Abstract
DNA transposition takes place within a higher-order complex known as the transpososome. Almost everything known about its assembly has been gleaned from bacterial transposons. Here we present a detailed analysis of transpososome assembly in the human Hsmar1 element. The transpososome is nominally symmetrical, consisting of two identical transposon ends and a dimer of transposase. However, after the transposase dimer has captured the first transposon end, an asymmetry is introduced, raising a barrier against recruitment of the second end. The barrier can be overcome by right-handed plectonemic intertwining of the transposon ends. This likely occurs mainly during transcription and episodes of nucleosome remodeling. Plectonemic intertwining favors only synapsis of closely linked transposon ends in the inverted-repeat configuration and therefore suppresses the promiscuous synapsis of distant transposon ends, which initiate McClintock's chromosomal breakage-fusion-bridge cycles in maize. We also show that synapsis of the transposon ends is a prerequisite for the first catalytic step. This provides constraints on the enzymatic mechanism of the double-strand breaks in mariner transposition, excluding the most prevalent of the current models.
Transposons are discrete segments of DNA that can move from one genomic location to another. These elements have profoundly affected the evolution of most, if not all, organisms by rearranging their genomes and modifying gene expression and function (16). Retrotransposons are the most numerous elements in mammals, in which their remnants constitute about 50% of the genome. DNA transposons, although generally less numerous, are almost ubiquitous and have colonized all branches of the tree of life.
The mariner family is probably the most successful group of DNA transposons if judged by the breadth and depth of its phylogenetic distribution. It is particularly widespread in multicellular animals, and some insects harbor very large numbers of members of this family. Mariner transposons are thought to require frequent horizontal transfer to new hosts (22). The prevailing model holds that a burst of transposition follows the successful infection of a new genome. This is subsequently dampened by the overproduction inhibition effect, a consequence of the increasing number of elements and the concomitant rise in transposase concentration. This phase is followed by a gradual decline in transposition as the pool of active transposase multimers is progressively poisoned by the accumulation of detrimental mutations that arise as the sequences of the different copies of the transposase genes begin to drift apart.
McClintock discovered transposons while investigating the causes of genetic instability in maize. She observed chromosomal breakage-fusion-bridge cycles caused by the aberrant transposition of the Activator (Ac) transposon. The aberrant transposition is caused by the promiscuous synapsis of transposon ends belonging to different copies of the element on the same chromosome or on different chromosomes (14). Promiscuous synapsis of transposon ends has been demonstrated for bacterial elements in vitro. Tn10 ends, for example, synapse almost equally well whether they are arranged as direct or inverted repeats or are located on different DNA molecules (6).
Such promiscuity is not an inherent property of all transposons or site-specific recombination systems. Many have topological filters and can distinguish the arrangement of their recombination sites (10). Examples include the phage Mu transposon, phage integrases, resolvases, and invertases. The selectivity in these systems is mediated by factors, such as enhancer sites and accessory proteins, which are lacking in structurally simple transposons such as Ac, mariner, and Tn10. Topological filters usually also rely on negative supercoiling in the DNA, which may drive assembly of a higher-order complex or provide directionality to the reaction.
In vitro transposition experiments are often performed using plasmid substrates purified from bacteria. Indeed, the effects of supercoiling on the rate of mariner transposition and target site selection have been previously noted (11, 18, 20). However, one might reasonably wonder about the biological significance of these observations. Bacteria and eukaryotes package supercoiling in different ways. Negatively supercoiled DNA in bacteria is mostly free to form right-handed plectosomes, as it does in vitro. In contrast, the DNA in eukaryotes is wrapped in a toroid around the nucleosome. Despite these general differences, both modes of packaging can be found in eukaryotes and bacteria (2, 27). In eukaryotes, several processes in particular produce high transient levels of free supercoiling, for example, DNA replication, transcription, and the displacement of nucleosomes associated with episodes of chromatin remodeling. Free supercoiling is probably therefore available in eukaryotes to any system able to use it as a mechanism of regulation.
Here we report that the synapsis of mariner transposon ends is accelerated by free negative supercoiling and inhibited by free positive supercoiling. Negative supercoiling can affect DNA transactions by a number of different mechanisms. Two of the most general mechanisms are that the relative concentrations of any two sites in a supercoiled domain increase by an order of magnitude as they are brought into juxtaposition in the plectosome and that their intertwining has a specific right-handed geometry. We show here that it is these two effects that stimulate mariner transposition by accelerating the rate at which the second transposon end is recruited into the developing transpososome. The effect constitutes a simple topological filter because it requires transposon ends in the inverted-repeat configuration and arises from the right-handed parallel interwrapping of the ends in the plectosome. We speculate that intertwining of the transposon ends disfavors synapsis of distant transposon ends and ends on different chromosomes. This would be advantageous to a transposon such as mariner, which is widespread in multicellular animals in which the genotoxic effects of breakage-fusion-bridge cycles have more serious consequences than in plants or microorganisms.
MATERIALS AND METHODS
Except as noted below, materials and reagents were from commercial sources. DNA-modifying enzymes were from New England Biolabs or Roche Applied Science and used according to the manufacturer's recommendations. All cloned PCR products were confirmed by nucleotide sequencing.
Plasmids.
Plasmid pRC880 is a pMAL-c2x-based expression vector for Hsmar1 transposase (8). Standard transposition reaction mixtures contained the inverted-repeat substrate pRC650. In pRC883, one transposon end is reversed to provide a direct-repeat substrate. The single-end substrate pRC919 has a single 30-bp Hsmar1 end inserted in the pBluescript polylinker. pRC704 carries a 2.3-kb Hsmar1 transposon with a 0.8-kb plasmid backbone. pRC1181 is based on pRC650 and contained phage lambda attP and attB sites in direct-repeat configuration, approximately on opposite sites of the map. pRC1105 is similar to pRC650 except that the sizes of the transposon and backbone segments were increased to 2.3 kb and 4.2 kb, respectively. This was necessary so that the two-dimensional (2D) gel analysis of the Hsmar1 reaction could be compared directly with similar gel analyses for Tn10, which were used as a yardstick. Open circular substrates were prepared by treatment with the Nb.BsrDI endonuclease, which nicks at a site(s) some distance from the transposon ends. Substrates with intermediate supercoiling densities were prepared by religation of nicked plasmids or by the treatment of supercoiled plasmids with topoisomerase I in the presence of ethidium bromide (25). Positively supercoiled substrate was generated by treating the substrates with reverse gyrase (28). After treatment, DNA was phenol-chloroform extracted, ethanol precipitated, and resuspended in Tris-EDTA (TE) buffer.
Protein expression, purification, and in vitro assays.
Purification of Hsmar1 transposase and the in vitro reaction conditions have been described previously (8). Briefly, the protein was expressed in Escherichia coli as a maltose binding protein fusion and purified by amylose resin and ion-exchange chromatography. Unless stated otherwise, a 50-μl reaction mixture contained 1 μg of the substrate plasmid and 0.1 μg of transposase in 20 mM Tris-HCl buffer, pH 8, 100 mM NaCl, 10% glycerol, 2 mM dithiothreitol (DTT), and 2.5 mM MgCl2. Reactions were stopped with EDTA and SDS treatment at 75°C for 30 min. DNA was recovered by ethanol precipitation, and 40% of each reaction mixture was loaded onto a Tris-borate-EDTA (TBE)-buffered 1.1% agarose gel. After electrophoresis, the gel was stained with ethidium bromide or SYBR green and photographed or recorded on a Fuji phosphorimager. Ethidium bromide staining provided a larger dynamic range than SYBR green. The conditions for two-dimensional gel electrophoresis were as previously described for Tn10 (6). Reverse gyrase, Tn10 transposase, lambda integrase, and integration host factor (IHF) were purified as described previously (3, 7, 15, 21, 28).
RESULTS
Transposon end synapsis precedes first-strand nicking.
There are about 200 defective copies of the Hsmar1 transposase in the human genome. The ancestral consensus sequence derived from these copies provides a highly active enzyme (23). Transposition of Hsmar1 follows the canonical pattern established for other cut-and-paste elements (8). The sequential cleavage of the DNA strands at each transposon end is followed by target site selection and integration (Fig. 1). In Tn5 and Tn10, cleavage is coupled to synapsis of the transposon ends by the trans architecture of the synaptic complex: the single active site of a transposase monomer bound to one transposon end cleaves both strands of DNA at the opposite transposon end via a hairpin intermediate (12). In contrast, mariner lacks a hairpin intermediate, and the mechanism remains unclear (13). The current model for mariner cleavage is that one of the transposon ends is nicked prior to synapsis (see references 13 and 19 and references therein). This is physically possible because the transposase is a dimer in solution prior to transposon end binding, thus making the trans subunit available to perform the first-strand nick. The model holds that second-strand nicking remains coupled to synapsis of the transposon ends by a conformational change or subunit exchange.
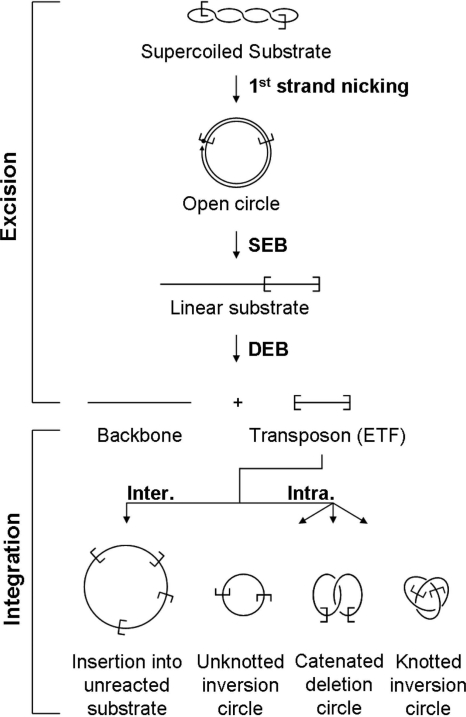
FIG. 1.
The Hsmar1 transposition reaction. A schematic representation of the different steps of a mariner transposition reaction using a supercoiled plasmid substrate. First-strand nicking at one transposon end generates an open circular product in which the 5′ end of the transposon is separated from the donor sequence. Second-strand nicking exposes the 3′ OH at the transposon end, yielding the single-end-break (SEB) product. A similar sequence of nicks at the other transposon end yields the double-end-break (DEB) products, which are the plasmid backbone plus the excised transposon fragment (ETF). Intermolecular (Inter.) insertions may target any DNA present in the reaction, such as an unreacted donor plasmid (as illustrated here). Intramolecular (Intra.) targets, within the transposon itself, generate a series of transposon circles, which may be knotted or catenated if supercoils have been trapped between the target site and the transposon end. For further details see references 6 and 9.
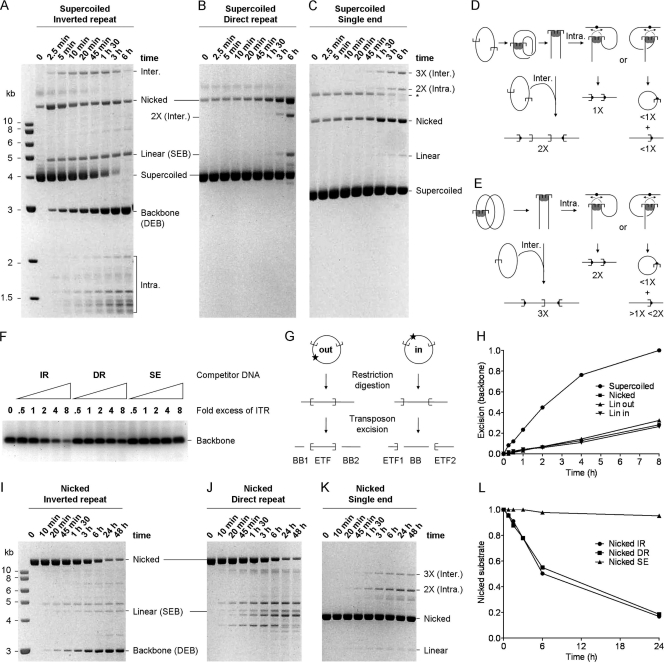
The unusually faithful and efficient in vitro system provided by Hsmar1 gave us the opportunity to test this model. We compared the reaction kinetics of the standard inverted-repeat substrate with those of plasmids carrying a direct repeat or a single transposon end (Fig. 2A, B, and C). A simple visual comparison of these reactions is difficult because the different arrangements of the transposon ends give rise to different spectrums of products (illustrated in Fig. 2D and E). The simplest comparison of the reactions is provided by the consumption of the supercoiled substrates, which shift position in the gel after the first transposon end is nicked. Since almost all of the substrate that achieves the first nick goes on to complete the reaction, the disappearance of the supercoiled substrate is almost as good a measure of transposon excision as the amount of plasmid backbone produced (Fig. 2A) (8).
FIG. 2.
The configuration of transposon ends influences the kinetics of cleavage. (A) The kinetics of transposition with the standard inverted-repeat substrate (pRC650). Excision of the transposon yields the plasmid backbone, which is an end product of the reaction. A photograph of an ethidium bromide-stained agarose gel is shown. (B and C) Cleavage with the direct-repeat (pRC883) (B) and single-end (pRC919) (C) substrates are difficult to quantify, because there is no product equivalent to the backbone in the inverted-repeat reactions. (The products of the reactions corresponding to panels B and C are shown in panels D and E, respectively.) The only unique products are from the integration reaction. The direct repeat yields an intermolecular product twice the size of the substrate (2X). The single-end substrate product yields a 2X intramolecular integration product and a 3X intermolecular integration product. The simplest estimate of cleavage is given by the rate at which the substrate disappears. * indicates the supercoiled dimer form of the substrate. A photograph of an ethidium bromide-stained agarose gel is shown. (D) With a direct-repeat substrate, cleavage yields two linear fragments that together equal the size of the substrate. Intermolecular integration into an unreacted substrate plasmid, which is the only target available, generates a linear product two times the size of the substrate (2X). Intramolecular events produce two alternative outcomes depending on orientation of the target site: either they generate a linear product the size of the substrate (1X) or they generate a pair of products, namely, an open-circle product plus a linear product that together equal the size of the plasmid. (E) With a single-ended substrate, cleavage at the transposon end yields a linear intermediate of the same size as the substrate (1X). Intermolecular integration into an unreacted substrate plasmid, which is the only target available in these reactions, generates a linear product three times the size of the substrate (3X). Intramolecular events produce two alternative outcomes depending on the orientation of the target site: either they generate a linear product twice the size of the substrate (2X) or they generate a pair of products, namely, an open-circle product (<1X substrate) plus a corresponding linear product between one and two times the size of the substrate (>1X <2X). (F) A competition assay was performed using a principal substrate (pRC704) yielding an 800-bp backbone fragment, which can be quantified easily because it runs in a clear area of the gel. Reaction mixtures contained 2 nM principal substrate and 10 nM transposase. They were titrated up to an 8-fold molar excess of the competitor transposon ends. This is equivalent to an 8-fold molar excess of the double-ended plasmids and a 16-fold molar excess of the single-ended plasmid. All of the reactions were adjusted to contain the same total amount of DNA by the addition of pBluescript, where required. The only difference between any of the reactions was therefore the concentration of the respective competitor transposon ends. Cleavage of the principal substrate was analyzed on an agarose gel stained with SYBR green. IR, inverted repeat; DR, direct repeat; SE, single end. (G) The inverted-repeat plasmid was linearized with XmnI and NruI to yield the LinOUT and LinIN substrates, respectively. The excision products for these substrates are illustrated. Transposon excision from the LinOUT substrate yields two backbone (BB) fragments. (H) The kinetics of the excision reactions with the indicated substrates were analyzed by ethidium bromide-stained agarose gel electrophoresis and quantified using a Fluorimager (Fujifilm). “SC” and “Nicked” refer to the supercoiled and open circular forms of the inverted-repeat substrate used in panel A. LinOUT and LinIN are illustrated in panel G. Reactions were normalized to the level of backbone produced by the SC substrate and plotted. (I, J, and K) The kinetics of transposition reactions using nicked forms of the inverted-repeat (I), direct-repeat (J), and single-end (K) substrates. These substrates were prepared by treatment with a nicking endonuclease (Nb.BsrDI). For each time point, 100 ng DNA was loaded on the gel. The reactivity of the substrates can be estimated by assessing the disappearance of the nicked donor. (L) The nicked substrates described for panels I, J, and K were quantified as described for panel H. Values were normalized to the signal at time zero for each substrate.
The supercoiled form of the inverted-repeat substrate was entirely consumed during the time course (Fig. 2A). The direct-repeat substrate was less efficient, and >50% of the supercoiled form remained unreacted after 6 h (Fig. 2B). The single-end substrate was the least reactive (Fig. 2C). The difference is particularly pronounced at 1.5 h, when reactions with supercoiled inverted-repeat substrate have almost reached completion. These results show that first-strand nicking cannot be independent of synapsis. If nicking were independent of synapsis, the direct-repeat and single-end substrates would be consumed as fast the standard inverted-repeat substrate. This is clearly not the case.
We have already shown that an event, most probably synapsis of the transposon ends, prior to the first nick is the rate-limiting step of the reaction (8). This means that the consumption of the substrate and the appearance of the plasmid backbone are both good proxies for the rate of synapsis. The higher reactivity of the inverted-repeat substrate therefore suggests that this configuration of transposon ends favors synapsis. This was unexpected, because the Tn10 transposon, which has a simple structure comparable to those of the mariner elements, works equally well with transposon ends in any topological arrangement.
We next performed a competition experiment to provide a more direct comparison of the three substrates (Fig. 2F). The principal substrate in this experiment included an inverted-repeat transposon, which had to compete for transposase against increasing amounts of the three substrates shown in Fig. 2A to C. Transposon excision from the principal substrate was measured by the release of an 800-bp backbone fragment, which migrated in a clear region of the gel, well separated from other products. The inverted-repeat substrate was the most effective competitor and acted almost stoichiometrically: an 8-fold molar excess of the competitor inhibited excision by about 8-fold (Fig. 2F). The direct-repeat substrate was less effective and provided about 50% inhibition at the highest concentration. The single-end plasmid had almost no effect on excision from the principal substrate, even when it was present at a 16-fold molar excess (equivalent to an 8-fold molar excess of transposon ends).
The lack of competition by the single-ended substrate was somewhat surprising. Even if this substrate did not compete for synapsis, one might have expected it to compete for transposase binding. The absence of competition therefore suggests that the interactions between transposase and the transposon ends are quite fluid and that they exist in a dynamic equilibrium until synapsis is achieved and the complex commits to the catalytic steps of the reaction.
Supercoiling favors inverted-repeat synapsis and accelerates the reaction.
Taken together, the results presented above suggest that synapsis is favored by the inverted-repeat configuration of the transposon ends. If true, this implies that the transposase protein has a mechanism to distinguish their configuration. One way this could be achieved is by a tracking mechanism in which the protein or complex finds the second binding site by a unidirectional search along the DNA. We tested this possibility by cutting the substrate either outside or inside the transposon to prevent tracking in either direction. These substrates are designated LinOUT and LinIN, respectively (Fig. 2G). We measured the kinetics of transposon excision with these substrates and included the supercoiled and nicked circular forms of the plasmid as controls (Fig. 2H). The results eliminate fixed-direction tracking mechanisms of synapsis, because there was no significant difference between the kinetics of the reactions with LinOUT, LinIN, and the nicked circular form of the substrate.
The supercoiled substrate reacted much faster than any of the others, suggesting that the preference for inverted-repeat synapsis may be peculiar to the supercoiled form of the substrate. If this is true, we would expect the inverted- and direct-repeat substrates to react at the same rate in the absence of supercoiling. This was the result obtained when we analyzed the reaction kinetics with nicked circular forms of the substrates. The rates of the reactions with the inverted and direct repeats were identical to one another and much faster than that with the single-end substrate (Fig. 2I, J, K, and L).
A concentration-dependent barrier to synapsis.
The reaction with the nicked single-end substrate being sluggish compared to reactions with the double-ended substrates was unexpected (Fig. 2I, J, K, and L). Nicking of the substrates releases the topological constraints, and one would therefore expect the single-end plasmid to react almost as fast as the double-ended plasmids. After nicking, the only difference between the single- and double-ended substrates was the relative concentrations of the transposon ends. The relative concentration of the transposon ends on the double-ended plasmids was about 100 nM, given by ring closure probability calculations (17). This is an order of magnitude greater than the 10 nM concentration of the single-end substrate in these reactions.
We have already shown that the rate-limiting step of the reaction is an event prior to the first nick (8). We assumed that this was synapsis of the ends because simple protein-DNA interactions, such as those between transposase and its binding site, take place very rapidly when the reactants are in the nM concentration range (see Discussion). We have been unable to address this question directly because the mariner paired-ends complex (PEC) does not survive in an electrophoretic mobility shift assay (EMSA) under noncatalytic conditions (1, 13, 19). We can, however, use an EMSA to rule out the possibility that the initial interaction between transposase and a single transposon end is very slow. To do this, we used an EMSA to analyze the kinetics of transposase binding to a transposon end carried on a linear DNA fragment. However, binding was too fast to measure and was complete within 30 s, which was the time required to mix and sample the reaction volume (not shown). This suggests that there is an asymmetry in the developing transpososome: interactions with the first transposon end are established quickly but raise a barrier against recruitment of the second end, which is slow.
Negative supercoiling, and not positive supercoiling, accelerates synapsis.
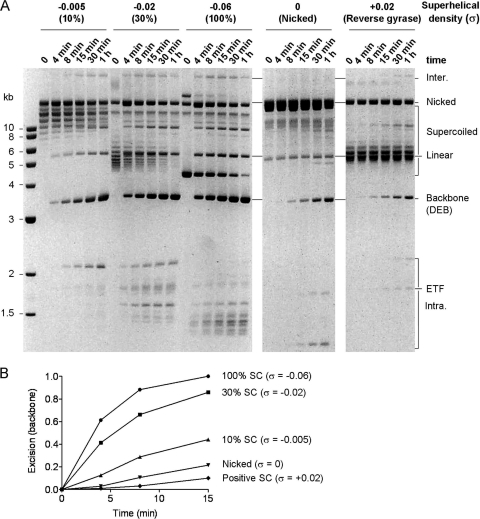
One way in which negative supercoiling could accelerate the rate of transposition is if synapsis is favored by the parallel juxtaposition of the transposon ends in the plectosome. To explore this issue, we prepared substrates with supercoiling densities (σ) ranging from values of −0.06 to +0.02 (Fig. 3A). In these experiments, we measured the production of plasmid backbone because it migrates at a fixed position in the gel and is easier to quantify at early time points than the consumption of the substrate. The results showed that the substrates with higher negative superhelicity reacted more quickly than their partially relaxed or nicked counterparts. The differences are most clearly evident at early time points (Fig. 3B).
FIG. 3.
Substrate supercoiling influences the kinetics of excision. (A) The kinetics of transposon excision were analyzed using the inverted-repeat substrate (pRC1181) with superhelical densities ranging from −0.06 to +0.02. A photograph of an ethidium bromide-stained agarose gel is shown. (B) The backbone product in panel A was quantified using a Fluorimager, normalized to the maximum level achieved with the substrate with a σ of −0.06, and plotted.
Of all the substrates, the positively supercoiled substrate was the least reactive (Fig. 3A and B). Furthermore, by examining the case at time zero, one can see that this substrate was contaminated with a significant amount of the nicked form of the plasmid (Fig. 3A). Since this would also contribute to the reaction, the low rate of backbone production observed is likely to be an overestimate. When reactions were performed with the single-end substrate, the kinetics with positive supercoiling and negative supercoiling were similar (not shown). This excludes the possibility that the overwinding of the helix in the positively supercoiled substrate is the inhibitory factor. If transposition is indeed stimulated by the juxtaposition of the ends when they meet in the plectosome, we can therefore conclude that it is the left-handed geometry provided by positive supercoiling that is unfavorable.
Synapsis in the plectosome.
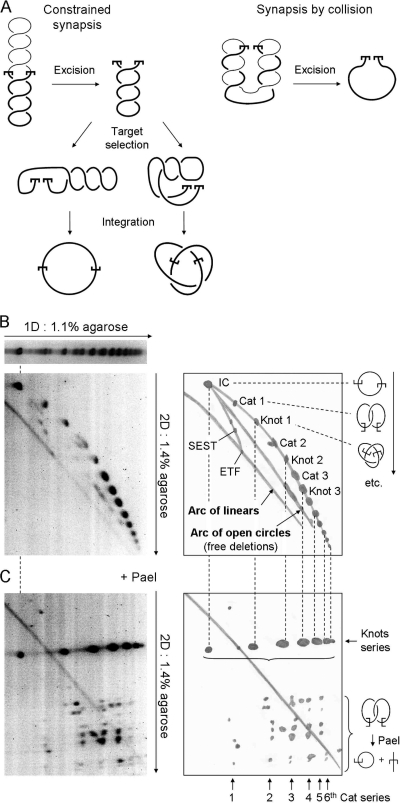
One way to decide whether the transposon ends achieve synapsis after meeting in the plectosome is to examine the structure of the intramolecular integration products. These products preserve a record of the topological relationship between the transposon ends immediately before cleavage and between the transpososome and the target site prior to integration. The relationships are revealed by the distribution of the numbers of knot and catenane nodes, which are derived from plectonemic nodes trapped during synapsis of the ends and integration at the target site (6). The principle is illustrated in Fig. 4A. If the transposon ends meet at a plectonemic node, the excised transposon will retain its full quota of supercoiling after excision (Fig. 4A, left panel, top illustration). If the transposon ends meet by random collision, the amount of supercoiling retained will depend on their relative position in the plectosome. If the ends are at their furthermost positions in the plectosome when they collide, all of the supercoils will be released by excision (Fig. 4A, right panel). Therefore, if the transposon ends meet by collision, the transposon will on average retain half of the supercoiling density present in the substrate.
FIG. 4.
Synapsis in the plectosome. (A) Two models for the assembly of the mariner synaptic complex are illustrated. (Left panel) When synapsis of the transposon ends is constrained by the plectosome, the ETF retains the full superhelical density of the substrate. If intramolecular target sites are selected by random collision, any number of nodes may be trapped, ranging from zero up to the total number present. However, on average, half of the nodes are trapped in the product. (Right panel) If the transposon ends meet by random collision, unconstrained by the plectosome, any number of nodes may be trapped, up to the maximum number present. The illustration shows the situation when the transposon ends are at opposite ends of the plectosome when they collide: excision releases all of the nodes. However, we would expect collision to trap half of the nodes on average. Half of these would go on to be trapped in the product if the target site is acquired by collision. (B) The topology of intramolecular integration products obtained from a reaction with a negatively supercoiled inverted-repeat substrate (pRC1105) was analyzed by two-dimensional (2D) gel electrophoresis using the conditions indicated beside the gels. A drawing of each gel is provided to indicate the identities of the products. The substrate includes a 2.3-kb transposon and a 4.2-kb backbone. The gel conditions and the respective sizes of the transposon and backbone fragments are identical to those previously used to characterize the products of Tn10 transposition, which were used as a yardstick (6). The gels were stained with SYBR green and recorded on a Fluorimager. Integration events that trap an even number of nodes between the transposon ends and the target site produce catenated deletion circles (Cat), while those that trap an odd number of nodes yield knotted inversion circles (Knot). IC, unknotted inversion circles; ETF, excised transposon fragment; SEST, single-ended-strand-transfer events, yielding lariat structures; 1D, first dimension. The identities of the products were determined by comparison with the gel shown in panel C. Further details are given in the text and reference 6. (C) The transposition reaction and the first dimension of the gel were identical to that in panel B. Annotations are also similar. After electrophoresis in the first dimension, the restriction endonuclease PaeI, which cuts at a single site within the transposon, was diffused into the gel. PaeI digestion converts knotted products into a linear species identical in size to the ETF. Upon digestion, catenated products yield one linear species and one open circular species that together equal the size of the ETF. The relative sizes of the linear and circular species depend on the site of intramolecular integration. Since the gels in panels B and C share a first dimension, the positions of the various products are the same and are indicated by the dashed vertical lines. Further details are given in the text and reference 6.
To characterize the intramolecular products, we used two different types of two-dimensional (2D) gel electrophoresis. In the first type of gel, the concentration of agarose is higher in the second dimension (Fig. 4B). This separates the DNA into three major arcs corresponding to linear, open circular, and topologically complex knotted and catenated products. In the second type of gel, the DNA is digested with a restriction endonuclease between the first and second dimensions (Fig. 4C). The enzyme cuts the substrate plasmid once at a site within the transposon. Digestion converts knotted transposon circles into linear molecules identical in size to the excised transposon fragment (ETF). The catenated products are converted into one linear species plus one open circular species, which together equal the size of the transposon. The identities of the various intramolecular products can be established by comparing their respective positions on the two gels, which share a first dimension.
The intramolecular product with the lowest electrophoretic mobility is the unknotted inversion circle (IC), which migrates on the arc of open circles before digestion and at the position of the ETF after digestion (Fig. 4B and C). The next product is the first catenane (Cat 1), which traps two nodes. Trapping one more node produces the first knotted transposon circle, which is linearized upon digestion and therefore migrates at the position of the ETF (Fig. 4C). Knots and catenanes of increasing complexity then alternate, producing the arc of topologically complex products. A total of 13 products can be distinguished on this arc, culminating in the sixth knot, which traps 13 nodes.
To deduce the topological relationship between the transposon ends during synapsis, we can assume that target sites are acquired by random collision. This is reasonable because it is the only mechanism consistent with the observed heterogeneity of the knotted and catenated products. The proportion of the transposon's supercoiling that becomes trapped in the intramolecular products will therefore depend on the location of the target site. Target sites furthermost from the transposon ends will trap the highest number of nodes, while those located close by may not trap any nodes (Fig. 4A, left panel, bottom illustration). It therefore follows that integration will on average trap approximately half of the nodes present in the excised transposon.
Since the superhelical density of the substrate is −0.06, a segment of DNA corresponding to a 2.3-kb transposon will have 14 nodes. If we first consider the case in which the transposon ends meet by random collision, the distribution of intramolecular products will peak at the first knot, which traps three nodes. This is because twice during the reaction, the transposon loses half of its superhelical density, once during synapsis and once during target acquisition. This was exactly the result obtained with Tn10, for which synapsis of the transposon ends and acquisition of the target are both by random collision (6). In contrast, the distribution of Hsmar1 products peaks at the third knot, which traps seven nodes (Fig. 4C). The excised transposon must therefore have contained 14 nodes. For a transposon of this size, this amount is equivalent to the full superhelical density of the substrate. The transposon ends must therefore have achieved synapsis while they were juxtaposed in the plectosome.
Catenation of single-ended circles stimulates excision.
The results presented thus far suggest that the right-handed parallel intertwining of the transposon ends in the plectosome accelerates the reaction kinetics. Other properties of the substrates, such as their covalent integrity or the actual orientation of the transposon ends, appear to be relatively unimportant. If this is true, we would expect the reaction with single-ended substrates to accelerate when they are catenated with a right-handed geometry. Such a substrate can be prepared by recombination using phage lambda integrase.
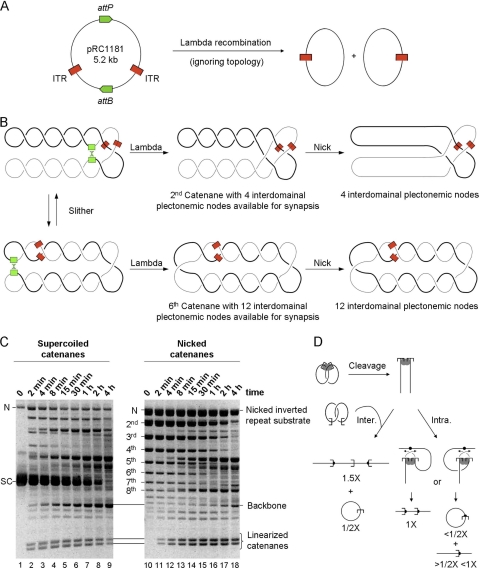
We constructed a recombination substrate with phage lambda attP and attB sites in direct-repeat configuration, opposite each other on the plasmid map. Treatment with lambda integrase generated a pair of catenated circles of almost equal size, each with a single transposon end (Fig. 5A and B). The extent of the lambda recombination reaction was estimated at about 65% from the amount of unreacted substrate present after nicking the catenanes (Fig. 5C) and by restriction endonuclease digestion of the catenanes (not shown).
FIG. 5.
DNA intertwining accelerates transposition. (A) An illustration of the phage lambda integrase reaction. Recombination between the phage lambda attP and attB sites separates the transposon ends onto two circles of almost identical size. Topological features of the reaction have been omitted but are shown in panel B. (B) An illustration of how phage lambda recombination can be used to generate catenated single-end transposition substrates. The lambda attP and attB sites (green) divide the plasmid into two equal-sized segments, indicated by the thick and thin lines. The att sites meet by random collision, trapping a number of plectonemic nodes. These are converted to catenane nodes by lambda recombination. The number of trapped nodes converted ranges from four, the minimum allowed by the mechanism of recombination, up to the maximum number of nodes present in the substrate. Lambda recombination preserves any supercoiling nodes that are not converted to catenane nodes (see the top element of the central panel). After recombination, each of the catenated circles contains a single transposon end (red). The catenated circles can slither past each other, providing opportunities for the transposon ends to meet in the right-handed crossing points. Treatment with the Nb.BsrDI endonuclease introduces a nick into each of the catenated circles, releasing any supercoiling that is present (rightmost panel). (C) The kinetics of transposition reactions with supercoiled and nicked catenane substrates. The absence of supercoiling in the nicked substrate reveals the distribution of the catenanes. The second catenane, with four nodes, is the most abundant product of lambda recombination. Its identity was confirmed in a high-resolution gel (not shown, but see reference 26). About 65% of the substrate was recombined by the lambda integrase, as revealed by the abundance of the unrecombined nicked inverted-repeat substrate in the rightmost panel (N). The backbone fragment produced from the 35% of the inverted-repeat substrate that remains in these reactions provides a useful internal control for the kinetic analysis (see the text for details). A photograph of an ethidium bromide-stained agarose gel is shown. (D) Products of transposition with catenated single-end substrates. Cleavage at the transposon ends yields two linear products of similar sizes. Intermolecular integration into an unreacted substrate plasmid, which is the only target available in these reactions, releases one of the half-circles and generates a linear product 1.5 times as large as the original plasmid used to generate the catenanes (i.e., 1.5X). Intramolecular events produce two alternative outcomes depending on orientation of the target site: a linear product the size of the original plasmid (1X) or a pair of products, namely, an open circle plus a linear product that together equal the size of the plasmid.
Lambda recombination maintains the superhelical density of the substrate. However, the number of supercoiling nodes converted into catenane nodes depends on the relative positions of the attP and attB sites in the plectosome when they collide (Fig. 5B). The distribution of the catenanes was revealed by treatment with the nicking endonuclease Nb.BsrDI, which introduces a nick in each of the catenated circles. Although the circles remain catenated, the nicks allow the supercoiling nodes to escape. This revealed that the most abundant product was the second catenane, with higher catenanes diminishing progressively (Fig. 5C, lane 10) (for further details, see reference 26).
Supercoiled and nicked catenanes were consumed rapidly by the transposition reaction (Fig. 5C). The spectrum of intermediates and products expected from the catenanes is illustrated in Fig. 5D. Transposon end cleavage yielded two linear fragments corresponding to the two catenated circles. Although this pair of fragments together is equivalent to the double-end-break product in a standard reaction, it is not an end product like the backbone but undergoes integration. Integration products can be seen appearing at numerous positions in the gel as the reactions progress. Since any DNA fragment in the reaction mixture can be used as a target, there is a large array of these products in addition to the major ones illustrated in Fig. 5D.
The reaction mixtures contained an internal control provided by the ∼35% of the inverted-repeat substrate that had failed to undergo lambda recombination. Excision from this substrate yields the standard backbone fragment, which accumulated during the time course. The backbone appeared only slowly in the reactions with the nicked substrate. This provides an important internal reference that greatly simplifies interpretation of the experiment. Despite the fact that the linearized catenanes are continually chased into integration products during the reaction, they are detected much earlier than the backbone in the time course with the nicked substrate. This shows that the nicked catenanes are reacting much faster than the nicked inverted-repeat substrate. Indeed, over 50% of the nicked inverted-repeat substrate remains uncleaved at the end of the time course when virtually all of the catenanes have been consumed. These results provide a very strong confirmation that it is the parallel right-handed intertwining of the transposon ends in the plectosome that drives synapsis and excision of the transposon.
DISCUSSION
Catalysis requires synapsis.
Transposition and site-specific recombination reactions are potentially genotoxic. Consequently, many recombinases have mechanisms to suppress noncanonical reactions. One common strategy is the trans architecture of the synaptic complex, in which recombining partners share the active sites and/or DNA binding domains of different subunits. This couples the initiation of catalysis to the synapsis of the recombining sites. This strategy is particularly powerful if the monomeric recombinase must bind to the recombination site before dimerization. This is the case for the phage Mu, Tn5, and Tn10 transposons, for example (10).
In contrast, the mariner transposase and the distantly related V(D)J recombinase bind the first recombination site as dimers from solution, making the trans subunit immediately available for catalysis. Experiments with an immobilized signal sequence demonstrated that V(D)J recombination can initiate on an unsynapsed site. However, it was later shown that the hairpin step, which completes cleavage, is coupled to synapsis by a conformational change in the complex (4, 5).
The detection of “self-inflicted wounds” in drosophilas gave the first indication that the first nick in mariner transposition may be independent of synapsis. This was supported by several lines of biochemical evidence that have, until now, been generally accepted. These observations form the basis for the current model for mariner transposition, which postulates that the first nick is followed by synapsis, which induces a conformational change, or subunit exchange, that licenses second-strand cleavage (13, 19).
This model is contradicted by the experiments with the single-ended plasmid (Fig. 2). The extremely low reactivity of this substrate is strong evidence against the idea that first-strand cleavage is independent of synapsis. If this was the case, we would expect first-strand nicking to be unaffected by the arrangement of the transposon ends. This was obviously not the case, as most of the single-end substrate remained unreacted after long incubation (compare Fig. 2A and C).
A kinetic barrier to synapsis.
Simple protein-DNA binding reactions take place very quickly when the reactants are in the low-nM concentration range used in our experiments. Indeed, a stopped-flow apparatus is usually required to study binding kinetics. Higher-order complexes may also form quickly. For example, the six components of the Tn10 transpososome are assembled within 30 s, which is the minimum time required to mix the reaction volume and take a sample (24). Our single-end substrate's lack of reactivity therefore suggests that there is a kinetic barrier to synapsis of Hsmar1 ends: that is, most collision events between the reactants are unproductive.
A kinetic barrier in a second-order reaction, such as that between transposon ends, should be ameliorated by increasing the concentration of the reactants. Our experiments are entirely consistent with this prediction. When two transposon ends are on the same plasmid, their effective concentration increases because they are constrained by covalent connections. Therefore, we expected that in the absence of supercoiling, the inverted- and direct-repeat substrates would react at the same rate, which is higher than that for the single-end substrate. This was exactly the result obtained (Fig. 2I, J, K, and L).
Supercoiling further increases the relative concentrations of any two sites on a plasmid by one to two orders of magnitude (29). This is because of constraints introduced as the random coil of the DNA backbone collapses down into a plectosome of much smaller volume. The acceleration of the inverted-repeat reaction by negative supercoiling can thus be explained by the increase in the relative concentrations of the transposon ends (Fig. 2I). However, an additional factor must be operating, since negative supercoiling does not stimulate the direct-repeat substrate, nor does positive supercoiling stimulate the inverted-repeat substrate (Fig. 2 and 3).
Reactions with the catenated single-end substrates suggest that it is the right-handed parallel intertwining of the transposon ends that accelerates synapsis. The simplest catenane produced by lambda recombination is the second member of the series with four nodes, which is equivalent to a σ of −0.0077 (Fig. 5B). This is not sufficient to collapse the random coil of the DNA backbones down into a compact plectosome (30). Nevertheless, it is sufficient to increase the relative concentrations of the ends substantially, and the right-handed intertwining of the catenanes would favor the parallel synapsis of the ends. We would therefore expect the nicked form of the second catenane to react much faster than the nicked inverted-repeat plasmid from which it is generated. This was precisely the result obtained (Fig. 5C, right panel). It therefore seems that the acceleration of the inverted-repeat reaction by negative supercoiling stems at least in part from the right-handed intertwining of the transposon ends in the plectosome. This is reminiscent of the topological filters used by some transposons and site-specific recombinases to distinguish the arrangements of their binding sites.
A topological filter.
Some recombination reactions are sensitive to the topological arrangement of their substrates. These systems have in common the fact that the synaptic complex involves an accessory protein-binding site in addition to the recombining sites. For example, assembly of the hin invertasome requires the FIS-bound enhancer, the phage Mu transpososome uses an IHF-bound enhancer, and the Tn3 resolvasome contains accessory binding sites for resolvase subunits which take no part in catalysis. Topological selectivity arises from negative supercoiling in the substrates, which promotes the interwrapping of the three respective sites in a favorable geometry. This mechanism is known as a topological filter because it disfavors recombination between sites on different molecules and between sites in the wrong orientation, e.g., between direct repeats in Mu and hin recombination and inverted repeats in the case of resolvase.
Supercoiling also serves other purposes in recombination reactions. In Tn10 transposition, for example, the energy of negative supercoiling is used to wrap one end of the transposon around the synaptic complex. However, this imparts no topological selectivity, and transposon ends recombine irrespective of whether they are in the inverted- or direct-repeat configuration or even on different molecules (6). It is intriguing to ask how, in the absence of a topological filter, simple transposons might regulate their activity to minimize the potential genotoxic results of unconstrained synapsis. For bacterial transposons, such as Tn10, the answer lies partly in the cis action of the transposase, which binds tightly to the first DNA it encounters before searching for nearby ends. The coupling of transcription and translation in bacteria thus greatly increases the probability of synapsing the ends of the encoding element.
In contrast, eukaryotic transposases are necessarily trans acting: when they enter the nucleus, they must search at random for transposon ends. If collision synapsis is efficient, there is a clear danger of noncanonical recombination events. These may arise between pairs of ends belonging to different copies of the element or between ends on different chromosomes. Such events are caused by the Ac transposon in maize and precipitate breakage-fusion-bridge cycles (14). These complicated genotoxic lesions are impossible to repair or certainly more difficult to repair than the simple double-strand break, which is the product of a canonical transposition reaction. Selection of the correct recombination partner is therefore an important decision.
The kinetic barrier to recruitment of the second transposon end into the developing transpososome seems to constitute a rudimentary topological filter. The barrier stems from the asymmetry of the complex after the transposase dimer has captured the first transposon end. We have shown that it is ameliorated by the increase in the effective concentrations of the reacting partners provided by the right-handed intertwining of the superhelix in negatively supercoiled DNA and the catenanes generated by lambda recombination. By favoring the parallel synapsis of appropriately oriented pairs of ends, the kinetic barrier thus serves the same purpose as the accessory binding sites in the topological filters discussed above.
We also note that the kinetic barrier to synapsis is increased by positive supercoiling, even though this also provides for the parallel synapsis of inverted repeats, and the same increase in effective concentration by the juxtaposition of reacting partners. Presumably, the penalty associated with positive supercoiling reflects an unfavorable angular distribution of the reacting partners in the left-handed superhelix.
Biological implications and conclusions.
This investigation into the effects of DNA supercoiling and the arrangement of mariner transposon ends has provided several insights into the mechanism of the reaction. The acceleration of the reaction in response to supercoiling is due to a right-handed intertwining of the transposon ends during synapsis. Synapsis is the rate-limiting step of the reaction, and its dependence on the concentration of the reacting partners disfavors the pairing of transposon ends on different DNA molecules and presumably also of ends on the same molecule when they are very distant. These constraints represent an important checkpoint, controlling the initiation of catalysis and promoting the canonical association of transposon ends at opposite ends of the same element.
Psoralen cross-linking experiments previously indicated that bulk eukaryotic DNA contained little free supercoiling. This view is challenged by recent genome-wide experiments showing significant regional differences in the level of free supercoiling (2). From a regulatory point of view, it is perhaps more important that mariner's sensitivity to free supercoiling allows the rate of transposition to respond to the dynamics of the nucleus. For example, transposition of elements ahead of an advancing replication fork will be blocked by positive supercoiling. This may help to amplify the number of transposons, because it will favor mobilization after replication when a sister chromosome is available for repair. Unconstrained negative supercoiling is also present behind an advancing transcription bubble. If this is sufficient to promote synapsis of mariner ends, it would allow a promoter, perhaps within the element, to influence the rate of transposition. Nucleosome remodeling, which accompanies transcription, is another source of unconstrained supercoiling. The loss of only one or two nucleosomes from a 1-kb transposon would provide a superhelical density of −0.02, sufficient for near-maximal stimulation of synapsis (Fig. 3). The levels of free supercoiling and its dynamics in eukaryotic cells are still poorly understood. However, it seems that mariner's sensitivity to supercoiling may be an important mechanism allowing transposition to synchronize with the cell cycle and helping to reduce the potentially genotoxic results of wayward recombination events.
Acknowledgments
We thank Art Landy for the gift of lambda integrase, Maria Ciaramella for the plasmid used to express reverse gyrase, Neil Walker for technical assistance throughout this work, and Paul O'Shea for advice about rate constants.
C.C.B. is supported by a BBSRC Doctoral Training Program studentship. This work was funded by a grant from The Wellcome Trust to R.C.
Footnotes
Published ahead of print on 1 November 2010.
REFERENCES
- 1.Augé-Gouillou, C., B. Brillet, M. H. Hamelin, and Y. Bigot. 2005. Assembly of the mariner MosI synaptic complex. Mol. Cell. Biol. 25:2861-2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bermúdez, I., J. Garcia-Martinez, J. E. Pérez-Ortín, and J. Roca. 2010. A method for genome-wide analysis of DNA helical tension by means of psoralen-DNA photobinding. Nucleic Acids Res. 38:e182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bischerour, J., and R. Chalmers. 2009. Base flipping in Tn10 transposition: an active flip and capture mechanism. PLoS One 4:e6201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bischerour, J., and R. Chalmers. 2007. Base-flipping dynamics in a DNA hairpin processing reaction. Nucleic Acids Res. 35:2584-2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bischerour, J., C. Lu, D. B. Roth, and R. Chalmers. 2009. Base flipping in V(D)J recombination: insights into the mechanism of hairpin formation, the 12/23 rule, and the coordination of double-strand breaks. Mol. Cell. Biol. 29:5889-5899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chalmers, R. M., and N. Kleckner. 1996. IS10/Tn10 transposition efficiently accommodates diverse transposon end configurations. EMBO J. 15:5112-5122. [PMC free article] [PubMed] [Google Scholar]
- 7.Chalmers, R. M., and N. Kleckner. 1994. Tn10/IS10 transposase purification, activation, and in vitro reaction. J. Biol. Chem. 269:8029-8035. [PubMed] [Google Scholar]
- 8.Claeys Bouuaert, C., and R. Chalmers. 2010. Transposition of the human Hsmar1 transposon: rate-limiting steps and the importance of the flanking TA dinucleotide in second strand cleavage. Nucleic Acids Res. 38:190-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Claeys Bouuaert, C., and R. M. Chalmers. 2010. Gene therapy vectors: the prospects and potentials of the cut-and-paste transposons. Genetica 138:473-484. [DOI] [PubMed] [Google Scholar]
- 10.Craig, N. L., R. Craigie, M. Gellert, and A. M. Lambowitz. 2002. Mobile DNA II. American Society for Microbiology, Washington, DC.
- 11.Crénès, G., D. Ivo, J. Herisson, S. Dion, S. Renault, Y. Bigot, and A. Petit. 2009. The bacterial Tn9 chloramphenicol resistance gene: an attractive DNA segment for MosI mariner insertions. Mol. Genet. Genomics 281:315-328. [DOI] [PubMed] [Google Scholar]
- 12.Davies, D. R., I. Y. Goryshin, W. S. Reznikoff, and I. Rayment. 2000. Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science 289:77-85. [DOI] [PubMed] [Google Scholar]
- 13.Dawson, A., and D. J. Finnegan. 2003. Excision of the Drosophila mariner transposon mos1. Comparison with bacterial transposition and v (d) j recombination. Mol. Cell 11:225-235. [DOI] [PubMed] [Google Scholar]
- 14.English, J. J., K. Harrison, and J. D. G. Jones. 1995. Aberrant transpositions of maize double Ds-like elements usually involve Ds ends on sister chromatids. Plant Cell 7:1235-1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hazelbaker, D., M. A. Azaro, and A. Landy. 2008. A biotin interference assay highlights two different asymmetric interaction profiles for lambda integrase arm-type binding sites in integrative versus excisive recombination. J. Biol. Chem. 283:12402-12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kazazian, H. H., Jr. 2004. Mobile elements: drivers of genome evolution. Science 303:1626-1632. [DOI] [PubMed] [Google Scholar]
- 17.Levene, S. D., and D. M. Crothers. 1986. Ring closure probabilities for DNA fragments by Monte Carlo simulation. J. Mol. Biol. 189:61-72. [DOI] [PubMed] [Google Scholar]
- 18.Lipkow, K., N. Buisine, and R. Chalmers. 2004. Promiscuous target interactions in the mariner transposon Himar1. J. Biol. Chem. 279:48569-48575. [DOI] [PubMed] [Google Scholar]
- 19.Lipkow, K., N. Buisine, D. J. Lampe, and R. Chalmers. 2004. Early intermediates of mariner transposition: catalysis without synapsis of the transposon ends suggests a novel architecture of the synaptic complex. Mol. Cell. Biol. 24:8301-8311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu, D., J. Bischerour, A. Siddique, N. Buisine, Y. Bigot, and R. Chalmers. 2007. The human SETMAR protein preserves most of the activities of the ancestral Hsmar1 transposase. Mol. Cell. Biol. 27:1125-1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu, D., P. Crellin, and R. Chalmers. 2005. Cyclic changes in the affinity of protein-DNA interactions drive the progression and regulate the outcome of the Tn10 transposition reaction. Nucleic Acids Res. 33:1982-1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lohe, A. R., E. N. Moriyama, D. A. Lidholm, and D. L. Hartl. 1995. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol. Biol. Evol. 12:62-72. [DOI] [PubMed] [Google Scholar]
- 23.Miskey, C., B. Papp, L. Mates, L. Sinzelle, H. Keller, Z. Izsvak, and Z. Ivics. 2007. The ancient mariner sails again: transposition of the human Hsmar1 element by a reconstructed transposase and activities of the SETMAR protein on transposon ends. Mol. Cell. Biol. 27:4589-4600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sakai, J., R. M. Chalmers, and N. Kleckner. 1995. Identification and characterization of a pre-cleavage synaptic complex that is an early intermediate in Tn10 transposition. EMBO J. 14:4374-4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singleton, C. K., and R. D. Wells. 1982. The facile generation of covalently closed, circular DNAs with defined negative superhelical densities. Anal. Biochem. 122:253-257. [DOI] [PubMed] [Google Scholar]
- 26.Spengler, S. J., A. Stasiak, and N. R. Cozzarelli. 1985. The stereostructure of knots and catenanes produced by phage lambda integrative recombination: implications for mechanism and DNA structure. Cell 42:325-334. [DOI] [PubMed] [Google Scholar]
- 27.Travers, A., and G. Muskhelishvili. 2007. A common topology for bacterial and eukaryotic transcription initiation? EMBO Rep. 8:147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Valenti, A., G. Perugino, A. D'Amaro, A. Cacace, A. Napoli, M. Rossi, and M. Ciaramella. 2008. Dissection of reverse gyrase activities: insight into the evolution of a thermostable molecular machine. Nucleic Acids Res. 36:4587-4597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vologodskii, A., and N. R. Cozzarelli. 1996. Effect of supercoiling on the juxtaposition and relative orientation of DNA sites. Biophys. J. 70:2548-2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vologodskii, A. V., and N. R. Cozzarelli. 1993. Monte Carlo analysis of the conformation of DNA catenanes. J. Mol. Biol. 232:1130-1140. [DOI] [PubMed] [Google Scholar]