Abstract
Proper gene expression relies on a class of ubiquitously expressed, uridine-rich small nuclear RNAs (snRNAs) transcribed by RNA polymerase II (RNAPII). Vertebrate snRNAs are transcribed from a unique promoter, which is required for proper 3′-end formation, and cleavage of the nascent transcript involves the activity of a poorly understood set of proteins called the Integrator complex. To examine 3′-end formation in Drosophila melanogaster, we developed a cell-based reporter that monitors aberrant 3′-end formation of snRNA through the gain in expression of green fluorescent protein (GFP). We used this reporter in Drosophila S2 cells to determine requirements for U7 snRNA 3′-end formation and found that processing was strongly dependent upon nucleotides located within the 3′ stem-loop as well as sequences likely to comprise the Drosophila equivalent of the vertebrate 3′ box. Substitution of the actin promoter for the snRNA promoter abolished proper 3′-end formation, demonstrating the conserved requirement for an snRNA promoter in Drosophila. We tested the requirement for all Drosophila Integrator subunits and found that Integrators 1, 4, 9, and 11 were essential for 3′-end formation and that Integrators 3 and 10 may be dispensable for processing. Depletion of cleavage and polyadenylation factors or of histone pre-mRNA processing factors did not affect U7 snRNA processing efficiency, demonstrating that the Integrator complex does not share components with the mRNA 3′-end processing machinery. Finally, flies harboring mutations in either Integrator 4 or 7 fail to complete development and accumulate significant levels of misprocessed snRNA in the larval stages.
In eukaryotes, the major transcripts produced by RNA polymerase II (RNAPII) include the polyadenylated [poly(A)+] mRNAs, the replication-dependent histone mRNAs, and the Sm class of small nuclear RNAs (snRNAs). The 3′ ends of these three general classes of RNAs are all formed by cotranscriptional cleavage, but each one has a distinct mechanism for 3′-end formation (for reviews, see references 29 and 32). In poly(A)+ and histone pre-mRNAs there are conserved upstream and downstream sequences that flank the cleavage site; factors bind to these sites and then recruit additional factors that initiate cleavage (53). In the case of poly(A)+ pre-mRNA, the upstream element is the canonical AAUAAA polyadenylation signal (PAS) and the downstream sequence is the G/U-rich downstream element (DSE). Recognition of the PAS is carried out by the cleavage and polyadenylation specificity complex (CPSF) component CPSF160 via its RNA recognition motifs (RRM) (36), whereas the DSE is bound by the RRM of the cleavage stimulation factor (CstF) component CstF64 (28). Subsequent to this recognition event is recruitment of additional factors that activate the endonucleolytic cleavage between the PAS and the DSE.
Histone pre-mRNA contains a distinct set of flanking elements. Upstream of the cleavage site is a conserved stem-loop structure (SL) and downstream a purine-rich element called the histone downstream element (HDE) (reviewed in reference 26). The SL is bound by the stem-loop binding protein (SLBP) (52), while the HDE base pairs with the U7 small nuclear RNA (35). Following these two recognition events, the same factors required for cleavage of poly(A)+ RNA, including a cleavage factor containing at least CPSF73, CPSF100, and a large scaffold protein called Symplekin, are recruited to cleave histone pre-mRNA. In Drosophila melanogaster, Symplekin, CPSF73, and CPSF100 form a stable complex which likely comprises the cores of both cleavage factors (49). CPSF73 is the catalytic component of the cleavage factor since it can be cross-linked to the cleavage site in histone pre-mRNA, and the recently published crystal structure of this protein definitively identifies it as a member of the zinc-dependent hydrolases within the β-lactamase family, containing a β-CASP motif, capable of cleaving the phosphodiester bond present in RNA (4, 7, 30).
In comparison to histone and poly(A)+ pre-mRNA processing, the mechanism of snRNA 3′-end formation is less well understood (reviewed in references 33, 19, and 11). Work in human cells and Xenopus laevis oocytes identified an AU-rich cis-acting element located 9 to 19 nucleotides (nt) downstream of the 3′ end of the mature transcript, which is termed the 3′ box (18). Mutations within the 3′ box of human snRNAs demonstrate a surprising degree of sequence flexibility in that no single point mutation results in a significant reduction in the efficiency of 3′-end formation (1). A unique property of vertebrate snRNA genes is that their 3′-end formation is dependent on the promoter driving transcription. Replacement of snRNA promoters with other RNAPII promoters results in a nearly total loss of proper 3′-end formation of the snRNA (6, 20). This suggests that the complex that carries out the 3′-end cleavage reaction is distinct from that used by other RNAPII genes and is loaded early in the transcription cycle.
Recently, the elusive complex responsible for snRNA 3′-end formation was biochemically purified from human cells and termed the “Integrator” complex (3). This complex consists of at least 12 separate Integrator subunits (IntS) in humans and 11 in Drosophila (reviewed in reference 5). Two subunits, the Integrator 9 (IntS9) and IntS11 proteins, are homologues of CPSF100 and CPSF73, respectively (10), suggesting that snRNA 3′ ends are formed by cotranscriptional cleavage. IntS9 and IntS11 exist as a heterodimer, with IntS11 likely to be the catalytic endonuclease responsible for cleaving the snRNA (10). Orthologues of the Integrator subunits have been identified in both metazoans and plants but are conspicuously absent in yeast, an observation that is consistent with the Nrd1/Nab3/Sen1 complex mediating the 3′-end formation of snRNA genes in those organisms (47, 48).
Here we investigate the function of Drosophila Integrator proteins in the 3′ processing of U7 snRNA and spliceosomal snRNAs. We developed a cell-based reporter that expresses green fluorescent protein (GFP) in response to misprocessing of U7 snRNA, allowing for facile and sensitive analysis of misprocessing in vivo. This reporter revealed novel cis-acting elements in the terminal stem-loop of U7 snRNA. Replacement of the U7 snRNA promoter with the actin 5 promoter abolished the processing of the reporter, indicating that the unique coupling of the promoter and the 3′ processing reaction has been conserved in evolution. We find that most, but not all, of the Drosophila homologs of the Integrator subunits are required for efficient U7 snRNA biosynthesis. Depletion of CPSF or CstF subunits or histone pre-mRNA processing factors did not affect U7 snRNA 3′-end formation, demonstrating that the Integrator complex does not share components with other processing factors. Finally, RNA interference (RNAi)-mediated depletion of Integrator proteins IntS1, -4, -9, and -11 in S2 cells results in accumulation of high levels of endogenous misprocessed snRNAs, and developing larvae harboring germ line mutations in IntS4 or IntS7 accumulate significant amounts of misprocessed snRNA and die at the mid-to-late larval stages.
MATERIALS AND METHODS
Cloning of the U7-GFP reporter and mutants.
The U7 reporter was cloned by replacing the histone promoter and coding region present in the histone H3-GFP reporter described previously with the U7 promoter, coding region, and putative 3′-box region (51). Briefly, we digested the H3-GFP reporter with HindIII and BamHI to remove the aforementioned sequences and subcloned into the same sites the U7 snRNA sequences using the primers U7F and U7R. U7-GFP mutants were generated by site-directed mutagenesis using the oligonucleotides listed in Table S1 in the supplemental material.
Northern blot analysis.
Northern blot analysis was performed on 3 μg of total RNA from S2 treated cells or 10 μg of total RNA from either wild-type or mutant larvae isolated at various time points of the Drosophila life cycle. After the RNA was dried by Speedvac, samples were dissolved in 10 μl of RNA loading buffer (7 M urea, 0.2% xylene cyanol FF, and 0.2% bromophenol blue) and boiled for 4 min. The RNA was resolved on 6% urea-polyacrylamide gels in 1× Tris-borate-EDTA (TBE) and transferred to Hybond N+ nitrocellulose (GE Healthcare Life Sciences, Piscataway, NJ) in 0.5× TBE for 60 min using the Genie Blotter transfer system (Idea Scientific Company, Minneapolis, MN). The membranes were probed in QuickHyb (Stratagene, La Jolla, CA) overnight at 50°C and washed with 1× SSC (0.15 M NaCl plus 0.015 M sodium citrate) plus 0.1% SDS for about 1 h. Probes were generated by randomly primed [α-32P]dCTP labeling (PrimeIt II kit; Stratagene) of PCR products generated to the complete coding region of snRNA U4:39B and the U7 snRNA genes.
Cell culture and RNAi treatment.
All templates used to create double-stranded RNA (dsRNA) were created through amplification from genomic DNA using primers containing T7 promoters at the 5′ end specifically targeting the Integrator genes (see Table S1 in the supplemental material for a complete oligonucleotide list). In all cases, oligonucleotides (Sigma, St. Louis, MO) were designed to target exons to preclude the inclusion of intronic sequences in the dsRNA. dsRNA was synthesized using T7 RNA polymerase from Ambion (Austin, Tx) and assessed for quality using gel electrophoresis. To perform RNAi, we plated 4 × 105 S2 cells into 24-well plates and treated them with 1 μg dsRNA each day for 3 or 4 days. Cells were harvested for analysis using radioimmunoprecipitation assay (RIPA) buffer (50 mM Tris-HCl [pH 8] 150 mM NaCl, 1% NP-40, 0.5% Na deoxycholate, 0.1% SDS) for protein lysate or TRIzol for total RNA extraction. All Western blotting was performed using standard techniques, and antibodies were raised against recombinant fragments of Drosophila Integrator proteins fused to glutathione S-transferase (GST).
Analysis of mutant Drosophila.
All flies were grown at 25°C on standard cornmeal-treacle media. The stocks w1118 and 11812 [w67c23 P(lacW)l(1)G0095G0095/FM7c] were obtained from the Bloomington Stock Center. The 11812 flies were rebalanced by crossing to an FM7c; Kr-GFP line to score the homozygous mutants. Genomic DNA was extracted from a single fly in 400 μl of lysis buffer (100 mM Tris-HCl [pH 7.5], 100 mM EDTA, 100 mM NaCl, and 0.5% SDS) at 65°C for 30 min and precipitated in 800 μl 1:2.5 (5 M) potassium acetate (KOAc)-6 M LiCl with 700 μl isopropanol, followed by phenol-chloroform extraction. The P element was detected by PCR using primers to the P element inverted repeat sequence (P-out) as well as a forward primer located in the genomic sequences 5′ of the P element (IntS4GF) and a second primer located downstream of the P element (IntS4GR). For reverse transcription-PCRs (RT-PCRs), P element-containing IntS4 homozygous mutant larvae were collected at the 1st-, 2nd-, and 3rd-instar larval stage using a Leica MZ16FA fluorescence stereomicroscope. Total RNA was extracted with TRIzol reagent (Invitrogen, Carlsbad, CA), and reverse transcription was performed using Moloney murine leukemia virus (MMLV) reverse transcriptase (Invitrogen). We measured the levels of IntS4 mRNA using the gene-specific primers IntS4F and IntS4R, and as a control we also amplified RpS17 using the gene-specific primers RpS17F and RpS17R.
Real-time RT-PCR (qRT-PCR) analysis.
Total RNA was isolated from Drosophila S2 cells using TRIzol reagent, and 2 μg was treated with DNase I (Fermentas, Burlington, ON, Canada) in accordance with the manufacturer's recommendations. Reverse transcription was performed on the 2 μg of RNA using random hexamer primers and MMLV reverse transcriptase (Invitrogen) in a total volume of 20 μl at 37°C for 1 h, followed by incubation at 95°C for 5 min. Reverse transcription reaction mixtures were then diluted to 50 μl with RNase-free water. In each case, 2 μl of the diluted reverse transcriptase was then used for real-time PCR using a SYBR green master mix and specific primers to each amplicon tested (for oligonucleotides, see Table S1 in the supplemental material). Data were acquired using a Stratagene Mx3000P real-time PCR machine, and data were calculated by the ΔΔCT (CT, threshold cycle) method using the equation  , where CTGOI is the CT for the gene of interest and CTnorm is the normalized CT. Triplicate experiments were performed for each Integrator knockdown, and data are plotted as fold increases in amplicon CT values normalized to the reference gene RpS17 versus those for the control dsRNA-treated cells, which were also normalized to RpS17.
, where CTGOI is the CT for the gene of interest and CTnorm is the normalized CT. Triplicate experiments were performed for each Integrator knockdown, and data are plotted as fold increases in amplicon CT values normalized to the reference gene RpS17 versus those for the control dsRNA-treated cells, which were also normalized to RpS17.
RESULTS
Construction and assessment of an snRNA misprocessing reporter.
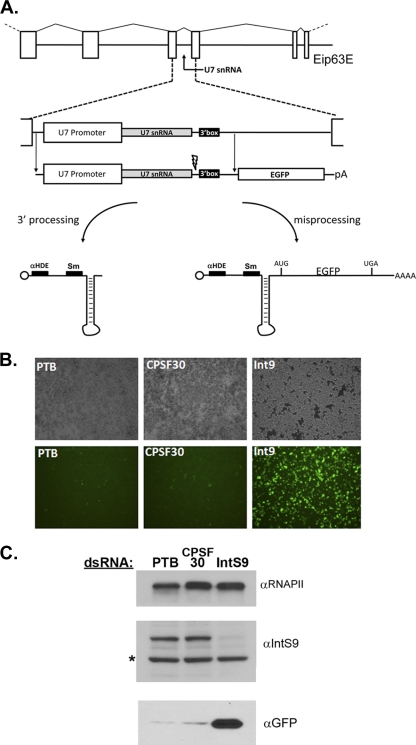
To measure snRNA misprocessing in vivo, we constructed a minigene reporter that expresses GFP under conditions of misprocessing resulting in read-through transcription. This enables a facile and semiquantitative fluorescence readout. We chose to use the Drosophila U7 snRNA as the model snRNA gene as it is the smallest snRNA and is predicted to have the least amount of secondary structure, which could dampen translation of the GFP reporter. The Drosophila genome contains a single copy of the U7 snRNA gene located within intron 4 of the ecdysone-induced protein 63E gene (Eip63E) on chromosome 3 (8). Surprisingly, a null mutation in the U7 snRNA gene is not lethal but instead results in misprocessing of histone mRNA and male/female sterility (14). This phenotype can be rescued by the expression of a genomic fragment containing only intron 4 of the Eip63 gene, indicating that all of the elements required for proper U7 snRNA biosynthesis are housed within this intron, including the promoter and putative 3′ box. Given this information, we cloned the Drosophila U7 snRNA gene, including the putative 3′ processing signals, upstream of an open reading frame (ORF) encoding GFP, which is itself followed by a strong canonical polyadenylation sequence downstream of the stop codon (Fig. 1A). Under normal conditions, the presence of this minigene should result in the formation of properly processed U7 snRNA and a failure to express GFP. In contrast, any impairment in the processing machinery should cause transcriptional read-through past the processing site, resulting in the use of a strong poly(A) site located downstream of the GFP ORF. Since the U7 snRNA sequences prior to the GFP ORF do not contain any AUG start codons, the resultant mRNA would be expected to become polyadenylated, encode GFP, and contain the U7 snRNA sequence as its 5′ untranslated region (UTR).
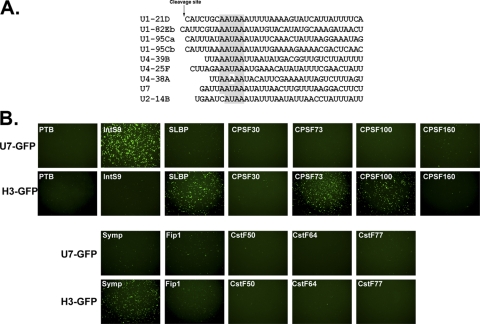
FIG. 1.
Design and assessment of a U7 snRNA misprocessing reporter. (A) Diagrammatic representation of the U7 snRNA gene, located within intron 4 of the Eip63E gene, which was cloned upstream of GFP. The endogenous U7 promoter, coding region, cleavage site, and 3′ box present in the minigene construct are depicted, as well as the predicted structures of both processed and misprocessed transcripts. αHDE, anti-HDE. (B) Drosophila S2 cells were treated with dsRNA (labeled in white), followed by transfection of the U7-GFP reporter. Bright-field images are in the top row, while corresponding GFP fluorescence is shown in the bottom row. (C) Western blot analysis of cell lysates treated with PTB, CPSF30 (negative controls) dsRNA, or dsRNA targeting Integrator 9 using anti-IntS9 (αIntS9) antibodies. The asterisk denotes a nonspecific cross-reacting band.
We performed a series of proof-of-principle experiments to determine if the U7-GFP minigene reporter is sensitive to known factors required for snRNA 3′-end formation. As controls, we treated Drosophila S2 cells with negative-control dsRNAs targeting production of either the 30-kDa subunit of the cleavage and polyadenylation factor (CPSF30) or the alternative splicing factor polypyrimidine tract binding protein (PTB). To determine if the U7-GFP reporter is sensitive to the levels of Integrator proteins, we used dsRNA to knock down production of the Drosophila homologue of IntS9 (CG5222), which is a component of the cleavage factor required to process the 3′ end of the snRNA. After several days of iterative dsRNA treatment, we saw no overt growth defect from any of the treatments, consistent with our previous results that revealed that there is a large pool of excess processing factors for poly(A)+ mRNAs in S2 cells (49, 51). Transfection of the U7-GFP reporter into cells treated with IntS9 dsRNA resulted in robust GFP expression relative to a near background level of fluorescence in the two control-treated cells (Fig. 1B). Western blotting of cell lysates using antibodies against GFP confirmed a strict correlation of GFP production only in cells displaying green fluorescence (Fig. 1C). To assess the knockdown of IntS9, we developed an antibody to this protein and used it to test the effect of dsRNA treatment on endogenous IntS9 accumulation. Treatment of cells with dsRNA targeting the IntS9 mRNA reduced IntS9 protein accumulation more than 90%, while treatment with dsRNA targeting two control mRNAs had no effect on the levels of IntS9 expression (Fig. 1C).
Drosophila U7 snRNA processing requires the snRNA promoter.
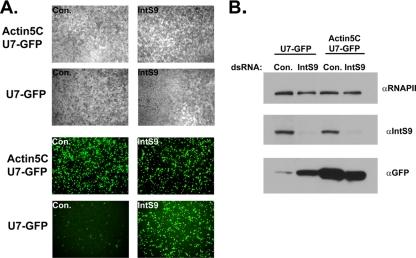
Like their vertebrate counterparts, snRNA promoters in Drosophila contain highly conserved elements not present in other Drosophila genes (17, 25, 34). To test whether Drosophila snRNA promoters are also necessary for correct 3′-end formation, as they are in vertebrates (6, 20), we removed the U7 snRNA promoter and replaced it with the actin 5C promoter, which normally drives the synthesis of a polyadenylated mRNA. When the U7-GFP reporter was driven by the actin 5C promoter, constitutive expression of GFP was observed in the absence of dsRNA targeting IntS9 (Fig. 2A). Moreover, the intensity did not significantly increase when IntS9 was knocked down in cells also transfected with actin 5C-driven U7-GFP. This suggests that there was little, if any, 3′-end processing of the U7 snRNA when driven by the actin 5C promoter. We found that the level of GFP fluorescence produced via the actin 5C promoter was essentially equivalent to that produced by the U7-GFP reporter when IntS9 was knocked down, suggesting that the relative strengths of these two promoters are comparable. Based upon this, we conclude that the coupling of the snRNA promoter with snRNA 3′-end formation has been conserved in Drosophila. Moreover, these data indicate that the U7 snRNA promoter is competent to produce poly(A)+ mRNA when Integrator 9 is knocked down.
FIG. 2.
A non-snRNA promoter causes constitutive misprocessing of U7 snRNA and insensitivity to Integrator knockdown. (A) Bright-field and fluorescence images of Drosophila S2 cells treated with either control (Con.) dsRNA or dsRNA targeting Integrator 9 and then transfected with either an actin 5C-driven U7-GFP reporter or with the native U7-GFP reporter. (B) Lysates from the cells in panel A were analyzed by Western blotting using anti-Integrator 9, anti-RNAPII (loading control), or anti-GFP antibodies.
Sequences in the 3′ stem-loop of U7 snRNA are required for snRNA processing.
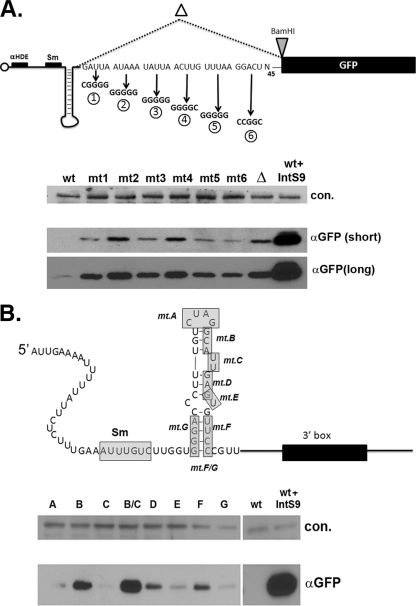
Much effort has been invested in defining the cis-acting elements required for pre-snRNA 3′-end processing in vertebrates (18, 54). Experiments in mammals and Xenopus located the sequences required to properly form the 3′ end of snRNA within an AT-rich core element located 9 to 19 nucleotides downstream of the cleavage site. A homologous region downstream of the 3′ end of Drosophila snRNAs is not conserved in vertebrates; however, we did note AT-rich stretches flanking all of the fly snRNA genes. Since the U7-GFP reporter exhibits very little misprocessing and subsequent GFP expression under normal conditions, the 30 nucleotides downstream of the cleavage site in the reporter are sufficient to mediate proper 3′-end formation. To identify critical nucleotides downstream of the 3′ end of the mature Drosophila U7 snRNA, we performed systematic mutagenesis of the region beginning with nucleotide +1 and continuing to and including nucleotide +30. We created six sets of 5-base mutations where any adenine or thymine was mutated to a guanine and any existing guanine was changed to a cytosine (Fig. 3A, schematic). In addition, we also deleted bases +1 to +30 (U7-GFPΔ), which effectively placed the GFP ORF directly downstream of the cleavage site. There are no other AU-rich sequences immediately 3′ of U7 snRNA in the minigene construct. We transfected each of the mutant reporters, the 30-base deletion, and the wild-type U7-GFP reporter (with and without dsRNA targeting to IntS9) into S2 cells, and lysates were prepared and probed for GFP expression. Nearly all of the mutants had some deleterious effect on 3′-end processing, although none of the mutants generated as much GFP expression as was achieved by the knockdown of Integrator 9 (Fig. 3A, bottom). Mutants 2 and 4 were reproducibly the most robust at increasing GFP expression. The positions mutated in mutants 2 and 4 constitute the bases located from +6 to +10 and +15 to +20 downstream of the cleavage site and likely represent the Drosophila 3′ box. The observation that none of the mutants were able to achieve the same degree of GFP expression as the wild type in the presence of Integrator 9 knockdown is consistent with previous evidence demonstrating that the human 3′ box is highly tolerant of mutation (1), and the results for the U7-GFPΔ mutant suggest that the 3′ box enhances cleavage but is not essential.
FIG. 3.
Characterization of the cis element requirements for U7 snRNA 3′-end processing. (A) Schematic of the scanning mutations within the 30 nucleotides downstream of the 3′ end of the mature U7 snRNA in the U7-GFP construct. Drosophila S2 cells were transfected with plasmids carrying mutations 1 to 6 (mt1 to mt6; top), and lysates were analyzed for snRNA misprocessing, as indicated by GFP expression, using anti-GFP antibodies or antibodies raised against RNAPII (con.). Both short and long exposures are shown for the GFP Western blot (bottom). (B) Schematic of the point mutations made within the 3′ terminal stem-loop of the U7 snRNA coding region (top) and Western blotting of cell extracts to quantitate GFP expression (bottom). The location of the mutated nucleotides (labeled mtA to mtG) are shaded. The lower blot represents an analysis similar to that described for panel A.
To determine the contribution of U7 snRNA coding sequences to 3′-end processing, we extended our mutational analysis into the coding region of the Drosophila U7 snRNA, testing whether the stem-loop at the 3′ end of the U7 snRNA is necessary for 3′-end formation. All snRNAs end in a 3′ stem-loop, and, although the sequence and length of the stem-loop vary greatly among snRNAs, whether the stem-loop has a function is not known. Previous studies in vertebrates measuring 3′-end formation required that the snRNA product be stable, making it difficult to address the role of coding region sequences in vertebrate snRNA 3′-end formation (18). Bioinformatic analysis of Drosophila U7 snRNA sequences (31) revealed that the 3′ stem-loop is markedly larger than its mammalian counterparts and in addition that there is a conserved bulge near the top of the stem-loop that is not found in mammals (Fig. 3B). As our assay does not rely on snRNA functionality or stability, we created a series of scanning mutations throughout the 3′ stem-loop of the Drosophila U7 snRNA, keeping sequences in the 3′ box intact (Fig. 3B, schematic). In all cases, mutations were substitutions that are predicted to disrupt the secondary structure present in the 3′ stem-loop. The majority of the mutations within the 3′ stem-loop did not result in GFP expression. However, mutations of the sequences in the top of the stem, but not in the loop or the bulge, caused significant GFP fluorescence and more GFP production than any of the 3′ box mutations (Fig. 3B, bottom). Combining a mutation of the stem at the top of the stem-loop with a bulge mutation resulted in an even larger increase in GFP than the individual mutations individually, suggesting that the stem and the four-base loop (size of the loop, not the sequence) were essential for efficient 3′-end processing.
Collectively, these data reveal the presence of a cis-acting element in the 3′ terminal stem-loop of Drosophila snRNAs and that flies, like humans, have an A/T-rich 3′ box that is moderately tolerant of mutations.
Knockdown of different Integrator components results in different amounts of readthrough of U7 snRNA.
The original study by Baillat et al. (3) in human cells identified 10 proteins that copurified with IntS9 and IntS11 and comprised the Integrator complex. In that study, RNAi-mediated depletion of IntS1 and IntS11 led to an accumulation of snRNA precursors, functionally implicating these two proteins as being required for processing. The other Integrator subunits, with the exception of IntS9, contain no obvious protein motifs or domain structures that would implicate them in an RNA-processing reaction. As no other Integrator subunits had been tested directly for their requirement for snRNA processing, we used our U7-GFP reporter system to assess each of the Drosophila Integrator subunits for their requirement in U7 snRNA biosynthesis.
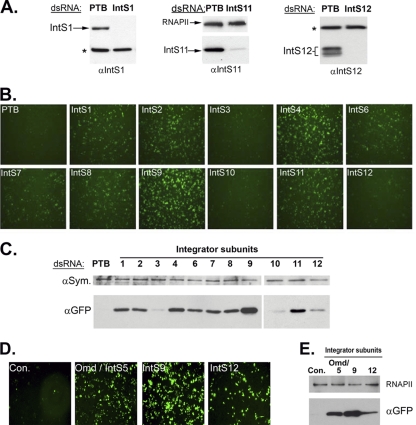
We created a series of dsRNAs, ranging from 400 to 500 nucleotides, to target all of the known Drosophila Integrator subunits. To minimize the potential off-target effects sometimes associated with using large dsRNAs, we used amplicons designed by the Drosophila RNAi Screening Center (www.flyrnai.org) which were predicted to have no other targets in the genome (2). As a negative control, we treated cells with dsRNA targeting PTB. S2 cells were treated with dsRNA for three consecutive days prior to transfection of the U7-GFP reporter. We observed no overt effect on cell growth in response to any of the dsRNAs used over a 120-h period. To assess the effect of knockdown efficiency on endogenous Integrator expression, we generated antibodies to several other Drosophila Integrator proteins in addition to IntS9 (Fig. 1C). We analyzed the effect of the dsRNA on protein expression using Western blotting with antibodies specific to Integrators 1, 11, and 12 (Fig. 4A). In all cases, the efficiency of knockdown was >90%, although we did observe some residual expression of each of these Integrator subunits on longer exposures (not shown). We assessed the effect of each knockdown on U7-GFP reporter activity by both fluorescence microscopy (Fig. 4B) and Western blotting using anti-GFP antibodies (Fig. 4C). We found that knockdown of nearly all of the Drosophila Integrator subunits resulted in various degrees of GFP expression from the U7-GFP reporter. Knockdown of Integrator 9 resulted in the highest level of GFP expression, and knockdown of IntS1, -2, -4, -6, -7, -8, and -11 resulted in robust GFP expression. The knockdown of Integrator 3 and Integrator 10 did not result in significant misprocessing of the reporter, and the knockdown of Integrator 12 resulted in only modest expression of GFP.
FIG. 4.
Knockdown of the Drosophila Integrator subunits causes misprocessing of the U7-GFP reporter. (A) Western blot analysis of lysates from Drosophila S2 cells treated with either control dsRNA targeting PTB or a specific dsRNA targeting production of Integrator 1, 11, or 12. RNAPII (center) or a cross-reacting band (asterisks) serves as a loading control. (B) Fluorescence images of cells treated with a specific dsRNA (labeled in white), then transfected with the U7-GFP reporter. (C) Western blot analysis of cell lysates from the cells shown in panel B. Lysates were probed with either anti-GFP antibodies or with anti-Symplekin (αSym.) antibodies as a control. (D) Fluorescence images of cells treated with dsRNA followed by transfection with the U7-GFP reporter. In this case the control dsRNA targeted LacZ and not PTB. (E) Western blot analysis of cell lysates from panel D.
The original report describing the identification of the Integrator complex did not predict the existence of a Drosophila IntS5 orthologue (3). We have identified a previously characterized gene in Drosophila called oocyte maintenance defects (omd) in the Ensembl database (www.ensembl.org) as a potential candidate, and Omd is 22% identical to human IntS5. Most of the Drosophila Integrator subunits are between 25% and 39% identical to their human orthologues with the exception of IntS9 and IntS11, which are 51% and 68% identical, respectively. To test whether Omd is functionally required for U7 snRNA 3′-end formation, we treated cells with dsRNA targeting Omd as well as dsRNA targeting IntS9 and IntS12. We observed significant levels of GFP expression in cells treated with Omd dsRNA compared to control-treated cells and found that the degree of fluorescence was between those seen after depletion of IntS9 and IntS12 (Fig. 4D and E). Together, these data and the bioinformatic analysis suggest that Omd is indeed the Drosophila orthologue of Integrator 5.
To rule out the possibility of ineffective dsRNA, we created a second set of dsRNAs for each Integrator subunit and treated S2 cells with these dsRNAs prior to transfection of the U7-GFP reporter. This second set of dsRNAs generated trends in the level of GFP expression very similar to that seen in Fig. 4 (see Fig. S1 in the supplemental material). We conclude that 10 of 12 Drosophila Integrator subunits tested play some role in U7 snRNA processing.
Cleavage and polyadenylation machinery is not required for U7 snRNA biosynthesis.
We and others have previously demonstrated that members of the cleavage and polyadenylation complex are involved in the processing of histone pre-mRNA in addition to their role in the cleavage and polyadenylation of cellular mRNA (7, 23, 40, 49, 51). Moreover, a recent proteomic analysis of the cleavage and polyadenylation complex found the presence of several Integrator proteins (45). There is a conserved AAUAAA sequence at the ends of all Drosophila snRNA genes (Fig. 5A), and mutation of these bases within the 3′ box of Drosophila U7 snRNA resulted in significant GFP expression of the U7-GFP minigene reporter (Fig. 3A, mt 2). Given this, we asked whether any cleavage and polyadenylation factors are involved in snRNA 3′-end formation by treating S2 cells with dsRNA targeting the primary subunits of the cleavage and polyadenylation machinery, including all five CPSF subunits (including Fip1), the three CstF subunits, and Symplekin. As a positive control, we also treated S2 cells with dsRNA targeting IntS9, and as a negative control, we treated cells with dsRNA targeting the splicing factor PTB or the histone pre-mRNA processing factor SLBP. Transfection of the U7 reporter into these dsRNA-treated cells resulted in robust GFP expression when Integrator 9 had been knocked down but failed to give significant GFP expression under any other dsRNA treatment (Fig. 5B, top row). In contrast, treatment of cells with dsRNA targeting SLBP, CPSF73, CPSF100, and Symplekin resulted in robust expression when the histone-GFP reporter (H3-GFP), which is a reporter capable of monitoring the 3′-end formation of histone pre-mRNA (51), was transfected. The histone-GFP data indicate that, for SLBP, CPSF73, CPSF100, and Symplekin, the dsRNA treatment was sufficient to affect RNA processing (Fig. 5B, bottom two rows). Given that the depletion of these factors had an effect on the H3-GFP reporter and not on the U7-GFP reporter, we conclude that the cleavage and polyadenylation machinery is not involved in Drosophila snRNA 3′-end formation. We also performed the reciprocal experiment by treating cells with dsRNA targeting Integrator subunits followed by transfection with the H3-GFP reporter and found no GFP expression, demonstrating that this complex is distinct from the histone pre-mRNA processing machinery (see Fig. S2 in the supplemental material). Collectively, these data demonstrate that snRNA 3′-end formation does not share components with the histone pre-mRNA processing or cleavage and polyadenylation machinery.
FIG. 5.
Cleavage and polyadenylation factors and histone pre-mRNA processing factors do not affect U7 snRNA 3′ processing. (A) Alignment of sequences downstream of several Drosophila snRNA mature 3′ ends reveals a well-conserved polyadenylation signal sequence present downstream of nearly all of the Drosophila snRNA genes (shaded). (B) Fluorescence images of Drosophila S2 cells treated with dsRNA to knock down specific cleavage and polyadenylation factors (labeled in white), followed by transfection with either the U7-GFP minigene reporter or the histone H3-GFP minigene reporter.
Knockdown of Drosophila Integrator proteins results in significant levels of misprocessed spliceosomal snRNA.
A significant advantage of the U7-GFP minigene reporter lies in its ability to detect snRNA misprocessing through the expression of a stable and visually observable product. However, it does not directly demonstrate that knockdown of Integrator complex subunits affects processing of endogenous Drosophila spliceosomal snRNAs. To address this, we used both real-time RT-PCR (qRT-PCR) and a Northern blot assay to determine whether misprocessed transcripts accumulate as a result of Drosophila Integrator subunit knockdown. We designed amplicons for each of the RNAPII-transcribed spliceosomal snRNAs, with the forward primers binding to sequences within the snRNA coding sequence and reverse primers binding approximately 50 nucleotides downstream of the 3′ cleavage site. These primer sets thus allowed specific detection of unprocessed or misprocessed snRNAs. All amplicons were confirmed by sequencing, and each primer set was found to generate a single band by electrophoresis through an ethidium-stained agarose gel (see Fig. S3 in the supplemental material). We then treated S2 cells with dsRNA targeting each Integrator subunit as well as PTB (negative control), extracted total RNA from these cells, and generated cDNA via reverse transcription. The levels of misprocessed snRNAs were then determined by qRT-PCR using primers for each misprocessed snRNA amplicon, relative to an internal control amplification of the RpS17 mRNA. We did not detect any significant changes in RpS17 expression after treatment of S2 cells with dsRNA targeting Integrator mRNAs (not shown). The data presented (Fig. 6A) are the averages of three independent RNAi experiments and are plotted as fold increases in the level of misprocessed snRNA relative to the level for control-treated cells. All values were normalized to RpS17 levels. With the exception of Integrator 3 dsRNA, which also did not score in the U7-GFP minigene assay, all knockdowns resulted in increased snRNA misprocessing (Fig. 6A). Integrator 10 displayed the smallest increase in misprocessing, consistent with very low GFP expression observed using the U7-GFP reporter (Fig. 4B). For each knockdown, the trends were consistent between the four different snRNA-misprocessed amplicons and correlated well with the level of GFP fluorescence achieved by the U7-GFP reporter. We find that knockdown of Integrators 1, 4, and 9 led to the largest amount of misprocessing (>50-fold increase), followed by the knockdowns of Integrators 11 and 7, which resulted in an ∼10-fold increase in misprocessed transcripts. With the exception of IntS3 and IntS10, knockdown of the remainder of the Integrator subunits resulted in a 2- to 10-fold increases in snRNA misprocessing.
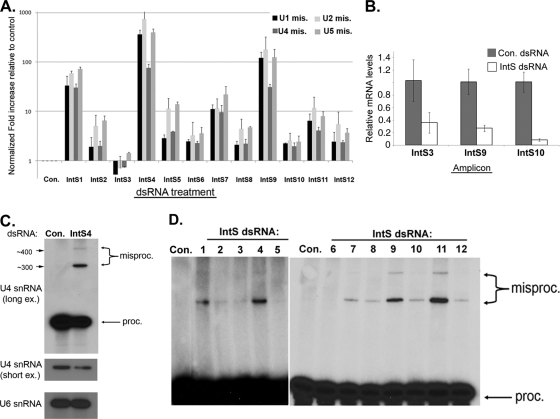
FIG. 6.
Knockdown of Integrator subunits causes various degrees of misprocessing of endogenous spliceosomal snRNAs. (A) Histogram of real-time PCR experimental data generated using primer pairs designed to detect the presence of misprocessed (mis.) spliceosomal U1, U2, U4, or U5 snRNAs. Results are plotted as fold increases relative to control-treated cells and reflect expression normalized to an internal control gene (RpS17). All results are derived from biological triplicates, with error bars indicative of the standard deviations of the triplicate quantification. (B) Histogram of qRT-PCR quantitation of IntS3, IntS9, and IntS10 mRNA following dsRNA treatment. Levels represented are the averages of triplicate experiments normalized to an internal control (RpS17) and then normalized to control-treated cells. (C) Northern blot analysis for endogenous U4 snRNA and control (U6) snRNA from Drosophila S2 cells treated with either control dsRNA or dsRNA targeting IntS4. Both short and long exposures (ex.) of the U4 snRNA blot are shown. misproc., misprocessed. (D) Northern blot analysis of endogenous U4 snRNA expression in S2 cells treated with dsRNAs to knock down specific Integrator subunits.
One possible explanation for the lack of a significant accumulation of misprocessed IntS3 or IntS10 snRNAs after dsRNA treatment is ineffective reduction in the levels of the target mRNA. To explore this possibility, we measured the levels of endogenous mRNA from control-treated S2 cells as well as those treated with dsRNA targeting IntS3, IntS9, or IntS10 (Fig. 6B). We observed a 65% reduction in the levels of IntS3 mRNA, a 75% reduction in the levels of IntS9 mRNA, and a >90% reduction in the levels of IntS10 mRNA. We conclude from these data that the dsRNA treatment was effective in reducing the levels of their respective target mRNAs and consequently reduced IntS3 and IntS10 protein expression, as a 75% reduction in IntS9 mRNA results in a >90% reduction in IntS9 protein levels (Fig. 1C).
To determine whether any specific misprocessed transcripts were formed, we performed a Northern blot analysis using a probe designed to hybridize within the coding region of U4 snRNA, thus enabling detection of both properly processed and misprocessed snRNA (Fig. 6B). Note that misprocessed snRNA will be detected by Northern blotting only if specific and discrete species are formed, and many misprocessed transcripts may have heterogenous 3′ ends. A short exposure demonstrated an only slight reduction in the levels of processed snRNA in response to Integrator 4 knockdown relative to control-treated cells, but a longer exposure revealed the presence of two discrete misprocessed bands that were present only when cells were treated with Integrator 4 dsRNA. We cannot distinguish whether the larger U4 bands are derived from ectopically cleaved (misprocessed) or unprocessed snRNA that is released due to downstream termination. The estimated sizes of these two bands are 300 and 400 nucleotides, and inspection of the sequences in this region downstream of the U4 snRNA cleavage site did not yield any obvious cis elements that might govern an alternative processing mechanism. Nevertheless, the two misprocessed bands were never observed in control cells and were present only when cells were treated with dsRNA targeting Integrator components. We extended our Northern blot analysis of U4 snRNA to examine the effects of knockdown of other Integrator subunits and found that the amount of the discrete misprocessed U4 snRNA varied between samples and, in general, showed a trend similar to that for the qRT-PCR analysis (Fig. 6C). We therefore conclude that most of the Integrator subunits are essential for spliceosomal snRNA biogenesis and that some yield more-severe phenotypes when depleted than others.
Analysis of Drosophila with mutations in Integrator 4.
Integrator 4 scores strongly in all the misprocessing assays in cultured cells, and dsRNA-treated cells showed no overt abnormalities. In contrast, the endogenous Drosophila Integrator 4 locus (CG12113) is annotated as an essential gene. To determine if IntS4 is essential for development, we acquired a candidate mutant called P(lacW)l(1)G0095 from the Bloomington Drosophila Stock Center; this mutant carries a P element insertion isolated in a screen for lethal insertions on the X chromosome using the P(LacW) enhancer trap (38) (Fig. 7A). The P(lacW)l(1)G0095 insertion was reported to be within the second exon, which we confirmed using PCR of genomic DNA isolated from mutant homozygous larvae (Fig. 7A, bottom). We found that the P element disrupts the conserved Armadillo-type fold housing the HEAT repeats and would be predicted to cause a C-terminal truncation of the protein if translated. The P(lacW)l(1)G0095 allele was propagated as a balanced heterozygote using the FM7 balancer chromosome, and no homozygous unbalanced flies were observed in the culture, consistent with this being a homozygously lethal P element insertion (38). We performed genetic crosses with these flies to create a line with the P allele chromosome balanced over an FM7 derivative that harbors a GFP cDNA under the control of the twist promoter (FM7c; Kr-GFP), which allows the detection of heterozygotes by GFP fluorescence early in development. We observed that homozygous P(lacW)l(1)G0095 larvae die at the junction of the 2nd- and 3rd-instar stages of development (with only a small percentage of homozygous larvae progressing to 3rd instar). Total RNA was isolated from homozygous late 2nd-instar larvae as well as from wild-type controls and analyzed using RT-PCR primers specific to the IntS4 cDNA (Fig. 7B). RNA from three different collections of mutant larvae was analyzed, and for each we observed negligible expression of IntS4 mRNA in the mutants while finding no difference in the level of RpS17 mRNA relative to that for wild-type larvae. These data suggest that the P(lacW)l(1)G0095 insertion is unlikely to produce any IntS4 protein.
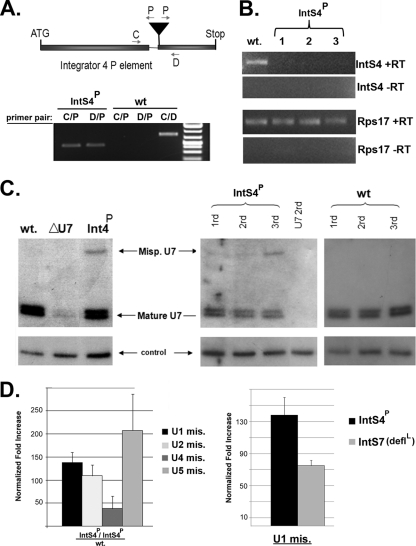
FIG. 7.
Drosophila carrying germ line mutations in Integrator subunits accumulates misprocessed snRNA and fails to complete development. (A, top) Schematic of the location of a P element within the second exon of the Integrator 4 gene in strain P(lacW)l(1)G0095, as well as the locations of PCR primers in exon 1 (C) and exon 2 (D) and within the inverted terminal repeat of the P element (P). (Bottom) PCR amplification of genomic DNA isolated from either wild-type (wt) larvae or larvae homozygous for the P element within the Integrator 4 gene. (B) RT-PCR analysis of RNA isolated from the larvae described in panel A using primers specific for the IntS4 mRNA or RpS17 mRNA (control). +RT indicates cDNA reaction mixtures containing reverse transcriptase, and −RT indicates mock cDNA reaction mixtures lacking reverse transcriptase. (C) Northern blot analysis of endogenous U7 snRNA from either wild-type, U7EY11305, or IntS4 P element-containing third-instar larvae showing accumulation of misprocessed snRNA in the IntS4 mutant. (D) Results from qRT-PCR analysis of misprocessed U1 snRNA or U7 snRNA isolated from third-instar larvae from either homozygous IntS4 [P(lacW)l(1)G0095] or IntS7 (deflL) null mutants.
We isolated RNA from the small percentage of larvae that developed to the 3rd-instar stage and analyzed the expression of U7 snRNA using a Northern blot analysis. As controls, we analyzed U7 snRNA expression from wild-type larvae as well as 3rd-instar larvae containing the U7 snRNA insertion mutant U7EY11305, which does not express any product (14). The wild-type U7 snRNA normally migrates as a doublet representing two differentially processed forms differing by 3 nucleotides. This doublet is significantly reduced in U7 mutant larvae (8) (Fig. 7C). The residual U7 snRNA observed in the U7 null larvae likely represents perdurance of the maternally derived product or possibly contamination from misgenotyped heterozygous larvae. The homozygous P(lacW)l(1)G0095 larvae surprisingly contain amounts of U7 snRNA comparable to that in the wild type but also contain a single additional band (∼150 to 200 nucleotides) above the properly processed product (Fig. 7C). A time course analysis of these larvae demonstrated that the larger U7 snRNA band increases in intensity as the larvae develop and is not detectable at any comparable time in the development of the wild-type fly (Fig. 7C). We also extended our analysis of misprocessing of the spliceosomal snRNAs by qRT-PCR and found the IntS4 mutant larvae display increased levels of misprocessed RNAPII-encoded spliceosomal snRNAs (Fig. 7D, left). Finally, we compared the levels of misprocessed U1 snRNA present in the IntS4 mutant larvae to that of a null allele of the deflated gene, which encodes the Drosophila homologue of Integrator 7. The deflated allele examined (deflL) was found also to be lethal at the second-instar larval stage and can be rescued through the expression of IntS7 cDNA (41). We found significant levels of misprocessed U1 snRNA in both Ints4 and IntS7 mutants, and, as observed in the RNAi experiments in cultured cells, higher levels of misprocessed U1 were observed when IntS4 was reduced/mutated than when IntS7 was reduced/mutated (Fig. 7D). We conclude from these experiments that germ line mutations in Integrator 4 perturb normal Drosophila larval development and that deficiencies in the expression of IntS4 or IntS7 lead to increased levels of misprocessed snRNAs.
DISCUSSION
The biochemical identification of the Integrator complex represents a significant advance in our understanding of the molecular mechanism of snRNA 3′-end formation. However, very little is known about the relative importance of these factors in snRNA 3′-end formation or whether all subunits are required for 3′-end formation. In this study, we demonstrate that the majority of the Integrator proteins are required functionally for efficient 3′-end formation of both spliceosomal snRNAs and U7 snRNA and that the processing reaction is highly dependent on 10 of the 12 subunits tested (Fig. 4C). The Drosophila snRNA promoters are distinct from the promoters of RNAPII-transcribed mRNAs (17), and the coupling of snRNA promoters to 3′-end formation observed for vertebrate spliceosomal snRNAs is conserved for Drosophila U7 snRNA 3′-end formation. This indicates that the 3′-end formation mechanism was already present early in metazoan evolution; this is supported by the observation of orthologues of Integrator subunits encoded in several plant genomes.
U7 snRNA 3′ stem-loop sequences are important for 3′-end formation.
The sequences of the elements that direct both the poly(A)+ mRNA and histone mRNA 3′-end processing have the common feature of being located on either side of the cleavage site. This is consistent with the fact that the molecular mechanism of pre-mRNA processing involves an interaction of the cleavage factor with factors bound on either side of the cleavage site. This conserved mechanism likely functions to enhance the specificity of the endonuclease and may also serve to regulate its activity, especially since the nuclease CPSF73 is one of the first factors recruited to the poly(A)+ pre-mRNA, and it is clearly advantageous to activate the nuclease only when it is at the cleavage site.
The primary element directing snRNA cleavage in vertebrates is the 3′ box located downstream of the cleavage site (1, 18, 54). We have shown here that the 3′ box defined in humans is present in Drosophila although the specific sequences are not well conserved. Moreover, we found that, similar to that of humans, the Drosophila 3′ box is tolerant of mutation, as neither any of the point mutations nor a full deletion of the 3′ box was able to produce the level of transcriptional readthrough that was attained when an Integrator component was knocked down by dsRNA treatment. This leads to the question of why a deletion of the presumed Integrator binding and cleavage element does not have an effect equal to the RNAi-mediated depletion of an Integrator subunit. There are two possible explanations for this observation. First, when the 3′ box is deleted, components of the Integrator complex are able to promiscuously bind to sequences in the 5′ end of the GFP ORF, allowing for a partial rescue of processing. The sequence of the new “3′ box” formed as a result of the deletion is totally dissimilar from the actual 3′ box sequence, given its enriched GC content; thus we disfavor this possibility. Alternatively, the deletion of the 3′ box is in fact equally deleterious to snRNA processing; however, this does not remove Integrator proteins from the elongation complex that is formed from the snRNA promoter. Consequently, the Integrator-RNAPII complex is unable to support the downstream cleavage and polyadenylation that are requisite for reporter expression.
Data presented here demonstrate that Drosophila snRNA processing is more dependent on the 3′ stem-loop present within the coding region of U7 snRNA than the sequences within the 3′ box. This is an unexpected observation given that the 3′ stem-loops present in all snRNAs are very different from each other and are unlikely to interact specifically with the same factor. It has been observed that in mammalian cultured cells and Xenopus, the histone stem-loop placed at the 3′ end of a transcript initiated from an snRNA promoter 5′ of the 3′ box functions efficiently in snRNA 3′-end formation (39; W. F. Marzluff, unpublished results). The dependency of the cleavage site on a 3′ stem-loop is not unique to snRNA processing. In humans, the position of the cleavage site within histone pre-mRNA is dictated by the HDE, and processing does not absolutely require the stem-loop or SLBP. Moving the HDE further downstream of the stem-loop shifts the cleavage site in turn (44, 43). This has led to the hypothesis that the U7 snRNP acts as a “molecular ruler,” dictating the cleavage site (44). The molecular details of the mechanism in Drosophila histone pre-mRNA processing are different, as the movement of the fly HDE further downstream of the stem-loop does not switch the cleavage site. In addition, the stem-loop and SLBP are absolutely required for processing in Drosophila in vitro, suggesting that the upstream stem-loop/SLBP complex is essential for determining where cleavage occurs (9). This Drosophila-specific dependence on the histone pre-mRNA stem-loop parallels our observations that the snRNA 3′ stem-loop located within the coding region is important for correct processing. We note that the previous studies on vertebrate snRNAs did not determine whether the stem-loop had an effect since removing the stem-loop would have destabilized the snRNA and their assays included determining the amount of processed snRNA (17). This suggests that the factors that dictate cleavage site choice rely on an upstream secondary structure as well as the downstream 3′ box and that the combination of these two elements directs the IntS11 nuclease to the proper target.
Differential requirements for Integrator subunits in Drosophila snRNA processing.
The molecular mechanism of Integrator action remains to be elucidated; however, we envision that it clearly has similarities to those of the other 3′-end formation complexes. The most striking of these similarities is in the mechanism of cleavage. In the cases of poly(A)+ and histone mRNA this is achieved through the endonuclease activity of the CPSF73/100 heterodimer while bound to Symplekin (7, 23, 24, 30, 49, 51). The signature the β-CASP domain found in CPSF73 is also present in IntS11, and the two proteins are highly similar, as are CPSF100 and IntS9 (4). It is not surprising that the knockdown of either IntS9 or IntS11 proteins causes significant readthrough of the U7-GFP reporter as well as the accumulation of high levels of misprocessed spliceosomal snRNA.
The cleavage of histone pre-mRNA requires a cleavage factor with a core of CPSF73/CPSF100 and Symplekin (24, 49). Symplekin is not required for snRNA formation since depletion of Symplekin does not affect snRNA 3′-end formation. We speculate that one of the other Integrator subunits, possibly IntS1 or IntS4, likely plays the role of a scaffold for assembly of the cleavage activity. It is noteworthy that IntS4 contains numerous HEAT repeats, which are also present in Symplekin (22), making it possible that IntS4 is a “Symplekin-like” factor in the snRNA cleavage complex. Alternatively, IntS1 and -4 could perform a function required for cleavage that does not include them as actual components of the core cleavage factor. For example, it was recently demonstrated that Integrator proteins can specifically bind to a unique phosphorylation pattern present on the C-terminal domain (CTD) of RNAPII (12). The identity of the specific Integrator subunit that binds this motif is not presently known, but it would be expected that RNAi-mediated depletion of this factor would result in significant misprocessing. In the case of both cleavage and polyadenylation and histone pre-mRNA processing, there exist components that bind the RNA in order to direct the cleavage event. That none of the Integrator subunits has a motif that is related to the known RNA binding domains suggests that additional Integrator subunits have yet to be identified or that the interaction with the 3′ stem-loop element and 3′ box is mediated by a novel domain within an existing member(s) of the complex. Depletion of such a protein would also be expected to dramatically impact processing.
In the U7-GFP minigene reporter assay, we observed that RNAi-mediated depletion of 10 of the 12 integrator subunits tested resulted in similar levels of GFP expression, whereas depletion of IntS3 or IntS10 generated little or no GFP production. We observed a small amount of misprocessing of endogenous snRNA in the IntS10 knockdowns but none in the IntS3 knockdowns (Fig. 6A). It is possible that IntS3 and IntS10 are not required for processing per se but may have other functions in snRNA biosynthesis (such as export). Alternatively, it is possible that they are not core members of the Integrator complex. Interestingly, during the course of this study, several reports have shown that Integrator 3 is also a member of the sensor of single-stranded DNA (SOSS) complex, involved in the maintenance of genome stability (21, 26, 46). None of the other Integrator subunits were implicated in this pathway, suggesting that IntS3 may have a function(s) distinct from snRNA processing.
Knockdown or mutation of Integrator subunits results in significant levels of misprocessed spliceosomal snRNA.
Failure to process snRNAs likely results in longer transcripts with heterogeneous 3′ ends. Mechanisms that could give rise to these transcripts include termination of transcription giving a defined product, ectopic processing events giving a defined 3′ end, and precocious polyadenylation, which would also generate length heterogeneity. In cells where Integrator subunits were knocked down, we readily detected misprocessed endogenous snRNAs via RT-PCR using primers that span the region 3′ to the cleavage site. We also detected misprocessed snRNAs in larvae homozygous for germ line mutations in genes encoding IntS4 or IntS7. Unexpectedly, we detected a longer defined U4 snRNA transcript in the cultured cells and a defined U7 snRNA transcript in larvae containing an Integrator 4 mutant. Each of these was about 200 to 300 nt longer than the normally processed snRNA. The origin of these transcripts is not clear, but they are unlikely to be polyadenylated given their discrete nature, and there are no obvious processing signals downstream of the 3′-end cleavage site.
Emerging data from other laboratories suggest a common developmentally lethal phenotype as a result of mutation or knockdown of Integrator proteins. P-element-mediated deletion mutations of the Drosophila gene deflated (encoding IntS7) result in lethality during late larval development (41). RNAi-mediated depletion of IntS6 in Caenorhabditis elegans results in embryonic lethality and mitochondrial defects (15), whereas depletion of IntS5 in zebrafish using antisense morpholino oligonucleotides also results in lethality and defects in hematopoiesis (50). Finally, targeted disruption of the gene encoding mouse IntS1 results in early embryonic lethality and the accumulation of snRNA precursors (16). Taken together, these studies suggest an evolutionarily conserved requirement for the Integrator complex. Here, we report that the Integrator 4 gene is also an essential gene and that homozygous larvae with defective IntS4 or IntS7 expression produce misprocessed snRNA. In mice, which make large amounts of snRNAs early in embryonic development (27), the early lethality of IntS1 knockout at the blastocyst stage correlates with the requirement for zygotic expression of snRNA at this stage. In contrast, Drosophila, sea urchins, Xenopus, and zebrafish contain large stores of maternal spliceosomal and U7 snRNAs (37, 42). There is a large synthesis of snRNAs at the midblastula transition in Xenopus (13) and by the morula stage of sea urchin development, as well as activation of the zygotic genome in the 2-cell stage in mice. The timing of zygotic expression of snRNAs in Drosophila is not known. However, the maternal U7 snRNA is sufficient for progression to the end of the 2nd-instar larval stage, and there is no defect in histone pre-mRNA processing until the 3rd-instar stage, at which point misprocessed histone RNA begins to accumulate (14). This timing correlates with our observation of lethality at the 2nd/3rd-instar larval border and suggests that lethality occurs once the maternal pool of spliceosomal snRNAs has been depleted below a critical threshold. Alternatively, the biosynthesis of a yet-to-be identified RNA may depend on Integrator function, and depletion of this RNA is primarily responsible for the stage-specific lethality.
The U7-GFP minigene reporter system described in this paper can likely be used to screen for all factors required for snRNA 3′-end formation in vivo; it was similarly used previously for histone mRNAs (51). These studies are in progress.
Supplementary Material
Acknowledgments
We thank Phillip Carpenter and Bob Duronio for helpful discussions and reviewing the manuscript.
J.C. acknowledges support from a GSBS training grant, W.F.M. acknowledges NIH grant GM58921, W.D.W. acknowledges the generous support of the Queensland Cancer Fund as well as the NHMRC through the Australian Drosophila Biomedical Research Support Facility at the ANU, and E.J.W. acknowledges support from the UTHSC startup funds and by NIH Pathway to Independence Award 5R00GM080447-03.
Footnotes
Published ahead of print on 15 November 2010.
Supplemental material for this article may be found at http://mcb.asm.org/.
REFERENCES
- 1.Ach, R. A., and A. M. Weiner. 1987. The highly conserved U small nuclear RNA 3′-end formation signal is quite tolerant to mutation. Mol. Cell. Biol. 7:2070-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Armknecht, S., et al. 2005. High-throughput RNA interference screens in Drosophila tissue culture cells. Methods Enzymol. 392:55-73. [DOI] [PubMed] [Google Scholar]
- 3.Baillat, D., et al. 2005. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell 123:265-276. [DOI] [PubMed] [Google Scholar]
- 4.Callebaut, I., D. Moshous, J. P. Mornon, and J. P. de Villartay. 2002. Metallo-beta-lactamase fold within nucleic acids processing enzymes: the beta-CASP family. Nucleic Acids Res. 30:3592-3601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J., and E. J. Wagner. 2010. snRNA 3′ end formation: the dawn of the Integrator complex. Biochem. Soc. Trans. 38:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Vegvar, H. E., E. Lund, and J. E. Dahlberg. 1986. 3′ end formation of U1 snRNA precursors is coupled to transcription from snRNA promoters. Cell 47:259-266. [DOI] [PubMed] [Google Scholar]
- 7.Dominski, Z., X. C. Yang, and W. F. Marzluff. 2005. The polyadenylation factor CPSF-73 is involved in histone-pre-mRNA processing. Cell 123:37-48. [DOI] [PubMed] [Google Scholar]
- 8.Dominski, Z., X. C. Yang, M. Purdy, and W. F. Marzluff. 2003. Cloning and characterization of the Drosophila U7 small nuclear RNA. Proc. Natl. Acad. Sci. U. S. A. 100:9422-9427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dominski, Z., X. C. Yang, M. Purdy, and W. F. Marzluff. 2005. Differences and similarities between Drosophila and mammalian 3′ end processing of histone pre-mRNAs. RNA 11:1835-1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dominski, Z., X. C. Yang, M. Purdy, E. J. Wagner, and W. F. Marzluff. 2005. A CPSF-73 homologue is required for cell cycle progression but not cell growth and interacts with a protein having features of CPSF-100. Mol. Cell. Biol. 25:1489-1500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egloff, S., D. O'Reilly, and S. Murphy. 2008. Expression of human snRNA genes from beginning to end. Biochem. Soc. Trans. 36:590-594. [DOI] [PubMed] [Google Scholar]
- 12.Egloff, S., et al. 2010. The integrator complex recognizes a new double mark on the RNA polymerase II carboxyl-terminal domain. J. Biol. Chem. 285:20564-20569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Forbes, D. J., T. B. Kornberg, and M. W. Kirschner. 1983. Small nuclear RNA transcription and ribonucleoprotein assembly in early Xenopus development. J. Cell Biol. 97:62-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Godfrey, A. C., et al. 2006. U7 snRNA mutations in Drosophila block histone pre-mRNA processing and disrupt oogenesis. RNA 12:396-409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Han, S. M., et al. 2006. Deleted in cancer 1 (DICE1) is an essential protein controlling the topology of the inner mitochondrial membrane in C. elegans. Development 133:3597-3606. [DOI] [PubMed] [Google Scholar]
- 16.Hata, T., and M. Nakayama. 2007. Targeted disruption of the murine large nuclear KIAA1440/Ints1 protein causes growth arrest in early blastocyst stage embryos and eventual apoptotic cell death. Biochim. Biophys. Acta 1773:1039-1051. [DOI] [PubMed] [Google Scholar]
- 17.Hernandez, G., Jr., F. Valafar, and W. E. Stumph. 2007. Insect small nuclear RNA gene promoters evolve rapidly yet retain conserved features involved in determining promoter activity and RNA polymerase specificity. Nucleic Acids Res. 35:21-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hernandez, N. 1985. Formation of the 3′ end of U1 snRNA is directed by a conserved sequence located downstream of the coding region. EMBO J. 4:1827-1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hernandez, N. 2001. Small nuclear RNA genes: a model system to study fundamental mechanisms of transcription. J. Biol. Chem. 276:26733-26736. [DOI] [PubMed] [Google Scholar]
- 20.Hernandez, N., and A. M. Weiner. 1986. Formation of the 3′ end of U1 snRNA requires compatible snRNA promoter elements. Cell 47:249-258. [DOI] [PubMed] [Google Scholar]
- 21.Huang, J., Z. Gong, G. Ghosal, and J. Chen. 2009. SOSS complexes participate in the maintenance of genomic stability. Mol. Cell 35:384-393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kennedy, S. A., et al. 2009. Crystal structure of the HEAT domain from the pre-mRNA processing factor Symplekin. J. Mol. Biol. 392:115-128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kolev, N. G., and J. A. Steitz. 2005. Symplekin and multiple other polyadenylation factors participate in 3′-end maturation of histone mRNAs. Genes Dev. 19:2583-2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kolev, N. G., T. A. Yario, E. Benson, and J. A. Steitz. 2008. Conserved motifs in both CPSF73 and CPSF100 are required to assemble the active endonuclease for histone mRNA 3′-end maturation. EMBO Rep. 9:1013-1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li, C., G. A. Harding, J. Parise, K. J. McNamara-Schroeder, and W. E. Stumph. 2004. Architectural arrangement of cloned proximal sequence element-binding protein subunits on Drosophila U1 and U6 snRNA gene promoters. Mol. Cell. Biol. 24:1897-1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li, Y., et al. 2009. HSSB1 and hSSB2 form similar multiprotein complexes that participate in DNA damage response. J. Biol. Chem. 284:23525-23531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lobo, S. M., et al. 1988. Localization and expression of U1 RNA in early mouse embryo development. Dev. Biol. 127:349-361. [DOI] [PubMed] [Google Scholar]
- 28.MacDonald, C. C., J. Wilusz, and T. Shenk. 1994. The 64-kilodalton subunit of the CstF polyadenylation factor binds to pre-mRNAs downstream of the cleavage site and influences cleavage site location. Mol. Cell. Biol. 14:6647-6654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mandel, C. R., Y. Bai, and L. Tong. 2008. Protein factors in pre-mRNA 3′-end processing. Cell. Mol. Life Sci. 65:1099-1122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mandel, C. R., et al. 2006. Polyadenylation factor CPSF-73 is the pre-mRNA 3′-end-processing endonuclease. Nature 444:953-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marz, M., A. Mosig, B. M. Stadler, and P. F. Stadler. 2007. U7 snRNAs: a computational survey. Genomics Proteomics Bioinformatics 5:187-195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Marzluff, W. F., E. J. Wagner, and R. J. Duronio. 2008. Metabolism and regulation of canonical histone mRNAs: life without a poly(A) tail. Nat. Rev. Genet. 9:843-854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Matera, A. G., R. M. Terns, and M. P. Terns. 2007. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat. Rev. Mol. Cell Biol. 8:209-220. [DOI] [PubMed] [Google Scholar]
- 34.McNamara-Schroeder, K. J., R. F. Hennessey, G. A. Harding, R. C. Jensen, and W. E. Stumph. 2001. The Drosophila U1 and U6 gene proximal sequence elements act as important determinants of the RNA polymerase specificity of small nuclear RNA gene promoters in vitro and in vivo. J. Biol. Chem. 276:31786-31792. [DOI] [PubMed] [Google Scholar]
- 35.Mowry, K. L., and J. A. Steitz. 1987. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science 238:1682-1687. [DOI] [PubMed] [Google Scholar]
- 36.Murthy, K. G., and J. L. Manley. 1995. The 160-kD subunit of human cleavage-polyadenylation specificity factor coordinates pre-mRNA 3′-end formation. Genes Dev. 9:2672-2683. [DOI] [PubMed] [Google Scholar]
- 37.Nijhawan, P., and W. F. Marzluff. 1979. Metabolism of low molecular weight ribonucleic acids in early sea urchin embryos. Biochemistry 18:1353-1360. [DOI] [PubMed] [Google Scholar]
- 38.Peter, A., et al. 2002. Mapping and identification of essential gene functions on the X chromosome of Drosophila. EMBO Rep. 3:34-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ramamurthy, L., T. C. Ingledue, D. R. Pilch, B. K. Kay, and W. F. Marzluff. 1996. Increasing the distance between the snRNA promoter and the 3′ box decreases the efficiency of snRNA 3′-end formation. Nucleic Acids Res. 24:4525-4534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruepp, M. D., et al. 2009. Mammalian pre-mRNA 3′ end processing factor CF I m 68 functions in mRNA export. Mol. Biol. Cell 20:5211-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutkowski, R. J., and W. D. Warren. 2009. Phenotypic analysis of deflated/Ints7 function in Drosophila development. Dev. Dyn. 238:1131-1139. [DOI] [PubMed] [Google Scholar]
- 42.Santiago, C., and W. F. Marzluff. 1989. Expression of the U1 RNA gene repeat during early sea urchin development: evidence for a switch in U1 RNA genes during development. Proc. Natl. Acad. Sci. U. S. A. 86:2572-2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scharl, E. C., and J. A. Steitz. 1996. Length suppression in histone messenger RNA 3′-end maturation: processing defects of insertion mutant premessenger RNAs can be compensated by insertions into the U7 small nuclear RNA. Proc. Natl. Acad. Sci. U. S. A. 93:14659-14664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Scharl, E. C., and J. A. Steitz. 1994. The site of 3′ end formation of histone messenger RNA is a fixed distance from the downstream element recognized by the U7 snRNP. EMBO J. 13:2432-2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shi, Y., et al. 2009. Molecular architecture of the human pre-mRNA 3′ processing complex. Mol. Cell 33:365-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Skaar, J. R., et al. 2009. INTS3 controls the hSSB1-mediated DNA damage response. J. Cell Biol. 187:25-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Steinmetz, E. J., and D. A. Brow. 1996. Repression of gene expression by an exogenous sequence element acting in concert with a heterogeneous nuclear ribonucleoprotein-like protein, Nrd1, and the putative helicase Sen1. Mol. Cell. Biol. 16:6993-7003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steinmetz, E. J., N. K. Conrad, D. A. Brow, and J. L. Corden. 2001. RNA-binding protein Nrd1 directs poly(A)-independent 3′-end formation of RNA polymerase II transcripts. Nature 413:327-331. [DOI] [PubMed] [Google Scholar]
- 49.Sullivan, K. D., M. Steiniger, and W. F. Marzluff. 2009. A core complex of CPSF73, CPSF100, and Symplekin may form two different cleavage factors for processing of poly(A) and histone mRNAs. Mol. Cell 34:322-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tao, S., Y. Cai, and K. Sampath. 2009. The Integrator subunits function in hematopoiesis by modulating Smad/BMP signaling. Development 136:2757-2765. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, E. J., et al. 2007. A genome-wide RNA interference screen reveals that variant histones are necessary for replication-dependent histone pre-mRNA processing. Mol. Cell 28:692-699. [DOI] [PubMed] [Google Scholar]
- 52.Wang, Z. F., M. L. Whitfield, T. C. Ingledue III, Z. Dominski, and W. F. Marzluff. 1996. The protein that binds the 3′ end of histone mRNA: a novel RNA-binding protein required for histone pre-mRNA processing. Genes Dev. 10:3028-3040. [DOI] [PubMed] [Google Scholar]
- 53.Weiner, A. M. 2005. E pluribus unum: 3′ end formation of polyadenylated mRNAs, histone mRNAs, and U snRNAs. Mol. Cell 20:168-170. [DOI] [PubMed] [Google Scholar]
- 54.Yuo, C. Y., M. Ares, Jr., and A. M. Weiner. 1985. Sequences required for 3′ end formation of human U2 small nuclear RNA. Cell 42:193-202. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.