Abstract
Box C/D ribonucleoprotein particles guide the 2′-O-ribose methylation of target nucleotides in both archaeal and eukaryotic RNAs. These complexes contain two functional centers, assembled around the C/D and C′/D′ motifs in the box C/D RNA. The C/D and C′/D′ RNPs of the archaeal snoRNA-like RNP (sRNP) are spatially and functionally coupled. Here, we show that similar coupling also occurs in eukaryotic box C/D snoRNPs. The C/D RNP guided 2′-O-methylation when the C′/D′ motif was either mutated or ablated. In contrast, the C′/D′ RNP was inactive as an independent complex. Additional experiments demonstrated that the internal C′/D′ RNP is spatially coupled to the terminal box C/D complex. Pulldown experiments also indicated that all four core proteins are independently recruited to the box C/D and C′/D′ motifs. Therefore, the spatial-functional coupling of box C/D and C′/D′ RNPs is an evolutionarily conserved feature of both archaeal and eukaryotic box C/D RNP complexes.
Eukaryotic rRNA nucleotides are extensively modified in the nucleolus as pre-rRNA transcripts are cleaved and bound with ribosomal proteins to assemble the large and small ribosome subunits (6, 22). The box C/D and box H/ACA small nucleolar RNAs (snoRNAs) guide nucleotide methylation (5, 18, 41) and pseudouridylation (12, 29), respectively. These snoRNAs possess guide sequences that base pair with complementary sequences in the rRNAs to determine the specific nucleotide for modification. Highly conserved box C/D and H/ACA sRNAs are also found in Archaea (7, 13, 32). The strong conservation of archaeal and eukaryotic guide RNAs and their core proteins indicates an ancient evolutionary origin for these nucleotide modification complexes prior to the divergence of Eukarya and Archaea some two billion years ago (38).
Box C/D RNAs are characterized by terminal box C and D sequences located at the 5′ and 3′ ends of the RNA. C′ and D′ sequences are located internally. The C and D boxes fold to form kink-turn (K-turn) motifs (34). K-turns are characterized by an asymmetric bulge flanked by two stems with tandem, sheared G:A base pairs hydrogen bonding across the asymmetric bulge (20, 44). In Archaea, the C′ and D′ sequences characteristically form K-loops, which are variant K-turns where one of the flanking stems is replaced by a loop (30). In Archaea, the box C/D and C′/D′ motifs are separated by a highly conserved distance (39). In contrast, the distances separating the eukaryotic box C/D and C′/D′ sequences are reported to be much larger and not well conserved in length (10).
The eukaryotic box C/D snoRNAs are associated with four proteins: the 15.5kD protein (Snu13p in Saccharomyces cerevisiae), nucleolar proteins Nop56 and Nop58, and the methyltransferase fibrillarin (Nop1 in yeast) (10, 34). Three highly conserved proteins, ribosomal protein L7Ae, Nop56/58 (Nop5), and fibrillarin bind the archaeal box C/D sRNAs to assemble the snoRNA-like RNP (sRNP) complex. In vitro assembly of the archaeal box C/D sRNP has revealed that a single copy of L7Ae, Nop56/58, and fibrillarin binds both the box C/D and the C′/D′ motifs, thus establishing a “symmetric” complex (31, 40). In apparent contrast, the eukaryotic Nop58 and Nop56 core proteins are asymmetrically cross-linked in vivo to the snoRNA box C/D and C′/D′ motifs, respectively (4). In vitro binding has also indicated that the Snu13p (15.5kD) core protein recognizes only the K-turn of the terminal box C/D motif (36, 43). Thus, only fibrillarin (Nop1) is clearly associated with both eukaryotic box C/D and C′/D′ RNPs.
An understanding of box C/D RNP-catalyzed nucleotide modification is emerging from investigations of the in vitro assembled archaeal sRNP complex. Efficient target RNA methylation directed by the terminal box C/D or internal C′/D′ RNPs requires symmetric assembly of the juxtaposed archaeal RNPs (40). Nucleotide methylation guided by each RNP requires maintenance of the highly conserved inter-RNP distance of 12 nucleotides (39). These studies have clearly demonstrated that the archaeal box C/D and C′/D′ RNPs are spatially and functionally coupled. The spatial-functional coupling of the eukaryotic box C/D and C′/D′ RNPs is not evident. The larger snoRNA size, greater inter-RNP distances, and apparent asymmetric distribution of core proteins suggest that the box C/D and C′/D′ RNPs may well function independently. However, we now demonstrate that the terminal box C/D and internal C′/D′ RNPs are indeed spatially and functionally coupled. Thus, the spatial-functional coupling of the box C/D and C′/D′ RNPs is an evolutionarily conserved feature of both archaeal and eukaryotic box C/D nucleotide modification complexes.
MATERIALS AND METHODS
Yeast strains and plasmid vectors.
Yeast strain YS625 (MATα ade2 trpl ura3 leu2 his3) (24) was provided by M. J. Fournier and used as the host strain for expressing engineered human U24 (hU24) and its mutants. Yeast strains YAF2 (MATα trpΔ his3Δ ura3-52 lys2-801 ade2-101 URA3::U24 NOP56::TAP::TRP1) (9) and YAF3 (MATα trpΔ his3Δ ura3-52 lys2-801 ade2-101 URA3::U24 NOP58::TAP::TRP1) (16) were provided by I. Bozzoni. Yeast strains LMA439 (MATa ade2 arg4 leu2-3,112 trp1-289 ura3-52 NOP1-CBP-TEV-protA::TRP1) (37) and YGALSNU13 (MATa his3Δ1 leu2Δ0 met15Δ0 LYS2 ura3Δ0 kanMX6:PGAL1-SNU13) (8) were provided by A. Jacquier and R. T. O'Keefe, respectively. Plasmids pBL150 (25) and pRS415 were provided by M. J. Fournier. Plasmid pRS413-snu13 (8) was provided by R. T. O'Keefe, and plasmid pFL45/snR5Nco (19) was provided by T. Kiss.
Recombinant plasmid construction.
Sequences of the number-designated DNA oligonucleotide primers used in this work are listed in the supplemental material. Plasmid pBL150/Sph was constructed for expression of human U24 snoRNA and its mutant constructs. DNA fragments corresponding to upstream and downstream regions of the snR38 coding sequence in the TEF4 gene were PCR amplified from plasmid pBL150 using oligonucleotide pairs 1 and 2 (1/2) and 3/4. These two DNA fragments were then ligated and PCR amplified using oligonucleotides 1 and 4. This PCR product lacked the intron-encoded snR38 coding sequence, which was replaced with a SphI restriction site to be used for insertion of the human U24 sequence and its various mutants. Plasmids pRS413-snR5Nco and pRS415-snR5Nco were constructed for expression of the chimeric human U24-yeast snR5 snoRNA and its mutants. The snR5 gene and its regulatory sequences were PCR amplified from plasmid pFL45/snR5Nco using the oligonucleotide primer pair 5/6 or 6/7. PCR fragments were digested with EcoRI and SacI or with SalI and SacI and inserted into plasmid pRS413-Snu13 or pRS415 digested with the same endonucleases. Plasmid pRS415-HASnu13 was constructed for making the hemagglutinin (HA)-tagged Snu13 yeast strain. The Snu13 gene with three HA tags (3×HA) at its N terminus along with flanking regulatory sequences was PCR amplified from plasmid pRS413-Snu13 using the primer pairs 8/9 and 10/11. These DNA fragments were ligated and PCR amplified using primer pair 8/10. This fragment was digested with SacI and SalI and inserted into plasmid pRS415 digested with the same enzymes. Engineered human U24 snoRNA (hU24) was PCR-amplified from plasmid pBS+hsc70Δs+hU24 using primer pair 12/13. The PCR product was digested with SphI and inserted into plasmid PBL150/Sph(PBL150/Sph/hU24). U24 snoRNA mutants were constructed by PCR amplification of this plasmid using appropriate oligonucleotide primer pairs followed by insertion into the SphI cloning site. The U24 mutants were constructed using the following primer pairs: mutC′, primer pair 14/15; mutD′, 16/17; FmutD′, 16/18; dmut, 14/19; delC′/D′, 14/20; 2L31, 21/22; 2L36, 23/24; 2L41, 25/26; 2L41V, 27/28; 2L41VD′, 14/28; 2L41VD, 16/27; 2S13, 29/30; hU24V, 31/32; 2S12, 33/34; 2S10, 35/36; SD′13, 16/29; LD, 14/26; SD′LD, 26/29; SD13, 14/30; LD′, 16/25; LD′SD, 25/30; SD′12, 32/33; SD′10, 32/35; SD12, 31/34; and SD10, 31/36. U24 snoRNA half-mers possessing box C/D motifs were constructed by direct insertion of oligonucleotide primer pairs into plasmid PBL150/Sph. The U24 C/D half-mers were constructed using the following primer pairs:: C/D, primer pair 37/38; C/D1, 39/40; and C/D2, 41/42. The U24 coding sequence in chimeric human U24-yeast snR5 snoRNA was PCR amplified from plasmid PBL150/Sph/hU24 using primer pair 43/44 and then inserted into the NcoI site of plasmid pRS413-snR5Nco or pRS415-snR5Nco. Mutant mutD was constructed by PCR amplification using primer pair 43/45. Chimeric box C/D and C′/D′ half-mers fused with downstream snR5 were constructed using the same method for making the U24 C/D half-mers. The chimeric C/D and C′/D′ half-mers were constructed using the following primer pairs: C/D, primer pair 46/47; C/D1, 48/49; C/D2, 50/51; C′/D′, 52/53; C′/D′1, 54/55; and C′/D′2, 56/57.
Yeast transformation and snoRNA expression.
Yeast transformations were performed using the lithium acetate (LiAc) method as previously described (15). LiAc-treated cells (50 μl) were transformed with approximately 1 μg of plasmid DNA mixed with 50 μg of salmon sperm DNA suspended in 200 μl of 40% polyethylene glycol (PEG) 3350. Cells were heat shocked at 42°C for 10 to 15 min and then spread on plates. After 2 to 3 days of growth, transformants were selected and transferred to selective medium for expression of the engineered snoRNAs. Yeast strain YHA-Snu13 was constructed by transformation of yeast strain YGALSNU13 with plasmid pRS415-HASnu13. PBL150-derived plasmids were transformed into yeast strain YS625. Transformants were screened on synthetic dextrose plates lacking Trp [SD(−Trp)], and selected clones were then grown in SG(−Trp) liquid medium to express the hU24 RNA variants. pRS413-snu13-derived plasmids were transformed into yeast strains YHASnu13, YAF2, and YAF3. pRS415-derived plasmids were transformed into yeast strain LMA439. Transformants were screened on SD(−His) or SD(−Leu) plates and then grown in the corresponding SD liquid medium to express the chimeric hU24-snR5 RNA variants. All yeast cultures were grown at 30°C with continuous shaking.
RNA analyses.
Yeast cells were grown in expression medium to mid-log phase (optical density at 600 nm [OD600] of 0.8 to 1.0), and total RNA was prepared using hot-phenol extraction (21). snoRNA expression was examined by Northern blot analysis as previously described (35). 5′-32P-labeled oligonucleotides hU24R and yU14R were used as probes for the human U24 and yeast U14 snoRNAs, respectively. Radiolabeled oligonucleotide ysnR5R was used to assess expression of both chimeric hU24-snR5 RNA transcripts and yeast snR5 snoRNA. Northern blots were revealed using a Molecular Dynamics PhosphorImager or with X-ray film. 2′-O-methylation of targeted yeast 18S rRNA nucleotides was assessed by primer extension analysis as previously described (26) with some modifications. Approximately 0.8 μg of total RNA was hybridized with approximately 0.15 pmol of 5′-labeled R576 primer at 55°C for 5 min. Primer extension was carried out at high (final concentration of 1 mM) and reduced (final concentration of 4 μM) deoxynucleoside triphosphate (dNTP) concentrations after adding 1 unit of avian myeloblastosis virus (AMV) reverse transcriptase (Promega). The cDNA extension products were separated on 8% polyacrylamide sequencing gels accompanied with RNA sequencing lanes of yeast 18S rRNA using the same oligonucleotide primer. Extension products were revealed using phosphorimaging or autoradiography. Quantification of A534 and U547 nucleotide methylation was accomplished by phosphorimaging and comparison with the wild-type (WT) U24 snoRNA.
Immunoprecipitation of box C/D snoRNPs.
Yeast cultures (60 ml) were grown in selective liquid medium and collected at mid-log phase, and cell extracts were prepared as described previously (42). Collected cells were washed with ice-cold water and then suspended in 600 μl of lysis buffer (150 mM KCl, 20 mM Tris-HCl [pH 7.5] 5 mM MgCl2, 2 mM phenylmethylsulfonyl fluoride [PMSF], 0.5% Triton X-100 [vol/vol], 10 mM vanadyl ribonucleoside complex [BRL]). An equal volume of acid-washed glass beads was added, and the cells were disrupted with rigorous vortexing for 1 h (vortexing for 5 min and then cooling on ice for 4 min [12 times] at 4°C). Cell lysates were cleared by centrifugation at 18,000 × g for 30 min at 4°C. Immunoprecipitation was performed as previously described (27). Antibody-agarose beads (rabbit IgG-agarose [ProtA tag] and mouse monoclonal anti-HA-agarose [HA tag]; Sigma) were washed (three times) in IPP buffer (150 or 500 mM NaCl, 10 mM Tris-HCl [pH 8.0], 0.1% NP-40, 0.1% NaN3). Approximately 25 μl of suspended antibody-agarose beads was incubated with 600 μl of cell extract and continuously rocked at 4°C for 3 to 18 h. Agarose beads were then washed (six times) with IPP buffer at 4°C. Washed beads were digested with proteinase K for 1 h, and the eluted RNAs were purified by phenol extraction and ethanol precipitation.
RESULTS
Construction of engineered human U24 box C/D snoRNA to guide the 2′-O-methylation of novel target nucleotides in yeast 18S rRNA.
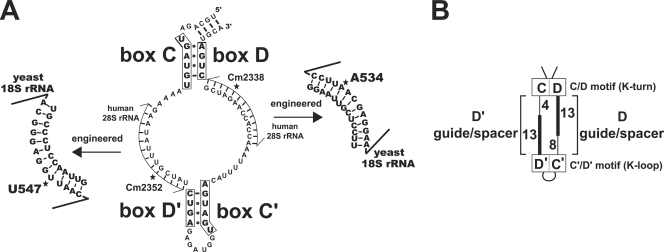
To test the potential spatial and functional coupling of the eukaryotic box C/D and C′/D′ RNPs for snoRNA-guided nucleotide modification, a human U24 box C/D snoRNA was engineered to guide the 2′-O-methylation of novel target nucleotides in yeast 18S rRNA (Fig. 1). This human snoRNA has been previously used in yeast as a model box C/D snoRNA for both structural and functional studies. Human U24 possesses well conserved C′ and D′ boxes and similarly sized C-D′ and C′-D guide/spacer regions. In addition, hybridization probes assessing human U24 transcription and stability resulted in no cross-hybridization with endogenous yeast snoRNAs.
FIG. 1.
Sequence and structure of engineered human U24 snoRNA (hU24). (A) Terminal C and D and internal C′ and D′ box sequences are in bold letters (enclosed) and folded to form the box C/D and C′/D′ motifs, respectively. Human D and D′ guide regions were replaced with the indicated sequences complementary to yeast 18S rRNA to target methylation of novel nucleotides A534 and U547, respectively, designated by asterisks. (B) Schematic presentation of engineered human U24 snoRNA. Box C, D, C′, and D′ sequences are indicated as boxes with the respective box C/D (K-turn) and C′/D′ (K-loop) motifs designated. Intermotif spacer regions are shown with the guide sequences complementary to 18S rRNA in bold lines and associated spacer regions in thin lines. The nucleotide lengths of the guide and associated spacer regions are indicated.
Modification of 18S rRNA nucleotides A534 and U547 was assessed by primer extension analysis using a common downstream primer. Nucleotide 2′-O-methylation was indicated by increasing primer extension termination one nucleotide before the modified target nucleotide when the concentration of nucleotides in the primer extension reaction was reduced (28). Selected target nucleotides were positioned not only near each other but also in regions of the 18S rRNA free of premature termination sites due to RNA folded structure. The occurrence of a naturally methylated site between these two target nucleotides (A541) helped serve as an internal control for assessing methylation levels in the different RNA preparations.
Box C/D RNP-guided nucleotide methylation is independent of the C′/D′ RNP.
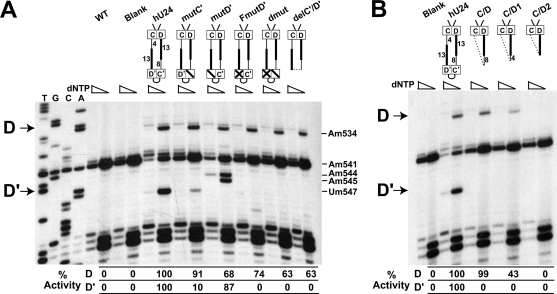
Initial experiments examined the functional interdependency of the terminal box C/D and internal C′/D′ RNPs (Fig. 2A). Primer extension analysis demonstrated strong methylation of the D guide (A534) and D′ guide (U547) target nucleotides for the wild-type U24 (hU24). Mutations were then made in the box C′ and D′ sequences, specifically altering the GA nucleotides critical for the tandem-sheared G-A base pairs that hydrogen bond across the asymmetric bulge of the K-loop motif (mutC′ and mutD′) (see Fig. S1 in the supplemental material for details of mutant constructs). These mutations severely reduced methylation of U547, whereas A534 was still strongly modified. Interestingly, mutation of the D′ box (mutD′) resulted in methylation of alternative target nucleotides, suggesting the creation of novel box D′ sequences (see Discussion). More severe mutation of the C′/D′ motif, which included alteration of 4 nucleotides of the loop region adjacent to the D′ box (FmutD′ and dmut), completely disrupted C′/D′-guided methylation (U547) while still permitting strong, albeit somewhat reduced, methylation of the box C/D-guided A534 nucleotide. Finally, complete loss of the C′/D′ motif and loop region (delC′/D′) also resulted in the complete loss of U547 modification but continued methylation of the A534 nucleotide. Importantly, all expressed U24 snoRNAs in this and subsequent analyses were stable in yeast as determined by Northern blot analyses (see Fig. S3 in the supplemental material). From these results, it was concluded that the box C/D RNP can function independently of the C′/D′ RNP in guiding the 2′-O-methylation of its target nucleotide.
FIG. 2.
The terminal box C/D RNP can guide nucleotide methylation independent of the internal C′/D′ RNP. (A) Box C/D RNP-guided nucleotide modification is independent of the C′/D′ RNP. Schematic presentations of hU24 and its mutant constructs are shown at the top of the appropriate gel lanes, with decreasing nucleotide concentrations in the primer extension assay indicated. Primer extension sequencing lanes are at the left, and the indicated primer extension control assays include cells transformed with no plasmid (WT) and empty plasmid (Blank). C′ and D′ mutations included alteration of the K-loop GA residues alone (indicated by a slash in the respective box) and in conjunction with loop mutations (indicated by an X in the respective box). The deleted C′/D′ motif is indicated by a dashed line. Termination sites due to box C/D RNP- and C′/D′ RNP-guided nucleotide modification are indicated on the left as D and D′, respectively. Altered methylation levels with respect to wild-type U24 are indicated below the respective gel lanes. A strong termination site seen between the hU24-targeted nucleotides is the naturally methylated A541 nucleotide. (B) The box C/D RNP requires a minimal size for box C/D RNP-guided nucleotide modification activity. Schematic presentations of full-length and shortened hU24 snoRNAs are shown at the top with targeted nucleotide methylation indicated on the left by D (A534) and D′ (U547). Altered methylation levels with respect to wild-type U24 are indicated below the respective gel lanes.
Subsequent experiments determined the minimal box C/D snoRNP capable of snoRNA-guided nucleotide modification. The U24 C/D RNA half-mer (delC′/D′) was further reduced in size by progressively removing spacer regions while maintaining the terminal box C/D motif (K-turn) essential for box C/D snoRNP assembly and snoRNA stability. The smallest of these constructs consisted of only the terminal box C/D motif and the 13-nucleotide D guide region (C/D2). Northern blot analysis revealed that the U24 half-mer snoRNAs were present at lower levels, indicating that their smaller size affected RNA stability, likely as a consequence of less efficiently assembled snoRNPs (see Fig. S3 in the supplemental material). Nevertheless, two of the three shortened U24 half-mers (C/D and C/D1) were still capable of guiding nucleotide methylation, albeit at reduced levels (Fig. 2B). Only the smallest snoRNA of 33 nucleotides (C/D2) was unable to guide modification of target nucleotide A534. We suspect that loss of activity was due to induced structural constraints that disrupted box C/D RNP-guided modification.
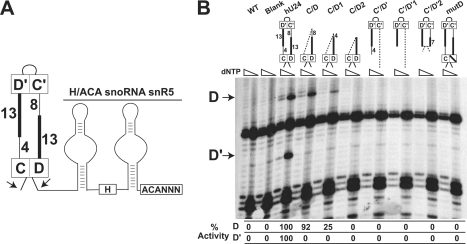
The terminal box C/D RNP is required for C′/D′ RNP-guided nucleotide methylation.
The ability of the internal C′/D′ RNP to independently guide nucleotide modification was examined next. Importantly, a terminal box C/D RNP is essential for snoRNP stability in vivo (3, 17). However, truncated box C/D snoRNAs can be stabilized by linkage to a downstream H/ACA snoRNA (Fig. 3A). Thus, the chimeric 5′-box C/D-box H/ACA-3′ construct previously established in the Kiss laboratory (19) was utilized to assess independent C′/D′ RNP methylation function. A chimeric U24-snR5 snoRNA possessing both terminal box C/D and internal C′/D′ RNPs (hU24) was able to guide methylation of both A534 (D guide) and U547 (D′ guide) target nucleotides (Fig. 3B). Chimeric snoRNAs possessing only the terminal box C/D RNP were also capable of methylating the D target nucleotide (C/D). The diminution and final loss of methylation activity as the terminal box C/D motif was reduced in size corresponded to the methylation capabilities of the box C/D snoRNA half-mers not linked to a downstream H/ACA snoRNA (Fig. 2B, mutants C/D1 and C/D2). Thus, the methylation capabilities of U24 snoRNA and the box C/D RNA half-mers were not altered when linked to the downstream snR5 H/ACA snoRNA. In contrast, chimeric RNAs possessing only the C′/D′ motif were inactive in modification of the target nucleotide (mutants C′/D′, C′/D′1, and C′/D′2). Similarly, an internal C′/D′ RNP linked to a terminal box C/D motif inactivated by mutation of the D box sequence (mutD) was also unable to methylate its target nucleotide. Supporting these results is our observation that utilization of an additional independent C′/D′ RNP which targets 18S rRNA U1758 does not 2′-O-methylate this target nucleotide (R. van Nues and N. Watkins, unpublished data). Collectively, these results demonstrated that the C′/D′ RNP cannot function independently and requires a coupled and functional box C/D RNP for nucleotide methylation.
FIG. 3.
Internal box C/D RNP-guided nucleotide methylation requires the terminal box C/D RNP. (A) Schematic presentation of the chimeric hU24 (box C/D)-snR5 (box H/ACA) snoRNA. The bipartite structure of snR5 with conserved H and ACA boxes (enclosed) is shown. Box C/D snoRNA inserts are positioned between two NcoI restriction sites indicated with arrows. (B) Schematic presentations of the box C/D portion of the chimeric hU24-snR5 snoRNAs are shown above the respective primer extension gel lanes. WT and blank lanes correspond to yeast cells transformed with no plasmid and plasmid lacking the hU24-snR5 chimeric insert, respectively. Primer extension stop sites for the box C/D (D) and C′/D′ (D′) RNP-guided nucleotide methylations are indicated on the left. Altered methylation levels with respect to wild-type U24 are indicated below the respective gel lanes.
The internal C′/D′ RNP is spatially coupled to the terminal box C/D RNP for nucleotide modification function.
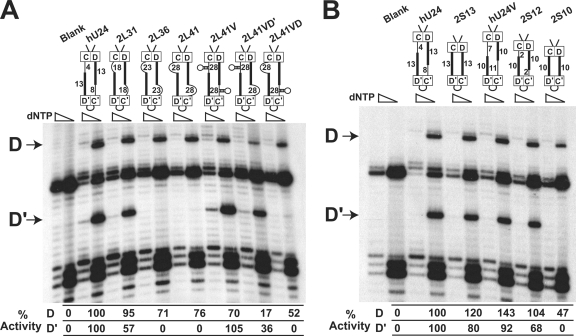
The potential spatial coupling of the internal C′/D′ RNP to the terminal box C/D RNP was examined next. The box C/D and C′/D′ RNPs of human U24 are separated by 21 (D guide/spacer) and 17 (D′ guide/spacer) nucleotides, with each region possessing a guide region of 13 nucleotides (Fig. 1B). Various U24 snoRNA constructs were engineered to possess longer spacer regions in a symmetric fashion (both D and D′ guide/spacer regions of equal length) while maintaining the 13-nucleotide length of D and D′ guide regions. These U24 snoRNAs were expressed in yeast, and the methylation of the respective D and D′ target nucleotides was assessed (Fig. 4A). Increasing each guide/spacer region to 31 nucleotides (Fig. 4A, 2L31) still permitted methylation of both target nucleotides. However, further increasing the guide/spacer regions to 36 or 41 nucleotides (2L36 and 2L41) disrupted C′/D′ RNP-guided nucleotide modification. Importantly, the terminal box C/D RNP continued to function independently and methylate its target A534 nucleotide. This is consistent with analysis demonstrating that the box C/D RNP is a minimal nucleotide modification complex that can function independently of the internal C′/D′ RNP (Fig. 2 and 3B).
FIG. 4.
The terminal box C/D and internal C′/D′ RNPs are spatially coupled for snoRNA-guided nucleotide methylation. Schematic presentations of engineered U24 snoRNA constructs with lengthened or shortened D and D′ spacer regions are shown above the respective primer extension gel lanes. Blank gel lanes correspond to transformed yeast cells with empty plasmid. Primer extension stop sites for the box C/D (D) and C′/D′ (D′) RNP-guided nucleotide methylations are indicated at the side. Altered methylation levels with respect to wild-type U24 are indicated below the respective gel lanes. (A) Box C/D and C′/D′ RNPs exhibit a maximal guide/spacer region length for spatially coupled C′/D′ RNP-guided nucleotide methylation. (B) Box C/D and C′/D′ RNPs exhibit a minimal guide/spacer region length for spatially coupled C′/D′ RNP-guided nucleotide methylation.
To demonstrate that the internal C′/D′ RNP was indeed spatially linked to the terminal box C/D RNP for methylation function, the U24 snoRNA possessing 41-nucleotide guide/spacer regions (2L41) was engineered to create two hairpin-loop regions such that the box C/D and C′/D′ RNPs should be drawn back together (see Fig. S2 in the supplemental material). These two hairpins should effectively reduce spacing between the box C/D and C′/D′ RNPs and thereby restore C′/D′ RNP-guided methylation of target nucleotide U537. Interestingly, insertion of this hairpin-loop structure in one guide/spacer region or the other resulted in differential effects upon nucleotide methylation. Insertion of the hairpin within the D′ guide/spacer region restored methylation of the C′/D′ RNP-guided target nucleotide U547 (2L41VD), whereas insertion of the hairpin within the D guide/spacer region (2L41VD′) did not. This indicated that it is particularly important for the D′ guide sequence to be within a guide/spacer region exhibiting the proper spatial context for methylation function (see below).
Similar experiments were then carried out to shorten the guide/spacer regions between the box C/D and C′/D′ RNPs (Fig. 4B). Removal of the D (8 nucleotides) and D′ (4 nucleotides) spacer regions leaving only the 13-nucleotide guide region resulted in methylation of both target nucleotides. Further reduction of the guide/spacer regions required reduction of the guide sequences themselves. Therefore, a control construct with D and D′ guide regions possessing only 10 nucleotides was first constructed in U24 snoRNA possessing native guide/spacer regions of 21 and 17 nucleotides (hU24V). This control U24 was still capable of methylating the respective target nucleotides. A U24 snoRNA with the guide/spacer region reduced to 12 nucleotides was capable of directing methylation from both box C/D and C′/D′ RNPs (2S12). However, when both guide/spacer regions were reduced to only the 10-nucleotide guide sequences (2S10), C′/D′ RNP-guided methylation of U547 was lost. Thus, a minimum as well as maximum distance was important for the spatial and functional coupling of the internal C′/D′ RNP with the terminal box C/D complex.
Asymmetric alteration of the D and D′ guide/spacer regions demonstrates the independent function of the box C/D RNP.
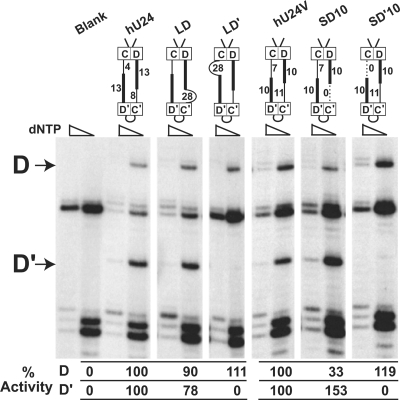
Asymmetric changes in the D and D′ guide/spacer regions were made, and the effect upon box C/D and C′/D′ RNP-guided nucleotide modification was assessed. Lengthening or shortening either the inter-RNP D or D′ spacer regions had differential effects upon guided nucleotide modification (Fig. 5). Increasing (LD) or decreasing (SD10) the D guide/spacer region resulted in both box C/D- or C′/D′-guided nucleotide modification although shortening this guide/spacer region altered the methylation levels of both target nucleotides. In contrast, increasing (LD′) or decreasing (SD′10) the D′ guide/spacer region disrupted C′/D′ RNP-guided nucleotide modification while leaving unaffected the target nucleotide modification guided by the box C/D RNP. These results indicated that the spatial positioning of the C′/D′ RNP with respect to its D′ guide region was particularly sensitive to linkage with the terminal box C/D RNP. Box C/D RNP activity was maintained when the length of either guide/spacer region was altered, reinforcing the notion that the terminal box C/D RNP functions independently of its internal partner RNP.
FIG. 5.
Asymmetric alteration of the D and D′ guide/spacer regions demonstrates the independent function of the box C/D RNP. Schematic presentations of engineered U24 snoRNA constructs with asymmetrically altered D or D′ spacer regions are shown above the respective primer extension gel lanes. The blank gel lane corresponds to transformed yeast cells with empty plasmid. Primer extension stop sites for the box C/D (D) and C′/D′ (D′) RNP-guided nucleotide methylations are indicated on the left. Altered methylation levels with respect to wild-type U24 are indicated below the respective gel lanes.
All four core proteins are recruited to the terminal box C/D and internal C′/D′ RNPs.
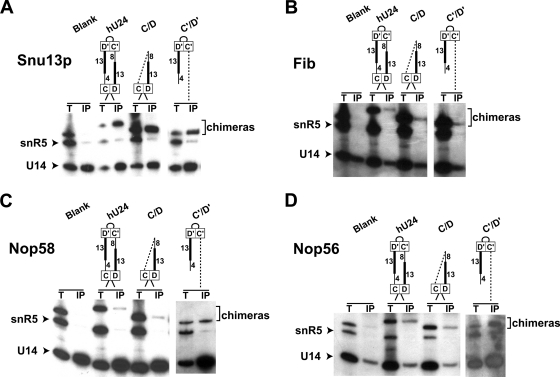
The core protein composition of the terminal box C/D and internal C′/D′ RNPs was examined in pulldown experiments using tagged Snu13p, Nop56, Nop58, and fibrillarin (Nop1) core proteins. These experiments utilized chimeric U24 box C/D-snR5 H/ACA snoRNAs, where individual box C/D and internal C′/D′ motifs were linked to the downstream snR5 H/ACA snoRNA. Coisolated snoRNAs associated with each tagged core protein were revealed in Northern blot analysis (Fig. 6). U14 box C/D snoRNA (positive control) was coisolated with Snu13p core protein, whereas endogenous snR5 (negative control) was not (Fig. 6A). Full-length U24 as well as the box C/D and C′/D′ motifs were pulled down with the tagged Snu13p, indicating the presence of this core protein in both box C/D and C′/D′ RNPs. The presence of Snu13p in the internal C′/D′ RNP was notable as in vitro binding experiments have shown that this core protein binds the K-turn (box C/D motif) but not the K-loop motif (C′/D′ motif) (see Discussion). Affinity isolation of tagged fibrillarin also revealed its association with both the box C/D and C′/D′ RNPs (Fig. 6B) as expected since both RNPs guide nucleotide methylation. Interestingly, the box C/D and C′/D′ motifs were pulled down with both Nop56 and Nop58 core proteins (Fig. 6C and D). We suspect that isolation of both Nop56 and Nop58 core proteins with both RNPs is a consequence of assembly of one protein in each of the box C/D and C′/D′ RNPs and coisolation of the second through protein-protein interactions via the coiled-coiled domain (see Discussion).
FIG. 6.
Box C/D and C′/D′ RNPs are symmetric in core protein composition. Chimeric U24 box C/D-snR5 box H/ACA snoRNAs were coexpressed with epitope-tagged Snu13p, Nop56, Nop58, or fibrillarin core proteins. Yeast extracts were prepared, and individual tagged core proteins were affinity purified from the extract. Coprecipitated RNAs were isolated, resolved on polyacrylamide gels, and revealed by Northern blot analysis. Hybridization probes included U14 snoRNA (box C/D snoRNA positive control) and snR5 (endogenous box H/ACA negative control). The snR5 probe also recognized the chimeric U24-snR5 snoRNAs coprecipitating with the individual core proteins. The box C/D snoRNA region of the U24-snR5 chimeric construct is shown above the respective gel lanes. T and IP indicate total RNA in the yeast extract and affinity-isolated snoRNAs, respectively. Blank lanes represent yeast not expressing a U24-snR5 chimeric snoRNA. Migration positions of U14, snR5, and chimeric U24-snR5 snoRNAs are indicated at the side. (A) snoRNAs coprecipitated with 3×HA-tagged affinity-isolated Snu13p core protein. (B) snoRNAs coprecipitated with TAP-tagged and affinity-isolated fibrillarin core protein. (C) snoRNAs coprecipitated with TAP-tagged and affinity-isolated Nop58 core protein. (D) snoRNAs coprecipitated with TAP-tagged and affinity-isolated Nop56 core protein.
DISCUSSION
Previous investigations have revealed the highly conserved inter-RNP spacing and functional interdependence of the box C/D and C′/D′ subcomplexes of the archaeal box C/D sRNP (39, 40). We have now demonstrated that the eukaryotic terminal box C/D and internal C′/D′ RNPs are also spatially and functionally constrained in the eukaryotic box C/D snoRNP. Like the archaeal complex, the terminal eukaryotic box C/D RNP can function independently of the C′/D′ complex. The internal eukaryotic C′/D′ RNP, also like its archaeal counterpart, requires a coupled box C/D RNP for guided 2′-O-methylation of its target nucleotide. This interdependency of methylation function implies inter-RNP protein-protein interactions. In summary, spatial-functional coupling of the box C/D and C′/D′ RNPs is an evolutionarily conserved feature of both archaeal and eukaryotic box C/D nucleotide modification complexes.
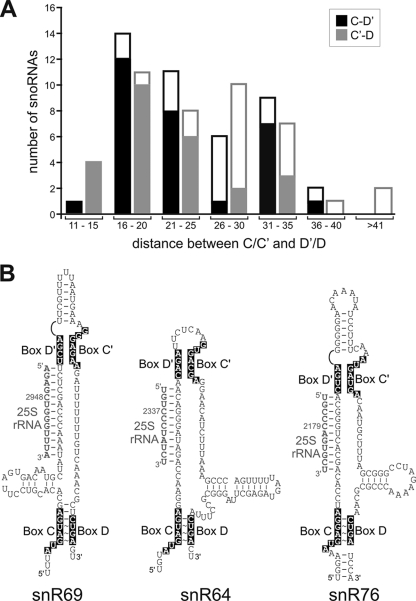
Internal C′/D′ RNP-guided nucleotide methylation function clearly requires a juxtaposed terminal box C/D RNP. Relative spatial positioning is important although not nearly as restricted as in the archaeal sRNP complex, where alteration of its highly conserved 12-nucleotide inter-RNP spacing by as few as 2 nucleotides disrupts C′/D′ RNP methylation activity (39). Analysis of the human U24 snoRNA revealed a range of inter-RNP spacing between 12 and 36 nucleotides necessary for coupled C′/D′ RNP methylation activity. This is consistent with the inter-RNP distancing observed for most yeast box C/D snoRNAs (Fig. 7A). The snoRNAs outside this range are likely to utilize secondary or tertiary structure(s) within the guide/spacer region to bring together their box C/D and C′/D′ RNP complexes for coupled function. This certainly appears to be the case for yeast snoRNAs such as snR69, snR64, and snR76, where evolutionarily conserved hairpins aid in folding each snoRNA to establish the requisite inter-RNP distance (Fig. 7B). Strikingly, as inter-RNP distancing increases in the yeast snoRNAs, so does the occurrence of potential hairpin structures within this region (Fig. 7A). Again, utilization of these hairpins to reduce inter-RNP distances to the acceptable range for inter-RNP interactions would promote snoRNA-guided nucleotide modification from both box C/D and C′/D′ RNPs. Alternatively, snoRNAs that do not guide nucleotide methylation from both RNP complexes may not require juxtaposed and coupled complexes. These RNAs would include those that function in pre-rRNA folding and cleavage. Finally, previous work had suggested that C′/D′ RNP function is independent of the terminal box C/D complex (19). We suspect that positioning of the novel target nucleotide close to a native methylation site complicated interpretation of these modification results. It was for this reason that we ultimately placed our D and D′ target nucleotides in yeast 18S rRNA well removed from native methylated nucleotides and internal rRNA folded structures.
FIG. 7.
Yeast box C/D and C′/D′ RNPs exhibit a defined range of inter-RNP spatial positioning assisted by spacer region folding. (A) Box C/D and C′/D′ inter-RNP spacing of yeast box C/D snoRNAs. Distances between the C-D′ boxes (black bars) and the C′-D boxes (gray bars) were determined and plotted versus the number of yeast box C/D snoRNAs in the indicated nucleotide spacing range. Regions of each bar represented in white are that fraction of snoRNAs which exhibit potential hairpin structures in their C-D′ or C′-D spacer regions. (B) Yeast box C/D snoRNAs utilizing spacer region folding to facilitate coupling of the box C/D and C′/D′ RNPs. Box C/D and C′/D′ sequences are blocked in black with the D′ guide regions base paired to their complementary sequences in 25S rRNA. Evolutionarily conserved hairpin structures which shorten the inter-RNP distances are indicated.
It is assumed that the observed requisite spatial positioning of the box C/D and C′/D′ RNPs facilitates required inter-RNP protein-protein interactions. The coiled-coil domain of Nop56 and Nop58 is the most likely candidate for this interaction(s). Precedence for this comes from the archaeal sRNP complex. Biophysical studies have demonstrated that the coiled-coil domain of archaeal Nop56/58 protein is responsible for this core protein's observed dimerization (1) and recent cryo-electron microscopy (cryo-EM) has confirmed this interaction (2). This domain of Nop56/58 is required for sRNP-guided nucleotide modification (46). Interestingly, results from our pulldown experiments utilizing epitope-tagged versions of all four core proteins are consistent with Nop56-Nop58 interaction via the coiled-coil domain. The observation that Nop56 and Nop58 are associated with both box C/D and C′/D′ RNPs suggests that dimerization of the coiled-coil domain of both proteins explains our observations, with a single protein of this homologous pair contained in each of the box C/D and C′/D′ RNPs (see below). Dimerization via the coiled-coil domain is indeed strong as pulldown experiments under elevated salt conditions (0.5 M) yielded the same results (data not shown).
Recently, Ye and coworkers presented a model where the guide/spacer regions span the two RNP complexes, suggesting critical interplay of their respective RNP-guided nucleotide modification functions (45). It is therefore also possible that lengthening the guide/spacer regions beyond the acceptable range may displace the catalytic site of the fibrillarin methylase from the target nucleotide, resulting in loss of target nucleotide methylation. Comparison of this crystal structure with the cryo-EM structure of the complete archaeal complex (2) raises some interesting questions. How are the conformational changes important for box C/D and C′/D′ methylation function proposed by the X-ray crystallographic model accommodated in the dimeric complex observed in the cryo-EM structure? Presently complicating this assessment is the inability to directly observe the sRNA in the cryo-EM structure and the partial nature of the sRNA in the crystal structure. An additional question is whether a dimeric complex is possible for the eukaryotic box C/D snoRNP. The permitted variation of inter-RNP spacing in the eukaryotic complex should make it difficult to assemble a dimeric snoRNP where individual monomeric snoRNPs can differ in overall length.
The archaeal box C/D sRNP is a symmetric complex in that the three box C/D RNP core proteins L7Ae, Nop56/58, and fibrillarin are found in both the terminal box C/D and internal C′/D′ RNPs (31, 33, 40). In contrast, the eukaryotic snoRNP complex has been described as asymmetric, with the Snu13p core protein found only in the terminal box C/D RNP. This proposed model is based upon the observations that the 15.5kD protein is unable to bind the C′/D′ motif (K-loop) in vitro (40, 43, 44) and is not cross-linked to the C′/D′ motif in vivo (36). Our results now demonstrate that Snu13p is indeed recruited to the C′/D′ RNP, despite its inability to recognize the K-loop motif. This would suggest that Snu13p presence in this complex likely occurs via protein-protein interactions. Precedence for this comes from the archaeal sRNP complex, where a mutated L7Ae core protein which is unable to recognize the K-loop motif is nevertheless incorporated into the C′/D′ RNP (11). This suggests that the eukaryotic Snu13p (15.5kD) core protein could also be incorporated into the internal C′/D′ RNP via protein-protein contacts.
Like archaeal L7Ae, the archaeal Nop56/58 core protein is also symmetrically distributed in the box C/D and C′/D′ RNPs. In eukaryotes, gene duplication has resulted in two homologous Nop56 and Nop58 core proteins. In vivo cross-linking indicated a differential distribution, with Nop58 and Nop56 binding the terminal box C/D and internal C′/D′ motifs, respectively (4). This is consistent with genetic analysis demonstrating the essentiality of Nop58 but not Nop56 for snoRNP stability (14, 23), presumably via Nop58 as a constituent of the terminal box C/D RNP preventing exonucleolytic turnover of the snoRNA. Our pulldown analyses revealing that both Nop56 and Nop58 are present in both the assembled box C/D and C′/D′ RNPs are likely due to dimerization of Nop56 (C′/D′ RNP) and Nop58 (box C/D RNP) via their coiled-coil domains. Coupled with the observation that Snu13p is found in both the box C/D and C′/D′ RNPs, this raises the possibility that, like the archaeal complex, the eukaryotic box C/D snoRNP is also symmetric in core protein composition.
Internal C′ and D′ box sequences, in contrast to terminal box C and D sequences, are often poorly conserved and sometimes difficult to identify by simple sequence inspection (19). This flexibility in sequence can result in the use of cryptic D′ box sequences when a native D′ is disrupted or ablated. The U24 mutD′ construct modified novel nucleotides corresponding to a cryptic D′ sequence (GAGA for Am545 and AGAG for Am544) (Fig. 2A). Previous work has even indicated the modification of successive nucleotides from a single D′ box as a consequence of D′ box “slide” (18). The independence of box C/D RNP-guided nucleotide modification may suggest that this terminal RNP assembles as the core complex upon which the internal C′/D′ RNP is constructed. While our pulldown experiments reveal that the C′/D′ RNP assembles with all core proteins in the absence of a box C/D RNP complex, this internal RNP nevertheless requires a juxtaposed box C/D RNP for methyltransferase function. An interesting observation is the “sidedness” of the guide-spacer length for methyltransferase function where C′/D′ RNP-guided methylation is particularly sensitive to the D′ guide-spacer distance. This again suggests critical core protein-protein interactions between the two RNPs spanning this region. The recent modeling of a terminal box C/D RNP crystal structure suggested dramatic changes in a dual guide complex directing alternating methyltransferase reactions from the terminal and internal RNPs (45). A crystal structure of the completely assembled snoRNP will ultimately reveal critical inter-RNP protein-protein interactions and explain the observed sidedness of guide/spacer region juxtapositioning. Finally, we imagine that lack of box C′ and D′ sequence conservation and the apparent flexibility in C′/D′ RNP assembly have contributed to the evolution of the snoRNP complex. The ability to utilize less conserved sequences for RNP assembly could lead to expanded snoRNA size and the resulting potential for snoRNP functional diversity, features which are found in the eukaryotic complex.
Supplementary Material
Acknowledgments
We thank Skip Fournier, Irene Bozzoni, Alain Jacquier, Raymond T. O'Keefe, and Tamas Kiss for generously providing yeast strains and/or plasmids.
This work was supported by NSF grant MCB 0543741 (E.S.M.) and Wellcome grant WT089378MA (N.J.W.).
Footnotes
Published ahead of print on 1 November 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Aittaleb, M., R. Rashid, Q. Chen, J. Almer, C. Daniels, and H. Lee. 2003. Structure and function of archaeal box C/D sRNP core proteins. Nat. Struct. Biol. 10:256-263. [DOI] [PubMed] [Google Scholar]
- 2.Bleichert, F., K. T. Gagnon, B. A. Brown II, E. S. Maxwell, A. E. Leschziner, V. M. Unger, and S. J. Baserga. 2009. A dimeric structure for archaeal box C/D small ribonucleoproteins. Science 325:1384-1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Caffarelli, E., A. Fatica, S. Prislei, E. De Gregorio, P. Fragapane, and I. Bozzoni. 1996. Processing of the intron-encoded U16 and U18 snoRNAs: the conserved C and D boxes control both the processing reaction and the stability of the mature snoRNA. EMBO J. 15:1121-1131. [PMC free article] [PubMed] [Google Scholar]
- 4.Cahill, N. M., K. Friend, W. Speckman, Z.H. Li, R. M. Terns, M. P. Terns, and J. A. Steitz. 2002. Site-specific cross-linking analyses reveal an asymmetric protein distribution for a box C/D snoRNP. EMBO J. 21:3816-3828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cavaillé, J., M. Nicoloso, and J. P. Bachellerie. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383:732-735. [DOI] [PubMed] [Google Scholar]
- 6.Decatur, W. A., and M. J. Fournier. 2002. rRNA modifications and ribosome function. Trends Biochem. Sci. 27:344-351. [DOI] [PubMed] [Google Scholar]
- 7.Dennis, P., A. Omer, and T. Lowe. 2001. A guided tour: small RNA function in Archaea. Mol. Microbiol. 40:509-519. [DOI] [PubMed] [Google Scholar]
- 8.Dobbyn, H. C., and R. T. O'Keefe. 2004. Analysis of Snu13p mutations reveals differential interactions with the U4 snRNA and U3 snoRNA. RNA 10:308-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fatica, A., M. Morlando, and I. Bozzoni. 2000. Yeast snoRNA accumulation relies on a cleavage-dependent/polyadenylation-independent 3′-processing apparatus. EMBO J. 19:6218-6229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gagnon, K. T., G. Qu, and E. S. Maxwell. Multicomponent 2′-O-ribose methylation machines: evolving box C/D RNP structure and function, p. 436-439. In H. Grosjean (ed.), DNA and RNA modification enzymes: comparative structure, mechanism, functions, cellular interactions and evolution. Landes Press, Austin, TX.
- 11.Gagnon, K. T., X. Zhang, G. Qu, S. Biswas, J. Suryadi, B. A. Brown II, and E. S. Maxwell. 2010. Signature amino acids enable the archaeal L7Ae box C/D RNP core protein to recognize and bind the K-loop RNA motif. RNA 16:79-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ganot, P., M. L. Bortolin, and T. Kiss. 1997. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89:799-809. [DOI] [PubMed] [Google Scholar]
- 13.Gaspin, C., J. Cavaillé, G. Erauso, and J. P. Bachellerie. 2000. Archaeal homologs of eukaryotic methylation guide small nucleolar RNAs: lessons from the Pyrococcus genomes. J. Mol. Biol. 297:895-906. [DOI] [PubMed] [Google Scholar]
- 14.Gautier, T., T Bergès, D. Tollervey, and E. Hurt. 1997. Nucleolar KKE/D repeat proteins Nop56p and Nop58p interact with Nop1p and are required for ribosome biogenesis. Mol. Cell. Biol. 17:7088-7098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gietz, R. D., and R. A. Woods. 1994. High efficiency transformation of yeast with lithium acetate, p. 121-134. In J. R. Johnston (ed.), Molecular genetics of yeast: a practical approach. Oxford University Press, Oxford, United Kingdom.
- 16.Giorgi, C., A. Fatica, R. Nagel, and I. Bozzoni. 2001. Release of U18 snoRNA from its host intron requires interaction of Nop1p with the Rnt1p endonuclease. EMBO J. 20:6856-6865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang, G. M., A. Jarmolowski, J. C. Struck, and M. J. Fournier. 1992. Accumulation of U14 small nuclear RNA in Saccharomyces cerevisiae requires box C, box D, and a 5′, 3′ terminal stem. Mol. Cell. Biol. 12:4456-4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kiss-Laszlo, Z., Y Henry, J. P. Bachellerie, M. Caizergues-Ferrer, and T. Kiss. 1996. Site-specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85:1077-1088. [DOI] [PubMed] [Google Scholar]
- 19.Kiss-Laszlo, Z., Y. Henry, and T. Kiss. 1998. Sequence and structural elements of methylation guide snoRNAs essential for site-specific ribose methylation of pre-rRNA. EMBO J. 17:797-807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klein, D. J., T. M. Schmeing, P. B. Moore, and T. A. Steitz. 2001. The kink-turn: a new RNA secondary structure motif. EMBO J. 20:4214-4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kohrer, K., and H. Domdey. 1991. Preparation of high molecular weight RNA. Methods Enzymol. 194:398-405. [DOI] [PubMed] [Google Scholar]
- 22.Kos, M., and D. Tollervey. 2010. Yeast pre-rRNA processing and modification occur cotranscriptionally. Mol. Cell 37:809-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lafontaine, D. L., and D. Tollervey. 1999. Nop58p is a common component of the box C+D snoRNPs that is required for snoRNA stability. RNA 5:455-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang, W. Q., and M. J. Fournier. 1995. U14 base-pairs with 18S rRNA: a novel snoRNA interaction required for rRNA processing. Genes Dev. 9:2433-2443. [DOI] [PubMed] [Google Scholar]
- 25.Liu, B., and M. J. Fournier. 2004. Interference probing of rRNA with snoRNPs: A novel approach for functional mapping of RNA in vivo. RNA 10:1130-1141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lowe, T. M., and S. R. Eddy. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283:1168-1171. [DOI] [PubMed] [Google Scholar]
- 27.Lygerou, Z., P. Mitchell, E. Petfalski, B. Séraphin, and D. Tollervey. 1994. The POP1 gene encodes a protein component common to the RNase MRP and RNase P ribonucleoproteins. Genes Dev. 8:1423-1433. [DOI] [PubMed] [Google Scholar]
- 28.Maden, B. E. 2001. Mapping 2′-O-methyl groups in ribosomal RNA. Methods 25:374-382. [DOI] [PubMed] [Google Scholar]
- 29.Ni, J., A. L. Tien, and M. J. Fournier. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89:565-573. [DOI] [PubMed] [Google Scholar]
- 30.Nolivos, S., A. J. Carpousis, and B. Clouet-d'Orval. 2005. The K-loop, a general feature of the Pyrococcus C/D guide RNAs, is an RNA structural motif related to the K-turn. Nucleic Acids Res. 33:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Omer, A., S. Ziesche, H. Ebhardt, and P. Dennis. 2002. In vitro reconstitution and activity of a C/D box methylation guide ribonucleoprotein complex. Proc. Natl. Acad. Sci. U. S. A. 99:5289-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Omer, A. D., T. M. Lowe, A. G. Russell, H. Ebhardt, S. Eddy, and P. Dennis. 2000. Homologs of small nucleolar RNAs in Archaea. Science 288:517-522. [DOI] [PubMed] [Google Scholar]
- 33.Rashid, R., M. Aittaleb, Q. Chen, K. Spiegel, B. Demeler, and H. Li. 2003. Functional requirement for symmetric assembly of archaeal box C/D small ribonucleoprotein particles. J. Mol. Biol. 33:295-306. [DOI] [PubMed] [Google Scholar]
- 34.Reichow, S. L., T. Hamma, A. R. Ferré-D'Amaré, and G. Varani. 2007. The structure and function of small nucleolar ribonucleoproteins. Nucleic Acids Res. 35:1452-1464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., and D. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- 36.Szewczak, L. B., S. J. DeGregorio, S. A. Strobel, and J. A. Steitz. 2002. Exclusive interaction of the 15.5 kD protein with the terminal box C/D motif of a methylation guide snoRNP. Chem. Biol. 9:1095-1107. [DOI] [PubMed] [Google Scholar]
- 37.Torchet, C., G. Badis, F. Devaux, G. Costanzo, M. Werner, and A. Jacquier. 2005. The complete set of H/ACA snoRNAs that guide rRNA pseudouridylations in Saccharomyces cerevisiae. RNA 11:928-938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tran, E., J. Brown, and E. S. Maxwell. 2004. Evolutionary origins of the RNA-guided nucleotide-modification complexes: from the primitive translation apparatus? Trends Biochem. Sci. 29:343-350. [DOI] [PubMed] [Google Scholar]
- 39.Tran, E., X. Zhang, L. Lackey, and E. S. Maxwell. 2005. Conserved spacing between the box C/D and C′/D′ RNPs of the archaeal box C/D sRNP complex is required for efficient 2′-O-methylation of target RNAs. RNA 11:285-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tran, E., X. Zhang, and E. S. Maxwell. 2003. Efficient RNA 2′-O-methylation requires juxtaposed and symmetrically assembled archaeal box C/D and C′/D′ RNPs. EMBO J. 22:3930-3940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tycowski, K. T., C. M. Smith, M. D. Shu, and J. A. Steitz. 1996. A small nucleolar RNA requirement for site-specific ribose methylation of rRNA in Xenopus. Proc. Natl. Acad. Sci. U. S. A. 93:14480-14485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Venema, J., H. R. Vos, A. W. Faber, W. J. van Venrooij, and H. A. Raué. 2000. Yeast Rrp9p is an evolutionarily conserved U3 snoRNP protein essential for early pre-rRNA processing cleavages and requires box C for its association. RNA 6:1660-1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Watkins, N. J., A. Dickmanns, and R. Luhrmann. 2002. Conserved stem II of the box C/D motif is essential for nucleolar localization and is required, along with the 15.5K protein, for the hierarchical assembly of the box C/D snoRNP. Mol. Cell. Biol. 22:8342-8352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Watkins, N. J., V. Segault, B. Charpentier, S. Nottrott, P. Fabrizio, A. Bachi, M. Wilm, M. Rosbash, C. Branlant, and R. Luhrmann. 2000. A common core RNP structure shared between the small nucleolar box C/D RNPs and the spliceosomal U4 snRNP. Cell 103:457-466. [DOI] [PubMed] [Google Scholar]
- 45.Ye, K., R. Jia, J. Lin, M. Ju, J. Peng, A. Xu, and L. Zhang. 2009. Structural organization of box C/D RNA-guided RNA methyltransferase. Proc. Natl. Acad. Sci. U. S. A. 106:13808-13813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang, X., E. A. Champion, E. J. Tran, B. A. Brown II, S. J. Baserga, and E. S. Maxwell. 2006. The coiled-coil domain of the Nop56/58 core protein is dispensable for sRNP assembly but is critical for archaeal box C/D sRNP-guided nucleotide methylation. RNA 12:1092-1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.