cartoon, UTRs are shown as thin black lines, ORFs are shown as gray boxes (marked ORF1 and ORF2), promoters are shown as small arrows, and target site duplications are shown as large black arrows. Each L1 produces two mRNAs using the sense promoter (gray curved line) or the antisense promoter (colored curved lines). Flanking genomic DNA is marked with a thin shadowed blue, red, orange, green, or purple line flanking the L1. (b) Total RNA was isolated from hESCs and primed with a RACE primer in an RT reaction to generate a library of cDNAs. (c) A fraction of these cDNAs were used in a PCR with primers Outer and ABIEL_library to generate double-stranded DNA products only from those cDNAs primed within the L1 antisense promoter. (d) PCR products were resolved on agarose gels and cloned according to size (see Materials and Methods), and approximately 30 clones from each fraction were sequenced. Then, sequences were mapped to the HGRS at http://genome.ucsc.edu using BLAT (51). (e) Schematic of the two possible outcomes of the sequence analysis. In both cases, a cartoon of a chromosome (blue) and the piece of DNA sequenced (marked with a broken green box) is shown. On the left is shown an example of a characterized clone corresponding to a previously annotated full-length L1 in the HGRS (blue shaded line with a full-length L1). On the right is shown an example of a polymorphic L1 that is not annotated in the HGRS (red shaded line).
We first conducted a mapping reaction using total RNA isolated from the hEC cell line 2102Ep and observed amplification products that ranged in size from ∼50 to 500 bp (data not shown). We cloned amplification products from this pool and sequenced randomly selected clones. We observed that the average size of inserts in the library was approximately 170 bp, likely due to bulk cloning (see Fig. 3a and b posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf) and identified several AS-L1 transcripts that could be precisely mapped to regions of the HGRS that contained an annotated full-length L1 element (Fig. 4a and Table 1). These results indicated that the antisense promoter of L1 can be used to identify the genomic loci from which individual L1s are expressed, at least by using their AS promoter as a proxy, in pluripotent cells. Notably, we also identified AS-L1 transcripts that precisely map to the HGRS but that did not have an annotated full-length L1 element upstream. These transcripts likely indicate the presence of novel dimorphic L1 insertions (see below).
FIG. 4.
LINE-1 elements expressed in hESC and hEC cell lines. (a to d) Each panel contains a cartoon of all of the cell line chromosomes, according to sex (a, 2102Ep; b, H13B; c, H7; d, H9). Each bar on the chromosome ideogram represents a sequenced transcript produced from a full-length (confirmed or inferred) LINE-1 element. We found that older subfamilies of LINE-1s are able to produce an antisense transcript, and the color of each line corresponds to a different L1 subfamily (green, L1Hs; red, L1PA3; orange, L1PA2; purple, L1PA8; blue, L1PA4; yellow, L1PA5; light blue, L1PA7; light orange, L1P1; purple, L1PA15; light brown, L1PA6). In panels b to d, it is indicated whether the sequence was obtained from a short- or a long-size pool (lines marked with _S_ and _L_, respectively). The chromosome band, if a full-length L1 was annotated in that position, the size of the transcript, and other details can be found in Tables 1 and 2 (for hEC cells and hESCs, respectively).
L1 elements expressed in hESCs.
We next conducted the L1 AS mapping procedure with total RNAs isolated from three undifferentiated hESC lines (H7, H9, and H13B). To avoid size enrichment artifacts, we size fractionated the amplified PCR products into two groups, <300 bp and 300 to 600 bp. As in hEC cells, we observed a range of amplification products in the three hESC lines. However, we obtained a better representation of the sizes of the AS-L1 transcripts in these libraries, likely due to the fractionation of amplified products, with maximum sizes of 300 and 595 bp in each size group (average sizes of 104 and 382 bp, respectively; see Fig. 3a and b posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf).
We next characterized approximately 30 clones per library (in triplicate) and then mapped the sequences to the HGRS using BLAT (51). An overview of the expressed AS-L1s in the three hESC lines is shown in Fig. 4b to d (see Table 2 for a detailed description of each annotated entry). These data indicate that active AS-L1 expression occurs from different chromosomal locations in hESCs. Notably, the majority of AS-L1 transcripts map to both human-specific and older L1 subfamilies in the HGRS (Fig. 4; see Fig. 6a), suggesting that the activity of the L1 antisense promoter is not restricted to L1Hs elements (see below). As we reasoned that those L1s showing AS transcription very likely would express sense L1 mRNA, we next analyzed whether L1Hs elements that generate an AS-L1 transcript in hEC cells and hESCs correspond to previously characterized active or hot L1 elements present in the HGRS (19). Indeed, among the L1Hs elements characterized in 2102Ep, H13B, H7, and H9 cells, some were previously demonstrated to be retrotransposition competent in a cell culture-based assay (19) and some correspond to elements that contain at least >20% of the activity of a hot reference L1 (see Table 2, last column). Remarkably, our results indicate that in some hESC lines, ∼30% of the expressed L1Hs elements (identified on the basis of active AS-L1 expression) correspond to known RC-L1Hs (19).
Identification of expressed polymorphic L1 elements in hESCs.
In analyzing L1 expression in hESCs, we found that almost half of the characterized L1Hs elements correspond to loci lacking an L1HS element in the HGRS. We reasoned that loci showing AS-L1 expression where a full-length L1Hs was absent from the HGRS represented polymorphic elements that are differentially present or absent in different hESC lines, consistent with previous findings (8, 15, 84). Notably, we did not find any polymorphic L1s in the set of identified older L1 elements (L1PA2 to L1PA15), consistent with their lack of activity during recent human evolution (14-16). To confirm that these loci contain polymorphic L1Hs elements, we genotyped a set of eight putatively polymorphic L1Hs elements isolated from H13B cells (Fig. 5a and Table 2). Two representative examples of the genotyping are shown in Fig. 5c and d, where the loci on chromosomes 10 and 3 (clones 13_F6 and 13_F10, respectively) produce amplification products consistent with the presence of an L1Hs insertion. Cloning and sequencing confirmed the presence of both L1Hs elements. We also confirmed the presence of the other six polymorphic L1Hs elements in the genome of H13B cells (data not shown), and in one case we found that the element is also present in the genome of HeLa cells (Fig. 5d). Thus, our results indicate that the L1 antisense promoter can be used to detect the presence of polymorphic L1 elements that are expressed in hESCs.
FIG. 5.
LINE-1 elements expressed in pluripotent cells are polymorphic with respect to their presence in the HGRS. (a) The graph represents the percentages of fixed (blue bars) and polymorphic (red bars) full-length L1Hs elements expressed in the indicated cell line (x axis). For the classification, we mapped L1Hs-derived antisense transcripts to the HGRS using BLAT and considered them fixed if a full-length L1Hs element was already annotated in the corresponding chromosomal position. (b) Rationale of the genotyping PCR. For genotyping, we designed a unique primer in a nonrepetitive region of the identified AS-L1 transcript and used it in conjunction with the L1 AS primer in a PCR. Genotyping primers are shown as blue arrows. (c and d) Genotyping PCR to confirm the presence of two polymorphic L1s in the genome of H13B cells. Shown are the results of the PCR conducted for two polymorphic full-length L1Hs elements using genomic DNA isolated from H13B cells or from HeLa cells as a control (c, 13_F6; d, 13_F10; see Table 2 for other details). A reaction was also conducted with water as an internal negative control (c− lane). MW, marker VIII (Roche). All of the other polymorphic full-length L1Hs elements identified in H13B cells were verified by the same procedure (data not shown).
LINE-1 antisense promoter activity is conserved through evolution.
In our analysis of the AS-L1 transcripts expressed in 2102Ep and hESCs, we observed that between 40 and 55% of the sequences correspond to older L1s, which suggests that the activity of the antisense L1 promoter is conserved through LINE-1 evolution. In hESCs, we found an overrepresentation of AS-L1 transcripts generated by L1PA2, L1PA3, and L1PA4 elements, although older subfamilies could also generate AS-L1 transcripts (Fig. 6a). Indeed, each clone containing an AS-L1 transcript originating from an old L1 (i.e., non-L1Hs elements) could be precisely mapped to an annotated repeat in the HGRS (Table 1 and 2). This suggests that they likely do not represent PCR recombination artifacts, although some of the older L1-containing AS-L1 transcripts may represent artifacts of priming within a longer transcript (generated upstream of the full-length L1 element [20, 48]).
FIG. 6.
LINE-1 elements expressed in pluripotent cells are produced from a range of L1s. (a) Pie charts showing the representations and percentages of L1 subfamilies expressing AS-L1 transcripts in hESCs and hEC cells (the cell line name is indicated above each pie chart). (b) Both sense and antisense promoters from the 5′ UTR of older LINE-1 elements are active in hEC cells. Each graph shows the quantification of the activity of the sense and antisense L1 promoters from L1PA1, L1PA2, L1PA3, L1PA4, L1PA6, L1PA7, L1PA8, and L1PA10 elements in PA-1 hEC cells. The graphs show the firefly activities (normalized to Renilla luciferase activity; see Materials and Methods) of the sense (S) and antisense (AS) promoters measured in triplicate (the standard deviation of the assay is indicated in each graph). As a control, PA-1 cells were transfected with a promoterless construct (pgl3basic). Similar data were obtained with 2102Ep and N-Tera2D1 cells (see Fig. 5 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf).
To unambiguously determine if the activity of the L1 antisense promoter is conserved through evolution, we cloned the first 900 bp of a cohort of old L1s (L1PA2, L1PA3, L1PA4, L1PA6, L1PA7, L1PA8, and L1PA10) into the vector pGL3Basic and determined their promoter strengths in both the sense and antisense orientations in hEC cells relative to those of the sense and antisense promoters of an L1Hs or L1PA1 element (L1.3, see above). Remarkably, we observed that both sense and antisense promoters from the cohort of old L1s were active in several hEC lines, including PA-1, 2102Ep, and N-Tera2D1 (Fig. 6b; see Fig. 4 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf), although their strength was always lower than that of an L1PA1 element in our experimental settings (see Fig. 5 posted at the above URL). This may mean that different TFBSs are differentially present in each promoter (see Document 1 posted at the above URL). In addition, technical limitations associated with the use of luciferase-based constructs to measure promoter strength may influence the reported level of activity of a promoter. Thus, these results indicate that the antisense promoter of L1 is conserved through evolution, at least up to L1PA10 elements.
Expressed L1 retrotransposons in hESCs are located mostly within genes.
When analyzing the expression of AS-L1s in hESCs, we identified genomic loci with active AS-L1 expression that were common to different hESC lines. Of these, five loci were shared by all of the hESC lines and seven were shared by at least two hESCs (Fig. 4c and d). Within these groups, four and six AS-L1 transcripts, respectively, are generated by full-length L1Hs elements, and the remainders are generated by L1PA3 and L1PA7 elements. Although the coverage of our procedure is unknown and may be biased toward shorter transcripts, it seems that active AS-L1 expression is largely confined to discrete loci in hESCs and that different hESC lines share some of these loci.
In addition, we found that most of the expressed AS-L1 transcripts characterized could be mapped to known/annotated genes and expressed sequence tags (ESTs; Table 2, 88%, 80%, and 81% in H13B, H7, and H9, respectively) and that their expression level appears to be independent of the L1's age. Notably, the proportion of expressed L1 elements that reside within genes in pluripotent cells seemed much higher than expected. To determine if expressed L1s in pluripotent cells are disproportionately located within genes, we compared our data to a nonredundant data set of human-specific full-length L1 sequences. These sequences were extracted from four large genomic DNA data sources (the GenBank nucleotide sequence database [April 2008], the HGRS [53], the Celera Genomics human genome sequence [81], and the HuRef diploid human assembly [54]). Of the 533 full-length L1Hs elements in these data sets, 164 are located within the transcription unit of Refseq genes (∼30%; see Materials and Methods for details). This starkly contrasts with the ∼80% of the expressed L1s reported here that are located within known genes. A hypergeometric analysis (see Materials and Methods) under the null hypothesis that the distribution of expressed L1s in genes is the same as their genomic distribution, irrespective of expression, confirmed that this null hypothesis can be robustly rejected (P = 4.25e-05, 1.70e-06, 8.84e-08, and 1.68e-09 in 2102Ep, H7, H9, and H13B cells, respectively). Assuming that full-length, human-specific L1s maintain a promoter activity similar to that of our data set of expressed L1s, these data strongly suggest that there is epigenetic regulation of the L1 antisense promoter in pluripotent cells.
In agreement with the above hypothesis, the expression of Alu elements correlated with the known number of Alu elements belonging to each subfamily in the human genome (Fig. 1a, see Table 1 posted at http://www.juntadeandalucia.es/fundacionprogresoysalud/repositorio/files/0000/0080/16._MCB00561_SupInfo_final.pdf) (9, 12, 62). However, we did detect a >2-fold enrichment of expressed Alu Y elements over Alu J elements (the average abundance across all three lines was 18% [Alu Y] and 7% [Alu J]), which is intriguing, as both subfamilies are present at similar copy numbers in the human genome (Fig. 1a and b) (9, 12, 62).
DISCUSSION
A recent study has determined that there are about 6,000 Alu active core elements per genome, including members of old and young subfamilies (12). Our results indicate that hESCs express a wide range of Alu elements, including both young and old subfamilies. These results are also consistent with previous L1 expression analyses where it was found that hESCs express a range of L1s of various ages (35). When corrected for the known copy number of Alu elements in the human genome, our results indicate that hESCs primarily express young Alu subfamily members (Y and S), which is in agreement with their recent evolutionary amplification in humans (9). To obtain an unbiased overview of expressed Alu elements that contain an active core, we analyzed the activity of a randomly selected cohort of Alu elements expressed in hESCs, and determined that, on average, 60% of them have >10% of the activity of a reference hot Alu element (12, 82). This result reflects the activity of the Alu cores and does not incorporate the influence of 5′ and 3′ flanking regions on the assayed Alu elements' activity (1, 22, 70). When combined with an in silico estimate of Alu activity, assuming that those elements with less than 10% variation with respect to the active core consensus represent active elements (12), approximately 40 to 50% of the core of Alu elements expressed in hESCs have a trans-retrotransposition potential of at least 10% of that of an active Alu element known to have caused a human disease (82).
To obtain a locus-specific census of expressed L1s in hESCs, we developed a method employing the antisense promoter contained within the L1 5′ UTR (76). First, we demonstrated that both sense (78) and antisense (76) L1Hs promoters are active in hEC cells and hESCs by using a plasmid reporter assay. Thus, it is likely that both promoters are active in genomic L1 copies, although our mapping protocol only captures transcription initiating from the L1 AS promoter. Indeed, we have confirmed that the mapping technique is useful in identifying L1Hs at various genomic loci that show active antisense transcription. Although we do not know the coverage of the transcriptome achieved by this procedure, the rate of false positives obtained is low, as we have never detected nonannotated old L1s generating AS-L1 transcripts (having analyzed >200 independent transcripts), and we have confirmed the existence of polymorphic L1Hs purely on the basis of their AS-L1 expression. It is worth mentioning that a significant proportion (∼50%) of AS-L1 transcripts originated from old L1 subfamilies. Thus, we analyzed a panel of old L1s for promoter activity and found that the activity of the L1 AS promoter is robust throughout L1 evolution (at least as far back as L1PA10 elements). These old L1s also contain active sense promoters and potentially can produce double-stranded RNAs that could trigger an RNAi response to regulate L1 activity (85). Due to its functional conservation during evolution, it will be interesting to elucidate if the AS L1 promoter serves as an autoregulatory signal or participates in any step of L1 retrotransposition.
We also have determined that some of the expressed AS-L1s correspond to previously identified active L1Hs elements. In agreement, recent reports on somatic human tissues revealed the presence of many L1 elements transcribed in those cells, but few of them are likely to be active (69). However, it should be noted that allelic heterogeneity could impact the activity of a given L1 allele, as previously reported (56, 73). Furthermore, we have determined that almost half of the identified L1Hs elements expressed in hESCs are polymorphic, consistent with previous reports (8, 15, 84). It was previously shown that de novo L1 insertions can accumulate during early human embryonic development in vivo (80). Very recently, in a mouse model of human L1 retrotransposition, it was found that most de novo insertions occur in early embryonic development and that insertions in germ cells are uncommon (47). Indeed, all known human mutagenic L1/Alu insertions could have occurred early in development, indicating that hESCs are a bona fide model to study the accumulation of new L1/Alu retrotransposition events in humans (47). Our results indicate that approximately 40% of all expressed Alu elements and up to 20% of expressed L1Hs elements in hESCs (on the basis of their AS promoter) may represent active elements with the potential to retrotranspose. This suggests that the degree of somatic mosaicism attributable to L1 insertions generated during early development may be much higher than previously anticipated (10, 27, 31, 42, 45).
Having captured a cohort of L1s actively expressed in hESCs, we have found that expression of L1s (at least from their antisense promoter) appears to be confined to discrete genomic loci and that some of these loci are shared by different hESC lines. Indeed, recent studies that analyzed the regulated retrotransposon transcriptome in a panel of human somatic tissues by using deep-sequencing cap analysis gene expression and other methods noted that, despite making up a third of our genome, retrotransposons are less expressed, on average, than nonrepetitive regions of the genome (32, 69). In hESCs, in contrast, the expression of L1s located in known or hypothetical human genes is readily detectable. This is reflected in a very significant enrichment of L1s expressed from within known genes, relative to their genomic distribution (∼80% of loci versus 30% expected). These data strongly suggest that while the L1 antisense promoter (and, by extrapolation, the sense promoter) is intrinsically active in human cells, there is apparently a relaxation in the control of L1 expression in pluripotent cells, most likely mediated by the widespread epigenetic remodeling typical of these cells. These results are also consistent with a recent report that demonstrated efficient epigenetic silencing and reactivation (by chromatin-modifying agents) of EGFP-marked de novo L1 retrotransposition insertion events in a panel of hEC cell lines (36). It remains to be seen whether the L1 expression detected in our study reflects programmed expression activation of hESC-specific genes or is a general feature of extensive epigenetic reprogramming. One model is that gene expression required to maintain the pluripotent state exposes L1 promoters within specific genes to chromatin contexts permissive for L1 expression. Indeed, it is tempting to speculate that active L1 elements present in expressed chromatin areas of human embryonic cells (i.e., ICM cells) would be more likely to generate copies of themselves that would be transmitted to new generations (Fig. 7).
FIG. 7.
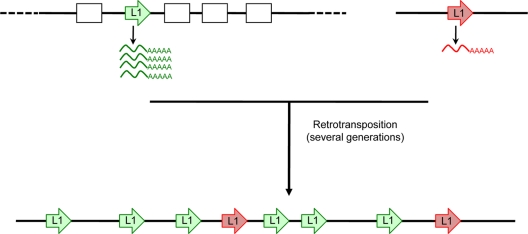
Epigenetic control of L1 expression in hESCs. The model shows retrotransposition events accumulating overtime as they arise from active L1s (depicted as arrows) located in different genomic contexts, either within genes (green arrow) or outside genes (red arrow). In the cartoon, noncoding regions are depicted as black lines and exons are depicted as white boxes. Active L1 elements located within genes (green arrow) would produce more L1 RNA and RNPs, and their subsequent insertion during early human development would result in an enrichment of this set of elements. In contrast, active elements located outside genes (red arrow) would produce less RNA and so produce fewer insertions over time.
Acknowledgments
We thank Thierry Heidmann (CNRS, France), Alicia Barroso-delJesus (Andalusian Stem Cell Bank, Spain), Peter W. Andrews (University of Sheffield, United Kingdom), and Astrid Roy-Engel (Tulane University) for providing the neo3 cassette, plasmid pRL-SV40, hEC cell lines, and HeLa-HA cells, respectively. We also thank Victoria Kelly (University of Michigan) for help with luciferase readings during early stages of this study, Sandra R. Richardson and John V. Moran (University of Michigan) for critically reading the manuscript, and Carolina Elosua (Andalusian Stem Cell Bank) and Alberto Labarga (Scientifik, Spain) for help with statistical analyses. J.L.C. thanks Gustavo Melen and Alicia Barroso-DelJesus (Andalusian Stem Cell Bank, Spain) for cloning advice and support. We are indebted to John V. Moran (Howard Hughes Medical Institute, University of Michigan), who enabled the proof-of-principle experiments for the antisense-based identification of expressed L1s using 2102Ep cells to be carried out in his laboratory when J.L.G.-P. was a postdoctoral fellow.
This work was supported by a Marie Curie IRG action (FP7-PEOPLE-2007-4-3-IRG to J.L.G.P.); by the Instituto de Salud Carlos III/FEDER, Spain (EMER07/056 to J.L.G.P., CP07/00065 to J.L.G.P., FIS PI08171 to J.L.G.P., and FIS PI070527 to J.A.M.); by the Junta de Andalucia, Spain (CICE P09-CTS-4980 to J.L.G.P., PeS PI-002 to J.L.G.P., and CICE P08-CTS-3678 to S.M.); by the Wellcome Trust (075163/Z/04/Z to R.M.B.); and by an MRC Capacity Building Ph.D. Studentship in Bioinformatics (to R.K.H.).
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Alemán, C., A. M. Roy-Engel, T. H. Shaikh, and P. Deininger. 2000. Cis-acting influences on Alu RNA levels. Nucleic Acids Res. 28:4755-4761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alisch, R. S., J. L. Garcia-Perez, A. R. Muotri, F. H. Gage, and J. V. Moran. 2006. Unconventional translation of mammalian LINE-1 retrotransposons. Genes Dev. 20:210-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.An, W., J. S. Han, S. J. Wheelan, E. S. Davis, C. E. Coombes, P. Ye, C. Triplett, and J. D. Boeke. 2006. Active retrotransposition by a synthetic L1 element in mice. Proc. Natl. Acad. Sci. U. S. A. 103:18662-18667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Andrews, P. W., P. N. Goodfellow, L. H. Shevinsky, D. L. Bronson, and B. B. Knowles. 1982. Cell-surface antigens of a clonal human embryonal carcinoma cell line: morphological and antigenic differentiation in culture. Int. J. Cancer 29:523-531. [DOI] [PubMed] [Google Scholar]
- 5.Andrews, P. W., M. M. Matin, A. R. Bahrami, I. Damjanov, P. Gokhale, and J. S. Draper. 2005. Embryonic stem (ES) cells and embryonal carcinoma (EC) cells: opposite sides of the same coin. Biochem. Soc. Trans. 33:1526-1530. [DOI] [PubMed] [Google Scholar]
- 6.Athanikar, J. N., R. M. Badge, and J. V. Moran. 2004. A YY1-binding site is required for accurate human LINE-1 transcription initiation. Nucleic Acids Res. 32:3846-3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Babushok, D. V., E. M. Ostertag, C. E. Courtney, J. M. Choi, and H. H. Kazazian, Jr. 2006. L1 integration in a transgenic mouse model. Genome Res. 16:240-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Badge, R. M., R. S. Alisch, and J. V. Moran. 2003. ATLAS: a system to selectively identify human-specific L1 insertions. Am. J. Hum. Genet. 72:823-838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Batzer, M. A., and P. L. Deininger. 2002. Alu repeats and human genomic diversity. Nat. Rev. Genet. 3:370-379. [DOI] [PubMed] [Google Scholar]
- 10.Beck, C. R., P. Collier, C. Macfarlane, M. Malig, J. M. Kidd, E. E. Eichler, R. M. Badge, and J. V. Moran. 2010. LINE-1 retrotransposition activity in human genomes. Cell 141:1159-1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Belancio, V. P., P. Deininger, and A. M. Roy-Engel. 2009. LINE dancing in the human genome: transposable elements and disease. Genome Med. 1:97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bennett, E. A., H. Keller, R. E. Mills, S. Schmidt, J. V. Moran, O. Weichenrieder, and S. E. Devine. 2008. Active Alu retrotransposons in the human genome. Genome Res. 18:1875-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bogerd, H. P., H. L. Wiegand, A. E. Hulme, J. L. Garcia-Perez, K. S. O'Shea, J. V. Moran, and B. R. Cullen. 2006. Cellular inhibitors of long interspersed element 1 and Alu retrotransposition. Proc. Natl. Acad. Sci. U. S. A. 103:8780-8785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boissinot, S., P. Chevret, and A. V. Furano. 2000. L1 (LINE-1) retrotransposon evolution and amplification in recent human history. Mol. Biol. Evol. 17:915-928. [DOI] [PubMed] [Google Scholar]
- 15.Boissinot, S., A. Entezam, L. Young, P. J. Munson, and A. V. Furano. 2004. The insertional history of an active family of L1 retrotransposons in humans. Genome Res. 14:1221-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boissinot, S., and A. V. Furano. 2001. Adaptive evolution in LINE-1 retrotransposons. Mol. Biol. Evol. 18:2186-2194. [DOI] [PubMed] [Google Scholar]
- 17.Bourc'his, D., and T. H. Bestor. 2004. Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L. Nature 431:96-99. [DOI] [PubMed] [Google Scholar]
- 18.Brouha, B., C. Meischl, E. Ostertag, M. de Boer, Y. Zhang, H. Neijens, D. Roos, and H. H. Kazazian, Jr. 2002. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am. J. Hum. Genet. 71:327-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brouha, B., J. Schustak, R. M. Badge, S. Lutz-Prigge, A. H. Farley, J. V. Moran, and H. H. Kazazian, Jr. 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U. S. A. 100:5280-5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carninci, P., T. Kasukawa, S. Katayama, J. Gough, M. C. Frith, N. Maeda, R. Oyama, T. Ravasi, B. Lenhard, C. Wells, R. Kodzius, K. Shimokawa, V. B. Bajic, S. E. Brenner, S. Batalov, A. R. Forrest, M. Zavolan, M. J. Davis, L. G. Wilming, V. Aidinis, J. E. Allen, A. Ambesi-Impiombato, R. Apweiler, R. N. Aturaliya, T. L. Bailey, M. Bansal, L. Baxter, K. W. Beisel, T. Bersano, H. Bono, A. M. Chalk, K. P. Chiu, V. Choudhary, A. Christoffels, D. R. Clutterbuck, M. L. Crowe, E. Dalla, B. P. Dalrymple, B. de Bono, G. Della Gatta, D. di Bernardo, T. Down, P. Engstrom, M. Fagiolini, G. Faulkner, C. F. Fletcher, T. Fukushima, M. Furuno, S. Futaki, M. Gariboldi, P. Georgii-Hemming, T. R. Gingeras, T. Gojobori, R. E. Green, S. Gustincich, M. Harbers, Y. Hayashi, T. K. Hensch, N. Hirokawa, D. Hill, L. Huminiecki, M. Iacono, K. Ikeo, A. Iwama, T. Ishikawa, M. Jakt, A. Kanapin, M. Katoh, Y. Kawasawa, J. Kelso, H. Kitamura, H. Kitano, G. Kollias, S. P. Krishnan, A. Kruger, S. K. Kummerfeld, I. V. Kurochkin, L. F. Lareau, D. Lazarevic, L. Lipovich, J. Liu, S. Liuni, S. McWilliam, M. Madan Babu, M. Madera, L. Marchionni, H. Matsuda, S. Matsuzawa, H. Miki, F. Mignone, S. Miyake, K. Morris, S. Mottagui-Tabar, N. Mulder, N. Nakano, H. Nakauchi, P. Ng, R. Nilsson, S. Nishiguchi, S. Nishikawa, et al. 2005. The transcriptional landscape of the mammalian genome. Science 309:1559-1563. [DOI] [PubMed] [Google Scholar]
- 21.Chiu, Y. L., and W. C. Greene. 2008. The APOBEC3 cytidine deaminases: an innate defensive network opposing exogenous retroviruses and endogenous retroelements. Annu. Rev. Immunol. 26:317-353. [DOI] [PubMed] [Google Scholar]
- 22.Comeaux, M. S., A. M. Roy-Engel, D. J. Hedges, and P. Deininger. 2009. Diverse cis factors controlling Alu retrotransposition: what causes Alu elements to die? Genome Res. 19:545-555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cordaux, R., and M. A. Batzer. 2009. The impact of retrotransposons on human genome evolution. Nat. Rev. Genet. 10:691-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cordaux, R., D. J. Hedges, S. W. Herke, and M. A. Batzer. 2006. Estimating the retrotransposition rate of human Alu elements. Gene 373:134-137. [DOI] [PubMed] [Google Scholar]
- 25.Cost, G. J., and J. D. Boeke. 1998. Targeting of human retrotransposon integration is directed by the specificity of the L1 endonuclease for regions of unusual DNA structure. Biochemistry 37:18081-18093. [DOI] [PubMed] [Google Scholar]
- 26.Cost, G. J., Q. Feng, A. Jacquier, and J. D. Boeke. 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J. 21:5899-5910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coufal, N. G., J. L. Garcia-Perez, G. E. Peng, G. W. Yeo, Y. Mu, M. T. Lovci, M. Morell, K. S. O'Shea, J. V. Moran, and F. H. Gage. 2009. L1 retrotransposition in human neural progenitor cells. Nature 460:1127-1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dewannieux, M., C. Esnault, and T. Heidmann. 2003. LINE-mediated retrotransposition of marked Alu sequences. Nat. Genet. 35:41-48. [DOI] [PubMed] [Google Scholar]
- 29.Dombroski, B. A., S. L. Mathias, E. Nanthakumar, A. F. Scott, and H. H. Kazazian, Jr. 1991. Isolation of an active human transposable element. Science 254:1805-1808. [DOI] [PubMed] [Google Scholar]
- 30.Esnault, C., J. F. Casella, and T. Heidmann. 2002. A Tetrahymena thermophila ribozyme-based indicator gene to detect transposition of marked retroelements in mammalian cells. Nucleic Acids Res. 30:e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ewing, A. D., and H. H. Kazazian. 2010. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res. 20:1262-1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Faulkner, G. J., Y. Kimura, C. O. Daub, S. Wani, C. Plessy, K. M. Irvine, K. Schroder, N. Cloonan, A. L. Steptoe, T. Lassmann, K. Waki, N. Hornig, T. Arakawa, H. Takahashi, J. Kawai, A. R. Forrest, H. Suzuki, Y. Hayashizaki, D. A. Hume, V. Orlando, S. M. Grimmond, and P. Carninci. 2009. The regulated retrotransposon transcriptome of mammalian cells. Nat. Genet. 41:563-571. [DOI] [PubMed] [Google Scholar]
- 33.Feng, Q., J. V. Moran, H. H. Kazazian, Jr., and J. D. Boeke. 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87:905-916. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Perez, J. L., A. J. Doucet, A. Bucheton, J. V. Moran, and N. Gilbert. 2007. Distinct mechanisms for trans-mediated mobilization of cellular RNAs by the LINE-1 reverse transcriptase. Genome Res. 17:602-611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garcia-Perez, J. L., M. C. Marchetto, A. R. Muotri, N. G. Coufal, F. H. Gage, K. S. O'Shea, and J. V. Moran. 2007. LINE-1 retrotransposition in human embryonic stem cells. Hum. Mol. Genet. 16:1569-1577. [DOI] [PubMed] [Google Scholar]
- 36.Garcia-Perez, J. L., M. Morell, J. O. Scheys, D. A. Kulpa, S. Morell, C. C. Carter, G. D. Hammer, K. L. Collins, K. S. O'Shea, P. Menendez, and J. V. Moran. 2010. Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466:769-773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goodier, J. L., and H. H. Kazazian. 2008. Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell 135:23-35. [DOI] [PubMed] [Google Scholar]
- 38.Hohjoh, H., and M. F. Singer. 1996. Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA. EMBO J. 15:630-639. [PMC free article] [PubMed] [Google Scholar]
- 39.Hohjoh, H., and M. F. Singer. 1997. Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon. J. Mol. Biol. 271:7-12. [DOI] [PubMed] [Google Scholar]
- 40.Hohjoh, H., and M. F. Singer. 1997. Sequence-specific single-strand RNA binding protein encoded by the human LINE-1 retrotransposon. EMBO J. 16:6034-6043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holmes, S. E., M. F. Singer, and G. D. Swergold. 1992. Studies on p40, the leucine zipper motif-containing protein encoded by the first open reading frame of an active human LINE-1 transposable element. J. Biol. Chem. 267:19765-19768. [PubMed] [Google Scholar]
- 42.Huang, C. R., A. M. Scheneider, Y. Lu, T. Niranjan, P. Shen, M. A. Robinson, J. P. Steranka, D. Valle, C. I. Civin, T. Wang, S. J. Wheelan, H. Ji, J. D. Boeke, and K. H. Burns. 2010. Mobile interspersed repeats are major structural variants in the human genome. Cell 141:1171-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hulme, A. E., H. P. Bogerd, B. R. Cullen, and J. V. Moran. 2007. Selective inhibition of Alu retrotransposition by APOBEC3G. Gene 390:199-205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hulme, A. E., D. A. Kulpa, J. L. Garcia-Perez, and J. V. Moran. 2006. The impact of LINE-1 retrotransposition on the human genome, p. 35-56. In J. Lupski and P. Stankiewicz (ed.), Genomic disorders: the genomic basis of disease. Humana Press, Totowa, NJ.
- 45.Iskow, R. C., M. T. McCabe, R. E. Mills, S. Torene, W. S. Pittard, A. F. Neuwald, E. G. Van Meir, P. M. Vertino, and S. E. Devine. 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141:1253-1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jurka, J. 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc. Natl. Acad. Sci. U. S. A. 94:1872-1877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kano, H., I. Godoy, C. Courtney, M. R. Vetter, G. L. Gerton, E. M. Ostertag, and H. H. Kazazian. 2009. L1 retrotransposition occurs mainly in embryogenesis and creates somatic mosaicism. Genes Dev. 23:1303-1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Katayama, S., Y. Tomaru, T. Kasukawa, K. Waki, M. Nakanishi, M. Nakamura, H. Nishida, C. C. Yap, M. Suzuki, J. Kawai, H. Suzuki, P. Carninci, Y. Hayashizaki, C. Wells, M. Frith, T. Ravasi, K. C. Pang, J. Hallinan, J. Mattick, D. A. Hume, L. Lipovich, S. Batalov, P. G. Engstrom, Y. Mizuno, M. A. Faghihi, A. Sandelin, A. M. Chalk, S. Mottagui-Tabar, Z. Liang, B. Lenhard, and C. Wahlestedt. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564-1566. [DOI] [PubMed] [Google Scholar]
- 49.Kazazian, H. H., Jr. 1999. An estimated frequency of endogenous insertional mutations in humans. Nat. Genet. 22:130. [DOI] [PubMed] [Google Scholar]
- 50.Kazazian, H. H., Jr. 2004. Mobile elements: drivers of genome evolution. Science 303:1626-1632. [DOI] [PubMed] [Google Scholar]
- 51.Kent, W. J. 2002. BLAT—the BLAST-like alignment tool. Genome Res. 12:656-664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kulpa, D. A., and J. V. Moran. 2005. Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition. Hum. Mol. Genet. 14:3237-3248. [DOI] [PubMed] [Google Scholar]
- 53.Lander, E. S., L. M. Linton, B. Birren, C. Nusbaum, M. C. Zody, J. Baldwin, K. Devon, K. Dewar, M. Doyle, W. FitzHugh, R. Funke, D. Gage, K. Harris, A. Heaford, J. Howland, L. Kann, J. Lehoczky, R. LeVine, P. McEwan, K. McKernan, J. Meldrim, J. P. Mesirov, C. Miranda, W. Morris, J. Naylor, C. Raymond, M. Rosetti, R. Santos, A. Sheridan, C. Sougnez, N. Stange-Thomann, N. Stojanovic, A. Subramanian, D. Wyman, J. Rogers, J. Sulston, R. Ainscough, S. Beck, D. Bentley, J. Burton, C. Clee, N. Carter, A. Coulson, R. Deadman, P. Deloukas, A. Dunham, I. Dunham, R. Durbin, L. French, D. Grafham, S. Gregory, T. Hubbard, S. Humphray, A. Hunt, M. Jones, C. Lloyd, A. McMurray, L. Matthews, S. Mercer, S. Milne, J. C. Mullikin, A. Mungall, R. Plumb, M. Ross, R. Shownkeen, S. Sims, R. H. Waterston, R. K. Wilson, L. W. Hillier, J. D. McPherson, M. A. Marra, E. R. Mardis, L. A. Fulton, A. T. Chinwalla, K. H. Pepin, W. R. Gish, S. L. Chissoe, M. C. Wendl, K. D. Delehaunty, T. L. Miner, A. Delehaunty, J. B. Kramer, L. L. Cook, R. S. Fulton, D. L. Johnson, P. J. Minx, S. W. Clifton, T. Hawkins, E. Branscomb, P. Predki, P. Richardson, S. Wenning, T. Slezak, N. Doggett, J. F. Cheng, A. Olsen, S. Lucas, C. Elkin, E. Uberbacher, M. Frazier, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409:860-921. [DOI] [PubMed] [Google Scholar]
- 54.Levy, S., G. Sutton, P. C. Ng, L. Feuk, A. L. Halpern, B. P. Walenz, N. Axelrod, J. Huang, E. F. Kirkness, G. Denisov, Y. Lin, J. R. MacDonald, A. W. Pang, M. Shago, T. B. Stockwell, A. Tsiamouri, V. Bafna, V. Bansal, S. A. Kravitz, D. A. Busam, K. Y. Beeson, T. C. McIntosh, K. A. Remington, J. F. Abril, J. Gill, J. Borman, Y. H. Rogers, M. E. Frazier, S. W. Scherer, R. L. Strausberg, and J. C. Venter. 2007. The diploid genome sequence of an individual human. PLoS Biol. 5:e254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Luan, D. D., M. H. Korman, J. L. Jakubczak, and T. H. Eickbush. 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72:595-605. [DOI] [PubMed] [Google Scholar]
- 56.Lutz, S. M., B. J. Vincent, H. H. Kazazian, Jr., M. A. Batzer, and J. V. Moran. 2003. Allelic heterogeneity in LINE-1 retrotransposition activity. Am. J. Hum. Genet. 73:1431-1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Malone, C. D., and G. J. Hannon. 2009. Small RNAs as guardians of the genome. Cell 136:656-668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin, S. L. 1991. Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells. Mol. Cell. Biol. 11:4804-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martin, S. L., and F. D. Bushman. 2001. Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon. Mol. Cell. Biol. 21:467-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Martin, S. L., M. Cruceanu, D. Branciforte, P. Wai-Lun Li, S. C. Kwok, R. S. Hodges, and M. C. Williams. 2005. LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein. J. Mol. Biol. 348:549-561. [DOI] [PubMed] [Google Scholar]
- 61.Mathias, S. L., A. F. Scott, H. H. Kazazian, Jr., J. D. Boeke, and A. Gabriel. 1991. Reverse transcriptase encoded by a human transposable element. Science 254:1808-1810. [DOI] [PubMed] [Google Scholar]
- 62.Mills, R. E., E. A. Bennett, R. C. Iskow, and S. E. Devine. 2007. Which transposable elements are active in the human genome? Trends Genet. 23:183-191. [DOI] [PubMed] [Google Scholar]
- 63.Montes, R., G. Ligero, L. Sanchez, P. Catalina, T. de la Cueva, A. Nieto, G. J. Melen, R. Rubio, J. Garcia-Castro, C. Bueno, and P. Menendez. 2009. Feeder-free maintenance of hESCs in mesenchymal stem cell-conditioned media: distinct requirements for TGF-beta and IGF-II. Cell Res. 19:698-709. [DOI] [PubMed] [Google Scholar]
- 64.Moran, J. V., and N. Gilbert. 2002. Mammalian LINE-1 retrotransposons and related elements, p. 836-869. In N. Craig, R. Craggie, M. Gellert, and A. Lambowitz (ed.), Mobile DNA II. ASM Press, Washington, DC.
- 65.Moran, J. V., S. E. Holmes, T. P. Naas, R. J. DeBerardinis, J. D. Boeke, and H. H. Kazazian, Jr. 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87:917-927. [DOI] [PubMed] [Google Scholar]
- 66.Muotri, A. R., V. T. Chu, M. C. Marchetto, W. Deng, J. V. Moran, and F. H. Gage. 2005. Somatic mosaicism in neuronal precursor cells mediated by L1 retrotransposition. Nature 435:903-910. [DOI] [PubMed] [Google Scholar]
- 67.Ostertag, E. M., R. J. DeBerardinis, J. L. Goodier, Y. Zhang, N. Yang, G. L. Gerton, and H. H. Kazazian, Jr. 2002. A mouse model of human L1 retrotransposition. Nat. Genet. 32:655-660. [DOI] [PubMed] [Google Scholar]
- 68.Prak, E. T., A. W. Dodson, E. A. Farkash, and H. H. Kazazian, Jr. 2003. Tracking an embryonic L1 retrotransposition event. Proc. Natl. Acad. Sci. U. S. A. 100:1832-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rangwala, S. H., L. Zhang, and H. H. Kazazian. 2009. Many LINE1 elements contribute to the transcriptome of human somatic cells. Genome Biol. 10:R100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roy-Engel, A. M., A. H. Salem, O. O. Oyeniran, L. Deininger, D. J. Hedges, G. E. Kilroy, M. A. Batzer, and P. L. Deininger. 2002. Active Alu element “A-tails”: size does matter. Genome Res. 12:1333-1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sassaman, D. M., B. A. Dombroski, J. V. Moran, M. L. Kimberland, T. P. Naas, R. J. DeBerardinis, A. Gabriel, G. D. Swergold, and H. H. Kazazian, Jr. 1997. Many human L1 elements are capable of retrotransposition. Nat. Genet. 16:37-43. [DOI] [PubMed] [Google Scholar]
- 72.Scott, A. F., B. J. Schmeckpeper, M. Abdelrazik, C. T. Comey, B. O'Hara, J. P. Rossiter, T. Cooley, P. Heath, K. D. Smith, and L. Margolet. 1987. Origin of the human L1 elements: proposed progenitor genes deduced from a consensus DNA sequence. Genomics 1:113-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Seleme, M. C., M. R. Vetter, R. Cordaux, L. Bastone, M. A. Batzer, and H. H. Kazazian. 2006. Extensive individual variation in L1 retrotransposition capability contributes to human genetic diversity. Proc. Natl. Acad. Sci. U. S. A. 103:6611-6616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Skowronski, J., T. G. Fanning, and M. F. Singer. 1988. Unit-length line-1 transcripts in human teratocarcinoma cells. Mol. Cell. Biol. 8:1385-1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Soifer, H. S., A. Zaragoza, M. Peyvan, M. A. Behlke, and J. J. Rossi. 2005. A potential role for RNA interference in controlling the activity of the human LINE-1 retrotransposon. Nucleic Acids Res. 33:846-856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Speek, M. 2001. Antisense promoter of human L1 retrotransposon drives transcription of adjacent cellular genes. Mol. Cell. Biol. 21:1973-1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Stetson, D. B., J. S. Ko, T. Heidmann, and R. Medzhitov. 2008. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell 134:587-598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swergold, G. D. 1990. Identification, characterization, and cell specificity of a human LINE-1 promoter. Mol. Cell. Biol. 10:6718-6729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thomson, J. A., J. Itskovitz-Eldor, S. S. Shapiro, M. A. Waknitz, J. J. Swiergiel, V. S. Marshall, and J. M. Jones. 1998. Embryonic stem cell lines derived from human blastocysts. Science 282:1145-1147. [DOI] [PubMed] [Google Scholar]
- 80.van den Hurk, J. A., I. C. Meij, M. C. Seleme, H. Kano, K. Nikopoulos, L. H. Hoefsloot, E. A. Sistermans, I. J. de Wijs, A. Mukhopadhyay, A. S. Plomp, P. T. de Jong, H. H. Kazazian, and F. P. Cremers. 2007. L1 retrotransposition can occur early in human embryonic development. Hum. Mol. Genet. 16:1587-1592. [DOI] [PubMed] [Google Scholar]
- 81.Venter, J. C., M. D. Adams, E. W. Myers, P. W. Li, R. J. Mural, G. G. Sutton, H. O. Smith, M. Yandell, C. A. Evans, R. A. Holt, J. D. Gocayne, P. Amanatides, R. M. Ballew, D. H. Huson, J. R. Wortman, Q. Zhang, C. D. Kodira, X. H. Zheng, L. Chen, M. Skupski, G. Subramanian, P. D. Thomas, J. Zhang, G. L. Gabor Miklos, C. Nelson, S. Broder, A. G. Clark, J. Nadeau, V. A. McKusick, N. Zinder, A. J. Levine, R. J. Roberts, M. Simon, C. Slayman, M. Hunkapiller, R. Bolanos, A. Delcher, I. Dew, D. Fasulo, M. Flanigan, L. Florea, A. Halpern, S. Hannenhalli, S. Kravitz, S. Levy, C. Mobarry, K. Reinert, K. Remington, J. Abu-Threideh, E. Beasley, K. Biddick, V. Bonazzi, R. Brandon, M. Cargill, I. Chandramouliswaran, R. Charlab, K. Chaturvedi, Z. Deng, V. Di Francesco, P. Dunn, K. Eilbeck, C. Evangelista, A. E. Gabrielian, W. Gan, W. Ge, F. Gong, Z. Gu, P. Guan, T. J. Heiman, M. E. Higgins, R. R. Ji, Z. Ke, K. A. Ketchum, Z. Lai, Y. Lei, Z. Li, J. Li, Y. Liang, X. Lin, F. Lu, G. V. Merkulov, N. Milshina, H. M. Moore, A. K. Naik, V. A. Narayan, B. Neelam, D. Nusskern, D. B. Rusch, S. Salzberg, W. Shao, B. Shue, J. Sun, Z. Wang, A. Wang, X. Wang, J. Wang, M. Wei, R. Wides, C. Xiao, C. Yan, et al. 2001. The sequence of the human genome. Science 291:1304-1351. [DOI] [PubMed] [Google Scholar]
- 82.Wallace, M. R., L. B. Andersen, A. M. Saulino, P. E. Gregory, T. W. Glover, and F. S. Collins. 1991. A de novo Alu insertion results in neurofibromatosis type 1. Nature 353:864-866. [DOI] [PubMed] [Google Scholar]
- 83.Wei, W., T. A. Morrish, R. S. Alisch, and J. V. Moran. 2000. A transient assay reveals that cultured human cells can accommodate multiple LINE-1 retrotransposition events. Anal. Biochem. 284:435-438. [DOI] [PubMed] [Google Scholar]
- 84.Witherspoon, D. J., E. E. Marchani, W. S. Watkins, C. T. Ostler, S. P. Wooding, B. A. Anders, J. D. Fowlkes, S. Boissinot, A. V. Furano, D. A. Ray, A. R. Rogers, M. A. Batzer, and L. B. Jorde. 2006. Human population genetic structure and diversity inferred from polymorphic L1(LINE-1) and Alu insertions. Hum. Hered. 62:30-46. [DOI] [PubMed] [Google Scholar]
- 85.Yang, N., and H. H. Kazazian, Jr. 2006. L1 retrotransposition is suppressed by endogenously encoded small interfering RNAs in human cultured cells. Nat. Struct. Mol. Biol. 13:763-771. [DOI] [PubMed] [Google Scholar]
- 86.Zeuthen, J., J. O. Norgaard, P. Avner, M. Fellous, J. Wartiovaara, A. Vaheri, A. Rosen, and B. C. Giovanella. 1980. Characterization of a human ovarian teratocarcinoma-derived cell line. Int. J. Cancer 25:19-32. [DOI] [PubMed] [Google Scholar]