Abstract
All gammaherpesviruses encode a glycoprotein positionally homologous to the Epstein-Barr virus gp350 and the Kaposi's sarcoma-associated herpesvirus (KSHV) K8.1. In this study, we characterized the positional homologous glycoprotein of bovine herpesvirus 4 (BoHV-4), encoded by the Bo10 gene. We identified a 180-kDa gene product, gp180, that was incorporated into the virion envelope. A Bo10 deletion virus was viable but showed a growth deficit associated with reduced binding to epithelial cells. This seemed to reflect an interaction of gp180 with glycosaminoglycans (GAGs), since compared to the wild-type virus, the Bo10 mutant virus was both less infectious for GAG-positive (GAG+) cells and more infectious for GAG-negative (GAG−) cells. However, we could not identify a direct interaction between gp180 and GAGs, implying that any direct interaction must be of low affinity. This function of gp180 was very similar to that previously identified for the murid herpesvirus 4 gp150 and also to that of the Epstein-Barr virus gp350 that promotes CD21+ cell infection and inhibits CD21− cell infection. We propose that such proteins generally regulate virion attachment both by binding to cells and by covering another receptor-binding protein until they are displaced. Thus, they regulate viral tropism both positively and negatively depending upon the presence or absence of their receptor.
Many viruses use a single glycoprotein for both cell binding and membrane fusion. Herpesviruses are more complex. Three proteins—gB, gH, and gL—form a core fusion machinery conserved in the Alpha-, Beta-, and Gammaherpesvirinae subfamilies (21). Most herpesviruses also encode at least one additional receptor-binding protein that is more specific for a given virus subfamily. For example, herpes simplex virus first attaches to cells by gB or gC binding to the heparan sulfate moieties of the cell surface proteoglycans. gD must then bind for fusion to occur (47).
Our understanding of gammaherpesvirus glycoprotein functions is more limited. This is due to the fact that the human gammaherpesviruses Epstein-Barr virus (EBV) and Kaposi's sarcoma-associated herpesvirus (KSHV) show limited lytic growth in vitro. Nevertheless, understanding how viral glycoproteins function is important for vaccination and for the development of neutralizing antibodies. While gB, gH, and gL are probably the key partners of the gammaherpesvirus membrane fusion machinery (22, 39, 41), the function of accessory entry proteins is less clear. EBV gp350/220 is a highly glycosylated membrane protein that adopts its 2 differently sized forms by alternative splicing (23). It is the most abundant protein in the virion envelope, binds to complement receptor 2 (CD21) on B cells (40, 49), and is a target for antibodies that neutralize B cell infection (53). Soluble recombinant gp350 can also inhibit EBV infection of CD21-positive (CD21+) cells (36). EBV lacking gp350/220 is poorly infectious for B cells (24, 46) yet infects CD21-negative (CD21−) epithelial cells better than the wild-type virus does (46). Virions treated with some antibodies against gp350/220 also show enhanced epithelial cell infection (54). The KSHV positional homolog of gp350/220, K8.1, also has 2 alternatively spliced forms (5). Both bind to cell surface heparan sulfate (HS) (2) and are thought to be involved in the initial steps of virion attachment. However, as for EBV gp350/220, K8.1 is dispensable for viral entry (33).
The difficulties of manipulating EBV and KSHV make related animal gammaherpesviruses important tools for deducing viral gene functions and their place in pathogenesis. Two of the best-established experimental models are provided by murid herpesvirus 4 (MuHV-4) and bovine herpesvirus 4 (BoHV-4). BoHV-4 infects healthy cattle worldwide and cattle with a variety of diseases (50). It readily propagates in cell culture and replicates in a broad range of host species both in vitro and in vivo. Recent viral genome sequencing (60) and bacterial artificial chromosome (BAC) cloning (12) have enhanced its value as an experimental model.
The positional homologs of K8.1 are M7 in MuHV-4, encoding gp150, and Bo10 in BoHV-4. The MuHV-4 gp150 is part of a multiprotein entry complex (19) and an immunodominant virion antibody target (14). However, gp150-specific antibodies do not neutralize. Instead, they strongly enhance Fc receptor-dependent infection (42). While gp150 seems to bind only weakly to glycosaminoglycans (GAGs) (6), the gp150− virus phenotype provides strong functional evidence for an important interaction (6, 10, 11, 15). While wild-type (WT) MuHV-4 infection is highly dependent on HS, gp150 knockouts are hardly dependent on HS at all, suggesting that a gp150-GAG interaction relieves a constitutive gp150-mediated inhibition of virion binding. The GAG-independent binding is presumably protected by gp150, thereby providing a mechanism of efficient virion release and antibody evasion. However, further studies are needed to determine whether this kind of binding regulation is a general feature of gammaherpesvirus biology.
The BoHV-4 Bo10 seems to be more closely related to the KSHV K8.1 than to the MuHV-4 M7. Sequence analysis shows that BoHV-4 Bo10 gene potentially comprises two exons that would together encode a protein with a length similar to that of K8.1 (30). In this study, Bo10 gene expression was characterized, and virus lacking this gene was produced, allowing us to analyze Bo10 gene function in the context of viral replication. Our results showed that Bo10 encodes a viral envelope protein involved in virion attachment. They suggested further that Bo10 regulates another viral binding interaction to make it essentially dependent on GAG.
MATERIALS AND METHODS
Cells and virus.
Madin-Darby bovine kidney (MDBK) cells (ATCC CCL-22), embryonic bovine trachea (EBTr) cells (ATCC CCL-44), CHO-K1 (ATCC CCL-61), and the glycosaminoglycan (GAG)-deficient mutant CHO-K1-pgs-A745 (ATCC CRL-2242) cells were cultured in Dulbecco's modified Eagle medium (Invitrogen) containing 10% fetal calf serum (FCS), 2% penicillin-streptomycin (Invitrogen), and 1% nonessential amino acids (Invitrogen). Bovine peripheral blood mononuclear cells (PBMC) were prepared as described elsewhere (17) and cultured in RPMI Glutamax medium containing 10% FCS, 2% penicillin-streptomycin (Invitrogen), 1% essential amino acids (Invitrogen), 1 mM sodium pyruvate, 25 mM HEPES, and 50 μM 2-mercaptoethanol. The BoHV-4 V. test strain initially isolated from a case of orchitis (51) and a derived recombinant strain, called V. test EGFP Xho I (EGFP stands for enhanced green fluorescent protein) (hereafter called WT eGFP) (16), were used throughout.
Determination of Bo10 kinetic class of transcription.
The experiments determining the kinetic class of transcription of Bo10 were performed as described elsewhere (34). Briefly, subconfluent monolayers of MDBK cells were infected with BoHV-4 V. test strain at a multiplicity of infection (MOI) of 1 PFU/cell. Four hours before infection, cycloheximide (CHX) (100 μg/ml) or phosphonoacetic acid (PAA) (300 μg/ml) was added to the culture medium to inhibit de novo protein synthesis or viral DNA polymerase activity, respectively. Eighteen hours after infection, cytoplasmic RNA was extracted, purified, and treated for reverse transcription-PCR (RT-PCR). The cDNA products were amplified by PCR using primers specific for Bo5 encoding BoHV-4 major immediate-early transcript (IE) (57), ORF21 encoding thymidine kinase expressed as an early gene (E) (27), ORF22 encoding glycoprotein H (gH) expressed as a late gene (L) (31) and Bo10-specific primers (Bo10 23-43 [5′-TCATACATTCAAATTGCATGC-3′] and Bo10 839-818 [5′-CATTGAATGAGAACAAACACG-3′]).
Production of rabbit polyclonal anti-Bo10 antibodies.
Anti-Bo10-c15 polyclonal mono-specific antibodies were produced by Sigma Genosys (Pampisford, United Kingdom). On day 0, equal volumes of diluted bovine serum albumin (BSA)-conjugated peptide (1 mg/ml) and Freund's complete adjuvant were emulsified and injected subcutaneously into the rabbits at three different sites (200 μg/rabbit). On days 14, 28, 42, 56, and 70, each rabbit was immunized again with 100 μg of peptide (1 mg/ml) emulsified with incomplete Freund's adjuvant. Serum samples were collected on day 77.
Western blotting.
Cells or virions were lysed and denatured by heating (95°C, 5 min) in SDS-PAGE sample buffer (31.25 mM Tris-HCl [pH 6.8], 1% [wt/vol] SDS, 12.5% [wt/vol] glycerol, 0.005% [wt/vol] bromophenol blue, 2.5% [vol/vol] 2-mercaptoethanol). Proteins were resolved by electrophoresis on Mini-PROTEAN TGX (Tris-glycine extended) precast 7.5% resolving gels (Bio-Rad) in SDS-PAGE running buffer (25 mM Tris base, 192 mM glycine, 0.1% [wt/vol] SDS) and transferred to polyvinylidene difluoride membranes (Immobilon-P transfer membrane with 0.45 μM pore size; Millipore). The membranes were blocked with 3% nonfat milk in phosphate-buffered saline (PBS) containing 0.1% Tween 20 (PBS-0.1% Tween 20), and then incubated with anti-Bo10-c15 rabbit antibodies or anti BoHV-4 polyserum in the same buffer. Bound antibodies were detected with horseradish peroxidase-conjugated goat anti-rabbit IgG polyclonal antibody (PAb) (Dako Corporation), followed by washing in PBS-0.1% Tween 20, development with enhanced chemiluminescence (ECL) substrate (GE Healthcare), and exposure to X-ray film.
5′ RACE.
The 5′ end of the Bo10 mRNA was mapped by rapid amplification of cDNA ends (RACE) (5′/3′ RACE kit second generation; Roche Diagnostics). The RACE primers were Bo10 900-878 (5′-TCATAATAAATTATATCCCTGAC-3′) for cDNA synthesis and Bo10 839-818 (5′-CATTGAATGAGAACAAACACG-3′) for PCR amplification [paired with a primer matching the 5′ poly(A) cDNA tail added by terminal deoxynucleotide transferase]. Both gene-specific primers were in exon 2, which encodes the amino acids recognized by the anti-Bo10-c15 immune serum. The PCR product was sequenced to locate the 5′ poly(A) tail.
Production of a BoHV-4 Bo10 deletion and revertant strains.
A BoHV-4 V. test strain deleted for nucleotides 43 to 608 of the Bo10 gene (Bo10 Del) was produced by homologous recombination (34). The restriction fragment EcoRI-B (containing Bo10) of the V. test strain genome was cloned into the pGEM-T Easy vector (Promega), resulting in pGEM-T Easy/EcoRI-B. This vector was then digested with HindIII and religated, resulting in pGEM-T Easy/EcoRI-B HinDIII. An enhanced green fluorescent protein (eGFP) expression cassette was then produced by PCR using the forward primer 5′-GCATGCACGCGTAATCAATTACGGGGTCATTAG-3′ with SphI-MluI sites (in italic type) and nucleotides 15 to 35 of the pEGFP-C1 vector (Clontech) (GenBank accession number U55763) and the reverse primer 5′-TTCGAACGCGTTAAGATACATTGATGAG-3′ with MluI-NspV sites (in italic type) and nucleotides 1640 to 1624 of pEGFP-C1. A modified version of the pEGFP-C1 vector in which most of the multiple cloning site had been deleted by digestion with BglII and BamHI was used as template. The PCR product was cloned into the pGEM-T Easy vector by TA cloning (Promega). The eGFP cassette released by NspV and SphI digestion was then cloned into pGEM-T Easy/EcoRI-B HinDIII, resulting in pGEM-T Easy/EcoRI-B HinDIII del Bo10 in which most of the Bo10 gene is replaced by the eGFP expression cassette. The latter plasmid was digested by MscI and XmnI, and the resulting fragment was used to generate the Bo10 Del V. test strain by homologous recombination in EBTr cells. Finally, a revertant strain (Bo10 Rev) was produced based on the same approach using the MscI-XmnI fragment of the pGEM-T Easy/EcoRI-B HinDIII vector as the recombination cassette.
Southern blotting.
Southern blot analysis of viral DNA digested with HindIII was performed (12) with probes corresponding to the entire Bo10 open reading frame (ORF) (exon I-intron-exon II) or to the deleted part of the V. test Bo10 ORF or the eGFP ORF.
Virus purification.
BoHV-4 strains grown on MDBK cells were purified as follows. Virions were harvested from infected MDBK cell supernatants by ultracentrifugation (100,000 × g, 2 h); infected-cell debris was then removed by low-speed centrifugation (1,000 × g, 10 min). Virions were then centrifuged through a 20 to 50% (wt/vol) potassium tartrate gradient in PBS (100,000 × g, 2 h). Virions were recovered from the gradient, washed, and concentrated in PBS (100,000 × g, 2 h).
Virion digestion with proteinase K.
Purified virions resuspended in PBS were subjected to proteinase K (Sigma) treatment (final concentration of 1 μg/μl) in the presence or absence of Triton X-100 (Sigma) (0.1% [vol/vol]) for 1 h at 56°C before Western blotting.
Growth curves.
The growth kinetics of mutant and revertant viruses were compared to those of the WT virus. Cell cultures were infected at a multiplicity of 0.05 (multistep assay) or 5 (one-step assay). After 1 h of adsorption, the cells were washed and then overlaid with minimal essential medium (MEM) containing 5% FCS. Supernatants of infected cultures and/or infected cells were harvested at successive intervals, and the amount of infectious virus was determined by plaque assay on MDBK cells (12).
Indirect immunofluorescence staining of adherent cells.
Cells were fixed in PBS containing 4% (wt/vol) paraformaldehyde (Merck) for 10 min on ice and then for 20 min at 20°C. After the cells were washed with PBS, they were permeabilized in PBS containing 0.1% (wt/vol) saponin (Sigma) at 37°C for 10 min. Immunofluorescence staining (incubation and washes) was performed in PBS containing 10% FCS (vol/vol) and 0.1% (wt/vol) saponin. Samples were incubated at 37°C for 45 min with mouse monoclonal antibody MAb 35 raised against BoHV-4 gB. After three washes, samples were incubated at 37°C for 30 min with Alexa Fluor 568-conjugated goat anti-mouse (GAM) IgG (2 μg/ml; Invitrogen). Fluorescence was then visualized with a Leica TCS/SP microscope and a Leica DC300F charge-coupled-device (CCD) camera system.
Plaque size.
MDBK cells grown on coverslips were infected with BoHV-4 and then overlaid with MEM containing 10% FCS and 0.6% (wt/vol) carboxymethylcellulose (CMC) (medium viscosity) (Sigma) in order to obtain isolated plaques (34). At successive intervals after infection, the plaques were stained by indirect immunofluorescence staining using MAb 35. Images were captured with a Leica TCS/SP microscope and a CCD camera system (DC 300F, IM50, version V1.20; Leica). For each virus, 30 plaques were measured, and the plaque area was determined with AnalySIS 3.2 software (Soft Imaging System).
Transmission electron microscopy.
Cells were washed with PBS and fixed directly in the dish in cacodylate buffer containing 2.5% glutaraldehyde and 2% paraformaldehyde. The cells were then scraped off and prepared for electron microscopy. Epon blocks and sections were prepared as described elsewhere (37). Sections were analyzed using a Technai Spirit transmission electron microscope (FEI, Eindhoven, Netherlands), and electron micrographs were taken using a bottom-mounted 4,000- by 4,000-pixel Eagle camera (FEI).
Enzymatic digestion of GAGs from cellular surface.
Adherent cells were washed two times with PBS and then incubated in 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 4 mM CaCl2, 0.01% BSA containing either heparinase II (5 U/ml) (Sigma) or chondroitinase ABC (5 U/ml) (Sigma) for 2 h at 37°C.
Sodium chlorate.
Sulfation was inhibited by sodium chlorate treatment (1, 26, 43). MDBK cells were cultured in MEM supplemented with 10% FCS, 2% penicillin-streptomycin (Invitrogen), 1% essential amino acids (Invitrogen), and different concentrations (0 to 100 mM) of sodium chlorate. In some experiments, 10 mM sodium sulfate was added to replenish sulfate to the cells. After overnight culture, cells were processed for virus infectivity.
Flow cytometry.
Cells exposed to eGFP+ viruses were washed in PBS and analyzed directly for green channel fluorescence (13). For intracellular staining, cells were fixed in 1% paraformaldehyde (30 min at room temperature) and then permeabilized with 0.1% saponin. The cells were incubated (1 h, 4°C) with either anti-Bo10-c15 rabbit antibodies or BoHV-4 gp8-specific MAb 103 (8) followed by Alexa Fluor 633-conjugated goat anti-rabbit PAb or Alexa Fluor 633-conjugated goat anti-mouse PAb (Invitrogen). The cells were then washed and analyzed in a FACSAria cell sorter (Becton Dickinson).
Silver staining.
Samples were electrophoresed on NuPAGE Novex 4 to 12% precast Bis-Tris polyacrylamide gels (Invitrogen) before silver staining (SilverXpress silver staining kit; Invitrogen).
RESULTS
Bo10 is expressed as a late gene during BoHV-4 infection.
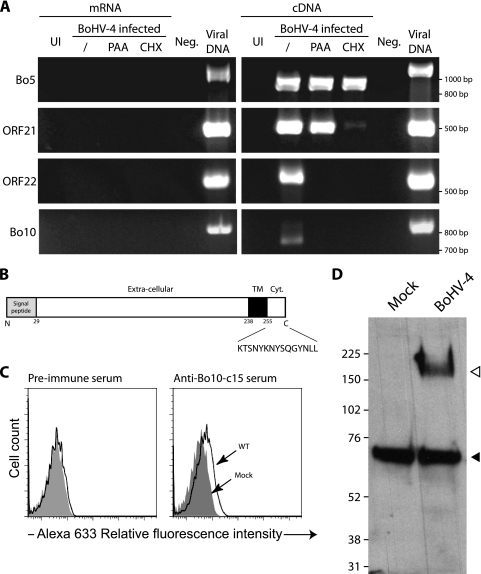
We first wanted to know how the Bo10 gene (GenBank accession number Z84818) was transcribed during BoHV-4 infection. The Bo10 gene is predicted to contain an intron (30), with the spliced mRNA encoding a 273-amino-acid (aa) protein with a signal sequence and membrane anchor. We used RT-PCR with primers spanning the putative intron to look for evidence of splicing. cDNA from BoHV-4-infected MDBK cells gave rise to a 738-bp PCR product (Fig. 1 A), corresponding to the expected size of the spliced mRNA. (An unspliced PCR product would be 816 bp.) No product was seen with mock-infected cells. DNA sequencing confirmed that the 738-bp product was from Bo10 with a splice as predicted (data not shown).
FIG. 1.
The Bo10 gene is expressed as a late gene and encodes a 180-kDa protein. (A) Determination of Bo10 kinetic class of transcription. MDBK cells were mock infected or infected with BoHV-4 V. test strain in the absence (/) or presence of CHX or PAA. Twenty-four hours postinfection, expression of immediate-early Bo5, early ORF21, and late ORF22 and Bo10 was studied using an RT-PCR approach as described in Materials and Methods. mRNA and cDNA represent PCR products generated when reverse transcriptase was omitted from the reaction mixtures and RT-PCR products, respectively. Amplification of viral genomic DNA is provided as control. UI and Neg. represent uninfected cell samples and PCR negative controls, respectively. The locations of the marker sizes (in base pairs) are indicated to the right of the gel. (B) Predicted product of expression of BoHV-4 Bo10 gene. Signal peptide and transmembrane (TM) regions were identified with SignalP and TMHMM software programs, respectively (http://www.expasy.ch/tools/). The sequence of the peptide used for rabbit immunization is shown below the map. Cyt., intracytoplasmic region. (C) Immunization with Bo10-c15 peptide elicited antibody response against the peptide. The antipeptide antibody responses were determined for two immunized rabbits by indirect immunofluorescence staining of permeabilized cells and FACS analysis as described in Materials and Methods. (D) Detection of specific BoHV-4 protein by the anti-Bo10-c15 serum. MDBK cells were left uninfected or infected with the WT V. test strain of BoHV-4 (1 PFU/cell). Forty-eight hours later, cells were scraped off the plate, and Western blotting was carried out as described in Materials and Methods. The positions of molecular mass (MM) standards (in kilodaltons) are shown to the left of the gel. The open and filled triangles indicate the specific 180-kDa protein and a background band, respectively.
Next, CHX and PAA were used to identify the kinetic class of Bo10 transcription. Bo10 expression was prevented by both CHX and PAA, indicating that it is a late gene. Bo5 (868 bp), ORF21 (499 bp), and ORF22 (564 bp) were used as controls; the results presented in Fig. 1A confirmed that they are immediate-early, early, and late genes, respectively. The absence of contaminant viral DNA in the mRNA preparations was confirmed by a lack of PCR product without reverse transcriptase. The size of the Bo5 RT-PCR product was also consistent with its known mRNA splicing (868 bp rather than 1,140 bp).
Detection of a BoHV-4 protein encoded by Bo10.
Bo10-specific polyclonal antibodies were generated by immunizing rabbits with a bovine serum albumin-conjugated peptide (KTSNYKNYSQGYNLL) from the C-terminal cytoplasmic tail of the predicted Bo10 gene product (Fig. 1B). To confirm that the serum recognized a viral gene product, we performed indirect immunofluorescence staining and fluorescence-activated cell sorting (FACS) analysis of MDBK cells either mock infected or infected with WT virus. Figure 1C shows positive albeit weak recognition of the WT virus-infected cells.
To identify the protein recognized by the anti-Bo10-c15 serum, Western blotting was performed on MDBK cells infected with WT virus. A protein with an apparent molecular mass (MM) of 180 kDa, hereafter called gp180, was detected under reducing conditions (Fig. 1D). No such protein was detected in mock-infected cells. The predicted MM of the Bo10 gene product is 24.9 kDa, so this result suggested that either the mRNA includes additional exons or that the protein runs aberrantly on SDS-polyacrylamide gels or that it undergoes extensive posttranslational modification. 5′ RACE identified the 5′ Bo10 AUG as that predicted by the sequence with GenBank accession number Z84818. Extensive glycosylation seemed the most likely explanation for the high MM of the Bo10 gene product. This hypothesis can be related to the sporadic detection of some proteins with MMs ranging from 63 kDa to 180 kDa, which likely represent some glycosylation intermediates (data not shown). It is noteworthy that this serum detected a cellular background band in both mock- and BoHV-4-infected cells that did not interfere with gp180 detection.
Bo10 is a structural protein that is nonessential for BoHV-4 replication in vitro.
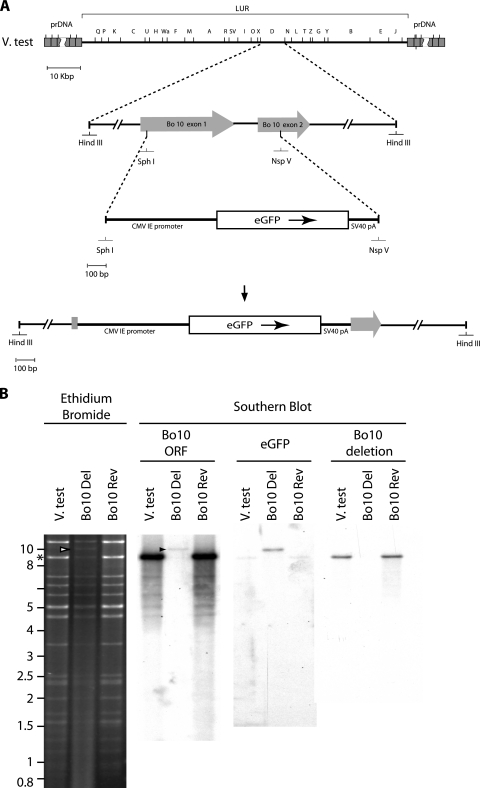
To establish the functional importance of Bo10 in virus replication, a mutant virus and revertant were produced (Fig. 2) as described in Materials and Methods. In the mutant, most of Bo10 exons 1 and 2 was replaced by an eGFP expression cassette (Fig. 2A and B). The predicted molecular structures of the recombinant strains were confirmed by restriction mapping and Southern blotting (Fig. 2B). The mutated region was also checked by DNA sequencing (data not shown) and found to be correct.
FIG. 2.
Production of a recombinant BoHV-4 strain deleted for Bo10. Using the BoHV-4 V. test strain as the parental strain, a recombinant BoHV-4 strain deleted for Bo10 and a derived revertant strain were produced by homologous recombination. (A) Recombination cassettes were constructed. The HindIII restriction map of the entire BoHV-4 V. test strain is shown at the top. Letters were assigned to the different restriction fragments according to their lengths, as described by Bublot et al. (4a). Most of Bo10 exons 1 and 2 were replaced by an eGFP expression cassette as described in Materials and Methods. prDNA, polyrepetitive DNA; CMV IE promoter, cytomegalovirus immediate-early promoter; SV40 pA, simian virus 40 pA. (B) Bo10 Del and Bo10 Rev strains were then characterized by a combined restriction endonuclease and Southern blotting approach. The DNA of the parental strain V. test and the derived recombinant strains Bo10 Del and Bo10 Rev were analyzed by HindIII restriction and further tested by Southern blotting using probes corresponding to the entire V. test Bo10 ORF or to the eGFP ORF or to the deleted part of the V. test Bo10 ORF. The asterisk and arrows indicate the restriction fragments containing the entire Bo10 and eGFP ORFs, respectively. Marker sizes (in kilobase pairs) are indicated on the left.
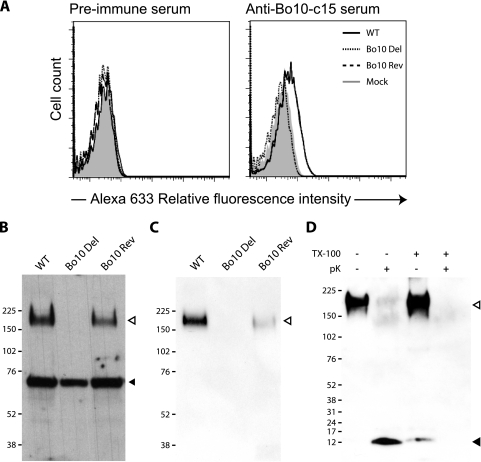
To confirm a lack of gp180 in cells infected with the Bo10 Del virus, we performed indirect immunofluorescence and Western blotting with the anti-Bo10-c15 serum. As seen in Fig. 3 A and B, both approaches confirmed that the mutant did not express gp180. By analogy with the positional homologs of other viruses, gp180 would be expected to be a virion component. To test this hypothesis, we purified virions on tartrate gradients before Western blotting. As with infected cells, we observed a band at 180 kDa for both WT and Bo10 Rev virions. In Fig. 3C, the cellular protein recognized by the Bo10 antiserum (Fig. 3B) was lacking, consistent with virion purification. To demonstrate that gp180 is in the viral envelope, we treated purified virions with proteinase K in the presence or absence of Triton X-100. A protected fragment was observed only without detergent, consistent with gp180 being a type I transmembrane protein. The weak band observed with Triton X-100 suggested only some nonspecific protein degradation and did not affect the conclusion that gp180 is a component of the BoHV-4 virion envelope.
FIG. 3.
gp180 detection in infected cells and virions. (A) MDBK cells were either mock infected or infected with the WT, Bo10 Del, or Bo10 Rev virus strain (1 PFU/cell). At 24 h postinfection, the cells were fixed and stained with either preimmune serum or anti-Bo10-c15 serum for FACS analysis as described in Materials and Methods. Alexa 633, Alexa Fluor 633. (B and C) Detection of specific BoHV-4 protein by the anti-Bo10-c15 serum. Infected MDBK cells (48 h, 1 PFU/cell) (B) or purified virions (5 × 105 virions per lane) (C) were subjected to Western blotting with anti-Bo10-c15 serum as described in Materials and Methods. The position of a MM standard is shown. Open and filled triangles indicate the specific 180-kDa protein and a background band, respectively. (D) BoHV-4 WT purified virions (5 × 105 virions per lane) were treated with proteinase K (pK) in the presence (+) or absence (−) of Triton X-100 (TX-100) (0.1% [vol/vol]) and then subjected to Western blotting with anti-Bo10-c15 serum as described in Materials and Methods. The open and filled triangles indicate the specific 180-kDa protein and the protected C-terminal end of gp180, respectively. The positions of MM standards (in kilodaltons) are shown to the left of the gels in panels B to D.
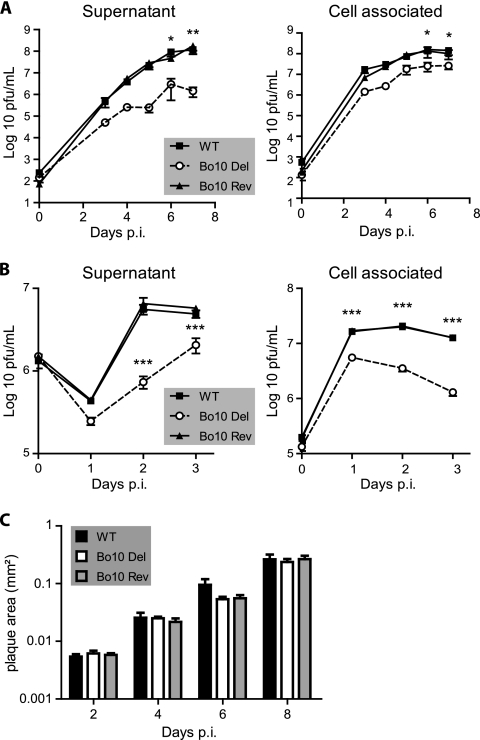
In order to address the role of Bo10 in BoHV-4 lytic infection, one-step and multistep growth assays were performed with WT, Bo10 Del, and Bo10 Rev viruses on MDBK cells. The Bo10 Del virus grew to lower titers than WT or Bo10 Rev virus did in both single-step and multistep growth assays (Fig. 4 A and B). The growth deficit was similar in supernatant and cell-associated virus fractions. Plaque sizes under semisolid medium were similar, suggesting that cell-to-cell virus spread was intact (Fig. 4C). Taken together, these results indicated that the Bo10 gene encodes a viral envelope protein that is not essential for BoHV-4 replication in vitro but that the absence of Bo10 impaired infection by cell-free virions.
FIG. 4.
Effect of Bo10 deletion on BoHV-4 replication in vitro. (A and B) MDBK cells grown in 6-well cluster dishes were infected at an MOI of 0.05 in a multistep assay (A) or at an MOI of 5 in a one-step assay (B) as described in Materials and Methods with BoHV-4 wild-type V. test, Bo10 Del, and Bo10 Rev. Supernatant of infected cultures and infected cells were harvested at different times (days) postinfection (p.i.), and the amount of infectious virus was determined by plaque assay on MDBK cells. For supernatants, time zero p.i. is retitration of the inoculums to ensure that similar amounts of virus were put on the cells. Plaques were visualized by immunofluorescence staining as described in Materials and Methods. The data presented are the averages ± standard errors of the means (SEMs) (error bars) for triplicate measurements. The data were analyzed by two-way analysis of variance (ANOVA) and Bonferroni posttests. Statistical significance is indicated as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001. (C) Bo10 deletion has no effect on BoHV-4 plaque size. MDBK cells grown on coverslips were infected with BoHV-4 wild type V. test, Bo10 Del, and Bo10 Rev strains and then overlaid with MEM containing CMC as described in Materials and Methods. At successive intervals after infection, plaques were revealed by indirect immunofluorescence staining using MAb 35 (recognizing gB) and Alexa Fluor 568-conjugated goat anti-mouse IgG as the primary and secondary antibodies, respectively. The pictures of plaques were captured with a CCD camera and analyzed with AnalySIS 3.2 software (Soft Imaging System) in order to determine plaque area. Each value presented is the average ± SEM (error bars) for the measurement of 30 plaques. The data were analyzed by two-way ANOVA and Bonferroni posttests, and no significant difference was observed.
The Bo10 deletion phenotype is not associated with a detectable deficit in virion release.
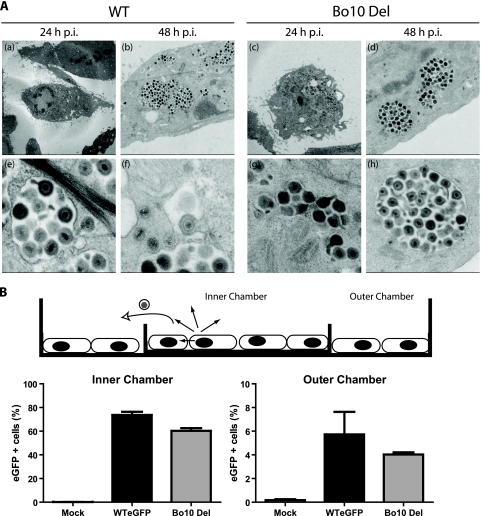
Because deletion of MuHV-4 gp150, one of the best-characterized positional homologs of Bo10, is associated with poor virion release (6), one possible explanation for the observed growth deficit of the Bo10 Del virus was poor virion release from infected cells. However, electron microscopy (Fig. 5 A) did not show Bo10 Del virions piled up on the plasma membrane as it had for MuHV-4.
FIG. 5.
The Bo10 deletion phenotype is not associated with a virion release deficit. (A) Electron microscopy of virus-infected cells. MDBK cells were infected (24 or 48 h; 1 PFU/cell) with WT or Bo10 Del virus as indicated. The pictures shown are representative of at least 10 sections per sample. For each time point, two different pictures at different magnifications are provided. (B) Fluid-phase virus spread. A total of 2 × 105 infected MDBK cells (MOI of 0.5) or 2 × 105 uninfected MDBK cells were seeded either on a 6-cm-diameter dish or in a 3.5-cm-diameter dish placed inside the first one. The cells were therefore separated by a physical barrier but connected by medium so that virus could not spread directly from the infected to the uninfected population but only via their common supernatant. After 48 h, the cells were harvested and analyzed for eGFP expression (eGFP + cells, eGFP-expressing cells). The data presented are the averages plus SEMs (error bars) for triplicate measurements and were analyzed by t test. The values for the WT eGFP and Bo10 Del virus strains were not significantly different.
We also conducted an experiment similar to that performed by de Lima et al. (6) to demonstrate the release deficit of gp150-deficient MuHV-4 (Fig. 5B). MDBK cells were seeded on either side of a separating barrier (a small “inner chamber” dish inside a larger “outer chamber”). Only the cells in the inner chamber were infected. After 2 h, the infected cells were washed with PBS, and medium was added to cover both monolayers and the divide between them. Thus, virus could spread from cell to cell within the infected population but could reach the uninfected population only via fluid-phase spread. In contrast to what was observed for MuHV-4, there was no difference in the spread of the WT and Bo10 Del viruses. A release deficit seemed therefore unlikely to account for the reduced growth of the Bo10 Del virus.
Kinetics of Bo10-deficient virion uptake.
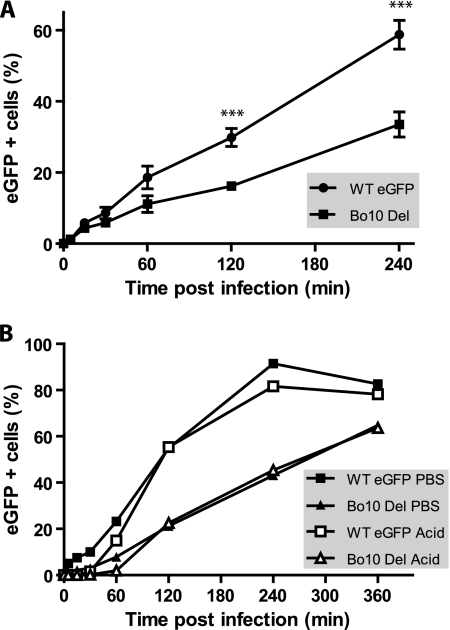
It was also possible that Bo10 Del replication deficit could be due to impaired virus binding. We tested this by incubating MDBK cells with eGFP-expressing WT virus or Bo10 Del BoHV-4 virus (0.5 PFU/cell) for various times before washing the input virus off with PBS. The number of infected cells (eGFP+) was determined by flow cytometry 18 h later (Fig. 6 A). It should be noted that in plaque assays of virus stocks, the viral inoculum was not removed so as to minimize any undertitering of Bo10-deficient viruses due to slow attachment. When the viral inoculum was removed, it was clear that the Bo10 Del virus was slower to attach than the WT virus. The deficit was equivalent whether the cells were washed with PBS (pH 7.4) to remove unbound virions or with acid (pH 3) (Fig. 6B) to also inactivate any virions that had not penetrated the plasma membrane. (BoHV-4 enters MDBK cells through endocytosis [C. Lété, unpublished data].) Thus, Bo10 Del virions appeared to be impaired in cell binding.
FIG. 6.
Infectivity assays with Bo10 Del BoHV-4 mutant. (A) MDBK cells were exposed to eGFP-expressing (eGFP +) wild-type virus (WT eGFP) or Bo10 Del BoHV-4 strain (0.5 PFU/cell) for the times indicated and then washed with PBS and cultured overnight. Viral infection was assayed 18 h later by flow cytometry for eGFP expression. The data presented are the average ± SEMs for triplicate measurements. The data were analyzed by two-way ANOVA and Bonferroni posttests. The values for WT eGFP-infected cells were significantly different (P < 0.001) from the values for the Bo10 Del strain as indicated (***). (B) MDBK cells were exposed to WT eGFP or Bo10 Del BoHV-4 strain (0.5 PFU/cell) for the times indicated and then washed either with PBS (pH 7.4) or with isotonic (pH 3) buffer (acid wash). Viral infection was assayed by measuring eGFP expression 18 h later as for panel A.
GAG interaction in BoHV-4 Bo10 Del infection.
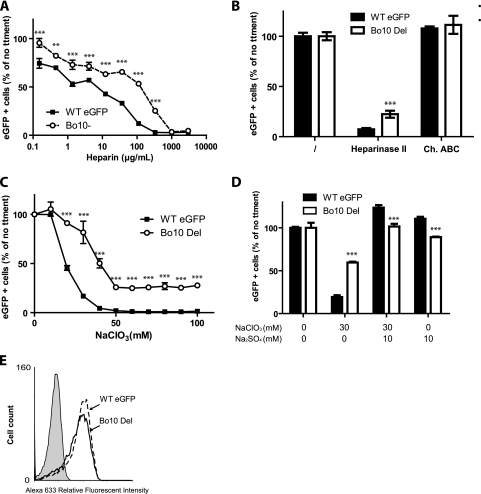
By analogy with HHV-8 K8.1 envelope glycoprotein, BoHV-4 gp180 could be involved in GAG interaction. Therefore, we tested the GAG dependence of WT virus and Bo10 Del BoHV-4 infections. As the Bo10 Del strain shows impaired binding, it was not possible to have equivalent percentages of eGFP-positive cells after 18 h of infection (one round of infection). We therefore present our results as the percentage of maximal infection with each virus. Compared to the WT virus, Bo10 Del viruses were approximately 10-fold-more resistant to inhibition by soluble heparin (Fig. 7 A). Heparinase treatment of cells similarly inhibited WT virus infection much more than Bo10 Del infection (Fig. 7B). In contrast, removing chondroitin sulfate did not affect MDBK infection by either WT or Bo10 Del viruses (Fig. 7B). Together, these results suggested that the defect of gp180-deficient BoHV-4 was in GAG binding.
FIG. 7.
gp180 is one of the BoHV-4 proteins interacting with heparan sulfate (HS). (A) WT eGFP or Bo10 Del BoHV-4 virions were preincubated with increasing amounts of soluble heparin for 2 h at 37°C. Viruses and heparin were then added to MDBK cells (MOI of 0.5 PFU/cell). Eighteen hours later, viral eGFP expression was assayed by flow cytometry. Each value is expressed as a percentage of the eGFP expression with untreated virus (no treatment [no ttment]). The data presented are the averages ± SEMs (error bars) for triplicate measurements and were analyzed by two-way ANOVA and Bonferroni posttests. Statistical significance was indicated as follows: **, P < 0.01; ***, P < 0.001. (B) Cell surface HS or chondroitin sulfate (CS) was enzymatically removed from MDBK cells after treatment with heparinase II or chondroitinase ABC (Ch. ABC), respectively. WT eGFP or Bo10 Del BoHV-4 was then added for 2 h on ice before extensive PBS washing to remove unbound virions. Eighteen hours later, viral eGFP expression was assayed by flow cytometry. Each value is expressed as a percentage of eGFP expression with untreated cells shown in the leftmost pair of bars. The data presented are the averages ± SEMs (error bars) for six measurements and were analyzed by two-way ANOVA and Bonferroni posttests. Heparinase II treatment of the cells resulted in a significantly lower level of infection by the B010 Del virus than by the WTeGFP virus (P < 0.001), as indicated (***). (C and D) Sulfation inhibition causes less entry of Bo10 Del virions than of WT virions. MDBK cells were cultured overnight in the presence of indicated amounts of sodium chlorate (NaClO3). The cells were then infected with WT eGFP or Bo10 Del BoHV-4 strain (MOI of 0.5 PFU/cell). Eighteen hours later, viral eGFP expression was assayed by flow cytometry. Each value is expressed as a percentage of the eGFP expression with untreated cells. The data presented are the averages ± SEMs (error bars) for four measurements and were analyzed by two-way ANOVA and Bonferroni posttests. Statistical significance is indicated as follows: ***, P < 0.001. (C) Addition of sodium sulfate (10 mM) restores WT eGFP and Bo10 Del infectivity. MDBK cells were cultured overnight with the indicated concentrations of sodium chlorate (NaClO3) and/or sodium sulfate (Na2SO4). The cells were then infected with WT eGFP or Bo10 Del BoHV-4 strain (MOI of 0.5 PFU/cell). Eighteen hours later, viral eGFP expression was assayed by flow cytometry. Each value is expressed as a percentage of the eGFP expression with untreated cells. The data presented are the averages ± SEMs for triplicate measurements and were analyzed by two-way ANOVA and Bonferroni posttests, Statistical significance is indicated as follows: ***, P < 0.001. (E) MDBK cells were left uninfected (filled histogram) or infected (1 PFU/cell) with either the WT eGFP or Bo10 Del strain of BoHV-4. Each population was then analyzed by flow cytometry for BoHV-4 gp8 (MAb 103) expression as described in Materials and Methods.
The specificity of GAG-protein interactions often depends on sulfation (20, 55). To clarify whether GAG sulfation is differentially required for WT or Bo10 Del virus infection, we treated MDBK cells with the reversible sulfation inhibitor sodium chlorate (1) (Fig. 7C). This did not affect cell viability as analyzed by propidium iodide exclusion (data not shown) but substantially reduced MDBK infection by WT BoHV-4. Bo10 Del virions were significantly less affected. Importantly, for both strains, the phenomenon was reversed by the addition of exogenous (10 nM) sodium sulfate (Fig. 7D). It is noteworthy that exogenous sodium sulfate increased MDBK infection by WT eGFP compared to no treatment but did not affect MDBK infection by the Bo10 Del strain (P < 0.01). This phenomenon possibly reflects better binding of the WT eGFP virions when sulfation of heparan sulfate is increased. These results established that GAG sulfation is critical for WT BoHV-4 infection and that Bo10 Del BoHV-4 is substantially less affected.
Since the BoHV-4 gp8 has been shown to bind heparinlike moieties (56), a possible explanation for the Bo10 Del phenotype was decreased gp8 expression. However, immunofluorescence staining showed that MDBK cells infected with either the WT or Bo10 Del virus strain expressed similar levels of gp8 (Fig. 7E).
Enhanced infection of GAG-deficient cells by the BoHV-4 Bo10 Del strain.
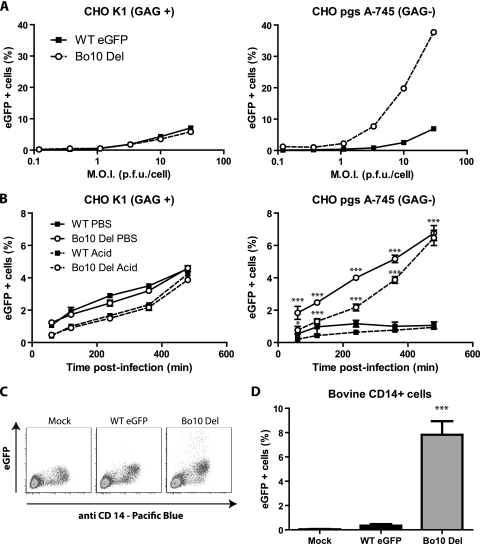
We further explored the apparently decreased GAG dependence of Bo10-deficient BoHV-4 by infecting CHO-K1 fibroblasts competent or not competent for GAG expression (Fig. 8 A). Bo10 Del viruses infected GAG+ CHO cells similarly to WT virus but infected GAG− CHO cells much better.
FIG. 8.
Bo10 deletion increases BoHV-4 entry in GAG-deficient (GAG−) cells. (A) CHO-K1 cells (CHO GAG+) and the GAG-deficient derivative CHO-pgsA-745 (CHO GAG−) were infected at the indicated MOI with the WT eGFP or Bo10 Del strain of BoHV-4. Twenty-four hours later, viral eGFP expression was assayed by flow cytometry. (B) CHO GAG+ and CHO GAG− cells were exposed to WT eGFP or Bo10 Del BoHV-4 strain (3 PFU/cell) for the times indicated and then washed either with PBS (pH 7.4) or with isotonic (pH 3) buffer (acid wash). Viral infection was assayed by measuring eGFP expression 18 h later as for panel A. The data presented are the averages ± SEMs (error bars) for three measurements. For the same treatment, WT eGFP and Bo10 del BoHV-4 strains were compared by two-way ANOVA and Bonferroni posttests. Statistical significance is indicated as follows: *, P < 0.05; ***, P < 0.001. (C and D) Bovine PBMC were isolated on Ficoll gradient and then left uninfected or infected with the WT eGFP or Bo10 Del strain of BoHV-4 (1 PFU/cell). (C) Twenty-four hours later, the cells were analyzed by flow cytometry for CD14 and viral eGFP expression as described in Materials and Methods. The data presented in panel D are the averages ± SEMs for six measurements and were analyzed by one-way ANOVA and Bonferroni posttests. Statistical significance is indicated as follows: ***, P < 0.001.
To discriminate between cell binding and penetration, we incubated CHO GAG+ and CHO GAG− cells with eGFP-expressing WT or Bo10 Del BoHV-4 (1 PFU/cell) for various times before PBS or acid washing. The number of infected cells (eGFP+) was determined by flow cytometry 18 h later (Fig. 8B). (The titers of the virus inputs were determined on MDBK cells, as CHO cells are not permissive for BoHV-4.) In contrast to CHO GAG+ cells, the mutant virus entered CHO GAG− cells better than the WT virus did. The ∼6 h for which the mutant virions remained susceptible to acid washing was comparable to the time taken for WT virions infecting CHO GAG+ cells to become acid wash resistant. Thus, the better infection of CHO GAG− cells by Bo10 Del virions appeared to reflect better cell binding, rather than better penetration.
In vivo, BoHV-4 infects monocytes, which are however relatively GAG deficient (28). We therefore compared the capacity of WT or Bo10 Del viruses to infect those cells ex vivo. While WT viruses infected bovine CD14+ cells very poorly, Bo10 Del viruses infected them much better (Fig. 8C and D).
Comparison of structural proteins of the different strains.
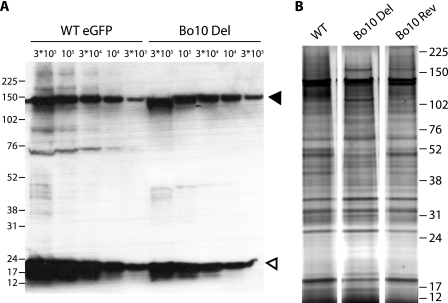
In view of the impaired capacity of Bo10 Del BoHV-4 to infect GAG+ cells, its enhanced capacity to infect GAG-deficient cells could potentially have been due in part to undertitration of virus stocks. However, immunoblots of virion lysates (Fig. 9 A) showed that Bo10 Del and WT eGFP virus stocks had equivalent protein contents for a given titer. We were also not able to see any difference in particle counts by electron microscopy (EM) on equivalent virus stocks.
FIG. 9.
Comparison of the structural proteins of the different strains. (A) WT eGFP and Bo10 Del concentrated stocks were compared for protein content by immunoblotting with anti-BoHV-4 rabbit polyserum. The black and white triangles indicate bands corresponding to VP7 and VP24 proteins, respectively; the two bands probably correspond to ORF25 and ORF65 encoding major capsid protein and capsid protein, respectively. 3*105, 3 × 105. (B) Tartrate-purified virions were loaded onto 4 to 12% Bis-Tris polyacrylamide gels and silver stained. The positions of molecular mass markers (in kilodaltons) are indicated to the left of the gel in panel A and to the right of the gel in panel B.
Some minor differences in proteins at 70 to 90 kDa were detected between WT and Bo10 Del virus stocks. Additional silver staining established that all the bands present in WT and Bo10 Rev stocks were also present in the Bo10 Del stock. Thus, while minor but systematic quantitative differences were hard to exclude, in our experience, the variation observed was no greater than the variation seen normally between different stocks of the same virus. Such differences can result from minor contamination of virion stocks with infected-cell debris. Possibly, it can also result from protein degradation. Certainly, it seemed clear that the enhanced infection of GAG-deficient cells by Bo10 Del BoHV-4 could not be attributed to an increased virus input relative to the WT virus.
DISCUSSION
In this study, we characterized the BoHV-4 Bo10 gene, a positional homolog of the EBV gp350 and KSHV K8.1 genes. Our results showed that Bo10 is expressed as a late gene and encodes a 180-kDa protein. A Bo10 mutant virus was viable but showed a growth deficit. This was not associated with a defect in cell-cell spread or viral release. Instead, gp180-deficient virions appeared slower than the WT virions to attach to GAG+ cells. Bo10 disruption also reduced the normal dependence of BoHV-4 on GAGs for efficient infection, such that gp180-deficient virions were more resistant to inhibition by soluble heparin and showed enhanced infection of GAG-deficient cells. Several hypotheses could explain these observations. One hypothesis is that some additional modifications on other structural proteins were induced by the Bo10 deletion. However, by analogy with MuHV-4 gp150, we propose that BoHV-4 gp180 regulates virion attachment to cells by covering a GAG-independent cell-binding epitope until displaced from that site by interaction with GAGs. Though the Bo10 Del virions would not necessarily bind more readily. Indeed, the cellular ligand might not become optimally available until heparan sulfate (HS) or other cellular ligand is engaged. For example, if gp180 binding signals to infected cells, it might lead to receptor recruitment/capping.
A possible rationale for this arrangement would be for gp180 to protect an otherwise vulnerable virion entry protein from antibody. Herpesviruses are generally transmitted from immunologically primed hosts as cell-free virions and so need to avoid neutralization by the host antibody response. The MuHV-4 gp150/BoHV-4 gp180 mechanism could allow key epitopes to be revealed only at the cell surface or even after endocytosis, thereby restricting the opportunities for neutralizing antibodies to bind. A very low affinity GAG interaction may be important for this, because it would help to avoid triggering by soluble GAGs. This seems to apply to gp150 and gp180, as neither binds to GAG+ cells as an Fc fusion protein (data not shown). A requirement for high avidity would limit gp150/gp180 displacement to after attachment, for example after BoHV-4 GAG binding via gp8. One prediction would be that BoHV-4 antibodies specific for the covered epitope should neutralize gp180-deficient virions but not the WT virions.
If BoHV-4 gp180 has to be displaced from its inhibitory site to reveal GAG-independent entry epitopes, this raises the question as to how BoHV-4 normally infects GAG-deficient cells. In vitro, this is generally inefficient (Fig. 8). However, in vivo BoHV-4 is readily recovered from monocytes (3, 9, 32) that are naturally HS deficient (6, 28). Direct cell-cell spread is a possible infection route, as observed for other herpesviruses (7, 35), that could be less dependent on GAG due to a lower need to attach cell-free virions. It is also possible that BoHV-4 produces gp180-deficient virions in vivo from some specific cell types, much as EBV from epithelial cells is more B cell tropic (4), perhaps by not splicing the Bo10 mRNA. BoHV-4 could also access GAG-deficient cells by transfer from the surfaces of GAG+ cells, similar to the epithelial infection route proposed for EBV (46). Finally, gp180-specific antibodies could make WT virions free of HS dependence by patching gp180 in the virus envelope as proposed for gp350/220 (54).
gp180 restricting GAG-independent virion binding could also be a mechanism of virion release. This appeared to operate for MuHV-4 by infected cells having decreased GAG expression (6). Herpesviruses could also do this by releasing GAG-binding glycoprotein fragments (10, 40, 43), including the BoHV-4 gp8 (8). This could explain why we did not detect a release deficit of gp180-deficient virions.
It is intriguing why the various gp180 positional homologs of different gammaherpesviruses are so diverse. Nevertheless, they all appear to be type I transmembrane proteins and have a heavily O-glycosylated stalk (Table 1). Our results suggest further that they are also related in function. Thus, they seem both to bind to a receptor and to block infection of cells that do not express this receptor (6, 48; this study). Therefore, we hypothesize that these proteins provide a glycan shield independent of cell binding (51) that could help to protect vulnerable virion epitopes as reported for the MuHV-4 gB N-terminal domain (18). Thus, their diversity could to a degree reflect immune selection. In conclusion, BoHV-4 Bo10 encodes a 180-kDa viral envelope protein that contributes to virus entry being dependent on GAG. We hypothesize that it acts by hiding a critical cell-binding epitope on virions until displaced by GAG, as proposed for MuHV-4 gp150. We hypothesize that all these homologs have an important role in regulating gammaherpesvirus tropism through both positive and negative effects.
TABLE 1.
Comparison of the different gammaherpesvirus K8.1 homologs
| Characteristic | Lymphocryptovirus HHV-4a | Rhadinovirusesa |
|||
|---|---|---|---|---|---|
| HHV-8 | MuHV-4 | SaHV-2 | BoHV-4 | ||
| Gene name | BLLF1 | K8.1 | M7 | ORF51 | Bo10 |
| Protein name | gp350 | K8.1 A | gp150 | ORF51 | gp180 |
| Protein characteristicsb | |||||
| Length (no. of aa) | 886 | 228 | 483 | 269 | 273 |
| Protein type | Type I | Type I | Type I | Type I | Type I |
| Predicted mass (kDa)c | 90 | 22.2 | 47.4 | 27.7 | 25 |
| Observed mass (kDa) | 350 (52) | 68-72 (59) | 150 (48) | 52 (38) | 180 |
| Glycosylationd | |||||
| N-glycosylation sites | 30 potential sites (confirmed)e (45) | 4 potential sites (confirmed) (58) | 3 potential sites | 7 potential sites (confirmed) (38) | 7 potential sites |
| O-glycosylation sites | 140 potential sites (confirmed) (45) | 12 potential sites (confirmed) (58) | 98 potential sites | 34 potential sites | 123 potential sites |
| Presence in virion | Yes (25) | Yes (29) | Yes (48) | Yes (38) | Yes |
| Neutralizing antibody target | Yes for B cell infection (53), no for epithelial cells (54) | Yes (in heterologous model) (44) | No (14) (immunodominant protein-immunological decoy) (14) | No (polyserum) (38) | No (polyserum) |
| Deleted strain viability | Viable (24) | Viable (33) | Viable (6) | Viable | |
| Putative function in virus entry | |||||
| Attachment of virion | Attachment of virion to B cell receptor CR2/CD21 (49) | Attachment of virion to heparan sulfate (2) | Attachment of virion to heparan sulfate (38) | ||
| Triggering cell entry | Triggers GAG+ cell entry through GAG interaction (6, 10) | Triggers GAG+ cell entry through GAG interaction | |||
| Inhibition of virus entry | Inhibits virus entry in CD21- cells (46) | Inhibits virus entry in GAG− cells (6, 10) | Inhibits virus entry in GAG− cells | ||
HHV-4 and HHV-8, human herpesvirus 4 and 8, respectively; MuHV-4, murid herpesvirus 4; SaHV-2, saimiriine herpesvirus 2; BoHV-4, bovine herpesvirus 4.
When 2 forms of the protein exist, only the characteristics of the predominant form are reported.
Predicted with the Compute pI/Mw tool on the ExPaSy proteomics server.
Predicted with NetNGlyc 1.0 and NetOGlyc 3.1 servers.
(confirmed), that type of glycosylation has been experimentally confirmed.
Acknowledgments
B.M., C.L., B.D., and L.G. are Research Fellow, Research Fellow, Postdoctoral Researcher, and Research Associate of the “Fonds de la Recherche Scientifique-Fonds National Belge de la Recherche Scientifique” (FRS-FNRS), respectively. P.G.S. is a Wellcome Trust Senior Clinical Fellow (GR076956MA). This work was supported by the following grants: starting grant (D-09/11) and GLYVIR ARC of the University of Liège and scientific impulse grant of the FRS-FNRS (grant F.4510.10).
Footnotes
Published ahead of print on 10 November 2010.
REFERENCES
- 1.Baeuerle, P. A., and W. B. Huttner. 1986. Chlorate-a potent inhibitor of protein sulfation in intact cells. Biochem. Biophys. Res. Commun. 141:870-877. [DOI] [PubMed] [Google Scholar]
- 2.Birkmann, A., K. Mahr, A. Ensser, S. Yaguboglu, F. Titgemeyer, B. Fleckenstein, and F. Neipel. 2001. Cell surface heparan sulfate is a receptor for human herpesvirus 8 and interacts with envelope glycoprotein K8.1. J. Virol. 75:11583-11593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boerner, B., W. Weigelt, H. J. Buhk, G. Castrucci, and H. Ludwig. 1999. A sensitive and specific PCR/Southern blot assay for detection of bovine herpesvirus 4 in calves infected experimentally. J. Virol. Methods 83:169-180. [DOI] [PubMed] [Google Scholar]
- 4.Borza, C. M., and L. M. Hutt-Fletcher. 2002. Alternate replication in B cells and epithelial cells switches tropism of Epstein-Barr virus. Nat. Med. 8:594-599. [DOI] [PubMed] [Google Scholar]
- 4a.Bublot, M., M. F. Van Bressem, E. Thiry, J. Dubuisson, and P. P. Pastoret. 1990. Bovine herpesvirus 4 genome: cloning, mapping and strain variation analysis. J. Gen. Virol. 71:133-142. [DOI] [PubMed] [Google Scholar]
- 5.Chandran, B., C. Bloomer, S. R. Chan, L. Zhu, E. Goldstein, and R. Horvat. 1998. Human herpesvirus-8 ORF K8.1 gene encodes immunogenic glycoproteins generated by spliced transcripts. Virology 249:140-149. [DOI] [PubMed] [Google Scholar]
- 6.de Lima, B. D., J. S. May, and P. G. Stevenson. 2004. Murine gammaherpesvirus 68 lacking gp150 shows defective virion release but establishes normal latency in vivo. J. Virol. 78:5103-5112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dingwell, K. S., C. R. Brunetti, R. L. Hendricks, Q. Tang, M. Tang, A. J. Rainbow, and D. C. Johnson. 1994. Herpes simplex virus glycoproteins E and I facilitate cell-to-cell spread in vivo and across junctions of cultured cells. J. Virol. 68:834-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dubuisson, J., I. Koromyslov, P. P. Pastoret, and E. Thiry. 1992. Proteins of bovine herpesvirus type 4 released into the culture medium of productively infected cells: identification of a 135K glycoprotein involved in viral attachment. J. Gen. Virol. 73:189-194. [DOI] [PubMed] [Google Scholar]
- 9.Fabian, K., R. Ivanics, M. Terenyi, and L. Egyed. 2005. Detection of bovine herpesvirus 4 in CD11b+ leukocytes of experimentally infected rabbits. Acta Vet. Hung. 53:265-273. [DOI] [PubMed] [Google Scholar]
- 10.Gillet, L., H. Adler, and P. G. Stevenson. 2007. Glycosaminoglycan interactions in murine gammaherpesvirus-68 infection. PLoS One 2:e347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillet, L., S. Colaco, and P. G. Stevenson. 2008. The murid herpesvirus-4 gH/gL binds to glycosaminoglycans. PLoS One 3:e1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gillet, L., V. Daix, G. Donofrio, M. Wagner, U. H. Koszinowski, B. China, M. Ackermann, N. Markine-Goriaynoff, and A. Vanderplasschen. 2005. Development of bovine herpesvirus 4 as an expression vector using bacterial artificial chromosome cloning. J. Gen. Virol. 86:907-917. [DOI] [PubMed] [Google Scholar]
- 13.Gillet, L., J. S. May, S. Colaco, and P. G. Stevenson. 2007. Glycoprotein L disruption reveals two functional forms of the murine gammaherpesvirus 68 glycoprotein H. J. Virol. 81:280-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gillet, L., J. S. May, S. Colaco, and P. G. Stevenson. 2007. The murine gammaherpesvirus-68 gp150 acts as an immunogenic decoy to limit virion neutralization. PLoS One 2:e705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gillet, L., J. S. May, and P. G. Stevenson. 2009. In vivo importance of heparan sulfate-binding glycoproteins for murid herpesvirus-4 infection. J. Gen. Virol. 90:602-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gillet, L., F. Minner, B. Detry, F. Farnir, L. Willems, M. Lambot, E. Thiry, P. P. Pastoret, F. Schynts, and A. Vanderplasschen. 2004. Investigation of the susceptibility of human cell lines to bovine herpesvirus 4 infection: demonstration that human cells can support a nonpermissive persistent infection which protects them against tumor necrosis factor alpha-induced apoptosis. J. Virol. 78:2336-2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gillet, L., H. Schroeder, J. Mast, M. Thirion, J. C. Renauld, B. Dewals, and A. Vanderplasschen. 2009. Anchoring tick salivary anti-complement proteins IRAC I and IRAC II to membrane increases their immunogenicity. Vet. Res. 40:51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gillet, L., and P. G. Stevenson. 2007. Antibody evasion by the N terminus of murid herpesvirus-4 glycoprotein B. EMBO J. 26:5131-5142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gillet, L., and P. G. Stevenson. 2007. Evidence for a multiprotein gamma-2 herpesvirus entry complex. J. Virol. 81:13082-13091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Habuchi, H., O. Habuchi, and K. Kimata. 2004. Sulfation pattern in glycosaminoglycan: does it have a code? Glycoconj. J. 21:47-52. [DOI] [PubMed] [Google Scholar]
- 21.Heldwein, E. E., and C. Krummenacher. 2008. Entry of herpesviruses into mammalian cells. Cell. Mol. Life Sci. 65:1653-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Herrold, R. E., A. Marchini, S. Fruehling, and R. Longnecker. 1996. Glycoprotein 110, the Epstein-Barr virus homolog of herpes simplex virus glycoprotein B, is essential for Epstein-Barr virus replication in vivo. J. Virol. 70:2049-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hutt-Fletcher, L. M. 2007. Epstein-Barr virus entry. J. Virol. 81:7825-7832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Janz, A., M. Oezel, C. Kurzeder, J. Mautner, D. Pich, M. Kost, W. Hammerschmidt, and H. J. Delecluse. 2000. Infectious Epstein-Barr virus lacking major glycoprotein BLLF1 (gp350/220) demonstrates the existence of additional viral ligands. J. Virol. 74:10142-10152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannsen, E., M. Luftig, M. R. Chase, S. Weicksel, E. Cahir-McFarland, D. Illanes, D. Sarracino, and E. Kieff. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286-16291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karger, A., U. Schmidt, and U. J. Buchholz. 2001. Recombinant bovine respiratory syncytial virus with deletions of the G or SH genes: G and F proteins bind heparin. J. Gen. Virol. 82:631-640. [DOI] [PubMed] [Google Scholar]
- 27.Kit, S., M. Kit, H. Ichimura, R. Crandell, and S. McConnell. 1986. Induction of thymidine kinase activity by viruses with group B DNA genomes: bovine cytomegalovirus (bovine herpesvirus 4). Virus Res. 4:197-212. [DOI] [PubMed] [Google Scholar]
- 28.Kolset, S. O. 1987. Proteoglycans in normal and neoplastic monocytes. Exp. Cell Res. 168:318-324. [DOI] [PubMed] [Google Scholar]
- 29.Li, M., J. MacKey, S. C. Czajak, R. C. Desrosiers, A. A. Lackner, and J. U. Jung. 1999. Identification and characterization of Kaposi's sarcoma-associated herpesvirus K8.1 virion glycoprotein. J. Virol. 73:1341-1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lomonte, P., M. Bublot, V. van Santen, G. M. Keil, P. P. Pastoret, and E. Thiry. 1995. Analysis of bovine herpesvirus 4 genomic regions located outside the conserved gammaherpesvirus gene blocks. J. Gen. Virol. 76:1835-1841. [DOI] [PubMed] [Google Scholar]
- 31.Lomonte, P., P. Filee, J. R. Lyaku, M. Bublot, P. P. Pastoret, and E. Thiry. 1997. Analysis of the biochemical properties of, and complex formation between, glycoproteins H and L of the gamma2 herpesvirus bovine herpesvirus-4. J. Gen. Virol. 78:2015-2023. [DOI] [PubMed] [Google Scholar]
- 32.Lopez, O. J., J. A. Galeota, and F. A. Osorio. 1996. Bovine herpesvirus type-4 (BHV-4) persistently infects cells of the marginal zone of spleen in cattle. Microb. Pathog. 21:47-58. [DOI] [PubMed] [Google Scholar]
- 33.Luna, R. E., F. Zhou, A. Baghian, V. Chouljenko, B. Forghani, S. J. Gao, and K. G. Kousoulas. 2004. Kaposi's sarcoma-associated herpesvirus glycoprotein K8.1 is dispensable for virus entry. J. Virol. 78:6389-6398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Markine-Goriaynoff, N., L. Gillet, O. A. Karlsen, L. Haarr, F. Minner, P. P. Pastoret, M. Fukuda, and A. Vanderplasschen. 2004. The core 2 beta-1,6-N-acetylglucosaminyltransferase-M encoded by bovine herpesvirus 4 is not essential for virus replication despite contributing to translational modifications of structural proteins. J. Gen. Virol. 85:355-367. [DOI] [PubMed] [Google Scholar]
- 35.Marsh, M., and A. Helenius. 2006. Virus entry: open sesame. Cell 124:729-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maruo, S., L. Yang, and K. Takada. 2001. Roles of Epstein-Barr virus glycoproteins gp350 and gp25 in the infection of human epithelial cells. J. Gen. Virol. 82:2373-2383. [DOI] [PubMed] [Google Scholar]
- 37.Mast, J., C. Nanbru, T. van den Berg, and G. Meulemans. 2005. Ultrastructural changes of the tracheal epithelium after vaccination of day-old chickens with the La Sota strain of Newcastle disease virus. Vet. Pathol. 42:559-565. [DOI] [PubMed] [Google Scholar]
- 38.Means, R. E. 2004. Characterization of the herpesvirus saimiri Orf51 protein. Virology 326:67-78. [DOI] [PubMed] [Google Scholar]
- 39.Molesworth, S. J., C. M. Lake, C. M. Borza, S. M. Turk, and L. M. Hutt-Fletcher. 2000. Epstein-Barr virus gH is essential for penetration of B cells but also plays a role in attachment of virus to epithelial cells. J. Virol. 74:6324-6332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nemerow, G. R., C. Mold, V. K. Schwend, V. Tollefson, and N. R. Cooper. 1987. Identification of gp350 as the viral glycoprotein mediating attachment of Epstein-Barr virus (EBV) to the EBV/C3d receptor of B cells: sequence homology of gp350 and C3 complement fragment C3d. J. Virol. 61:1416-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pertel, P. E. 2002. Human herpesvirus 8 glycoprotein B (gB), gH, and gL can mediate cell fusion. J. Virol. 76:4390-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rosa, G. T., L. Gillet, C. M. Smith, B. D. de Lima, and P. G. Stevenson. 2007. IgG Fc receptors provide an alternative infection route for murine gamma-herpesvirus-68. PLoS One 2:e560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Safaiyan, F., S. O. Kolset, K. Prydz, E. Gottfridsson, U. Lindahl, and M. Salmivirta. 1999. Selective effects of sodium chlorate treatment on the sulfation of heparan sulfate. J. Biol. Chem. 274:36267-36273. [DOI] [PubMed] [Google Scholar]
- 44.Sakamoto, K., H. Asanuma, T. Nakamura, T. Kanno, T. Sata, and H. Katano. 2010. Immune response to intranasal and intraperitoneal immunization with Kaposi's sarcoma-associated herpesvirus in mice. Vaccine 28:3325-3332. [DOI] [PubMed] [Google Scholar]
- 45.Serafini-Cessi, F., N. Malagolini, M. Nanni, F. Dall'Olio, G. Campadelli-Fiume, J. Tanner, and E. Kieff. 1989. Characterization of N- and O-linked oligosaccharides of glycoprotein 350 from Epstein-Barr virus. Virology 170:1-10. [DOI] [PubMed] [Google Scholar]
- 46.Shannon-Lowe, C. D., B. Neuhierl, G. Baldwin, A. B. Rickinson, and H. J. Delecluse. 2006. Resting B cells as a transfer vehicle for Epstein-Barr virus infection of epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 103:7065-7070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Spear, P. G., and R. Longnecker. 2003. Herpesvirus entry: an update. J. Virol. 77:10179-10185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, J. P., N. J. Janjua, S. D. Pepper, G. Bennion, M. Mackett, T. Allen, A. A. Nash, and J. R. Arrand. 1996. Identification and characterization of murine gammaherpesvirus 68 gp150: a virion membrane glycoprotein. J. Virol. 70:3528-3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanner, J., J. Weis, D. Fearon, Y. Whang, and E. Kieff. 1987. Epstein-Barr virus gp350/220 binding to the B lymphocyte C3d receptor mediates adsorption, capping, and endocytosis. Cell 50:203-213. [DOI] [PubMed] [Google Scholar]
- 50.Thiry, E., M. Bublot, J. Dubuisson, M. F. Van Bressem, A. S. Lequarre, P. Lomonte, A. Vanderplasschen, and P. P. Pastoret. 1992. Molecular biology of bovine herpesvirus type 4. Vet. Microbiol. 33:79-92. [DOI] [PubMed] [Google Scholar]
- 51.Thiry, E., P. P. Pastoret, C. Dessy-Doizé, C. Hanzen, and C. M. Calberg-Bacq. 1981. Herpesvirus in infertile bull's testicle. Vet. Rec. 108:426. [DOI] [PubMed] [Google Scholar]
- 52.Thorley-Lawson, D. A., and C. M. Edson. 1979. Polypeptides of the Epstein-Barr virus membrane antigen complex. J. Virol. 32:458-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thorley-Lawson, D. A., and C. A. Poodry. 1982. Identification and isolation of the main component (gp350-gp220) of Epstein-Barr virus responsible for generating neutralizing antibodies in vivo. J. Virol. 43:730-736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Turk, S. M., R. Jiang, L. S. Chesnokova, and L. M. Hutt-Fletcher. 2006. Antibodies to gp350/220 enhance the ability of Epstein-Barr virus to infect epithelial cells. J. Virol. 80:9628-9633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Turnbull, J., A. Powell, and S. Guimond. 2001. Heparan sulfate: decoding a dynamic multifunctional cell regulator. Trends Cell Biol. 11:75-82. [DOI] [PubMed] [Google Scholar]
- 56.Vanderplasschen, A., M. Bublot, J. Dubuisson, P. P. Pastoret, and E. Thiry. 1993. Attachment of the gammaherpesvirus bovine herpesvirus 4 is mediated by the interaction of gp8 glycoprotein with heparinlike moieties on the cell surface. Virology 196:232-240. [DOI] [PubMed] [Google Scholar]
- 57.van Santen, V. L. 1991. Characterization of the bovine herpesvirus 4 major immediate-early transcript. J. Virol. 65:5211-5224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wu, L., R. Renne, D. Ganem, and B. Forghani. 2000. Human herpesvirus 8 glycoprotein K8.1: expression, post-translational modification and localization analyzed by monoclonal antibody. J. Clin. Virol. 17:127-136. [DOI] [PubMed] [Google Scholar]
- 59.Zhu, L., V. Puri, and B. Chandran. 1999. Characterization of human herpesvirus-8 K8.1A/B glycoproteins by monoclonal antibodies. Virology 262:237-249. [DOI] [PubMed] [Google Scholar]
- 60.Zimmermann, W., H. Broll, B. Ehlers, H. J. Buhk, A. Rosenthal, and M. Goltz. 2001. Genome sequence of bovine herpesvirus 4, a bovine Rhadinovirus, and identification of an origin of DNA replication. J. Virol. 75:1186-1194. [DOI] [PMC free article] [PubMed] [Google Scholar]