Abstract
The Rb/E2F pathway has long been appreciated for its role in regulating cell cycle progression. Emerging evidence indicates that it also influences physiological events beyond regulation of the cell cycle. We have previously described a requirement for Rb/E2F mediating neuronal migration; however, the molecular mechanisms remain unknown, making this an ideal system to identify Rb/E2F-mediated atypical gene regulation in vivo. Here, we report that Rb regulates the expression of neogenin, a gene encoding a receptor involved in cell migration and axon guidance. Rb is capable of repressing E2F-mediated neogenin expression while E2F3 occupies a region containing E2F consensus sites on the neogenin promoter in native chromatin. Absence of Rb results in aberrant neuronal migration and adhesion in response to netrin-1, a known ligand for neogenin. Increased expression of neogenin through ex vivo electroporation results in impaired neuronal migration similar to that detected in forebrain-specific Rb deficiency. These findings show direct regulation of neogenin by the Rb/E2F pathway and demonstrate that regulation of neogenin expression is required for neural precursor migration. These studies identify a novel mechanism through which Rb regulates transcription of a gene beyond the classical E2F targets to regulate events distinct from cell cycle progression.
The Rb pathway is best characterized for its role in regulating cell cycle progression through E2F-mediated transcriptional regulation of classical cell cycle machinery target genes. Recently, however, accumulating in vivo and in vitro evidence is emerging to suggest that Rb and E2F are capable of regulating expression of atypical target genes with functions other than cell cycle regulation in cell-type-specific manners (reviewed in reference 35). In vivo, several studies have emerged that implicate Rb and E2F interaction in novel processes beyond well-characterized roles in cell cycle regulation (10; for a review, see reference 6). In the nervous system, in particular, we have recently shown that an Rb-E2F3 interaction mediates migration of a subpopulation of GABAergic interneurons (34). In the same study, we also observed deregulation of a number of genes with known roles in neuronal migration in cell populations lacking Rb, suggesting a role for E2F3 in regulating transcription of novel targets (34). A second cell cycle-independent role for E2F3a in regulating Rb-mediated interneuron differentiation was also reported in the retina (9). Thus far, in vivo studies have failed to identify the mechanism through which these cell cycle-independent processes occur.
In parallel, in vitro several microarray studies examining changes in gene expression in response to various models of deregulated E2F expression have each identified groups of overlapping novel target genes with well-characterized roles in differentiation, development, and migration (5, 15, 25, 31, 39, 41, 60). More recently, chromatin immunoprecipitation (ChIP)-on-chip studies have identified putative E2F binding sites within the promoters of a number of genes unrelated to the cell cycle (3, 4, 7, 28, 46, 56, 57). Finally, by using an approach whereby novel genes induced by E2F1 are identified based on subtraction screening, genes with known roles in differentiation and migration were identified as being directly induced by E2F1 in a cell cycle-independent manner (26). Thus, these data provide evidence that our understanding of the significance of Rb/E2F function should be expanded to include transcriptional regulation of genes beyond the well-characterized subset of targets that regulate the cell cycle.
Our identification of a role for Rb/E2F3 in mediating neuronal migration represents an attractive model to identify novel cell cycle-independent E2F target genes in the context of an in vivo physiological function (16, 34). Given our previous observations revealing (i) deregulation of a number of genes in families of known chemotactic ligands and receptors implicated in neuronal migration in the absence of Rb; and (ii) the cell-autonomous requirement for Rb in neuronal migration, we hypothesized that Rb/E2F may modulate the transcription of novel target genes involved in neuronal migration. We focused our efforts on neogenin, a receptor for the netrin and repulsive guidance molecule (RGM) families of chemotropic ligands (reviewed in reference 14). Notably, neogenin is highly expressed by a subpopulation of interneurons migrating from the ventral forebrain and has been independently identified, in an in vitro overexpression system, as an E2F-regulated gene (26, 34). Here, we report that Rb directly regulates the expression of a nontraditional target, neogenin. Rb is capable of repressing E2F-mediated transcription of neogenin while E2F3 binds to a region containing a conserved E2F consensus site on the neogenin promoter in native chromatin. The absence of Rb results in aberrant neuronal migration and adhesion in response to the neogenin ligand, netrin-1. Finally, increased expression of neogenin through ex vivo electroporation results in impaired neural precursor migration similar to that observed in forebrain-specific Rb deficiency. From these findings, we show direct regulation of neogenin by the Rb/E2F pathway and demonstrate that correct regulation of neogenin expression is required for neural precursor cell migration. Through these studies we identify a novel mechanism through which Rb interacts with E2F to regulate transcription of genes beyond the classical E2F targets to influence biologically relevant events distinct from cell cycle progression.
MATERIALS AND METHODS
Mice.
Telencephalon-specific Rb-deficient mice were generated by crossing floxed Rb-F19 (33, 53) and Foxg1-cre mice (23), and mice were genotyped according to standard protocols with previously published primers (16, 17). For embryonic time points, the time of plug identification was counted as embryonic day 0.5 (E0.5). For all experiments littermate Rb conditional mutants (Rbflox/flox Foxg1-cre+/−) and double heterozygous controls (Rbflox/+ Foxg1-cre+/−) were compared. Due to Rb autoregulation (49), Rb expression in heterozygous mice is similar to that of wild-type controls. All experiments were approved by the University of Ottawa's Animal Care ethics committee, adhering to the Guidelines of the Canadian Council on Animal Care.
Western blotting.
Protein was isolated from neurospheres by treating cells with lysis buffer (10 mM Tris, 0.15 M NaCl, 1 mM EDTA, 0.4 mM sodium vanadate, and 0.5% Triton-X). Cells were incubated on ice for 20 min, followed by a 10-min centrifugation at maximum speed to remove debris. Western blotting was performed as previously described (16) with antibodies directed toward neogenin (H-175; Santa Cruz), Rb (Pharmingen), and beta-actin (Sigma). Immunoblotting was performed on three independent samples, and results were quantified using ImageJ software.
Tissue preparation and in situ hybridization.
Tissue was dissected, fixed, cryoprotected, and sectioned as previously described (34). Nonradioactive in situ hybridization and digoxigenin probe labeling were performed according to previously described protocols (54). Neogenin, deleted in colorectal cancer (DCC), netrin, and RGMa, were generous gifts of Helen Cooper of the University of Queensland (20), Elke Stein of Yale University, and Silvia Arber of the University of Basel (40). All results shown are representative of those obtained with a minimum of three independent animals.
Transcription factor binding sites.
The 5′ region of the mouse Neo1 locus containing the intergenic region, the untranslated region, and exon 1 was analyzed for putative E2F binding sites using the TRANSFAC Professional Library, version 10.2, through Mulan/MultiTF (http://rvista.dcode.org). All sites identified in Mulan were manually examined for their similarity to the consensus (TTTSSCGC) and nonconsensus (BKTSSCGS) E2F motifs.
Chromatin immunoprecipitation.
Neurosphere cultures were prepared from CD1 embryos (Charles River) at embryonic day 14.5. Proliferating neurospheres were triturated, cross-linked with formaldehyde, lysed, sonicated, and centrifuged at 14,000 × g to remove cellular debris. Each immunoprecipitation was performed using 2 μg of antibody. Antibodies against E2F3 (sc-878), E2F1 (sc-193), and normal rabbit immunoglobulin G (sc-2027) were obtained from Santa Cruz Biotechnology. Immunocomplexes were captured using protein A/G9-Sepharose beads and washed extensively, and cross-links were reversed overnight, followed by treatment with RNase A at 37°C for 1 h and proteinase K at 65°C for 30 min. The purified DNA was examined by PCR using primers designed around the E2F consensus sites at bp −44 and bp −821 in the 5′ region of the neogenin gene. Immunoprecipitations were performed from three cultures obtained from three independent animals.
DNA constructs.
The neogenin construct was PCR amplified from pSecTagA-Neogenin (rat) using the following primers: forward, GAACTGCAGACCATGGAAGAAAGA; reverse, CTTCTGCAGGTTGCCCTCTAGCTAG. The PCR fragment was then subcloned into pCIG2. The 5′ regulatory region of neogenin was PCR amplified from embryonic genomic DNA. Primers were designed as follows with flanking XhoI restriction sites inserted: full-length forward, AGACTCGAGGAGGTGCAGAGGAGTCGC; full-length reverse, TGTCTCGAGGTTGAAAAACCAATTCCCG. To create 5′ truncations the following primers were used with full-length reverse primer: FTrunc247, AGACTCGAGAGCCGGGGGGTGG; FTrunc618, AGACTCGAGAAGCGATCCGCCTCCT. To create the 3′ truncation the following primer was used with full-length forward primer: RTrunc 247np, TGTCTCGAGCCACCCCCCGGCT. The construct consisting of bp 247 to 618 was made using the FTrunc247 and RTrunc 618bp (TGTCTCGAGGCGGATCGCTTCTCC) primers. PCR products were digested with XhoI and ligated into pGL4.24 (Promega). Sequence was confirmed by DNA sequencing (StemCore Laboratories, University of Ottawa).
Luciferase reporter assays.
HEK293T cells were transfected using Lipofectamine (Invitrogen) as per the manufacturer's protocol. Briefly, cells were transfected with 500 ng of pGL4.24, pGL4.24-Neogenin, or pGL4.24-NeoTruncations, 10 ng of E2F3, 300 ng of Rb, and total transfected plasmid normalized with pcDNA3.1. Transfection efficiency was normalized using 10 ng of pRL Renilla-expressing vector. Cells were lysed at 24 h posttransfection and examined by spectrophotometer (LMaxII; Molecular Devices) for luciferase expression by a Dual-Glo luciferase kit (Promega).
In vitro explant cultures.
In vitro explant cultures were performed as described previously (13, 29, 38, 43, 44) with some modifications. Briefly, brains were removed from E14.5 embryos in L-15 (Gibco) medium, and medial ganglionic eminence (MGE) was dissected as described previously (16); explants were subsequently divided into pieces approximately 200 μm in diameter by using sharpened tungsten needles. MGE explants were then transferred into collagen (PureCol, catalog number 5409; Inamed BioMaterials) inside culture dishes and allowed to solidify for 40 min prior to addition of Neurobasal medium (Gibco) supplemented with fetal bovine serum (FBS). Purified netrin-1 was added to the explant culture medium at a final concentration of 200 ng/ml.
Explants cultured alone were grown in vitro for 24 h. Images were captured with a Zeiss Axioscope microscope. For quantification of cell migration in collagen, the total number of cell bodies migrating from explants was counted. Two to four explants per embryo were measured, and values were averaged. Two-tailed t tests were performed to compare mean migration between genotypes or treatment groups. Differences were considered significant at a P value of <0.05.
Neural progenitor cultures.
Pregnant mice were euthanized at gestation day 14.5, embryos were removed, and the ganglionic eminences were isolated by microdissection. For determination of cell proliferation and cell death in single-cell preparations, ganglionic eminences were dissociated, and equal cell numbers were plated on poly-d-lysine- and laminin-1-coated dishes in duplicate. Cells were cultured in Neurobasal medium supplemented with 0.5 mM l-glutamine, 1% N-2, 2% B-27, 10 ng/ml fibroblast growth factor 2 (FGF-2), and 20 ng/ml epidermal growth factor (EGF) in either the presence or absence of netrin-1 at 200 ng/ml. Cells were treated with bromodeoxyuridine (BrdU) at a final concentration of 10 μg/ml for 45 min prior to fixation. After 24 h, cells were fixed for 15 min in 4% paraformaldehyde (PFA) and then treated sequentially with 2 N HCl and 0.01 M NaB4O7, followed by BrdU immunohistochemistry (anti-BrdU at 1:100; BD Biosciences, San Jose, CA) and Hoechst nuclear staining. The total cells and BrdU-positive cells were counted in three microscope fields per duplicate well. Rates of proliferation were obtained by calculating the proportion of BrdU-positive cells relative to the total cell number. Fold increase in proliferation in response to netrin-1 was calculated for each genotype by dividing the percentage of proliferating cells in the presence of netrin-1 by the percentage of proliferating cells in the absence of netrin-1. (Three separate embryos were analyzed in quadruplicate for both control and conditional mutant embryos). For Hoechst labeling, dead cells were identified by the characteristic condensation of chromatin. Fold increase in apoptotic nuclei was calculated in an analogous manner to cell proliferation. (Three separate embryos were analyzed in quadruplicate for both control and conditional mutant embryos).
Substrate-bound adhesion assay.
To assess cell adhesion in the presence of netrin-1, a substrate-bound adhesion assay was performed as described previously (50). Briefly, 20 μl of 0.1% nitrocellulose (Hybond ECL; Amersham Biosciences) dissolved in methanol was dried on the bottom of a four-well plate. Plates were then incubated with either Hanks' balanced salt solution (HBSS) or 2 μg/ml netrin-1 in HBSS for 2 h at room temperature. Wells were then blocked for 1 h with 1% bovine serum albumin (BSA; Fisher Scientific) in HBSS and then again with 1% heparin (Sigma) in HBSS. A total of 2.5 × 105 cells from dissociated ganglionic eminences were plated in Neurobasal medium (Gibco) supplemented with 2% B-27 and 2 mM glutamine. Cells were cultured for 2 h at 37°C in 5% CO2, gently washed once with phosphate-buffered saline (PBS), and fixed with 4% PFA in PBS overnight. Nuclei were labeled with 0.5 μg/ml Hoechst 33258 (Sigma) in PBS for 30 min. Experiments were performed on four wild-type and three mutant embryos. Paired, two-tailed t tests were performed to compare genotypes, with differences considered significant at a P value of <0.05.
Ex vivo cortical electroporation.
Cortical electroporation and ex vivo slice culture were performed as described previously (16, 22, 42), with some modifications. Briefly, pregnant female mice (Charles River) were euthanized at E15 with a lethal injection of sodium pentobarbital. Embryos were removed and decapitated, and a 2 μg/μl solution of pCIG-Neogenin or empty pCIG vector (supplemented with 0.5% Brilliant Blue FCF for visualization) was injected, using a Picospritzer II (General Valve Corporation), into the lateral ventricles. Brains were subjected to 10 pulses at 70 V using an ElectroSquarePorator ECM830 (BTX/Genetronics San Diego, CA). Brains were then isolated and embedded in low-melting-point agarose. Agarose-embedded brains were sectioned coronally into 250-μm sections on a Leica VT1000S vibratome. Brain sections were collected and plated on poly-l-lysine-laminin-coated filter membrane inserts placed on top of the culture medium in each well of a six-well dish, as described previously (16, 42). Slices were then cultured for 72 h to assess the degree of migration. Two sections from five to six embryos from three litters were measured, and values were averaged. Migration was assessed by measuring the total area occupied by green fluorescent protein (GFP)-positive cells at both 24-h and 72-h time points. Degree of migration under each condition was assessed by subtracting the area occupied at 24 h from the area occupied at 72 h and then dividing this value by the initial area to obtain a percentage of migration over time. Degree of migration in neogenin-expressing samples relative to the control was obtained by dividing percent migration in neogenin by percent migration of the control. Two-tailed t tests were performed to compare migration between control and neogenin overexpression, with differences considered significant at a P value of <0.05.
RESULTS
Rb deficiency results in specific deregulation of neogenin expression in vivo.
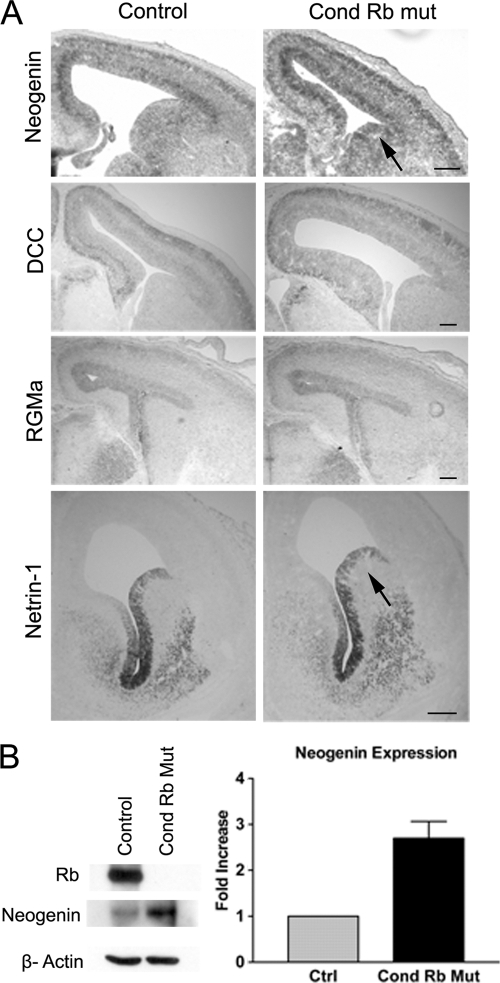
Our previous studies have described a physiological requirement for Rb interacting through E2F to mediate nervous system development (34). Furthermore, we reported that Rb deficiency results in increased expression of mRNA encoding neogenin, a receptor involved in regulating axon guidance and cell migration during neural development (59; for a review, see reference 14).The physiological significance of Rb-mediated regulation of neogenin expression on nervous system development, however, is unknown. To assess the contribution of neogenin to Rb-mediated nervous system development, we first asked if deregulation is unique to neogenin or extends across the family of neogenin ligands and related receptors. To address this question in the telencephalon, sections from Foxg1-cre conditional Rb mutants were subjected to in situ hybridization to examine the expression profiles of neogenin, the closely related receptor deleted in colorectal cancer (DCC), and the known neogenin ligands repulsive guidance molecule (RGMa) and netrin-1 (Fig. 1A). Consistent with our previous microarray and in situ hybridization findings (34), we detected increased neogenin expression throughout the ventral and dorsal telencephalon in conditional Rb mutants. In the ganglionic eminence, a source of migrating interneurons, no difference was detected in the expression patterns of DCC, netrin-1, and RGMa between control and conditional Rb mutants (n = 3) (Fig. 1A). These expression patterns parallel results of other studies that have shown neogenin and netrin-1 overlapping protein expression in the ganglionic eminence (19, 52). Thus, of the members of the netrin signaling pathway, a significant change was detected in the expression only of neogenin within the ventrally derived population of neural precursor cells. To validate increased neogenin levels identified by in situ hybridization, we assessed expression at the protein level within the migrating cell population. Total protein was extracted from the population of ventrally derived neural progenitor cells from three separate embryos for each genotype, and a similar increase in neogenin protein was identified in conditional Rb mutants (Fig. 1B). Efficient excision of the Rb allele in the context of primary ventral neural precursors was confirmed by protein levels (Fig. 1B). Together, these data support the hypothesis that Rb may play an important role in regulating expression of neogenin, a nontraditional E2F target gene.
FIG. 1.
Neogenin is upregulated in the absence of Rb in the developing forebrain. (A) In the absence of Rb (Foxg1-Cre/+; RbloxP/loxP) increased neogenin expression is detected in the subventricular zone, cortex, and striatum compared with results in controls (Foxg1-Cre/+; RbloxP/+). In situ hybridizations were performed on three or more independent samples for DCC, RGMa, netrin-1, and neogenin. Arrows indicate regions of overlapping expression between neogenin and netrin-1 in the ganglionic eminence. (B) Western blot analysis was performed on neural precursor cells isolated from E14.5 Rb conditional mutants and control embryos. Note efficient recombination of the floxed Rb allele, showing no detectable Rb. Rb mutants showed an upregulation of neogenin compared to levels in the control. Densitometric quantification of neogenin expression detected by Western blotting was performed using ImageJ analysis of three independent experiments. Significance was determined through a paired two-tailed t test for the control and conditional Rb mutant (P < 0.05).
E2F3 binds the 5′ locus of neogenin in vivo.
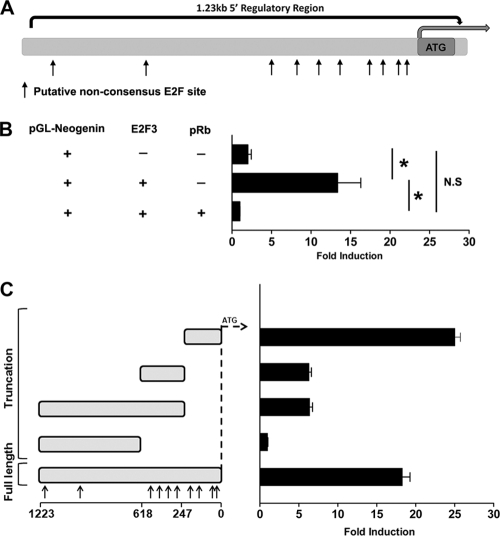
Recent studies have reported that neogenin is among a novel class of atypical E2F target genes regulated in a cell cycle-independent manner (26). E2F1 was shown to be capable of directly inducing neogenin expression independent of growth stimulation; however, no classical E2F binding (TTTSSCGC) site was observed within the 5′ region (26). With the implementation of new bioinformatic techniques, a new broad E2F consensus binding site(s) (BKTSSCGS) has recently been characterized (45). We asked whether E2F could be mediating neogenin expression through one of these newly defined E2F sites contained in its 5′ regulatory region. Using Mulan/rVista software, the 5′ region of the mouse neogenin gene was examined for the presence of broad-spectrum BKTSSCGS E2F motifs (45). Consistent with previous reports, no classical E2F binding sites could be found in the mouse neogenin promoter (26). Closer examination revealed 10 broad E2F binding sites located within 1.23 kb upstream of the translational start site. The majority of these putative E2F sites are clustered within the first 600 bp upstream of the ATG (Fig. 2A).
FIG. 2.
E2F3 interacts with a region containing multiple putative E2F sites within the 5′ regulatory region of neogenin. (A) Putative E2F sites were identified in the 5′ region of the mouse (mm9) Neo1 gene using Mulan/rVista software and confirmed by manual sequence analysis (BKTSSCGS). (B) ChIP was performed on neurospheres isolated from the ganglionic eminence of E14.5 wild-type embryos. Immunoprecipitation was performed using an antibody specific to E2F3 or E2F1, followed by PCR amplification of the indicated regions in the 5′ regulatory region of the neogenin gene. ChIP was performed on three independent cultures per condition. An interaction was detected only with E2F3 at the region containing multiple clustered sites; the putative site at 852 bp showed no specific binding. Immunoprecipitations were performed on independent cultures from three animals.
Prior studies examined the regulation of neogenin expression by E2F1; however, as we have previously described unique roles for E2F3 in nervous system development (34, 36), we asked whether E2F3 interacted with the 5′ regulatory region of the neogenin gene in the context of native chromatin. To address this question, we performed chromatin immunoprecipitation (ChIP) in primary neural progenitor cultures to see if E2F3 associated with the regions containing atypical E2F sites. Chromatin was immunoprecipitated with antibodies to E2F3, followed by PCR with primers designed around the cluster of E2F sites immediately upstream of the translation start and the region containing only a single site 852 bp upstream (Fig. 2). ChIP with the primer set surrounding the cluster demonstrated enrichment of E2F3 binding, with no detectable E2F3 binding at the site at −852 bp (Fig. 2B). Consistent results were obtained from three independent primary cultures. Previous studies using a subtractive microarray analysis revealed that E2F1 could induce neogenin expression in rat embryonic fibroblasts; however, a direct interaction in the neogenin regulatory regions was not shown (26). We therefore asked if E2F1 might physically interact with the putative promoter region of neogenin. To this end we performed ChIP with an antibody directed to E2F1 and examined the region in which E2F3 binding was detected. No enrichment was found in this region (Fig. 2B), suggesting that E2F1 does not interact with the neogenin promoter in precursors from the ventral forebrain. These results demonstrate that, in the context of neural precursor cells, E2F3 specifically binds the neogenin promoter at the region (−600 to ATG) encompassing the multiple putative E2F consensus sites in an in vivo context, and this is consistent with the hypothesis that E2F3 is capable of modulating neogenin gene expression.
Rb/E2F regulates transcriptional activity at the 5′ neogenin-regulatory region.
Given that we observe deregulated neogenin expression in the absence of Rb, we next asked whether Rb regulates this expression through its interaction with E2F. To address this question, we performed in vitro luciferase reporter assays in HEK293T cells. Ideally, these studies should be performed in primary systems; however, overexpression of E2F1 or E2F3 induces a rapid and robust apoptotic response in embryonic tissue and in primary neural precursor cells. As HEK293T cell lines express E1B preventing apoptosis (58), they withstand overexpression of “activating” E2F constructs without undergoing cell death. Thus, all reporter assays were performed in HEK293T cell lines. The neogenin promoter region was amplified from embryonic genomic DNA with primers designed to flank a 1.23-kb region containing the putative E2F binding sites (Fig. 3A). The fragment was then subcloned into a luciferase reporter vector (pGL4.24) which was subsequently transfected into HEK293 cells.
FIG. 3.
The 5′ neogenin promoter is responsive to Rb/E2F regulation. (A) Schematic of the 5′ region of the neogenin gene. The 1.23-kb region isolated and cloned into the pGL4.24 vector contains 10 putative E2F binding sites identified using Dcode/Mulan software. (B) Dual-Glo luciferase (Promega) promoter assays in HEK293T cells utilizing the neogenin promoter reveal that the addition of E2F3 induces a 13-fold induction of neogenin promoter activity. When cells are stimulated with Rb and E2F3, the activation is eliminated, and luciferase levels return to that of promoter alone. (C) Luciferase analysis of truncations of the 5′ region of the neogenin gene. Activity is ablated upon removal of the initial 618 bp. The region between bp 247 and 618 results in slight E2F3 responsiveness; however, the first 247 bp recapitulates full-length promoter activity. *, P < 0.05; N.S., nonsignificant difference.
In the absence of exogenous E2F, the neogenin promoter displayed a minimal level of activation (Fig. 3B). Upon addition of E2F3, however, we observed a strong 13.3-fold increase in luciferase activity, demonstrating that E2F3 is capable of transcriptionally activating the neogenin promoter. We next asked if Rb is capable of repressing E2F-mediated activation of the neogenin promoter. Upon cotransfection of E2F3 and Rb expression plasmids, E2F3-mediated activation of the neogenin promoter was repressed back to basal levels (Fig. 3B). Together these results demonstrate that Rb acts to repress E2F3-mediated activation of the neogenin promoter through interaction in the 5′ regulatory region.
We next sought to determine the specific region in the neogenin promoter in which E2F3 was binding to activate transcription. Examination of the 5′-proximal promoter for both classical/nonclassical E2F binding motifs revealed 10 putative sites within the 1.23 kb examined. To define the essential regions required for E2F-mediated activation, we created multiple truncation constructs at roughly 250-bp intervals from both the 5′ and 3′ ends of the 1.23-kb promoter construct (Fig. 3C). While constructs lacking the regions upstream of bp −247 had no effect on E2F responsiveness, absence of the region from the ATG to bp −618 abolished the ability of E2F to activate transcription (Fig. 3C). This region was most enriched with eight putative E2F sites, and the results suggest that E2F3 is binding one or more of the several clustered sites in this region. To more precisely identify the sites, two further truncation constructs (bp 0 to 247 and bp 247 to 618) were made lacking each of the two clusters of E2F sites within the first 618 bp of the promoter (Fig. 3C). Introduction of the construct containing only the region encompassing bp 247 to 618 upstream of the promoter (Fig. 3C) resulted in slight activation in response to E2F (Fig. 3C). This result recapitulated the slight E2F responsiveness observed in the construct lacking the first 247 bp upstream of the ATG. To determine if the first 247 bp could confer E2F-mediated activation, this 247-bp fragment alone was ligated into the reporter construct, and full E2F responsiveness, equivalent to that of the full-length construct, was obtained. These results demonstrate that the cluster of E2F sites contained in the 247 bp upstream of the ATG are essential for neogenin promoter activation, consistent with the region identified by our E2F3 ChIP (Fig. 2B). These findings support the conclusion that Rb, acting through E2F3, directs the expression of neogenin, an atypical E2F target gene, that functions outside cell cycle progression.
Rb deficiency results in aberrant neuronal migration in the presence of netrin-1.
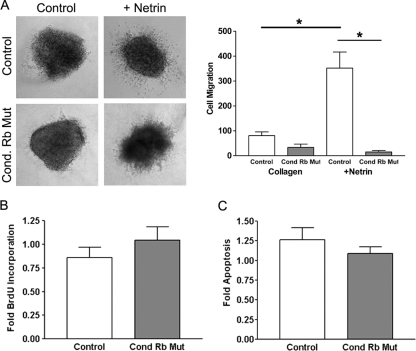
We next sought to determine the functional consequences of deregulation of neogenin expression as a result of the ablation of the Rb gene. We hypothesized that if deregulated expression of neogenin contributes to the aberrant migration of ventrally derived neurons in the conditional Rb mutant, then neural precursor cells should elicit an aberrant response in the presence of neogenin ligand. During mammary gland development, netrin-1-neogenin interactions have been shown to be crucial for proper stabilization of the multipotent progenitor cell layer (19, 51). This interaction may play an analogous role during tangential migration in the developing forebrain. We therefore determined if netrin-1 is capable of influencing migration of MGE-derived cells under wild-type conditions. To effectively address this question, we employed a reductionist in vitro approach using primary neural precursor explants cultured in a collagen matrix. This approach created a defined extracellular environment containing netrin-1 alone and allowed us to determine the effect of the neogenin ligand, netrin, in the absence of other competing signals known to influence migration (32). Explants of ventral ganglionic eminence were microdissected from control and mutant E14.5 cerebral hemispheres and then cultured for 24 h supplemented with netrin-1, after which cell migration from the explant was quantified. We assessed the relative contribution of netrin-1, a ligand demonstrated to elicit neogenin-dependent chemoattractant responses in the developing nervous system. Explants were cultured in collagen, a matrix suitable for assessing chemoattractant responses (29, 38). Both control and conditional Rb mutant explants cultured in collagen alone exhibited modest numbers of cells migrating, with no appreciable difference in migration from either type of explant. In the presence of netrin-1, however, a clear difference was observed (Fig. 4A). While control explants exhibited a 4-fold increase in migration in the presence of netrin-1, there was no significant difference in the number of cells migrating from conditional Rb mutant explants cultured in the presence or absence of netrin-1 (Fig. 4A).
FIG. 4.
Conditional Rb mutants display a defective migratory response to netrin-1. (A) Control (Foxg1-Cre/+; RbloxP/+) and conditional Rb mutant (Foxg1-Cre/+; RbloxP/loxP) MGE explants were cultured in collagen in the absence or presence of recombinant netrin-1. Migration was quantified by counting the individual cell bodies migrating from each explant. Bars in the graph at left represent the mean of the average number of cells migrating from an individual explant ± standard error of the mean. While control cells exhibit a nearly 4-fold increase in migration in the presence of netrin, no difference was observed in conditional Rb mutants between the presence and absence of netrin. Significance was determined through a paired two-tailed t test for explants of the same genotype and a two-tailed t test for explants of different genotypes. *, P < 0.05 (n = 4 embryos per treatment, per genotype; two to three explants were examined per embryo). (B and C) Cells from the ganglionic eminence of control and conditional Rb mutants were dissected and cultured as single-cell preparations in the presence or absence of netrin-1. Quantification of the proportion of cells in S phase (BrdU) or dying (Hoechst) reveals no change upon addition of netrin-1 in either genotype. Three separate embryos were analyzed in quadruplicate for both the control and conditional Rb mutant.
Netrin-1 has been hypothesized to mediate cell proliferation and cell death (37; for a review, see reference 11). We therefore verified that the differences detected in migration were not due to either of these processes. To dissect the potential contribution of altered proliferation or cell death, netrin-1 treatment was performed under conditions that recapitulate those used in our in vitro explant culture in order to ensure that there are no changes in proliferation and cell death under those specific conditions. To examine proliferation, cultures were treated with bromodeoxyuridine (BrdU), and the proportion of cells in S phase of the cell cycle was counted. In three independent control and mutant cultures, netrin-1 treatment did not significantly impact the number of proliferating cells (Fig. 4B). Assessment of chromatin condensation revealed no significant change in cell death between control and mutant cultures upon addition of netrin-1 (n = 3) (Fig. 4C). These results suggest that the increased number of cells migrating in response to netrin-1 is not a consequence of increased cell proliferation, nor can the absence of migration in the conditional Rb mutant be attributed to increased cell death. Thus, these data support a model whereby netrin-1 is capable of influencing migration of ventrally derived progenitors, an effect that is not observed in the conditional Rb mutants. These results suggest that ventrally derived progenitors from Rb mutants are inherently unable to elicit the appropriate migratory response to netrin-1 itself.
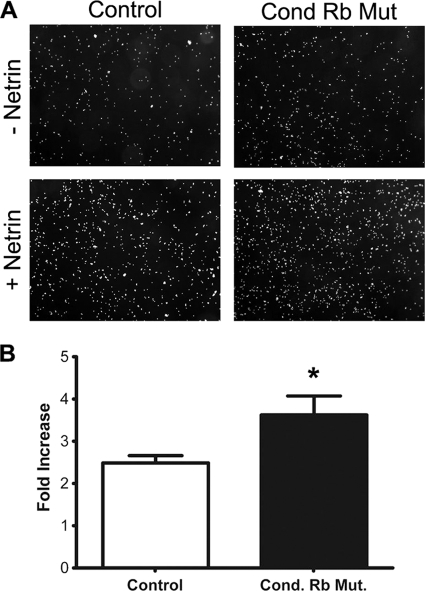
Increased adhesion to substrate-bound netrin-1 in conditional Rb mutants.
A previous study demonstrated that netrin-1 and neogenin interact to mediate adhesion in the mammary gland (51). Given that we observe reduced migration in response to netrin-1 in the conditional Rb mutant, where neogenin expression is increased, we determined if the increased amount of neogenin present would increase adhesion of neural precursor cells. To address this, adhesion assays were performed which have been previously used to assess netrin-neogenin-mediated adhesion in fibroblasts (51) and adapted the assay for neural precursor cells (50). Using this assay, we examined the capacity of ventrally derived precursors from control and conditional Rb mutants to adhere specifically to immobilized netrin-1. The ventral telencephalon from E14.5 control and conditional Rb mutants was dissected and dissociated into single-cell suspensions. Cells were quantified, and then equal numbers were plated and allowed to adhere to culture dishes preadsorbed with nitrocellulose alone or with netrin-1 and nitrocellulose. After 2 h, cells were washed and fixed, and cell adhesion was quantified. Data were represented as the fold increase in adhesion upon netrin-1 treatment to eliminate the experimental variability observed from each independent assay. Independent experiments, however, produced highly consistent results (Table 1). In the presence of netrin-1, conditional Rb mutants displayed a 3.6-fold increase in adhesion, whereas control littermates displayed a significantly smaller 2.5-fold increase (Fig. 5). Our findings suggest that Rb-deficient neural precursor cells have increased adhesive properties, consistent with previous findings revealing a role for netrin-neogenin in mediating cellular adhesion (50, 51). Given the elevated levels of neogenin expression detected, increased adhesion in response to netrin-1 may be a key contributing factor to the migration defect present in Rb-deficient brains.
TABLE 1.
Rb-deficient neural precursors show an increased propensity to adhere to substrate bound netrin-1
| Expta | Embryo | Genotype | Cell adhesion (avg no. of cells/field)b |
Fold increasec | |
|---|---|---|---|---|---|
| Without netrin | With netrin | ||||
| CTL | MGA57 | Foxg1-Cre/+; RbloxP/+ | 382 | 925 | 2.421466 |
| CTL | MGA62 | Foxg1-Cre/+; RbloxP/+ | 235.5 | 513 | 2.178344 |
| CTL | MGA132 | Foxg1-Cre/+; RbloxP/+ | 121.5 | 285.5 | 2.349794 |
| CTL | MGA133 | Foxg1-Cre/+; RbloxP/+ | 127.75 | 291.4 | 2.281018 |
| Rb−/− | MGA134 | Foxg1-Cre/+; RbloxP/RbloxP | 100 | 316.25 | 3.1625 |
| Rb−/− | MGA136 | Foxg1-Cre/+; RbloxP/RbloxP | 113.5 | 504.75 | 4.447137 |
| Rb−/− | MGA59 | Foxg1-Cre/+; RbloxP/RbloxP | 464 | 1635 | 3.523707 |
CTL, control.
Cells from the ganglionic eminence of control and conditional Rb mutants were dissected at E14.5. Cells were allowed to adhere to netrin-1 or noncoated wells for 2 h before they were fixed and stained. Cells were then imaged, and nuclei were counted. The total number of nuclei per field was averaged for each condition. In the absence of Rb, cells from the ganglionic eminence display more significant adherence to netrin-1 than cells from control littermates.
Fold increase represents the increase in adhesion from noncoated to netrin-1-coated wells.
FIG. 5.
Rb-deficient neural precursors show an increased propensity to adhere to substrate-bound netrin-1. (A) Cells from the ganglionic eminence of control (Foxg1-Cre/+; RbloxP/+) and conditional Rb mutants (Foxg1-Cre/+; RbloxP/loxP) were dissected at E14.5. Cells were allowed to adhere to netrin-1 or noncoated wells for 2 h and were then fixed and stained. Cells were then imaged, and nuclei were counted. In the absence of Rb, cells from the ganglionic eminence display more significant adherence to netrin-1 than cells from control littermates. (B) Fold increase represents the increase in adhesion from noncoated to netrin-1-coated wells. Error bars represent standard error of the mean (n = 4 for the controls and n = 3 for conditional Rb mutants). Significance was determined through a paired two-tailed t test for control and conditional Rb mutant cultures. *, P < 0.05.
Increased neogenin impedes neuronal migration.
While we have demonstrated that Rb is capable of regulating neogenin transcription through E2F in the developing nervous system, the consequence of increased neogenin expression remains unknown. We therefore asked if upregulation of neogenin as found in the Rb-deficient forebrain was sufficient to disrupt the migration of MGE-derived neurons. To determine whether increased neogenin expression could perturb neuronal migration, we performed ex vivo cortical electroporation of the full-length neogenin or a control internal ribosome entry site (IRES)-GFP vector into the ventral telencephalon of wild-type E15.5 embryonic brains (22). Following electroporation, brains were cultured as slices for 72 h to observe migration. Expression of the plasmid carrying GFP-positive cells was first observed at 24 h postelectroporation and subsequently at 72 h. At 24 h, brains electroporated with either control or neogenin-containing plasmids displayed GFP-positive cells lining the ventricular zone of the ventral forebrain, with no difference observed between controls and neogenin-electroporated cells (Fig. 6A, red). At 72 h, numerous GFP-positive cells from control slices were observed to have migrated considerably from their initial position within the ventricular zone. GFP-positive cells from neogenin slices, however, remained clustered within a similar band along the ventricular zone (Fig. 6A, green). Migration was quantified by measuring the total area occupied by GFP-positive cells at the endpoint, subtracting the initial area, and then dividing this value by the initial area to obtain the percent increase in migration [(total migration − initial migration)/total migration], and values were normalized to the percentage of the control. Upregulation of neogenin resulted in a 77% decrease in migration (P < 0.05) compared to that of control-electroporated embryos (Fig. 6B). We conclude that increased expression of neogenin by cells in the ganglionic eminence results in reduced migration of precursors away from the ventricular zone, paralleling the migration defect observed in the Rb-deficient forebrain.
FIG. 6.
Increased neogenin expression impairs migration of neuroblasts from the subventricular zone. (A) Ex vivo overexpression of control IRES-GFP- or neogenin-IRES-GFP-expressing plasmids in E15 embryos. Embryos were sectioned at 250 μm and plated on poly-l-lysine-laminin-coated inserts. Cells were visualized at 24 h postelectroporation (red) to determine their baseline vector expression and migration. Cells were imaged again at 72 h postelectroporation to assess the degree to which they migrated (green). Under both control and neogenin-overexpressing conditions cells expressing the plasmids initially line the ventricle (red, arrows). After migration (lower panels) the cells move away from the ventricular zone into the striatum (green, arrows) in the control; however, in the mutant they fail to shift position. (B) Quantification of the capacity of cells to migrate depicted in panel A. In order to quantify migration, fold increase was obtained by calculating (total migration − initial migration)/total migration and then normalizing values to wild-type migration levels. Error bars represent standard error of the mean (n = 5 for the control and n = 6 for the mutant, with two sections per embryo). Significance was determined through a paired two-tailed t test of control versus neogenin-overexpressing slices. *, P < 0.05.
Taken together, our results demonstrate a function for the Rb pathway in regulating expression of a nontraditional E2F target gene, neogenin, during neuronal migration. Furthermore, we demonstrate that aberrant neogenin expression, similar to that found in conditional Rb mutants, leads to impaired migration. Overall, these finding support the conclusion that Rb/E2F regulation of neogenin expression, an atypical target, influences appropriate neural development in a manner beyond traditional regulation of the cell cycle.
DISCUSSION
Here, we demonstrate the existence of an Rb/E2F-mediated molecular mechanism regulating expression of an atypical E2F target gene, neogenin. First, we have shown that neogenin expression is deregulated in the absence of Rb at the mRNA and protein levels in neural precursor cells. While neogenin has previously been shown to be an E2F-regulated target gene in vitro, here we complement previous findings by demonstrating that E2F3 is capable of activating neogenin expression, and we extend these findings by demonstrating the binding of E2F3 to the 5′ regulatory region of the neogenin promoter in neural precursor cells. It is possible that other E2Fs are contributing to regulation of neogenin expression in different biological contexts; however, given that E2F3 has previously been implicated in multiple aspects of nervous system development in vivo (9, 34, 36), these observations lend further support to the idea that E2F3 regulation is significant in this context. Finally, we demonstrate that E2F transcriptional regulation of neogenin is, in turn, strongly repressed by Rb activity. These results, along with our data regarding increased neogenin expression among migrating neurons in the absence of Rb, suggest a direct role for the regulation of neogenin by the Rb/E2F pathway in the developing forebrain.
Previously, we have shown that the Rb/E2F pathway mediates migration of a population of precursors from the ventral telencephalon during nervous system development (16, 34). Here, we provide mechanistic insight into this process, showing that in the absence of Rb, migrating ventral precursors exhibit a decreased response to the neogenin ligand, netrin-1. Consistent with decreased migration, we observe increased adhesion of ventrally derived Rb-deficient precursors to substrate-bound netrin-1. This suggests a mechanism in which increased neogenin expression results in augmented neuronal adhesion leading to the decreased migration. Our ex vivo manipulation of neogenin expression resulted in a defect in neuronal migration similar to that seen in the conditional Rb mutant (16). While we favor our model as a hypothesis to explain how Rb/E2F regulates migration, we note that neogenin is likely only one of many factors contributing to Rb-mediated neuronal migration. Indeed, neuronal migration is a complex phenomenon involving multiple genes and genetic pathways (24). Through our previous microarray analysis we identified several known genes that regulate migration in the central nervous system, and therefore dysfunction in their expression could also be contributing to several facets of the observed migration defect in the conditional Rb mutants (34). While the extent to which deregulated neogenin contributes to the migration defect in Rb mutants is unknown, our studies reveal that overexpression of neogenin perturbs neuroblast migration in wild-type tissue (Fig. 6). Indeed, a rescue experiment in our conditional Rb mutants would be challenging as reducing neogenin expression to physiological levels without causing a complete knockdown would likely lead to variable results. As presented, our results provide strong evidence that, by regulating neogenin expression, Rb/E2F has an important physiological role beyond regulation of the cell cycle machinery, a phenomenon that has not yet been reported. It is probable that Rb is involved in the regulation of multiple genes, which through distinct mechanisms contribute to the regulation of neuronal migration.
The idea that the Rb/E2F pathway can regulate genes outside the prototypical cell cycle machinery in the context of nervous system development may also broaden its role in tumorigenesis. As Rb is the first identified tumor suppressor, intense interest has been focused on defining the molecular mechanisms through which it mediates tumor suppression. While early studies established the model that Rb-mediated tumor suppression is the result of its restraint of E2F transcription factors at the G1/S transition, (reviewed in reference 55), more recent studies suggest that the role of Rb as a tumor suppressor is more complex than originally hypothesized. Indeed, roles for Rb in maintaining genome stability and promoting senescence have broadened the scope and complexity of Rb-mediated tumor suppression (reviewed in references 21 and 30). Further deregulation of the Rb pathway in cancer has been traditionally associated with sustained proliferation; however, Rb mutations are frequently found in metastatic cancers, including small-cell lung carcinoma and osteosarcoma, as well as invasive poor-prognosis glioblastomas (reviewed in reference 12).
Having demonstrated a novel role for the Rb/E2F pathway in mediating expression of a specific gene involved in neuronal migration, the data presented here raise the possibility that Rb activity could contribute to the regulation of other cellular processes involved in cancer beyond regulation of cell division. Recent studies employing conditional transgenic alleles to remove tumor suppressor genes specifically in adult neural precursor cells have shown the important nonoverlapping roles for Rb, PTEN, Nf1, and p53 (1, 27). The study of Jacques et al. correlated the ablation of Rb gene expression in the adult subventricular zone with the appearance of primitive neuroectodermal tumors (pNETS). These tumors display significant differentiation across all three neural lineages and ectopic infiltration of surrounding brain tissue. This lends itself to the hypothesis that Rb may also regulate the differentiation and localization of these tumor cells. Many families of genes which mediate neuronal migration, such as the netrin signaling axis (18, 48), have been implicated in multiple aspects of cancer and tumorigenesis. Ligands and receptors from these migration pathways are frequently found deregulated or are lost altogether in numerous cancers (reviewed in references 2 and 8). Our findings demonstrate a key role for Rb/E2F-regulated expression of neogenin. Contributing to neuronal migration gives rise to the possibility that Rb-mediated mechanisms may regulate expression of migration-related genes during steady-state events such as neurogenesis. The deregulation of these processes may contribute to facets of tumor progression that expand from the typical aberrant S-phase entry associated with Rb loss of function. Further exploration of this hypothesis in the context of tumorigenesis could lend new insight into our understanding of the mechanisms of Rb-mediated tumor suppression.
In conclusion, our results suggest that Rb/E2F is required for the regulation of neogenin during neuronal migration. Further, these results provide strong support to our overall hypothesis that Rb acts through E2F to mediate events distinct from cell cycle progression by regulating transcription of genes that are not classical E2F targets.
Acknowledgments
We thank Philippe Monnier, Carol Schuurmans, Silvia Arber, Helen Cooper, and Elke Stein for providing valuable reagents. We thank Vladimir Ruzhynsky, Angela Nguyen, Jason G. MacLaurin, and David Douda for excellent technical assistance.
This work was supported by a CIHR grant to R.S.S. M.G.A. is supported by awards from OGSST and HSFO, K.A.M. and L.M.J. are recipients of a CIHR Canada Graduate Doctoral Research Award, D.D.T. holds an OGSST award, and T.E.K. holds an FRSQ Chercheur National Award and is a Killam Foundation Scholar.
Footnotes
Published ahead of print on 8 November 2010.
REFERENCES
- 1.Alcantara Llaguno, S., J. Chen, C. H. Kwon, E. L. Jackson, Y. Li, D. K. Burns, A. Alvarez-Buylla, and L. F. Parada. 2009. Malignant astrocytomas originate from neural stem/progenitor cells in a somatic tumor suppressor mouse model. Cancer Cell 15:45-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arakawa, H. 2004. Netrin-1 and its receptors in tumorigenesis. Nat. Rev. Cancer. 4:978-987. [DOI] [PubMed] [Google Scholar]
- 3.Balciunaite, E., A. Spektor, N. H. Lents, H. Cam, H. Te Riele, A. Scime, M. A. Rudnicki, R. Young, and B. D. Dynlacht. 2005. Pocket protein complexes are recruited to distinct targets in quiescent and proliferating cells. Mol. Cell. Biol. 25:8166-8178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bieda, M., X. Xu, M. A. Singer, R. Green, and P. J. Farnham. 2006. Unbiased location analysis of E2F1-binding sites suggests a widespread role for E2F1 in the human genome. Genome Res. 16:595-605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Black, E. P., T. Hallstrom, H. K. Dressman, M. West, and J. R. Nevins. 2005. Distinctions in the specificity of E2F function revealed by gene expression signatures. Proc. Natl. Acad. Sci. U. S. A. 102:15948-15953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burkhart, D. L., and J. Sage. 2008. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer. 8:671-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cam, H., E. Balciunaite, A. Blais, A. Spektor, R. C. Scarpulla, R. Young, Y. Kluger, and B. D. Dynlacht. 2004. A common set of gene regulatory networks links metabolism and growth inhibition. Mol. Cell 16:399-411. [DOI] [PubMed] [Google Scholar]
- 8.Chedotal, A., G. Kerjan, and C. Moreau-Fauvarque. 2005. The brain within the tumor: new roles for axon guidance molecules in cancers. Cell Death Differ. 12:1044-1056. [DOI] [PubMed] [Google Scholar]
- 9.Chen, D., R. Opavsky, M. Pacal, N. Tanimoto, P. Wenzel, M. W. Seeliger, G. Leone, and R. Bremner. 2007. Rb-mediated neuronal differentiation through cell-cycle-independent regulation of E2f3a. PLoS Biol. 5:e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen, H. Z., S. Y. Tsai, and G. Leone. 2009. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nat. Rev. Cancer 9:785-797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cirulli, V., and M. Yebra. 2007. Netrins: beyond the brain. Nat. Rev. Mol. Cell Biol. 8:296-306. [DOI] [PubMed] [Google Scholar]
- 12.Classon, M., and E. Harlow. 2002. The retinoblastoma tumour suppressor in development and cancer. Nat. Rev. Cancer 2:910-917. [DOI] [PubMed] [Google Scholar]
- 13.Colombo, E., P. Collombat, G. Colasante, M. Bianchi, J. Long, A. Mansouri, J. L. Rubenstein, and V. Broccoli. 2007. Inactivation of Arx, the murine ortholog of the X-linked lissencephaly with ambiguous genitalia gene, leads to severe disorganization of the ventral telencephalon with impaired neuronal migration and differentiation. J. Neurosci. 27:4786-4798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Vries, M., and H. M. Cooper. 2008. Emerging roles for neogenin and its ligands in CNS development. J. Neurochem. 106:1483-1492. [DOI] [PubMed] [Google Scholar]
- 15.Dimova, D. K., O. Stevaux, M. V. Frolov, and N. J. Dyson. 2003. Cell cycle-dependent and cell cycle-independent control of transcription by the Drosophila E2F/RB pathway. Genes Dev. 17:2308-2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferguson, K. L., K. A. McClellan, J. L. Vanderluit, W. C. McIntosh, C. Schuurmans, F. Polleux, and R. S. Slack. 2005. A cell-autonomous requirement for the cell cycle regulatory protein, Rb, in neuronal migration. EMBO J. 24:4381-4391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ferguson, K. L., J. L. Vanderluit, J. M. Hebert, W. C. McIntosh, E. Tibbo, J. G. MacLaurin, D. S. Park, V. A. Wallace, M. Vooijs, S. K. McConnell, and R. S. Slack. 2002. Telencephalon-specific Rb knockouts reveal enhanced neurogenesis, survival and abnormal cortical development. EMBO J. 21:3337-3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fitamant, J., C. Guenebeaud, M. M. Coissieux, C. Guix, I. Treilleux, J. Y. Scoazec, T. Bachelot, A. Bernet, and P. Mehlen. 2008. Netrin-1 expression confers a selective advantage for tumor cell survival in metastatic breast cancer. Proc. Natl. Acad. Sci. U. S. A. 105:4850-4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitzgerald, D. P., S. J. Cole, A. Hammond, C. Seaman, and H. M. Cooper. 2006. Characterization of neogenin-expressing neural progenitor populations and migrating neuroblasts in the embryonic mouse forebrain. Neuroscience 142:703-716. [DOI] [PubMed] [Google Scholar]
- 20.Gad, J. M., S. L. Keeling, A. F. Wilks, S. S. Tan, and H. M. Cooper. 1997. The expression patterns of guidance receptors, DCC and neogenin, are spatially and temporally distinct throughout mouse embryogenesis. Dev. Biol. 192:258-273. [DOI] [PubMed] [Google Scholar]
- 21.Goodrich, D. W. 2006. The retinoblastoma tumor-suppressor gene, the exception that proves the rule. Oncogene 25:5233-5243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hand, R., D. Bortone, P. Mattar, L. Nguyen, J. I. Heng, S. Guerrier, E. Boutt, E. Peters, A. P. Barnes, C. Parras, C. Schuurmans, F. Guillemot, and F. Polleux. 2005. Phosphorylation of Neurogenin2 specifies the migration properties and the dendritic morphology of pyramidal neurons in the neocortex. Neuron 48:45-62. [DOI] [PubMed] [Google Scholar]
- 23.Hebert, J. M., and S. K. McConnell. 2000. Targeting of cre to the Foxg1 (BF-1) locus mediates loxP recombination in the telencephalon and other developing head structures. Dev. Biol. 222:296-306. [DOI] [PubMed] [Google Scholar]
- 24.Huang, Z. 2009. Molecular regulation of neuronal migration during neocortical development. Mol. Cell Neurosci. 42:11-22. [DOI] [PubMed] [Google Scholar]
- 25.Ishida, S., E. Huang, H. Zuzan, R. Spang, G. Leone, M. West, and J. R. Nevins. 2001. Role for E2F in control of both DNA replication and mitotic functions as revealed from DNA microarray analysis. Mol. Cell. Biol. 21:4684-4699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Iwanaga, R., H. Komori, S. Ishida, N. Okamura, K. Nakayama, K. I. Nakayama, and K. Ohtani. 2006. Identification of novel E2F1 target genes regulated in cell cycle-dependent and independent manners. Oncogene 25:1786-1798. [DOI] [PubMed] [Google Scholar]
- 27.Jacques, T. S., A. Swales, M. J. Brzozowski, N. V. Henriquez, J. M. Linehan, Z. Mirzadeh, C. O'Malley, H. Naumann, A. Alvarez-Buylla, and S. Brandner. 2010. Combinations of genetic mutations in the adult neural stem cell compartment determine brain tumour phenotypes. EMBO J. 29:222-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jin, V. X., A. Rabinovich, S. L. Squazzo, R. Green, and P. J. Farnham. 2006. A computational genomics approach to identify cis-regulatory modules from chromatin immunoprecipitation microarray data—a case study using E2F1. Genome Res. 16:1585-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy, T. E., T. Serafini, J. R. de la Torre, and M. Tessier-Lavigne. 1994. Netrins are diffusible chemotropic factors for commissural axons in the embryonic spinal cord. Cell 78:425-435. [DOI] [PubMed] [Google Scholar]
- 30.Liu, H., B. Dibling, B. Spike, A. Dirlam, and K. Macleod. 2004. New roles for the RB tumor suppressor protein. Curr. Opin. Genet. Dev. 14:55-64. [DOI] [PubMed] [Google Scholar]
- 31.Ma, Y., R. Croxton, R. L. Moorer, Jr., and W. D. Cress. 2002. Identification of novel E2F1-regulated genes by microarray. Arch. Biochem. Biophys. 399:212-224. [DOI] [PubMed] [Google Scholar]
- 32.Marin, O., and J. L. Rubenstein. 2001. A long, remarkable journey: tangential migration in the telencephalon. Nat. Rev. Neurosci. 2:780-790. [DOI] [PubMed] [Google Scholar]
- 33.Marino, S., M. Vooijs, H. van Dergulden, J. Jonkers, and A. Berns. 2000. Induction of medulloblastomas in p53-null mutant mice by somatic inactivation of Rb in the external granular layer cells of the cerebellum. Genes Dev. 14:994-1004. [PMC free article] [PubMed] [Google Scholar]
- 34.McClellan, K. A., V. A. Ruzhynsky, D. N. Douda, J. L. Vanderluit, K. L. Ferguson, D. Chen, R. Bremner, D. S. Park, G. Leone, and R. S. Slack. 2007. Unique requirement for Rb/E2F3 in neuronal migration: evidence for cell cycle-independent functions. Mol. Cell. Biol. 27:4825-4843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.McClellan, K. A., and R. S. Slack. 2007. Specific in vivo roles for E2Fs in differentiation and development. Cell Cycle 6:2917-2927. [DOI] [PubMed] [Google Scholar]
- 36.McClellan, K. A., J. L. Vanderluit, L. M. Julian, M. G. Andrusiak, D. Dugal-Tessier, D. S. Park, and R. S. Slack. 2009. The p107/E2F pathway regulates fibroblast growth factor 2 responsiveness in neural precursor cells. Mol. Cell. Biol. 29:4701-4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mehlen, P., and C. Furne. 2005. Netrin-1: when a neuronal guidance cue turns out to be a regulator of tumorigenesis. Cell Mol. Life Sci. 62:2599-2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Metin, C., D. Deleglise, T. Serafini, T. E. Kennedy, and M. Tessier-Lavigne. 1997. A role for netrin-1 in the guidance of cortical efferents. Development 124:5063-5074. [DOI] [PubMed] [Google Scholar]
- 39.Muller, H., A. P. Bracken, R. Vernell, M. C. Moroni, F. Christians, E. Grassilli, E. Prosperini, E. Vigo, J. D. Oliner, and K. Helin. 2001. E2Fs regulate the expression of genes involved in differentiation, development, proliferation, and apoptosis. Genes Dev. 15:267-285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Niederkofler, V., R. Salie, M. Sigrist, and S. Arber. 2004. Repulsive guidance molecule (RGM) gene function is required for neural tube closure but not retinal topography in the mouse visual system. J. Neurosci. 24:808-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Polager, S., Y. Kalma, E. Berkovich, and D. Ginsberg. 2002. E2Fs up-regulate expression of genes involved in DNA replication, DNA repair and mitosis. Oncogene 21:437-446. [DOI] [PubMed] [Google Scholar]
- 42.Polleux, F., and A. Ghosh. 2002. The slice overlay assay: a versatile tool to study the influence of extracellular signals on neuronal development. Sci. STKE 2002:pl9. [DOI] [PubMed] [Google Scholar]
- 43.Pozas, E., and C. F. Ibanez. 2005. GDNF and GFRalpha1 promote differentiation and tangential migration of cortical GABAergic neurons. Neuron 45:701-713. [DOI] [PubMed] [Google Scholar]
- 44.Pozas, E., M. Pascual, K. T. Nguyen Ba-Charvet, P. Guijarro, C. Sotelo, A. Chedotal, J. A. Del Rio, and E. Soriano. 2001. Age-dependent effects of secreted Semaphorins 3A, 3F, and 3E on developing hippocampal axons: in vitro effects and phenotype of Semaphorin 3A (−/−) mice. Mol. Cell Neurosci. 18:26-43. [DOI] [PubMed] [Google Scholar]
- 45.Rabinovich, A., V. X. Jin, R. Rabinovich, X. Xu, and P. J. Farnham. 2008. E2F in vivo binding specificity: comparison of consensus versus nonconsensus binding sites. Genome Res. 18:1763-1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ren, B., H. Cam, Y. Takahashi, T. Volkert, J. Terragni, R. A. Young, and B. D. Dynlacht. 2002. E2F integrates cell cycle progression with DNA repair, replication, and G2/M checkpoints. Genes Dev. 16:245-256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Reference deleted.
- 48.Rodrigues, S., O. De Wever, E. Bruyneel, R. J. Rooney, and C. Gespach. 2007. Opposing roles of netrin-1 and the dependence receptor DCC in cancer cell invasion, tumor growth and metastasis. Oncogene 26:5615-5625. [DOI] [PubMed] [Google Scholar]
- 49.Shan, B., C. Y. Chang, D. Jones, and W. H. Lee. 1994. The transcription factor E2F-1 mediates the autoregulation of RB gene expression. Mol. Cell. Biol. 14:299-309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shekarabi, M., S. W. Moore, N. X. Tritsch, S. J. Morris, J. F. Bouchard, and T. E. Kennedy. 2005. Deleted in colorectal cancer binding netrin-1 mediates cell substrate adhesion and recruits Cdc42, Rac1, Pak1, and N-WASP into an intracellular signaling complex that promotes growth cone expansion. J. Neurosci. 25:3132-3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Srinivasan, K., P. Strickland, A. Valdes, G. C. Shin, and L. Hinck. 2003. Netrin-1/neogenin interaction stabilizes multipotent progenitor cap cells during mammary gland morphogenesis. Dev. Cell 4:371-382. [DOI] [PubMed] [Google Scholar]
- 52.Stanco, A., C. Szekeres, N. Patel, S. Rao, K. Campbell, J. A. Kreidberg, F. Polleux, and E. S. Anton. 2009. Netrin-1-α3β1 integrin interactions regulate the migration of interneurons through the cortical marginal zone. Proc. Natl. Acad. Sci. U. S. A. 106:7595-7600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vooijs, M., M. van der Valk, H. te Riele, and A. Berns. 1998. Flp-mediated tissue-specific inactivation of the retinoblastoma tumor suppressor gene in the mouse. Oncogene 17:1-12. [DOI] [PubMed] [Google Scholar]
- 54.Wallace, V. A., and M. C. Raff. 1999. A role for Sonic hedgehog in axon-to-astrocyte signalling in the rodent optic nerve. Development 126:2901-2909. [DOI] [PubMed] [Google Scholar]
- 55.Weinberg, R. A. 1995. The retinoblastoma protein and cell cycle control. Cell 81:323-330. [DOI] [PubMed] [Google Scholar]
- 56.Weinmann, A. S., S. M. Bartley, T. Zhang, M. Q. Zhang, and P. J. Farnham. 2001. Use of chromatin immunoprecipitation to clone novel E2F target promoters. Mol. Cell. Biol. 21:6820-6832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Weinmann, A. S., P. S. Yan, M. J. Oberley, T. H. Huang, and P. J. Farnham. 2002. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 16:235-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.White, E., P. Sabbatini, M. Debbas, W. S. Wold, D. I. Kusher, and L. R. Gooding. 1992. The 19-kilodalton adenovirus E1B transforming protein inhibits programmed cell death and prevents cytolysis by tumor necrosis factor alpha. Mol. Cell. Biol. 12:2570-2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yamashita, T., B. K. Mueller, and K. Hata. 2007. Neogenin and repulsive guidance molecule signaling in the central nervous system. Curr. Opin. Neurobiol. 17:29-34. [DOI] [PubMed] [Google Scholar]
- 60.Young, A. P., R. Nagarajan, and G. D. Longmore. 2003. Mechanisms of transcriptional regulation by Rb-E2F segregate by biological pathway. Oncogene 22:7209-7217. [DOI] [PubMed] [Google Scholar]